Abstract
We report a biomarker-based non-endoscopic method for detecting Barrett’s esophagus (BE), based on detecting methylated DNAs retrieved via a swallowable balloon-based esophageal sampling device. BE is the precursor of, and a major recognized risk factor for, developing esophageal adenocarcinoma (EAC). Endoscopy, the current standard for BE detection, is not cost-effective for population screening. We performed genome-wide screening to ascertain regions targeted for recurrent aberrant cytosine methylation in BE, identifying high-frequency methylation within the CCNA1 locus. We tested CCNA1 DNA methylation as a BE biomarker in cytology brushings of the distal esophagus from 173 individuals with or without BE. CCNA1 DNA methylation demonstrated an area under the curve (AUC)=0.95 for discriminating BE-related metaplasia and neoplasia cases versus normal individuals, performing identically to methylation of VIM DNA, an established BE biomarker. When combined, the resulting two biomarker panel was 95% sensitive and 91% specific. These results were replicated in an independent validation cohort of 149 individuals, who were assayed using the same cutoff values for test positivity established in the training population. To progress toward non-endoscopic esophageal screening, we engineered a well-tolerated, swallowable, encapsulated balloon device able to selectively sample the distal esophagus within 5 minutes. In balloon samples from 86 individuals, tests of CCNA1 plus VIM DNA methylation detected BE metaplasia with 90.3% sensitivity and 91.7% specificity. Combining the balloon sampling device with molecular assays of CCNA1 plus VIM DNA methylation enables an efficient, well-tolerated, sensitive, and specific method of screening at-risk populations for BE.
Introduction
The incidence of esophageal adenocarcinoma (EAC) has more than quadrupled in the past thirty years (1–4), and the prognosis for EAC patients remains poor, with a less than 20% survival at 5 years (4, 5). Barrett’s esophagus (BE), a pre-malignant intestinal-type columnar metaplasia that replaces the normal squamous mucosa of the distal esophagus, is the only known precursor for EAC, but its detection currently requires performing esophagogastroduodenoscopy (EGD). Due to the high cost of EGD and the lack of a randomized controlled trial demonstrating cost-effective reduction in EAC, endoscopy screening for BE has not been routinely recommended (6, 7). Thus, in approximately 95% of cases of EAC, the presence of the antecedent BE remains undetected and unknown (8). Hence, there is a need for additional methods for BE detection that are less expensive than EGD and can be widely and readily implemented in an at-risk population. Molecular biomarkers for detecting BE are potentially valuable in this regard.
Acquisition of aberrant cytosine methylation within CpG-rich genomic islands is a common accompaniment of many cancers and can serve as a neoplasia biomarker (9–13). We have previously reported that de novo DNA methylation of the CpG island overlapping the first exon of the vimentin gene (mVIM) is a highly sensitive BE biomarker that is present in biopsies of approximately 90% of BE patients, suggesting utility of mVIM as a potential biomarker for BE screening (14). In an initial pilot study, we further found that mVIM could be detected in esophageal brushings, suggesting that mVIM can serve as a molecular cytology biomarker to enable non-endoscopic detection of BE (14). Reduced Representation Bisulfite Sequencing (RRBS) is a whole-genome approach that allows for unbiased assessment of DNA methylation within CpG-dense regions of the human genome, which encompass the majority of promoters and other regulatory regions (15, 16). RRBS thus provides the potential to identify an extended biomarker panel with increased effectiveness for detecting BE.
The aims of this study were hence to identify methylated DNA biomarkers of esophageal neoplasia, to define the sensitivity and specificity of such markers in comparison to and/or in combination with mVIM for detecting BE metaplasias and related neoplasias, and to further test the efficacy of these biomarkers for detecting BE in samples obtained via a non-endoscopic swallowable balloon-based esophageal sampling device as compared to samples obtained via endoscopy directed brushings.
Results
RRBS discovery of DNA methylation biomarkers of esophageal neoplasia
We performed RRBS analysis on a set of 26 esophageal cancer (EAC) biopsies and their respective matched normal squamous biopsies, 15 biopsy or brushing samples of Barrett’s esophagus (BE), and 5 esophageal cancer cell lines (fig. S1). Out of 3,091,193 analyzable CpGs, 26,601 CpGs showed methylation below 10% in all the informative normal squamous samples (requiring at least 4 informative normal samples, each having sequencing depth of at least 20X). When compared in BE, 1,970 of these CpGs showed > 30% methylation in at least 4 samples comprising 45% or more of informative cases (all having sequencing depth of >10X). Of these 1,970 CpGs methylated in BE, 1011 CpGs additionally demonstrated >30% methylation in at least 8 informative EAC biopsies (all having sequencing depth of >10X).
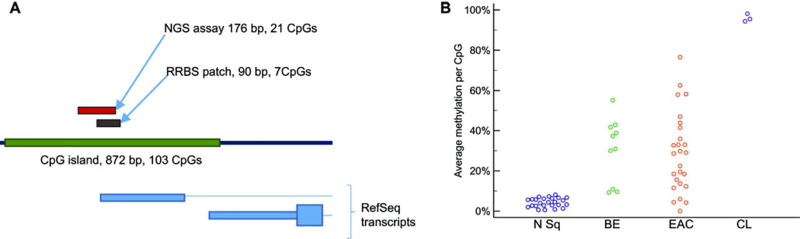
These 1011 CpGs that were differentially methylated between normal squamous versus BE and EAC samples were clustered into 412 differentially methylated CpG patches, defined as clusters of differentially methylated CpGs each less than 200 bp apart. The best 26 of these patches were selected for further inspection (table S1). Of these, the best candidate for discriminating BE-related lesions from normal esophagus was a patch of 7 CpGs located on chromosome 13, in the CpG island spanning the promoter and 5’ UTR of CCNA1 (Fig. 1A). 70% of the BEs characterized by RRBS showed average methylation exceeding 30% across this patch (Fig. 1B). Moreover, all three informative esophageal cancer cell lines showed nearly 100% methylation across this patch, suggesting that both CCNA1 alleles were densely methylated, and that the methylation signal in the biopsies was derived from the abnormal Barrett’s epithelial cells (Fig. 1B).
Figure 1. CCNA1 region methylation in esophageal neoplasia.
(A) Location of the differentially methylated region in the CCNA1 promoter on chromosome 13. The patch of CpGs found to be differentially methylated in reduced representation bisulfite sequencing (RRBS) and the amplicon assayed by next-generation sequencing are indicated above the map of the CpG island (green), with structures of CCNA1 RefSeq transcripts indicated below. (B) Average methylation of CpGs in the CCNA1 RRBS-defined patch of 7 CpGs in: biopsies of normal squamous mucosa (N Sq), Barrett’s esophagus (BE), esophageal adenocarcinoma (EAC), and in esophageal cell lines (CL).
CCNA1 and VIM DNA methylation in esophageal biopsies
To further interrogate the CCNA1-associated DNA methylation patch, we designed a next-generation sequencing (NGS)-based assay for targeted resequencing of this differentially-methylated region (Fig. 1A). We targeted a cluster of 21 consecutive CpG residues that overlap with the methylated patch identified by RRBS (Fig. 1A). The NGS approach allows for determining the methylation status for each of these 21 CpGs across a single individual DNA read, and thereby calculating the percent of CpG methylation present across this region for each individual CCNA1 locus DNA molecule, thus enabling each DNA read to be classified as “methylated” versus “unmethylated”. This approach further provides a metric of tissue methylation, corresponding to the percent of all the individual CCNA1 DNA molecules identified as methylated (percent of CCNA1 “methylated reads”), that can further be referenced as exceeding or not exceeding the upper limit of the normal range. This patch-based algorithm suppresses background from random methylation of individual CpGs and provides enhanced discrimination of normal versus diseased tissue (fig. S2).
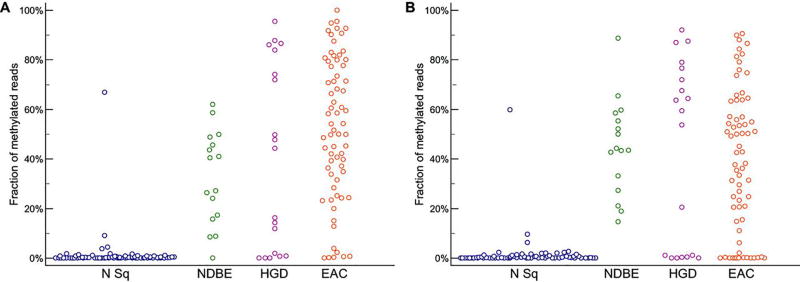
We used this method to characterize an expanded set of esophageal biopsies (fig. S1) and to compare the performance of DNA methylation at the CCNA1 versus VIM loci, reflecting our previous identification of VIM region methylation as a high-performance biomarker of esophageal neoplasia (14). CCNA1 locus methylation (mCCNA1) was significantly increased in all BE-related lesions versus normal (P<0.001, Fig. 2A), with mCCNA1 detected in 81% percent of nondysplastic BE, 68% percent of BE with high-grade dysplasia, and 90% percent of EAC, but in only 1% of normal squamous samples (classifying a tissue as methylated if it had >10% of methylated CCNA1 reads, and classifying a CCNA1 read as methylated when methylation was detected at ≥ 16 out of target 21 target CpG positions). These results were similar to those obtained by bisulfite-sequencing analysis of these samples for VIM locus methylation (mVIM) (Fig. 2B), in which > 10% methylation was demonstrated by 100% percent of nondysplastic BE, 63% percent of BE with high-grade dysplasia, and 76.5% percent of EAC, but by only 1% of normal squamous samples (classifying VIM reads as methylated when methylated at ≥ 8 out of 10 target CpG positions).
