Abstract
Despite approaches in regenerative medicine using stem cells, bio‐engineered scaffolds, and targeted drug delivery to enhance human tissue repair, clinicians remain unable to regenerate large‐scale, multi‐tissue defects in situ. The study of regenerative biology using mammalian models of complex tissue regeneration offers an opportunity to discover key factors that stimulate a regenerative rather than fibrotic response to injury. For example, although primates and rodents can regenerate their distal digit tips, they heal more proximal amputations with scar tissue. Rabbits and African spiny mice re‐grow tissue to fill large musculoskeletal defects through their ear pinna, while other mammals fail to regenerate identical defects and instead heal ear holes through fibrotic repair. This Review explores the utility of these comparative healing models using the spiny mouse ear pinna and the mouse digit tip to consider how mechanistic insight into reparative regeneration might serve to advance regenerative medicine. Specifically, we consider how inflammation and immunity, extracellular matrix composition, and controlled cell proliferation intersect to establish a pro‐regenerative microenvironment in response to injuries. Understanding how some mammals naturally regenerate complex tissue can provide a blueprint for how we might manipulate the injury microenvironment to enhance regenerative abilities in humans. Stem Cells Translational Medicine 2018;7:220–231
Keywords: Animal models, Tissue regeneration, Tissue‐specific stem cells, Monocyte
Significance Statement.
Continued study using mammalian models of regeneration will undoubtedly deepen our understanding of how the injury microenvironment stimulates blastema formation in lieu of fibrotic repair. The challenge ahead is how to translate basic biological understanding into meaningful clinical advances that manifest into tractable regenerative therapies. A key problem to solve is the extent to which the local injury microenvironment can be manipulated to induce blastema formation and subsequent regeneration.
Introduction
For most of us, the sight of an amputated digit is a reminder that humans cannot regenerate organs in response to severe trauma and prompts the question, why not us? Throughout the animal kingdom, we observe a panoply of animals that can regenerate limbs, spinal cords, hearts and even entire bodies from a small fragment, which begets the question, how do they do it? Historically, scientists have approached the how through careful description of regenerative phenomena in animals at the genomic, molecular, cellular, and tissue level of organization, and by inhibiting the regenerative process at various stages. Many such studies promoted the idea that understanding the various mechanisms regulating regeneration in animals could provide a pathway toward stimulating regeneration in humans 1.
In an unlucky twist of fate, the ability to genetically and transgenically modify certain organisms to study embryonic development left classic animal models of regeneration on the sidelines. Focus shifted toward stem cell biology and tissue engineering, which ultimately produced the modern field of regenerative medicine. The progression of regenerative medicine coincided with rapid technological advances in genomic sequencing, computational genomics, gene manipulation, cellular re‐programming, and the production of tissue scaffolds and bioreactors. The result is that scientists are now able to reprogram adult somatic cells into multipotent and totipotent stem cells 2 and subsequently differentiate these cells into defined cell types 3, build complex tissue scaffolds with three‐dimensional printing technology to incorporate stem cells (reviewed in 4), and construct simplistic organs ex vivo for transplantation 5. And yet, despite conceptual and technological advances, we still cannot faithfully induce a digit or other complex organs to naturally regenerate in humans. A reckoning suggests that a path forward for regenerative medicine is to directly re‐engage with regenerative biologists to understand how animals regulate the injury environment to create local bioreactors in situ that can organize cells to faithfully replace damaged tissue.
Being mindful of a species sampling bias and confounding traits such as age, size, and life‐stage 6, regenerative ability appears to be unevenly distributed among adult vertebrates (reviewed in 7). Generally speaking, fishes exhibit extensive regenerative ability 8, 9 and among tetrapods, Urodele amphibians stand as outliers given the extent of their regenerative abilities 10. Beyond these species, some frogs, lizards, and mammals show enhanced regenerative capacity of complex tissues as adults suggesting either, regenerative ability is broadly suppressed in vertebrates and has re‐emerged in some species, or regenerative ability has been broadly lost and subsequently re‐evolved in some instances. In spite of the interesting evolutionary questions these comparisons raise, scientists have tended to focus on those vertebrates with the most extensive powers of regeneration. Using a few key species, the hope was that discovering the underlying mechanisms in these models might stimulate new approaches or insight into developing regenerative therapies for humans 1, 11.
In particular vertebrates, appendage amputation triggers cellular reactions—activated cell‐cycling, developmental signaling, morphogenesis, and differentiation—and studies in these animal models provide a basic blueprint for how tissues can naturally regenerate (Fig. 1). While studies in fish and salamanders continue to provide resolution at the molecular level for how vertebrate regeneration occurs, lack of closely related nonregenerating species makes it difficult to disentangle the mechanisms differentially driving a regenerative or fibrotic response to injury 12. Important genomic, cellular, and physiological differences exist between vertebrates necessitating a broader expansion of regenerative animal models. In this light, adult mammalian models of regeneration are poised to make a unique contribution to regenerative medicine. Adult mammals more closely mimic the human condition with respect to genomic architecture, metabolic rate, immunity, and homeothermy. Moreover, mammalian models of regeneration can provide a comparative system to study regeneration and scar formation between species (e.g., ear holes, skin, etc.) or in the same tissue (e.g., distal digit tip vs. middle phalanx), and thus studies can uncover the switches regulating a fibrotic or regenerative response to injury. A similar paradigm has been exploited to compare embryonic scar‐free healing to adult fibrotic repair 13, 14. While this body of work has contributed much to our understanding of skin healing and regeneration, the confounding factors of developmental stage (e.g., incomplete state of tissue development, cellular differentiation, immune system maturation, etc.) make it difficult to determine the extent to which embryonic scar‐free healing mimics instances of naturally occurring adult regeneration. Thus, the focus of this review is directed toward complex tissue regeneration in adult mammals.
Figure 1.
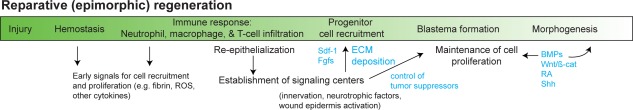
An outline of the events during reparative (epimorphic) regeneration in mammals. Injury initiates hemostasis, an immune response and re‐epithelialization. These early events help to establish key signaling centers and a regenerative microenvironment that recruits progenitor cells. Subsequent progenitor cell proliferation forms a blastema, a defining characteristic of epimorphic regeneration. The blastema undergoes morphogenesis to restore the missing tissue, and subsequent growth will ensure a functional replacement. Vertebrate models have shed light on specific growth factors, chemokines, and extracellular matrix proteins that control reparative regeneration and include: SDF1, FGFs, BMPs, Wnt/ß‐catenin signaling, RA, and SHH. Abbreviations: BMPs, bone morphogenetic proteins; ECM, extracellular matrix; FGFs, fibroblast growth factors; RA, retinoic acid signaling; SDF1, stromal derived factor 1; SHH, sonic hedgehog signaling; ROS, reactive oxygen species.
Epimorphic Regeneration and Fibrotic Healing
Definitions of regeneration create an important foundation for studying regenerative phenomena. One can investigate regeneration in the context of homeostatic regeneration (i.e., regular and repeated turnover of a single cell type or multiple cell types within a tissue), or in the context of reparative regeneration (i.e., replacement of complex tissue in response to injury). Although these two broad classes of regeneration are distinct, in many cases, homeostatic regeneration and reparative regeneration can occur within the same tissue. For example, the mammalian epidermis undergoes continuous replacement during the majority of a normal lifetime (reviewed in 15). Thus, epidermal cells exhibit homeostatic regeneration. However, if a large portion of epidermis is lost along with the underlying dermis, as in full‐thickness skin injuries, although keratinocytes migrate and proliferate to re‐epithelialize the wound and replace the lost section of tissue, the epidermally‐derived hair follicles fail to regenerate. The distinction between the two types of regeneration is important because it underscores the idea that regenerating organs require the coordinated activity of multiple cell types and that regenerative ability at the single cell level does not necessarily translate into tissue‐level regeneration. On the other hand, the fact that all animals exhibit homeostatic regeneration of some tissues is promising from a clinical perspective because it suggests there is some regenerative ability intrinsic to all vertebrates, including humans. In our mind, studying the mechanisms that control reparative regeneration is the key to understanding how to stimulate regeneration.
