Abstract
IMPORTANCE
Methotrexate is a first-line systemic agent for treating of psoriasis, although its onset of effects is slower and overall it is less effective than tumor necrosis factor blockers.
OBJECTIVE
To differentiate the response of psoriatic disease to adalimumab and methotrexate sodium.
DESIGN, SETTING, AND PARTICIPANTS
Single-center, randomized, assessor-blind, 2-arm clinical trial of 30 patients from the outpatient dermatology center of Tufts Medical Center, enrolled from August 18, 2009, to October 11, 2011. Patients aged 18 to 85 years with chronic plaque-type psoriasis, a minimum Physician Global Assessment score of 3 (higher scores indicate more severe disease), and a psoriatic plaque of at least 2 cm were randomized in a 1:1 fashion to receive subcutaneous adalimumab or oral methotrexate. Skin biopsy specimens obtained at baseline and weeks 1, 2, 4, and 16 were given a histologic grade by blinded assessors to evaluate treatment response. Analyses were conducted from April 16, 2013, to January 5, 2015.
INTERVENTIONS
A 16-week course of subcutaneous adalimumab (40 mg every 2 weeks after a loading dose) or low-dosage oral methotrexate sodium (7.5–25 mg/wk).
MAIN OUTCOMES AND MEASURES
Changes in genomic, immunohistochemical, and messenger RNA (mRNA) profiles.
RESULTS
Methotrexate responders experienced significant downregulation of helper T-cell– related (TH1, TH17, and TH22) mRNA expression compared with methotrexate nonresponders. Comparisons among adalimumab-treated patients were limited by the number of nonresponders (n = 1). Between adalimumab and methotrexate responders, we found no significant differences in gene expression at any study point or in the expression of T-cell–related mRNA at week 16. Adalimumab responders demonstrated early downregulation of chemokine (C-C motif) ligand 20 (CCL20) mRNA (mean [SE] at week 2, −1.83 [0.52], P < .001; week 16, −3.55 [0.54], P < .001) compared with late downregulation for methotrexate responders (week 2, 0.02 [0.51], P = .96; week 16, −2.96 [0.51], P < .001). Similar differences were observed with interleukin 22 (IL22) mRNA showing early downregulation for adalimumab responders (week 2, −3.17 [1.00], P < .001; week 16, −3.58 [1.00], P < .001) compared with late downregulation for methotrexate responders (week 2, −0.44 [0.68], P = .64; week 16, −5.14 [0.68], P < .001). Analysis of variance findings for key mRNA and immunohistochemical marker expression over the study course were significant only for CCL20 (P = .03) and IL22 (P = .006) mRNA comparing adalimumab and methotrexate responders.
CONCLUSIONS AND RELEVANCE
Methotrexate is an immunomodulator with effects on helper T-cell signaling in psoriasis. Similar genomic and immunohistochemical response signatures and levels of mRNA downregulation at study completion among adalimumab and methotrexate responders suggest a disease-driven instead of therapeutic-driven pathway regulation. Adalimumab and methotrexate responses are differentiated by patterns of normalization of CCL20 and IL22 mRNA expression and may explain the varied onset and degree of clinical responses by each treatment.
TRIAL REGISTRATION
clinicaltrials.gov Identifier: NCT00932113
Psoriasis is a chronic immune-mediated disorder of dysregulated T-cell signaling that affects approximately 3% of the global population. Methotrexate sodium was first evaluated as a treatment for psoriasis in the 1950s, whereas newer biological agents, including adalimumab, were introduced in the 1990s. Studies have demonstrated the superior clinical efficacy and faster onset of the clinical response of adalimumab in the treatment of psoriasis compared with methotrexate.1,2 However, to our knowledge, no investigations have profiled the features of genomic, immunohistochemical, and messenger RNA (mRNA) expression that determine and differentiate responses of psoriatic disease to methotrexate and adalimumab. The primary objective of our study was to characterize the genomic, immunohistochemical, and mRNA signatures associated with methotrexate and adalimumab in the treatment of plaque-type psoriasis.
Methods
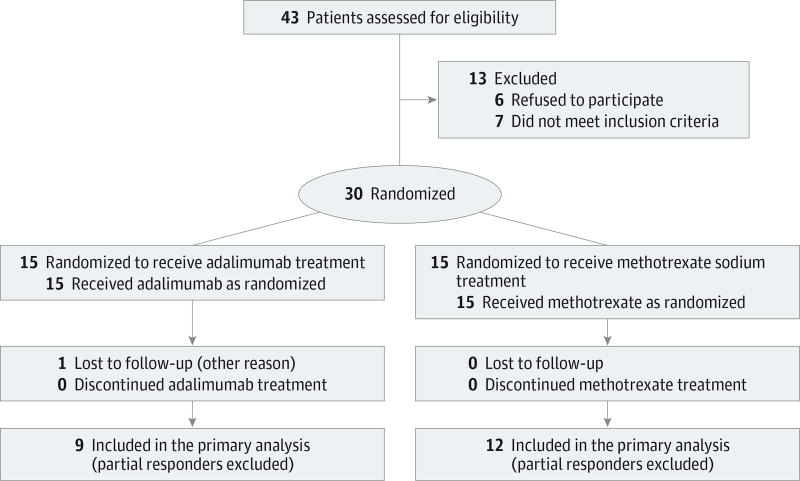
We performed a single-center, randomized, assessor-blind, 2-arm clinical trial. We consecutively enrolled patients attending the tertiary-referral, academic dermatology clinic at Tufts Medical Center from August 18, 2009, through October 11, 2011. Samples were sent to The Rockefeller University in New York, New York, for further analysis. The study received approval from the internal review boards at both institutions and followed CONSORT recommendations for reporting of randomized clinical trials (Figure 1). The study protocol is found in the Supplement 1.
Figure 1.
CONSORT Flow Diagram
Patients
Each patient provided written informed consent for the study before initiation of study participation. Eligible patients were men or women aged 18 through 85 years with chronic plaque-type psoriasis. A minimum Physician Global Assessment (PGA)3 score of 3, based on erythema, induration, and scale (composite score range, 0–5; higher scores indicate more severe disease), and a psoriatic plaque of at least 2 cm in an area that could undergo repeated biopsy were also required for enrollment.
