Abstract
Poorly differentiated cell populations including tumor-initiating stem cells have been demonstrated to display a unique ability to natively internalize fragmented double-stranded DNA. Using this feature as a marker, we show that 0.1% to 6% of human glioblastoma cells from the bioptates can effectively internalize a fluorescently labeled DNA probe. Of these, using samples from 3 patients, 66% to 100% cells are also positive for CD133, a well-established surface marker of tumor-initiating glioma stem cells. Using the samples from primary malignant brain lesions (33 patients), we demonstrate that tumor grading significantly correlates (R = .71) with the percentage of DNA-internalizing cells. No such correlation is observed for relapse samples (18 patients).
Keywords: glioblastoma, tumor-initiating stem cells, TAMRA-DNA, CD133, neurosphere
Introduction
Gliomas are brain tumors originating from neuroglial cells. Star-shaped glial cells are referred to as astrocytes, and so tumors derived from such cells are known as astrocytic tumors. The 2007 World Health Organization classification of astrocytic tumors groups them into 4 grades differing in the aggressiveness:1
Grade I, pilocytic astrocytoma—a benign, noninvasive, slow-growing tumor with clear borders.
Grade II, diffuse astrocytoma—a slow-growing tumor lacking clear borders and with marked cellular atypia.
Grade III, anaplastic astrocytoma—a malignant, aggressive tumor infiltrating the brain parenchyma and lacking clear borders. Such tumors display cellular atypia and high mitotic activity.
Grade IV, glioblastoma multiforme (hereafter referred to as glioblastoma) is the most aggressive malignant tumor lacking clear borders and rapidly spreading throughout the brain. These tumors display highly heterogeneous cell composition, rapid proliferation, neovascularization, extensive invasion, areas of hypoxia, and necrosis.
Modern diagnostics of brain tumors relies on the methods of computer-aided tomography (computed tomography scans) and magnetic resonance imaging . These techniques are indispensable for establishing the localization and size of the lesion(s), yet in most cases, they are poorly informative on the tumor histology.2 Cytology and histopathology analyses of bioptates collected during brain surgery have long remained the only option to reliably establish glioma grade and stage.3 Recently, microRNA (miRNA) profiling of glioma biopsies via quantitative PCR has been proposed to aid in glioma grading.4,5 Also, a number of noninvasive techniques have been developed to inform on the status of glioma and its grade.6,7 Modern diagnostic tools are all geared toward accurate assessment of the disease stage, which is clearly required for appropriate choice of the therapeutic options. To our knowledge, though, none of the existing diagnostic methods provide prognostic information on the disease progression.
Tumor-initiating stem cells (TISCs) were first described by Lapidot and Bonnet and Dick.8,9 These cells are primarily responsible for tumor engraftment, and they provide high proliferative capacity of cancer cell population, contributing to the tumor structure10,11,12 and metastasis.13–15 Our recent studies have shown that eliminating such TISCs from the cancer cell population leads to tumor eradication and full recovery of tumor-engrafted mice (ascites and solid forms of murine Krebs-2 adenocarcinomas).12 Based on this finding, we hypothesized that the percentage of cells capable of internalizing TAMRA-labeled (carboxy tetramethyl-rhodamine) DNA probe may serve as a valuable measure of the tumor aggressiveness and grade, thereby indicating the disease stage. In a clinical setting, this would translate into an opportunity to simultaneously assess the malignant potential of the tumor and to stage the progression of disease. Clearly then, this would help understand whether the chosen course of therapy is effective and provide the prognosis for the patient.
In the present work, we analyzed the percentage of TAMRA-positive cells in the biopsies of 51 patients with glioma (33 primary cases and 18 recurrent gliomas). We show that this parameter is positively correlated (R = .71) with the tumor grade. Further, using 3 patient samples, we show that 66% to 100% of human glioblastoma cells that internalize TAMRA-labeled DNA probe are also positive for the glial TISC marker CD133.16
Materials and Methods
Preparation of Glioma Cell Suspension and Establishment of Primary Cell Cultures Collected at Surgical Resection
Patient’s written consent for material collection and all the downstream procedures was obtained prior to the surgery for brain cancer. All investigations were approved by local ethics committee of Ya. L. Tsivian Novosibirsk Research Institute of Traumatology and Orthopaedics (protocol #003/15-1, 17/02/2015). It should be noted that patients with primary tumors did not receive any treatment before the operation.