Figure 2. NGS bisulfite sequencing assay of DNA methylation in esophageal biopsies.
(A) CCNA1 locus methylation (mCCNA1) in esophageal neoplasia and control patients. N Sq (normal squamous biopsies); NDBE (non-dysplastic Barrett’s esophagus); HGD (Barrett’s esophagus with high-grade dysplasia); EAC (esophageal adenocarcinoma). Fraction of methylated reads in each sample is indicated on the Y-axis. P-value <0.001 for one way ANOVA comparison, and p<0.001 for post-hoc Student-Newman-Keuls test of N Sq versus BE, HGD, and EAC.
(B) VIM locus methylation (mVIM) in esophageal neoplasia and control patients. N Sq (normal squamous biopsies); NDBE (non-dysplastic Barrett’s esophagus); HGD (Barrett’s esophagus with high-grade dysplasia); EAC (esophageal adenocarcinoma). Fraction of methylated reads in each sample is indicated on the Y-axis. P-value <0.001 for one way ANOVA comparison, and p<0.001 for post-hoc Student-Newman-Keuls test of N Sq vs BE, HGD, and EAC.
CCNA1 and VIM DNA methylation for detecting BE and EAC in cytology brushings
To model a “molecular cytology” assay for detecting BE, we used the NGS bisulfite-sequencing assay to measure CCNA1 DNA methylation (mCCNA1) in a training set of esophageal cytology brushings (fig. S1). Brushings of the gastroesophageal (GE) junction were obtained from 62 control subjects with or without symptomatic gastroesophageal reflux disease (GERD), but without BE. These controls included persons with normal endoscopic findings (n=54) plus individuals with erosive esophagitis (n=8). Brushings were also obtained from 111 patients including 62 individuals with cancer, either EAC (n=48) or GE junction adenocarcinoma (n=14), and 49 individuals with BE. Of BE cases, 12 had non-dysplastic short-segment BE (SSBE, < 3 cm), 19 had nondysplastic long segment BE (LSBE ≥3 cm), 8 had low-grade dysplasia (LGD), and 10 had high-grade dysplasia (HGD). Eighty-three percent of study participants were white. The disease group was older than controls (P < 0.0001) and included more males, reflecting the epidemiology of Barrett’s esophagus. There was no significant difference in the prevalence of smoking between cases and controls (table S2).
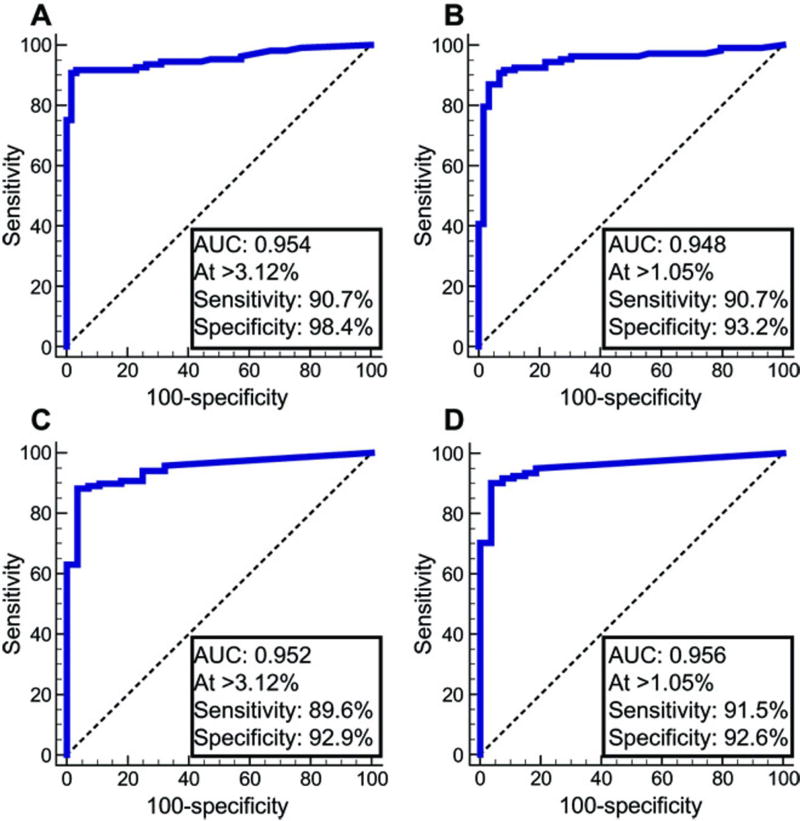
Figure 3A shows the receiver operating characteristic (ROC) curve for mCCNA1 in these training set samples, in which the assay demonstrated an area under the curve (AUC) of 0.95. An optimal cutpoint, in which a sample was detected as positive if it had >3.12% methylated CCNA1 templates, maximized the sum of sensitivity plus specificity. At this cutpoint, mCCNA1 demonstrated 90.7% sensitivity for detecting BE or cancer with 98.4% specificity. As a comparator, Figure 3B shows the ROC curve for the same samples assayed for mVIM by bisulfite-sequencing analysis. Similar to mCCNA1, the mVIM assay showed an AUC=0.95. The optimal cutpoint for this assay, of 1.05% mVIM content, provided sensitivity of 90.7% and specificity of 93.2% (Fig. 3B). These mVIM results are consistent with our previous observation in a pilot set of 34 esophageal brushings that were analyzed for mVIM by qMS-PCR (14).
Figure 3. ROC curves of mCCNA1 and mVIM assayed in esophageal cytology brushings from control normal-appearing GE junctions versus BE and EAC cases.
(A) and (B) Training samples. (A) mCCNA1, n= 61 controls, 108 cases. (B) mVIM, n= 59 controls and 107 cases. (C) and (D) Validation samples. (C) mCCNA1, n= 28 controls, 115 cases. (D) mVIM, n= 27 controls and 117 cases. Area under the curve (AUC) and the sensitivity and specificity of the assays at the indicated cutpoint are listed for each graph, with the cutpoint value of percent methylation that defines a positive test denoted by “At >”.
To validate the performance of mCCNA1 and mVIM in molecular cytology applications, we examined a second independent set of esophageal cytology brushings from 149 new individuals recruited at five different cancer centers from across the United States (fig. S1). These individuals included 30 controls with normal esophageal morphology and 119 cases with BE or cancer (table S2). Similarly to the training set, and reflecting the biology of BE, participants in the validation set were 93% white. Training and validation populations did not significantly differ by gender or smoking history, but controls were older in the validation than in training set (table S2). As shown in Figure 3C and 3D, the validation population again showed an AUC of 0.95 for mCCNA1 and 0.96 for mVIM. Moreover, sensitivity and specificity of mCCNA1 and mVIM in the validation population replicated those of the training population, using the same cutoffs for test positivity as pre-specified by the training dataset (Figures 3C, 3D versus 3A, 3B and table S3).
BE progresses from non-dysplastic metaplasias to low and then high grade dysplasias that ultimately give rise to EAC (17, 18). Given the equivalent performance of mVIM and mCCNA1 in the training and validation populations, we performed a pooled analysis to examine these markers’ performance at each of the stages of progression of BE-related neoplasias. As shown in Table 1, mCCNA1 and mVIM markers were both highly sensitive for detecting each of the stages of BE-related disease, including early non-dysplastic BE metaplasias (NDBE), low and the high-grade dysplasias (LGD and HGD), and cancers. mVIM performed better than mCCNA1 in detecting non-dysplastic BE (91.5% vs 79.7%), whereas mCCNA1 showed a slightly higher sensitivity for dysplastic BE (94.5% vs 91.1%) and cancer (94.9% vs 90.7%). When considered in combination, mVIM and mCCNA1 jointly detected 92% of NDBE, 96% of dysplastic BE, and 96% of cancers, while maintaining specificity above 90% for negatively classifying brushings from the normal GE junction (Table 1). The combination of mVIM and mCCNA1 was more sensitive than either marker individually, even when the specificity of each individual marker was adjusted to match that of the combination (table S4).
Table 1. mVIM and mCCNA1 performance in the combined set of all distal esophagus brushings.