Before study initiation, patients were required to receive (1) no investigational drugs or biological agents within the previous 12 weeks; (2) no methotrexate within the previous 6 weeks; (3) no psoralen–UV-A or oral systemic treatments within the previous 4 weeks; and (4) no UV-B or topical therapies within the previous 2 weeks. However, a continued, stable regimen of classes I or II topical corticosteroids applied to the scalp, axilla, or groin was permitted during study participation. We excluded patients who were previously treated with adalimumab, who had a primary nonresponse to methotrexate, infliximab, or etanercept, or who had experienced safety-related issues secondary to treatment with methotrexate or anti–tumor necrosis factor (TNF) agents. The remaining exclusionary criteria consisted of a history of internal malignant neoplastic disease within 5 years, alcohol or other drug abuse, pregnancy or active nursing, known seropositivity for human immunodeficiency virus, an untreated positive tuberculosis test result, chronic hepatitis B or C virus infection, multiple sclerosis, transverse myelitis, optic neuritis, epilepsy, or a severe infection or receipt of a live vaccine within the previous 30 days.
Study Design
Thirty patients were enrolled in the trial. Through restricted randomization, the patients were distributed in a 1:1 fashion by the study pharmacist to receive adalimumab or methotrexate. The role of the study pharmacist was limited to randomizing patients to treatment and dispensing the study drug; the pharmacist otherwise had no access to patient information or study data. Methotrexate sodium was given as an oral dosage starting at 7.5 mg/wk that was increased based on published protocols.1 Folic acid was also administered as a 1-mg oral dose, 5 days per week, during study participation. Adalimumab was given as an 80-mg subcutaneous loading dose at baseline and then administered as a 40-mg dose at week 1 and every 2 weeks thereafter. The total treatment course was 16 weeks for each group.
Biopsy specimens of lesional and nonlesional skin (LS and NLS, respectively) were obtained before the first dose of each study drug (baseline). During the course of treatment, LS biopsy specimens were also obtained at weeks 1, 2, 4, and 16, from a single area of psoriatic skin selected at baseline (the target lesion). Each patient was scheduled to undergo a total of 6 biopsies.
Clinical Efficacy and Safety Monitoring
The primary clinical efficacy end points included the proportion of patients achieving a Psoriasis Area Severity Index (PASI)3 of 75, defined as a 75% improvement in PASI from baseline. The PASI values range from 0 (no disease) to 72 (severe disease) and measure the severity of psoriasis (erythema, induration, and scale) and the total body surface area of disease involvement. Other clinical end points included improvement in body surface area (calculated using the patient’s palm size as a 1% equivalent), PGA score, and the target lesion site (the PGA score for a single plaque selected at baseline for each patient). Routine laboratory tests and monitoring of vital signs and adverse events were performed throughout the study.
Statistical Analysis
Sample Size Calculation for Microarray Experiments
The R package ssize.fdr4 was used to calculate the sample size needed for a microarray experiment powered to detect differentially expressed genes among patients who responded to treatment (responders) at week 16 (end of treatment).5 Estimates for sample size calculations were based on previous experiments with anti-TNF agents that used a similar study design.6 A sample size larger than 30 guaranteed that we could detect differentially expressed genes at a false discovery rate (FDR) of less than 0.05 and a fold change (FCH) of greater than 2.0 with a power higher than 80% for week 16.
Assessor Blinding
Assessors of clinical (A.B.G.) and histologic responses (M.S.-F. and J.G.K.) were blinded. All nonclinical assessments were performed after study completion.
Microarray Analysis
Preprocessing and Illumina-Based Psoriasis Transcriptome
Classic quality control steps excluded 3 samples (patient 18 at week 16, patient 13 at baseline [NLS], and patient 11 at baseline [LS]). Data were preprocessed using R package lumi,7 quantile normalization, and variance stabilization (log2) transformation. Expression values were filtered to eliminate probe sets with low variation or intensity. The disease profile (psoriasis transcriptome) was defined by the differentially expressed genes between LS and NLS samples from each patient with an FDR of less than 0.05 and an FCH of greater than 1.5.
Modeling Response-Profile to Treatment
Response to treatment was determined by assessor-blinded, histologic scores of keratin 16 expression and epidermal thickness compared with LS.8,9 Each patient was classified as a responder, a nonresponder, or a partial responder by these criteria.10 Histologic response was characterized by elimination of suprabasal keratin 16 staining, normalization of epidermal differentiation, and reduced acanthosis. Nonresponse was characterized by retained acanthosis, abnormal epidermal differentiation, and positive keratin 16 staining among suprabasal keratinocytes. Epidermal, dermal, and overall expression of CD3 and CD11c were also evaluated, and other immunohistochemical markers were replaced in favor of additional array analyses. Complete data analysis was performed on the patients from each treatment group who were classified as responders or nonresponders. A mixed-effect linear model was used to model gene expression for the clinical trial’s time course design. This model estimated the variables of interest even with 3 samples excluded based on poor quality control. Model fitting and hypothesis testing were conducted using the limma package from Bioconductor.11 We used a moderated t statistic for differences in time and calculated a moderated F score for composite comparisons. P values were calculated with adjustment for multiple hypotheses using the Benjamini-Hochberg approach.12
Reverse Transcription–Polymerase Chain Reaction Analysis
Expression of mRNA was measured for chemokines, cytokines, and antimicrobial peptides, which are known to be altered among patients with psoriasis and in response to treatment.13,14 These included mRNA expression related to the following sets of helper T cells: TH1 (interferon γ [IFNG; OMIM 147570] and myxovirus resistance 1 [MX1; OMIM 147150]), TH17 (cathelicidin antimicrobial peptide [CAMP; OMIM 600474], chemokine [C-C motif] ligand 20 [CCL20; OMIM 601960], chemokine [C-X-C motif] ligand 1 [CXCL1; OMIM 155730], defensin β4A [DEFB4A; OMIM 602215], interleukin 17A [IL17A; OMIM 603149], IL17F [OMIM 606496]; and IL23A [OMIM 605580]), and TH22 (IL22; OMIM 605330). Values were normalized to the housekeeping gene hARP, log2-transformed for statistical analysis, and modeled using mixed-effect models. Several other mRNAs of interest were not evaluated by reverse transcription–polymerase chain reaction analysis owing to their high sensitivity on gene arrays. Analyses were conducted from April 16, 2013, to January 5, 2015.
