Abstract
Paired box 7 (PAX7) gene regulates the conversion of muscle satellite cells into myogenic cells and participates in multi-step processes in myogenesis. Expression levels of PAX7 are decisive for its regulatory function. Previous reports revealed that PAX7 were responsible for the developmental traits of muscle. The relationship of the PAX7 promoter variants and livestock phenotypic traits has not been fully elucidated. We detected a novel 10-bp insertion/deletion (indel) polymorphism in the bovine PAX7 promoter and revealed that the indel altered the binding of the transcriptional factor ZNF219. Luciferase reporter assay showed that deletion-deletion (Del-Del) genotype of the PAX7 gene showed 2.79-fold higher promoter activity than the insertion-insertion (Ins-Ins) genotype (P < 0.05), and ZNF219 overexpression significantly diminished the luciferase activity in Ins-Ins groups. Moreover, the expression of PAX7 and its down-stream genes were detected in fetal skeletal muscle of cattle with different PAX7 genotypes, where the Del-Del genotype also displayed high expression levels. Statistical association analysis demonstrated that this indel had significant effects on early growth traits in cattle. These findings provide a complete overview of the function of the PAX7 10-bp variant, which may have potential as a genetic marker for marker-assisted selection in improving economically significant traits of cattle.
Introduction
Cattle phenotypes of economic significance, such as growth rates, carcass, and meat quality, have attracted much attention from breeders in the beef cattle industry. Such phenotypes constitute a multifactorial background that arises from interactions between environmental and genetic factors1. In marker-assisted selection programs, the efficiency of selection is associated with the identification of candidate genes, as well as the exploration of genome-wide variation markers that are responsible for the complex quantitative traits2. The establishment of genetic association approach provides a unique opportunity to study genotype-phenotype relationships3. Therefore, the integration of screening for variations and gene association programs can improve assessments of the genetic effects of candidate genes, thus providing promising materials for successful genetic selection in cattle breeding.
Muscle satellite cells, known as muscle-specific committed progenitors, are located under the basal lamina of muscle fibres and play important roles in the growth, maintenance, homeostasis, repair, and regeneration of skeletal muscle4–6. Paired box 7 (PAX7) gene, a member of the paired box gene family, is the marker gene of myogenic satellite cells7. PAX7 is specifically expressed in quiescent, activated, and proliferating satellite cells, and PAX7-expressing satellite cells are indispensable for the postnatal muscle development and regeneration after the injury of skeletal muscle, however, their regenerative function can not be performed by other endogenous cell types. In addition, the deficiency of the myogenic cells in skeletal muscle of the PAX7 knockout mice demonstrated that the PAX7 was required for specification of the satellite cell lineage8. As a transcription factor, PAX7 affects the conversion of the myogenic progenitors entry into skeletal myoblast program by regulating the down-stream myogenic determination genes9,10. For example, Kumar et al.11 revealed that PAX7 can block premature differentiation and maintain stem cell status of quiescent satellite cells by inducing the expression of the inhibitor of DNA binding 2 (ID2) and ID3. Previous studies reported that PAX7 knockout mice exhibited muscle malformations, and the body sizes of the homozygous deletion mice were significantly smaller as compared with the wildtype and heterozygous counterparts12,13. Therefore, given its fundamental roles in satellite cell differentiation and muscle development, we hypothesized that the PAX7 could be considered as a selection marker gene for muscle involving phenotypic traits.
Previous association studies have identified substantial genetic variations in the coding regions of candidate genes that are responsible for economical traits of cattle14–16. In addition, regulatory promoter elements contribute to the promotion and suppression of gene transcription, thus, the exploration of the variants in the promoter region is also of the utmost importance. Promoter polymorphisms may change the binding sites of transcriptional factors that could completely alter the inducibility of the promoter17 or significantly influence the transcriptional activity18. The promoter sequence of PAX7 comprises multiple regulatory elements19. In humans, a (CCT)n microsatellite polymorphism, located in the specificity protein 1 (SP1) binding site, was detected in the PAX7 promoter, and the (CCT)11 variant showed significantly higher transcriptional efficiency in comparison to the (CCT)8 and (CCT)10 genotypes20. Similarly, a G/C single nucleotide polymorphism (SNP) and a (CA)n microsatellite variant have been identified in the promoter region of PAX3 gene (a paralogue of PAX7 gene)21. Our previous studies have reported that seven SNPs in the exon and intron regions of PAX7 were significantly associated with growth traits of Chinese cattle breeds22,23. However, to our knowledge, there have been no reports about the PAX7 promoter polymorphisms and their functional effects on economically significant traits in cattle.
The bovine PAX7 gene is located on chromosome 2 and underlies the quantitative trait locus (QTL) for body weight24. The objectives of this study were to detect genetic polymorphism in the PAX7 promoter among five Chinese indigenous cattle breeds, determine the relationship of the variation with growth traits, and further explore the biological effects of the PAX7 gene with different allelic variants. These results may provide evidence for further investigation on exploiting the significant polymorphisms as molecular markers in cattle breeding programs.
Results
Identification of a novel PAX7 10-bp indel polymorphism
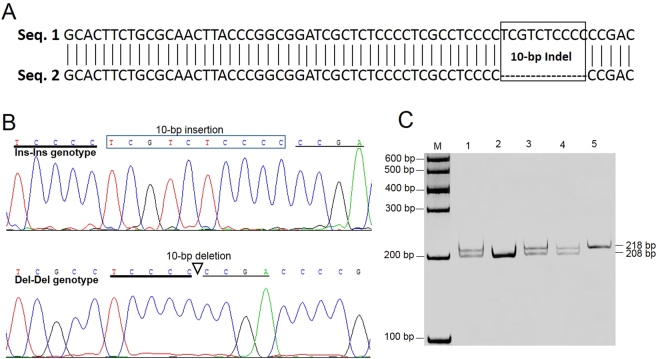
We sequenced the 1868-bp promoter region of the bovine PAX7 gene and revealed a 10-bp (TCGTCTCCCC) indel polymorphism between nucleotide position −633 and −643 (Fig. 1A,B). Using the P2 primer (Supplementary Table S1), the 10-bp indel variant was genotyped by polyacrylamide gel electrophoresis (PAGE) in five Chinese cattle breeds. As illustrated in Fig. 1C, the genotypic patterns were determined with a 208-bp fragment for the Del-Del genotype, a 218-bp fragment for the Ins-Ins genotype, and 208-bp and 218-bp fragments for the Ins-Del genotype. This polymorphic sequence has been deposited in the National Center for Biotechnology Information (NCBI accession number: ss 831883063).