VIM and CCNA1 gene methylation was assayed in DNA samples from cytology brushings of the distal esophagus from: Unaffected controls brushed at the gastroesophageal junction (control GEJ); cases of nondysplastic Barrett’s esophagus (NDBE), further subclassified as short-segment Barrett’s esophagus of 1–3 cm (SSBE) or long-segment Barrett’s esophagus of ≥ 3 cm (LSBE); Barret’s esophagus with low-grade dysplasia (LGD); Barrett’s esophagus with high-grade dysplasia (HGD); esophageal adenocarcinoma and/or junctional cancer of the esophagus (EAC). Samples were scored as VIM methylated for mVIM >1.05% and as CCNA1 methylated for mCCNA1 >3.12% (using ROC defined cutpoints from Fig. 3A, B). Cases were positive for the panel of mCCNA1 plus mVIM if either marker tested positive. Controls were negative for the panel when both mCCNA1 and mVIM were negative. Controls with one negative marker and one marker with assay failure were excluded. Entries indicate percent sensitivity or specificity (%) and total number of individuals tested (n).
| mVIM | mCCNA1 | Either mVIM or mCCNA1 |
||||
|---|---|---|---|---|---|---|
| % | n | % | n | % | n | |
| Specificity control GEJ | 93.0% | 86 | 96.6% | 89 | 90.5% | 84 |
| Sensitivity all cases | 91.1% | 224 | 90.1% | 223 | 94.8% | 229 |
| Sensitivity all NDBE | 91.5% | 71 | 79.7% | 69 | 91.7% | 72 |
| Sensitivity SSBE | 87.1% | 31 | 76.7% | 30 | 87.1% | 31 |
| Sensitivity LSBE | 95.0% | 40 | 82.1% | 39 | 95.1% | 41 |
| Sensitivity all dysplastic BE | 91.1% | 56 | 94.5% | 55 | 96.5% | 57 |
| Sensitivity LGD | 93.9% | 33 | 90.6% | 32 | 94.1% | 34 |
| Sensitivity HGD | 87.0% | 23 | 100.0% | 23 | 100.0% | 23 |
| Sensitivity EAC | 90.7% | 97 | 94.9% | 99 | 96.0% | 100 |
Smoking-induced methylation in the upper esophagus
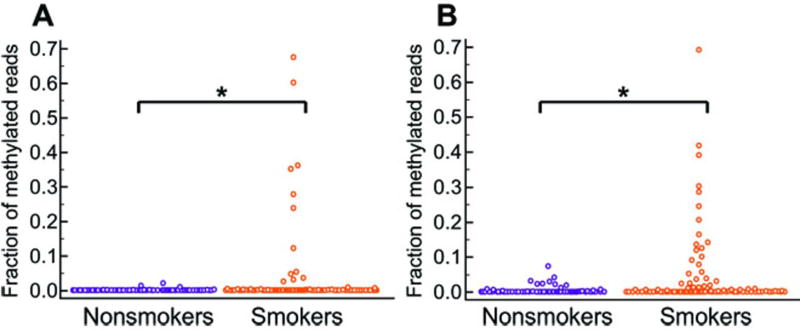
Esophageal brushings under endoscopic guidance provided for directed sampling of the distal esophagus. However, certain non-endoscopic approaches to esophageal sampling, for example using sponge-based devices, require sampling the entire esophagus. To simulate this process, we obtained additional cytology brushings of the proximal normal squamous esophagus from all participants in our training population. To avoid contamination, these proximal brushings were obtained before any brushings of the GE junction. Unexpectedly, a subset of these proximal squamous mucosa brushings tested positive for mVIM or mCCNA1, demonstrating methylation values above the cutpoint for normal GE junction as determined by the training set of brushings. Positive tests were obtained in 21% of proximal squamous samples assayed by mVIM and 6% assayed by CCNA1 (Fig. 4A, 4B). Insight into the basis for methylation in the proximal esophagus came from noting that the great majority of methylation-positive samples were obtained from current or former smokers, with smokers accounting for 75% of samples testing mVIM positive (p=0.0155) and 100% of samples testing mCCNA1 positive (p=0.0094). Further review determined that most of the methylation-positive normal squamous biopsies from our initial experiments (Fig. 2) also came from smokers, though the numbers were too few to support statistical analyses. These findings suggest that using methylated DNA markers for non-endoscopic detection of BE will be enhanced by device designs that allow for selective sampling of the distal esophagus and for protection of samples from contamination by the proximal esophagus.
Figure 4. DNA methylation in the proximal squamous esophagus of smokers versus non-smokers.
(A) mCCNA1, *p=0.0094; (B) mVIM, *p=0.0155. Patients were classified as smokers if they had any history of ever smoking. P values for differences between smokers and non-smokers were computed using the Mann-Whitney rank sum test.
Reassuringly for such a selective sampling approach, reanalysis of brushings from the distal esophagus showed smoking to have no significant effect on the extent of VIM and of CCNA1 methylation detectable in either the normal GE junction of control individuals or in cases with BE or Barrett’s related neoplasia (table S5).
Detecting BE and EAC via non-endoscopic balloon sampling
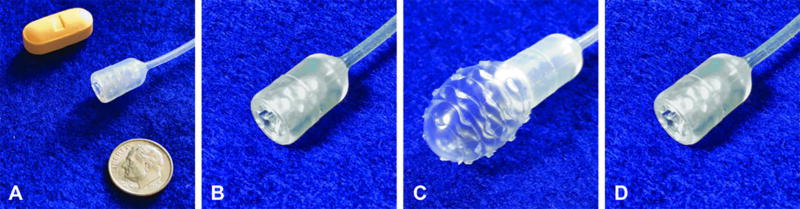
To enable non-endoscopic targeted sampling of the distal esophagus, we designed and built an encapsulated, inflatable, surface-textured balloon. The device is swallowed in a pill-sized 16×9 mm capsule attached to a thin 2.16 mm silicone catheter (Fig. 5A, 5B). After delivery to the stomach, the balloon is inflated by injecting 5 cc of air through the catheter (Fig. 5C) and then gently withdrawn 3 to 6 cm back through the distal esophagus to sample the luminal epithelial surface. The balloon is then deflated and inverted back into the capsule (Fig. 5D), thus protecting the acquired biosample from further dilution or contamination in the proximal esophagus and the oropharynx. After retrieval of the capsule through the mouth, DNA is extracted from the balloon surface for molecular analysis.
Figure 5. Non-endoscopic balloon device.
(A) Device capsule and catheter in comparison to a vitamin pill and a dime. (B) Capsule containing inverted balloon in configuration for swallowing. (C) Capsule with inflated balloon in configuration for esophageal sampling. (D) Capsule containing inverted balloon in configuration for device and biospecimen retrieval.
One hundred fifty six patients underwent unsedated distal esophageal sampling with the balloon device before scheduled EGD. The majority of participants were male (71%) and white (83%), with an average age of 64.1 (SD = 12.5) years. Twenty-eight (18%) were unable to swallow the device. However, the 128 successful participants in trial reported little to no anxiety, pain, or choking, and only low to intermediate gagging (table S6). On average, the balloon reached the stomach in 3.3 min (range 1.0–7.7 min) with excellent tolerance in 72% of cases (table S6). Ninety-three percent of individuals who participated in esophageal balloon testing would repeat the procedure again if necessary while 95% would recommend it to others (table S6).
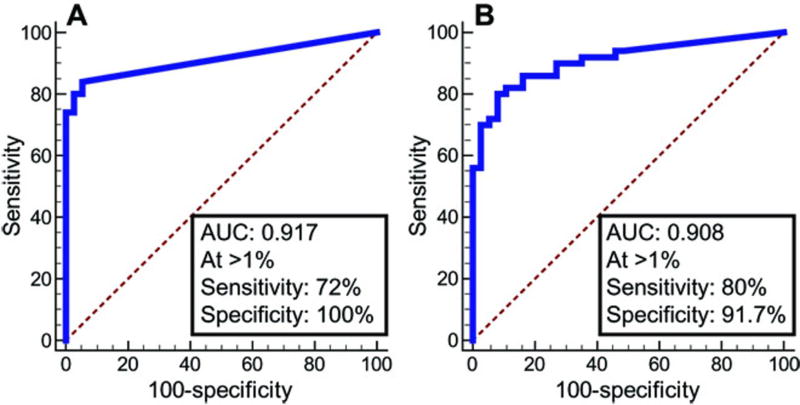
From the 128 balloons processed, an adequate DNA amount of at least 60 ng was obtained in 116 instances (91%). Thirty individuals were additionally excluded from further analysis due to either having a history of prior esophageal ablation for Barrett’s dysplasia (n=23), having gastric intestinal metaplasia (n=4), or having intestinal metaplasia (IM) of <1 cm in length (ultrashort BE, n=3). The 86 evaluable individuals (table S7) included 36 controls free of IM, but with erosive esophagitis (n=3) or other indications for upper endoscopy (n=33). The evaluable individuals also included 50 cases, 42 with BE (31 non-dysplastic, 6 LGD, 4 HGD, 1 indefinite for dysplasia), and 8 with EAC or junctional cancers (Table 2). In these balloon samples, mVIM and mCCNA1 discriminated cases and controls with performance near identical to that found in the cytology brushing samples, with ROC curves for balloon samples demonstrating AUC=0.92 for mCCNA1 and AUC =0.91 for mVIM (Fig. 6A versus Fig. 3A, and Fig. 6B versus Fig. 3B). Balloon samples of the normal GE junction showed less background methylation of CCNA1 than did cytology brushings, and at a cutpoint of 1% for mCCNA1, the assay detected 72% of cases while retaining 100% specificity (Table 2). mVIM demonstrated essentially the same cutpoint in balloon samples (1%) as in cytology brushings, and the mVIM assay detected 80% of cases with specificity of 91.7% (Table 2). When the two markers were jointly applied to the balloon samples, the panel detected 90.3% of nondysplastic BE cases with a specificity of 91.7% (Table 2). The combination had a slightly higher sensitivity of 94.4% for detecting long-segment BE (≥3 cm) but still detected short segment BE of 1–3 cm length with 84.6% sensitivity. Although the study included only a few cases of dysplasia or cancer, the combination of mVIM and mCCNA1 did detect 9 of 11 dysplasias and 7 of 8 cancers, yielding sensitivity of 88% for detection of all BE, dysplasias, and cancers studied (Table 2). Overall, the encapsulated balloon device successfully sampled the distal esophagus with excellent tolerability and acceptability and, when combined with bisulfite sequencing for mVIM plus mCCNA1, demonstrated high sensitivity and specificity for detecting BE.