When studying tissue regeneration in vertebrates, reparative regeneration is defined as epimorphic regeneration (i.e., regenerative morphogenesis involving cell proliferation) 11, 16 (Fig. 1). Epimorphosis is initiated by amputation or direct injury and leads to a hemostatic response including the recruitment of neutrophils, macrophages, osteoclasts, lymphocytes, and other hematopoietic cells 17, 18, 19, 20, 21, 22. Re‐epithelialization is initiated in damaged epidermis and occurs when keratinocytes migrate to cover the injured tissue. Meanwhile, enzymatic activity from resident cells contributes to tissue histolysis in the damaged area 23, 24, 25, 26, 27, 28. During these initial events, inflammatory cells set the stage for local cell proliferation 18, 20, 21, 29 and in cases of appendage regeneration, resident cells transition to form a blastema, a recruited heterogeneous mass of cells that undergo morphogenesis to replace the missing tissue. Blastemal cells respond to signals from regenerating axons, Schwann cells, and the wound epidermis to remain in a proliferative state (reviewed in 30). Ultimately, blastema cells will differentiate into a functional replacement of the lost organ, and regeneration of many vertebrate tissues (i.e., fin, limb, digit, ear pinna, etc.) occurs through a blastema intermediate. Some complex tissues can regenerate without the formation of a blastema (e.g., heart, skin, bone fracture, etc.) 19, 31, 32 and in these cases resident cells that accumulated in response to injury differentiate directly into the replacement tissue.
Notably, epimorphic regeneration involves several processes that also occur during scar formation 19. Specifically, both epimorphic regeneration and fibrotic repair are necessarily triggered by injury and a hemostatic event 10, 33. Leukocytes infiltrate the damaged tissue and produce chemotactic signals for fibroblasts, endothelial cells, and axons, while also releasing proliferative signals for local fibroblasts (reviewed in 34). Re‐epithelialization seals the wounded area from the environment and new matrix is laid down to support cell infiltration in both injury responses 19, 35, 36, 37. Beyond similarities, important differences exist in the response to injury between fibrotic healing and tissue regeneration 38, 39, 40, 41 and ultimately, a regenerative microenvironment gives rise to a regeneration blastema, which can self‐regulate in size, pattern, and tissue complexity 42. With this in mind, a major goal of regenerative medicine should be to stimulate formation of a regeneration blastema 12. If the local microenvironment can be manipulated to shift the injury response, the self‐sustaining nature of a blastema should be capable of morphogenesis (Fig. 2A).
Figure 2.
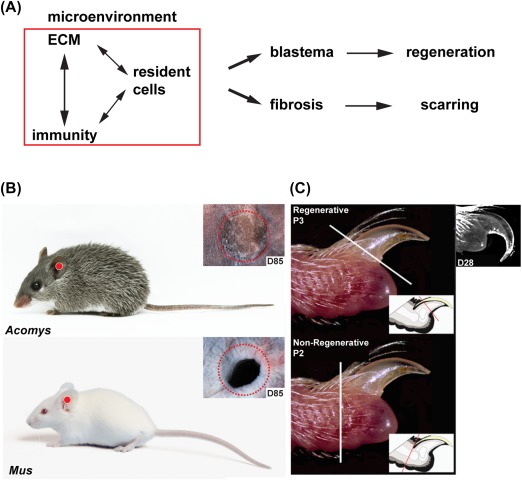
The injury microenvironment differentially regulates healing outcomes in mammalian models of regeneration and fibrotic repair. (A): The composition of the local microenvironment is determined by crosstalk among the ECM, cells of the immune system, and resident tissue progenitor cells. Comparative studies suggest a pro‐regenerative microenvironment stimulates blastema formation in lieu of fibrosis. Combinatorial therapies that target these three facets could dictate the future of regenerative medicine. Comparative models of natural regeneration and fibrotic repair include a 4‐mm ear punch assay (B) and digit tip amputations (C). These models inform how the local tissue environment functions during both tissue repair processes. (B) A 4‐mm biopsy punch through the external ear pinna of the African spiny mouse (Acomys) results in epimorphic regeneration, whereas the same injury through the ear pinna of outbred lab mice (Mus) results in a scar. (C) Amputation of the third phalangeal element (P3) results in epimorphic regeneration, whereas amputation through the second phalangeal element (P2) results in scar formation. Image in (C) adapted with permission from 43. Abbreviation: ECM, extracellular matrix.
Adult Mammalian Models of Epimorphic Regeneration
Bonafide examples of epimorphic regeneration in adult mammals do exist, and these can occur with or without formation of a blastema. Regeneration of the costal cartilage in mice and humans 44 and the annual replacement of deer antlers occur without a blastema intermediate, relying instead on dedicated stem cells from the perichondrium and periosteum respectively 45, 46. Likewise, full‐thickness skin regeneration occurs through direct differentiation and remodeling of granulation tissue similar to heart regeneration in zebrafish 9, 47, 48. In contrast, regeneration of ear pinna holes and distal digit tips proceed through a blastema before replacing the missing tissue 37, 39, 47, 49, 50, 51, 52, 53. An additional strength of mammalian regeneration models lies in the ability to directly compare the differential response to injury that leads to fibrotic repair or regeneration (Fig. 2A).
A biopsy punch through the external ear pinnae removes cartilage, connective tissue dermis, muscle, adipose tissue, epidermis, and hair follicles with associated sebaceous glands. Using this ear punch assay, researchers have determined that rabbits 50, 52, 54 and African spiny mice 39, 47, 53 are capable of epimorphic regeneration. In contrast, an identical injury undergoes fibrotic repair and scar formation, remaining as an open hole in other rodent species including Rattus norvegicus, Myomyscus brockmani, outbred Mus musculus, and inbred Mus such as the MRL/MpJ strain 1, 39, 47, 53, 55 (Figure 2B). Interestingly, anecdotal reports of ear pinna regeneration in cats, pikas, and hares suggest that additional mammalian species may exhibit enhanced regenerative ability including others to be discovered 1. It should be noted that the size of the hole in the ear punch assay matters in so far as holes 2mm or smaller can be closed with scar tissue. Thus, a ≥4‐mm ear punch assay provides a model to compare epimorphic regeneration and fibrotic repair in the same tissue across closely related species.
Amputation through the distal digit tip (third phalanx—P3) removes bone, bone marrow, connective tissue, dermis, epidermis, and nail (Fig. 2C). Regeneration of the distal digit tip has been documented in young humans 56, 57, 58, primates 59, neonatal mice 51 and adult mice 38, 49. Digit tip regeneration, however, is amputation level dependent. A distal amputation through the P3 element results in complete structural and functional replacement, whereas an amputation through the second phalangeal element (P2) results in scar formation and loss of original structure 37, 38, 49 (Fig. 2C). Comparing regeneration competent (P3) and incompetent (P2) amputations provides a model for studying fibrotic repair and regeneration within the same tissue, and thus the same individual.
How Do Mammals Establish a Regenerative Microenvironment?
Comparative models of epimorphic regeneration and fibrotic repair highlight a major question for regenerative medicine: is a regenerative response to injury driven by intrinsic cellular differences (i.e., response competency) or variation in extrinsic signals (i.e., signal variability)? While not new, this question has been addressed almost exclusively during cellular regeneration using skeletal muscle in the context of aging. Transplantation experiments with rodent skeletal muscle have addressed potential differences in intrinsic and extrinsic controls of regeneration 60, 61. For instance, muscle satellite cells in older rodents show a reduced ability to self‐renew when compared with those in young animals, and thus older animals lose muscle mass over time 62. This impaired self‐renewal in aged cells has been linked to over‐activation of the p38α/β MAPK pathway and this defect is sustained in older cells when they are transplanted into young muscle fibers 60. These and other studies suggest that a progressive decline in the capacity for satellite cell self‐renewal has an intrinsic component. However, when older satellite cells are cultured in vitro with serum from young mice, self‐renewal capacity seen in younger muscle is restored 63 suggesting that extrinsic inputs are capable of inducing cellular regeneration. Furthermore, transplantation of whole muscle fibers, which normally lose muscle mass, from old animals into young animals restores the ability of the older transplanted muscles to gain mass over time 61 supporting extrinsic regulation of the stem cell niche. Together, these studies suggest that intrinsic and extrinsic factors are required to regulate progenitor contribution to tissue regeneration. Do intrinsic or extrinsic differences in cellular phenotype equally contribute to the differential response to injury when comparing epimorphic regeneration and fibrotic repair?
Resident Cells: Strict Maintenance of Cell Proliferation
Evidence from newts and salamanders suggests that blastema cells are derived from within 500 μm of the initial wound site 64, 65 supporting local cells as essential building blocks to re‐create lost tissue. In mammalian digits, lineage tracing studies have shown that blastema cells are derived from lineage‐restricted cells insofar as lineages can be accurately labeled 66. What is less clear is the extent to which blastema cells represent a population of quiescent tissue‐specific progenitor cells or local cells that have de‐differentiated from mature tissue. Tissue‐specific progenitor cells have been discovered throughout many mammalian tissues 67, and these cells are essential for repairing damaged muscle 68, 69, 70, cartilage 44, and bone 71, 72. On the other hand, stem cells with varying plasticity have been isolated from a wide array of adult tissues 73, 74 suggesting that inherently plastic cells like connective tissue fibroblasts may contribute a majority of the starting material for tissue regeneration 75. The tricky problem is identifying stable markers for inherently plastic cells 66, 76, 77. Nonetheless, the available evidence supports the idea that multiple lineage‐restricted cell types contribute to the blastema during complex tissue regeneration rather than formation of a pluripotent cell mass 75, 78, 79, 80, 81, 82. Coupled with existing abilities to reprogram cells, perhaps the more pertinent question is not one of origin, but figuring out how resident cells are activated to proliferate and how proliferation is controlled.