Figure 1.
Identification of the 10-bp indel polymorphism in the bovine PAX7 gene. (A) BLAST results for two different partial promoter sequences of PAX7. Seq. 1 was the sequences from NCBI (accession no. NC_007300); Seq. 2 was obtained by DNA sequencing of the bovine PAX7 promoter with the 10-bp deletion. (B) Sequencing maps of the 10-bp indel polymorphism. Top: the rectangular frame contains the inserted nucleotides; bottom: the triangle shows the position of the 10-bp deletion. (C) Genotyping patterns of the 10-bp indel polymorphism obtained by polyacrylamide gels electrophoresis. Lanes 1, 3, 4: Ins-Del genotype; lane 2: Del-Del genotype; lane 5: Ins-Ins genotype.
Genetic parameter analysis of the indel in five cattle breeds
Distributions of genotypic and allelic frequencies of the 10-bp indel, as well as its genetic diversity, were given in Table 1. The Del was the predominant allele in Nanyang (NY) breed, however, the Ins allele was predominant in Jiaxian (JX), Qinchuan (QC), Luxi (LX), and Chinese Caoyuan CY) populations. Interestingly, the results of a χ2 test showed that the 10-bp indel was in Hardy-Weinberg equilibrium (P > 0.05) in the NY breed, whereas the deviations from the Hardy-Weinberg equilibrium (P < 0.01) were detected in JX, QC, LX, and CY populations, which may be attributed to the selection and the population history, specifically, the degree of selection, small population size, and population mixture. The values of genetic heterozygosity (He) and effective allele numbers (Ne) were 0.460–0.550 and 1.853–2.000, respectively. According to the classification of polymorphism information content (PIC), all of the five cattle breeds exhibited moderate polymorphism (0.250 < PIC < 0.500) at the 10-bp indel locus, suggesting that the five cattle breeds may undergo the similarly continued selection pressure in evolutionary history.
Table 1.
Genetic parameters of the PAX7 10-bp indel polymorphism in five cattle breeds.
| Breeds | Sex | Genotypic frequency | Allelic frequency | He | Ne | PIC | HWE (P)1 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Ins-Ins | Ins-Del | Del-Del | Ins | Del | ||||||
| NY (220) | Female | 0.245 | 0.505 | 0.250 | 0.498 | 0.502 | 0.500 | 2.000 | 0.375 | > 0.05a |
| JX (395) | Female | 0.400 | 0.215 | 0.385 | 0.508 | 0.492 | 0.500 | 2.000 | 0.375 | < 0.01 |
| QC (224) | Female | 0.513 | 0.254 | 0.232 | 0.641 | 0.359 | 0.460 | 1.853 | 0.354 | < 0.01 |
| LX (161) | Female | 0.317 | 0.385 | 0.298 | 0.509 | 0.491 | 0.500 | 1.999 | 0.375 | < 0.01 |
| CY (233) | Female | 0.468 | 0.129 | 0.403 | 0.532 | 0.468 | 0.498 | 1.992 | 0.374 | < 0.01 |
1P-value with “a” indicates that the breed was in Hardy-Weinberg equilibrium (HWE). He, heterozygosity. Ne, effective allele numbers. PIC, polymorphism information content.
Association analysis of the indel with cattle growth traits
As shown in Table 2, the 10-bp indel of the PAX7 gene revealed significant associations with body weight (P = 0.0004), body height (P = 0.0248), body length (P = 0.0043), heart girth (P = 0.0001), hucklebone width (P = 0.0128) and average daily gain (P = 0.0006) in NY cattle with an age of 6 months, where the Del-Del genotype showed higher values than the Ins-Ins and Ins-Del genotypes. Regarding the individuals aged 12 months old, this indel was associated with body weight (P = 0.014), body length (P = 0.0035), and heart girth (P = 0.0001). Consistently, the cattle with Del-Del genotype also had significantly improved traits in comparison to those with Ins-Ins and Ins-Del genotypes. However, no associations were found between the indel locus and growth traits of cattle with ages of 18 and 24 months (P > 0.05, Supplementary Table S2).
Table 2.
Association analysis of the PAX7 10-bp indel with growth traits in NY cattle aged 6/12 months.
| Age | Growth traits | PAX7-10-bp indel genotypes (Mean ± SE) | P-value | ||
|---|---|---|---|---|---|
| Ins-Ins(n = 54) | Ins-Del(n = 111) | Del-Del(n = 55) | |||
| 6 months | Body weight, kg | 144.00 ± 4.61B | 159.24 ± 2.44A,b | 167.48 ± 3.32A,a | 0.0004 |
| Body height, cm | 102.71 ± 1.31b | 106.42 ± 0.69a | 106.96 ± 0.94a | 0.0248 | |
| Body length, cm | 101.14 ± 1.46B | 106.06 ± 0.77A | 107.07 ± 1.05A | 0.0043 | |
| Heart girth, cm | 121.86 ± 1.78B | 128.82 ± 0.94A,b | 132.15 ± 1.28A,a | 0.0001 | |
| Hucklebone width, cm | 17.64 ± 0.34B | 18.21 ± 0.18b | 18.87 ± 0.25A,a | 0.0128 | |
| Average daily gain, kg | 0.64 ± 0.02B | 0.72 ± 0.01A | 0.76 ± 0.02A | 0.0006 | |
| 12 months | Body weight, kg | 210.71 ± 6.04B | 220.86 ± 3.19b | 232.11 ± 4.35A,a | 0.0140 |
| Body length, cm | 112.07 ± 1.91B,b | 116.88 ± 1.01a | 120.22 ± 1.38A | 0.0035 | |
| Heart girth, cm | 134.50 ± 1.89 C | 140.62 ± 1.00B | 145.44 ± 1.36A | 0.0001 | |
Within the same row, values with different lowercase letters (a,b) differ significantly at P < 0.05, and those with different uppercase letters (A, B, C) differ significantly at P < 0.01.
