Abstract
Selenoproteins are essential components of antioxidant defense, redox homeostasis, and cell signaling in mammals, where selenium is found in the form of a rare amino acid, selenocysteine. Selenium, which is often limited both in food intake and cell culture media, is a strong regulator of selenoprotein expression and selenoenzyme activity. Aging is a slow, complex, and multifactorial process, resulting in a gradual and irreversible decline of various functions of the body. Several cellular aspects of organismal aging are recapitulated in the replicative senescence of cultured human diploid fibroblasts, such as embryonic lung fibroblast WI-38 cells. We previously reported that the long-term growth of young WI-38 cells with high (supplemented), moderate (control), or low (depleted) concentrations of selenium in the culture medium impacts their replicative lifespan, due to rapid changes in replicative senescence-associated markers and signaling pathways. In order to gain insight into the molecular link between selenium levels and replicative senescence, in the present work, we have applied a quantitative proteomic approach based on 2-Dimensional Differential in-Gel Electrophoresis (2D-DIGE) to the study of young and presenescent cells grown in selenium-supplemented, control, or depleted media. Applying a restrictive cut-off (spot intensity ±50% and a p value < 0.05) to the 2D-DIGE analyses revealed 81 differentially expressed protein spots, from which 123 proteins of interest were identified by mass spectrometry. We compared the changes in protein abundance for three different conditions: (i) spots varying between young and presenescent cells, (ii) spots varying in response to selenium concentration in young cells, and (iii) spots varying in response to selenium concentration in presenescent cells. Interestingly, a 72% overlap between the impact of senescence and selenium was observed in our proteomic results, demonstrating a strong interplay between selenium, selenoproteins, and replicative senescence.
Keywords: proteomics, 2-Dimensional Differential in-Gel Electrophoresis (2D-DIGE), selenium, protein abundance, selenoprotein, replicative senescence, WI-38 cells
1. Introduction
Selenium has been shown to be an essential trace element in all animals. Deficiency of selenium has been linked to several diseases and disorders, including an increased risk of cancer and of cardiovascular and neurological diseases, as well as a decrease in immune function [1,2,3]. Most of the beneficial effects of selenium are due to the existence of a pool of selenoproteins, which are involved in redox biology and homeostasis. In effect, the selenoproteins have the ability to translationally incorporate a rare amino acid residue, selenocysteine (Sec, U), using a specific and complex machinery (for details, see [4]). The selenocysteine residue is more reactive and less sensitive to oxidation than its sulfur analog cysteine. At the catalytic site of enzymes, selenocysteine participates in redox reactions, which are essential for antioxidant defense, redox homeostasis, and cell signaling [5,6,7,8]. Gene inactivation in mice of certain selenoproteins, such as glutathione peroxidase 4 (Gpx4), thioredoxin reductase 1 (Txnrd1), or SelenoT, leads to embryonic lethality. Therefore, it is comprehensible that gene inactivation of Sec-tRNA[Ser]Sec or Selenocysteine insertion sequence binding protein-2 SECISBP2 [9,10], altering selenoprotein synthesis, is also fatal in mice. The level of selenoprotein expression in the organism is tightly regulated not only by the bioavailability of selenium, but also by exogenous stimuli [11]. For in vitro cultured cells, the level of selenium dictates the range of selenoprotein expression in accordance with a cell-line-specific hierarchy [4]. The interplay between selenium levels and exogenous stimuli, such as aging or oxidative stress, has only been studied in specific cases [11,12,13,14].
Aging is a slow, complex, and multifactorial process, resulting in a gradual and irreversible decline of various functions of the body, making organisms more vulnerable and more likely to die [11]. Several cellular aspects of organismal aging are recapitulated in the replicative senescence of cultured human diploid fibroblasts, such as embryonic lung fibroblast WI-38 cells, which thus offer a commonly used cellular model to study in vitro features of the aging phenomenon [15]. Cellular senescence, also referred to as replicative senescence, was initially described by Hayflick and Moorehead in 1961 [16]. Senescence is defined by the finding that diploid cells go through a finite number of divisions—also referred to as the Hayflick limit [16]. Cellular senescence is characterized by an irreversible cell cycle arrest via the p53, pRb, p16, and p21 signaling pathways [17,18,19]; telomere shortening; accumulation of oxidative damage; and several senescence-associated markers, including β-galactosidase (SABG) and heterochromatin foci (SAHF). Interestingly, the number of senescent cells increases with age and senescent-associated phenotypes are considered to be predictive features of age-related phenotypes. In various cell types, premature senescence can be induced by exogenous stimuli. For example, repeated exposure to mild oxidative stress (utraviolet radiation (UV) or tert-Butyl hydroperoxide) provokes a stress-induced premature senescence (SIPS) phenotype [20,21,22] in human diploid fibroblasts. Additionally, in cancer cells, the use of drug therapy can also lead to premature senescence, namely therapy-induced senescence (TIS) [23,24]. Although, in every case, a G0 cell cycle arrest is observed, in general, the molecular mechanisms involved and intracellular targets differ between the different senescence phenotypes.
Even though redox status is implicated in the aging phenotype, the link between selenium and aging or replicative senescence has, so far, been poorly investigated (reviewed in [11]). Primarily, it was reported that selenium levels and selenium-dependent glutathione peroxidase activity decreased in the serum of healthy Italian subjects [25]. Interestingly, it was observed in this study that the selenium levels in the elderly were much more scattered than those in young persons. It has been suggested that the decrease of selenium status in aging could lead to a weakening of the antioxidant defense and a decrease in longevity, although the molecular mechanisms and selenoproteins involved remain to be identified. At the cellular level, a few studies have reported extension of the replicative lifespan of cultured cells with selenium supplementation [13,26]. In the most recent study, a 30 nM selenium supplementation of the culture medium significantly reduced the level of senescence markers, including signaling molecules (p16, p21, p53), telomere shortening, SABG and SAHF [13]. As a corollary, the expression of several selenoproteins was altered by replicative senescence. In this study, it was found that selenoproteins were regulated at both the transcriptional and translational levels.
In the present study, we have further investigated the interrelation between selenium and replicative senescence at the molecular level using a proteomic approach. We have compared the response to senescence with long-term growth in culture media containing various levels of selenium, previously found to modulate the replicative capacity of WI-38 cells. Due to a large overlap in protein targets, our data favor an action of selenium in the generic replicative senescence pathway, rather than a premature senescence similar to SIPS.
2. Materials and Methods
2.1. Cell Growth
Human embryonic fibroblasts WI-38 were purchased from SIGMA (Chicago, IL, United States of America) (passage 15, estimated cumulative population doublings (CPD) 30), grown and maintained in 75 cm2 plates in Dulbecco’s Modified Eagle Medium (D-MEM) supplemented with 10% fetal calf serum (FCS), 100 µg mL−1 streptomycin, 100 UI mL−1 penicillin, 1 mM sodium pyruvate, and 2 mM l-glutamine. Cells were cultivated in 5% CO2 at 37 °C and a humidified atmosphere. The different culture media referred to as depleted (Dpl), control (Ctl), and supplemented (Sup) were prepared according to [13] and contained, respectively, 3, 15, and 45 nM of selenium, as determined by Inductively coupled plasma-mass spectrometry ICP-MS [12,13,27,28]. The quantification of the selenium levels in the raw FCS (150 nM) was obtained with a limit of quantification (LOQ) of 2.5 nM. Although much lower than selenium levels in serum, the concentration of 45 nM in the culture medium yielded a strong increase in protein expression for several selenoproteins when selenite was used to supplement the medium [13]. When cells reached confluence (approximately at 100,000 cells per cm2), they were passaged in a new flask at 10,000 cells per cm2 density. WI-38 cells were counted at each passage to calculate the Population Doubling (PD) using the equation ΔPD = log ((Final Cell Number)/(Initial Cell Number))/log (2). Cumulative PD was plotted as a function of time.
2.2. Protein Extraction. 2D Clean-Up Kit. Protein Quantification. Verification of Protein Quality by SDS PAGE
All chemical reagents purchased were 2D-DIGE grade (i.e., PlusOne, GEHealthcare, Little Chalfont, United Kingdom). Whole protein extracts were obtained from 75 cm2 plates using cell lysis buffer composed of 40 mM Tris-HCl pH 7.8, 8 M urea, 2 M thiourea, 4% (w/v) 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate (CHAPS), 50 mM DL-Dithiothreitol DTT, and protease inhibitor (Complete Mini, Roche Diagnostics). Cells were disrupted by passing them through a 23 gauge syringe 10 times. The homogenates were centrifuged at 20,000 g for 1 h at 4 °C. The supernatant was collected and conserved at −20 °C. An aliquot of crude protein extract was precipitated and washed according to the 2D clean-up kit procedure (GEHealthcare). Pellets were resuspended in 50 µL of Urea, Thiourea, CHAPS (UTC) buffer (8 M urea, 2 M thiourea, 4% CHAPS). Protein concentrations were measured using the Detergent compatible (DC)kit protein assay kit (Biorad) in microplate assays (BMG Labtech GmbH, Ortenberg, Germany). To verify the integrity of protein extracts and correct quantification of protein concentrations, 5 µg of each sample was separated in Bis-Tris NuPAGE Novex Midi Gels (Life Technologies, Carlsbad, CA, United States of America, under 3-(N-Morpholino)propanesulfonic acid, 4-Morpholinepropanesulfonic acid (MOPS) conditions. The gel was then stained using Coomassie staining solution.