The ability for cells to re‐enter the cell cycle and commit to cell proliferation is paramount for blastema formation. Early thymidine tracing experiments in newt limbs showed that the only actively dividing cells prior to amputation were found in the epidermis and blood, and neither population contributed to the actively dividing cells of the blastema 64. Surveying adult tissues from mammalian regeneration models demonstrates that the number of actively dividing cells is negligible prior to injury, specifically when compared with tissues with high rates of cell turnover (e.g., small intestine) in the same animal (Fig. 3). Interestingly, actively dividing cells are found throughout uninjured adult tissues in the highly regenerative axolotl (Fig. 3). These data suggest quiescent cell populations in adult mammalian tissues are re‐activated to undergo cell division after injury and make up the bulk of the blastema rather than existing as a permanent slow‐cycling population.
Figure 3.
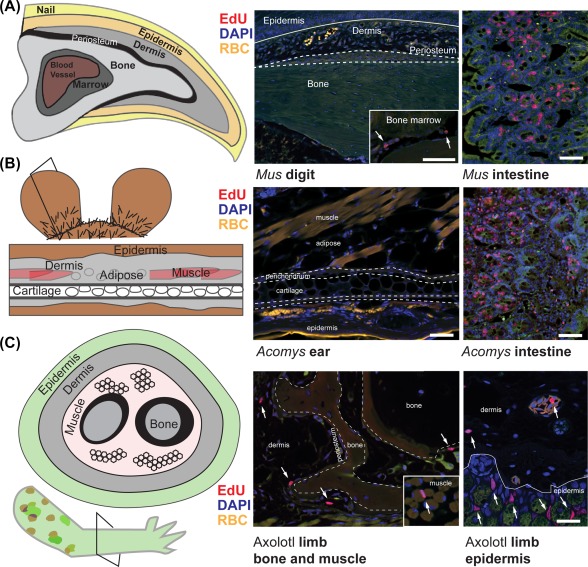
Tissue analysis of the S‐phase marker EdU in uninjured mouse digit tip, spiny mouse ear pinna and salamander limb. (A): Proximal to distal (longitudinal) section of the third phalangeal element (P3) from an adult Mus shows very few EdU+ (red) cells which are restricted to the bone marrow (inset, white arrows). EdU staining of adult Mus small intestine is displayed for comparison. (B): Proximal to distal (longitudinal section) section through an adult spiny mouse ear pinna shows very few EdU+ (red) cells. EdU staining of adult spiny mouse small intestine is displayed for comparison. (C): In contrast to mammalian regeneration models, cross‐section of an uninjured 1‐year old axolotl limb shows many EdU+ cells (white arrows) in the dermis, periosteum, connective tissue surrounding muscle (inset), epidermis, and blood vessels. Scale bars = 50 μm. EdU+ cells = red; DAPI (4′,6‐Diamidino‐2‐phenylindole) nuclear stain = blue; autofluorescent RBC = yellow. Abbreviation: RBC, red blood cells.
An outstanding hypothesis concerns the degree to which resident cells from regenerating species may be primed and possess a lower barrier to cell‐cycle re‐entry or whether differential regulation of tumor suppressor proteins regulates activation of progenitor cells 83. Experiments with newt and mammalian myotubes in culture showed that newt, but not mammalian myotubes, could re‐enter the cell cycle in response to serum 84. Compared to the newt muscle cells in culture, mammalian primary and established myoblast cultures displayed a differential regulation of several tumor suppressor genes and differential control of certain tumor suppressor proteins, including retinoblastoma protein (pRb) and p53, to initiate cell cycle arrest 84, 85, 86, 87. Inactivating pRb alone in mammalian C2C12 myotubes failed to initiate cell cycle re‐entry 84. However, combined inactivation of both pRb and the Ink4a isoform Arf was able to drive C2C12 de‐differentiation, and inactivation of p53 was able to induce cell‐cycle re‐entry in primary myotubes 86, 88. Additionally, tumor suppressors such as Arf are specific to mammalian cells 89 and expression of human Arf in regenerating axolotl or zebrafish tissues inhibits regeneration 84, 90, 91.
Mammalian models of epimorphic regeneration shed further light on cell cycle control in vivo, and suggest differential regulation of tumor suppressor pathways during regenerative or fibrotic healing 39. In ear punches from regenerating and nonregenerating mammals, injury appears to similarly activate resident cells to re‐enter the cell cycle 39. In the early stages of regeneration and fibrotic repair, perichondrial cells and dermal fibroblasts are positive for cell cycle proteins Ki67, pRb, pHH3, and they incorporate EdU. However, during fibrotic repair in Mus, resident cells lose these cell proliferation markers coincident with nuclear localization of the tumor suppressors, p21 and p27 39. In contrast, during regeneration in Acomys, resident cells continue to proliferate as a blastema forms to produce new tissue to close ear holes. Importantly, nuclear localization of p21 and p27 did not occur in blastema cells, although it was observed in cells proximal to the injury. This local control of cell proliferation appears key to ensuring new tissue is appropriately produced and does not lead to unregulated growth. In line with observations from the spiny mouse ear, differential regulation of tumor suppressor pathways also occurs when comparing digit cells from regenerative and nonregenerative amputations. When cells were cultured from the nonregenerative P2 region of the Mus digit they divided more slowly and upregulated Cdkn2a (p16) and Cdkn2b (p15) when compared with P3 cells on plastic 92. Together, these studies support regulation of tumor suppressor pathway activation, whether extrinsically or intrinsically controlled, as a key feature of cell proliferation during epimorphic regeneration.
Extrinsic Inputs and Developmental Signals
Although injury promotes formation of a new bone callus in nonregenerative P2 amputations, and new cartilage nodules in the nonregenerative Mus ear pinna, in both cases these new tissues are usually disorganized and mispatterned 39, 47, 53, 71. With respect to bone and cartilage, these observations support direct differentiation of periosteal and perichondrial cells into new tissue, but underscores the failure to organize a multicellular response typical of epimorphic regeneration. A failure to simulate epimorphosis in these mammalian tissues may result from different signaling cues produced in response to injury. Fibroblasts isolated from the P3 and P2 region of mouse digits display distinct differences in their ability to signal to the epidermis; P3 fibroblasts induce formation of a mucosal‐type epidermis while P2 cells induce a stratified epidermis in keratinocyte cocultures 93. These findings support fibroblast identity as a key component of regeneration, similar to how fibroblasts from different dermal layers or anatomical regions differentially contribute to wound healing 66, 76. Similarly, in most mammals the ability to regenerate hair follicles in full‐thickness skin wounds is lost during ontogeny and can be traced to changes in fibroblast signaling patterns, specifically a downregulation of Wnt signals and an upregulation of inflammatory cytokines in mature fibroblast populations 94. However, studies using the Mus digit tip model suggest that fibroblasts from normally nonregenerative amputation levels maintain the ability to respond to appropriate patterning signals as both P2 and P3 digit cells show similar expression of BMP receptors and SMAD activation 95, and addition of exogenous BMP2 and BMP7 in vivo is able to induce skeletal patterning in nonregenerative amputations of neonatal and adult mice 40, 96, 97, 98. While these studies suggest patterning can be rescued if the proper signaling environment is restored, there appears to be a temporal component as well. Treatment with BMP2 induces skeletal patterning of adult P2 only at specific time points after amputation 98. BMP2 treatment 9 days after adult P2 injury induces segmental regeneration and patterned bone formation, but BMP2 treatment 24 days after injury fails to induce a cellular response. A re‐injury to the P2 bone 24 days after injury rescues the ability of cells to respond to BMP2 and create new patterned bone 98. Together, these examples support specific and appropriately timed molecular signals within the regenerative microenvironment as important extrinsic cues regulating the injury response (Fig. 2A).