Table 2. mVIM and mCCNA1 performance in esophageal balloon samples.
VIM and CCNA1 gene methylation was assayed in DNA samples from non-endoscopic balloon sampling of the distal esophagus from: Unaffected controls (individuals with GERD, erosive esophagitis, or no pathology detected during endoscopy); cases of nondysplastic Barrett’s esophagus (NDBE), further subclassified as short-segment Barrett’s esophagus of 1–3 cm (SSBE) or long-segment Barrett’s esophagus of ≥ 3 cm (LSBE); Barret’s esophagus with low-grade dysplasia (LGD); Barrett’s esophagus with high-grade dysplasia (HGD); esophageal adenocarcinoma and/or junctional cancer of the esophagus (EAC). Samples were scored as VIM methylated for mVIM >1.0%, and as CCNA1 methylated for mCCNA1 >1.0%, (using ROC defined cutpoints from Fig. 6). Samples were positive for the panel of mCCNA1 plus mVIM if either marker tested positive. Entries indicate percent sensitivity or specificity (%) and total number of individuals tested (n). We note that only 4 HGD were studied, and in this small sample size differences in the rate of mVIM and mCCNA1 detection of HGD versus detection of NBDE or EAC are not statistically significant (P>0.088 for any between group comparison).
| mVIM | mCCNA1 | Either mVIM or mCCNA1 |
||||
|---|---|---|---|---|---|---|
| % | n | % | n | % | n | |
| Specificity unaffected controls | 91.7% | 36 | 100.0% | 36 | 91.7% | 36 |
| Sensitivity all cases | 80.0% | 50 | 72.0% | 50 | 88.0% | 50 |
| Sensitivity all NDBE | 80.6% | 31 | 71.0% | 31 | 90.3% | 31 |
| Sensitivity SSBE | 69.2% | 13 | 53.8% | 13 | 84.6% | 13 |
| Sensitivity LSBE | 88.9% | 18 | 83.3% | 18 | 94.4% | 18 |
| Sensitivity all dysplastic BE | 72.7% | 11 | 72.7% | 11 | 81.8% | 11 |
| Sensitivity LGD | 83.3% | 6 | 100.0% | 6 | 100.0% | 6 |
| Sensitivity HGD | 50.0% | 4 | 50.0% | 4 | 50.0% | 4 |
| Sensitivity EAC | 87.5% | 8 | 75.0% | 8 | 87.5% | 8 |
Figure 6. ROC curves of mCCNA1 and mVIM assayed on esophageal balloon samplings of the distal esophagus.
(A) mCCNA1, n=36 controls, 50 cases. (B) mVIM, n=36 controls, 50 cases. Area under the curve (AUC), and the sensitivity and specificity of the assays at the indicated cutpoints are listed for each graph, with the cutpoint value of percent methylation that defines a positive test denoted by “At >”.
Methylation in other upper GI tract pathologies
To identify potential confounding sources of VIM and CCNA1 DNA methylation, we performed a retrospective analysis of archival formalin fixed paraffin embedded (FFPE) esophageal and gastric specimens that captured common pathologies of the upper GI tract (Table 3). As expected, FFPE biopsies of BE were highly methylated, with 90% testing positive for mVIM, 75% testing positive for mCCNA1, and 90% testing positive for the two marker panel. Clinical criteria for BE require identifying 1 cm or more of esophageal intestinal metaplasia (IM). However, the two marker panel additionally detected 80% of early esophageal intestinal metaplasias, identifying 80% of IM lesions <1 cm and still confined to the endoscopic GE junction (70% positive for mVIM and 67% positive for mCCNA1). Moreover, among individuals with BE, mVIM or mCCNA1 methylation could also be detected in 30% of biopsy samples of columnar mucosa that had no histologic IM (regions of gastric metaplasia) (Table 3), suggesting that the IM in these BE cases arose in an antecedent field of methylation-positive columnar cells. In contrast, among individuals without BE or IM, assays for mVIM and for mCCNA1 were negative in all GE junction samples (n=55), whichincluded 15 cases with chronic carditis typified by columnar mucosa without IM (Table 3), and methylation was additionally negativein all distal esophagus samples (n=24).Together, these findings suggest that aberrant methylation occurs at the earliest stages of BE development, in those columnar mucosae that are actively evolving toward IM. Methylation is not simply a consequence of inflammation, because mVIM and mCCNA1 tests were all negative in each of 15 cases of eosinophilic esophagitis, as well as in the above-mentioned chronic carditis samples. Methylation was, however, additionally detected in a subset of gastric IM lesions, 22% of which were positive for at least 1 of the two markers. Moreover, 2 of 13 individuals with H. pylori gastritis, who were at increased risk for but did not have gastric IM, were also positive for VIM or CCNA1 methylation. However, 24 normal gastric fundic mucosa without IM were negative for mVIM in all samples and negative for mCCNA1 in all but one sample. Positive tests for mVIM and mCCNA1 are thus highly associated with IM, predominantly IM of the esophagus, but sometimes IM of the stomach.
Table 3. mVIM and mCCNA1 Detection in FFPE biopsies of upper GI tract pathologies.
mVIM and mCCNA1 were assayed in microdissected FFPE biopsies that captured each of the histologies shown. BE (Barrett’s esophagus); IM (intestinal metaplasia); GEJ (gastroesophageal junction). Samples were scored as methylated for mVIM >1.05%, mCCNA1>3.12% (the cutpoints established in ROC analysis of esophageal brushings assayed for each marker). Samples were positive for the panel of mCCNA1 plus mVIM if either marker tested positive. Entries indicate percent positive samples (%) and total number of individuals tested (n).
| mCCNA1 | mVIM | Either mVIM or mCCNA1 |
||||
|---|---|---|---|---|---|---|
| % | n | % | n | % | n | |
| BE (IM) | 75% | 20 | 90% | 21 | 90% | 21 |
| GEJ/cardia with IM (<1cm extent) | 67% | 9 | 70% | 10 | 80% | 10 |
| Non-IM columnar metaplasia concurrent with BE | 11% | 9 | 30% | 10 | 30% | 10 |
| GEJ/cardia without IM (including 15 cases with chronic carditis) | 0% | 53 | 0% | 54 | 0% | 55 |
| Distal normal squamous esophagus from control patients without glandular metaplasia | 0% | 24 | 0% | 24 | 0% | 24 |
| Eosinophilic esophagitis | 0% | 12 | 0% | 15 | 0% | 15 |
| Gastric fundic mucosa without IM | 4% | 24 | 0% | 24 | 4% | 24 |
| Intestinal metaplasia of stomach | 14% | 7 | 11% | 9 | 22% | 9 |
| Helicobacter pylori gastritis without IM | 15% | 13 | 8% | 13 | 15% | 13 |
Consistent with the predictions from this FFPE archive, positive methylation was detected on 2 of 3 balloon samples from cases of ultrashort BE (<1 cm), which were unevaluable in the primary analysis (2 positive for mVIM and none for mCCNA1). Similarly, positive methylation was detected in balloon samples from 4 of 4 cases of gastric intestinal metaplasia that were also unevaluable for the primary analysis (4 positive for mVIM and 2 for mCCNA1). This finding of DNA methylation markers that are shared between gastric metaplasias and very early esophageal metaplasias is consistent with other genomic and cell biology lines of evidence that also point to a common origin between esophageal adenocarcinomas and intestinal type gastric cancers (19, 20)
VIM methylation was additionally identifiable in esophageal balloon samples from certain individuals in whom dysplastic BE had been previously ablated. Specifically, analysis of balloon samples from 23 endoscopically normal individuals who were unevaluable for the primary study, due to their having had ablation of prior dysplastic BE lesions, also demonstrated positive mVIM signals present in 8 (35%) patients (P < 0.017 for increased mVIM positivity in post-ablation versus evaluable control individuals) (table S8). This finding suggests that the endoscopically normal post-ablation GE junction, and/or post-ablation neosquamous epithelium, may retain molecular abnormalities associated with prior IM and may hence harbor continued cellular precursors of BE. It will be intriguing to determine whether, on longitudinal follow-up, these individuals who retain VIM methylation after BE ablation will demonstrate a higher risk for BE recurrence. Curiously, only 2 of these 23 post-ablation individuals were positive for CCNA1 methylation.