2.3. 2-Dimensional Differential in-Gel Electrophoresis (2D-DIGE) Labeling, Isoelectric Focusing Electrophoresis (IEF), and Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE ) of Protein Samples
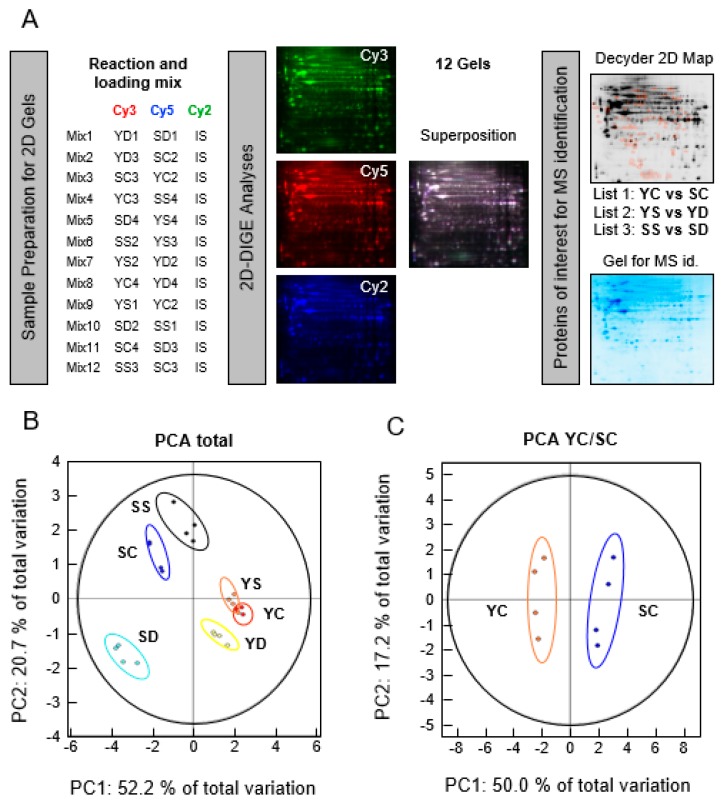
Four experimental replicates for each set of conditions were collected from cells grown in four individual flasks. As shown in Figure 1, six growth conditions were generated, i.e., young Se-depleted (YD), young Se-control (YC), young Se-supplemented (YS), senescent Se-depleted (SD), senescent Se-control (SC), and senescent Se-supplemented (SS). In total, 24 different protein extracts were analyzed using the 2D-DIGE strategy. A quantity of 50 µg of protein from each sample was transferred to a different tube (i.e., 24 in total), and the volume adjusted to 11 µL with UTC buffer. An internal standard (IS) was made by mixing 25 µg of each sample in another tube, with the volume completed to 132 µL with UTC buffer. CyDye DIGE Fluor minimal dyes (GEHealthcare, Little Chalfont, United Kingdom), which react with lysine residues, were resuspended at 400 pM/µL concentration in dimethylformamide (DMF). A quantity of 1 µL of a specific CyDye solution (i.e., Cy3 or Cy5) was add to the 11 µL of protein sample according to the schematic in Figure 2A. Biological replicates were randomly labelled to prevent dye bias. On the other hand, 12 µL of Cy2 was added to the 132 µL of internal standard. The labeling reaction was left to occur for 30 min on ice, protected from light. To the Cy3 and Cy5 reactions, 1 µL of DIGE stop solution (10 mM Lysine) was added. On the other hand, 12 µL of DIGE stop solution was added to the Cy2 (IS) reaction. The stop reaction was left to occur for 10 min protected from light at 4 °C. Twelve mixes composed of one Cy3 (13 µL), one Cy5 (13 µL), and 13 µL of the IS (Cy2) reaction were generated according to the table in Figure 2A. The volume was completed with 311 µL of isoelectric focusing electrophoresis (IEF) rehydration buffer composed of 8 M urea, 2 M thiourea, 2% (w/v) CHAPS, 12 µL/mL DeStreak reagent (GEHealthcare, Little Chalfont, United Kingdom), 0.5% (v/v) immobilized pH gradient (IPG) Buffer (3–10 non-linear (NL)), and traces of Bromophenol blue. This solution was loaded onto an 18 cm Immobiline DryStrip (3–10 NL) by passive rehydration under a layer of DryStrip Cover Fluid (GEHealthcare, Little Chalfont, United Kingdom) overnight. Then, IEF was performed using IPGPhor2 (GEHealthcare, Little Chalfont, United Kingdom) with the following run: 150 V for 1 h, 200 V for 1 h, gradient 200–1000 V for 9 h, gradient 1000–8000 V for 3 h, and 8000 V until total volt hours (Vh) reached 35,000. At this stage, the strips were kept frozen at −20 °C until further use.
Figure 1.
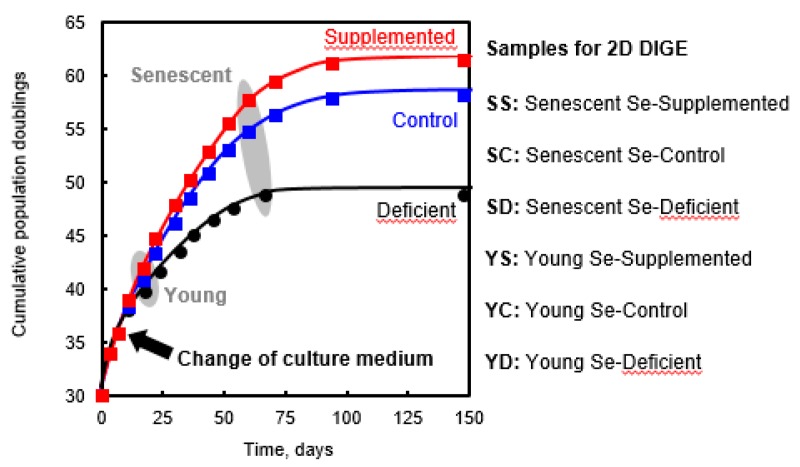
The replicative life span of WI-38 cells is affected by the selenium level of the culture medium. At cumulative population doublings (CPD) 35.8, WI-38 cells were plated in three different conditions, namely, Se-deficient (Dpl, black dots), Se-control (Ctl, blue square), and Se-supplemented (Sup, red square) for two passages and until they reached the senescent stage. The gray ellipses illustrate the time and stage the cells were harvested for 2-Dimensional Differential in-Gel Electrophoresis (2D-DIGE) analyses.
Figure 2.
Experimental design and statistical validation of our 2D-DIGE analyses. YD: young Se-depleted; SD: senescent Se-depleted; SC: senescent Se-control; YC: young Se-control; SS: senescent Se-supplemented; YS: young Se-supplemented; MS: mass spectrometry; PCA: principal component analysis; PC1: principal component 1; PC2: principal component 2.
The strips were thawed and incubated for 12 min in equilibration buffer 1 composed of 50 mM Tris-HCl pH 8.6, 6 M urea, 2% (w/v) SDS, 30% (v/v) glycerol, and 65 mM DTT. Then, the strips were transferred for 12 min to equilibration buffer 2, composed of 50 mM Tris-HCl pH 8.6, 6 M urea, 2% (w/v) SDS, 30% (v/v) glycerol, and 250 mM Iodoacteamide. After the equilibration stage, each strip was positioned on top of a 12.5% acrylamide (acrylamide/bis acrylamide, 37.5:1) SDS-PAGE and run overnight using the Ettan DALTII system (GEHealthcare, Little Chalfont, United Kingdom), allowing the simultaneous migration of six gels. Migration conditions were as follows: 15 W per gel for 30 min, and 25 W per gel for 16 h, under a water cooling system. It should be noted that the gels were cast in low-fluorescence glass plates to allow a direct reading with a Typhoon fluorescence scanner.
2.4. Fluorescence Scanning of the Gel and Decyder Analysis
After the second-dimension electrophoresis run, the plates containing the gels were carefully cleaned and directly scanned with a Typhoon FLA 9500 (GEHealthcare, Little Chalfont, United Kingdom), without uncasting the gel. The orientation of the plates was kept identical for the 12 gels. Every gel was scanned three times with the following parameters: for Cy2, laser 488 nm, emission filter 520BP40, photomultiplier voltage setting (PMT) 490; for Cy3, laser 532 nm, emission filter 580BP30, PMT 520; for Cy3, laser 633 nm, emission filter 670BP30, PMT 470. Importantly, the scanning parameters were kept identical for the 12 gels with a resolution set at 100 µm. The data analysis, including the principal component analysis (PCA), was performed with Decyder 7.0 software (GEHealthcare, Little Chalfont, United Kingdom) according to the manufacturer’s instructions.
2.5. Mass Spectrometry Protein Analysis
2.5.1. Sample Preparation
A preparative gel was formed with 500 µg of protein completed with rehydration buffer to a total of 340 µL. Rehydration, migration of the strips, and SDS-PAGE were performed as described earlier for the fluorescence-labeled samples. Each spot of interest was localized in the Coomassie-stained 2D-gel, excised, and subjected to enzymatic digestion. Trypsin digestion of selected spots was performed following reduction/alkylation of cysteine residues as described previously [29] with the addition of 10 µL of trypsin (12.5 ng/µL) per spot and 20 µL of extraction solvent for peptide extraction. Tryptic peptides were vacuum-dried and finally resuspended in 0.1% formic acid and 5% acetonitrile before nanoscale liquid chromatography coupled to tandem mass spectrometry (nanoLC-MS/MS) analyses.
2.5.2. NanoLC-MS/MS Analyses
Tryptic peptides were analyzed by nanoLC-MS/MS analysis using first a nanoAcquity liquid chromatograph (Waters, Milford, MA, United States of America) coupled to a quadrupole-time-of-flight (Q-TOF) Premier mass spectrometer (Waters, Milford, MA, United States of America), and secondly an EASY-nLC II HPLC system (Proxeon, ThermoScientific, Waltham, MA, United States of America) coupled to a nanoESI-Linear Trap Quadropole (LTQ)-Orbitrap Velos mass spectrometer (Thermo Scientific) in case no significant protein identification was obtained in the first analysis. Nano-liquid chromatography elution conditions were similar for both systems, operated with a flow rate of 300 nL/min and an acetonitrile gradient of 5–40% (v/v) over 20 min. The nanoLC system (Waters, Milford, MA, United States of America) coupled to the Q-TOF Premier was equipped with a trapping column (C18 symmetry, 180 μm × 20 mm, particle size 5 μm, from Waters) and an analytical column (BEH 130 C18, 75 μm × 100 mm, particle size 1.7 μm, from Waters). In the Q-TOF analysis, MS/MS spectra were acquired by a data-dependent acquisition method involving selection of the three precursors giving the most intense signals. Raw data acquired on the Q-TOF were processed with a ProteinLynx Global Server (Waters). For proteins identified with less than two peptides, the tryptic peptide samples were further analyzed on the more sensitive LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). In the nanoLC system coupled to the LTQ-Orbitrap, peptide separation was performed on a reversed-phase C18 nanoHPLC column (100 µm inner diameter, 5 µm C18 particles, 15 cm length, NTCC-360/100-5, from NikkyoTechnos (NikkyoTechnos Co., Tokyo, Japan). For the LTQ-Orbitrap, MS/MS spectra were acquired using the data-dependent acquisition mode operating with a Top20 collision induced dissociation (CID) method: the mass of the precursors was measured with high resolution (60,000 full width at half maximum (FWHM)) in the Orbitrap and the twenty most intense ions, above an intensity threshold of 5000 counts, were selected for Collision-Induced Dissociation (CID) fragmentation and analysis in the LTQ. Raw data acquired with the LTQ-Orbitrap were processed with Proteome Discoverer 1.3 software (Thermo Fisher Scientific, Waltham, MA, United States of America).
2.5.3. Protein Identification
Proteins were identified using the MASCOT search engine (Matrix Science, London, UK) against the Swissprot 0112 database with carbamidomethylation of cysteines set as a fixed modification, and oxidation of methionines as variable modifications. Peptide and fragment mass tolerances were set at 20 ppm and 0.1 Da, respectively, for Q-TOF data, and at 5 ppm and 0.6 Da, respectively, for Orbitrap data. Only proteins identified with at least two peptides with a score higher than the identity threshold calculated at a false discovery rate of less than 1% (Mascot decoy option) were considered. The generated protein lists were analyzed with the use of Ingenuity Pathways Analysis™ software (QIAGEN Inc., Available Online: https://www.qiagenbioinformatics.com/).