Tumor was first minced with a scalpel and tumor pieces were washed twice with phosphate buffered saline. Next, the tumor material was treated for 30 minutes with 0.1% type IA collagenase (Sigma-Aldrich, USA) at 37°C. To quench collagenase activity, DMEM (Dulbecco's Modified Eagle Medium; Gibco, USA) + 10% fetal bovine serum (FBS; HyClone, USA) was added, and the cells were washed with DMEM + FBS 2 more times. An aliquot of cell suspension was taken for the analysis of TAMRA positivity and CD133 expression. The remaining cells were left in the culture flasks. Five to 7 days later, floating cells were transferred into a new flask and cultivated for 3 to 5 days in DMEM + 10% FBS. This time, all the floating cells were removed and adherent cells were kept in the culture with regular passaging once or twice a week, until 70% to 80% confluency was achieved. Cell passaging was done using trypsin/EDTA treatment for 5 to 10 minutes.
TAMRA DNA Labeling
Fluorescent labeling of human Alu repeat DNA using Polymerase chain reaction-based incorporation of TAMRA-5′-dUTP (deoxyuridine triphosphate) was performed exactly as described by Dolgova et al.10
Analysis of TAMRA+ DNA Internalization by Primary Tumor cells
To analyze the percentage and distribution of TAMRA+ cells in human gliomas, 1 million freshly isolated cells were incubated in 1 mL DMEM premixed with 0.5 μg TAMRA-labeled AluDNA for 1 hour at room temperature in the dark. The cells were spun down by brief centrifugation, washed once with the medium, and resuspended in the final volume of 200 μL DMEM. Cell suspensions were layered onto the glass slides using cytospin apparatus (1000 rpm, 1 minutes) and mounted in a droplet of ∼10 μL antifade (DABCO) +0.5 μg/mL DAPI. The slides were covered with coverslips and analyzed under fluorescent microscope AxioVision (Zeiss, Germany) using ISIS V. 5.4.9 (MetaSystems) software for imaging or under LSM 780 NLO confocal laser scanning microscope (Zeiss, Germany) and ZEN 2010 B SP1 software.
Analysis of CD133 Expression and TAMRA Positivity by the Primary Tumor Cells, Percoll Density Gradient
To understand whether TAMRA+ and CD133+ cell populations from glioma samples display any overlap, we modified a Percoll density gradient–based protocol to enrich for the mononuclear cells from the collagenase-treated total cell suspensions. First, the stock isotonic percoll (SIP) solution was prepared by mixing 9 volumes of Percoll (MP Biomedicals, USA) and 1 volume of 10 × Hank’s balanced salt solution (HBSS). Total cell suspension was centrifuged, resuspended in 1 × HBSS, and adjusted with SIP to 30%. Cell suspension obtained was carefully layered on top of the 70% SIP made on HBSS and centrifuged at 500g for 10 minutes at 18°C. In order not to disturb the gradient, centrifuge deceleration rate was set to minimum. Following centrifugation, cell debris remained on top of the gradient, red blood cells dropped to the bottom of the tube, and mononuclear cells formed a circle in the middle. The collected cells were washed with 7 to 8 mL of DMEM and counted using hemocytometer. Next, the cells were either incubated with TAMRA-labeled Alu DNA probe or stained with CD133-specific FITC conjugates (Miltenyi Biotec, Germany). Alternatively, the cells were first incubated with the TAMRA DNA probe, washed several times, and processed for immunostaining using CD133-FITC conjugates. Cells were then placed on glass slides and analyzed using fluorescence microscopy to calculate the percentages of TAMRA+, CD133+, and TAMRA+/CD133+ cells. For each tumor sample, at least 2 slides were analyzed and 2000 to 4000 cells were scored. Whenever possible, CD133 expression was analyzed using FACSAriaflow cytometer (Becton Dickinson, USA).
Neurosphere Formation in the Primary Human Glioblastoma Cell Cultures and Analysis of TAMRA Positivity in the Neurosphere Cells
Neurospheres were observed to form in the primary glioblastoma cell cultures after the third passage, which typically corresponded to week 6 of the cell cultivation. Prior to TAMRA incorporation, neurospheres were grown at a density of 6 × 104 cells/mL in α-MEM supplemented with 10 U/mL heparin, bFGF (basic fibroblast growth factor)(20 ng/mL), EGF (epidermal growth factor) (50 ng/mL), and 1% B27 supplements. Neurospheres were treated with TAMRA-labeled DNA probe and monitored for probe internalization in real time using confocal laser scanning microscope LSM 780 NLO (Zeiss) and ZEN software (Core Facility Center for microscopy analysis of biological samples of the SB RAS). TAMRA signal intensity in each neurosphere was measured every 10 minutes for a total period of 70 minutes.
