Significance
The human face is extraordinarily variable, and the extreme similarity of the faces of identical twins indicates that most of this variability is genetically determined. We have devised an approach to increase the chance of identifying specific large genetic effects on particular facial features, by choosing features with high heritability and selecting individuals with relatively extreme facial phenotypes for comparison with a control population. This has yielded three specific and replicated genetic variants, two for features of facial profiles, and one for the region around the eyes. Further application of these methods should enable the understanding, eventually at the molecular level, of the nature of this extraordinary genetic variability, which is such an important feature of our everyday human interactions.
Keywords: human genetics, facial features, SNPs, 3D imaging, additive genetic value
Abstract
To discover specific variants with relatively large effects on the human face, we have devised an approach to identifying facial features with high heritability. This is based on using twin data to estimate the additive genetic value of each point on a face, as provided by a 3D camera system. In addition, we have used the ethnic difference between East Asian and European faces as a further source of face genetic variation. We use principal components (PCs) analysis to provide a fine definition of the surface features of human faces around the eyes and of the profile, and chose upper and lower 10% extremes of the most heritable PCs for looking for genetic associations. Using this strategy for the analysis of 3D images of 1,832 unique volunteers from the well-characterized People of the British Isles study and 1,567 unique twin images from the TwinsUK cohort, together with genetic data for 500,000 SNPs, we have identified three specific genetic variants with notable effects on facial profiles and eyes.
Face to face—that is how we mostly recognize each other, and we can do that because the human face is so hugely variable. Galton (1) pioneered the idea that twins could be used to study the different effects of “nature and nurture” (a term he created) on various human traits. In particular, having noticed that twins fell into different categories, one of which was determined by their extreme similarity, which we now recognize as identical or monozygous (MZ) twins, he argued that this suggested that the resemblance was due much more to nature than nurture. This foreshadowed our current recognition of the fact that, because the faces of genetically identical MZ twins are extraordinarily difficult to distinguish, especially at first sight, the varying facial features by which we recognize people are almost totally genetically determined. The added fact that the facial features of identical twins raised apart are as similar to each other as those raised together (2) strongly supports the view that normal environmental effects on facial features as we recognize them are usually very limited.
There is strong anecdotal evidence that similar facial features often tend to occur in families and follow on from one parent or recent ancestor to the next generation. This suggests the existence of single gene variants for such facial features, with relatively large effects. There are evolutionary arguments to support this expectation. The extent of genetic variation that must exist to explain the high level of variation for facial features has most probably been driven by natural selection, possibly connected with individual recognition and recognition of group membership. In addition, there are special regions of the brain for face recognition in humans (3), as well as in other primates (4). The evolutionary arguments suggest that our minds are ingrained to perceive those features of a face that are likely to be strongly genetically determined.
Recently published studies on normal facial variation, using population-based genome-wide association studies (GWAS), have only found variants with quite small effects on facial features that have mostly only moderate heritabilities or have no associated heritability data (5–9). When the effect of a genetic variant on a particular phenotype is estimated to be small, it is generally very difficult, if not impossible, to establish whether there is any direct functional effect of the genetic variant, or a variant closely linked to it, on the associated phenotype. With large enough samples, it is possible to find highly significantly associated variants whose effects are so small that they cannot distinguish particular individuals who have the variant from those who do not by the way they look. Such small effects are unlikely to have any biologically interpretable function. Our aim is to discover, and replicate, genetic variants with sufficiently large effects on facial phenotypes to allow individual characterization and potential identification of the molecular basis of action of the discovered variant or one closely linked to it.
Our approach was based on the premise that the nature of facial individuality and its recognition means that the face must be treated effectively as a discrete characteristic that is quite different from quantitative characteristics, such as height. We cannot recognize individuals by their height or by any single quantitative facial feature, such as the distance between the eyes or the height-to-width ratio of the face. We proposed that the key to identifying single-locus SNPs with detectable effects on particular individual facial features lies in the careful definition of the phenotype chosen for a genetic association analysis. Our approach to this was to choose phenotypes, based on a principal components analysis (PCA), that have the maximum possible heritability and by selecting individuals with relatively extreme principal component (PC) scores for comparison with a control population. We identified PCs with high heritability by using data on MZ and dizygous (DZ) twins to estimate the additive genetic value (AGV), as described below, of each of the three measurements that define a point on the surface of the face, for each of the ∼30,000 points identified using the three-dimensional (3D) 3dMD imaging system, and then performed the PCA on these AGVs.
The AGV is a concept devised by animal breeders (see, e.g., ref. 10) that is defined to be the expected measurement for each individual, conditioned on their genetic information. In other words, it is what we expect an individual’s phenotype to be if the environmental variance and measurement error were eliminated. Using AGVs should then give rise to measurements of facial features that are more heritable than those based on the original measurements provided by use of the 3dMD imaging system. The prediction of AGVs uses the genetic variances and covariances between measurements that are obtained from the known relationships between MZ and DZ twins, and so takes into account the correlation structure of the face—namely, that each point on the face is correlated to an appreciable extent with other points on the surface of the face. It is this correlation structure of faces that makes it difficult to obtain single SNP associations with relatively large effects on any particular measured quantitative facial feature, such as the distance between the eyes or some measure of local facial curvature. The estimation of the AGVs was done without any reference to the SNP-determined genotypes, to avoid any possibility of biasing the choice of phenotype by any aspect of genotype. The PCA values with highest heritability were then dichotomized into sets of upper and lower values to turn them into discrete characteristics, and it is this definition of phenotypes—namely, comparing individuals who lie in the extremes of the selected phenotypic PC distributions with those who do not—that was used for the genetic association analysis.
We used a portable version of the 3dMD camera system to obtain 3D images of 1,832 unique volunteers from the very-well-characterized People of the British Isles (PoBI) study (11, 12) and 1,567 unique twin images from the TwinsUK cohort, previously used to analyze the heritability of facial phenotypes derived in various ways from the camera measurements (13) (www.twinsuk.ac.uk). For most of these individuals, we had genetic data for at least 500,000 SNPs. In addition, we had 33 images of East Asian (mainly Chinese) volunteers for comparison with the UK images. This enabled us to look for features in the UK faces that overlapped those in East Asians, which presumably must have a genetic basis.
Each image modeled the surface of the face with ∼30,000 points. By using an approach that harks back to Galton’s (14) method for obtaining average faces, 14 landmark points, such as the tip of the nose, were marked on each facial image, which, through a process of mesh registration, enabled the overlaying of all images in relation to each other. This then made it possible to estimate the AGVs of each position on a face for subsets of the data within a facial region around the eyes and for a profile. The two resulting sets of values were then subjected to a PCA. We then searched for those PCs that had high heritability or were notably associated with the overlap of UK and East Asian faces. At this stage, the top and bottom 10% of PoBI individuals for each of the chosen PCs were used for a genome-wide association analysis. Discovery SNPs based on the UK PoBI data were then replicated by using the TwinsUK data. This procedure has so far yielded three convincing associations for SNPs with minor allele frequencies (MAFs) ∼10% in the UK population and with odds ratios (ORs) >7 based on recessive models for the minor frequency alleles.
Results
The 3dMD Images, Landmarking, and Face Registration.
The primary data for our analysis of facial features were 3D images of 1,832 volunteers from the PoBI project, 1,567 imaged individuals from the TwinsUK cohort, and 33 East Asian volunteers, taken by using a 3dMD camera (www.3dmd.com). The images produced by the 3dMD proprietary software defined the surface of each face in terms of a mesh over 50,000–150,000 surface points in 3D space. To perform meaningful calculations on this collection of facial images, it was necessary to align the faces with each other so that any given surface point on a face could be matched with the corresponding point on any other face. This was done by a combination of manually annotating each face with 14 well-defined landmarks (for example, the tip of nose or corners of eyes) and then using a series of algorithms that registered each face against a generic model face (see Materials and Methods and SI Appendix for the details of these processes). The raw data from the camera software were transformed via landmarking and registration into a set of 29,658 surface points for each individual’s face, where each point had an identity corresponding to a biologically equivalent point on every other face. Each point had three coordinate variables describing its 3D position, giving 29,658 × 3 = 88,974 measurements per face.
AGV Prediction and Choice of Facial Subregions.