3. Results
3.1. 2-Dimensional Differential in-Gel Electrophoresis (2D-DIGE) and Mass Spectrometry Identification of Protein Spots
Replicative senescence is characterized by a finite number of cell divisions under defined growth conditions. We have recently shown that selenium levels in the culture medium modulated the proliferative capacity of WI-38 [13]. In other words, selenium supplementation (Sup) significantly increased the number of cell divisions and decreased the senescence-associated markers and phenotypes, in comparison with control (Ctl) conditions. On the other hand, selenium deficiency (Dpl) accelerated entry into senescence and reduced the proliferative capacity of WI-38 cells, measured by the population doubling at each passage. This acceleration of the senescence-associated phenotype coincides with the decrease in selenoproteins involved in antioxidant defense, glutathione peroxidase 1 (Gpx1) and glutathione peroxidase 4 (Gpx4). Experimentally, at CPD 35.8, mitotically active WI-38 cells, initially grown in Ctl medium, were plated in three different conditions, namely Dpl, Ctl, and Sup. After two passages, young WI-38 cells grown in the different culture media were harvested, at CPD 39.7, 40.4, and 41.9, respectively, from four experimental replicates, and referred to as Young Dpl, Ctl, and Sup (YD, YC, and YS, respectively). As previously observed, even after two passages of young cells in various selenium-containing media, a difference of cell proliferation could be noticed. In parallel, additional flasks were maintained in respective media and harvested (again in four experimental replicates) when they reached presenescent stage, at CPD 48.7, 54.7, and 57.6, respectively, and were referred to as senescent Dpl, Ctl, and Sup (SD, SC, and SS, respectively), see Figure 1.
A proteomic study was then applied to our samples. Comparison of expression patterns between our six different cell growth conditions was performed using 2D-DIGE methodology (Figure 2A), which displays additional advantages compared with classical 2D-gel proteomic analyses. We took advantage of the high resolution of the 2D-gel electrophoresis that allows the separation of proteins as spots according to their isoelectric point (pI) in the first dimension, and their molecular weight in the second dimension. This technique is particularly efficient to separate protein isoforms with different posttranslational modifications (PTMs). The CyDye fluorescent minimal labeling provides at least the same sensitivity as protein silver staining, but with a much higher dynamic range for quantification. Usually, 2500 protein spots can be mapped in a typical 2D-gel from a whole cell extract. An important feature of the 2D-DIGE methodology is the use of an internal standard in all gels, which allowed a reliable spot assignment between the 12 different gels and a precise inter-gel spot quantification. The scanned images were analyzed with Decyder 7.0 software (GEHealthcare). Six groups, namely YD, YC, YS, SD, SC, and SS were assigned to the various scans, each group being composed of four replicates. The gel images were treated with an exclusion volume filter of 30,000 to remove artifacts from the analyses. A master gel was identified by the software, with 2499 spots. Among this number of spots in the master gel, 1155 spots were found in 80% of the gel maps (i.e., 32 out of 36), and were used for further analyses. In order to validate the quality of the replicates, and, if necessary, remove an outlier replicate from the analysis, we performed a principal component analysis (PCA). In effect, when we applied a statistical filter to the spot maps (one-way analysis of variance (ANOVA) < 0.01), we were able to cluster the 2D protein profiles into six groups of four replicates (Figure 2B) corresponding to our six experimental conditions. The first component separated the protein profiles according to the number of passages (young vs. senescent cells, principal component 1 (PC1): 52% of total variation), while the second dimension differentiated the sample according to the selenium variation (Dpl, Ctl vs. Sup, principal component 2 (PC2): 20.7% of total variation). This statistical analysis indicated that protein expression was more impacted by senescence phenotype than by selenium variation, which seemed to be more subtle. In addition, it appeared that SD, SC, and SS were much more segregated than YD, YC, and YS, indicating that the selenium levels of the culture medium induced more differences in protein expression in senescent than in young cells. When considering only young and senescent cells grown in control medium, YC and SC, respectively, the segregation between the two groups was also obvious with PCA (Figure 2C).
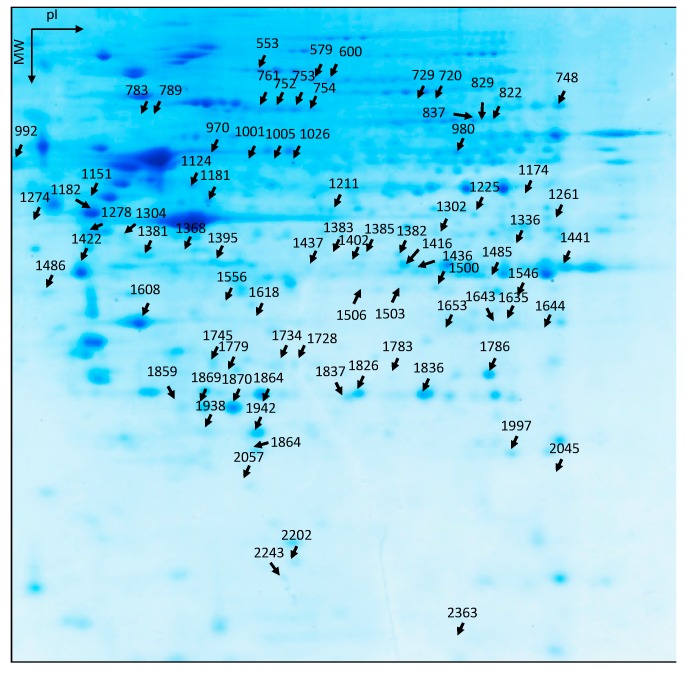
When we applied specific filters for differentially and significantly expressed proteins (±50% and t-test < 0.05), we found 136 spots among the 1155 common to 80% of the spot maps. In other words, the expression of almost 12% of the proteome was affected by either replicative senescence and/or selenium levels of the culture media. In the preparative gel stained with Coomassie staining, only 81 spots were noticeable (see Figure 3). These protein spots were collected and sent for mass spectrometry protein identification and peptide match search with the human proteome databank as described in Materials and Methods. Basically, protein identification was performed after trypsin digestion with a Q-TOF mass spectrometer, except when two or less peptides were identified by MS/MS. In these cases, the remaining tryptic digestion was analyzed with an Orbitrap mass spectrometer, which provided more peptide sequences than the Q-TOF analysis. Among the 81 spots, 123 proteins were identified. The list of identified proteins together with the mass spectrometer used are indicated in Table 1. Three lists of proteins of interest were generated: (i) proteins differentially regulated between YC and SC (±50% and t-test < 0.05, see Table S1); (ii) proteins differentially regulated between YD and YS (same cut-off parameters, see Table S2); (iii) proteins differentially regulated between SD and SS (same cut-off parameters, see Table S3). The proteins common to at least two lists are presented in Table S4. Typical examples of differentially regulated proteins are shown in Figure 4, for spots number 2202, 1734, and 1869, respectively.
Figure 3.
Spot position of the proteins of interest in the spot map of a blue-stained preparative 2D-gel. A quantity of 500 µg of protein sample was separated on a 2D-gel. The identified spots were excised and sent for mass spectrometry identification.
Table 1.
List of proteins identified by tandem mass spectrometry (MS/MS) (either by quadrupole-time-of-flight (Q-TOF) or Orbitrap) from the coomassie blue-stained gel (presented in Figure 3).
| Master Spot Number | Targets of Senescence | Targets of Selenium in Young Cells | Targets of Selenium in Senescent Cells | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Gene Name | MS Ident | Score | Nb | Nb acc | Average Ratio (S/Y) | p Value | Average Ratio (Sup/Dpl) | p Value | Average Ratio (Sup/Dpl) | p Value | |
| Mascot | Peptides | |||||||||||
| Proteins Targets Common to Three Conditions | ||||||||||||
| 754 | Vinculin | VCL | Orbitrap | 452 | 15 | P18206 | 2.40 | 0.01800 | 2.68 | 0.01800 | 3.06 | 0.00540 |
| 2057 | Ferritin light chain | FTL | Orbitrap | 422 | 21 | P02792 | 1.91 | 0.00260 | −1.78 | 0.00046 | −3.05 | 0.00000 |
| 1870 | Superoxide dismutase [Mn], mitochondrial | SOD2 | Orbitrap | 165 | 5 | P04179 | 1.58 | 0.00016 | −1.64 | 0.00057 | −2.42 | 0.00086 |
| 1870 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | Orbitrap | 197 | 12 | P09936 | 1.58 | 0.00016 | −1.64 | 0.00057 | −2.42 | 0.00086 |