Influence of the indel on the binding of the zinc finger transcription factor 219 (ZNF219)
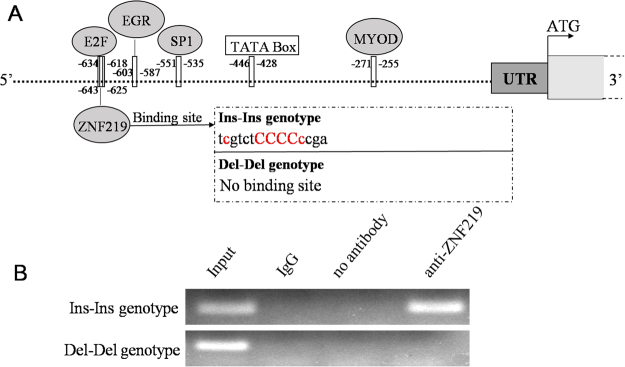
Given the significant association between the PAX7 10-bp indel and cattle growth traits, the mechanism that contributed to these phenotypic variations remained to be determined. We hypothesized that the observed differences in genotype may reflect differences in the activity of the PAX7 promoter. Therefore, the PAX7 promoter region adjacent to the 10-bp indel was analyzed, and the results were shown in Fig. 2A. The binding sites of many transcriptional factors (TFs), such as myoblast determination protein (MYOD), SP1, early growth response (EGR), and E2F, were found in the analyzed sequences. Specifically, in the presence of the Ins-Ins genotype, the binding site of ZNF219 is created, while this site was lost in the presence of the Del-Del genotype (Fig. 2A). Correspondingly, previous studies have reported that ZNF219 functioned as a transcriptional regulator and was a sequence-specific DNA binding protein in nucleus25. In addition, we used chromatin immunoprecipitation (ChIP) assay to further test the binding of the ZNF219 to the 10-bp indel locus. The results indicated that ZNF219 bound to the Ins-Ins genotype of the PAX7 promoter, while no binding was detected in the Del-Del genotype (Fig. 2B).
Figure 2.
The PAX7 10-bp indel affects the binding site of ZNF219. (A) Schematic representation of the putative binding sites of TFs in the presence of Ins-Ins or Del-Del genotypes of the 10-bp indel polymorphism, as predicted by MatInspector Release professional 8.0.4. The sequence of the core binding site of ZNF219 transcription factor was shown, with ci-values of >60 in red and the core sequence in red capitals. Grey ellipses: predicted TFs; white box: TATA box element; dark-gray box: 5′-untranslated region (UTR); light gray box: coding region. (B) ChIP assay of ZNF219 binding to the PAX7 gene promoter with Ins-Ins or Del-Del genotypes. The images were cropped from the same gel, and the full-length gel was shown in Supplementary Fig. S1.
Detection of the PAX7 promoter activity
In order to confirm whether the 10-bp indel was located in the core active region of the PAX7 promoter, pGL3-pro1, pGL3-pro2, pGL3-pro3, pGL3-pro4, and pGL3-pro5 plasmids were co-transfected with pRL-TK into C2C12 cells, respectively. The results showed that the pGL3-pro2 vector yielded a significantly stronger fluorescence than the other vectors (P < 0.01 or P < 0.05 Fig. 3A). Notably, the pGL3-pro1 construct, which contained a larger portion of the PAX7 promoter, displayed significantly lower promoter activity than the pGL3-pro2 construct (P < 0.05), suggesting that there may be an inhibitor binding site existing in the promoter region −1603~−1279 of the PAX7 gene. In fact, the 10-bp Ins-Ins genotype was included in the pGL3-pro2 plasmid, denoted as pGL3-pro2InsIns. Similarly, the vector pGL3-pro2DelDel (including 10-bp Del-Del genotype) was constructed, and its transcription activity was also detected in C2C12 cells. As shown in Fig. 3B, the Del-Del genotype showed 2.79-fold higher luciferase activity than the Ins-Ins genotype (P < 0.05). Luciferase reporter assay was performed to determine whether ZNF219 regulated PAX7 promoter activity (Fig. 3B). The results demonstrated that overexpression of ZNF219 significantly diminished the luciferase activity in the Ins-Ins groups (P < 0.05), while ZNF219 overexpression had no effects on the promoter activity in the Del-Del groups (P > 0.05).
Figure 3.
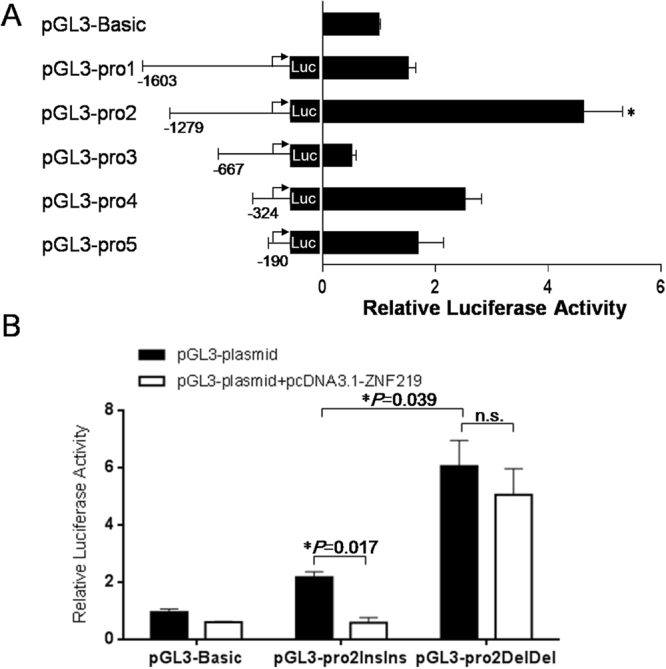
PAX7 promoter activity modulated by the 10-bp indel polymorphism. (A) Luciferase activity in C2C12 cells transfected with recombinant plasmids containing serial promoter fragments (pGL3-pro1, pGL3-pro2, pGL3-pro3, pGL3-pro4, and pGL3-pro5) of the Pax7 promoter. The backbone vector pGL3-Basic was used as a negative control. PpGL3-pro2 vs. pGL3-pro1 = 0.0118, PpGL3-pro2 vs. pGL3-pro3 = 0.0042, PpGL3-pro2 vs. pGL3-pro4 = 0.0495, PpGL3-pro2 vs. pGL3-pro5 = 0.0241. (B) Luciferase activity in C2C12 cells transfected with recombinant plasmids containing the Ins-Ins or Del-Del genotype. pcDNA3.1-ZNF219 was transiently co-transfected into C2C12 cells together with the reporter vectors. Each experiment was repeated at least three times. The data were mean ± standard error (S.E.) of the normalized luciferase activity. *P < 0.05 represents a significant difference.