As already mentioned, to help identify facial features with high heritability, we devised a method for obtaining the AGVs of every surface point on a face. These are known as “breeding values” in the quantitative genetics literature. The position of each surface point correlated with that of many other points, particularly those immediately surrounding it. Much of this correlation structure will be genetic, and it is this that represents the biological phenotype of interest. Therefore, we exploited the correlation structure to further transform each original registered variable into its corresponding AGV. We did this by using the MZ and DZ twin data to estimate the additive genetic variances of each surface point, and the additive genetic covariances between points, which are needed to estimate a set of coefficients θjk using a least-squares minimization. The predicted AGV for each individual i at variable j is then given by the equation
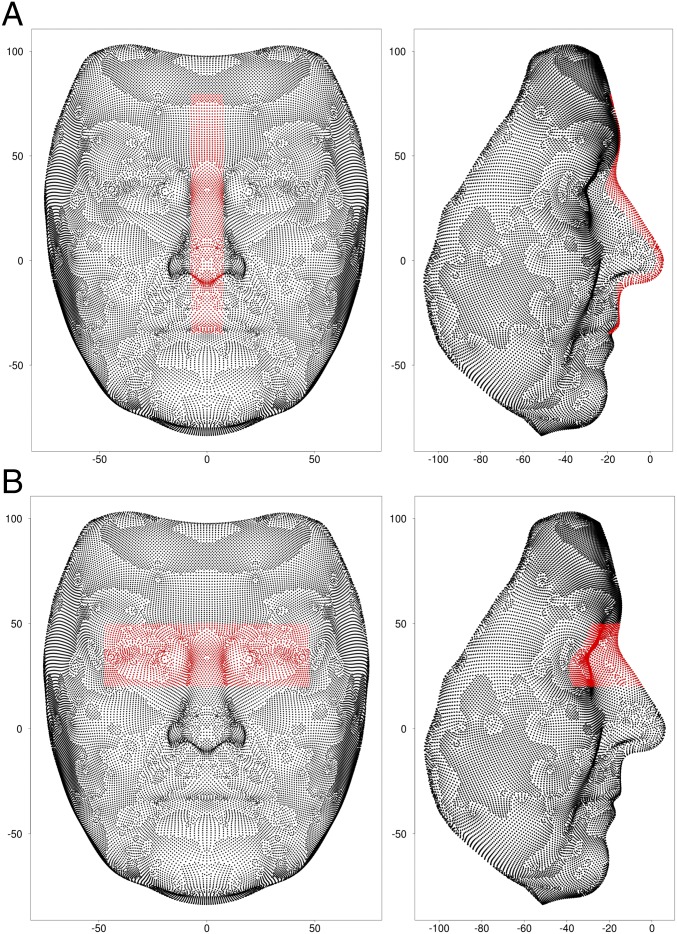
where xik are the observed values of the coordinates of the kth original variable (an x, y, or z axis position for a particular registered surface point) for individual i and the summation is over the total, m, of variables per face (three times the number of surface points) (see Materials and Methods and SI Appendix for further details of the procedures for estimating the AGVs). Estimation of each θjk was performed, without any use of genetic SNP data, merely by knowing the relationships between MZ and DZ pairs. This allowed individuals from the twin dataset to be reused for subsequent genetic association analysis, without having unwittingly introduced associations between SNPs and AGV phenotypes. Ideally, the estimation process should have been carried out by using all of the 88,974 variables that were used to define the surface of each face. Computational limitations so far limited us to carrying out this AGV estimation for two subregions of the face (eyes and profile) chosen by visual inspection (see Materials and Methods and Fig. 1 for further details).
Fig. 1.
Points, highlighted in red, constituting the profile (A) and eyes (B) subregions, with the scale for both axes in millimeters.
The eyes subregion included 2,736 points, giving 3 × 2,763 = 8,289 variables for further analysis. Only the height (y) and depth (z) dimensions were used for the profile subregion, as the width (x) dimension was much less heritable (SI Appendix, Fig. S2A). There were 1,646 points in the profile subregion, and so 2 × 1,646 = 3,292 variables were used for further analysis.
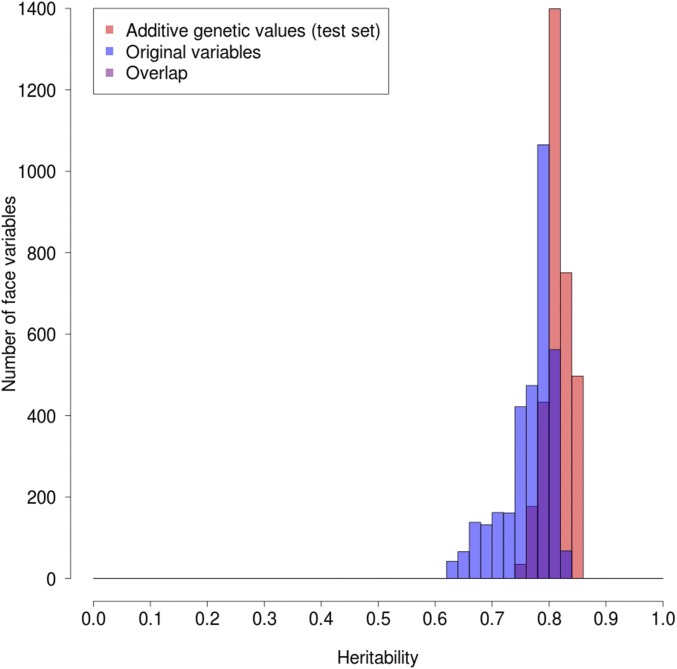
The mean heritability of the AGVs for eyes was 76.1% and for the profile 81.5%, compared with 69.8% and 76.6%, respectively, for the original variables. The gain in heritability for the AGVs for the profile is shown more directly in Fig. 2, which plots for all of the variables the histograms for both the original data and the AGVs as determined from a subset of the TwinsUK data not used for the estimation of the AGVs. This clearly shows the significant increase in the AGV heritabilities and also, perhaps more importantly, the substantial reduction in the variance of the AGV heritabilities. This is due to much-improved AGV heritabilities for variables with previously low heritabilities (see also SI Appendix, Fig. S3 for the eyes subregion). There are also many more AGVs than original variables with heritabilities >0.8.
Fig. 2.
Comparison of profile heritabilities for original values vs. AGVs.
Shape Translation and PCA.
As described in Materials and Methods, PCAs were carried out on the AGVs after a Procrustes-like transformation that centered both profile and eyes subregions on the origin to avoid any influence of large-scale craniofacial morphology causing differences between individuals in the positions of regions. The PCAs were then carried out separately for the eyes and profile facial subregions, using all of the PoBI, TwinsUK, and the 33 East Asian individuals’ images combined. The resulting eigenvectors were used as the PC scores.
Selection of Facial PC Axes for Further Analysis.
Five PC axes were chosen for genetic association analysis from the largest 50 PC axes, for each of the eyes and profile subregions (PCs 1, 2, 3, 5, and 6 in the former and PCs 1, 2, 3, 5 and 7 in the latter; marked by red circles in SI Appendix, Fig. S4). The choices were based on PC scores having a heritability of >75% as determined from the TwinsUK data or having a heritability >65% and a relatively large difference between the mean PoBI and East Asian scores (SI Appendix, Fig. S4). This latter criterion was based on the fact that major ethnic groups generally have certain distinct types of facial features, which must be largely genetically determined. A large difference in the PC phenotype between populations implies that there are facial features common in East Asian populations which are much less common in European populations, or vice versa, and that this difference is due to one or more face-controlling genetic variants that are significantly more or less frequent in East Asian than in European populations. These comparisons were made by using the set of 33 East Asian images. Although this was a relatively small sample, we found several PCs with large between-group differences, suggesting the influences of genetic variants that differ in frequency between the two populations. The East Asian images were only used as a contribution to the choice of PCs in the PoBI data for further analysis. They were not used directly for any genetic analysis.
Discovery Genetic Association Analysis.
To investigate the hypothesis that there are genetic variants conferring large effects on facial morphology, and taking into account the discrete nature of facial features, we focused on individuals with facial features that could, in some sense, be considered “extreme” relative to the general population. We therefore dichotomized the PC scores for the facial phenotypes into subsets of upper and lower extremes, chosen to be the top and bottom 10% of individuals when ranked according to their PC scores. This meant that the five PCs chosen for further analysis from each of the eyes and profile subregions gave rise to a total of 20 extreme phenotypes to be tested for genetic associations.
Only the PoBI data were used for the SNP association discovery analysis, leaving the TwinsUK data for subsequent replication of those SNPs chosen for further analysis. As described in Materials and Methods and SI Appendix, after quality control (QC) procedures, there were 3,161 PoBI individuals (1,532 males and 1,629 females) genotyped for 524,576 SNPs, of which 512,181 were non-sex-linked and were used for the association analysis. Of the 3,161 genotyped PoBI individuals, only 1,423 (652 males and 771 females) had both good-quality facial images and genotypes. The remaining 1,738 individuals with genotype data but no image data were used as controls. A further 409 individuals with image data but no genotype data were also available, giving a total of 1,832 imaged individuals, and all of these were used for the definition of the phenotypes, as previously described.