| 1869 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | Q-TOF | 251 | 9 | P09936 | 1.54 | 0.00010 | −1.51 | 0.00013 | −2.34 | 0.00003 |
| Proteins Targets Common to Two Conditions | ||||||||||||
| 600 | Glycyl-tRNA synthetase | GARS | Q-TOF | 88 | 4 | P41250 | 1.90 | 0.00059 | −2.09 | 0.00150 | ||
| 600 | Mitochondrial inner membrane protein | IMMT | Q-TOF | 72 | 2 | Q16891 | 1.90 | 0.00059 | −2.09 | 0.00150 | ||
| 579 | Glycyl-tRNA synthetase | GARS | Orbitrap | 1047 | 47 | P41250 | 1.64 | 0.00300 | −1.63 | 0.00130 | ||
| 579 | Mitochondrial inner membrane protein | IMMT | Orbitrap | 1000 | 45 | Q16891 | 1.64 | 0.00300 | −1.63 | 0.00130 | ||
| 579 | Vinculin | VCL | Orbitrap | 940 | 31 | P18206 | 1.64 | 0.00300 | −1.63 | 0.00130 | ||
| 1546 | Prohibitin-2 | PHB2 | Q-TOF | 191 | 6 | Q99623 | 4.48 | 0.00078 | 2.69 | 0.00140 | ||
| 789 | 78 kDa glucose-regulated protein | HSPA5 | Orbitrap | 660 | 22 | P11021 | 2.67 | 0.00280 | 4.47 | 0.00073 | ||
| 1500 | L-lactate dehydrogenase B chain | LDHB | Orbitrap | 235 | 6 | P07195 | 2.61 | 0.00029 | 2.33 | 0.00010 | ||
| 1500 | Malate dehydrogenase, cytoplasmic | MDH1 | Orbitrap | 380 | 15 | P40925 | −2.61 | 0.00029 | 2.33 | 0.00010 | ||
| 761 | Heat shock cognate 71 kDa protein | HSPA8 | Orbitrap | 557 | 20 | P11142 | 2.60 | 0.00540 | 1.79 | 0.01200 | ||
| 761 | Stress-70 protein, mitochondrial | HSPA9 | Orbitrap | 573 | 20 | P38646 | 2.60 | 0.00540 | 1.79 | 0.01200 | ||
| 753 | Gelsolin | GSN | Orbitrap | 252 | 10 | P06396 | 2.40 | 0.01100 | 2.80 | 0.00230 | ||
| 1385 | Actin, cytoplasmic 1 | ACTB | Orbitrap | 184 | 6 | P60709 | 2.33 | 0.00099 | 1.96 | 0.01500 | ||
| 1385 | Translation initiation factor eIF-2B subunit beta | EIF2B2 | Orbitrap | 302 | 9 | P49770 | 2.33 | 0.00099 | 1.96 | 0.01500 | ||
| 1382 | Biliverdin reductase A | BLVRA | Q-TOF | 97 | 5 | P53004 | 2.27 | 0.00130 | 2.14 | 0.00510 | ||
| 753 | Heat shock protein 70 kDa | HSPA1A | Orbitrap | 252 | 7 | P08107 | 2.21 | 0.01100 | 2.80 | 0.00230 | ||
| 752 | Heat shock protein 70 kDa | HSPA1A | Orbitrap | 687 | 24 | P08107 | 2.21 | 0.01500 | 2.50 | 0.00300 | ||
| 1503 | Desmoplakin | DSP | Orbitrap | 198 | 10 | P15924 | 2.21 | 0.00190 | 1.90 | 0.00095 | ||
| 1503 | L-lactate dehydrogenase B chain | LDHB | Orbitrap | 225 | 5 | P07195 | 2.21 | 0.00190 | 1.90 | 0.00095 | ||
| 1503 | Malate dehydrogenase, mitochondrial | MDH2 | Orbitrap | 172 | 6 | P40926 | 2.21 | 0.00190 | 1.90 | 0.00095 | ||
| 1383 | Elongation factor 2 | EEF2 | Orbitrap | 260 | 11 | P13639 | 2.15 | 0.00120 | 2.42 | 0.01200 | ||
| 1383 | PDZ domain-containing protein GIPC1 | GIPC1 | Orbitrap | 244 | 7 | O14908 | 2.15 | 0.00120 | 2.42 | 0.01200 | ||
| 1383 | Transaldolase | TALDO1 | Orbitrap | 164 | 5 | P37837 | 2.15 | 0.00120 | 2.42 | 0.01200 | ||
| 1837 | Peroxiredoxin-6 | PRDX6 | Q-TOF | 214 | 11 | P30041 | 2.04 | 0.00020 | −1.54 | 0.01300 | ||
| 553 | Neutral alpha-glucosidase | GANAB | Orbitrap | 416 | 15 | Q14697 | 1.85 | 0.00007 | −1.64 | 0.00140 | ||
| 553 | Programmed cell death 6-interacting protein | PDCD6IP | Orbitrap | 482 | 18 | Q8WUM4 | 1.85 | 0.00007 | −1.64 | 0.00140 | ||
| 553 | Caldesmon | CALD1 | Q-TOF | 365 | 20 | Q05682 | 1.85 | 0.00007 | −1.64 | 0.00140 | ||
| 553 | Neutral alpha-glucosidase AB | GANAB | Q-TOF | 422 | 20 | Q14697 | 1.85 | 0.00007 | −1.64 | 0.00140 | ||
| 1635 | Annexin A1 | ANXA1 | Orbitrap | 486 | 12 | P04083 | 1.84 | 0.00096 | −2.54 | 0.00460 | ||
| 1635 | Guanine nucleotide-binding protein subunit beta-2-like 1 | GNB2L1 | Orbitrap | 647 | 22 | P63244 | 1.84 | 0.00096 | −2.54 | 0.00460 | ||
| 1635 | Syntenin-1 | SDCBP | Orbitrap | 747 | 22 | O00560 | 1.84 | 0.00096 | −2.54 | 0.00460 | ||
| 1211 | Actin, cytoplasmic 2 | ACTG1 | Q-TOF | 63 | 4 | P63261 | 1.83 | 0.02500 | 1.81 | 0.00014 | ||
| 1734 | Prohibitin | PHB | Q-TOF | 249 | 10 | P35232 | 1.76 | 0.00053 | −1.63 | 0.00160 | ||
| 1859 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | Q-TOF | 119 | 5 | P09936 | 1.69 | 0.03500 | −2.18 | 0.00023 | ||
| 1336 | 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial | HIBCH | Orbitrap | 181 | 5 | Q6NVY1 | 1.62 | 0.00014 | −1.69 | 0.01700 | ||
| 1336 | Methionine adenosyltransferase 2 subunit beta | MAT2B | Orbitrap | 186 | 4 | Q9NZL9 | 1.62 | 0.00014 | −1.69 | 0.01700 | ||
| 1336 | Phosphoserine aminotransferase | PSAT1 | Orbitrap | 261 | 11 | Q9Y617 | 1.62 | 0.00014 | −1.69 | 0.01700 | ||
| 1644 | Annexin A1 | ANXA1 | Q-TOF | 198 | 5 | P04083 | 1.52 | 0.00520 | −1.91 | 0.00600 | ||
| 1644 | Guanine nucleotide-binding protein subunit beta-2-like 1 | GNB2L1 | Q-TOF | 110 | 4 | P63244 | 1.52 | 0.00520 | −1.91 | 0.00600 | ||
| 829 | Pyruvate kinase isozymes M1/M2 | PKM2 | Q-TOF | 299 | 14 | P14618 | −1.54 | 0.00200 | 1.70 | 0.00280 | ||
| 1486 | Actin, alpha skeletal muscle | ACTA1 | Q-TOF | 259 | 5 | P68133 | −1.60 | 0.00190 | 1.61 | 0.00044 | ||
| 1486 | Actin, cytoplasmic 2 | ACTG1 | Q-TOF | 259 | 12 | P63261 | −1.60 | 0.00190 | 1.61 | 0.00044 | ||
| 822 | Pyruvate kinase isozymes M1/M2 | PKM2 | Q-TOF | 152 | 9 | P14618 | −1.78 | 0.00130 | 2.59 | 0.00069 | ||
| 1381 | Serine-threonine kinase receptor-associated protein | STRAP | Orbitrap | 667 | 17 | Q9Y3F4 | −1.87 | 0.00004 | 1.65 | 0.02300 | ||
| 1381 | Vimentin | VIM | Orbitrap | 269 | 13 | P08670 | −1.87 | 0.00004 | 1.65 | 0.02300 | ||
| 720 | Heat shock protein 75 kDa, mitochondrial | TRAP1 | Q-TOF | 143 | 6 | Q12931 | −1.89 | 0.00056 | 2.23 | 0.00120 | ||
| 729 | Heat shock protein 71 kDa | HSPA8 | Orbitrap | 702 | 24 | P11142 | −2.01 | 0.00046 | 2.54 | 0.00110 | ||
| 729 | Prelamin-A/C | LMNA | Orbitrap | 577 | 23 | P02545 | −2.01 | 0.00046 | 2.54 | 0.00110 | ||
| 729 | Heat shock protein 75 kDa, mitochondrial | TRAP1 | Orbitrap | 783 | 32 | Q12931 | −2.01 | 0.00046 | 2.54 | 0.00110 | ||
| 2202 | ADP-ribosylation factor 1 | ARF1 | Orbitrap | 298 | 9 | P00441 | −2.18 | 0.00093 | 2.74 | 0.00160 | ||
| 2202 | Cofilin-1 | CFL1 | Orbitrap | 193 | 5 | P23528 | −2.18 | 0.00093 | 2.74 | 0.00160 | ||
| 2202 | Superoxide dismutase [Cu-Zn] | SOD1 | Orbitrap | 399 | 13 | P00441 | −2.18 | 0.00093 | 2.74 | 0.00160 | ||
| Proteins Targets of One Condition | ||||||||||||
| 1836 | Peroxiredoxin-6 | PRDX6 | Q-TOF | 197 | 11 | P30041 | 1.80 | 0.00001 | ||||
| 1826 | Heat shock protein beta-1 | HSPB1 | Q-TOF | 324 | 11 | P04792 | 1.74 | 0.00031 | ||||
| 1506 | 26S proteasome non-ATPase regulatory subunit 14 | PSMD14 | Q-TOF | 139 | 6 | O00487 | 1.73 | 0.02200 | ||||
| 1745 | Cathepsin D | CTSD | Q-TOF | 195 | 8 | P07339 | 1.68 | 0.00002 | ||||
| 1422 | Reticulocalbin-1 | RCN1 | Q-TOF | 432 | 19 | Q15293 | 1.67 | 0.00027 | ||||
| 1422 | Vimentin | VIM | Q-TOF | 68 | 3 | P08670 | 1.67 | 0.00027 | ||||
| 1643 | Electron transfer flavoprotein subunit alpha, mitochondrial | ETFA | Orbitrap | 1130 | 47 | P13804 | 1.62 | 0.00035 | ||||
| 1181 | Eukaryotic initiation factor 4A-I | EIF4A1 | Q-TOF | 138 | 5 | P60842 | −1.53 | 0.00300 | ||||
| 748 | Pyruvate kinase isozymes M1/M2 | PKM2 | Q-TOF | 236 | 7 | P14618 | −1.59 | 0.00074 | ||||
| 1304 | Spermine synthase | SMS | Q-TOF | 116 | 5 | P52788 | −1.62 | 0.00049 | ||||
| 1304 | Vimentin | VIM | Q-TOF | 83 | 5 | P08670 | −1.62 | 0.00049 | ||||
| 1402 | Transaldolase | TALDO1 | Q-TOF | 279 | 7 | P37837 | −1.66 | 0.01100 | ||||
| 1274 | Actin, cytoplasmic 2 | ACTG1 | Q-TOF | 296 | 9 | P63261 | −1.76 | 0.00100 | ||||
| 1395 | Annexin A2 | ANXA2 | Q-TOF | 122 | 4 | P07355 | 2.16 | 0.00300 | 2.13 | 0.00930 | ||
| 2243 | Actin-related protein 2/3 complex subunit 5 | ARPC5 | Q-TOF | 175 | 5 | O15511 | 1.91 | 0.00230 | 1.57 | 0.02100 | ||
| 1653 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1 | Orbitrap | 820 | 21 | O15144 | 1.53 | 0.00001 | ||||
| 1278 | Protein SET | SET | Q-TOF | 116 | 3 | Q01105 | −1.54 | 0.00620 | ||||
| 980 | Glucose-6-phosphate 1-dehydrogenase | G6PD | Q-TOF | 279 | 14 | P11413 | −1.57 | 0.00000 | ||||