Biological Implications
These studies in the esophagus help to inform the larger understanding of the genesis of aberrant DNA methylation in human neoplasias. Our data strongly point to epithelial cells as the source of both VIM and CCNA1 methylation signals. First, VIM and CCNA1 methylation were both detected in epithelial fractions from microdissected FFPE samples of early and late BE lesions (Table 3). Second, EAC cell lines demonstrated nearly complete biallelic methylation (>97% of alleles methylated) in 5 of 5 cell lines for CCNA1 and in 4 of 5 cell lines for VIM (fig. S3). Third, testing in whole organ gastroesophagus porcine explants showed that the balloon device samples a nearly exclusively epithelial cell population (fig. S4). Consistent with this, no breaches of the surface epithelium (no abrasions) were detected in the human participants in this study, all of whom underwent upper endoscopic examination immediately after the balloon procedure. The basis for the aberrant methylation of VIM and CCNA1 genomic loci that we commonly found in BE-related lesions remains obscure. As noted above, smoking was associated with increased mVIM and mCCNA1 only in the proximal (upper) but not in the distal esophagus (Fig. 4, table S5). Moreover, there is no clear functional implication of CCNA1 methylation in BE, because expression of this cyclin family gene is normally restricted primarily to the testis, with lesser expression in the brain, and because CCNA1 expression is not present in either unmethylated normal squamous esophagus or in methylated EAC cells (fig. S3). Similarly, VIM expression is also absent in the unmethylated normal esophagus (fig. S3). However, VIM expression is associated with epithelial to mesenchymal transition (EMT) of cancer cells (21), and although VIM expression was absent in all 4 VIM methylated EAC cell lines, VIM expression was detected in unmethylated FLO1 EAC cells (fig. S3), suggesting that VIM methylation may provide a mechanism for cancer cells to suppress EMT.
Discussion
This study demonstrates the feasibility of non-endoscopic office-based molecular cytology screening for BE and EAC. First, we have identified cytosine methylation of the CCNA1 locus as a methylation marker of Barrett’s esophagus. Secondly, we have shown that molecular cytology assays of distal esophageal brushings, by bisulfite-sequencing detection of the 2-marker panel of mVIM and mCCNA1 DNAs, detects BE and EAC with sensitivity and specificity both greater than 90%. Thirdly, we have shown that a swallowable balloon device can obtain DNA samples from the distal esophagus in a rapid simple unsedated outpatient examination, and, when combined with bisulfite sequencing for detecting DNA methylation, maintains both sensitivity and specificity for detecting BE and EAC at close to 90%. The combination of this sampling device and molecular diagnostic assay demonstrates the feasibility of non-endoscopic molecular cytology screening for BE as a method for ultimately preventing EAC development.
EAC has steadily increased in incidence over recent decades. With an 82% five year mortality rate, this cancer is the most rapidly increasing cause of cancer mortality from solid tumors in the American population (5). BE is a precursor lesion from which EAC develops, and ablation of BE with HGD and/or LGD is recommended to prevent EAC (22, 23). However, the great majority of EACs are diagnosed in patients who have never had prior BE screening. This reflects that most patients with GERD symptoms do not undergo upper endoscopy, and, moreover, that 40% of EACs develop in patients with no prior symptoms (24, 25). Thus, the success of ablation approaches in preventing EAC will remain limited without an acceptable and cost-effective method to effectively screen at-risk populations and detect those individuals who are harboring BE. Non-endoscopic balloon sampling paired with molecular assay for mVIM and mCCNA1 offers a technology to address this need for simple non-invasive BE screening.
Given the need, other investigators have also advanced alternative approaches for BE screening. One such approach uses sponge-based devices for esophageal sampling (26, 27). One advantage of the balloon device is that it deploys rapidly by inflation, eliminating the waiting time for the coating on typical sponge devices to dissolve. Additionally, as the balloon retracts into its capsule after sampling, it enables directed sampling of the distal esophagus plus provides protection of the sample from dilution or contamination from the proximal esophagus or oral cavity. Moreover, the smaller dimensions of the retracted balloon provide for easier and more comfortable retrieval. Balloon sampling of the esophagus was first demonstrated by Falk and colleagues (28), with the current balloon based device now incorporating additional design enhancements that include optimized surface texturing and the addition of a protective capsule for delivery and retrieval.
Immunohistochemical detection of trefoil factor 3 has also been proposed as a biomarker for detection of BE, with a reported sensitivity of 79.9% when used with a cytosponge collection device (29). Attractions of DNA-based biomarkers include absence of subjectivity in interpretation and ease of automation for processing large sample numbers. Although direct comparison between the approaches will be of interest, the finding of 91% sensitivity for detecting BE metaplasia by mVIM plus mCCNA1 assays performed on balloon samples suggests this approach is likely at least as robust and may offer ease of examination and easier scalability to large populations. The lower background of CCNA1 methylation in control individuals sampled by esophageal balloons, as compared to cytology brushings, is intriguing, and perhaps reflects a useful advantage arising from the somewhat more superficial cell layer sampled by the balloon versus by cytology brushings.
We note that this study does have some limitations. This investigation was conducted at a single tertiary care institution, and establishing generality will require replication at other centers and in community-based populations. Our study population is also predominantly male Caucasians, suggesting caution in extrapolating these results to females and other ethnic groups, in which BE is less common. Additionally, we suffered failures of some participants to swallow the balloon device or of the device to obtain adequate sample. Remedying this limitation will need to be a focus of future enhancements to the device design. Last, we lack longitudinal follow-up to be able to interpret implications of finding positive tests for mVIM and mCCNA1 in endoscopically normal individuals without IM of stomach or esophagus or in higher risk individuals who are post ablation of dysplastic BE.
In summary, this study suggests that the combination of a balloon-based sampling device with bisulfite sequencing of the VIM and CCNA1 loci provides a highly sensitive and specific yet minimally invasive screening procedure that could be clinically useful for detection and screening of BE.
Materials and Methods
Study Design
The study protocol was approved by University Hospitals Case Medical Center and Cleveland Clinic Institutional Review Boards for Human Subject Investigation. Clinical trial registration numbers at ClinicalTrials.gov are NCT02451124 for the non-endoscopic balloon trial and NCT00288119 for the endoscopic cytology brushings study. Subjects referred for outpatient EGD were approached for study participation. Consent for obtaining esophageal brushings and biopsies for research was obtained from subjects before their EGD. Cases were classified as subjects with newly diagnosed BE, those undergoing surveillance of BE, or those with a new diagnosis of esophageal or gastroesophageal junctional adenocarcinoma undergoing an endoscopic procedure. BE was defined according to current ACG guidelines as at least one cm of endoscopically visible columnar mucosa in the distal esophagus with intestinal metaplasia confirmed on histology (7). Lesions < 1 cm were classified as ultrashort BE and analyzed separately. Control subjects had no endoscopic evidence of BE and no histological evidence of intestinal metaplasia if a clinical biopsy was obtained from either the distal esophagus or gastroesophageal junction.
The overall study was a non-randomized observational study. Esophageal brushings were first used to validate findings from esophageal biopsies of methylated vimentin as a biomarker for detection of Barrett’s esophagus. Balloon-based esophageal samples were then obtained as a second validation sample. Study size was not pre-specified, and results are reported for esophageal brushing samples accrued from June 2011 to February of 2017 and for all balloon samples accrued from July 2015 to August of 2016. No subjects were excluded from reporting. The primary endpoint of detection of Barrett’s esophagus and related progressed lesions was pre-specified before study initiation. All laboratory samples were assayed by investigators blinded to the clinical status of the subjects.