Influence of the indel on gene expression in the muscle of cattle
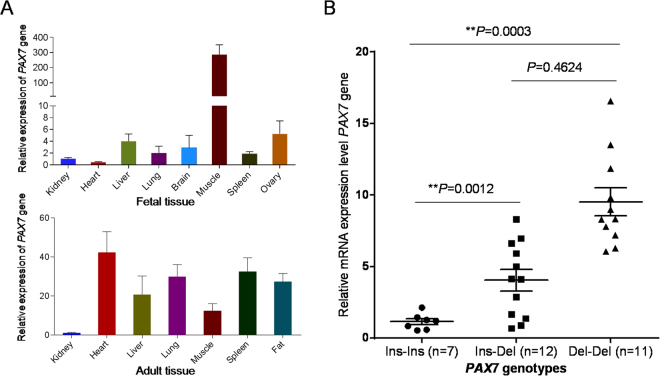
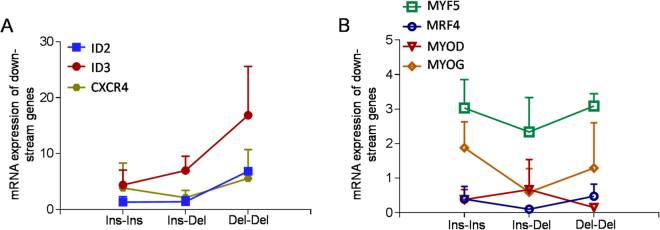
Expression pattern analysis revealed that the PAX7 gene was highly expressed in the skeletal muscle of cattle embryos, whereas it displayed lower expression levels in other tissues (Fig. 4A). Thus, we collected the muscle tissues from 30 cattle embryos, in which 7, 12, and 11 individuals were genotyped as Ins-Ins, Ins-Del, and Del-Del, respectively. Next we detected the PAX7 expression in the skeletal muscle of fetal cattle with different genotypes, and found that the Del-Del genotype showed 8.30-fold higher expression than the Ins-Ins group (P < 0.01, Fig. 4B). In addition, the expression of the PAX7 down-stream genes were also investigated in the different genotypic groups. As shown in Fig. 5A, we found the same tendency of the higher expression of ID2, ID3 and chemokine receptor 4 (CXCR4) in the individuals with the Del-Del genotype than the Ins-Ins and Ins-Del groups, which was in parallel with enhancive PAX7 expression. However, the other four genes, myogenic factor 5 (MYF5), myogenic regulatory factor 4 (MRF4), myoblast determination protein (MYOD) and myogenin (MYOG), displayed non-significant differences among the three genotypic individuals (Fig. 5B).
Figure 4.
Expression levels of the PAX7 gene in tissues of QC cattle. (A) Expression profiling of the PAX7 gene in fetal and adult tissues. (B) Expression levels of the PAX7 gene in skeletal muscles in cattle embryo with different genotypes. The relative expression levels of PAX7 normalized that of bovine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown for all animals with the genotypes Ins-Ins (n = 7), Ins-Del (n = 12), and Del-Del (n = 11). The data are mean ± S.E. for each genotype group. **P < 0.01 represents a highly significant difference.
Figure 5.
Expression patterns of the PAX7 down-stream genes in fetal skeletal muscles of cattle with different genotypes. (A) Relative expression levels of genes expressed in myogenic satellite cells: ID2, ID3, and CXCR4. (B) Relative expression levels of genes involved in myogenic determination and differentiation: MYF5, MRF4, MYOD, and MYOG. The mRNA expressions were normalized to GAPDH. Each experiment was repeated at least three times. The data are the mean ± S.E. for each genotype group.
Discussion
The identification of DNA markers that contributes to phenotypic evolution is a powerful aid to animal breeding, importantly, thorough functional research of the causative markers should be explored before their application in breeding projects26. In cattle, previous studies have identified substantial indel variants acrossing the whole genome in different species27,28, however, there have been few attempts to associate indels with functional effects. In this study, we firstly detected a novel 10-bp indel variant in the promoter region of the PAX7 gene in five Chinese cattle breeds, and further investigated the genetic diversity, characterization of genetic properties, and the determination of functional impacts.
Paired box (PAX) genes, termed as the PAX gene family, encode for specific DNA-binding transcription factors, which play critical roles in early development29. The PAX family includes nine genes that are assigned to four subgroups based on conservation of their primary structure: (1) PAX1/PAX9, (2) PAX2/PAX5/PAX8, (3) PAX3/PAX7, and (4) PAX4/PAX630. PAX3 and PAX7 participate in the specification, survival, proliferation and self-renewal of muscle progenitor cells, and are required for skeletal muscle development13. The sequences of PAX genes are evolutionarily conserved among various species, suggesting that the genetic mutation may cause remarkable phenotypic alteration, even result in serious disease. For example, SNPs of the PAX3 and PAX7 have been associated with human alveolar rhabdomyosarcoma31. Our previous study reported that the SNPs in PAX3 (promoter, exon and intron) and PAX7 (exons and introns) showed significant associations with growth traits in Chinese cattle22,23,32. In the present study, we further detected the 10-bp indel in the PAX7 promoter, and revealed that the indel significantly affect growth traits by regulating expression effects. Similarly, Zhang et al.33 found a novel 31-bp indel in intron 3 of the PAX7 gene that was associated with chicken growth, carcass and meat quality traits. These results may be attributed to the location of the PAX7 gene in QTL region that is linked to economic traits24.