| 1302 | Sialic acid synthase | NANS | Q-TOF | 208 | 6 | Q9NR45 | −1.60 | 0.00000 | ||||
| 783 | 78 kDa glucose-regulated protein | HSPA5 | Orbitrap | 887 | 31 | P11021 | 3.00 | 0.00100 | ||||
| 2045 | Desmoplakin | DSP | Q-TOF | 504 | 18 | P15924 | 2.87 | 0.00460 | ||||
| 1124 | Actin, cytoplasmic 2 | ACTG1 | Q-TOF | 383 | 14 | P63261 | 1.75 | 0.00380 | ||||
| 1124 | POTE ankyrin domain family member E | POTEE | Q-TOF | 269 | 7 | Q6S8J3 | 1.75 | 0.00380 | ||||
| 2363 | Actin-related protein 2/3 complex subunit 5-like protein | ARPC5L | Orbitrap | 961 | 36 | Q4R5P2 | 1.73 | 0.00860 | ||||
| 2363 | Heat shock protein beta-6 | HSPB6 | Orbitrap | 554 | 28 | O14558 | 1.73 | 0.00860 | ||||
| 837 | Catalase | CAT | Q-TOF | 176 | 9 | P04040 | 1.68 | 0.00940 | ||||
| 837 | Pyruvate kinase isozymes M1/M2 | PKM2 | Q-TOF | 151 | 6 | P14618 | 1.68 | 0.00940 | ||||
| 1174 | Fumarate hydratase, mitochondrial | FH | Q-TOF | 244 | 5 | P07954 | 1.64 | 0.03400 | ||||
| 1783 | Endoplasmic reticulum resident protein 29 | ERP29 | Q-TOF | 238 | 7 | P30040 | 1.55 | 0.05000 | ||||
| 1864 | Ubiquitin carboxyl-terminal hydrolase isozyme L1 | UCHL1 | Q-TOF | 121 | 6 | P09936 | −1.50 | 0.02900 | ||||
| 1786 | Phosphoglycerate mutase 1 | PGAM1 | Q-TOF | 65 | 3 | P18669 | −1.53 | 0.00009 | ||||
| 1779 | Endoplasmic reticulum resident protein 29 | ERP29 | Orbitrap | 447 | 14 | P30040 | −1.55 | 0.01100 | ||||
| 1779 | Beta-hexosaminidase subunit beta | HEXB | Orbitrap | 533 | 21 | P07686 | −1.55 | 0.01100 | ||||
| 1779 | Nicotinamide N-methyltransferase | NNMT | Orbitrap | 415 | 19 | P40261 | −1.55 | 0.01100 | ||||
| 1005 | Tubulin beta chain | TUBB | Q-TOF | 311 | 10 | P07437 | −1.56 | 0.00035 | ||||
| 1416 | Annexin A1 | ANXA1 | Q-TOF | 343 | 13 | P04083 | −1.56 | 0.00500 | ||||
| 1728 | Cathepsin D | CTSD | Q-TOF | 175 | 8 | P07339 | −1.56 | 0.00900 | ||||
| 1261 | Fructose-bisphosphate aldolase A | ALDOA | Q-TOF | 196 | 8 | P04075 | −1.57 | 0.00380 | ||||
| 1437 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Q-TOF | 222 | 7 | P04406 | −1.61 | 0.01100 | ||||
| 1225 | Isocitrate dehydrogenase [NADP] cytoplasmic | IDH1 | Q-TOF | 375 | 14 | O75874 | −1.63 | 0.02300 | ||||
| 1026 | Aldehyde dehydrogenase, mitochondrial | ALDH2 | Q-TOF | 185 | 5 | P05091 | −1.65 | 0.00004 | ||||
| 1026 | Xaa-Pro dipeptidase | PEPD | Q-TOF | 184 | 5 | P12955 | −1.65 | 0.00004 | ||||
| 1026 | Tubulin beta-4B chain | TUBB4B | Q-TOF | 140 | 6 | P68371 | −1.65 | 0.00004 | ||||
| 1985 | Peroxiredoxin-2 | PRDX2 | Q-TOF | 201 | 10 | P32119 | −1.65 | 0.00380 | ||||
| 970 | Protein disulfide-isomerase A3 | PDIA3 | Q-TOF | 409 | 19 | P30101 | −1.70 | 0.00200 | ||||
| 970 | Vimentin | VIM | Q-TOF | 158 | 4 | P08670 | −1.70 | 0.00200 | ||||
| 1618 | Annexin A5 | ANXA5 | Q-TOF | 175 | 11 | P08758 | −1.71 | 0.00002 | ||||
| 1182 | Desmin | DES | Q-TOF | 415 | 18 | P17661 | −1.77 | 0.01400 | ||||
| 1182 | Vimentin | VIM | Q-TOF | 415 | 18 | P08670 | −1.77 | 0.01400 | ||||
| 1441 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Q-TOF | 246 | 6 | P04406 | −1.77 | 0.00650 | ||||
| 1942 | Glutathione S-transferase P | GSTP1 | Q-TOF | 329 | 10 | P09211 | −1.82 | 0.00094 | ||||
| 1938 | Glutathione S-transferase P | GSTP1 | Orbitrap | 1357 | 40 | P09211 | −1.85 | 0.00000 | ||||
| 992 | Vimentin | VIM | Orbitrap | 2287 | 97 | P08670 | −1.87 | 0.00400 | ||||
| 1368 | Actin, cytoplasmic 1 | ACTB | Q-TOF | 215 | 8 | P60709 | −2.02 | 0.00056 | ||||
| 1001 | Tubulin beta chain | TUBB | Q-TOF | 213 | 6 | P07437 | −2.03 | 0.00010 | ||||
| 1151 | Protein disulfide-isomerase | P4HB | Q-TOF | 121 | 3 | P07237 | −2.04 | 0.00270 | ||||
| 1151 | Ribonuclease inhibitor | RNH1 | Q-TOF | 86 | 3 | P13489 | −2.04 | 0.00270 | ||||
| 1151 | Vimentin | VIM | Q-TOF | 40 | 2 | P08670 | −2.04 | 0.00270 | ||||
| 1556 | l-lactate dehydrogenase B | LDHB | Orbitrap | 234 | 7 | P07195 | −2.05 | 0.00310 | ||||
| 1556 | Inorganic pyrophosphatase | PPA1 | Orbitrap | 376 | 21 | Q15181 | −2.05 | 0.00310 | ||||
| 1556 | Tubulin beta-6 chain | TUBB6 | Orbitrap | 239 | 8 | Q9BUF5 | −2.05 | 0.00310 | ||||
| 1436 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | Q-TOF | 206 | 6 | P04406 | −2.07 | 0.00900 | ||||
| 1608 | Annexin A4 | ANXA4 | Q-TOF | 481 | 28 | P09525 | −2.17 | 0.00011 | ||||
| 1485 | Annexin A1 | ANXA1 | Q-TOF | 108 | 4 | P04083 | −2.26 | 0.00032 | ||||
| 1485 | Malate dehydrogenase, cytoplasmic | MDH1 | Q-TOF | 99 | 3 | P40925 | −2.26 | 0.00032 | ||||
Nb: number; Sup/Dpl: Supplemented vs. Depleted.
Figure 4.
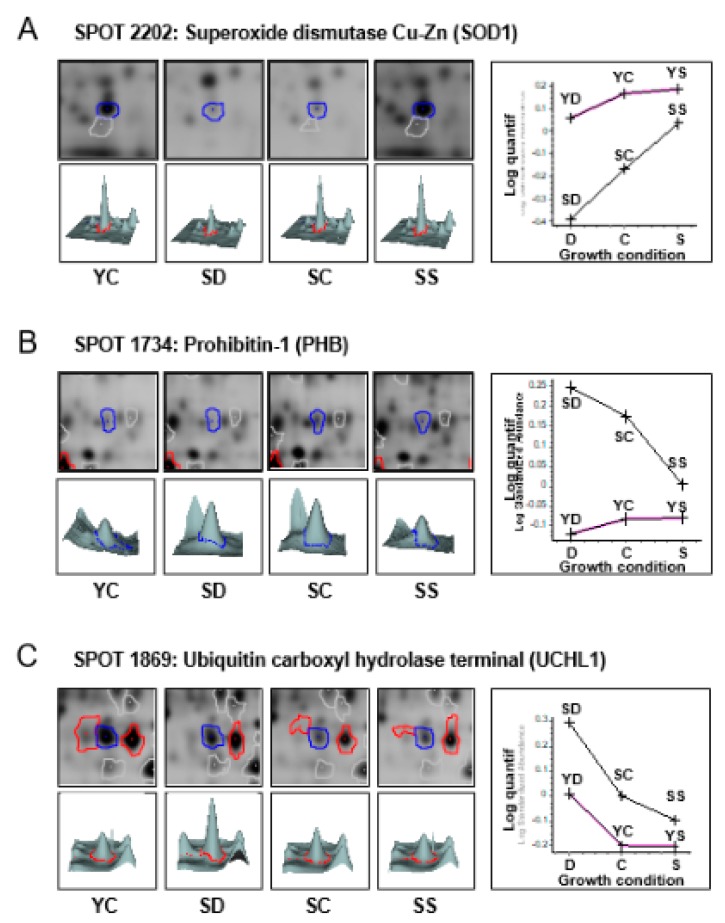
Typical examples of 2D spots differentially regulated by senescence and selenium levels of the culture media. Respective spot maps in 2D (top panels) and 3D (bottom panel) and average quantification (right graphs) are shown for spots number 2202 (A), 1734 (B), and 1869 (C).
3.2. Effect of Senescence in Ctl Conditions
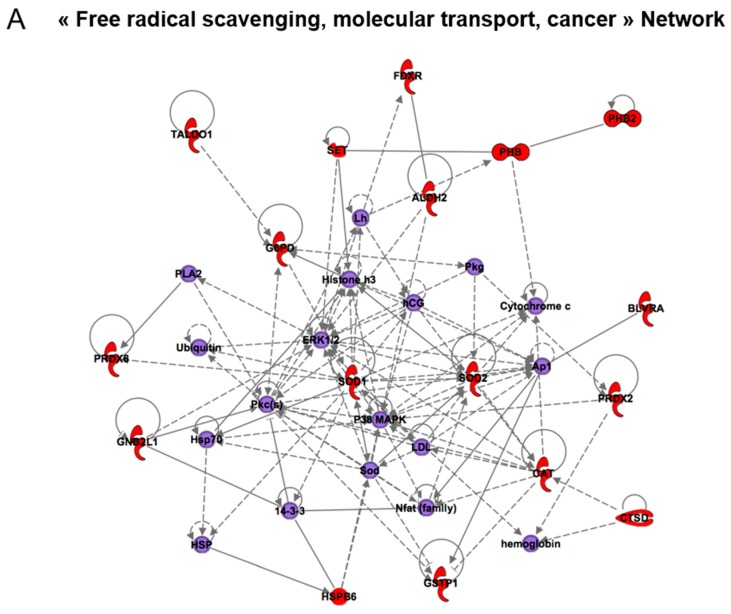
Many proteomic studies have investigated the phenotype of premature senescence, induced either by drug therapy (TIS) or by repeated exposure to mild oxidative stress (SIPS), but very few have investigated the replicative senescence of human diploid cells, such as WI-38 [22,30,31]. From a comparative study, it appeared that SIPS, TIS, and replicative senescence have distinct phenotypes but limited protein targets and pathways in common [22]. In this study, the authors found 50 spots with significant differences (±30% and t-test < 0.05) between young and senescent cells. Among these 50 spots, 12 were common with two different models of SIPS, suggesting that partially distinct mechanisms are involved. Unfortunately, in this study, the proteins were not identified by mass spectrometry. Here, our analysis—performed with slightly different parameters (±50% and t-test < 0.05)—allowed the detection of 43 spots, differentially regulated between young and senescent cell extracts, from which 71 proteins were identified by mass spectrometry (see Table 1 and Table S1). Interestingly, we found three times more proteins whose expression was stimulated in senescent compared to young cells than we did downregulated proteins. Among the most upregulated proteins, we found prohibitin-2 (PHB2), 78 kDa glucose-regulated protein (HSP5A), the cytoplasmic malate dehydrogenase (MDH1), heat shock cognate 71 kDa protein (HSPA8), and Vinculin (VCL), in spots 1546, 789, 1500, 761, and 754, respectively (see Figure 3 for spot location). In contrast, several proteins were found to be downregulated during replicative senescence, including the cytosolic superoxide dismutase (SOD1), the mitochondrial heat shock protein 75 kDa (TRAP1), the serine-threonine kinase receptor-associated protein (STRAP), and Vimentin (VIM) (Table 1 and Table S1). In principle, 2D DIGE allowed the detection of approximately 2500 spots which represent at maximum only 5 to 10% of the proteome, typically the most abundant proteins. It follows that most of the information regarding low abundance proteins is missing. To uncover the molecular pathways that are affected by replicative senescence of other potentially identifiable targets of this phenotype, we performed an Ingenuity® Pathway Analysis (IPA®) on the list of significantly differentially expressed proteins (Table 2). This analysis proposed the three most relevant networks, which were (i) free radical scavenging, molecular transport, cancer; (ii) developmental disorders, and neurological and inherited diseases; and (iii) cancer, neurological disease, and cell signaling. The best-represented network in terms of number of members identified by mass spectrometry is shown in Figure 5A. These data suggest that the free radical scavenging pathway is a target of replicative senescence in WI-38 cells, making relevant the impact of selenium in slowing down the senescence phenotype by improving antioxidant status and redox homeostasis.
Table 2.