Statistical Analysis
Statistical analysis was performed using Statistica 10 software. In the figure, bars show 0.95 confidence interval. The level of significance was estimated using Mann-Whitney U test. Correlation coefficient was performed using Microsoft Excel software. Differences were considered “statistically significant” when P < .1 (*), P < .05 (**).
Results
Internalization of Extracellular Double-Stranded DNA By The Neurosphere-Forming Cultured Primary Glioblastoma Cells
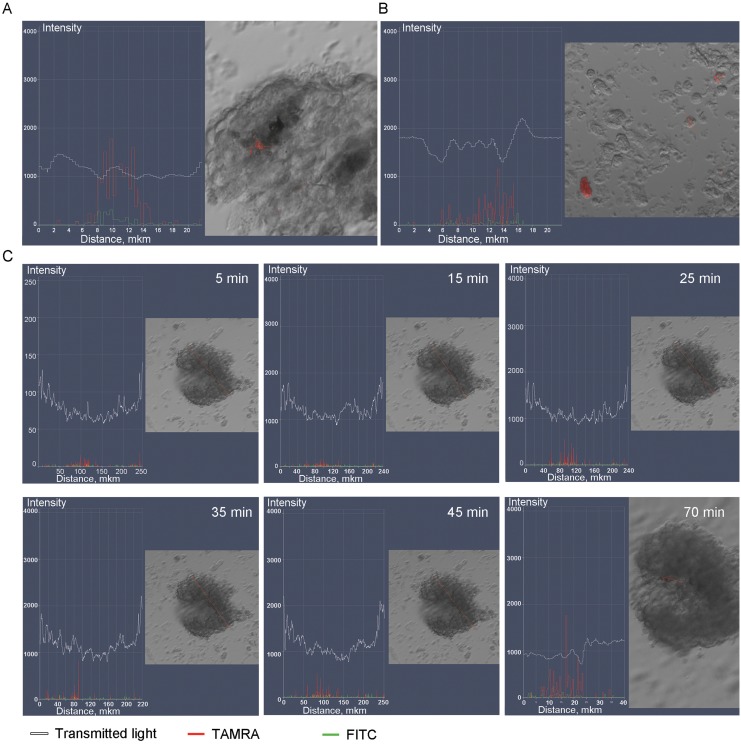
Neurospheres were obtained from adherent primary glioma cell culture (resection material from the patient K). Neurospheres were incubated with TAMRA-labeled DNA probe and monitored in real time for TAMRA signal intensity at 10-minute intervals for 70 minutes. Weak TAMRA signal was detectable as soon as 5 minutes following incubation (Figure 1B). Notably, some of the cells lying outside of neurospheres were also becoming TAMRA-positive (Figure 1C).
Figure 1.
Analysis of TAMRA-labeled Alu DNA probe accumulation in the neurospheres. A, Fragment of a neurosphere with a single TAMRA+ cells. B, Individual cells scattered through the slide are also TAMRA+. C, Time-lapse imaging of progressive TAMRA signal accumulation in the neurospheres. White curve denotes a transmitted light profile of the neurosphere, red and green graphs correspond to the TAMRA (specific) and FITC (nonspecific fluorescence) channels. The signal magnitude increases until 25 minutes and plateaus thereafter; at 70 minutes, individual cells may show even stronger specific fluorescence.
On average, neurospheres that formed in the cell culture of glioblastoma material from the patient K encompassed ∼30 cells, of which 1 to 2 cells were TAMRA+ (4.2%). In total, 27 neurospheres were subjected to microscopy analysis, of these 10 had TAMRA+ cells (∼37%). Analysis of mixed cell suspensions containing both neurosphere cells and glial cells (adherent fraction) indicated that 1.2% cells were TAMRA+.