Inflammation and Immunity
Prior to blastema formation, cellular interactions with the immune environment partially determine if a blastema will form 18, 20, 21, 29, 99, 100, 101. In adult tissues, the immediate response to injury includes the infiltration of a diverse subset of leukocytes including neutrophils, macrophages, and T cells. While intense interest has focused on macrophages and T‐lymphocytes during mammalian wound healing 102, 103, 104, 105, their importance in blastema‐based regeneration is just beginning to be understood 18, 20, 21, 29. Macrophages are essential to the mammalian regeneration process and depletion studies in the spiny mouse ear show that loss of early macrophage populations delays regeneration until macrophages re‐populate the tissue to stimulate blastema formation 18. Similarly, depletion studies in the mouse digit tip show loss of early macrophage populations inhibits re‐epithelialization, tissue histolysis, blastema formation and differentiation 20. Rescuing re‐epithelialization can promote blastema initiation, but without macrophages the blastema regresses and fails to differentiate into new tissue 20. Together with studies in salamanders 21 and zebrafish 29, 106, these studies show that macrophages are a key initiator of blastema‐based regeneration in adult vertebrates. Identifying specific macrophage signaling products will be essential for determining how local immunity regulates the tissue microenvironment to promote regeneration.
While the critical macrophage signaling pathways are not yet known, timed depletions studies show that early infiltrating macrophage populations are essential for regeneration 20, 21, 29. The timing of macrophage infiltration is closely linked to their phenotype and activity. Studies during skeletal muscle repair 102, liver repair 107, spinal cord injury 108, myocardial infarction 109, and skin healing 110, 111 document changes in macrophage phenotype with time. In these wound healing models, the initial macrophage populations are dominated by pro‐inflammatory (classically activated M1) phenotypes while later repair stages are dominated by anti‐inflammatory (alternatively activated M2) resolving phenotypes 111. Characteristic differences include cytokine expression and metabolic activity between these macrophage subtypes 112, 113. These observations of changing macrophage phenotypes in wound healing models combined with the timed depletion studies in regeneration models would lead one to hypothesize that initial pro‐inflammatory signals from macrophages are required for mammalian tissue regeneration.
Comparing the early microenvironments of spiny mice and laboratory mice ear injuries has revealed important similarities and differences in the initial immune environment. First, the initial pro‐inflammatory responses are distinct: early stages of regeneration are dominated by high NADPH‐oxidase derived reactive oxygen species (ROS) production and low myeloperoxidase activity, whereas early stages proceeding fibrotic repair exhibit low NADPH‐oxidase derived ROS production and high myeloperoxidase activity 18. Second, compared with fibrotic repair in lab mice, the local inflammatory cytokine milieu is unique during regeneration in the spiny mouse (unpublished data) and digit tip studies provide evidence that specific cytokines, for example, oncostatin M, tumor necrosis factor alpha (TNFα), and stimulation of lymphotoxin beta receptor, support blastema formation in vivo and in vitro 114, 115. These cytokines are produced by macrophages, Schwann cells 114, the epidermis 116, and potentially T cells (unpublished data). Last, macrophages exhibit a distinct spatial distribution in vivo. CD206+ cells (M2) infiltrate scar‐forming and regenerating wounds early after injury whereas CD86+ cells (M1) appeared to primarily infiltrate the injury microenvironment in scar‐forming wounds 18. Together, these observations demonstrate distinct inflammatory environments are present prior to a regenerative or fibrotic healing outcome and point toward the need for a more detailed phenotyping to determine the functional requirement for specific macrophages and cytokine pathways during regeneration.
Extracellular Matrix Composition
In addition to developmental signaling pathways and the immune environment, blastema organization, maintenance and morphogenesis is governed by the extracellular matrix (ECM). Far from a passive support structure, the ECM is capable of providing cues for cell migration, proliferation and differentiation (reviewed in 117, 118). A surge in bioengineered scaffolds has capitalized on the instructive abilities of the ECM and de‐cellularized scaffolds derived from natural tissues can promote tissue regeneration in bone, cartilage, muscle, and skin (reviewed in 119). Synthetic and natural scaffolds have been modified to mimic conditions observed during embryonic development using developmental matrix molecules to guide cell differentiation. For instance, scaffolds rich in the ECM fiber tenascin‐C have been shown to promote chondrocyte differentiation in vitro 120, 121. While this can be beneficial in certain contexts, other studies have shown that treating cartilage explants with tenascin‐C can increase the loss of proteoglycans and exacerbate tissue histolysis 122, suggesting that trying to direct cellular activity by enriching for single matrix components can have antagonistic outcomes as well. These studies underscore the need for understanding the composition and dynamic nature of naturally occurring regenerative matrices.
The mammalian regenerative matrix environment appears to superficially resemble the ECM present during embryonic development which is composed of matrix proteins that stimulate proliferation (tenascin‐C), cell migration (fibronectin) and are more easily remodeled (collagen type 3) 28, 39, 47, 115, 123, 124, 125, 126. Low resolution comparison of the regenerative ECM suggests this matrix environment may be conserved across regenerative species 39, 47, 123, 124, 127, 128. In adult tissues, tenascin‐C and fibronectin contribute to the periosteum, perichondrium, tendon, and muscle where they surround progenitor cells 39, 124, 127, and perhaps play a role in supporting the progenitor cell niche 129, 130. In vitro studies have demonstrated a direct positive effect of tenascin‐C and fibronectin on salamander and mouse myotube proliferation 124. Despite parity across regeneration models, comparative studies in mammalian models of regeneration and scarring redirect our understanding of regenerative matrices. Healing Ear punches in Acomys and Mus revealed similar levels of tenascin‐C during the first fifteen days post amputation suggesting that traditional components of a regenerative ECM are deposited during fibrotic repair as well 39. Despite early expression and deposition of tenascin‐C during fibrotic healing in Mus, proportionally higher expression of Col1a1, Col3a1 and chondroitin sulfate proteoglycans occurred prior to, and during scar formation. In contrast, persistently high levels of tenascin‐C were maintained throughout the regeneration process in Acomys, and proportionately higher levels of matrix fibers involved in nerve guidance proceeded blastema formation 39. These comparisons indicate that relative composition and timing of ECM deposition is critical toward healing outcomes.
Far from a static structure, the ECM is dynamic, undergoing active remodeling as some components are digested and others deposited. Functional studies in newts have shown that active remodeling of the ECM by matrix remodeling enzymes is required for blastema formation 27. Although remodeling enzymes such as matrix metalloproteinases 9 and 13 (MMP9 and MMP13) are upregulated in response to injury regardless of the outcome, timing of expression is, again, a prominent difference between regeneration and scarring. In regenerating systems, these enzymes remain significantly upregulated during blastema formation and morphogenesis when compared with fibrotic repair 25, 27, 39, 124, 131. While the exact role of remodeling enzymes during regeneration remains obscure, a few studies suggest that degraded ECM components, as well as growth factors and chemokines released by histolysis, are able to promote cell migration 132, 133. As we continue to discover the extent to which the ECM facilitates regeneration, a more comprehensive understanding of its temporal composition will guide attempts to design next‐generation bio‐reactive scaffolds for clinical applications in vivo and ex vivo.
Current Regenerative Approaches to Complex Tissue Injury
The clinical challenge of regenerating organs and complex, multi‐tissue injuries is immense. In cases of severe organ damage or dysfunction, the gold standard remains whole organ transplantation from a matched donor 134, 135 and in cases of severe tissue damage, autologous tissue transplant. While the long‐term prognosis for transplant patients can be quite good, especially for certain procedures, the availability of suitable organs for transplant is relatively low and even when a match is made, there is still a need for lifetime immunosuppressive therapy 135. It is no surprise then that there is a ravenous appetite from the patient and clinical communities to see basic scientific advances transformed into useful regenerative therapies. Unfortunately, although advances in regenerative biology continue to be made, very few regenerative technologies have either proved their efficacy in humans or significantly outperformed surgical alternatives 134, 136. The failure to create new, useful, and tractable regenerative therapies for complex tissue trauma or loss can be traced to inherent design problems with ex vivo constructs (i.e., vascular integration, innervation, mechanical properties, etc.), an incomplete understanding of how tissues naturally develop and regenerate, and a poor understanding of how the immune system positively/negatively interacts with local cells to regulate tissue morphogenesis during natural instances of tissue regeneration.
Current approaches in regenerative medicine attempt to leverage cell‐based therapies, scaffolds (natural or synthetic) or a combination of cells and scaffolds to stimulate tissue repair in vivo or build tissues ex vivo for implantation. Targeted drug delivery is a third approach, typified by the application of demineralized bone matrix (DBM) or isolated bioactive molecules to regenerate skeletal injuries 137. Where damage or dysfunction can be traced to a single cell type or simple tissue (e.g., epidermis, corneal epithelium) available stem‐cell therapies have shown encouraging success in humans. For instance, hematopoietic stem cell transplants (HSCT) have been used to successfully treat a range of leukemias and lymphoproliferative disorders by regenerating a patient's blood system following chemotherapy 138, although like whole organ transplantation, HSCT does carry long‐term complications 139. Limbal stem cell transplants have proven successful in regenerating the corneal epithelium in humans 140 and a recent report has demonstrated proof‐of‐principle success using transgenically altered autologous keratinocytes to treat junctional epidermolysis bullosa through re‐population of a significant portion of the patient's epidermis 141, 142. In the case of critical size defects in the skeleton (craniofacial, axial or appendicular), autogenous bone grafts are the gold standard and most widely used clinical treatment 137. However, the osteoinductive properties of demineralized bone matrix (DBM) 143, 144 which have been used clinically for over thirty years, have made DBM products, alone or in combination with autogenous grafts, a viable option to stimulate the intrinsic regenerative properties of bone 137. Importantly, attempts to bridge a critically‐sized fibular defect in humans using the osteoinductive molecule BMP7 (OP1) were not as effective as DBM after one year 145. This and other attempts to stimulate regeneration using single bioactive molecules reinforces the importance of designing therapies with the complexity of natural tissue development in mind where a multitude of signaling pathways act synergistically and antagonistically to precisely control tissue morphogenesis.