Results
Forty-three patients underwent assessment for eligibility, and 30 were randomized to each treatment group (Figure 1). Thirteen patients were excluded, including 6 lost to or unavailable for follow-up after screening, 4 with an inadequate PGA score or target lesion site at baseline, 2 who were seropositive for hepatitis B or C virus, and 1 with a history of clinical nonresponse to methotrexate. No significant difference in baseline demographic data was found between treatment groups (Table 1). All patients underwent evaluation at each designated study visit except 1 adalimumab-treated patient who was unavailable for follow-up at week 12. The mean maximum dosage of methotrexate sodium was 22 (range, 15–25) mg/wk.
Table 1.
Baseline Characteristics for Methotrexate Sodium– and Adalimumab-Treated Patientsa
| Characteristic | Study Treatment | P Value | |
|---|---|---|---|
| Methotrexate (n = 15) |
Adalimumab (n = 15) |
||
| Male sex | 13 | 11 | .36 |
| Age, mean (range), y | 50.3 (22–69) | 50.5 (22–69) | .97 |
| Race, white | 14 | 12 | .28 |
| Mean PASIb | 15.9 | 16.8 | .79 |
| Target lesion location | |||
| Upper extremity | 3 | 3 | .98 |
| Lower extremity | 6 | 5 | |
| Trunk | 5 | 6 | |
| Buttock | 1 | 1 | |
| History of psoriatic arthritis | 3 | 2 | .62 |
| Previous systemic therapy | 4 | 6 | .44 |
| Mean BMI | 35.0 | 32.1 | .62 |
| Disease duration, mean (range), y | 21.5 (0–47) | 17.3 (1–45) | .41 |
| Age at onset, mean (range), y | 29.1 (5–68) | 33.3 (4–56) | .54 |
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); PASI, Psoriasis Area Severity Index.
Unless otherwise indicated, data are expressed as number of patients.
Values range from 0 (no disease) to 72 (severe disease).
Clinical and Histologic Efficacy
At weeks 8 and 16, 5 (33%) and 10 (67%) adalimumab-treated patients, respectively, achieved a PASI of 75 compared with 1 (7%) and 4 (27%) methotrexate-treated patients, respectively. Among adalimumab-treated patients, 11 (73%) achieved a PGA score of clear or almost clear (0–1) at week 16 compared with 4 (27%) methotrexate-treated patients. Adalimumab responders had a faster reduction in PASI from baseline and a greater percentage reduction in PASI at week 16 (mean, 84.5% [interquartile range, 76.8%–94.7%]) than methotrexate responders (mean, 64.4% [interquartile range, 58.0%–80.7%]) (eFigure, A in Supplement 2). Percentage of improvement in PASI and in target lesion site were strongly correlated for adalimumab-treated (Pearson r = 0.81) and methotrexate-treated (Pearson r = 0.75) patients. Based on histologic criteria, adalimumab-treated patients included 8 responders (53%), 6 partial responders (40%), and 1 nonresponder (7%) compared with 7 responders (47%), 3 partial responders (20%), and 5 nonresponders (33%) among methotrexate-treated patients (eFigure, B and C, Supplement 2).
The adverse events in order of frequency included upper respiratory tract infection (n = 8), flulike illness (n = 2), dizziness (n = 2), and injection site reaction (n = 2) for adalimumab-treated patients. They included upper respiratory tract infection (n = 11), gastrointestinal tract upset (n = 8), and minor infection (n = 4) for methotrexate-treated patients.
Psoriasis Transcriptome
A psoriasis transcriptome of 671 upregulated and 624 down-regulated transcripts (representing 526 and 521 genes, respectively) was generated to compare baseline gene expression among LS and NLS samples from all 30 patients before treatment (FDR, <0.05; FCH, >1.5). This transcriptome was consistent with study findings among patients with psoriasis described previously.6,15 Before initiation of therapy, no differentially expressed genes were found between the 2 treatment groups (LS-to-NLS sample comparison).
Methotrexate Responders and Nonresponders
Genomic Profile
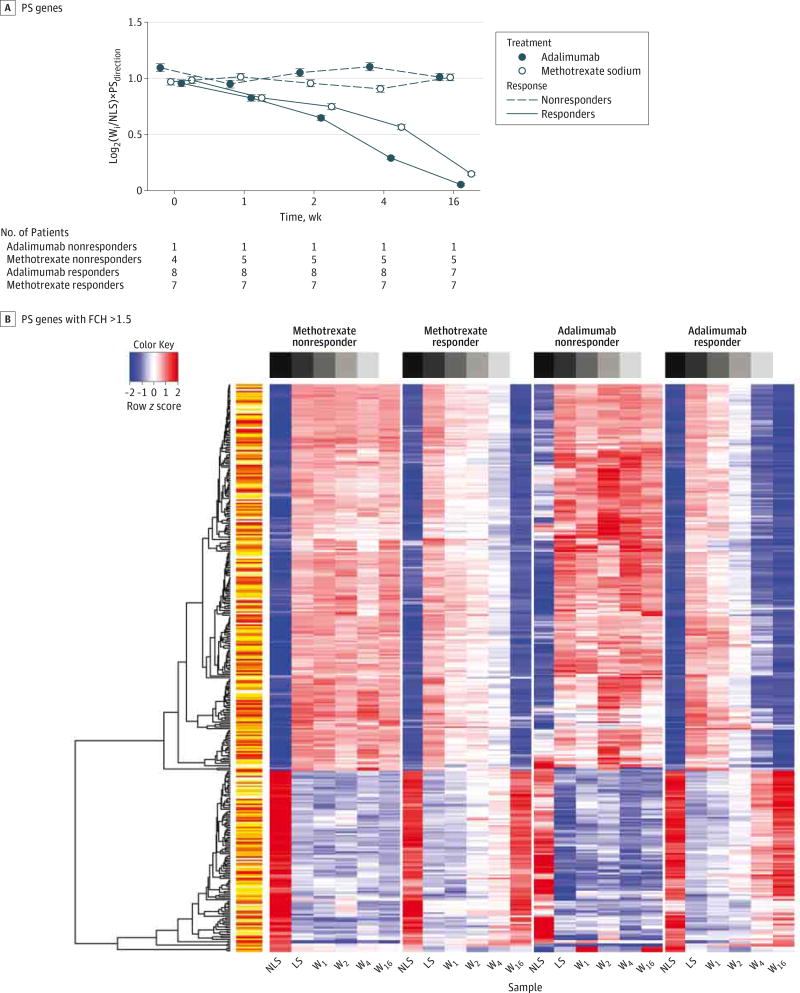
Overall gene expression was plotted as a GG plot of log2-transformed gene expression against time (Figure 2A). Methotrexate responders demonstrated greater normalization of psoriasis transcriptome gene expression compared with nonresponders at all study points after baseline. A heat map of gene expression showed normalization of psoriasis transcriptome genes during the 16-week study for methotrexate responders but not for nonresponders (Figure 2B). The differential treatment effect (response to methotrexate therapy) was defined as genes in which the treatment effect (weeki compared with LS at each time point i) was significantly different between responders compared with nonresponders at the set FDR and FCH criteria of less than 0.05 and greater than 1.5, respectively. A differential treatment effect was observed only for the comparison of week 16 with LS.