Ingenuity pathway analysis of functional networks affected by replicative senescence.
| Network | Members | Molecules | Associated Groups or Molecules |
|---|---|---|---|
| Free radical scavenging, molecular transport, cancer | 20 | ACTA1, ACTG1, ANXA1, ANXA5, CASP8AP2, CTSD, FDXR, HSPB1, LMNA, PHB2, PHB, PRDX2, PRDX6, PSMD14, SMS, SOD1, SOD2, STRAP, TRAP1, VIM | 14-3-3, 26S Proteasome, caspase, CD3, Cytochrome c, estrogen receptor, F actin, glutathione peroxidase, hemoglobin, Insulin, Lh, NFkB (complex), Rb, Ubiquitin |
| Developmental disorder, neurological and inherited diseases | 13 | BLVRA, EIF4A1, ETFA, GARS, GNB2L1, IMMT, PKM2, RAB11A, RCN1, TALDO1, TMEM132D, UCHL1, VSP13D | AkT, Ap1, CENPI, ERK1/2, FAM189B, FRY, FSH, Histone h3, Jnk, LEMD2, Mapk, NUDT6, P38 MAPK, PDGFB, PI3K (complex), Pkc(s), PTCD2, SLC9A6, SPRED3, UBC, USP35, USP40 |
| Cancer, neurological disease and cell signaling | 1 | CEP104 | NR2F6 |
Figure 5.
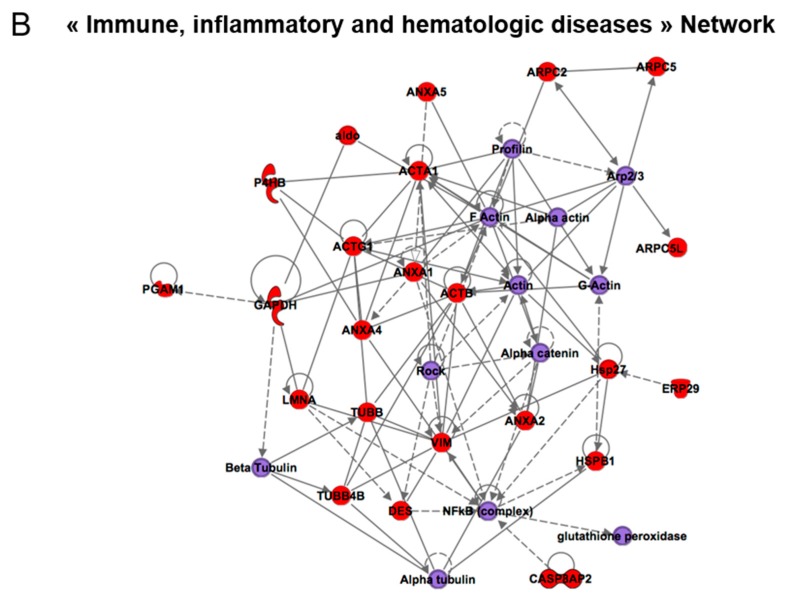
Illustration of the top networks identified by Ingenuity Pathway Analysis for the response to senescence (A) and selenium level variation (B). ACTA1: Actin, alpha skeletal muscle; ACTG1: Actin, cytoplasmic 2; ALDH2: Aldehyde dehydrogenase, mitochondrial; aldo: aldolase; ANXA1: Annexin A1; ANXA4: Annexin A4; ANXA5: Annexin A5; APA: Aminopeptidase A; Arp2/3: Actin-related protein 2/3; ARPC2: Actin-related protein 2/3 complex subunit 2; ARPC5: Actin-related protein 2/3 complex subunit 5; ARPC5L: Actin-related protein 2/3 complex subunit 5-like protein; BLVRA: Biliverdin reductase A; CASP8AP2: Caspase-8-associated protein 2; CAT: Catalase; CTSD: Cathepsin D; DES: Desmin; ERK1/2: Extracellular signal-regulated kinase 1/2; ERP29: Endoplasmic reticulum resident protein 29; FDXR: Ferredoxin reductase; G6PD: Glucose-6-phosphate 1-dehydrogenase; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GNB2L1: Guanine nucleotide-binding protein subunit beta-2-like 1; GSTP1: Glutathione S-transferase P; hCG: Human chorionic gonadotropin; HSP: Heat shock protein; HSP27: Heat shock protein beta-1; HSPB1: Heat shock protein beta-1; HSPB6: Heat shock protein beta-6; LDL: Beta Lipoprotein; Lh: Light-harvesting complex; LMNA: Prelamin-A/C; Nfat: Nuclear factor of activated T-cells; NKkB: Nuclear factor NF-kappa-B; P38 MAPK: Mitogen-activated protein kinase 14; P4HB: Protein disulfide-isomerase; PBH: Prohibitin; PGAM1: Phosphoglycerate mutase 1; PHB2: Prohibitin-2; pkg: protein KINASE G; PLA2: Phospholipases A2; PRDX2: Peroxiredoxin-2; PRDX6: Peroxiredoxin-6; SET: Protein SET; SOD1: Superoxide dismutase [Cu-Zn]; SOD2: Superoxide dismutase [Mn], mitochondrial; TALDO1: Transaldolase; TUBB: Tubulin beta chain; TUBB4B: Tubulin beta-4B chain; VIM: Vimentin.
3.3. Effect of Selenium on Young and Presenescent Cells
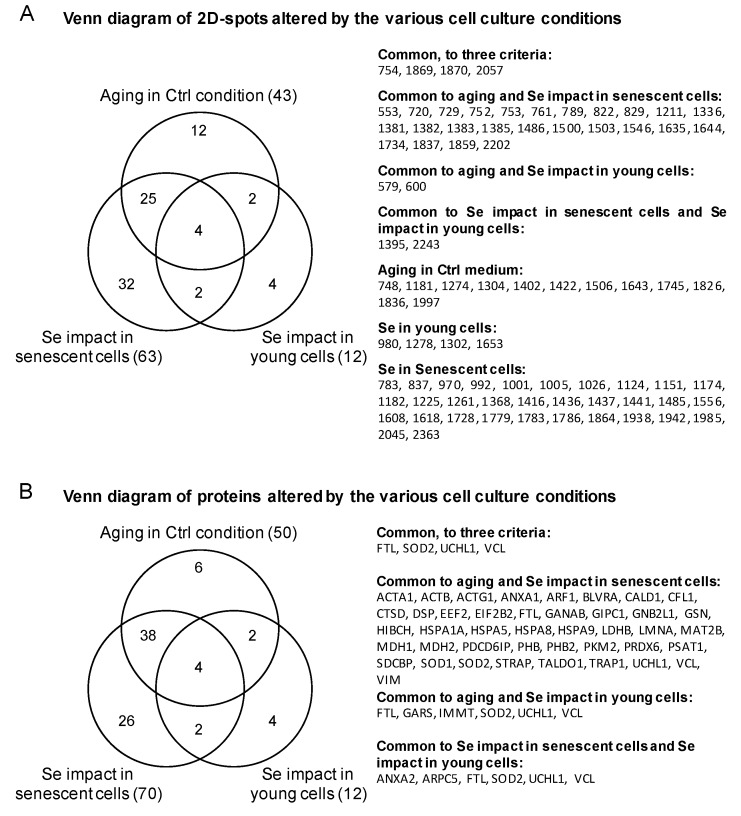
A similar analysis was performed to identify the proteins that were affected by selenium levels in either young or senescent cells. A cut-off of ±50% change in protein expression between Sup and Dpl with a t-test < 0.05 was applied. Spots 12 and 63 fulfilled these criteria in young and senescent conditions, respectively. The proteins identified by MS are listed in Table 1 and in Tables S2 and S3. Among these differentially expressed spots, 12 (young) and 70 (senescent) proteins were identified by mass spectrometry. It appears that selenium levels have much more impact in senescent than in young cells. In addition, the overlap of selenium-dependent variations between young and senescent cells is rather limited, as illustrated in the Venn diagrams in Figure 6. In the list of identified proteins regulated by selenium levels in senescent cells, we found 78 kDa glucose-regulated protein (HSPA5), Vinculin (VCL), Desmoplakin (DSP), Gelsolin (GSN), and Heat shock protein 70 kDa (HSPA1A) among the most upregulated proteins in Sup conditions. Among the most downregulated proteins in Sup conditions, we identified Ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1), Phosphoglycerate mutase 1 (PGAM1), Peroxiredoxin-6 (PRDX6), and Endoplasmic reticulum resident protein 29 (ERP29). An Ingenuity® Pathway Analysis was then performed on the list of significantly differentially expressed proteins upon selenium variation (Table 3). This analysis proposed the three most relevant networks, namely: (i) immune, inflammatory, and hematologic diseases; (ii) cellular function and maintenance, energy production, lipid metabolism; and (iii) cancer, neurological diseases, and cell signaling. The first network, which is composed of 21 identified members, is shown in Figure 5B to illustrate the various connections of the network with associated groups or molecules.
Figure 6.
Venn diagram analyses for the differentially expressed spots (A) and identified proteins (B) in response to senescence and/or selenium level variation in young or senescent cells.
Table 3.
Ingenuity Pathway Analysis of functional networks affected by selenium in young or senescent cells.
| Network | Members | Molecules | Associated Groups or Molecules |
|---|---|---|---|
| Immune, inflammatory and hematologic diseases | 21 | ACTA1, ACTB, ACTG1, ALDH2, ANXA1, ANXA2, ANXA4, ANXA5, DES, FDXR, GADPH, LMNA, P4HB, PHB2, PPA1, RNH1, STRAP, TUBB, TUBB4B, VIM | Actin, alpha actin, alpha tubulin, Ap1, Beta tubulin, CD3, estrogen receptor, F actin, FSH, Insulin, Lh, NFkB (complex), Profilin, Rock |
| Cellular Function and Maintenance, energy production, lipid metabolism | 12 | ALDOA, BLVRA, CAT, FH, IDH1, MDH1, PEPD, PKM2, POTEE/POTEF, PSMD14, TRAP1, VPS13D | 26S proteasome, Akt, BLVRB, ERK1/2, Histone H4, IL22R1-IL10R2, INPP4B, Jnk, Jun-GABP, LEMD2, Mapk, N-arachidonylglycine, NUDT6, P38 MAPK, Pkc(s), PSME4, S100A16, SBSN, SLC26A6, SPRED3, TBCE, UBC, Ybx1-ps3 |
| Cancer, neurological disease and cell signaling | 1 | CEP104 | NR2F6 |
3.4. Intersection between Selenium and Senescence
In order to determine whether the impact of selenium levels on the replicative potential of WI-38 fibroblasts involves similar pathways to regular senescence as suggested in [13], we investigated the overlap between the differentially expressed proteins listed in Tables S1–S3. As inferred by the Venn diagrams, the common targets encompassed 31 spots (Figure 6A), from which 44 proteins were identified by mass spectrometry (Figure 6B and Table S4). Our data show that 72% of the variation observed between young and senescent cells (31 spots over 43) are common with the response to selenium level variation. This value strongly suggests that the change of selenium levels in the culture affects predominantly molecular pathways linked to replicative senescence and may not be related to a premature senescence phenotype like TIS or SIPS. The link between selenium and senescence is illustrated by several selected examples in Figure 6, namely, spots 2202, 1734, and 1869, identified as Superoxide dismutase Cu-Zn (SOD1), Prohibitin-1 (PHB), and Ubiquitin carboxyl hydrolase terminal (UCHL1), respectively. Spot 1869 (UCHL1) is a common target of senescence, selenium in young cells, and selenium in senescent cells. As shown in Figure 4C and Table 1, this protein is upregulated by selenium depletion (+51% in young and +134% in senescent cells) and in response to senescence (+54%). Interestingly, selenium supplementation seemed to counteract this upregulation in senescent cells. It is tempting to speculate that UCHL1 and thence the connected pathway of proteasome protein degradation were affected immediately after selenium changes in the culture media, and that this effect lasted up to the senescence stage. Interestingly, the level of spot 1869 in SS conditions was almost identical to that in young cells (YC). On the other hand, spot 2202 (SOD1) exemplifies the opposite situation (Figure 4A), where a downregulation of the protein occurred during replicative senescence (−118%) and selenium deficiency (−174% in senescent cells). This effect was counteracted in senescent cells by selenium supplementation to recover an almost identical level as that of young cells (compare SS with YC conditions in Figure 4A). Interestingly, the downregulation of this spot seemed to also occur at early passages in Dpl medium, suggesting an early event that could initiate the senescence phenotype. Another kind of regulation is exemplified by spot 1734 (PHB, Figure 4B), which is upregulated (+76%) during the senescence process and by selenium deficiency in senescent cells (63%). Similar to UCHL1 and SOD1, the level of PHB in selenium-supplemented senescent cells (SS) almost reached that of the young conditions (YC), again suggesting a mechanism for the selenium-dependent slow-down of the senescence phenotype. It is important to bear in mind that protein content does not always correlate with enzyme activity. Most enzymes are subjected to PTMs that can alter catalytic activity. Such PTMs are often visible on 2D-Gel electrophoresis. It will be necessary to further characterize PTM changes in identified targets of selenium and/or senescence.