TAMRA+ and CD133 Cell Populations In Primary Human Gliomas Display Significant Overlap
CD133 (Prominin-1) is a transmembrane glycoprotein encoded by the PROM1 gene.17–19 CD133 localizes to the membrane of poorly differentiated cells of various origin, including glial TISCs present in human glioblastomas.20,21 Our previous work firmly established that the ability of cells to internalize fragmented extracellular double-stranded DNA is also a marker of poorly differentiated cells,10–12,22–25 although the exact mechanism of this internalization is currently unknown. We asked whether these 2 markers may be present on the same cells in human glioblastomas, as this could translate into our better control of the patient response to therapy. Also, this question is also interesting from the biological standpoint, as presence of both or either of the markers on the glioma stem cells may be important for understanding whether such cells display similar or distinct degrees of differentiation.
Analysis of TAMRA and CD133 positivity was performed either separately (microscopy analysis of TAMRA+ signal, rhodamine channel, and FACS analysis of CD133 expression [CD133-Allophycocyanin channel] or in exactly the same cells (microscopy analysis). This was due to the high background fluorescence levels that could not be completely eliminated even after extensive cell purification using Percoll density gradient (Supplemental Figure 1).
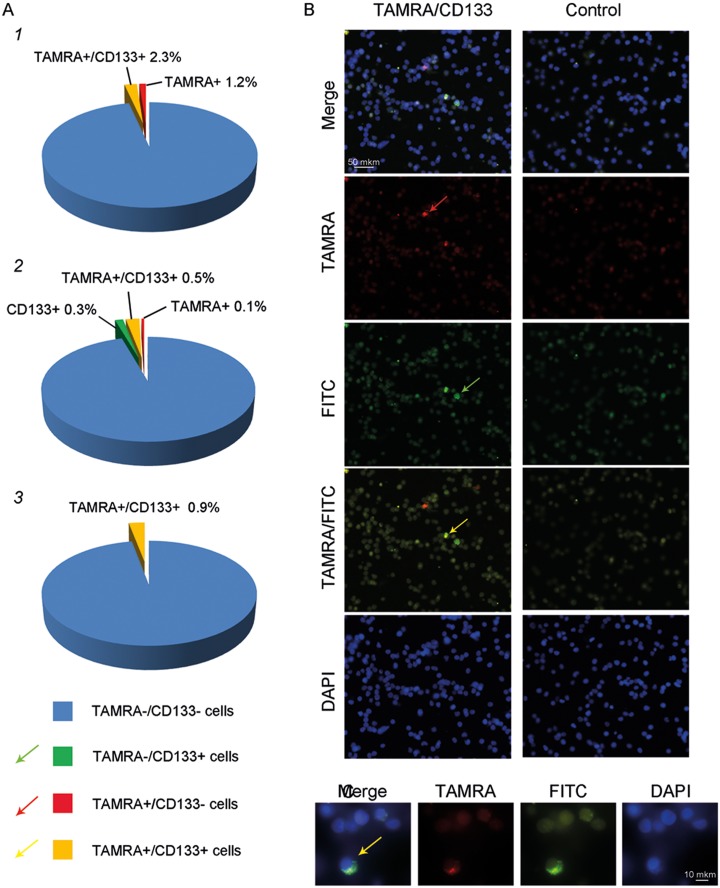
Our analysis of 3 patient samples (patients #51—grade III, #47—grade II, and #48—grade II) indicates that 66%, 83%, and 100% TAMRA+ cells are also positive for CD133 expression (this corresponds to 2.3%/3.5%, 0.5%/0.6%, and 0.9%/0.9% total cells; CD133+%/TAMRA+%; Figure 2). One of the prominent histomorphological features of grade IV gliomas is the presence of necrotic tissue, which produces significant nonspecific fluorescence upon microscopy analysis. Hence, our failure to quantify the percentage of double-positive cells in the samples from grade IV gliomas is likely attributable to this technical issue (Figure 2).
Figure 2.
Analysis of TAMRA and CD133 double staining in glioma cells. A, Pie chart showing the percentage of CD133+, TAMRA+, and CD133+/TAMRA+ cells in 3 patient samples (patients #51, #47, and 48),10,22,23 as assayed by microscopy analysis of 7000 and 500 cells, respectively. B, Cytology images showing TAMRA+ signal (red arrow), CD133+ signal (green arrow), and colocalization of both TAMRA+ and CD133+ signals in the cells (yellow arrow, merged image). C, Zoom in of a TAMRA+/CD133+ double-positive cell.