For each PC, the individuals in the 10% extreme (upper or lower) were tested against all those not in the extreme together with unphenotyped individuals. The latter will include some individuals with the extreme phenotype being tested, which will dilute any suggested association but cannot bias the result toward an association. We assumed that the relatively large number of PoBI controls who were not phenotyped offset any slight loss of power for detecting an association due to the presence of a relatively small number of people with the extreme phenotype in the controls.
To allow for male vs. female differences, all association analyses were performed for female upper and lower extremes (incorporating all males into the control samples and comparing 77 individuals in the upper extremes against 3,084 = 3,161 − 77 controls, and 76 individuals in lower extremes against 3,085 = 3,161 − 76 controls) and combined male and female upper and lower extremes (142 upper-extreme individuals against 3,019 = 3,161 − 142 controls and 140 lower-extreme individuals against 3,021 = 3,161 − 140 controls). Male extremes were not studied at this stage because the TwinsUK data to be used for replication hardly included any males. Males were, however, included in the controls, as no association analyses were done using sex-linked SNPs. In total, there were therefore four association analyses performed for each of the 10 PCs under consideration—namely, 10 × 2 = 20, as above, taking into account upper and lower extremes for each of the 10 PCs, and 20 × 2 = 40, taking into account that each of these 20 phenotypes was tested separately in females and in combined sexes. Association analyses were therefore effectively done on 40 different facial variables.
It is important to emphasize that none of the steps taken to reach this definition of phenotypes involved any relationship to the SNP genotyping. Thus, each of these phenotypes was initially tested independently for genome-wide associations, using only the number of SNPs used for the association analysis to take into account multiple comparisons for assessing significance, just as is done for standard GWAS for diseases.
For each of the selected 40 PC extremes, assessment of the statistical significance of SNP associations was based on analysis of the 3 × 2 contingency tables of genotypes (aa/Aa/AA, where a represents the minor frequency allele) vs. extreme/control status, for all 512,181 non-sex-linked SNPs. Dominant (A) and recessive (aa) models were both tested for each SNP and the P value for the best fit of the two then used, multiplied by 2 for the extra comparison being made. The relatively small number of extreme individuals motivated a careful selection of the appropriate statistical test for contingency table significance. Most tests—for example, the Pearson’s χ2 test—rely on assumptions that usually only hold in large samples (15). This is especially pertinent when examining models of inheritance involving the effects of minor allele homozygotes, which can be at quite low frequencies among the extremes for MAFs of ∼10%. In practice, we therefore only considered SNPs with MAFs ≥10%. To establish the most appropriate test, null hypothesis simulations of 3 × 2 tables were performed, and the type-I error rates were quantified (SI Appendix).
The best-performing approximate test, showing only mild deflation of P values within the borderline genome-wide significance range (10−5 to 10−6), was an implementation of a Wald test (SI Appendix, Fig. S5A), in which standardized log ORs under recessive and dominant models were tested against an N(0, 1) distribution for significant departures from 0 (16) (see SI Appendix for further details). Pearson’s χ2 test showed serious inflation of statistical significance, even when applying Yates’ continuity correction (SI Appendix, Fig. S5B).
Variants were deemed to be sufficiently associated with an extreme facial feature for taking forward for replication, by passing one of three thresholds: (i) having a P value below the standard level of genome-wide significance (5 × 10−8); (ii) belonging to the candidate SNP set and having a P value <500,000 × 5 × 10−8/66,769 = 3.7 × 10−7, where 66,769 is the number of tested SNPs in the candidate list (SI Appendix); or (iii) having a P value <10−4 and an OR >9. The rationale for this threshold was that, at the cost of a greater number of false positives, variants could be discovered with (i) biologically significant consequences and (ii) high power for replication, conditional on them being true positives. An OR lower limit of 9 gave a suitable balance between having either too few or too many SNPs to take forward for replication.
There were, for the females, 15 associations for the profile and 10 for the eyes that passed the threshold of choice for taking forward for further analysis. There were, in addition, for combined males and females, two SNP associations for the profile and two for the eyes that passed this threshold. A total of 17 SNPs for the profile and 12 SNPs for the eyes were therefore taken forward for replication in the TwinsUK data. For the profile, three of the five PC phenotypes (accounting for 9 of 17 associated SNPs to be taken forward for further analysis), and for the eyes ,four of five PC phenotypes (accounting for 10 of 12 associated SNPs to be taken forward for further analysis) showed a substantial difference between East Asian and European individuals. Interestingly, all of the discovery associations were statistically more significant under a recessive rather than a dominant inheritance model.
Replication in TwinsUK Data.
The TwinsUK dataset provides the basis for valid and unbiased replication since, although it was used for the AGV estimation, this process makes no reference to DNA data. Although a combination of high OR and low P value was used to determine whether a SNP should be followed up for replication, statistical significance alone was used to determine whether replication had been achieved.
Of the 12 eye-associated discovery SNPs, 1 (rs2039473, eyes PC3 associated in females) was not present on the TwinsUK genotyping platform and so could not easily be pursued further. One SNP of the 17 selected for replication for the profile had an r2 > 0.1 with another discovery hit. Thus, retaining the SNP with the highest OR in the discovery analysis, there remained 16 SNPs with profile-associated phenotypes and 11 eyes-associated SNPs, making a total of 27 SNPs to be taken forward as candidates for replication.
For each of these 27 replication SNPs, association was only tested for the PC extreme with which it was associated in the discovery analysis. To increase power, control samples from the appropriate discovery analyses were incorporated into the replication contingency tables before testing. A Wald test (one-tailed) was then performed under the appropriate inheritance model (recessive in all cases) for each of the 27 replication SNPs.
A complication in using the TwinsUK cohort as a replication dataset was the high degree of relatedness between MZ and DZ twins. Removing all related individuals satisfied the requirements for independent samples required by standard statistical analyses, but also lost information and resulted in an arbitrarily chosen set of unrelated samples. To address these issues, we carried out a randomization and permutation testing analysis using a limited number of the DZ twins. Thus, noting that statistical power might be gained by utilizing more than half of the DZ twins per analysis, and that a pair of DZ twins contained the genetic information equivalent to 1.5 unrelated samples, we performed 10,000 random selections of one from each pair of DZ twins plus, for a random half of those DZ twins, their twin relative.
Each random selection thus included 75% of the available 654 DZ individuals (327 pairs) with genetic data plus 246 composite MZ imaged individuals (formed by averaging the data for the two MZ twins and treating them as a single individual) and 129 remaining individuals with no relative available, all after QC. The total of ∼865 (654 × 0.75 + 246 + 129) is approximately equivalent to the full set of independent genetic information available from the TwinsUK data. The appropriate genotype distributions for the extremes and nonextremes from this dataset were then produced by using the mean cell counts over the random selections, rounded to the nearest integer. For the controls, the PoBI controls (nonextreme phenotyped PoBI individuals and all genotyped but not phenotyped PoBI individuals) were combined with the nonextreme twins’ data. A random permutation test was then used to assess the significance of the difference between the extremes and the controls, and the OR was calculated from the appropriate recessive 2 × 2 table (see Materials and Methods and SI Appendix for further details of these procedures).
Of the 27 discovery SNPs taken forward for replication analysis in this way in the TwinsUK dataset, 3 replicated with 5% or lower false discovery rate (FDR; calculated on the basis of having used 27 SNPs) and with ORs >4: rs2045145 (in PCDH15), profile PC2 associated in females; rs11642644 (in MBTPS1), profile PC7 associated in females; and rs7560738 (in TMEM163), eyes PC1 associated in the combined sexes. None of these SNPs were in any of our candidate gene regions.
Detailed Analysis of the Three Replicated SNP Associations.
For each of these SNPs, we show the 3 × 2 tables for the discovery analysis, replication analysis, and combined TwinsUK and PoBI data. The combined data tables were formed by adding the extremes results for the PoBI discovery and the TwinsUK replication, and testing the combination for significance against the same controls used in the replication analysis (namely, the combined controls from the PoBI and TwinsUK sets as described in Replication in TwinsUK Data) using a Wald test. The overall OR was estimated from the 2 × 2 table for a recessive model, as was appropriate for each of the three replicated associations, and the P values were for one-sided tests, given the expected direction of a SNP effect in the replication.
rs2045145 (PCDH15) association with the more European extreme profile PC2 in females.
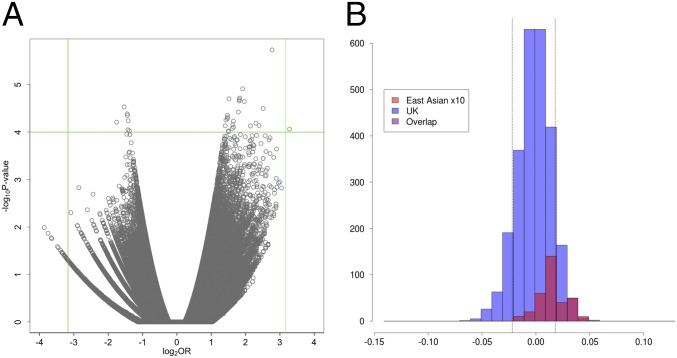
The volcano plot of −log10 P values against log2 ORs for the profile PC2 phenotype in females for the PoBI discovery analysis is shown in Fig. 3A, indicating in green the rs2045145 SNP that replicated. The green lines are drawn to show the P < 10−4 (horizontal) and OR > 9 (vertical) thresholds above and beyond which SNPs were taken forward for replication.