4. Discussion
Our work has consisted of further investigating the initial findings of [13], who reported an acceleration of replicative senescence in WI-38 cells due to selenium deficiency. However, selenium supplementation of the culture media was found to improve the replicative potential of WI-38 cells via the p21, p16, p53, and Rb pathways and to reduce senescence-associated markers (SABG, SAHF, telomere shortening). Here, we have used a proteomic approach to study whether this phenotype was relevant to mechanisms implied in premature senescence, induced by repeated stress (SIPS) or by therapy (TIS), or shared common features with the generic replicative senescence phenotype. The proteomics approach used here is based on separation of the cellular proteome in 2D-gels, fluorescent labelling of the proteins allowing a high sensitivity, and reliable data for inter-gel quantitative comparison. In this work, we detected 43 spots differentially and significantly regulated between young and senescent WI-38 cell extracts grown in Ctl conditions. From these spots, 71 proteins were identified by mass spectrometry (Q-TOF or Orbitrap). We compared this set of proteomic data to that obtained by growing WI-38 in various selenium-containing media for two passages or until they reached the senescence stage. We found that the 2D spots differentially expressed in response to replicative senescence variation share a 72% overlap with the spots of interest (31 spots over 43) identified in response to selenium level variation. Our data strongly suggest that selenium variation interferes with the generic senescence program but not with the premature senescence induced by exogenous stimuli. These data further confirm the strong interplay between selenium level and replicative senescence. This is in agreement with the finding that reactive oxygen species (ROS) are increased in senescent cells, together with a reduction of antioxidant defense and cell metabolism [4]. We posit a hypothesis stipulating that most of the physiological effect of selenium is due to the activity of selenoproteins involved in redox cellular homeostasis and intracellular ROS levels. Selenoproteins are indeed highly responsive to selenium variation following a gene-specific prioritization of selenium incorporation in selenocysteine, also referred to as the selenoprotein hierarchy [32]. This phenomenon has been described in WI-38 cells and found to be involved in transcriptional and translational regulatory mechanisms. To further understand the causal role of selenoproteins in replicative senescence, it will be crucial to follow the impact of gene inactivation of each member of this group of proteins in the acceleration of the senescence phenotype as has been done for several other markers of senescence. For example, the gene inactivation of SOD1, which we found to be downregulated in senescent WI-38 cells, leads to induction of the senescence phenotype [33]. Similar strategies could be employed for selenoproteins by targeting individual members or factors involved in their biogenesis [4]. To date, a single work has reported the impact of selenoprotein knockdown, in this case, SelenoH (also known as SelH), in human MRC-5 fibroblasts [34]. The authors describe a rapid enhancement of senescence-associated markers—including SABG ROS levels and cell autofluorescence—and a decrease in proliferative capacity. The fact that selenoproteins were not found in this proteomic analysis is rather surprising, since they were found to be highly sensitive to selenium variation in a previous study using this cell line. A reasonable explanation could be that the selenoproteome is present at trace levels within the overall proteome. Even the most abundant selenoproteins such as Gpx1 and Gpx4 seemed to be expressed below the detection limits of the CyDye strategy. We would suggest the use of a selenium-specific detection strategy such as (i) radioactive 75Se labelling, followed by autoradiography [35] or (ii) nonradioactive selenium labeling and detection by elemental mass spectrometry [36,37]. Alternatively, a state-of-the-art label-free proteomic strategy may also offer a better proteome recovery. Interestingly, glutathione peroxidases were found in the Ingenuity Pathway Analysis, as illustrated in Figure 5B.
5. Conclusions
Our present work further specifies the action of selenium in replicative senescence of human diploid cells. Our data, from a proteomic perspective, indicate that a significant proportion of the proteins altered by senescence are at least partially counteracted by selenium supplementation, which could explain the improvement of the replicative potential and the reduction of senescence-associated markers observed in our previous work [13]. This study opens the path to selective characterization of selenoproteins in the mechanism of replicative senescence and, more globally, the mechanism of aging.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS) (ATIP program to LC), Institut National de la Santé Et de la Recherche Médicale (INSERM), and Ecole Normale Supérieure (ENS) de Lyon “Emerging Project” (LC), the Fondation pour la Recherche Médicale (LC), the Ligue Contre le Cancer (Comité de l’Essonne, LC), the programme interdisciplinaire de recherche du CNRS longévité et vieillissement (LC), the Association pour la Recherche sur le Cancer [grant number 4849, LC] and the Agence Nationale de la Recherche [grant number ANR-09-BLAN-0048 to LC]. YL was a Recipient of a fellowship from the Ministère Français de l’Enseignement Supérieur et de la Recherche. We thank Manuela Argentini and Willy Bienvenut for assistance in mass spectrometry using the proteomic platform SICaPS (http://www.i2bc.paris-saclay.fr/spip.php?article201) and Virginie Redeker (CNRS, Gif-sur-Yvette) for helpful discussions and comments on the manuscript. We thank the CNRS, the Fondation pour la Recherche Médicale (FRM), the Région Ile-de-France, and IBiSA (Infrastructures en Biologie Santé et Agronomie) for financial support of the mass spectrometers that have been acquired for the SICaPS Proteomic Platform and used in this work.
Abbreviations
ACTA1: Actin, alpha skeletal muscle; ACTB: Actin, cytoplasmic 1; ACTG1: Actin, cytoplasmic 2; AkT: RAC serine/threonine-protein kinase; ALDH2: Aldehyde dehydrogenase, mitochondrial; ALDOA: Fructose-bisphosphate aldolase A; ANXA1: Annexin A1; ANXA2: Annexin A2; ANXA4: Annexin A4; ANXA5: Annexin A5; Ap1: Activator protein 1; ARF1: ADP-ribosylation factor 1; ARPC5: Actin-related protein 2/3 complex subunit 5; BLVRA: Biliverdin reductase A; BLVRB: Biliverdin reductase B; CALD1: Caldesmon; CAT: Catalase; CD3: T-cell surface glycoprotein CD3; CENPI: Centromere protein I; CEP104: Centrosomal protein of 104 kDa; CFL1: Cofilin-1; CTSD: Cathepsin D; DES: Desmin; DSP: Desmoplakin; ECH1: Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial; EEF2: Elongation factor 2; EIF2B2: Translation initiation factor eIF-2B subunit beta; EIF4A1: Eukaryotic initiation factor 4A-I; ERK1/2: Extracellular signal-regulated kinase 1/2; ERP29: Endoplasmic reticulum resident protein 29; ETFA: Electron transfer flavoprotein subunit alpha, mitochondrial; FAM189B: Family with sequence similarity 189, member B; FH: Fumarate hydratase, mitochondrial; FRY: Furry homolog; FSH: follicle- stimulating hormone; FTL: Ferritin light chain; G6PD: Glucose-6-phosphate 1-dehydrogenase; GANAB: Neutral alpha-glucosidase AB; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; GARS: Glycyl-tRNA synthetase; GARS: Glycyl-tRNA synthetase; GIPC1: PDZ domain-containing protein GIPC1; GNB2L1: Guanine nucleotide-binding protein subunit beta-2-like 1; GSN : Gelsolin; GSTP1: Glutathione S-transferase P; HEXB: Beta-hexosaminidase subunit beta; HIBCH: 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial; HSPA1A: Heat shock protein 70 kDa; HSPA5: 78 kDa glucose-regulated protein; HSPA8: Heat shock protein 71 kDa; HSPA9: Stress-70 protein, mitochondrial; HSPB1: Heat shock protein beta-1; HSPB6: Heat shock protein beta-6; IDH1: Isocitrate dehydrogenase [NADP] cytoplasmic; IL22R1-IL10R2: Interleukin-22 receptor subunit alpha-1, Interleukin-10 receptor subunit beta; IMMT: Mitochondrial inner membrane protein; INPP4B: Type II inositol 3,4-bisphosphate 4-phosphatase; Jun-GABP: Jun, GA-binding protein subunit beta-1; LDHB: L-lactate dehydrogenase B; LEMD2: LEM (lamina-associated polypeptide-emerin-MAN1) domain containing 2; Lh complex: Light-harvesting complex; LMNA: Prelamin-A/C; Mapk: Mitogen-activated protein kinase; MAT2B: Methionine adenosyltransferase 2 subunit beta; MDH1: Malate dehydrogenase, cytoplasmic; MDH2: Malate dehydrogenase, mitochondrial; NANS: Sialic acid synthase; NFkB: Nuclear factor NF-kappa-B; NNMT: Nicotinamide N-methyltransferase; NR2F6: Nuclear receptor subfamily 2 group F member 6; NUDT6: Nucleoside diphosphate-linked moiety X motif 6; P38 MAPK: Mitogen-activated protein kinase 14; P4HB: Protein disulfide-isomerase; PDCD6IP: Programmed cell death 6-interacting protein; PDGF BB: Platelet-derived growth factor subunit B; PDIA3: Protein disulfide-isomerase A3; PEPD: Xaa-Pro dipeptidase; PGAM1: Phosphoglycerate mutase 1; PHB: Prohibitin; PHB2: Prohibitin-2; Pkc: Protein kinase C; PKM2: Pyruvate kinase isozymes M1/M2; POTEE: POTE ankyrin domain family member E; PPA1: Inorganic pyrophosphatase; PRDX2: Peroxiredoxin-2; PRDX6: Peroxiredoxin-6; PSAT1: Phosphoserine aminotransferase; PSMD14: 26S proteasome non-ATPase regulatory subunit 14; PSME4: Proteasome activator complex subunit 4; PTCD2: Pentatricopeptide repeat-containing protein 2, mitochondrial; Rb: Retinoblastoma; RCN1: Reticulocalbin-1; RNH1: Ribonuclease inhibitor; S100A16: Protein S100-A16; SBSN: Suprabasin; SDCBP: Syntenin-1; SET: Protein SET; SLC26A6: Solute carrier family 26 member 6; SLC9A6: Sodium/hydrogen exchanger 6; SMS: Spermine synthase; SOD1: Superoxide dismutase [Cu-Zn]; SOD2: Superoxide dismutase [Mn], mitochondrial; SPRED3: Sprouty-related, EVH1 domain-containing protein 3; STRAP: Serine-threonine kinase receptor-associated protein; TALDO1: Transaldolase; TBCE: Tubulin-specific chaperone E; TRAP1: Heat shock protein 75 kDa, mitochondrial; TUBB: Tubulin beta chain; TUBB4B: Tubulin beta-4B chain; TUBB6: Tubulin beta-6 chain; UBC: Ubiquitin C; UCHL1: Ubiquitin carboxyl-terminal hydrolase isozyme L1; USP35: Ubiquitin carboxyl-terminal hydrolase 35; USP40: Ubiquitin carboxyl-terminal hydrolase 40; VCL: Vinculin; VIM: Vimentin; Ybx1-ps3: Nuclease-sensitive element-binding protein 1-like
Supplementary Materials
The following are available online at http://www.mdpi.com/2076-3921/7/1/19/s1, Table S1: Proteins that are differentially expressed between young (Y) and senescent (S) WI-38 grown in Ctl media; Table S2: Proteins that are differentially expressed between Sup and Dpl conditions in Young WI-38 cells; Table S3: Proteins that are differentially expressed between Sup and Dpl conditions in Senescent WI-38 cells; Table S4: List of proteins that are affected by senescence and by selenium (either in young or senescent cells).