As injuries increase in complexity involving multiple cell types and tissue compartments, regenerative therapies have seen far less clinical success. Replacement of so‐called hollow organs (e.g., bladder, blood vessels, urethra, etc.) with ex vivo constructs from scaffolds alone or cell‐seeded scaffolds have achieved a measure of reported success 146, 147, but these approaches are used infrequently and present short and long‐term complications 134, 148. Among complex tissues, the skin is injured more frequently than any other organ. The primary approach to treating acute wounds (small or large) and burns remains the split‐thickness autograft 149. In an attempt to create an artificial skin substitute that could stimulate natural tissue regeneration, Integra was developed as a composite of naturally derived collagen, chondroitin 6‐sulphate and Silastic 150. Integra is used in two stages. First, when placed in a wound or burn, the collagen compartment encourages cell infiltration and neo‐dermis production as it is slowly degraded during healing. Subsequently, the Silastic is removed and replaced by a meshed epidermal autograft which restores the epidermal skin compartment 150. Integra represents one of the true successes for regenerative engineering and has inspired many competing products to facilitate skin regeneration and repair, including de‐cellularized extracellular matrices from human and animal dermis 119, 149, 151. Despite the success of artificial skin and natural scaffolds in treating burns and full‐thickness skin wounds, comparative human studies show a long‐term clinical outcome no different from split‐thickness skin grafts 152, 153. As outlined above, the available human data supports the need to refine (or reconstruct) current regenerative therapies in light of our expanding knowledge of natural regeneration in animal models. Appreciating the dynamic interaction of cells, the ECM and immune system in the context of adult tissue should provide guidance on optimizing new approaches to regenerative medicine.
Future Clinical Practices: Balancing Immunity, Cell Proliferation, and Fibrosis
Continued study using comparative mammalian models of regeneration will undoubtedly deepen our understanding of how the injury microenvironment stimulates blastema formation in lieu of fibrotic repair. The challenge is how to translate basic biological understanding into meaningful clinical advances that manifest into tractable regenerative therapies. In our view, a key problem to solve in the treatment of complex tissue injuries is the extent to which the local microenvironment can be altered to induce blastema formation. If injured tissue can transition to a blastema then the self‐sustaining nature of the blastema will facilitate morphogenesis and regeneration. This same principle can be applied to tissue injuries where a blastema is not required (e.g., skin, heart, lung, etc.). In these cases, the local microenvironment still controls the cellular response to injury and as such, can polarize the healing response.
Current tissue engineering techniques modulate the tissue microenvironment and promote endogenous repair through the creation of scaffolds (reviewed in 119), and through the modulation of the immune response (reviewed in 154). It is notable that these current therapies are designed to target individual tissues, that is, collagen‐glycosaminoglycan scaffolds for skin (Integra) 150, 155, silk scaffolds for ligament 156, 157, organic scaffolds for bone 158, 159 or hyaluronic acid infused scaffolds for cartilage repair 160. Similarly, immune‐modulatory therapies are designed to target single specific arms of the immune cell response; that is, targeted dampening of interleukin 1 beta pro‐inflammatory cascades to improve healing in skin 161, 162, 163, calvarial bone defects 164, and arthritic cartilage 165. Yet the ultimate goal of regenerative medicine is to stimulate integrated multi‐tissue regeneration, and the extent to which current scaffold designs and immune targeting drugs induce complex tissue regeneration is unknown. Is it possible to create scaffolds that target multiple tissues with a therapy that stimulates evolution of the immune cell response across healing phases? What would these therapies look like? We can benefit from comparative analyses of regeneration and scarring to quickly guide new therapy designs and help answer these questions. For example, we have learned from comparative analysis in mammals that regenerating systems deposit ECM fibers for nerve guidance 39, that BMP treatment is more successful when combined with factors that mobilize periosteal cells 98, and that early inflammatory events promoting high NADPH‐oxidase derived ROS production and lower H2O2 stimulate blastema formation 39. As engineering techniques become more sophisticated enabling duel growth factor delivery 166, guided nerve growth 167, and cytokine capture 168, it should be possible to design dynamic scaffolds to simultaneously incorporate early control of the immune system, guidance of nerve growth, and timed release of growth factors to promote blastema formation.
Conclusion
Although many yearn for a single factor or scaffold that will magically jumpstart a regenerative response, it has yet to be discovered, and is more likely not to exist at all. Instead, the complexity of establishing and maintaining a controlled regenerative response in adult tissue will require combinatorial therapies delivered in a progressive timeline to manipulate the local tissue microenvironment. Future goals for designing new approaches in regenerative medicine should embrace an understanding of how to integrate immune modulation, scaffold design, and controlled cell cycle activation to enhance intrinsic regenerative abilities in humans. In our view, a productive path to discovering new regenerative therapies will arise from comparative studies of regeneration and fibrotic repair, and active engagement between biologists, engineers, and clinician‐scientists.
Author Contributions
J.S. and A.S.: wrote and approved the final manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Note Added in Proof
This article was published online on 22 December 2017. Minor edits have been made that do not affect data. This notice is included in the online and print versions to indicate that both have been corrected 29 January 2018.
Acknowledgments
A.W.S. is supported by the National Science Foundation (NSF) and the Office for International Science and Engineering (OISE) (IOS‐1353713) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS—NIH) (R01AR070313).
Contributor Information
Jennifer Simkin, Email: jennifer.simkin@uky.edu.
Ashley W. Seifert, Email: awseifert@uky.edu.