Fig. 3.
(A) Volcano plot for female lower (more European) extreme PC2 profile. Green lines show the upper and lower thresholds for ORs and the upper threshold for the P values. Only the green dot satisfies the criteria for further analysis. (B) Distribution of PC values for the PC2 profile in UK (PoBI) and East Asian individuals, with the East Asian distribution magnified in size by 10 for visualization purposes. Dotted lines show the thresholds for the upper and lower 10% of individuals.
The distribution of the PC values for the PC2 profile phenotype in UK and East Asian individuals is shown in Fig. 3B. This clearly demonstrates the difference between the UK and East Asian individuals for this PC2 phenotype, which was expected based on the choice of this PC for analysis, including an overall difference for this PC between the UK and East Asian individuals (Materials and Methods and SI Appendix, Fig. S4B). In this case, it was the lower extreme, being the more characteristically UK face, that showed the genetic association. The 3 × 2 tables for the rs2045145 SNP for the discovery analysis, the replication analysis, and the combined TwinsUK and PoBI (i.e., replication and discovery) data are shown in Table 1.
Table 1.
rs2045145: Association with female profile PC2 European-like extreme
| Discovery | aa | Aa | AA | P value | OR |
| Extremes | 6 | 7 | 63 | 8.65 × 10−5 | 9.70 |
| Controls* | 27 | 564 | 2,492 | ||
| Replication | |||||
| Extremes | 3 | 9 | 70 | 2.85 × 10−3 | 4.67 |
| Controls | 31 | 727 | 3,089 | FDR = 0.026 | |
| Combined | |||||
| Extremes | 9 | 16 | 133 | 1.11 × 10−6 | 7.44 |
| Controls | 31 | 727 | 3,089 |
The 3 × 2 tables for PoBI discovery, TwinsUK replication, and the combined data. Minor allele (a) frequency 0.103. P values and OR are for the minor allele (aa) recessive homozygote; FDR by Benjamini and Hochberg procedure (37). Male imaged individuals were excluded from the discovery analysis. The number of controls for discovery is the total number of genotyped individuals, 3,161, minus the number of extremes, 76, minus 2 missing genotypes. The number of controls for replication and combined is the number for discovery (3,083) plus the number of genotyped and phenotyped twins (∼865) minus the number of extreme twins (82) minus 18 twins not typed for this SNP.
Two genotypes missing for technical reasons.
The gene frequency of the a allele in a representative Chinese population [301 Chinese from the 1,000 Genomes Project (17)] was 0.06, and so, as expected based on the direction of the phenotypic effect, lower than in the UK population.
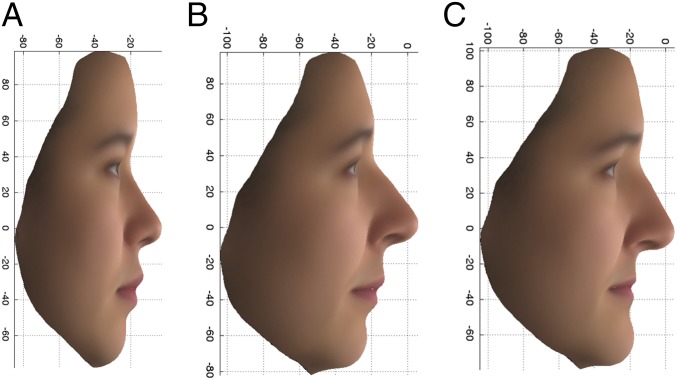
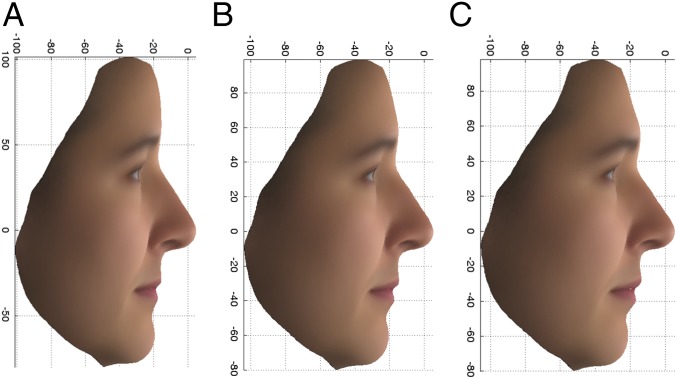
For visualization of the difference between the extreme facial phenotypes, average faces within each extreme (upper and lower 10%) were produced by plotting the arithmetic mean for each coordinate measurement in each vertex, among all individuals falling into the designated extreme, and overlaying with a surface texture. Only females were used to produce average faces, so as not to obscure the genetic difference by sex differences. Average faces were produced by using all East Asian females and all PoBI females separately, and from the original variables rather than the AGVs. The average profiles for the East Asian females and the upper and lower 10% of the PoBI females for the PC2 profile are given in Fig. 4. It is the lower, more European-looking extreme profile of Fig. 4C that was significantly associated with the recessive (aa) genotype of the rs2045145 SNP with an overall OR of 7.44. It is, however, notable that the East Asian-like PoBI female face (Fig. 4B) has an upturned nose and upper lip and a receded chin, similar to the average East Asian female face. Although extreme phenotypic status was determined by comparing individuals’ PC scores derived from AGV values, the phenotypes depicted in the average faces were calculated directly from original variables and are an appropriate representation of the associated phenotypes.
Fig. 4.
PC2 profile: Average faces, using the original variables, for 14 East Asian females (A) and the upper 10% (more East Asian) (B) and lower 10% (more European) (C) extremes of the PoBI females.
The rs2045145 SNP is in an intron of two transcripts (PCDH15-220 and PCDH15-225), 670 Kb downstream from the first exon of PCDH15-225 and 580 Kb upstream from its terminus, and so itself has no obvious functional effect. By using the approach to functional analysis given in Materials and Methods, three potentially functionally relevant variants, but with relatively low ORs, were found to be in linkage disequilibrium (LD) with rs2045145 (r2 = 0.53, 0.53, and 0.55). These were within 5 Kb upstream of a long noncoding RNA (lncRNA; 207-bp mature transcript) situated in an intron of PCDH15. PCDH15 is one of several genes for which loss-of-function recessive variants cause one of the forms of Usher syndrome, which involves hearing and balance malfunction in addition to retinal defects. Usher syndrome is not usually associated with a distinctive facial appearance, although dysmorphology has sometimes been seen (18). PCDH15 is expressed in the nose (olfactory epithelium and nasal cartilage) of developing mice (19), perhaps fitting in with the observation that the minor homozygote is associated with a straight rather than upturned nose (Fig. 4C). An additional tagged SNP with an OR of 4.65 in the sequenced TwinsUK data (rs61850893, r2 = 0.72) is situated 229 bp upstream of a conserved intron region within PCDH15, but its possible functional significance is not clear. As shown in SI Appendix, Fig. S9A, the discovery SNP lies in a region of sequence conserved between primates at a position where the common allele (G) is the common allele in humans. This suggests that, in this case, the discovery allele could itself be functional with respect to the facial phenotype.
rs11642644 (MBTPS1) association with upper extreme profile PC7 in females.
Table 2 shows the 3 × 2 associations for rs11642644 for the discovery, replication, and combined data analyses and SI Appendix, Fig. S6A, the volcano plot that led to the choice of this SNP for replication.
Table 2.
rs11642644 (MBTPS1) associations with profile PC7 upper extreme in females
| Discovery | aa | Aa | AA | P value | OR |
| Extremes | 6 | 15 | 56 | 7.38 × 10−5 | 9.57 |
| Controls | 27 | 578 | 2,479 | ||
| Replication | |||||
| Extremes | 3 | 13 | 69 | 2.7 × 10−3 | 4.48 |
| Controls | 31 | 688 | 3,110 | FDR = 0.026 | |
| Combined | |||||
| Extremes | 9 | 28 | 125 | 1.79 × 10−6 | 7.21 |
| Controls | 31 | 688 | 3,110 |
Minor allele (a) frequency 0.097. P values and OR are for the minor allele (aa) recessive homozygote; FDR Benjamini and Hochberg procedure (37). Male imaged individuals were excluded from the discovery analysis. The number of controls for Discovery is the total number of genotyped individuals, 3,161, minus the number of extremes, 77. The number of controls for replication and combined is the number for discovery (3,084) plus the number of genotyped and phenotyped twins (∼865) minus the number of extreme twins (85) minus 35 twins not typed for this SNP.