Quantitative Analysis Of TAMRA+ And Cd133+ Cells In Primary Human Glioma Samples
Glioma bioptates or resection material were collected from 51 patients with different disease stages; of these, TAMRA+ cells were analyzed for 47 patient samples, whereas CD133 profiling was done for 33 patient samples (Supplemental Table 1).
When we turned to the analysis of these data, notable differences were observed in the percentage of TAMRA+ and CD133+ cells in the resection samples from patients with primary tumors and with tumor relapse that had identical histopathology tumor grades. This is not surprising, as cancer stem cells that largely contribute to the disease relapse are also known to display stronger invasiveness and more pronounced malignant features.13,15 These cells have survived the treatment and have likely acquired resistance to the drugs administered to the patient. Further, such cells have activated the epithelial-to-mesenchymal transition program, which helps them circumvent the control by various molecular and surveillance systems in the body. Finally, they are largely resistant to apoptotic stimuli and secrete multiple molecules that help establish appropriate stromal microenvironment. For this reason, patients with grade III (relapse) and grade IV (relapse) tumors were grouped in a separate category.
The distribution of patients according to the categories (primary lesion/relapse) and disease stages was as follows (Table 1).
Table 1.
Distribution of Patients According to the Categories (Primary Lesion/Relapse) and Disease Stages.
| Grade | Designation | Number of Patients | |
|---|---|---|---|
| I | Pilocytic astrocytoma | 1 | |
| II | Diffuse astrocytoma | 3 | |
| II progressing into III | 5 | ||
| III | Anaplastic astrocytoma | Primary tumor | 8 |
| Relapse | 3 | ||
| IV | Glioblastoma multiforme | Primary tumor | 22 |
| Relapse | 8 | ||
| Breast cancer metastasis in the brain | 1 | ||
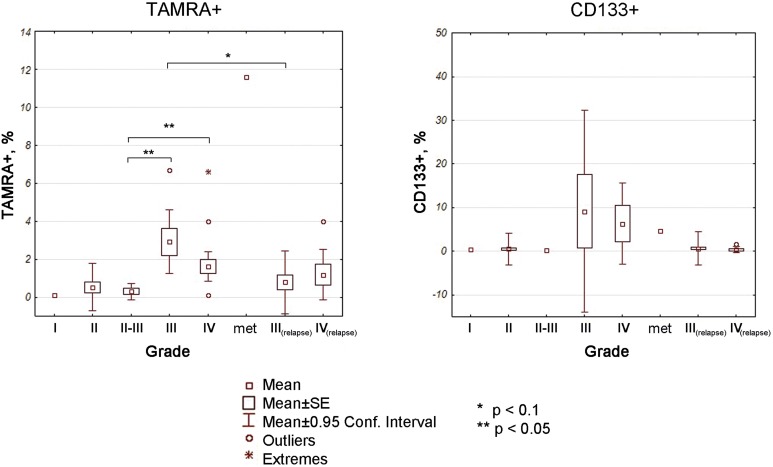
We show that primary high-grade gliomas (grades III and IV) have 0.10% to 6.70% TAMRA+ cells (Figure 3). One grade I tumor sample had 0.10% TAMRA+ cells, three grade II samples had 0%, 0.60%, and 0.99% TAMRA+ cells (Supplemental Table 1). Tumor samples classified as grade II progressing into grade III (n = 5) had a range from 0% to 0.89% TAMRA+ cells. We observed significant difference (P < .05) in the percentage of TAMRA+ cells between grade II progressing into grade III tumors and grade III as well as grade IV samples. No significant differences were observed between grades I, II and grades III, IV due to the limited number of grade I/II samples (n = 1 and 3) available (Figure 3).
Figure 3.
Percentage of TAMRA+ and CD133+ cells in fresh glioma samples, grades I to IV. The more malignant the tumor, the more TAMRA+ cells it has. Significance of differences between different grades was analyzed using Mann-Whitney U test in Statistica 10 package. *P < .1; **P < .05; met—breast cancer metastasis in the brain (n = 1).
Although CD133+ percentage is positively associated with the percentage of TAMRA+ cells, and both TAMRA and CD133 positivity tend to increase in higher grade tumors, the percentage of CD133+ varies in extremely wide range, which precludes statistically meaningful comparisons for this marker from being made (for instance, the grade III samples show 0% to 40.30% CD133+ cells).