The promoter, where multiple cis-transcriptional elements exist in, plays critical roles in modulating gene expression. Polymorphisms in the promoter region of a gene may affect its gene product by altering transcription factor binding sites or RNA stability34. Numerous studies have revealed that promoter variants can cause significantly potential phenotype diversity, and most of the regulatory mechanisms are associated with the change of transcription factor binding and promoter activity35. For example, a single mutation at position −1,687 in human L-plastin gene affected the binding strength of the transcriptional suppressor NKX3.1, and further reduced the expression of L-plastin and potentially decreased the tumorigenesis and progression of prostate cancer36. Dominquez et al. indicated that a 23-bp indel in porcine TLR5 gene, creating an additional STAT binding site, is associated with an increase of the promoter activity37. We found a 10-bp indel variant in the promoter region of the bovine PAX7 gene that the 10-bp insertion can reduce promoter activity and PAX7 expression, and negatively associate the cattle growth traits by creating a transcriptional suppressor ZNF219 binding site, while the 10-bp deletion can enhance the effects. This 10-bp indel could be a potential selection marker for superior muscle involving traits in cattle breeding industry.
The results of promoter activity assay and association analysis demonstrated that ZNF219 negatively regulated the expression of the PAX7 gene by binding to its promoter in the presence of the 10-bp indel. The ZNF219 gene is a member of the Kruppel-like zinc finger gene family that are involved in many biological processes, such as cell growth, differentiation, embryogenesis and tumorigenesis, and ZNF219 is often depleted in the early stage of tumor progression38. Loss of ZNF219 expression is correlated with high oncogene level and the presence of some metastatic diseases39. There was evidence in the literature to support the claim that ZNF219 is a nuclear transcriptional factor that regulates the expression of its downstream target genes. For example, Sakai et al.25 reported that ZNF219 could modulate the expression of the HMGN1 gene directly by binding its upstream DNA sequence. Recently, Liu et al.40 showed that the expression of ZNF219 was associated with skeletal muscle reduction in cancer cachexia. In our association analysis, the Ins-Ins genotype with the ZNF219 binding presented remarkably lower body weight, body height, body length, heart girth, hucklebone width and average daily gain than Del-Del genotype. This finding provided a foundation for further studies of the regulatory mechanisms and roles of ZNF219 on PAX7 gene expression in the development of skeletal muscle.
Although it is not fully elucidated if the decreased PAX7 expression is directly defined by the 10-bp insertion or is the result of interaction with other genetic pathways, it is interesting to note that the homozygous genotype (Ins-Ins) of the 10-bp indel, which reduced the promoter activity of PAX7 by creating the ZNF219 binding site, was related to decreased phenotypic traits of cattle. Furthermore, we found that the significant associations of the 10-bp indel with growth traits were detected in the early stage (6 and 12 months old) of cattle. Consistently, the PAX7 gene with Del-Del genotype showed higher expression than the Ins-Ins and Ins-Del genotypes in skeletal muscle of fetal cattle. These results may be attributed to the regulatory function of PAX7 in early muscle development. PAX7 participates in successive phases of embryonic and post-natal myogenesis that lead to the formation and growth of skeletal muscles41. Previous study reported that the body weight of PAX7 knockout mice markedly decreased from birth to the age of two weeks, which was due to the absence of PAX7 in the progenitor satellite cells12. During myogenesis, PAX7 is considered as an upstream regulators that can regulate the satellite cells entry into the myogenic programme by stimulating transcriptional activation of target genes42. For instance, ID2 and ID3 are two notable PAX7 targets that are coordinately expressed with PAX7 in quiescent satellite cells and can be induced by ectopic expression of PAX711. CXCR4 is another PAX7 target in satellite cells43. Herein, we found the same tendency of the high expression of ID2, ID3 and CXCR4 in the individuals with Del-Del genotype that was accompanied by PAX7 level. However, the expression of other four genes, MYF5, MYOD, MRF4 and MYOG, were not associated with the genotypes of the PAX7 10-bp indel, which may be explained by the reason that the four genes play crucial roles in proliferation and differentiation of myoblast cells, especially, the MRF4 and MYOG are expressed in a later stage of myogenesis44. Therefore, the PAX7 10-bp indel could be utilized in early selection project for cattle breeding.
In summary, we identified a 10-bp indel in the PAX7 promoter and demonstrated that this indel, located in the ZNF219 binding site, affected the promoter activity and expression of the PAX7 gene, which in turn was associated with phenotypic traits in the early stages of cattle. The regulatory mechanism of the 10-bp indel and the validation of the indel locus in other populations are need to be explored in further study. Taken together, this study revealed the functional role of the PAX7 10-bp indel and provided promising insight into its further exploitation in molecular breeding of cattle.
Methods
Ethics statement
The experiments and animal care were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2004) and approved by the Institutional Animal Care and Use Committee (College of Animal Science and Technology, Northwest A&F University, China). Embryos of slaughtered cattle were collected from a local slaughterhouse, Tumen Abattoir (Xi’an, China). Cattle were allowed access to feed and water ad libitum under normal conditions. All animal experiments and methods were carried out in accordance with the approved guidelines and all efforts were made to minimize suffering.