References
- 1. Goss RJ. Prospects for regeneration in man. Clin Orthop Relat Res 1980;151:270–282. [PubMed] [Google Scholar]
- 2. Takahashi K, Tanabe K, Ohnuki M et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872. [DOI] [PubMed] [Google Scholar]
- 3. Pagliuca FW, Millman JR, Gurtler M et al. Generation of functional human pancreatic beta cells in vitro. Cell 2014;159:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Derby B. Printing and prototyping of tissues and scaffolds. Science 2012;338:921–926. [DOI] [PubMed] [Google Scholar]
- 5. Oberpenning F, Meng J, Yoo JJ et al. De novo reconstitution of a functional mammalian urinary bladder by tissue engineering. Nat Biotechnol 1999;17:149–155. [DOI] [PubMed] [Google Scholar]
- 6. Seifert AW, Monaghan JR, Smith MD et al. The influence of fundamental traits on mechanisms controlling appendage regeneration. Biol Rev 2012;87:330–345. [DOI] [PubMed] [Google Scholar]
- 7. Bely AE, Nyberg KG. Evolution of animal regeneration: Re‐emergence of a field. Trends Ecol Evol 2010;25:161–170. [DOI] [PubMed] [Google Scholar]
- 8. Wagner GP, Misof BY. Evolutionary modification of regenerative capability in vertebrates: A comparative study on teleost pectoral fin regeneration. J Exp Zool 1992;261:62–78. [DOI] [PubMed] [Google Scholar]
- 9. Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188–2190. [DOI] [PubMed] [Google Scholar]
- 10. Carlson BM. Principles of Regenerative Biology. Burlington, NJ: Academic Press, 2007. [Google Scholar]
- 11. Carlson BM. Some principles of regeneration in mammalian systems. Anat Rec B New Anat 2005;287:4–13. [DOI] [PubMed] [Google Scholar]
- 12. Seifert AW, Muneoka K. The blastema and epimorphic regeneration in mammals. Dev Biol 2017;433:190–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adzick NS, Longaker MT. Fetal wound healing. New York: Elsevier Publishing Company, 1992. [Google Scholar]
- 14. Bullard KM, Longaker MT, Lorenz HP. Fetal wound healing: Current biology. World J Surg 2003;27:54–61. [DOI] [PubMed] [Google Scholar]
- 15. Blanpain C, Fuchs E. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 2009;10:207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morgan TH. Regeneration, by Thomas Hunt Morgan. New York: The Macmillan Company, 1901. [Google Scholar]
- 17. Li L, Yan B, Shi Y‐Q et al. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem 2012;287:25353–25360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simkin J, Gawriluk TR, Gensel JC et al. Macrophages are necessary for epimorphic regeneration in African spiny mice. eLife 2017;6:e24623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seifert AW, Monaghan JR, Voss SR et al. Skin regeneration in adult axolotls: A blueprint for scar‐free healing in vertebrates. PLoS One 2012;7:e32875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simkin J, Sammarco MC, Marrero L et al. Macrophages are required to coordinate mouse digit tip regeneration. Development 2017;144:3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proc Natil Acad Sci USA 2013;110:9415–9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goode RP. The regeneration of limbs in adult anurans. Development 1967;18:259–267. [PubMed] [Google Scholar]
- 23. Kato T, Miyazaki K, Shimizu‐Nishikawa K et al. Unique expression patterns of matrix metalloproteinases in regenerating newt limbs. Dev Dyn 2003;226:366–376. [DOI] [PubMed] [Google Scholar]
- 24. Yang EV, Bryant SV. Developmental regulation of a matrix metalloproteinase during regeneration of axolotl appendages. Dev Biol 1994;166:696–703. [DOI] [PubMed] [Google Scholar]
- 25. Grillo HC, Lapie CM, Dresden MH et al. Collagenolytic activity in regenerating forelimbs of the adult newt (Triturus viridescens). Dev Biol 1968;17:571–583. [DOI] [PubMed] [Google Scholar]
- 26. Yang EV, Gardiner DM, Carlson MR et al. Expression of Mmp‐9 and related matrix metalloproteinase genes during axolotl limb regeneration. Dev Dyn 1999;216:2–9. [DOI] [PubMed] [Google Scholar]
- 27. Vinarsky V, Atkinson DL, Stevenson TJ et al. Normal newt limb regeneration requires matrix metalloproteinase function. Dev Biol 2005;279:86–98. [DOI] [PubMed] [Google Scholar]
- 28. Simkin J, Sammarco MC, Dawson LA et al. Epidermal closure regulates histolysis during mammalian (Mus) digit regeneration. Regeneration (Oxf) 2015;2:106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petrie TA, Strand NS, Yang CT et al. Macrophages modulate adult zebrafish tail fin regeneration. Development 2014;141:2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stocum DL, Cameron JA. Looking proximally and distally: 100 years of limb regeneration and beyond. Dev Dyn 2011;240:943–968. [DOI] [PubMed] [Google Scholar]
- 31. Yin VP, Lepilina A, Smith A et al. Regulation of zebrafish heart regeneration by miR‐133. Dev Biol 2012;365:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. I‐Aql ZA, Alagl A, Graves D et al. Molecular mechanisms controlling bone formation during fracture healing and distraction osteogenesis. J Dent Res 2008;87:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singer AJ, Clark RA. Cutaneous wound healing. New Engl J Med 1999;341:738–746. [DOI] [PubMed] [Google Scholar]
- 34. Shaw TJ, Martin P. Wound repair: A showcase for cell plasticity and migration. Curr Opin Cell Biol 2016;42:29–37. [DOI] [PubMed] [Google Scholar]
- 35. Seifert AW, Maden M. New insights into vertebrate skin regeneration. Int Rev Cell Mol Biol 2014;310:129–169. [DOI] [PubMed] [Google Scholar]
- 36. Xue M, Jackson CJ. Extracellular matrix reorganization during wound healing and its impact on abnormal scarring. Adv Wound Care (New Rochelle) 2015;4:119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernando WA, Leininger E, Simkin J et al. Wound healing and blastema formation in regenerating digit tips of adult mice. Dev Biol 2011;350:301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neufeld DA, Zhao W. Phalangeal regrowth in rodents: Postamputational bone regrowth depends upon the level of amputation. Prog Clin Biol Res 1993;383a:243–252. [PubMed] [Google Scholar]
- 39. Gawriluk TR, Simkin J, Thompson KL et al. Comparative analysis of ear‐hole closure identifies epimorphic regeneration as a discrete trait in mammals. Nat Commun 2016;7:11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu L, Han M, Yan M et al. BMP signaling induces digit regeneration in neonatal mice. Development 2010;137:551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reginelli AD, Wang YQ, Sassoon D et al. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development 1995;121:1065–1076. [DOI] [PubMed] [Google Scholar]
- 42. Stocum DL. The urodele limb regeneration blastema: Determination and organization of the morphogenetic field. Differentiation 1984;27:13–28. [DOI] [PubMed] [Google Scholar]
- 43. Simkin J, Han M, Yu L et al. The mouse digit tip: From wound healing to regeneration. Methods Mol Biol 2013;1037:419–435. [DOI] [PubMed] [Google Scholar]
- 44. Srour MK, Fogel JL, Yamaguchi KT et al. Natural large‐scale regeneration of rib cartilage in a mouse model. J Bone Miner Res 2015;30:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kierdorf U, Li C, Price JS. Improbable appendages: Deer antler renewal as a unique case of mammalian regeneration. Semin Cell Dev Biol 2009;20:535–542. [DOI] [PubMed] [Google Scholar]
- 46. Kierdorf U, Kierdorf H, Szuwart T. Deer antler regeneration: Cells, concepts, and controversies. J Morphol 2007;268:726–738. [DOI] [PubMed] [Google Scholar]
- 47. Seifert AW, Kiama SG, Seifert MG et al. Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 2012;489:561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brant JO, Yoon JH, Polvadore T et al. Cellular events during scar‐free skin regeneration in the spiny mouse, Acomys. Wound Repair Regen 2016;24:75–88. [DOI] [PubMed] [Google Scholar]
- 49. Borgens RB. Mice regrow the tips of their foretoes. Science 1982;217:747–750. [DOI] [PubMed] [Google Scholar]
- 50. Goss RJ, Grimes LN. Tissue interactions in regeneration of rabbit ear holes. Am Zool 1972;12:151–157. [Google Scholar]
- 51. Han M, Yang X, Lee J et al. Development and regeneration of the neonatal digit tip in mice. Dev Biol 2008;315:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Joseph J, Dyson M. Tissue replacement in the rabbit's ear. Br J Surg 1966;53:372–380. [DOI] [PubMed] [Google Scholar]
- 53. Matias Santos D, Rita AM, Casanellas I et al. Ear wound regeneration in the African spiny mouse Acomys cahirinus. Regeneration 2016;3:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goss RJ, Grimes LN. Epidermal downgrowths in regenerating rabbit ear holes. J Morphol 1975;146:533–542. [DOI] [PubMed] [Google Scholar]
- 55. Williams‐Boyce PK, Daniel JC. Jr. Comparison of ear tissue regeneration in mammals. J Anat 1986;149:55–63. [PMC free article] [PubMed] [Google Scholar]
- 56. McKim LH. Regeneration of the distal phalanx. Can Med Assoc J 1932;26:549–550. [PMC free article] [PubMed] [Google Scholar]
- 57. Illingworth CM. Trapped fingers and amputated finger tips in children. J Pediatr Surg 1974;9:853–858. [DOI] [PubMed] [Google Scholar]
- 58. Douglas BS. Conservative management of guillotine amputation of the finger in children. J Paediatr Child Health 1972;8:86–89. [DOI] [PubMed] [Google Scholar]
- 59. Singer M, Weckesser EC, Geraudie J et al. Open finger tip healing and replacement after distal amputation in rhesus monkey with comparison to limb regeneration in lower vertebrates. Anat Embryol 1987;177:29–36. [DOI] [PubMed] [Google Scholar]
- 60. Bernet JD, Doles JD, Hall JK et al. p38 MAPK signaling underlies a cell‐autonomous loss of stem cell self‐renewal in skeletal muscle of aged mice. Nat Med 2014;20:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carlson B, Faulkner J. Muscle transplantation between young and old rats: Age of host determines recovery. Am J Physiol Cell Physiol 1989;256:C1262–C1266. [DOI] [PubMed] [Google Scholar]