Discussion
Our earlier studies have established that TAMRA+ cells display a number of features characteristic of poorly differentiated cell types and are typically present in various cell populations. Of note, in the context of murine Krebs-2 carcinoma, TAMRA+ marker is characteristic of TISC subpopulation.10,12,22,23 Furthermore, targeting of these cells was shown to have a strong therapeutic effect on mice engrafted with one of the most aggressive forms of transplantable cancer (Krebs-2 ascites).12
TAMRA+ cells were also detected in neurospheres obtained from passaged primary human glioblastomas. The presence of such cells was directly correlated with graft tumorigenicity.10 This observation also led us speculate that human glioblastomas (at least those cultured ex vivo) may have the same key cell subpopulation that can be and should be targeted to eliminate these tumors in the patients, that is, this represents a viable treatment modality to be explored in more detail.
In the present study, we show that TAMRA+ cells are present in the primary glioblastoma neurospheres as well as in the primary patient bioptates of patients with glioblastoma. In other words, specific cell targets apparently exist in primary tumors, and killing these cells may be necessary to eradicate brain cancers.
There are 2 basic parameters central to the target therapy: first, it is the number of cells to be selectively targeted, and second, it is the marker that helps trace the fate of such cells. We took advantage of a feature of TISCs to internalize TAMRA-DNA probe and used it as a marker. With this approach in hands, we quantified the numbers of TISC-like cells in primary human glioblastoma samples and related them with the disease progression (correlation between the disease stage [grades I-IV] and TAMRA positivity [which we assume translates into percentage of TISCs] was R = .71). This pronounced correlation between the numbers of such TAMRA-DNA internalizing cells and the tumor grade may indicate that declining TAMRA+ cell numbers in course of therapy should be viewed as a positive prognostic feature.
Our data provide a unique opportunity to discriminate between tumor grades II and III versus grades III and IV tumors on the day of brain surgery. This information can be instrumental for characterizing the disease progression and adjusting the therapeutic strategy before the histology report is obtained. To further increase the diagnostic power, we expect that our approach can be safely combined with a number of express tests available, such as those based on qPCR analysis of miRNA expression4,5,26,27 or on cytotoxicity of peripheral dendritic cells.6
In our future studies, we plan to unequivocally establish whether TAMRA+ glioblastoma cells are robust cell targets, and so the data presented in our current report will constitute the first quantitative analysis of the presence of target cells in primary glioblastomas. In other words, these numbers will help address the question of how many cells need to be targeted to achieve remission. Furthermore, TAMRA-DNA internalization may become a useful marker for measuring the numbers of key tumorigenic cells present in gliomas, as well as the response to therapy.
Multiple studies have reported the identification and validation of glioma TISC markers, which include CD133, CD44, L1CAM (CD171), β-tubulin, nestin, GFAP, A2B5, CD15, ALDH1 activity.28–32 In the present study, CD133 was used as an independent “control” TISC marker among other options. This surface glycoprotein has been characterized as a marker of neural TISCs20,33–35 that may be informative of the disease aggressiveness. Nonetheless, data are also available indicating that CD133-negative cells may have TISC-like features and that tumors lacking CD133+ cells can engraft in immunodeficient mice.36 Noteworthy, conflicting reports have been published on the glioma grade versus CD133+ correlation. One recent study37 demonstrated that no such correlation exists, whereas Zhang with coauthors,38 in contrast, provided evidence for the strong positive correlation between CD133 expression and glioma grading.
In our work, 2 directions to relate TAMRA and CD133 positivity with tumor grades were taken. First, we measured the percentage of TAMRA+ or CD133+ cells in primary glioma samples (51 patients in total, with 47 and 33 samples characterized for TAMRA and CD133 markers, respectively). Our data are consistent with the idea that higher CD133+ cell counts are observed in higher grade gliomas. Second, we performed pilot experiments to detect both markers simultaneously (Figure 2). The degree of overlap between TAMRA+ and CD133+ cell populations was only partial (66%, 83%, and 100%); in other words, the positive correlation was clearly present, yet the overlap was incomplete. These data indicate that TAMRA+ and CD133+ cell populations form a common tumorigenic core and that their numbers are correlated with tumor progression. Thus, immunostaining of glioma cell samples with CD133-specific antibodies may aid in more accurate disease staging.