Animals and sample preparation
Blood samples were obtained from a multi-breed panel of up to 1233 adult individuals including five Chinese cattle breeds: NY (n = 220), JX (n = 395), QC (n = 224), LX (n = 161) and CY (n = 233). These breeds represent the main populations of P. R. China and are reared in the Henan, Henan, Shaanxi, Shandong and Jilin province, respectively. Calves were weaned at six months of age on average and raised from weaning to slaughter on a diet of corn and corn silage. Genomic DNA was isolated from blood samples according to the procedure used by Sambrock, J. & Russell, D. W.45. Records of growth traits in NY breed were collected for further statistical analysis, including body weight, body height, body length, heart girth, hucklebone width and average daily gain at different growth periods (6, 12, 18, and 24 months).
The muscle samples (taken from the longissimus thoracis) were collected from 30 cattle embryos (gestation 90 days) in the QC breed within 10 min after slaughter. Total RNA was prepared from the muscles with Trizol reagent (Takara, Japan) according to the manufacturer’s protocol. RNA integrity was monitored by denaturing 1% agarose gel electrophoresis. Concentrations and purities of RNA were measured by spectrophotometry. Reverse transcription into cDNA was performed using a PrimeScript® RT reagent Kit (Perfect Real Time) (Takara, Japan) with 2 μg total RNA in a 20 μl reaction. Genomic DNA was extracted from the same muscles (10 mg) by two rounds of proteinase K digestion and phenol-chloroform extraction.
DNA pool sequencing and genotyping
DNA pool sequencing has been explored as an efficient strategy to increase the detection throughput of SNPs and small indels. In this study, five DNA pools were constructed and each pool contained 80–100 individuals that were randomly chosen from the five cattle breeds, respectively. All DNA samples was dissolved to 50 ng/μl, and then contributed same volume to their respective pool46.
Based on the sequence of the bovine PAX7 gene (GenBank accession no. NC_007300), the primer P1 (Table S1) was designed to amplify a 1868-bp fragment, which encompassed the bovine PAX7 promoter region and a part of the exon 1. PCR amplification was carried out using the genomic DNA pool as template, and PCR product was sequenced on an automated sequencer (ABI PRISM 3730 DNA analyzer). Sequence polymorphisms were identified in silico using BLASTN and sequencing maps results. In fact, a 10-bp insertion or deletion (10-bp indel) was identified at the position of −633 and −643 of the PAX7 promoter region. Correspondingly, another primer pair P2 (Table S1) was designed for genotyping the 10-bp indel variant. The different genotypic fragments were separated on 10% PAGE in 1 × TBE buffer with ethidium bromide staining.
Analysis of transcription factor binding sites in PAX7 promoter region
The bovine PAX7 promoter sequence was analyzed to identify putative regulatory regions in silico using TFSEARCH (www.cbrc.jp/research /db/TFSEARCH.html) and Genomatix MatInspector Release professional 8.0.4, to make sequence-based prediction of TF binding sites47.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was strictly operated according the protocol of SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology). Cells were fixed with 1% formaldehyde for 10 min to cross-link DNA-protein complexes and quenched with glycine for 5 min. After washing with ice-cold PBS for three times, cells were re-suspended in lysis buffer. Then the lysis was digested by micrococcal nuclease at 37 °C for 20 min to length of approximately 150–900 bp. Digestion was terminated by the addition of 0.5 M EDTA. Chromatin was sonicated at 30% output for 6 × 10 sec. Clarify lysates by centrifugation at 10,000 rpm for 10 min at 4 °C. The supernatant was diluted with ChIP buffer (1:9). Add the immunoprecipitating anti-ZNF219 antibody (Atlas antibodies) into 500 μl of the diluted chromatin. IgG (Santa cruz) was used as the negative control. Reverse cross-linked DNA was purified by spin columns. The DNA was used as a template for PCR, and the products were separated on 1% agarose gel. The PAX7 promoter region primers: Forward: 5′- GTTACAACCAGCACTTCTGC -3′, Reverse: 5′- TCTGGGGAGGGAAGAAGGAA -3′.
Plasmid construction
According to the TF binding sites prediction results, five fragments of the PAX7 promoter region were amplified by PCR using different primer pairs (Table S1). The large 1671-bp fragment spanning −1603 to +60, was cloned into the pGEM-T vector (Promega, Madison, WI). For luciferase reporter construction, the insert was released by NheI and HindIII digestion, and then subcloned into the luciferase reporter vector pGL3-Basic (Promega, Madison, WI). This plasmid was designated as pGL3-pro1. Serial deletion vectors, pGL3-pro2 (−1279 to +60), pGL3-pro3 (−667 to +60), pGL3-pro4 (−324 to +60), pGL3-pro5 (−190 to +60) were constructed from pGL3-pro1 using the primers including KpnI and HindIII recognized sites, respectively. Additionally, the two vectors, termed as pGL3-pro2InsIns (10-bp Ins-Ins genotype) and pGL3-pro2DelDel (10-bp Del-Del genotype), were constructed for detecting the effects of the PAX7 indel. Their inserted sequences were obtained from the individuals with homozygous locus. The vector pRL-TK (Promega, Madison, WI) was used as internal reference in the luciferase reporter system.
To generate ZNF219 overexpressing vector, the full length of ZNF219 coding sequence was amplified using a pair of specific primer with BamHI and EcoRI, respectively (Table S1), and were cloned into the plasmid pcDNA3.1( + ). All recombinants were verified by DNA sequencing to proof correct insertion and proper orientation.
Cell culture
A skeletal muscle cell line of C2C12 cells was grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), and 100 units/ml penicillin with 100 μg/ml streptomycin at 37 °C in a humidified 5% CO2 atmosphere.