- 62. Barani AE, Durieux A‐C, Sabido O et al. Age‐related changes in the mitotic and metabolic characteristics of muscle‐derived cells. J Appl Physiol 2003;95:2089–2098. [DOI] [PubMed] [Google Scholar]
- 63. Carlson ME, Conboy IM. Loss of stem cell regenerative capacity within aged niches. Aging Cell 2007;6:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hay ED, Fischman DA. Origin of the blastema in regenerating limbs of the newt Triturus viridescens. An autoradiographic study using tritiated thymidine to follow cell proliferation and migration. Dev Biol 1961;3:26–59. [DOI] [PubMed] [Google Scholar]
- 65. Currie JD, Kawaguchi A, Traspas RM et al. Live imaging of axolotl digit regeneration reveals spatiotemporal choreography of diverse connective tissue progenitor pools. Dev Cell 2016;39:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rinkevich Y, Walmsley GG, Hu MS et al. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 2015;348:aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Young HE, Duplaa C, Romero‐Ramos M et al. Adult reserve stem cells and their potential for tissue engineering. Cell Biochem Biophys 2004;40:1–80. [DOI] [PubMed] [Google Scholar]
- 68. Lepper C, Partridge TA, Fan C‐M. An absolute requirement for Pax7‐positive satellite cells in acute injury‐induced skeletal muscle regeneration. Development 2011;138:3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sambasivan R, Yao R, Kissenpfennig A et al. Pax7‐expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 2011;138:3647–3656. [DOI] [PubMed] [Google Scholar]
- 70. Bischoff R. Regeneration of single skeletal muscle fibers in vitro. Anat Rec 1975;182:215–235. [DOI] [PubMed] [Google Scholar]
- 71. Dawson LA, Simkin J, Sauque M et al. Analogous cellular contribution and healing mechanisms following digit amputation and phalangeal fracture in mice. Regen (Oxf) 2016;3:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J Bone Miner Res 2009;24:274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Abreu SC, Antunes MA, Xisto DG et al. Bone marrow, adipose, and lung tissue‐derived murine mesenchymal stromal cells release different mediators and differentially affect airway and lung parenchyma in experimental asthma. Stem Cells Translational Medicine 2017;6:1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pachon‐Pena G, Yu G, Tucker A et al. Stromal stem cells from adipose tissue and bone marrow of age‐matched female donors display distinct immunophenotypic profiles. J Cell Physiol 2011;226:843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev Biol 1986;116:256–260. [DOI] [PubMed] [Google Scholar]
- 76. Driskell RR, Lichtenberger BM, Hoste E et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 2013;504:277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Plikus MV, Guerrero‐Juarez CF, Ito M et al. Regeneration of fat cells from myofibroblasts during wound healing. Science 2017;355:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kragl M, Knapp D, Nacu E et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 2009;460:60–65. [DOI] [PubMed] [Google Scholar]
- 79. Tu S, Johnson SL. Fate restriction in the growing and regenerating zebrafish fin. Dev Cell 2011;20:725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Stewart S, Stankunas K. Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev Biol 2012;365:339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lehoczky JA, Robert B, Tabin CJ. Mouse digit tip regeneration is mediated by fate‐restricted progenitor cells. Proc Natl Acad Sci USA 2011;108:20609–20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rinkevich Y, Lindau P, Ueno H et al. Germ‐layer and lineage‐restricted stem/progenitors regenerate the mouse digit tip. Nature 2011;476:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pomerantz JH, Blau HM. Tumor suppressors: Enhancers or suppressors of regeneration? Development 2013;140:2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tanaka EM, Gann AA, Gates PB et al. Newt myotubes reenter the cell cycle by phosphorylation of the retinoblastoma protein. J Cell Biol 1997;136:155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yun MH, Gates PB, Brockes JP. Regulation of p53 is critical for vertebrate limb regeneration. Proc Natl Acad Sci USA 2013;110:17392–17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang H, Loof S, Borg P et al. Turning terminally differentiated skeletal muscle cells into regenerative progenitors. Nat Commun 2015;6:7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lööf S, Straube WL, Drechsel D et al. Plasticity of mammalian myotubes upon stimulation with a thrombin‐activated serum factor. Cell Cycle 2007;6:1096–1101. [DOI] [PubMed] [Google Scholar]
- 88. Pajcini KV, Corbel SY, Sage J et al. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 2010;7:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kim S‐H, Mitchell M, Fujii H et al. Absence of p16INK4a and truncation of ARF tumor suppressors in chickens. Proc Natl Acad Sci USA 2003;100:211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hesse RG, Kouklis GK, Ahituv N et al. The human ARF tumor suppressor senses blastema activity and suppresses epimorphic tissue regeneration. eLife 2015;4:e07702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Khattak S, Schuez M, Richter T et al. Germline transgenic methods for tracking cells and testing gene function during regeneration in the axolotl. Stem Cell Reports 2013;1:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lynch KM, Ahsan T. Modulating the physical microenvironment to study regenerative processes in vitro using cells from mouse phalangeal elements. Tissue Eng Part A 2013;19:1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wu Y, Wang K, Karapetyan A et al. Connective tissue fibroblast properties are position‐dependent during mouse digit tip regeneration. PLoS One 2013;8:e54764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rognoni E, Gomez C, Pisco AO et al. Inhibition of beta‐catenin signalling in dermal fibroblasts enhances hair follicle regeneration during wound healing. Development 2016;143:2522–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lynch KM, Ahsan T. Correlating the effects of bone morphogenic protein to secreted soluble factors from fibroblasts and mesenchymal stem cells in regulating regenerative processes in vitro. Tissue Eng Part A 2014;20:3122–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yu L, Han M, Yan M et al. BMP2 induces segment‐specific skeletal regeneration from digit and limb amputations by establishing a new endochondral ossification center. Dev Biol 2012;372:263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee J, Marrero L, Yu L et al. SDF‐1alpha/CXCR4 signaling mediates digit tip regeneration promoted by BMP‐2. Dev Biol 2013;382:98–109. [DOI] [PubMed] [Google Scholar]
- 98. Dawson LA, Yu L, Yan M et al. The periosteal requirement and temporal dynamics of BMP2‐induced middle phalanx regeneration in the adult mouse. Regeneration 2017;4:140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Love NR. Amputation‐induced reactive oxygen species (ROS) are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol 2013;15:222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hameed LS, Berg DA, Belnoue L et al. Environmental changes in oxygen tension reveal ROS‐dependent neurogenesis and regeneration in the adult newt brain. eLife 2015;4:e08422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang Q, Wang Y, Man L et al. Reactive oxygen species generated from skeletal muscles are required for gecko tail regeneration. Sci Rep 2016;6:20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Novak ML, Weinheimer‐Haus EM, Koh TJ. Macrophage activation and skeletal muscle healing following traumatic injury. J Pathol 2014;232:344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mirza R, DiPietro LA, Koh TJ. Selective and specific macrophage ablation is detrimental to wound healing in mice. Am J Pathol 2009;175:2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Arnold L, Henry A, Poron F et al. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 2007;204:1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Alexander KA, Chang MK, Maylin ER et al. Osteal macrophages promote in vivo intramembranous bone healing in a mouse tibial injury model. J Bone Miner Res 2011;26:1517–1532. [DOI] [PubMed] [Google Scholar]
- 106. Kyritsis N, Kizil C, Zocher S et al. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 2012;338:1353–1356. [DOI] [PubMed] [Google Scholar]
- 107. Duffield JS, Forbes SJ, Constandinou CM et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest 2005;115:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gensel JC, Zhang B. Macrophage activation and its role in repair and pathology after spinal cord injury. Brain Res 2015;1619:1–11. [DOI] [PubMed] [Google Scholar]
- 109. Nahrendorf M, Swirski FK, Aikawa E et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 2007;204:3037–3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lucas T, Waisman A, Ranjan R et al. Differential roles of macrophages in diverse phases of skin repair. J Immunol 2010;184:3964–3977. [DOI] [PubMed] [Google Scholar]
- 111. Varga T, Mounier R, Horvath A et al. Highly dynamic transcriptional signature of distinct macrophage subsets during sterile inflammation, resolution, and tissue repair. J Immunol 2016;196:4771–4782. [DOI] [PubMed] [Google Scholar]
- 112. Mantovani A, Biswas SK, Galdiero MR et al. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 2013;229:176–185. [DOI] [PubMed] [Google Scholar]
- 113. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008;8:958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Johnston AP, Yuzwa SA, Carr MJ et al. Dedifferentiated Schwann cell precursors secreting paracrine factors are required for regeneration of the mammalian digit tip. Cell Stem Cell 2016;19:433–448. [DOI] [PubMed] [Google Scholar]