Figure 2. Changes in Genomic Data by Disease Response and Treatment Arm.
A, gg plot showing variance stabilization (log2) transformation of gene expression for psoriasis transcriptome (PS) genes (1295 probes/1047 genes) among patients who responded to adalimumab treatment (responders), patients who did not respond to adalimumab treatment (nonresponders), methotrexate sodium–treated responders, and methotrexate nonresponders at week (W) 1, 2, 4, and 16 compared with baseline expression in nonlesional skin (NLS). B, Heat map representing expression of PS genes for methotrexate nonresponders, methotrexate responders, adalimumab nonresponders, and adalimumab responders with fold change (FCH) of greater than 1.5 for baseline lesional and nonlesional skin (LS and NLS, respectively) samples and samples at weeks 1, 2, 4, and 16.Wi indicates expression at any given week (weeki).
mRNA Response Signatures
Among methotrexate responders, our study showed significant downregulation of TH1, TH17, and TH22 pathway mRNA expression compared with nonresponders at week 16 (Figure 3). To further assess the role of TH-specific cell lineages, we performed repeated-measures analysis of variance (ANOVA) on mRNA expression and demonstrated significance among TH17 (P < .01 for all except CAMP [P = .10]) and TH22 (P < .001) axis expression (Table 2). For the TH1 axis, results of the ANOVA for MX1 (P = .03), but not IFNG (P = .09) mRNA expression was statistically significant. Results of the ANOVA were not significant for immunohistochemical markers, including epidermal, dermal, and overall expression of CD3 and of CD11c (P > .05 for all) when we compared methotrexate responders and nonresponders.
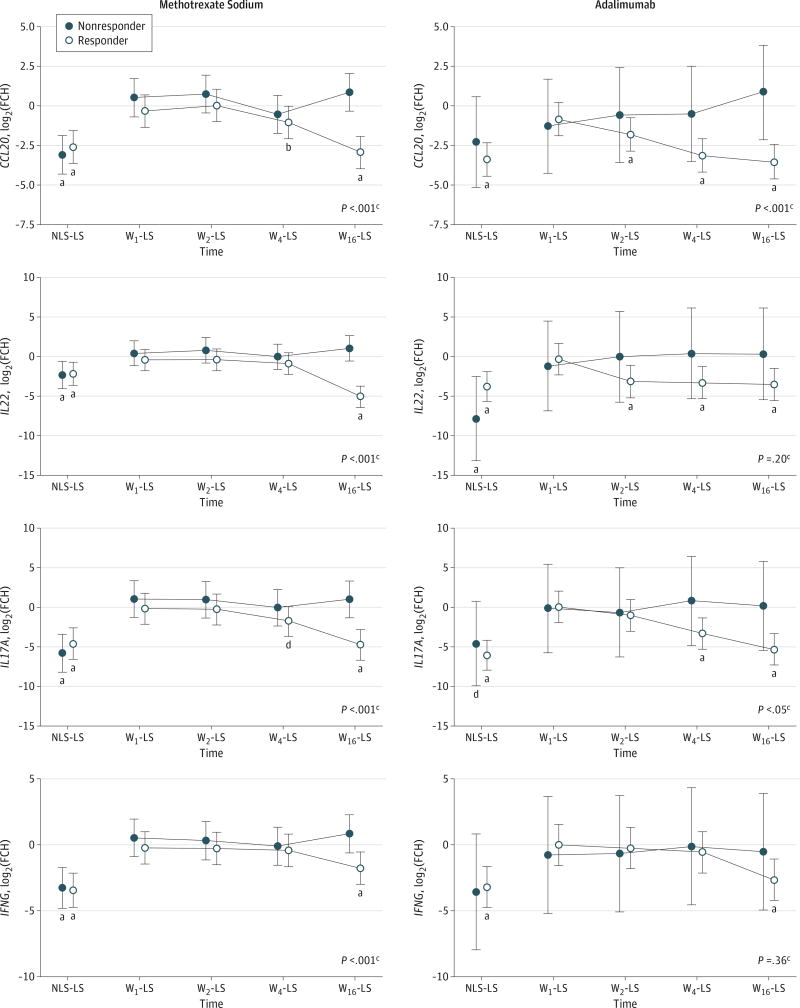
Figure 3. Changes in Select Messenger RNA (mRNA) Expression by Disease Response and Treatment Arm.
Variance stabilization (log2) transformation fold change (FCH) expression of mRNA for patients who responded (responders) and did not respond (nonresponders) to study treatments. Data points indicate mean; error bars, 95%CI. Chemokine (C-C motif) ligand 20 (CCL20), interleukin 22 (IL22), IL17A, and interferon-γ (IFNG) mRNA expression at baseline are compared for baseline lesional and nonlesional skin (LS and NLS, respectively), and week (W) 1, 2, 4, and 16 for methotrexate sodium (left column) and adalimumab (right column).
a P < .001.
b P < .01.
c P value for responders vs nonresponders, W16-LS.
d P < .05.
Table 2.