Author Contributions
Ghania Hammad, Zahia Touat-Hamici and Laurent Chavatte designed the study; Ghania Hammad, Yona Legrain, Zahia Touat-Hamici, Stéphane Duhieu, David Cornu, Anne-Laure Bulteau and Laurent Chavatte performed the experiments and analyzed the results; Laurent Chavatte prepared the draft manuscript; all authors contributed to discussion of the results and the editing and approval of the final version.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Papp L.V., Holmgren A., Khanna K.K. Selenium and selenoproteins in health and disease. Antioxid. Redox Signal. 2010;12:793–795. doi: 10.1089/ars.2009.2973. [DOI] [PubMed] [Google Scholar]
- 2.Latrèche L., Chavatte L. Selenium incorporation into selenoproteins, implications in human health. Metal Ions in Biol. Med. X. 2008;10:731–737. [Google Scholar]
- 3.Whanger P.D. Selenium and its relationship to cancer: An update. Br. J. Nutr. 2004;91:11–28. doi: 10.1079/BJN20031015. [DOI] [PubMed] [Google Scholar]
- 4.Bulteau A.L., Chavatte L. Update on selenoprotein biosynthesis. Antioxid. Redox Signal. 2015;23:775–794. doi: 10.1089/ars.2015.6391. [DOI] [PubMed] [Google Scholar]
- 5.Gladyshev V.N., Arner E.S., Berry M.J., Brigelius-Flohe R., Bruford E.A., Burk R.F., Carlson B.A., Castellano S., Chavatte L., Conrad M., et al. Selenoprotein gene nomenclature. J. Biol. Chem. 2016;291:24036–24040. doi: 10.1074/jbc.M116.756155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labunskyy V.M., Hatfield D.L., Gladyshev V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobanov A.V., Hatfield D.L., Gladyshev V.N. Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta. 2009;1790:1424–1428. doi: 10.1016/j.bbagen.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 9.Seeher S., Atassi T., Mahdi Y., Carlson B.A., Braun D., Wirth E.K., Klein M.O., Reix N., Miniard A.C., Schomburg L., et al. Secisbp2 is essential for embryonic development and enhances selenoprotein expression. Antioxid. Redox Signal. 2014;21:835–849. doi: 10.1089/ars.2013.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosl M.R., Takaku K., Oshima M., Nishimura S., Taketo M.M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc. Natl. Acad. Sci. USA. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Touat-Hamici Z., Legrain Y., Sonet J., Bulteau A.-L., Chavatte L. Alteration of selenoprotein expression during stress and in aging. In: Hatfield D.L., Schweizer S.U., Tsuji P.A., Gladyshev V.N., editors. Selenium: Its Molecular Biology and Role in Human Health. 4th ed. Springer Science + Business Media, LLC; New York, NY, USA: 2016. pp. 539–551. [Google Scholar]
- 12.Touat-Hamici Z., Legrain Y., Bulteau A.L., Chavatte L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014;289:14750–14761. doi: 10.1074/jbc.M114.551994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legrain Y., Touat-Hamici Z., Chavatte L. Interplay between selenium levels, selenoprotein expression, and replicative senescence in wi-38 human fibroblasts. J. Biol. Chem. 2014;289:6299–6310. doi: 10.1074/jbc.M113.526863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papp L.V., Lu J., Striebel F., Kennedy D., Holmgren A., Khanna K.K. The redox state of secis binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell. Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs B.G., Durik M., Baker D.J., van Deursen J.M. Cellular senescence in aging and age-related disease: From mechanisms to therapy. Nat. Med. 2015;21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 17.Campisi J., d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 18.Herbig U., Jobling W.A., Chen B.P., Chen D.J., Sedivy J.M. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol. Cell. 2004;14:501–513. doi: 10.1016/S1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 19.D’Adda di Fagagna F., Reaper P.M., Clay-Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N.P., Jackson S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 20.Debacq-Chainiaux F., Borlon C., Pascal T., Royer V., Eliaers F., Ninane N., Carrard G., Friguet B., de Longueville F., Boffe S., et al. Repeated exposure of human skin fibroblasts to UVB at subcytotoxic level triggers premature senescence through the TGF-beta1 signaling pathway. J. Cell Sci. 2005;118:743–758. doi: 10.1242/jcs.01651. [DOI] [PubMed] [Google Scholar]
- 21.Toussaint O., Royer V., Salmon M., Remacle J. Stress-induced premature senescence and tissue ageing. Biochem. Pharmacol. 2002;64:1007–1009. doi: 10.1016/S0006-2952(02)01170-X. [DOI] [PubMed] [Google Scholar]
- 22.Dierick J.F., Eliaers F., Remacle J., Raes M., Fey S.J., Larsen P.M., Toussaint O. Stress-induced premature senescence and replicative senescence are different phenotypes, proteomic evidence. Biochem. Pharmacol. 2002;64:1011–1017. doi: 10.1016/S0006-2952(02)01171-1. [DOI] [PubMed] [Google Scholar]
- 23.Flor A.C., Wolfgeher D., Wu D., Kron S.J. A signature of enhanced lipid metabolism, lipid peroxidation and aldehyde stress in therapy-induced senescence. Cell Death Discov. 2017;3:17075. doi: 10.1038/cddiscovery.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M., Ye H., Shao C., Zheng X., Li Q., Wang L., Zhao M., Lu G., Chen B., Zhang J., et al. Metabolomics-proteomics combined approach identifies differential metabolism-associated molecular events between senescence and apoptosis. J. Proteome Res. 2017;16:2250–2261. doi: 10.1021/acs.jproteome.7b00111. [DOI] [PubMed] [Google Scholar]
- 25.Olivieri O., Stanzial A.M., Girelli D., Trevisan M.T., Guarini P., Terzi M., Caffi S., Fontana F., Casaril M., Ferrari S., et al. Selenium status, fatty acids, vitamins A and E, and aging: The nove study. Am. J. Clin. Nutr. 1994;60:510–517. doi: 10.1093/ajcn/60.4.510. [DOI] [PubMed] [Google Scholar]
- 26.Hornsby P.J., Harris S.E. Oxidative damage to DNA and replicative lifespan in cultured adrenocortical cells. Exp. Cell Res. 1987;168:203–217. doi: 10.1016/0014-4827(87)90429-0. [DOI] [PubMed] [Google Scholar]
- 27.Vacchina V., Dumont J. Total selenium quantification in biological samples by inductively coupled plasma mass spectrometry (ICP-MS) Methods Mol. Biol. 2018;1661:145–152. doi: 10.1007/978-1-4939-7258-6_10. [DOI] [PubMed] [Google Scholar]
- 28.Latreche L., Duhieu S., Touat-Hamici Z., Jean-Jean O., Chavatte L. The differential expression of glutathione peroxidase 1 and 4 depends on the nature of the SECIS element. RNA Biol. 2012;9:681–690. doi: 10.4161/rna.20147. [DOI] [PubMed] [Google Scholar]
- 29.Redeker V., Bonnefoy J., Le Caer J.P., Pemberton S., Laprevote O., Melki R. A region within the C-terminal domain of Ure2p is shown to interact with the molecular chaperone Ssa1p by the use of cross-linkers and mass spectrometry. FEBS J. 2010;277:5112–5123. doi: 10.1111/j.1742-4658.2010.07915.x. [DOI] [PubMed] [Google Scholar]
- 30.Dierick J.F., Kalume D.E., Wenders F., Salmon M., Dieu M., Raes M., Roepstorff P., Toussaint O. Identification of 30 protein species involved in replicative senescence and stress-induced premature senescence. FEBS Lett. 2002;531:499–504. doi: 10.1016/S0014-5793(02)03604-9. [DOI] [PubMed] [Google Scholar]
- 31.Toussaint O., Medrano E.E., von Zglinicki T. Cellular and molecular mechanisms of stress-induced premature senescence (SIPS) of human diploid fibroblasts and melanocytes. Exp. Gerontol. 2000;35:927–945. doi: 10.1016/S0531-5565(00)00180-7. [DOI] [PubMed] [Google Scholar]
- 32.Driscoll D.M., Copeland P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 33.Blander G., de Oliveira R.M., Conboy C.M., Haigis M., Guarente L. Superoxide dismutase 1 knock-down induces senescence in human fibroblasts. J. Biol. Chem. 2003;278:38966–38969. doi: 10.1074/jbc.M307146200. [DOI] [PubMed] [Google Scholar]
- 34.Wu R.T., Cao L., Chen B.P., Cheng W.H. Selenoprotein H suppresses cellular senescence through genome maintenance and redox regulation. J. Biol. Chem. 2014;289:34378–34388. doi: 10.1074/jbc.M114.611970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yim S.H., Tobe R., Turanov A.A., Carlson B.A. Radioactive 75Se Labeling and Detection of Selenoproteins. Methods Mol. Biol. 2018;1661:177–192. doi: 10.1007/978-1-4939-7258-6_13. [DOI] [PubMed] [Google Scholar]
- 36.Sonet J., Mounicou S., Chavatte L. Nonradioactive isotopic labeling and tracing of selenoproteins in cultured cell lines. Methods Mol. Biol. 2018;1661:193–203. doi: 10.1007/978-1-4939-7258-6_14. [DOI] [PubMed] [Google Scholar]
- 37.Sonet J., Mounicou S., Chavatte L. Detection of selenoproteins by laser ablation inductively coupled plasma mass spectrometry (LA-ICP MS) in immobilized pH gradient (IPG) strips. Methods Mol. Biol. 2018;1661:205–217. doi: 10.1007/978-1-4939-7258-6_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.