The average female PC7 profiles for the upper (Fig. 5A) and lower (Fig. 5C) extremes, as well as the overall average (Fig. 5B), calculated from original variables, are shown in Fig. 5.
Fig. 5.
Average profile PC7 female faces, using the original variables, for the upper 10% (A) and lower 10% (C) extremes and the overall average (B).
In this case, it is the upper extreme, Fig. 5A, that is associated with the recessive homozygote (aa) for the rs11642644 SNP, and this does not show any obvious association with the East Asian faces (SI Appendix, Fig. S6B), as expected from the data in SI Appendix, Fig. S4B, on which the choice of profile PC7 was made for further analysis.
The SNP rs11642644 is in the gene MBTPS1 and is located in an exon of a 543-bp transcript (MBTPS1-016), which is within the MBTPS1 gene. This exon has no ORF and is likely to be part of a lncRNA. Two of the transcript’s three exons overlap with those of the protein-coding MBTPS1 transcript. rs11642644 also lies in a region of open chromatin. The minor allele, associated with the upper 10% extreme phenotype as a recessive genotype, is present in African green monkey, macaque, and olive baboon, while the major allele (T) is present in orangutan, gorilla, chimpanzee, and marmoset (SI Appendix, Fig. S9B). This suggests that the minor allele is itself a functional candidate, as these species differ considerably in their craniofacial morphology.
A major function of MBTPS1, also known as site-1 protease, is to cleave SREP proteins in the endoplasmic reticulum (20). The SREP proteins are transcription factors encoded by two genes, SREBF1 and SREBF2, which regulate production of enzymes responsible for steroid biosynthesis. SREBF1 is in a 3.7-Mb region of 17p11.2, which is reciprocally deleted and duplicated in Smith–Magenis and Potocki–Lupski Syndromes (21), both of which have accompanying facial dysmorphias. SREBF2 is within the 22q13 deletions in Phelan–McDermid syndrome (also known as 22q13 deletion syndrome), also with a dysmorphic facial appearance. Both proteins are strongly craniofacially expressed in mice at 10.5 d after conception, and MBTPS1 also seems to be expressed in a smaller region at the tip of the snout at this stage of development (22). Together, this evidence provides a possible explanation for the functional effect of the rs11642644 SNP, either through an effect on the open chromatin or through an effect of the presumed lncRNA on the expression of the MBTPS1 gene.
rs7560738 (in TMEM163) association with the upper extreme PC1 eyes phenotype in the combined sexes.
Table 3 shows the data on the rs7560738 association with the upper extreme PC1 eyes phenotype in the combined sexes for the discovery, replication, and overall analyses. The volcano plot that led to the selection of this SNP for further analysis is shown in SI Appendix, Fig. S7 A and B, which shows that there is no East Asian association for this SNP, as expected from SI Appendix, Fig. S4A.
Table 3.
rs7560738 (in TMEM163) associations with the eyes phenotype PC1 upper extreme in the combined sexes
| Discovery (both sexes) | aa | Aa | AA | P value | OR |
| Extremes | 9 | 30 | 103 | 4.39 × 10−7 | 10.15 |
| Controls | 20 | 577 | 2,422 | ||
| Replication (female only) | |||||
| Extremes | 3 | 23 | 57 | 2.50 × 10−3 | 5.12 |
| Controls | 29 | 729 | 3,040 | FDR = 0.026 | |
| Combined | |||||
| Extremes | 12 | 53 | 160 | 4.00 × 10−8 | 7.32 |
| Controls | 29 | 729 | 3,040 |
Minor allele (a) frequency 0.107. P values and OR are for the minor allele (aa) recessive homozygote; FDR by Benjamini and Hochberg procedure (37); All imaged individuals were used in the discovery analysis. The number of controls for discovery is the total number of genotyped individuals, 3,161, minus the number of extremes, 142. The number of controls for replication and combined is the number for discovery (3,019) plus the number of genotyped and phenotyped twins (∼865) minus the number of extreme twins (83). The discrepancy of 3 is a function of the way the twins are analyzed using random permutation.
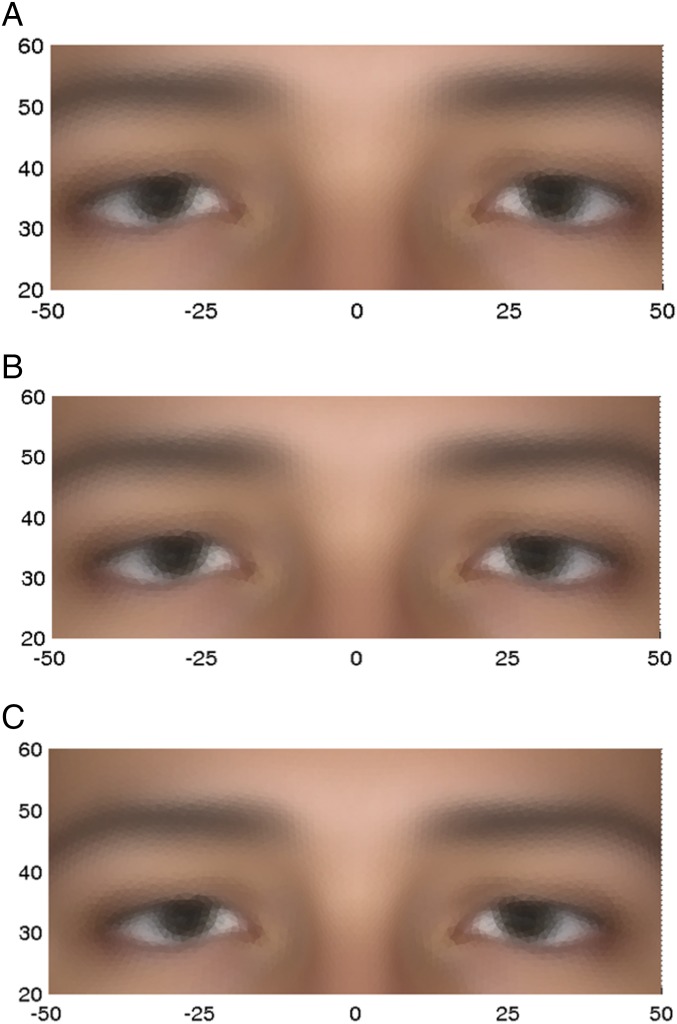
The average faces for the PC1 eyes phenotype upper and lower extremes and the overall average, calculated from original variables in females only, are shown in Fig. 6.
Fig. 6.
Average eyes phenotype, using the original variables, for PC1 (females only) for the upper 10% (A) and lower 10% (C) extremes and the overall average (B).
It is the upper extreme (Fig. 6A) that is associated with the minor allele homozygote (aa). Note the differences in eye width and eye height, against the x and y scale, respectively, which are both greater in the upper extreme.
rs7560738 lies in TMEM163, which is a highly conserved gene in mammals coding for a transmembrane protein that is a putative zinc transporter expressed in the brain and retina, as well as in a limited number of other tissues. Recent work (23) shows that TMEM163 is a binding partner of MCOLN1, for which loss-of-function mutations (including deletions and point mutations) cause Mucolipidosis type IV (MLIV) (24), a lysosomal storage disorder. Although MLIV is not commonly thought of as having distinctive facial features (25), several cases have been identified which share particular facial dysmorphias, especially around the eyelids (26, 27). Photographs of these cases document some atypical eye shapes and positions (26). TMEM163 may be involved in the pathology of the disorder through its influence on cellular zinc levels, which are elevated in MLIV (28) and increased when TMEM163 is knocked down (23). Two orthologs of MCOLN1 are expressed in zebra fish eyes during development, supporting the case for the TMEM163–MCOLN1 interaction playing some role in the determination of eye morphology (29). These data suggest that TMEM163 could be a plausible functional candidate for the eyes PC1 phenotype. This is supported by the data on the DNA sequences around the discovery SNP in a range of primates (SI Appendix, Fig. S9C), which shows that the discovery SNP is a variant in the midst of a conserved region. It is the derived allele (A) that has an approximate frequency of 10% in Europeans.