Reduced Representation Bisulfite Sequencing
Reduced representation bisulfite sequencing was performed as previously described (15). Genomic DNA isolated from frozen esophageal biopsies or cell lines was digested with MspI (New England Biolabs # R0106T). End repair, A-tailing, and adapter ligation were carried out using the NEXTFlex Bisulfite-sequencing library prep kit (Bioo Scientific, cat# NOVA-5119-02) with additional use of methylated NEXTFlex bisulfite-sequencing barcoded adapters (Bioo Scientific, cat# NOVA-511912) to allow for multiplexing multiple samples on a single sequencing lane. After adapter ligation, the library was size fractionated on an agarose gel, and DNA fragments between 170–350 bps (corresponding to the initial unligated 40–220 bp MspI digest fragments) were isolated and purified from the gel using Macherey-Nagel gel extraction and PCR purification kit (Cat#740609.250). After elution from the columns, DNA was bisulfite converted using the EpiTect Bisulfite Conversion Kit (QIAGEN, cat# 59104). Adapter-ligated and converted libraries were then PCR amplified and purified using Macherey-Nagel gel extraction and PCR purification kit.
Sequencing was performed on an Illumina HiSeq 2500 using paired-end 100 bp reads. Individual BAM files for each sample were generated for alignment. DNA sequencing reads from each RRBS experiment were aligned to bisulfite-converted and unconverted versions of the human reference genome (hg18) using Bowtie2, and percent methylation for each CpG was calculated by dividing the number of methylated Cs by the total coverage of that base. These analyses were facilitated by the Bismark software (32), which was specifically developed for processing RRBS data. The overall pipeline converts raw RRBS fastq files to tables of read depth and percent methylation at each individual CpG site for each patient sample.
Endoscopic Sampling
During the endoscopy, one cytology brushing (US Endoscopy) was obtained from the proximal squamous esophagus, 20–25 cm from the incisors, as soon as the esophagus was intubated. The second cytology brushing was obtained from endoscopic BE or cancer lesions. In endoscopic normal controls, the gastroesophageal junction was brushed to sample the glandular mucosa as well as distal squamous mucosa. Standard of care clinical biopsies were obtained from suspected BE and cancer cases. Data were collected from subjects regarding demographics, indications for EGD, GERD symptoms, exposures, and past medical history. All diagnoses (Table 1) were established by endoscopic report and histopathology review of clinical biopsy samples. Demographic data for all patients are provided in table S2. Brushes were immediately clipped with wire cutters into empty nuclease-free 0.5 ml cryo-safe tubes and immediately snap frozen on dry ice for transport to storage at −80°C until use. Genomic DNA was extracted from endoscopic brushes using the DNeasy blood and tissue kit (Qiagen #69504). The protocol for the purification of Total DNA from Animal Tissues (Spin-Column Protocol) was used with the modification of increasing the digestion time to overnight. Final elution volume was 100 µl. DNA yields from clinical samples were quantitated using the Qubit fluorometer (Invitrogen).
FFPE Tissues Specimens
Archival normal and neoplastic tissue specimens were obtained from the Department of Pathology at University Hospitals Case Medical Center under a tissue procurement protocol approved by University Hospitals Case Medical Center Institutional Review Board. Before use, diagnostic slides of all samples were reviewed by a gastrointestinal pathologist (J.W.) for confirmation of the recorded diagnoses. After confirmation of diagnoses, specimens for this study were prepared via punch biopsies of tissue blocks. The presence of intestinal metaplasia in designated esophageal biopsies was required for a diagnosis of Barrett’s esophagus according to published guidelines.(7)
DNA was purified using QIAamp DNA micro kit (QIAGEN) according to the manufacturer’s protocol with the following modifications: The starting extraction volume was increased to 200 µl ATL buffer and 50 µl proteinase K. The cores were digested for four days at 60 degrees. An additional 6 µl of Proteinase K was added on days 2 and 3 of incubation. The DNA was eluted from columns in 100 µl of low-TE elution buffer and used immediately for bisulfite conversion, or frozen at −80°C until use.
Non-endoscopic esophageal brushing via a balloon device
We designed an encapsulated, inflatable, surface-featured balloon for targeted sampling of the distal esophagus. The device was delivered in a 16 by 9 mm capsule that also protected the acquired biospecimen from potential contamination with proximal esophagus material during withdrawal. Subjects referred for outpatient EGD were approached for study participation. Patients underwent un-sedated distal esophageal sampling with the balloon device before scheduled EGD. After a patient swallowed the capsule with the balloon and the capsule was allowed to reach the stomach, the balloon was inflated with 5–5.5 cc of air using the attached tubing and syringe. On average, the balloon reached the stomach in 3:21 minutes (range, 1 to 7:39 minutes). The inflated balloon was pulled slowly for 3–6 cm from the point where the constriction of the esophageal sphincter was felt by the endoscopist, and then deflated using an attached syringe, thus retracting the balloon inside the capsule and protecting the sample. After the capsule was removed, the balloon was re-inflated, cut off from the capsule, and immediately frozen. DNA was extracted from balloons using the same protocol as for endoscopic brushings, but with the modification of increasing the volumes of kit buffers ATL and AL to 1 ml, in order to completely cover the balloons. After the procedure, each patient filled out a standardized tolerance and acceptance questionnaire (table S9) (30).
Bisulfite conversion of the genomic DNA
To create a template for PCR and then DNA methylation analysis, DNA samples were subjected to treatment with sodium bisulfite, which converts unmethylated cytosine bases into uracil, while leaving methylated cytosines intact, using an Epitect kit (QIAGEN) according to the manufacturer’s protocol. The person performing the methylation analysis was blinded to the clinical history of the samples until after all procedures and calculations were completed.
Bisulfite-Sequencing-based methylation detection (Bisulfite-Seq
Bisulfite-converted DNA samples from cytology brushings were analyzed by NGS. Bisulfite-specific, methylation-indifferent PCR primers were constructed as a mixture of primers against converted products of fully methylated or fully unmethylated templates and were used to amplify a differentially-methylated region of the vimentin exon 1 CpG island (previously described, (31)) or CCNA1 (table S10). Platinum Taq reaction mix (Invitrogen) was supplemented with 1 mM MgCl2, 0.2 mM dNTP mix (New England Biolabs), 0.5 M Betaine (Sigma), and a mix of the 4 primers, each at 0.1 µM final concentration. PCR was performed using a touchdown protocol where after the activation of Taq polymerase at 95°C for 5 min, the initial cycling conditions were: 95°C for 45 sec, 67°C for 45 sec, 72°C for 45 sec. The annealing temperature was decreased by 3°C every 3 cycles, to a final of 55°C. An additional 33 cycles of PCR were performed at the annealing temperature of 55°C. Successful amplification was confirmed by agarose gel electrophoresis. PCR products were purified using NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel), and quantitated by Qubit. The NEXTflex Rapid DNA-seq kit (BIOO Scientific) was used to prepare indexed libraries for NGS sequencing (Illumina-compatible), and NGS was performed using a MiSeq platform at the McGill University and Génome Québec Innovation Centre, Montréal, Canada. FFPE DNA samples were bisulfite-converted and sequenced by Epiquest (Zymo Research), using the same protocol as above.
Analysis of non-endoscopic balloon DNA samples was done the same way as for brushing samples above, except that the PCR amplification primers (table S10) were indexed by adding 96 different 7 bp index tags to the 5’ end of both forward and reverse primers (table S11). The amplification with indexed primers was carried out under the same conditions as for non-indexed primers, and the PCR products were subsequently mixed together, before preparing a library for NGS using a non-indexed library adapter.
DNA sequencing reads from each sample were aligned to bisulfite converted and unconverted versions of the human reference genome (hg18) using Bowtie2. VIM aligned reads were classified as methylated if they indicated that 8 or more CpG dinucleotides were methylated (out of total of 10 CpGs present between the primers in the VIM PCR fragment). CCNA1 aligned reads were classified as methylated if they indicated that 16 or more CpG dinucleotides were methylated (out of total of 21 CpGs present between the primers in the CCNA1 PCR fragment). These analyses were facilitated by use of Bismark software (32), which was specifically developed for processing bisulfite-sequencing data.
Statistical methods
Between-group comparisons of continuous variables were performed using unpaired t test (for two groups) or one-way ANOVA for groups of 3 or more, followed by the post-hoc Student-Newman-Keuls test for all pairwise comparisons. The Mann-Whitney rank sum test was used to determine the p-value for comparison of smokers and non-smokers. Fisher’s exact test was used for comparison of demographic composition of cases/controls in brushing and balloon studies. ROC curves, and all associated statistics were generated using MedCalc software. The optimal cut-points were calculated as maximizing the sum of sensitivity plus specificity.
Supplementary Material
Supplementary figure 1. Flowchart of study analyses.
Supplementary figure 2. Comparison of methylation of individual CpGs versus CpG patches in discriminating esophageal lesions.
Supplementary figure 3. Comparing VIM and CCNA1 Expression versus methylation.
Supplementary figure 4. Morphology of touch preps from balloon brushings of three intact porcine esophagus samples, from esophagogastric organ explants.
Supplementary table 1. Location of 26 differentially methylated patches identified by RRBS comparison of normal esophageal squamous mucosa versus Barrett’s lesions.
Supplementary table 2. Demographic characteristics of training and validation esophageal brushings populations.