Transient transfection and luciferase reporter assay
Transfections were conducted using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. Before transfection, C2C12 cells were seeded into 48-well plate at a density of 2 × 104 cells/well in 1 ml of complete medium and cultured overnight. Luciferase reporter constructs containing the PAX7 promoter with 10-bp insertion (pGL3-pro2InsIns) or 10-bp deletion (pGL3-pro2DelDel) sites and/or pcDNA3.1-ZNF219 plasmid were temporarily transfected into C2C12 cells using Lipofectamine 3000. To normalize the transfection efficiency, 40 ng of the pRL-TK Renilla transfection control plasmid was co-transfected into the cells. After 48 hr of serum starvation, the cells were lysed, and the luciferase activity was measured using the Dual Luciferase Assay System (Promega, Heidelberg, Germany) and the SpectraMax M5 reader (Molecular Devices, CA). The luciferase signal from the PAX7 promoter reporter constructs were calculated and normalized to the Renilla luciferase activity48. All transfections were carried out in triplicate.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed to detect the expression of the PAX7 and its down-stream genes on a Bio-Rad CFX 96TM Real Time Detection System (Bio-Rad, Hercules, CA). The bovine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control to normalize the differences in the amount of total cDNA added to each reaction. Gene-specific qPCR primer pairs were designed using the Beacon DesignerTM software (version 8.1) (Table S1). The reaction contained 100 ng of cDNA, 10 μl SYBR® Premix Ex Taq TM II (TaKaRa, Japan), and 10 pmol of primers in a volume totaling 20 μl. The mixture was denatured for 30 s at 95 °C and was followed by 40 cycles of 5 s at 95 °C and 30 s at 60 °C.
Statistical analysis
The genotypic and allelic frequencies of the 10-bp indel within the PAX7 gene were estimated by the standard counting method. Each breed and the overall dataset were tested for deviation from Hardy-Weinberg equilibrium by χ2-test performed by the POPGENE software23 (version 3.2). PIC, He, and Ne were determined using the methods of Nei and Roychoudhury49. The association analysis with the growth traits was established using the General Linear Model (GLM) procedure as implemented in the SPSS software50 (version 18.0). The traits and PAX7 genotypes were statistically analyzed according to our previously reported statistical model23: Yijkl = μ + BFi + Aj + Gk + eijkl, where Yijkl is the observation of the growth traits; μ is the overall mean of each trait, BFi is the fixed effect associated with ith breed and farm, Aj is the effect due to jth age, Gk is the fixed effect of kth genotype of the 10-bp indel and eijkl is the random residual error.
Where appropriate, data were expressed as mean value ± standard error of duplicates. For comparison of the mean of two groups, statistical significance was assessed by Student’s t-test51. Calculations were performed with the STATA Statistical Software (StataCorp, College Station, TX, USA). Statistical significance was defined as P < 0.05.
Electronic supplementary material
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 31600617), the Program of National Beef Cattle and yak Industrial Technology System (CARS-37), Natural Science Foundation of Hubei Province (2016CFB171), Bio-breeding capacity-building and industry specific projects from National Development and Reform Commission (2014–2573), Specific Projects of Science and Technology in Henan Province (141100110200), Science and Technology Co-ordinator Innovative engineering projects of Shaanxi Province (No. 2015KTCL02-08).
Author Contributions
Y.X., H.C. designed the study and Y.X. wrote the manuscript; Y.X., T.S. and Y.Z. performed the study; Y.Z. and M.L. participated in data analysis; S.K. and X.L. contributed to the revision of the manuscript; C.L. provided valuable assistance with the experimental procedures and for collecting the tissues of the adult cattle.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20177-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bolormaa S, et al. Non-additive genetic variation in growth, carcass and fertility traits of beef cattle. Genet Sel Evol. 2015;47:015–0114. doi: 10.1186/s12711-015-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haskell, M. J., Simm, G. & Turner, S. P. Genetic selection for temperament traits in dairy and beef cattle. Front Genet5 (2014). [DOI] [PMC free article] [PubMed]
- 3.Wientjes YC, Calus MP, Goddard ME, Hayes BJ. Impact of QTL properties on the accuracy of multi-breed genomic prediction. Genet Sel Evol. 2015;47:015–0124. doi: 10.1186/s12711-014-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laumonier T, Bermont F, Hoffmeyer P, Kindler V, Menetrey J. Human myogenic reserve cells are quiescent stem cells that contribute to muscle regeneration after intramuscular transplantation in immunodeficient mice. Sci Rep. 2017;7:017–03703. doi: 10.1038/s41598-017-03703-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumont NA, Bentzinger CF, Sincennes MC, Rudnicki MA. Satellite Cells and Skeletal Muscle Regeneration. Compr Physiol. 2015;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 7.von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sambasivan R, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 9.Gunther S, et al. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calhabeu F, Hayashi S, Morgan JE, Relaix F, Zammit PS. Alveolar rhabdomyosarcoma-associated proteins PAX3/FOXO1A and PAX7/FOXO1A suppress the transcriptional activity of MyoD-target genes in muscle stem cells. Oncogene. 2013;32:651–662. doi: 10.1038/onc.2012.73. [DOI] [PubMed] [Google Scholar]
- 11.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell. 2009;20:3170–3177. doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seale P, et al. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 13.Zalc A, Rattenbach R, Aurade F, Cadot B, Relaix F. Pax3 and Pax7 play essential safeguard functions against environmental stress-induced birth defects. Dev Cell. 2015;33:56–66. doi: 10.1016/j.devcel.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Deiner C, et al. Allelic variations in coding regions of the vitamin D receptor gene in dairy cows and potential susceptibility to periparturient hypocalcaemia. J Dairy Res. 2012;79:423. doi: 10.1017/S0022029912000465. [DOI] [PubMed] [Google Scholar]