Discussion
Our success in finding specific single SNPs with relatively large effects on facial features depends on our ability to identify facial phenotypes that have the high heritability expected from the extraordinary facial similarity between MZ twins. Key to this has been the estimation of the AGVs of points on the surface of the face using data from the 3dMD camera system’s 3D facial scans of MZ and DZ twins. Tsagkrasoulis et al. (13) have recently used the same cohort of images to accurately characterize heritabilities of facial geometry measurements, demonstrating the efficacy of the 3D technology for representing heritable aspects of faces, though they have not looked for genetic associations. The objective behind estimating additive genetic values is to maximize the heritability captured by each facial surface measurement in order to increase the strength of genetic associations. Another contribution to the phenotype heritability is the use of ethnic differences, in our case between the UK and East Asian populations, which played a significant role in the discovery of the rs2045145 (PCDH15) association with the more European extreme PC2 profile in females. These differences are necessarily largely genetic and must be due to differences between ethnic groups in the frequencies of genetic variants with defined effects on facial features. The final contribution to our success has been the use of dichotomized variables for the genetic association analysis rather than a traditional quantitative variable approach. Basing our analysis on facial variation within the relatively homogeneous UK population has necessarily, to some extent, limited the range of facial genetic variability available for study. However, the use of the well-characterized and carefully sampled PoBI population as our control for genetic association studies has minimized the possibility of population stratification effects that are especially troublesome for studies on mixed ethnic populations. It is important to emphasize that none of the approaches to the choice of phenotypes to study were in any way dependent on the genetic information. This prevented any bias of the chosen phenotype toward having significant associations with SNPs, so that the correction for multiple testing of each chosen phenotype will therefore only depend on the number of SNPs tested for.
The choice of extreme phenotypes for study to maximize the chance of finding variants with relatively large effects inevitably limited the number of individuals available for a genetic association analysis. This problem turned out to be particularly relevant given that the genetic associations we have found involve homozygotes for relatively low-frequency variants. This led to a careful consideration of the appropriate statistical test to use for selection of variants for subsequent replication. Simulations of expected distributions using random permutations between controls and cases strongly suggested that the conventional χ2 tests were too permissive for the balance of cases and controls in our study (SI Appendix, Fig. S5B). This led us to adopt a Wald test based on standardized log ORs. We also decided to take into account the estimated magnitude of the effect of a potential discovery SNP, choosing a lower threshold of an OR of ∼9 as a criterion for taking forward a SNP for replication, while allowing for a less stringent P value threshold. Once a criterion has been set for the choice of SNPs to be taken forward for replication, it is then only the number of these chosen SNPs, effectively 27 candidate SNPs, that needs to be taken into account in assessing an acceptable P value for replication.
Another limitation of our study has been the resource-limited availability of a suitable sample of phenotyped and genotyped individuals for replication. We were fortunate to have the TwinsUK samples for this, but that created the problem of effectively only having females for replication. It also posed the challenge of how best to use the information from the DZ twins without losing information due to using only one member for each pair in the replication analysis.
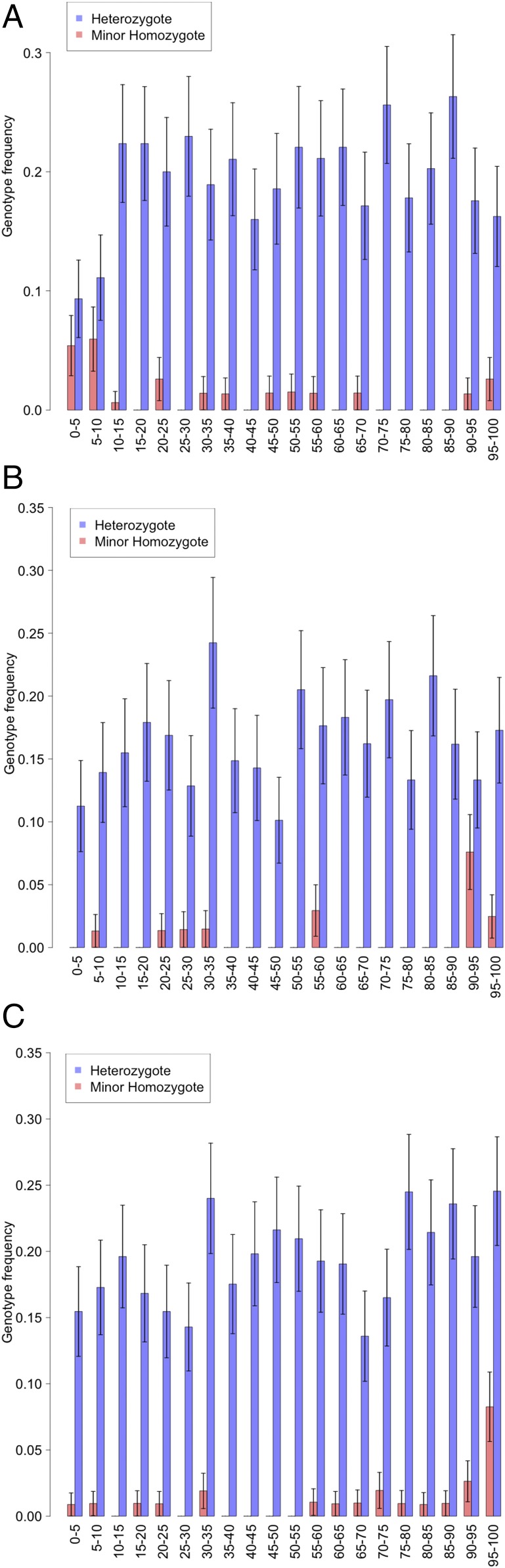
The three replicated SNPs were identified as significant by an association with individuals in a 10% extreme of selected PC distributions. We sought for more subtle effects by looking at the frequencies of the associated homozygotes (aa) and heterozygotes (Aa) in successive 5% quantiles across the appropriate PC distributions for combined PoBI and TwinsUK data (Materials and Methods), as shown in Fig. 7. This shows that in each case the frequency of the aa homozygotes is largely concentrated in the extreme 5% or 10% of the PC distribution with no obvious variation in the (Aa) heterozygote frequencies over the different levels of the PC7 profile and PC1 eyes phenotypes. This is consistent with the lack of heterozygote association with these phenotypes in the data given in Tables 2 and 3, and so a clearly recessive control of the extreme phenotypes. Each variant has no obvious quantitative effect on the trait value outside of the extreme group, which is consistent with our choice of a 10% threshold for dichotomization of the phenotype. It is questionable whether these associations could be detected by using a quantitative analysis comparing the average phenotype values within each genotype. For the PC2 profile phenotype (Fig. 7A), there is a remarkable association between the presence of aa homozygotes and the absence of Aa heterozygotes in the lower SNP associated extreme (OR 13.2, Yates corrected χ2 44.5, using data from Table 1). This implies that the presence of a single A allele in this case positively inhibits the expression of the lower extreme phenotype. This is consistent with the observed negative correlation between heterozygotes Aa and the lower extreme phenotype shown in Table 1 (OR 0.51, P = 0.01, Fisher’s exact test).
Fig. 7.
Bar graphs of frequencies of minor allele homozygotes and heterozygotes at 5% quantiles for the three replicated SNP associations. Percentage quantiles are labeled on the x axis. (A) rs2045145 profile lower extreme PC2 association. (B) rs11642644 profile upper extreme PC7 association. (C) rs7560738 eyes upper extreme PC1 association.
Despite the strong recessive effects of the three replicated SNPs on their associated extreme phenotypes, there are in each case many extreme phenotypes that are not recessive homozygotes for these SNPs. While some of these may be due to measurement error in the phenotype, it seems more probable, given the high heritabilities involved, that most are likely to be associated with other sources of genetic variation that our analysis was too weakly powered to detect. These could include lesser effects of either homozygotes or heterozygotes for other SNPs or epistatic effects. The preponderance of recessive phenotypes may be due to the fact that we are looking for unusual phenotypic differences, with overall dominant effects controlling the corresponding commoner facial features. It may seem curious that recessive effects are imposed on phenotypes that were initially produced via maximization of the additive genetic variance via prediction of additive genetic values. However, the extent of additive genetic variation, calculated under the assumption of no dominance variation, can be affected by the extent of dominant variation that is actually present. In addition, it is possible that additive genetic variation could be the most important factor across the continuous range of values for a given PC phenotype, while recessive genotypes become accumulated in the extremes.
The familial pattern of apparent parent to offspring passing on of particular facial features will be expected to be seen largely in the matings between the strongly associated homozygotes (aa) and the heterozygotes (Aa). This is because the probability of the Aa heterozygotes expressing the associated feature is 5- to 10-fold less than for the aa homozygotes. Since the proportion of aa homozygotes in these meetings will be 0.5, this gives the appearance of an apparent dominantly determined phenotype.
In our small sample of three replicated SNPs, there was no evidence of any particular pattern of gene functions. This is in marked contrast, for example, to the genetic control of tissue type incompatibility by the HLA system in humans or of taste and smell differences by the extraordinary complex of olfactory receptors. Selection for genetic variation in facial features connected, for example, with recognition of group membership, and mate choice preferences is likely to be frequency-dependent. This can explain high levels of polymorphism, as was suggested many years ago for the HLA system (30). However, in the case of the face, the genetic variation selected for may be adventitious with respect to gene function in the sense that there may be very many different ways in which variation in particular genes can influence the inheritance of facial features.