It must be noted that in its present form, our approach is not applicable to the samples from patients with glioma having disease relapse, as correlations between the percentage of TAMRA+ cells and disease stage are not as pronounced as in the primary glioma cases. Nonetheless, whenever several biopsies become available during the therapy, TAMRA positivity may inform on the efficacy of the strategy chosen. Should such TAMRA+ cells be found in or near the tumor site following the surgery, chemotherapy, or radiation therapy, this may indicate that disease relapse is highly probable.
To conclude, the phenomenon of DNA internalization by poorly differentiated cells of different origin, including TISCs, appears to have broad biological significance and may provide important insights into the biology of stem cells. Whenever DNA is delivered into the cell interior, it is by no means an inert material, instead it launches multiple molecular cascades, in a cell-type and cell state–dependent manner. For instance, in normal stem cells, such internalized DNA molecules undergo partial hydrolysis and ligate to form closed circles.25,39 In cells that have been irradiated or pretreated with cytostatic drugs and which are undergoing DNA repair, the internalized DNA fragments actively participate in and interfere with nucleotide excision repair, homologous recombination, and nonhomologous end joining pathways.25,39,40 Thus, this interesting phenomenon can be used for targeting various cancer stem cell–like populations and has a clear translational potential.
Supplementary Material
Abbreviations
- FBS
fetal bovine serum
- HBSS
Hank’s balanced salt solution
- miRNA
microRNA
- TISCs
tumor-initiating stem cells
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the State Project of the Institute of Cytology and Genetics No 0324-2018-0019 and the grant No 15-04-03386 from the Russian Fund for Basic Research.
Supplemental Material: Supplementary material for this article is available online
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hashemi RN, Bradley WG, Lisanti CJ. MRI: The Basics. Philadelphia, PL: Lippincott Williams & Wilkins; 2004:353. [Google Scholar]
- 3. Lozano AM, Gildenberg Philip L, eds. Textbook of Stereotactic and Functional Neurosurgery. 2nd ed Berlin: Springer; 2009:679–698. [Google Scholar]
- 4. Han IB, Kim M, Lee SH, et al. Down-regulation of microRNA-126 in glioblastoma and its correlation with patient prognosis: a pilot study. Anticancer Res. 2016;36(12):6691–6697. [DOI] [PubMed] [Google Scholar]
- 5. Titov SE, Ivanov MK, Karpinskaya EV, et al. miRNA profiling, detection of BRAF V600E mutation and RET-PTC1 translocation in patients from Novosibirsk oblast (Russia) with different types of thyroid tumors. BMC Cancer. 2016;16:201 doi:10.1186/s12885-016-2240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tyrinova TV, Leplina OY, Mishinov SV, et al. Cytotoxic activity of ex-vivo generated IFNα-induced monocyte-derived dendritic cells in brain glioma patients. Cell Immunol. 2013;284(1-2):146–153. doi:10.1016/j.cellimm.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 7. Syatkin SP, Frolov VA, Gridina NY, Draguntseva NG, Skorik AS. Differential diagnostics of neoplastic and inflammatory processes in the brain by modifications NMDA receptor activity in blood cells with verapamil and ketamine. Bull Exp Biol Med. 2016;161(5):703–705. [DOI] [PubMed] [Google Scholar]
- 8. Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. [DOI] [PubMed] [Google Scholar]
- 9. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. [DOI] [PubMed] [Google Scholar]
- 10. Dolgova EV, Alyamkina EA, Efremov YR, et al. Identification of cancer stem cells and a strategy for their elimination. Cancer Biol Ther. 2014;15(10):1378–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Potter EA, Dolgova EV, Proskurina AS, et al. Gene expression profiling of tumor-initiating stem cells from mouse Krebs-2 carcinoma using a novel marker of poorly differentiated cells. Oncotarget. 2017;8(6):9425–9441. doi:10.18632/oncotarget.14116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Potter EA, Dolgova EV, Proskurina AS, et al. A strategy to eradicate well-developed Krebs-2 ascites in mice. Oncotarget. 2016;7(10):11580–11594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ebben JD, Treisman DM, Zorniak M, Kutty RG, Clark PA, Kuo JS. The cancer stem cell paradigm: a new understanding of tumor development and treatment. Expert Opin Ther Targets. 2010;14(6):621–632. doi:10.1517/14712598.2010.485186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavelic SK, Sedic M, Bosnjak H, Spaventi S, Pavelic K. Metastasis: new perspectives on an old problem. Mol Cancer. 2011;10:22 doi:10.1186/1476-4598-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198(3):281–293. doi:10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010;16(12):3113–3120. doi:10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 17. Yin AH, Miraglia S, Zanjani ED, et al. AC133, is a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–5012. [PubMed] [Google Scholar]