- 15.Szewczuk M, Zych S, Wójcik J, Czerniawska-Piątkowska E. Association of two SNPs in the coding region of the insulin-like growth factor 1 receptor (IGF1R) gene with growth-related traits in Angus cattle. J Appl Genet. 2013;54:305–308. doi: 10.1007/s13353-013-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, et al. Associations between variants of the HAL gene and milk production traits in Chinese Holstein cows. BMC Genet. 2014;15:014–0125. doi: 10.1186/s12863-014-0125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo XM, et al. Characterization of the NLRC5 promoter in chicken: SNPs, regulatory elements and CpG islands. Anim Genet. 2016;47:579–587. doi: 10.1111/age.12450. [DOI] [PubMed] [Google Scholar]
- 18.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinnell IW, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2007;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syagailo YV, et al. Structural and functional characterization of the human PAX7 5′-flanking regulatory region. Gene. 2002;294:259–268. doi: 10.1016/S0378-1119(02)00798-9. [DOI] [PubMed] [Google Scholar]
- 21.Okladnova O, et al. Functional Characterization of the Human PAX3 Gene Regulatory Region. Genomics. 1999;57:110–119. doi: 10.1006/geno.1998.5711. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y, et al. Consistent effects of single and combined SNP (s) within bovine paired box 7 gene (Pax7) on growth traits. J Genet. 2011;90:e53–e57. [PubMed] [Google Scholar]
- 23.Xu Y, et al. Integrating haplotypes and single genetic variability effects of the Pax7 gene on growth traits in two cattle breeds. Genome. 2012;56:9–15. doi: 10.1139/gen-2012-0124. [DOI] [PubMed] [Google Scholar]
- 24.McClure M, et al. A genome scan for quantitative trait loci influencing carcass, post-natal growth and reproductive traits in commercial Angus cattle. Anim Genet. 2010;41:597–607. doi: 10.1111/j.1365-2052.2010.02063.x. [DOI] [PubMed] [Google Scholar]
- 25.Sakai T, Hino K, Wada S, Maeda H. Identification of the DNA binding specificity of the human ZNF219 protein and its function as a transcriptional repressor. DNA research. 2003;10:155–165. doi: 10.1093/dnares/10.4.155. [DOI] [PubMed] [Google Scholar]
- 26.Andersson L. Molecular consequences of animal breeding. Curr Opin Genet Dev. 2013;23:295–301. doi: 10.1016/j.gde.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Mei C, et al. Whole-genome sequencing of the endangered bovine species Gayal (Bos frontalis) provides new insights into its genetic features. Sci Rep. 2016;6:19787. doi: 10.1038/srep19787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Msalya G, et al. Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet. 2010;41:218–221. doi: 10.1111/j.1365-2052.2009.01983.x. [DOI] [PubMed] [Google Scholar]
- 29.Miller DJ, et al. Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): implications for the evolution of the Pax gene family. Proc Natl Acad Sci USA. 2000;97:4475–4480. doi: 10.1073/pnas.97.9.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Underhill DA. Genetic and biochemical diversity in the Pax gene family. Biochem Cell Biol. 2000;78:629–638. doi: 10.1139/o00-077. [DOI] [PubMed] [Google Scholar]
- 31.Davicioni E, et al. Molecular classification of rhabdomyosarcoma—genotypic and phenotypic determinants of diagnosis: a report from the Children’s Oncology Group. Am J Pathol. 2009;174:550–564. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, et al. SNP and haplotype analysis of paired box 3 (PAX3) gene provide evidence for association with growth traits in Chinese cattle. Mol Biol Rep. 2014;41:4295–4303. doi: 10.1007/s11033-014-3300-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhang S, et al. A novel 31-bp indel in the paired box 7 (PAX7) gene is associated with chicken performance traits. Br Poult Sci. 2014;55:31–36. doi: 10.1080/00071668.2013.860215. [DOI] [PubMed] [Google Scholar]
- 34.Ponomarenko JV, et al. rSNP_Guide: an integrated database-tools system for studying SNPs and site-directed mutations in transcription factor binding sites. Hum Mutat. 2002;20:239–248. doi: 10.1002/humu.10116. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Tomso DJ, Liu X, Bell DA. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol Appl Pharmacol. 2005;207:84–90. doi: 10.1016/j.taap.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, et al. An NKX3.1 binding site polymorphism in the l-plastin promoter leads to differential gene expression in human prostate cancer. Int J Cancer. 2016;138:74–86. doi: 10.1002/ijc.29677. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez MA, Landi V, Martinez A, Garrido JJ. Identification and functional characterization of novel genetic variations in porcine TLR5 promoter. Dna Cell Biol. 2014;33:469–476. doi: 10.1089/dna.2013.2318. [DOI] [PubMed] [Google Scholar]
- 38.Sakai T, Toyoda A, Hashimoto K, Maeda H. Isolation and characterization of a novel zinc finger gene, ZNF219, and mapping to the human chromosome 14q11 region. DNA research: an international journal for rapid publication of reports on genes and genomes. 2000;7:137–141. doi: 10.1093/dnares/7.2.137. [DOI] [PubMed] [Google Scholar]
- 39.Takigawa Y, et al. The transcription factor Znf219 regulates chondrocyte differentiation by assembling a transcription factory with Sox9. J Cell Sci. 2010;123:3780–3788. doi: 10.1242/jcs.071373. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, et al. Identification and functional analysis of a potential key lncRNA involved in fat loss of cancer cachexia. 2017 doi: 10.1002/jcb.26328. [DOI] [PubMed] [Google Scholar]
- 41.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 42.Buckingham M, Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin Cell Dev Biol. 2015;44:115–125. doi: 10.1016/j.semcdb.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 43.Bosnakovski D, et al. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandurangan M, et al. Effects of stress hormone cortisol on the mRNA expression of myogenenin, MyoD, Myf5, PAX3 and PAX7. Cytotechnology. 2014;66:839–844. doi: 10.1007/s10616-013-9635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrock, J. & Russell, D. W. Molecular cloning: a laboratory manual. 3rd edn, 49-56 (Cold Spring Harbor Laboratory Press, 2001).
- 46.Sham P, Bader JS, Craig I, O’Donovan M, Owen M. DNA pooling: a tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- 47.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 48.Solberg N, Krauss S. Luciferase assay to study the activity of a cloned promoter DNA fragment. Methods in molecular biology (Clifton, N.J.) 2013;977:65–78. doi: 10.1007/978-1-62703-284-1_6. [DOI] [PubMed] [Google Scholar]
- 49.Nei M, Roychoudhury AK. Sampling variances of heterozygosity and genetic distance. Genetics. 1974;76:379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver B, Wuensch KL. SPSS and SAS programs for comparing Pearson correlations and OLS regression coefficients. Behavior research methods. 2013;45:880–895. doi: 10.3758/s13428-012-0289-7. [DOI] [PubMed] [Google Scholar]
- 51.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.