The ultimate test for the validity of any particular SNP found to be strongly associated with a particular human facial feature will be to verify the functional effect associated with that SNP. This can be done, for example, by evolutionary comparisons relating SNP variability to facial features, such as for the differences between primate species (as we have done for our replicating SNPs) or in the variety of dog breeds, and by exploring the effects of specific gene mouse knockouts or knockins on snout morphology.
Our studies as described here have been limited by:
-
i)
The need for observations on larger panels of individuals, both for discovery and replication, to increase the power for detection of variants with lower gene frequencies than 0.1 and also somewhat lower ORs. It would be possible to define the phenotypes in precisely the same way in a separate cohort of individuals as a means of further replicating our results, by registering facial images and using the relevant coefficients for producing AGVs and PC scores;
-
ii)
The need to extend the computational tractability of the analysis to process the whole face; and
-
iii)
The possibility of studying a wider range of ethnic differences.
We suggest that many more specific and relatively large genetic variant effects on human facial features will be found in the future using approaches such as we have described.
Materials and Methods
Sources of Volunteers.
The PoBI individuals were sampled from throughout the United Kingdom, primarily from rural areas and from individuals all of whose grandparents came from approximately the same area, as described by Winney et al. (12). The majority of the samples (2,039) have been subject to a detailed population genetic structure analysis (11), which, while revealing extraordinary genetic structure related to geography, showed that the average FST between regional groups was only 0.0007. Thus, for the purpose of genetic association studies, this is an ideal population. MZ and DZ twins were from the TwinsUK cohort (www.twinsuk.ac.uk).
Image Phenotyping.
All facial images were taken by using a portable version of the static 3dMDface System3dMD camera and its accompanying dedicated software (www.3dMD.com and SI Appendix). Where possible, three images were taken of each participant, to allow averaging of measurements over multiple images, with the aim of reducing technical noise. Volunteers were asked to assume a neutral, relaxed facial expression. A total of 2,556 images of PoBI individuals and 2,025 images of TwinsUK volunteers were collected. Of the 2,556 images of PoBI volunteers, 1,832 were of unique individuals (819 male and 1,005 female and 8 for whom sex was not correctly recorded), while for the twins, 1,567 were of unique individuals (1,551 females and 16 males), not all of whom were satisfactorily genotyped (see below). In addition, single images were taken of 33 East Asian Chinese origin volunteers (14 females and 19 males). Images from all datasets were registered at 29,658 locations on the facial surface, each referred to as a vertex or surface point. Every vertex has three coordinate positions describing its x, y, and z dimensional positions, yielding 88,974 phenotypic variables per individual.
Informed consent was obtained from all subjects, and the whole project conformed to the UK standard research ethical consent procedures with approval granted by the National Research Ethics Service Committee, Yorkshire and the Humber–Leeds West, United Kingdom (Reference 05/Q1205/35).
Facial Registration.
The 3D face images generated by the 3dMD camera system are provided in the form of a triangulated mesh (SI Appendix, Fig. S1A). Each vertex has associated with it a 3D location and an RGB (red green blue) appearance value. However, the identity of each mesh vertex or point at this stage is unknown, and the number of points varies from image to image. This was dealt with by mesh registration, which involved fitting the mesh of a generic face model to the unknown mesh to produce a standardized triangulated mesh in which the identity of each node in the mesh is known (SI Appendix, Fig. S1B; refs. 31 and 32). It is this registration process that enables meaningful calculations on the collection of facial surfaces to be made (for further details of the process, see SI Appendix).
AGV Prediction.
The original facial surface measurements xij are continuous random variables describing a position in one-dimensional space (three measurements for each vertex), where i and j are indices, respectively, for individuals (total n) and variables (total m = 88,974). We aim to estimate the unobserved values Yj = Ee[Xj|g], where the expectation is taken over the stochastic environmental effects (Ee), for a given individual, for each measurement, j, where Xj is the random variable of which xij is a realization, and g represents a random vector of genotypes belonging to the individual. In quantitative genetics, the departure of Yj from its population mean would be termed the genetic value. Under purely additive genetic effects (no dominance or epistasis), the assumption we rely on below, this is the AGV. The objective is to predict yij, the realized AGV for individual i, at measurement j, using the complete set of measurements taken on the same individual: . This is performed for each j in turn. Each Xk is modeled as
| [1] |
for all k = 1,2,3...m, where εk represents the effects of the environment. We assume that Ee[εk|g] = 0, equivalent to there being no gene–environment interactions. To estimate the coefficients θjk, which effectively measure the predictive influence of variable Xk on Yj, we minimize the expected least-squares error,
| [2] |
with respect to each coefficient θjk. This double expectation is taken with respect to the stochastic environmental and genetic effects (Ee and Eg, respectively). After applying constraints to avoid bias and so introducing a Lagrangian parameter, and estimating genetic variances and covariances from the twins’ data assuming only additive genetic effects, we obtain a matrix equation whose solution gives the estimates of the coefficients θjk. The predicted AGVs, for each individual i at measurement j, are then
| [3] |
For further details of the estimation procedure, see SI Appendix.
Fitting Process Using Facial Subsets.
Ideally the linear predictor uses all facial variables as described in AGV Prediction, where m (=88,974) is the total number processed and registered from the camera system. However, computational limitations have so far prevented this, and so we used a workaround whereby analysis was restricted to two rectangular subregions of the face, which we call “profile” and “eyes.” These were selected based on visual inspection of an image of the average face (Fig. 1). Vertices that fell within the subregion on the average face were taken forward for AGV prediction analysis in all individuals (SI Appendix). All subsequent statistical analyses were performed on the predicted AGVs.
Shape Translation and PCA.
Using a Procrustes transformation approach, we translated each individual’s subregion by subtracting the mean x coordinate for their own subregion (taken across all vertices in that region for the particular individual) from the x coordinates of all variables in the subregion, and similarly for the y and z coordinates for both eyes and profile subregions. This has the effect of centering all individuals’ subregions on the origin. Downstream statistical analyses using these centered values are not then unduly influenced by large-scale craniofacial morphology. To avoid losing information that could be of genetic interest, no other Procrustes transformations—for example, matching subregions by rotation or scaling by size—were applied.
PCA was carried out in the standard way, performing eigenvalue decomposition on the nxn covariance matrix of scaled (by population SD) and centered (on population mean) facial variables for both subregions separately, using all TwinsUK, East Asian, and PoBI images combined. The resulting eigenvectors were used as the PC scores.
Projecting TwinsUK individuals onto PoBI-fitted PC axes showed that PC scores for the largest five axes had r > 0.9 with the scores produced using the combined data, for both subregions. This relatively high correlation between the two sets of PC scores justified the use of the combined PCA analysis. PC scores were averaged over all TwinsUK and PoBI individuals for whom multiple images were available, to reduce sources of environmental variation such as differences in facial expression and small measurement errors.
PC Subset Selection.
The largest 50 PC axes were inspected for promising phenotypes using two criteria: (i) heritability (33) of each PC axis as determined by using the PC scores of the 1,567 TwinsUK individuals; and (ii) the squared difference between the mean PoBI (European) PC score and the mean East Asian score, taken as a ratio against the within-population variance. Plots of these statistics are shown in SI Appendix, Fig. S4. PCs with heritability >0.75 or heritability >0.65 and a between/within population variance ratio >1 were taken forward for genetic association analysis.
Genotype QC and Genotyping.
Genotyping of the PoBI discovery samples was performed on two separate Illumina platforms. As part of the Wellcome Trust Case Control Consortium 2 study (34), 2,912 samples were typed on the Illumina Human 1.2M-Duo genotyping chip, 2,039 of which were previously used to analyze the population structure of the British population (11). Since 2011, a total of 823 newly recruited samples were genotyped on the Illumina Infinium OmniExpress-24 BeadChip (750K) platform. The intersection between the two platforms was 547,863 SNPs. Before QC, there were 3,735 genotyped DNA samples, constituting 3,616 unique individuals.
Standard genotype QC procedures were performed by using PLINK [Versions 1.07 (35) and 1.90 (36)] in combination with R scripts, after which there remained 3,161 individuals (1,532 male and 1,629 female) genotyped at 524,576 SNPs.
TwinsUK genotype data were available on two platforms; 1,278 samples represented by 2,287,998 array-typed SNPs and 612 whole-genome sequenced samples (19,725,734 autosomal variants). Separate QC procedures were performed for the two platforms before merging. After merging the array and sequencing data and retaining variants common to both sets, there were 1,887,250 variants typed on 1,794 samples, 1,275 of whom were unique individuals.
For more details of QC procedures, see SI Appendix.
Procedure for Dealing with the Relatedness Between MZ and DZ Twins.