Supplementary Table 3. mVIM and mCCNA1 performance in training and validation esophageal brushing samples.
Supplementary Table 4. Comparison of sensitivities of mVIM plus mCCNA1, versus mVIM, and versus mCCNA1, all at equal specificities.
Supplementary table 5. Influence of smoking on VIM and CCNA1 methylation in proximal versus distal esophagus.
Supplementary table 6. Participant evaluation of the non-endoscopic balloon sampling of the esophagus.
Supplementary table 7. Demographic characteristics of subjects in non-endoscopic balloon study.
Supplementary table 8. Methylation in post-ablation subjects.
Supplementary table 9. Post-examination questionnaire.
Supplementary table 10. Bisulfite-specific methylation-independent PCR primer sequences.
Supplementary table 11. Index tags for bisulfite-specific methylation-independent PCR primer sequences.
Acknowledgments
The authors wish to acknowledge the contribution of staff of the McGill University and Génome Québec Innovation Centre, Montréal, Canada for NGS sequencing and to thank Nottingham Spirk, North Coast Medical Development, Michael Haag and Stephanie Weidenbecher for support and assistance in development of the balloon device.
Funding: Supported by NIH grants P50CA150964, U54CA163060, UO1CA152756, UL1TR000439, UH2CA205105, American Cancer Society grant 123436-RSG-12-159-01-DMC, and awards from the Case-Coulter Foundation Partnership for Translational Research and the Ohio Third Frontier Technology Validation and Start-Up Fund.
Footnotes
Author contributions
S.D.M. and A.C. designed the study; A.C., J.D., P.N.T, A.F., and J.B.S., wrote and submitted the clinical protocol and treated the patients; P.G.I., M.I.C., J.S.W., and N.J.S. recruited patients and obtained research tissue samples; A.K.C. and W.B. archived and managed the clinical samples; J.E.W. performed pathology review of FFPE samples; J.L. prepared and archived DNA for all samples used in the study; H.R.M. and K.G. performed the experiments; O. dl C.C. and T.L. performed the NGS analyses; H.R.M. and S.D.M. analyzed the data; H.R.M., S.D.M., and A.C. wrote the manuscript. All authors reviewed and approved the final manuscript.
Competing interests: Drs. Chak, Willis, and Markowitz have awarded patents on the use of methylated Vimentin for detection of Barrett’s esophagus and other GI cancers and have also pending patent on a balloon based device for non-endoscopic sampling of the esophagus. Drs. Chak, Willis, Markowitz, LaFramboise, de la Cruz Cabrera, and Moinova have pending patents on methylated CCNA1. Patent rights have been assigned to Case Western Reserve University and are managed under institutional conflict of interest policies. Awarded patents include: U.S. Patent 9580754, Methods and Compositions for Detecting Gastrointestinal and Other Cancers; U.S. Patent 8415100, Methods and Compositions for Detecting Gastrointestinal and Other Cancers; U.S. Patent 8221977, Methods and Compositions for Detecting Colon Cancers. Pending patents include: PCT/US2014/070060, Device for collecting a biological sample; PCT/US2010/030084, Digital quantification of DNA methylation; PCT/US2015/068131, Methods and compositions for detecting esophageal neoplasias and metaplasias; PCT/US2017/040708, Methods and compositions for detecting esophageal neoplasias and/or metaplasias in the esophagus. S.D.M. has consulting relationships with Rodeo Therapeutics, Jannsen Pharmaceuticals, and GlaxoSmithKline. A.C. has consulting relationships with U.S. Endoscopy and Coldplay Therapeutics. N.J.S. consults for Shire, Ambu, and Boston Scientific and has research funding from Medtronic, C2 Therapeutics, CSA Medical, EndoStim, CDx Medical, and Interpace Diagnostics. P.G.I. consults for Medtronic and has research funding from Exact Sciences, Intromedic, and C2 Therapeutics. M.I.C. consults for Pentax Medical Corporation and Cook Medical and has research funding from C2 Therapeutics, Inc. J.D. consults for US Endoscopy and CSA Medical and has research funding from C2 Therapeutics. No other financial conflicts of interest pertain to the authors of this paper.
References and notes
- 1.Blot WJ, Devesa SS, Kneller RW, Fraumeni JF., Jr Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA. 1991;265:1287–1289. [PubMed] [Google Scholar]
- 2.Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. [PubMed] [Google Scholar]
- 3.Pera M, Cameron AJ, Trastek VF, Carpenter HA, Zinsmeister AR. Increasing incidence of adenocarcinoma of the esophagus and esophagogastric junction. Gastroenterology. 1993;104:510–513. doi: 10.1016/0016-5085(93)90420-h. [DOI] [PubMed] [Google Scholar]
- 4.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.American Gastroenterological A, et al. American Gastroenterological Association medical position statement on the management of Barrett's esophagus. Gastroenterology. 2011;140:1084–1091. doi: 10.1053/j.gastro.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 7.Shaheen NJ, Falk GW, Iyer PG, Gerson LB. ACG Clinical Guideline: Diagnosis and Management of Barrett's Esophagus. Am J Gastroenterol. 2016;111:30–50. doi: 10.1038/ajg.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dulai GS, Guha S, Kahn KL, Gornbein J, Weinstein WM. Preoperative prevalence of Barrett's esophagus in esophageal adenocarcinoma: a systematic review. Gastroenterology. 2002;122:26–33. doi: 10.1053/gast.2002.30297. [DOI] [PubMed] [Google Scholar]
- 9.Chen WD, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 10.Grady WM, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 11.Marzese DM, Hoon DS. Emerging technologies for studying DNA methylation for the molecular diagnosis of cancer. Expert Rev Mol Diagn. 2015;15:647–664. doi: 10.1586/14737159.2015.1027194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moinova HR, et al. HLTF gene silencing in human colon cancer. Proc Natl Acad Sci U S A. 2002;99:4562–4567. doi: 10.1073/pnas.062459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noehammer C, et al. Strategies for validation and testing of DNA methylation biomarkers. Epigenomics. 2014;6:603–622. doi: 10.2217/epi.14.43. [DOI] [PubMed] [Google Scholar]
- 14.Moinova H, et al. Aberrant vimentin methylation is characteristic of upper gastrointestinal pathologies. Cancer Epidemiol Biomarkers Prev. 2012;21:594–600. doi: 10.1158/1055-9965.EPI-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 16.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richmond E, Umar A. Mechanisms of esophageal adenocarcinoma formation and approaches to chemopreventive intervention. Semin Oncol. 2016;43:78–85. doi: 10.1053/j.seminoncol.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Singh A, Chak A. Advances in the management of Barrett's esophagus and early esophageal adenocarcinoma. Gastroenterol Rep (Oxf) 2015;3:303–315. doi: 10.1093/gastro/gov048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayakawa Y, Sethi N, Sepulveda AR, Bass AJ, Wang TC. Oesophageal adenocarcinoma and gastric cancer: should we mind the gap? Nat Rev Cancer. 2016;16:305–318. doi: 10.1038/nrc.2016.24. [DOI] [PubMed] [Google Scholar]
- 20.N. Cancer Genome Atlas Research et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene CL, et al. Emerging Concepts for the Endoscopic Management of Superficial Esophageal Adenocarcinoma. J Gastrointest Surg. 2016;20:851–860. doi: 10.1007/s11605-015-3056-0. [DOI] [PubMed] [Google Scholar]
- 23.Small AJ, et al. Radiofrequency Ablation Is Associated With Decreased Neoplastic Progression in Patients With Barrett's Esophagus and Confirmed Low-Grade Dysplasia. Gastroenterology. 2015;149:567–576. e563. doi: 10.1053/j.gastro.2015.04.013. quiz e513-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chak A, et al. Gastroesophageal reflux symptoms in patients with adenocarcinoma of the esophagus or cardia. Cancer. 2006;107:2160–2166. doi: 10.1002/cncr.22245. [DOI] [PubMed] [Google Scholar]
- 25.Lagergren J, Bergstrom R, Lindgren A, Nyren O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340:825–831. doi: 10.1056/NEJM199903183401101. [DOI] [PubMed] [Google Scholar]
- 26.Kadri S, Lao-Sirieix P, Fitzgerald RC. Developing a nonendoscopic screening test for Barrett's esophagus. Biomark Med. 2011;5:397–404. doi: 10.2217/bmm.11.40. [DOI] [PubMed] [Google Scholar]
- 27.Kadri SR, et al. Acceptability and accuracy of a non-endoscopic screening test for Barrett's oesophagus in primary care: cohort study. BMJ. 2010;341:c4372. doi: 10.1136/bmj.c4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falk GW, et al. Surveillance of patients with Barrett's esophagus for dysplasia and cancer with balloon cytology. Gastroenterology. 1997;112:1787–1797. doi: 10.1053/gast.1997.v112.pm9178668. [DOI] [PubMed] [Google Scholar]