- 18. Corbeil D, Fargeas CA, Huttner WB. Rat prominin, like its mouse and human orthologues, is a pentaspan membrane glycoprotein. Biochem Biophys Res Commun. 2001;285(4):939–944. doi:10.1006/bbrc.2001.5271. [DOI] [PubMed] [Google Scholar]
- 19. Irollo E, Pirozzi G. CD133: to be or not to be, is this the real question? Am J Transl Res. 2013;5(6):563–581. [PMC free article] [PubMed] [Google Scholar]
- 20. Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(1):5821–5828. [PubMed] [Google Scholar]
- 21. Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. doi:10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 22. Dolgova EV, Potter EA, Proskurina AS, et al. Properties of internalization factors contributing to the uptake of extracellular DNA into tumor-initiating stem cells of mouse Krebs-2 cell line. Stem Cell Res Ther. 2016;7(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dolgova EV, Shevela EY, Tyrinova TV, et al. Non-adherent spheres with multiple myeloma surface markers contain cells that contribute to the sphere formation and are capable of internalizing extracellular double-stranded DNA. Clin Lymphoma Myeloma Leuk. 2016;16(10):563–576. [DOI] [PubMed] [Google Scholar]
- 24. Dolgova EV, Proskurina AS, Nikolin VP, et al. “Delayed death” phenomenon: a synergistic action of cyclophosphamide and exogenous DNA. Gene. 2012;495:134–145. [DOI] [PubMed] [Google Scholar]
- 25. Dolgova EV, Efremov YR, Orishchenko KE, et al. Delivery and processing of exogenous double-stranded DNA in mouse CD34+ hematopoietic progenitor cells and their cell cycle changes upon combined treatment with cyclophosphamide and double-stranded DNA. Gene. 2013;528(2):74–83. [DOI] [PubMed] [Google Scholar]
- 26. Somasundaram K, Mahabala Rao SA, Santosh V. MicroRNAa (miRNA) as biomarkers for diagnosing different grades of gliomas and pathways of glioma progression. Patent US 8637241 B2. [Google Scholar]
- 27. Berger F, Issartel JP, Atifi-Borel ME, Lages E, Guttin A. Use of Mirnas as biomarkers in glioma diagnosis. Patent US 20120322680 A1. [Google Scholar]
- 28. Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23(58):9392–9400. [DOI] [PubMed] [Google Scholar]
- 29. Wei KC, Huang CY, Chen PY, et al. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30(1):253–259. [PubMed] [Google Scholar]
- 30. Brescia P, Richichi C, Pelicci G. Current strategies for identification of glioma stem cells: adequate or unsatisfactory? J Oncol. 2012;2012:376894 doi:10.1155/2012/376894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dahlrot RH, Hermansen SK, Hansen S, Kristensen BW. What is the clinical value of cancer stem cell markers in gliomas? Int J Clin Exp Pathol. 2013;6(3):334–348. [PMC free article] [PubMed] [Google Scholar]
- 32. Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–512. doi:10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uchida N, Buck DW, He D, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci U S A. 2000;97(26):14720–14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007. 67(9):4010–4015. [DOI] [PubMed] [Google Scholar]
- 35. Wang J, Sakariassen PØ, Tsinkalovsky O, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122(4):761–768. [DOI] [PubMed] [Google Scholar]
- 36. Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31(5):857–869. doi:10.1002/stem.1317. [DOI] [PubMed] [Google Scholar]
- 37. Dahlrot RH, Hansen S, Jensen SS, Schrøder HD, Hjelmborg J, Kristensen BW. Clinical value of CD133 and nestin in patients with glioma: a population-based study. Int J Clin Exp Pathol. 2014;7(7):3739–3751. [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang M, Song T, Yang L, et al. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27:85 doi:10.1186/1756-9966-27-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rogachev VA, Likhacheva A, Vratskikh O, et al. Qualitative and quantitative characteristics of the extracellular DNA delivered to the nucleus of a living cell. Cancer Cell Int. 2006;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Likhacheva AS, Nikolin VP, Popova NA, et al. Exogenous DNA can be captured by stem cells and be involved in their rescue from death after lethal-dose γ-radiation. Gene Ther Mol Biol. 2007;11(2):305–314. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.