The MZ twins were dealt with by averaging phenotypes (PC scores) over each member of the MZ pair and taking the average as a single individual, which reduced the influence of environmental variation in the associated facial phenotype.
We dealt with the presence of DZ twins by performing 10,000 random selections of a list of unrelated DZ twins plus, for a random half of those individuals, their twin relative, as already described. The most significant FDR-adjusted P value (37) over the 10,000 selections was then taken as the overall observed P value, and this was compared against null hypothesis permutations to obtain an empirical P value. While there may be more efficient ways of dealing with this problem of the use of the twin data, our approach is certainly more efficient than just using one member of each twin pair.
Frequency Trend Analysis.
Combined TwinsUK and PoBI data were used to inspect for evidence for trends in genotype frequency across the quantitative PC phenotypes. This was done for each of the three replicating SNPs in the PC phenotype with which they were associated. Individuals were sorted into 5% quantiles based on their ranked PC values. This was performed within the TwinsUK data by using a randomization scheme similar to that described in Procedure for Dealing with the Relatedness Between MZ and DZ Twins and in SI Appendix, in which a random set of unrelated DZ pair members was retained and each time combined with the full set of phenotyped individuals from the PoBI discovery set, giving ∼1,467 individuals when analyzing females only and 2,118 individuals when analyzing combined sexes. The mean frequency of genotypes in each bin for the combination of PoBI and TwinsUK data, the latter taken over the set of randomizations for DZ twins, was taken as the observed data for plotting.
Function Analysis of Replicating SNPs.
By using the TwinsUK sequence data (600 individuals), replicated variants were assessed for high r2 (as a measure of LD) with novel variants in the sequence data that were not present among the 524,576 SNPs used for the association analysis and which were within 2 Mb upstream or downstream of the physical location of the replicated variant. Variants satisfying this requirement and with r2 > 0.2 were screened for functional information by using SnpEff (38) and ensembl’s online tools (www.ensembl.org). Variants were determined to be putatively functional if they were (i) exonic; (ii) located within 5 Kb upstream or downstream of a gene; (iii) within a UTR; (iv) within a splice site; (v) in a highly conserved region, notably that could be coding for a functional RNA sequence; or (vi) in a regulatory region, possibly a known enhancer.
Interspecies conservation, another possible clue to function, was assessed by using ensembl’s online tools by comparing the 10 base pair flanking regions of variants among eight primate species (human, chimpanzee, gorilla, orangutan, green monkey, macaque, olive baboon, and marmoset).
Supplementary Material
Acknowledgments
We thank the many researchers and volunteers who helped us at data collection events; in particular, Ellen Royrvik, Helen Bodmer, Stephen Day, Stephen Leslie, and Ann Ganesan. We also thank the many volunteers who donated their 3D images and genetic data. This work was supported by Wellcome Trust Grant 088262/Z/09/Z and Engineering and Physical Sciences Research Council Grant EP/N007743/1. The TwinsUK study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme FP7/2007-2013. The TwinsUK study also receives support from the National Institute for Health Research-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service Foundation Trust in partnership with King’s College London. TwinsUK SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via the NIH/Center for Inherited Disease Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708207114/-/DCSupplemental.
References
- 1.Galton F. The history of twins, as a criterion of the relative powers of nature and nurture. J Anthropol Inst. 1875;5:391–406. doi: 10.1093/ije/dys097. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard TJ. Individuality and Determinism. Springer; New York: 1984. Twins reared together and apart: What they tell us about human diversity. [Google Scholar]
- 3.Nelson C. The development and neural bases of face recognition. Infant Child Dev. 2001;10:3–18. [Google Scholar]
- 4.Leopold DA, Bondar IV, Giese MA. Norm-based face encoding by single neurons in the monkey inferotemporal cortex. Nature. 2006;442:572–575. doi: 10.1038/nature04951. [DOI] [PubMed] [Google Scholar]
- 5.Adhikari K, et al. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nat Commun. 2016;7:11616. doi: 10.1038/ncomms11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claes P, et al. Modeling 3D facial shape from DNA. PLoS Genet. 2014;10:e1004224. doi: 10.1371/journal.pgen.1004224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coussens AK, van Daal A. Linkage disequilibrium analysis identifies an FGFR1 haplotype-tag SNP associated with normal variation in craniofacial shape. Genomics. 2005;85:563–573. doi: 10.1016/j.ygeno.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, et al. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet. 2012;8:e1002932. doi: 10.1371/journal.pgen.1002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paternoster L, et al. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am J Hum Genet. 2012;90:478–485. doi: 10.1016/j.ajhg.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th Ed, xv. Longman; Harlow, UK: 1996. p. 464. [Google Scholar]
- 11.Leslie S, et al. Wellcome Trust Case Control Consortium 2; International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winney B, et al. People of the British Isles: Preliminary analysis of genotypes and surnames in a UK-control population. Eur J Hum Genet. 2012;20:203–210. doi: 10.1038/ejhg.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsagkrasoulis D, Hysi P, Spector T, Montana G. Heritability maps of human face morphology through large-scale automated three-dimensional phenotyping. Sci Rep. 2017;7:45885. doi: 10.1038/srep45885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galton F. Composite portraits, made by combining those of many different persons into a single resultant figure. J Anthropol Inst G B Irel. 1879;8:132–144. [Google Scholar]
- 15.Bigdeli TB, Neale BM, Neale MC. Statistical properties of single-marker tests for rare variants. Twin Res Hum Genet. 2014;17:143–150. doi: 10.1017/thg.2014.17. [DOI] [PubMed] [Google Scholar]
- 16.Agresti A. Categorical Data Analysis. 2nd Ed. Wiley; Hoboken, NJ: 2002. p. 710. [Google Scholar]
- 17.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimberling WJ, et al. Usher syndrome: Clinical findings and gene localization studies. Laryngoscope. 1989;99:66–72. doi: 10.1288/00005537-198901000-00013. [DOI] [PubMed] [Google Scholar]
- 19.Murcia CL, Woychik RP. Expression of Pcdh15 in the inner ear, nervous system and various epithelia of the developing embryo. Mech Dev. 2001;105:163–166. doi: 10.1016/s0925-4773(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 20.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee CG, Park SJ, Yun JN, Yim SY, Sohn YB. Reciprocal deletion and duplication of 17p11.2-11.2: Korean patients with Smith-Magenis syndrome and Potocki-Lupski syndrome. J Korean Med Sci. 2012;27:1586–1590. doi: 10.3346/jkms.2012.27.12.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 23.Cuajungco MP, et al. Cellular zinc levels are modulated by TRPML1-TMEM163 interaction. Traffic. 2014;15:1247–1265. doi: 10.1111/tra.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altarescu G, et al. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59:306–313. doi: 10.1212/wnl.59.3.306. [DOI] [PubMed] [Google Scholar]
- 25.Wakabayashi K, Gustafson AM, Sidransky E, Goldin E. Mucolipidosis type IV: An update. Mol Genet Metab. 2011;104:206–213. doi: 10.1016/j.ymgme.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chitayat D, et al. Mucolipidosis type IV: Clinical manifestations and natural history. Am J Med Genet. 1991;41:313–318. doi: 10.1002/ajmg.1320410310. [DOI] [PubMed] [Google Scholar]
- 27.Bindu PS, et al. A variant form of mucolipidosis IV: Report on 4 patients from the Indian subcontinent. J Child Neurol. 2008;23:1443–1446. doi: 10.1177/0883073808318537. [DOI] [PubMed] [Google Scholar]
- 28.Eichelsdoerfer JL, Evans JA, Slaugenhaupt SA, Cuajungco MP. Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J Biol Chem. 2010;285:34304–34308. doi: 10.1074/jbc.C110.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benini A, et al. Characterization and expression analysis of mcoln1.1 and mcoln1.2, the putative zebrafish co-orthologs of the gene responsible for human mucolipidosis type IV. Int J Dev Biol. 2013;57:85–93. doi: 10.1387/ijdb.120033gb. [DOI] [PubMed] [Google Scholar]
- 30.Bodmer WF. Evolutionary significance of the HL-A system. Nature. 1972;237:139–145, passim. doi: 10.1038/237139a0. [DOI] [PubMed] [Google Scholar]
- 31.Tena JR, Hamouz M, Hilton A, Illingworth JA. Proceedings of the International Conference on Video and Signal Based Surveillance. IEEE; New York: 2006. A validated method for dense non-rigid 3D face registration. [Google Scholar]
- 32.Tena Rodriguez JR. 2007. 3D face modelling for 2D+3D face recognition. PhD thesis (University of Surrey, Guildford, UK)
- 33.Cavalli-Sforza LL, Bodmer WF. 1971. The Genetics of Human Populations, Series of Biology Books (W.H. Freeman, San Francisco), p 582.
- 34.Strange A, et al. Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate–A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 38.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.