- 29.Ross-Innes CS, et al. Evaluation of a minimally invasive cell sampling device coupled with assessment of trefoil factor 3 expression for diagnosing Barrett's esophagus: a multi-center case-control study. PLoS Med. 2015;12:e1001780. doi: 10.1371/journal.pmed.1001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peery AF, et al. Feasibility, safety, acceptability, and yield of office-based, screening transnasal esophagoscopy (with video) Gastrointest Endosc. 2012;75:945–953. e942. doi: 10.1016/j.gie.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooki A, et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 34.Shen S, et al. Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenetics. 2017;9:88. doi: 10.1186/s13148-017-0392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka H, et al. Adherens junctions associated protein 1 serves as a predictor of recurrence of squamous cell carcinoma of the esophagus. Int J Oncol. 2015;47:1811–1818. doi: 10.3892/ijo.2015.3167. [DOI] [PubMed] [Google Scholar]
- 36.Cogdell D, et al. Tumor-associated methylation of the putative tumor suppressor AJAP1 gene and association between decreased AJAP1 expression and shorter survival in patients with glioma. Chin J Cancer. 2011;30:247–253. doi: 10.5732/cjc.011.10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EJ, et al. Identification of Global DNA Methylation Signatures in Glioblastoma-Derived Cancer Stem Cells. J Genet Genomics. 2015;42:355–371. doi: 10.1016/j.jgg.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsusaka K, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–7197. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 39.Lai HC, et al. DNA methylation as a biomarker for the detection of hidden carcinoma in endometrial atypical hyperplasia. Gynecol Oncol. 2014;135:552–559. doi: 10.1016/j.ygyno.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Chen YC, et al. Methylomics analysis identifies epigenetically silenced genes and implies an activation of beta-catenin signaling in cervical cancer. Int J Cancer. 2014;135:117–127. doi: 10.1002/ijc.28658. [DOI] [PubMed] [Google Scholar]
- 41.Kitchen MO, et al. Quantitative genome-wide methylation analysis of high-grade non-muscle invasive bladder cancer. Epigenetics. 2016;11:237–246. doi: 10.1080/15592294.2016.1154246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami M, et al. Integration of transcriptome and methylome analysis of aldosterone-producing adenomas. Eur J Endocrinol. 2015;173:185–195. doi: 10.1530/EJE-15-0148. [DOI] [PubMed] [Google Scholar]
- 43.Kostareli E, et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J Clin Invest. 2013;123:2488–2501. doi: 10.1172/JCI67010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange CP, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:e50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kloten V, et al. Epigenetic inactivation of the novel candidate tumor suppressor gene ITIH5 in colon cancer predicts unfavorable overall survival in the CpG island methylator phenotype. Epigenetics. 2014;9:1290–1301. doi: 10.4161/epi.32089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose M, et al. Epigenetic inactivation of ITIH5 promotes bladder cancer progression and predicts early relapse of pT1 high-grade urothelial tumours. Carcinogenesis. 2014;35:727–736. doi: 10.1093/carcin/bgt375. [DOI] [PubMed] [Google Scholar]
- 47.Kloten V, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15:R4. doi: 10.1186/bcr3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dittmann J, et al. Gene expression analysis combined with functional genomics approach identifies ITIH5 as tumor suppressor gene in cervical carcinogenesis. Mol Carcinog. 2017;56:1578–1589. doi: 10.1002/mc.22613. [DOI] [PubMed] [Google Scholar]
- 49.Zhang S, et al. Genome-wide analysis of DNA methylation in tongue squamous cell carcinoma. Oncol Rep. 2013;29:1819–1826. doi: 10.3892/or.2013.2309. [DOI] [PubMed] [Google Scholar]
- 50.Ma G, et al. KCNMA1 cooperating with PTK2 is a novel tumor suppressor in gastric cancer and is associated with disease outcome. Mol Cancer. 2017;16:46. doi: 10.1186/s12943-017-0613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanaja DK, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27:549–560. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi JM, et al. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol. 2012;33:363–372. doi: 10.1007/s13277-011-0302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae JH, et al. Detection of DNA hypermethylation in sera of patients with Crohn's disease. Mol Med Rep. 2014;9:725–729. doi: 10.3892/mmr.2013.1840. [DOI] [PubMed] [Google Scholar]
- 54.Li D, et al. [Restriction landmark genomic scanning for screening aberrant CpG methylations in prostate cancer] Nan Fang Yi Ke Da Xue Xue Bao. 2016;36:103–108. [PubMed] [Google Scholar]
- 55.Arantes LM, et al. Validation of methylation markers for diagnosis of oral cavity cancer. Eur J Cancer. 2015;51:632–641. doi: 10.1016/j.ejca.2015.01.060. [DOI] [PubMed] [Google Scholar]
- 56.Chalertpet K, Pakdeechaidan W, Patel V, Mutirangura A, Yanatatsaneejit P. Human papillomavirus type 16 E7 oncoprotein mediates CCNA1 promoter methylation. Cancer Sci. 2015;106:1333–1340. doi: 10.1111/cas.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klajic J, et al. DNA methylation status of key cell-cycle regulators such as CDKNA2/p16 and CCNA1 correlates with treatment response to doxorubicin and 5-fluorouracil in locally advanced breast tumors. Clin Cancer Res. 2014;20:6357–6366. doi: 10.1158/1078-0432.CCR-14-0297. [DOI] [PubMed] [Google Scholar]
- 58.Maldonado L, et al. An epigenetic marker panel for recurrence risk prediction of low grade papillary urothelial cell carcinoma (LGPUCC) and its potential use for surveillance after transurethral resection using urine. Oncotarget. 2014;5:5218–5233. doi: 10.18632/oncotarget.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, et al. A network-based, integrative approach to identify genes with aberrant co-methylation in colorectal cancer. Mol Biosyst. 2014;10:180–190. doi: 10.1039/c3mb70270g. [DOI] [PubMed] [Google Scholar]
- 60.Bae H, et al. Epigenetically regulated Fibronectin leucine rich transmembrane protein 2 (FLRT2) shows tumor suppressor activity in breast cancer cells. Sci Rep. 2017;7:272. doi: 10.1038/s41598-017-00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kusakabe M, et al. Identification of G0S2 as a gene frequently methylated in squamous lung cancer by combination of in silico and experimental approaches. Int J Cancer. 2010;126:1895–1902. doi: 10.1002/ijc.24947. [DOI] [PubMed] [Google Scholar]
- 62.Wu Y, et al. Methylation profiling identified novel differentially methylated markers including OPCML and FLRT2 in prostate cancer. Epigenetics. 2016;11:247–258. doi: 10.1080/15592294.2016.1148867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Windhorst S, Song K, Gazdar AF. Inositol-1,4,5-trisphosphate 3-kinase-A (ITPKA) is frequently over-expressed and functions as an oncogene in several tumor types. Biochem Pharmacol. 2017;137:1–9. doi: 10.1016/j.bcp.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang YW, et al. ITPKA Gene Body Methylation Regulates Gene Expression and Serves as an Early Diagnostic Marker in Lung and Other Cancers. J Thorac Oncol. 2016;11:1469–1481. doi: 10.1016/j.jtho.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takamaru H, et al. Aberrant methylation of RASGRF1 is associated with an epigenetic field defect and increased risk of gastric cancer. Cancer Prev Res (Phila) 2012;5:1203–1212. doi: 10.1158/1940-6207.CAPR-12-0056. [DOI] [PubMed] [Google Scholar]
- 66.Uehiro N, et al. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 2016;18:129. doi: 10.1186/s13058-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1. Flowchart of study analyses.
Supplementary figure 2. Comparison of methylation of individual CpGs versus CpG patches in discriminating esophageal lesions.
Supplementary figure 3. Comparing VIM and CCNA1 Expression versus methylation.
Supplementary figure 4. Morphology of touch preps from balloon brushings of three intact porcine esophagus samples, from esophagogastric organ explants.
Supplementary table 1. Location of 26 differentially methylated patches identified by RRBS comparison of normal esophageal squamous mucosa versus Barrett’s lesions.
Supplementary table 2. Demographic characteristics of training and validation esophageal brushings populations.
Supplementary Table 3. mVIM and mCCNA1 performance in training and validation esophageal brushing samples.
Supplementary Table 4. Comparison of sensitivities of mVIM plus mCCNA1, versus mVIM, and versus mCCNA1, all at equal specificities.
Supplementary table 5. Influence of smoking on VIM and CCNA1 methylation in proximal versus distal esophagus.
Supplementary table 6. Participant evaluation of the non-endoscopic balloon sampling of the esophagus.
Supplementary table 7. Demographic characteristics of subjects in non-endoscopic balloon study.
Supplementary table 8. Methylation in post-ablation subjects.
Supplementary table 9. Post-examination questionnaire.
Supplementary table 10. Bisulfite-specific methylation-independent PCR primer sequences.
Supplementary table 11. Index tags for bisulfite-specific methylation-independent PCR primer sequences.