Reverse Transcription–Polymerase Chain Reaction Data for Key TH17, TH22, and TH1 Mediatorsa
| Pathway and Gene |
Methotrexate Sodium | Adalimumab | Methotrexate Responders vs Adalimumab Responders, P Valuec |
||||
|---|---|---|---|---|---|---|---|
| Responders | Nonresponders | P Valueb | Responders | Nonresponders | P Valueb | ||
| TH17 | |||||||
| CAMP | −2.83 (0.82) [.001] | 0.87 (0.97) [.38] | .10 | −3.85 (0.79) [<.001] | 0.05 (2.2) [.98] | .03 | .69 |
| CCL20 | −2.96 (0.51) [<.001] | 0.84 (0.60) [.17] | <.001 | −3.55 (0.54) [<.001] | 0.84 (1.5) [.57] | .052 | .03 |
| CXCL1 | −3.18 (0.70) [<.001] | 1.22 (0.83) [.15] | <.001 | −3.01 (0.61) [.002] | 1.03 (1.6) [.48] | .02 | .58 |
| DEFB4A | −6.00 (1.1) [<.001] | 1.83 (1.2) [.15] | .001 | −6.96 (0.96) [<.001] | −0.07 (2.6) [.98] | .03 | .50 |
| IL17F | −3.24 (0.74) [<.001] | 1.02 (0.87) [.25] | .009 | −1.97 (0.99) [.054] | 0.79 (2.8) [.78] | .87 | .68 |
| IL17A | −4.75 (0.96) [<.001] | 1.00 (1.1) [.39] | <.001 | −5.31 (0.98) [<.001] | 0.18 (2.8) [.95] | .31 | .74 |
| IL23A | −2.72 (0.56) [<.001] | 0.89 (0.67) [.19] | .003 | −3.90 (0.79) [<.001] | −0.81 (2.2) [.72] | .36 | .65 |
| TH22 | |||||||
| IL22 | −5.14 (0.68) [<.001] | 1.00 (0.80) [.22] | <.001 | −3.58 (1.0) [.001] | 0.25 (2.8) [.93] | .10 | .006 |
| TH1 | |||||||
| IFNG | −1.77 (0.61) [.005] | 0.82 (0.72) [.26] | .09 | −2.66 (0.77) [.002] | −0.52 (2.2) [.81] | .84 | .87 |
| MX1 | −2.00 (0.46) [<.001] | 0.47 (0.54) [.39] | .03 | −1.90 (0.31) [<.001] | 0.04 (0.86) [.96] | .001 | .10 |
Abbreviations: CAMP, cathelicidin antimicrobial peptide; CCL20, chemokine (C-C motif) ligand 20; CXCL1, chemokine (C-X-C motif) ligand 1; DEFB4A, defensin β4A; FCH, fold change; IFNG, interferon-γ; IL, interleukin; mRNA,messenger RNA; MX1,myxovirus resistance 1; TH, helper T cell.
Unless otherwise indicated, data are given as mean (SE) [P value] for the treatment effect at the week 16 study end point.
P value for the interaction effect time × response comparing responders and nonresponders.
P value for the interaction effect time × treatment.
Genomic Profile and mRNA Response Signatures
Adalimumab Responders and Nonresponders
No statistically significant differences between adalimumab responders and nonresponders in individual gene expression during the study course were found through microarray analysis. A GG plot and heat map of gene expression demonstrated normalization of psoriasis transcriptome genes during the 16-week study course among adalimumab responders but not among nonresponders (Figure 2). Compared with adalimumab nonresponders, responders demonstrated significant downregulation of CAMP, CXCL1, DEFB4A, and MX1 mRNA expression during the study course (Table 2). Results of the ANOVA were also significant for immunohistochemical markers, including epidermal, dermal, and overall CD11c expression (P < .001 for all) and epidermal (P < .001) but not dermal or overall CD3 expression (P > .70). We observed greater downregulation among adalimumab responders than nonresponders.
Adalimumab and Methotrexate Responders
Adalimumab demonstrated greater normalization of psoriasis transcriptome gene expression compared with methotrexate, with the largest difference observed at week 4 (Figure 2A). From weeks 4 to 16, the rate of change for methotrexate surpassed that for adalimumab, although overall normalization of gene expression favored adalimumab at study completion. A heat map similarly showed faster and more complete normalization of gene expression for adalimumab responders compared with methotrexate responders (Figure 2B). During the entire study, however, we observed no differential response effect genes, defined as individual genes with a treatment effect significantly different when we compared the responders of each study group (weeki compared with LS; FDR, <0.05; FCH, >1.5).
Log2-transformed FCH mRNA expression, including TH1-, TH17-, and TH22-related mRNA, was evaluated for adalimumab and methotrexate responders. The comparison of mRNA levels at week 16 with those of LS reached pretreatment NLS-to-LS expression levels for all transcripts studied (Figure 3). Adalimumab responders demonstrated early downregulation of CCL20 mRNA (mean [SE] at week 2, −1.83 [0.52], P < .001; week 16, −3.55 [0.54], P < .001) compared with late downregulation for methotrexate responders (week 2, 0.02 [0.51], P = .96; week 16, −2.96 [0.51], P < .001). Similar differences were observed with interleukin 22 (IL22) mRNA showing early down-regulation for adalimumab responders (week 2, −3.17 [1.00], P < .001; week 16, −3.58 [1.00], P < .001) compared with late downregulation for methotrexate responders (week 2, −0.44 [0.68], P = .64; week 16, −5.14 [0.68], P < .001). Results of the ANOVA for mRNA expression during the 16-week study were significant only for CCL20 (P = .03) and IL22 (P = .006) (Table 2). Results of the ANOVA were not significant for immunohistochemical markers, including epidermal, dermal, and overall expression of CD3 and of CD11c (all P > .40), when we compared adalimumab and methotrexate responders.
Discussion
This assessor-blinded, 2-arm study investigates the effects of adalimumab and methotrexate in the treatment of psoriasis through clinical, genomic, immunohistochemical, and mRNA analyses. Similar to the findings of previous studies,1,2 we demonstrate faster and more complete clinical disease responses among adalimumab-treated than methotrexate-treated patients. We show that methotrexate responders compared with methotrexate nonresponders experience significant down-regulation of mRNAs in the TH1-, TH17-, and TH22-related signaling pathways and normalization of psoriasis transcriptome genes. Among adalimumab responders, normalization of CCL20 and IL22 mRNA expression is faster than among methotrexate responders, but both groups share similar genomic and mRNA profiles at week 16, the study end point. Our findings highlight methotrexate’s role as an immunomodulator and suggest that further investigations of CCL20-and IL22-related signaling in psoriasis are warranted.