- 115. Marrero L, Simkin J, Sammarco M et al. Fibroblast reticular cells engineer a blastema extracellular network during digit tip regeneration in mice. Regeneration 2017;4:69–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Piran R, Lee SH, Kuss P et al. PAR2 regulates regeneration, transdifferentiation, and death. Cell Death Dis 2016;7:e2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Watt FM, Huck WTS. Role of the extracellular matrix in regulating stem cell fate. Nat Rev Mol Cell Biol 2013;14:467–473. [DOI] [PubMed] [Google Scholar]
- 118. Hynes RO. The extracellular matrix: Not just pretty fibrils. Science 2009;326:1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Swinehart IT, Badylak SF. Extracellular matrix bioscaffolds in tissue remodeling and morphogenesis. Dev Dyn 2016;245:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mackie EJ, Thesleff I, Chiquet‐Ehrismann R. Tenascin is associated with chondrogenic and osteogenic differentiation in vivo and promotes chondrogenesis in vitro. J Cell Biol 1987;105:2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mackie EJ, Murphy LI. The role of tenascin‐C and related glycoproteins in early chondrogenesis. Microsc Res Tech 1998;43:102. [DOI] [PubMed] [Google Scholar]
- 122. Patel L, Sun W, Glasson SS et al. Tenascin‐C induces inflammatory mediators and matrix degradation in osteoarthritic cartilage. BMC Musculoskeletal Disord 2011;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature 2003;423:876–881. [DOI] [PubMed] [Google Scholar]
- 124. Calve S, Odelberg SJ, Simon HG. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev Biol 2010;344:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem 2009;78:929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Giblin SP, Midwood KS. Tenascin‐C: Form versus function. Cell Adh Migr 2015;9:48–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nace JD, Tassava RA. Examination of fibronectin distribution and its sources in the regenerating newt limb by immunocytochemistry and in situ hybridization. Dev Dyn 1995;202:153–164. [DOI] [PubMed] [Google Scholar]
- 128. Onda H, Poulin ML, Tassava RA et al. Characterization of a newt tenascin cDNA and localization of tenascin mRNA during newt limb regeneration by in situ hybridization. Dev Biol 1991;148:219–232. [DOI] [PubMed] [Google Scholar]
- 129. Chiquet‐Ehrismann R, Orend G, Chiquet M et al. Tenascins in stem cell niches. Matrix 2014;37:112–123. [DOI] [PubMed] [Google Scholar]
- 130. Singh P, Schwarzbauer JE. Fibronectin and stem cell differentiation – lessons from chondrogenesis. J Cell Sci 2012;125:3703–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Miyazaki K, Uchiyama K, Imokawa Y et al. Cloning and characterization of cDNAs for matrix metalloproteinases of regenerating newt limbs. Proc Natl Acad Sci USA 1996;93:6819–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tang Y, Wu X, Lei W et al. TGF‐β1–induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med 2009;15:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Agrawal V, Johnson SA, Reing J et al. Epimorphic regeneration approach to tissue replacement in adult mammals. Proc Natl Acad Sci USA 2010;107:3351–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Orlando G, Wood KJ, De Coppi P et al. Regenerative medicine as applied to general surgery. Ann Surg 2012;255:867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Orlando G, Soker S, Stratta RJ et al. Will regenerative medicine replace transplantation? Cold Spring Harb Perspect Med 2013;3:a015693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Robey P. “Mesenchymal stem cells”: Fact or fiction, and implications in their therapeutic use. F1000Res 2017;6:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gruskin E, Doll BA, Futrell FW et al. Demineralized bone matrix in bone repair: History and use. Adv Drug Deliv Rev 2012;64:1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Gratwohl A, Pasquini MC, Aljurf M et al. One million haemopoietic stem‐cell transplants: A retrospective observational study. Lancet Haematol 2015;2:e91–100. [DOI] [PubMed] [Google Scholar]
- 139. Inamoto Y, Lee SJ. Late effects of blood and marrow transplantation. Haematologica 2017;102:614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rama P, Matuska S, Paganoni G et al. Limbal stem‐cell therapy and long‐term corneal regeneration. N Engl J Med 2010;363:147–155. [DOI] [PubMed] [Google Scholar]
- 141. Hirsch T, Rothoeft T, Teig N et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature 2017;551:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Mavilio F, Pellegrini G, Ferrari S et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med 2006;12:1397–1402. [DOI] [PubMed] [Google Scholar]
- 143. Urist MR. Bone: Formation by autoinduction. Science 1965;150:893–899. [DOI] [PubMed] [Google Scholar]
- 144. Sampath TK, Reddi AH. Dissociative extraction and reconstitution of extracellular matrix components involved in local bone differentiation. Proc Natl Acad Sci USA 1981;78:7599–7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Geesink RG, Hoefnagels NH, Bulstra SK. Osteogenic activity of OP‐1 bone morphogenetic protein (BMP‐7) in a human fibular defect. J Bone Joint Surg Br 1999;81:710–718. [DOI] [PubMed] [Google Scholar]
- 146. Atala A, Bauer SB, Soker S et al. Tissue‐engineered autologous bladders for patients needing cystoplasty. Lancet 2006;367:1241–1246. [DOI] [PubMed] [Google Scholar]
- 147. Palminteri E, Berdondini E, Fusco F et al. Long‐term results of small intestinal submucosa graft in bulbar urethral reconstruction. Urology 2012;79:695–701. [DOI] [PubMed] [Google Scholar]
- 148. Mangera A, Chapple CR. Tissue engineering in urethral reconstruction–an update. Asian J Androl 2013;15:89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Schulz JT, 3rd , Tompkins RG, Burke JF. Artificial skin. Annu Rev Med 2000;51:231–244. [DOI] [PubMed] [Google Scholar]
- 150. Burke JF, Yannas IV, Quinby WC, Jr et al. Successful use of a physiologically acceptable artificial skin in the treatment of extensive burn injury. Ann Surg 1981;194:413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yannas IV. Emerging rules for inducing organ regeneration. Biomaterials 2013;34:321–330. [DOI] [PubMed] [Google Scholar]
- 152. Lagus H, Sarlomo‐Rikala M, Bohling T et al. Prospective study on burns treated with Integra(R), a cellulose sponge and split thickness skin graft: Comparative clinical and histological study–randomized controlled trial. Burns 2013;39:1577–1587. [DOI] [PubMed] [Google Scholar]
- 153. Philandrianos C, Andrac‐Meyer L, Mordon S et al. Comparison of five dermal substitutes in full‐thickness skin wound healing in a porcine model. Burns 2012;38:820–829. [DOI] [PubMed] [Google Scholar]
- 154. Zakrzewski JL, van den Brink MRM, Hubbell JA. Overcoming immunological barriers in regenerative medicine. Nat Biotechnol 2014;32:786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Yannas IV, Tzeranis DS, Harley BA et al. Biologically active collagen‐based scaffolds: Advances in processing and characterization. Philos Trans Ser A Math Phys Eng Sci 2010;368:2123–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ran J, Hu Y, Le H et al. Ectopic tissue engineered ligament with silk collagen scaffold for ACL regeneration: A preliminary study. Acta Biomater 2017;53:307–317. [DOI] [PubMed] [Google Scholar]
- 157. Fan H, Liu H, Wong EJ et al. In vivo study of anterior cruciate ligament regeneration using mesenchymal stem cells and silk scaffold. Biomaterials 2008;29:3324–3337. [DOI] [PubMed] [Google Scholar]
- 158. Keller L, Regiel‐Futyra A, Gimeno M et al. Chitosan‐based nanocomposites for the repair of bone defects. Nanomedicine 2017;13:2231–2240. [DOI] [PubMed] [Google Scholar]
- 159. Petite H, Viateau V, Bensaid W et al. Tissue‐engineered bone regeneration. Nat Biotechnol 2000;18:959. [DOI] [PubMed] [Google Scholar]
- 160. Huey DJ, Hu JC, Athanasiou KA. Unlike bone, cartilage regeneration remains elusive. Science 2012;338:917–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Mirza RE, Fang MM, Ennis WJ et al. Blocking interleukin‐1beta induces a healing‐associated wound macrophage phenotype and improves healing in type 2 diabetes. Diabetes 2013;62:2579–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Mirza RE, Fang MM, Novak ML et al. Macrophage PPARgamma and impaired wound healing in type 2 diabetes. J Pathol 2015;236:433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Thomay AA, Daley JM, Sabo E et al. Disruption of interleukin‐1 signaling improves the quality of wound healing. Am J Pathol 2009;174:2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Martino MM, Maruyama K, Kuhn GA et al. Inhibition of IL‐1R1/MyD88 signalling promotes mesenchymal stem cell‐driven tissue regeneration. Nat Commun 2016;7:11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Giacomelli R, Ruscitti P, Alvaro S et al. IL‐1beta at the crossroad between rheumatoid arthritis and type 2 diabetes: May we kill two birds with one stone? Exp Rev Clin Immunol 2016;12:849–855. [DOI] [PubMed] [Google Scholar]
- 166. Richardson TP, Peters MC, Ennett AB et al. Polymeric system for dual growth factor delivery. Nat Biotechnol 2001;19:1029–1034. [DOI] [PubMed] [Google Scholar]
- 167. Curley JL, Moore MJ. Facile micropatterning of dual hydrogel systems for 3D models of neurite outgrowth. J Biomed Mater Res Part A 2011;99:532–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lohmann N, Schirmer L, Atallah P et al. Glycosaminoglycan‐based hydrogels capture inflammatory chemokines and rescue defective wound healing in mice. Sci Transl Med 2017;9:pii: eaai9044. [DOI] [PubMed] [Google Scholar]