Methotrexate is a first-line, systemic therapy in the treatment of psoriasis, and its use is typically required before prescription of biological agents. Despite methotrexate’s significantly lower cost, current evidence suggests that biological agents are safe, have a faster onset of clinical response, and demonstrate an overall higher efficacy than methotrexate.1,2 However, no known studies have evaluated methotrexate’s effects on pathway regulation in psoriasis or compared the changes induced by methotrexate and biological agents at the genomic, immunohistochemical, and mRNA levels.
Our study shows that methotrexate is an immunomodulator with significant downregulatory effects on TH1-, TH17-, and TH22-related signaling pathways in diseased psoriatic skin. Current research suggests that an aberrant TH17 axis is a key component in the process leading to and maintaining inflammatory dysregulation in psoriasis.16 In susceptible hosts, dendritic cell–driven IL23A production leads to activation of TH17 and TH22 subsets. Activated TH17 produces IL17A and IL17F, which stimulate keratinocyte production of inflammatory mediators, including CCL20, CXCL1, and DEFB4A. Interleukin 22–producing TH22 also plays a key role in mediating the epidermal changes observed in psoriasis.17,18 As we demonstrated with methotrexate, effective treatment of psoriasis may be expected to normalize these aberrant signaling pathways. This hypothesis is supported by similarities between adalimumab and methotrexate responders in the differential response effect genes and in mRNA expression levels at week 16, suggesting disease-specific instead of treatment-specific pathway regulation. However, although quantitative gene and mRNA expression for responding patients appears to converge at week 16, adalimumab responders experienced faster downregulation of CCL20 and IL22 mRNA than methotrexate responders.
As a member of the IL10 cytokine family, IL22 is primarily produced by T lymphocytes, particularly TH22 lymphocytes.19 Interleukin 22 stimulation of barrier tissues, such as the skin, joints, and gastrointestinal tract, leads to regulation of chemokine and cytokine production within these tissues. Downstream processes, including cellular differentiation and proliferation, are essential for the maintenance of these physiologic barriers subject to frequent external insults.20 However, dysregulated activity of these pathways can be pathogenic in disease states such as psoriasis. Psoriatic skin is characterized by increased expression of IL22 and other factors (eg, TNF, IFNγ) that increase keratinocyte and fibroblast expression of the IL22 receptor complex 1.19 Within psoriatic skin, increased IL22 pathway signaling promotes characteristic findings, such as acanthosis and keratinocyte dedifferentiation.21 Mutations in the IL22 promoter site are known to lead to T-cell–mediated upregulation of IL22 expression and are associated with early-onset psoriasis.22 In addition, the chronic course of psoriasis may in part be linked to persistent expression of IL22-producing, CD4+, tissue-resident, memory T cells within the normal-appearing (NLS) psoriatic epidermis.23 In patients with psoriatic arthritis, IL22 levels are significantly elevated in the synovial fluid and can regulate fibroblastlike synoviocyte proliferation.24 Osteoblast-dependent bone remodeling, a characteristic finding among patients with inflammatory arthritis, is also mediated in part by IL22 signaling.25 Therefore, IL22-related signaling participates in the pathogenic inflammatory processes occurring in the skin and joints of patients with psoriasis.
The chemokine CCL20 is produced by multiple cell lines, including keratinocytes, neutrophils, and T cells, particularly TH17 lymphocytes. Its chemokine receptor, C-C receptor 6 (CCR6), is strongly expressed by T cells and immature dendritic cells and therefore promotes the migration and entry of key psoriatic disease effector cells into target tissues. One of the immediate response psoriasis factors, CCL20, is upregulated by synergistic IL17/TNF signaling and attracts clusters of CCR6+ dendritic cells and T cells to the psoriatic epidermis.26,27 Administration of anti-CCL20 antibodies in a murine model reduce IL23-induced, psoriasiform inflammation and limit epidermal infiltration of IL22-producing CCR6+ T cells, thereby disrupting the psoriatic phenotype.28 The CCR6-knockout mice also experience significant reductions in IL23-induced IL22 production compared with wild-type mice.
Two previous single-arm trials29,30 have investigated the ability of adalimumab to regulate key inflammatory pathways in psoriasis on the mRNA level. In one study by Hendriks et al,29 10 clinically responding, adalimumab-treated patients demonstrated early downregulation of innate immune responses and epidermal complex–proliferation makers but late downregulation of adaptive immune responses. A study by Balato et al30 showed significant reductions in CCL20 and IL22 mRNA expression, among others, in the skin and peripheral blood mononuclear cells isolated from 18 adalimumab-treated patients with psoriasis who had histologic evidence of response. In the study by Hendriks et al,29 CCL20 and IL22 expression was not evaluated. Because both studies had a single arm, they were unable to evaluate qualitative differences between disease response (improvement in pathogenic axes associated with psoriasis) and medication response (how pathways are regulated by the therapy administered). Other single-arm studies have also demonstrated significant downregulation of CCL20 and IL22 lesional mRNA among treatment responders with psoriasis, including patients who receive cyclosporine or etanercept.14,31 However, potential comparative evaluations of pathway regulation between these trials are limited by varied study methods.
Our trial suggests that CCL20 and IL22 signaling may be key determinants of the faster and more complete clinical responses observed with adalimumab compared with methotrexate in the treatment of psoriasis. Previous studies have investigated the anti-IL22 monoclonal antibody, fezakinumab, for the treatment of psoriasis (phase 1)32 and rheumatoid arthritis (phase 2).33 At present, no known, active clinical studies in psoriasis are ongoing, but targeting of IL22 alone may be too far downstream in the pathogenic mechanism of psoriasis to be an effective monotherapy. Although IL22 is a key regulator of epidermal hyperplasia and keratinocyte differentiation, other central effector cells and signaling path-ways in the disease process would not be targeted directly through IL22 inhibition. For example, blockade of IL22 receptor complex 1 may interfere with additional ligands and lead to more effective signaling disruption.19 Signaling of CCL20/ CCR6 may also present an effective target in drug development because the signal links the innate (dendritic cells) and adaptive (T cells) immune responses and is involved in chemotaxis and effector cell trafficking. Preclinical murine models investigating the blockade of these pathways have promising results, although no clinical studies have been initiated, to our knowledge.28,34
Our study has several limitations. Patterns of genomic and mRNA regulation may not reflect changes seen at the post-translational levels, including protein expression and post-translational modification. Additional studies investigating such expression may expand on the findings reported here. In addition, differences observed between adalimumab responders (n = 8) and nonresponders (n = 1) require further investigation because of the limited sample size. However, this study is one of the larger ones to characterize genomic and mRNA data from psoriatic skin samples. This study also represents, to our knowledge, the first known evaluation of methotrexate and comparison of methotrexate with an anti-TNF agent by these methods. Finally, methotrexate was dosed in a graduated fashion; therefore, delayed responses in clinical and mRNA expression may to some degree be expected. However, unlike CCL20 and IL22, all other mRNA transcripts studied among methotrexate responders mirrored those of adalimumab responders in rates of normalization. Dose sensitivity in CCL20 and IL22 expression may be greater than those of the other transcripts studied, although this possibility is less likely, and would support the role of CCL20 and IL22 above the other chemokines and cytokines studied in the differential rates of disease response between adalimumab and methotrexate.
Conclusions
Clinical response to adalimumab in psoriasis is faster, more complete, and characterized by faster downregulation of CCL20 and IL22 mRNA compared with methotrexate. These cytokines are important disease mediators with downstream effects, including inflammatory cell migration, keratinocyte dysregulation, and joint space remodeling. Given the expression of CCL20 and CCR6 by the innate (dendritic cells) and adaptive (T cells) immune system and the role of IL22 in mediating crosstalk between keratinocytes and other immune cells, targets within these pathways may disrupt the psoriatic disease process at multiple critical points.35 Therefore, IL22-and CCL20-related signaling pathways are candidates for further investigation to understand disease mechanisms and drug development in psoriasis.28,35
Supplementary Material
Acknowledgments
Funding/Support: This study was supported in part by an investigator-initiated grant from AbbVie (Dr Gottlieb); by the National Psoriasis Foundation; by Aram Sogomonian; and by grant UL1 TR000043 from the National Center for Advancing Translational Sciences, National Institutes of Health Clinical and Translational Science Award program (Dr Suárez-Fariñas).
Role of the Funder/Support: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Supplemental content at jamadermatology.com
Author Contributions: Drs Goldminz and Suárez-Fariñas had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Krueger, Gottlieb.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Goldminz, Suárez-Fariñas, Krueger, Gottlieb.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Goldminz, Suárez-Fariñas.
Obtained funding: Gottlieb.
Administrative, technical, or material support: Wang, Dumont, Krueger.
Study supervision: Dumont, Gottlieb.
Conflict of Interest Disclosures: Dr Krueger is a consultant for Janssen, Lilly, and Pfizer and has grant agreements with Amgen, Janssen, Lilly, Merck, and Pfizer. Dr Gottlieb has consulting/ advisory board agreements with AbbVie, Actelion, Akros, Amgen, Astellas, Beiersdorf, Bristol-Myers Squibb, Canfite, Catabasis, Celgene, Coronado, CSL Behring, Dermipsor, Ltd, GlaxoSmithKline, Incyte, Janssen, Karyopharm, Lilly, Novartis, Novo Nordisk, Pfizer, TEVA, UCB, Vertex, Dusa, and Xenoport and receives research/educational grants (paid to Tufts Medical Center) from AbbVie, Amgen, Celgene, Coronado, Janssen, Levia, Lilly, Novartis, Merck, and Pfizer. No other disclosures were reported.
Additional Contributions: Biljana Bazdar-Vinovrski, MS, PharmD, acted as the study pharmacist. No financial compensation was given for these services.
References
- 1.Saurat JH, Stingl G, Dubertret L, et al. CHAMPION Study Investigators. Efficacy and safety results from the randomized controlled comparative study of adalimumab vs methotrexate vs placebo in patients with psoriasis (CHAMPION) Br J Dermatol. 2008;158(3):558–566. doi: 10.1111/j.1365-2133.2007.08315.x. [DOI] [PubMed] [Google Scholar]
- 2.Nast A, Sporbeck B, Rosumeck S, et al. Which antipsoriatic drug has the fastest onset of action? systematic review on the rapidity of the onset of action. J Invest Dermatol. 2013;133(8):1963–1970. doi: 10.1038/jid.2013.78. [DOI] [PubMed] [Google Scholar]
- 3.Spuls PI, Lecluse LL, Poulsen ML, Bos JD, Stern RS, Nijsten T. How good are clinical severity and outcome measures for psoriasis? quantitative evaluation in a systematic review. J Invest Dermatol. 2010;130(4):933–943. doi: 10.1038/jid.2009.391. [DOI] [PubMed] [Google Scholar]
- 4.Orr M, Liu P. Sample size estimation while controlling false discovery rate for microarray experiments using the ssize.fdr package. R J. 2009;1:47–53. [Google Scholar]
- 5.Liu P, Hwang JT. Quick calculation for sample size while controlling false discovery rate with application to microarray analysis. Bioinformatics. 2007;23(6):739–746. doi: 10.1093/bioinformatics/btl664. [DOI] [PubMed] [Google Scholar]
- 6.Zaba LC, Suárez-Fariñas M, Fuentes-Duculan J, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol. 2009;124(5):1022–10.e1. 395. doi: 10.1016/j.jaci.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du P, Kibbe WA, Lin S. [Accessed March 16, 2015];Using lumi, a package processing Illumina Microarray. http://bioconductor.org/packages/2.0/bioc/vignettes/lumi/inst/doc/lumi.pdf. Published April 25, 2007.
- 8.Krueger JG, Wolfe JT, Nabeya RT, et al. Successful ultraviolet B treatment of psoriasis is accompanied by a reversal of keratinocyte pathology and by selective depletion of intraepidermal T cells. J Exp Med. 1995;182(6):2057–2068. doi: 10.1084/jem.182.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb AB, Krueger JG, Wittkowski K, Dedrick R, Walicke PA, Garovoy M. Psoriasis as a model for T-cell–mediated disease: immunobiologic and clinical effects of treatment with multiple doses of efalizumab, an anti-CD11a antibody. Arch Dermatol. 2002;138(5):591–600. doi: 10.1001/archderm.138.5.591. [DOI] [PubMed] [Google Scholar]
- 10.Wittkowski KM, Lee E, Nussbaum R, Chamian FN, Krueger JG. Combining several ordinal measures in clinical studies. Stat Med. 2004;23(10):1579–1592. doi: 10.1002/sim.1778. [DOI] [PubMed] [Google Scholar]
- 11.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005. [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 13.Gottlieb AB, Chamian F, Masud S, et al. TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol. 2005;175(4):2721–2729. doi: 10.4049/jimmunol.175.4.2721. [DOI] [PubMed] [Google Scholar]
- 14.Zaba LC, Cardinale I, Gilleaudeau P, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced TH17 responses. J Exp Med. 2007;204(13):3183–3194. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suárez-Fariñas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger JG. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132(11):2552–2564. doi: 10.1038/jid.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowes MA, Russell CB, Martin DA, Towne JE, Krueger JG. The IL-23/T17 pathogenic axis in psoriasis is amplified by keratinocyte responses. Trends Immunol. 2013;34(4):174–181. doi: 10.1016/j.it.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolk K, Witte E, Wallace E, et al. IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes: a potential role in psoriasis. Eur J Immunol. 2006;36(5):1309–1323. doi: 10.1002/eji.200535503. [DOI] [PubMed] [Google Scholar]
- 18.Wolk K, Haugen HS, Xu W, et al. IL-22 and IL-20 are key mediators of the epidermal alterations in psoriasis while IL-17 and IFN-γ are not. J Mol Med (Berl) 2009;87(5):523–536. doi: 10.1007/s00109-009-0457-0. [DOI] [PubMed] [Google Scholar]
- 19.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22–IL-22R1 system. Nat Rev Drug Discov. 2014;13(1):21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy-Crispin M, Billick E, Mitsui H, et al. Human keratinocytes’ response to injury upregulates CCL20 and other genes linking innate and adaptive immunity. J Invest Dermatol. 2012;132(1):105–113. doi: 10.1038/jid.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Danilenko DM, Valdez P, et al. Interleukin-22, a TH17 cytokine, mediates IL-23–induced dermal inflammation and acanthosis. Nature. 2007;445(7128):648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 22.Nikamo P, Cheuk S, Lysell J, et al. Genetic variants of the IL22 promoter associate to onset of psoriasis before puberty and increased IL-22 production in T cells. J Invest Dermatol. 2014;134(6):1535–1541. doi: 10.1038/jid.2014.5. [DOI] [PubMed] [Google Scholar]
- 23.Cheuk S, Wikén M, Blomqvist L, et al. Epidermal TH22 and Tc17 cells form a localized disease memory in clinically healed psoriasis. J Immunol. 2014;192(7):3111–3120. doi: 10.4049/jimmunol.1302313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra A, Raychaudhuri SK, Raychaudhuri SP. Functional role of IL-22 in psoriatic arthritis. Arthritis Res Ther. 2012;14(2):R65. doi: 10.1186/ar3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherlock JP, Joyce-Shaikh B, Turner SP, et al. IL-23 induces spondyloarthropathy by acting on ROR-γt+ CD3+CD4−CD8− entheseal resident T cells. Nat Med. 2012;18(7):1069–1076. doi: 10.1038/nm.2817. [DOI] [PubMed] [Google Scholar]
- 26.Chiricozzi A, Guttman-Yassky E, Suárez-Fariñas M, et al. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131(3):677–687. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 27.Kim TG, Jee H, Fuentes-Duculan J, et al. Dermal clusters of mature dendritic cells and T cells are associated with the CCL20/CCR6 chemokine system in chronic psoriasis. J Invest Dermatol. 2014;134(5):1462–1465. doi: 10.1038/jid.2013.534. [DOI] [PubMed] [Google Scholar]
- 28.Mabuchi T, Singh TP, Takekoshi T, et al. CCR6 is required for epidermal trafficking of γδ-T cells in an IL-23–induced model of psoriasiform dermatitis. J Invest Dermatol. 2013;133(1):164–171. doi: 10.1038/jid.2012.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendriks AG, van der Velden HM, Wolberink EA, et al. The effect of adalimumab on key drivers in the pathogenesis of psoriasis. Br J Dermatol. 2014;170(3):571–580. doi: 10.1111/bjd.12705. [DOI] [PubMed] [Google Scholar]
- 30.Balato A, Schiattarella M, Di Caprio R, et al. Effects of adalimumab therapy in adult subjects with moderate-to-severe psoriasis on TH17 pathway. J Eur Acad Dermatol Venereol. 2014;28(8):1016–1024. doi: 10.1111/jdv.12240. [DOI] [PubMed] [Google Scholar]
- 31.Haider AS, Lowes MA, Suárez-Fariñas M, et al. Identification of cellular pathways of “type 1,” TH17 T cells, and TNF- and inducible nitric oxide synthase–producing dendritic cells in autoimmune inflammation through pharmacogenomic study of cyclosporine A in psoriasis. J Immunol. 2008;180(3):1913–1920. doi: 10.4049/jimmunol.180.3.1913. [DOI] [PubMed] [Google Scholar]
- 32.ClinicalTrials.gov. [Accessed January 6, 2015];Study Evaluating the Safety and Tolerability of ILV-094 in Subjects With Psoriasis. NCT00563524. https://clinicaltrials.gov/ct2/show/NCT00563524.
- 33.ClinicalTrials.gov. [Accessed January 6, 2015];Study to Evaluate the Safety and Efficacy of ILV-094 in Subjects With Rheumatoid Arthritis. NCT00883896. https://clinicaltrials.gov/ct2/show/NCT00883896.
- 34.Hedrick MN, Lonsdorf AS, Hwang ST, Farber JM. CCR6 as a possible therapeutic target in psoriasis. Expert Opin Ther Targets. 2010;14(9):911–922. doi: 10.1517/14728222.2010.504716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AY, Körner H. CCR6 and CCL20: emerging players in the pathogenesis of rheumatoid arthritis. Immunol Cell Biol. 2014;92(4):354–358. doi: 10.1038/icb.2013.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.