Significance
A timely diagnosis is key for both survival and quality of life of children with CHARGE syndrome (coloboma, heart defects, atresia of choanae, retardation of growth/development, genital abnormalities, and ear anomalies). Such diagnosis is often difficult to establish, in part because many patients test negative for mutation of CHD7, the only gene associated with this condition to date. Identifying additional CHARGE-associated genes would not only help resolve diagnosis issues but could also help in identifying common pathogenic mechanisms, which in turn could lead to desirable curative interventions for all patients. Here, FAM172A is reported as a new candidate gene for CHARGE syndrome. This discovery has allowed us to reveal a molecular process that is dysregulated in both CHD7 mutation-positive and -negative cases, such defect being correctable in vitro with rapamycin.
Keywords: alternative splicing, CHARGE syndrome, neural crest cells, sex reversal, Fam172a
Abstract
CHARGE syndrome—which stands for coloboma of the eye, heart defects, atresia of choanae, retardation of growth/development, genital abnormalities, and ear anomalies—is a severe developmental disorder with wide phenotypic variability, caused mainly by mutations in CHD7 (chromodomain helicase DNA-binding protein 7), known to encode a chromatin remodeler. The genetic lesions responsible for CHD7 mutation-negative cases are unknown, at least in part because the pathogenic mechanisms underlying CHARGE syndrome remain poorly defined. Here, we report the characterization of a mouse model for CHD7 mutation-negative cases of CHARGE syndrome generated by insertional mutagenesis of Fam172a (family with sequence similarity 172, member A). We show that Fam172a plays a key role in the regulation of cotranscriptional alternative splicing, notably by interacting with Ago2 (Argonaute-2) and Chd7. Validation studies in a human cohort allow us to propose that dysregulation of cotranscriptional alternative splicing is a unifying pathogenic mechanism for both CHD7 mutation-positive and CHD7 mutation-negative cases. We also present evidence that such splicing defects can be corrected in vitro by acute rapamycin treatment.
CHARGE syndrome affects ∼1/10,000 newborns worldwide and has a very complex clinical presentation (1). This phenotypic complexity is notably highlighted by the acronym CHARGE, which stands for coloboma of the eye, heart defects, atresia of choanae, retardation of growth/development, genital abnormalities, and ear anomalies. However, diagnosis of CHARGE syndrome does not depend on the concomitant presence of all these characteristics, as each one varies from severe to absent in affected children. It is thought that CHARGE syndrome is an underdiagnosed condition with the mildest forms presenting with hypogonadotropic hypogonadism and additional features such as cleft palate, characteristic craniofacial dysmorphisms, inner ear dysplasia, and intellectual disability (2, 3). Familial cases have also been reported and are characterized by extensive clinical variability with the transmitting parent often being very mildly affected or even asymptomatic (4–6). Such a wide range of phenotypic presentations has resulted in multiple revisions of the diagnostic criteria over the past decade (2, 3). These criteria have been subdivided into “major” and “minor” features based on their predictive value, with the most recent inclusion rule being two major and an unlimited number of minor features (2). It is noteworthy that the multiple anomalies in CHARGE syndrome can be life-threatening and, consequently, about 30% of affected children die before their fifth birthday (7). Survival and quality of life of these children are tightly linked to age of diagnosis, which is hard to establish not only because of the variable clinical presentation but also because an important subset of cases remains genetically unexplained (1, 2).
Heterozygous mutation of CHD7 (chromodomain helicase DNA-binding protein 7) is currently the only known genetic cause of CHARGE syndrome (8). However, depending on the diagnostic criteria used, up to ∼30% of patients do not test positive for CHD7 mutations (2). Based on high levels of CHD7 gene expression in neural crest derivatives and proposed roles for CHD7 in neural crest development, we hypothesized that genetically undefined CHARGE patients may harbor pathogenic variants that affect the integrity of the neural crest cell (NCC) transcriptome. Indeed, previous studies with cellular and mouse models revealed that CHD7 is a chromatin remodeler that interacts with the SWI/SNF complex for fine-tuning the expression levels of multiple genes at the heart of the NCC gene regulatory network such as SOX9, TWIST1, and SNAI1 (9–12). It is also important to note that activation of p53 appears as another relevant event in the pathogenic cascade initiated by CHD7 deficiency (13).
Intriguingly, CHD7 has recently been proposed to be one of several chromatin factors that might influence alternative splicing in mammalian cell lines of non-NCC origin (14). Many studies in such cell lines have shown that chromatin remodeling and histone modifications not only can regulate transcription but also impact alternative splicing by modulating the elongation rate of RNA polymerase II and/or by participating to the recruitment of splicing factors (15–17). In this regard, it is noteworthy that one of the preferred binding partners of CHD7 appears to be PARP1 [poly(ADP ribose) polymerase 1] (9, 18), which has been recently proposed to directly influence alternative splicing by interacting with chromatin-associated proteins, pre-mRNAs, and splicing factors (19). There is also compelling evidence that the interplay between chromatin structure and alternative splicing in mammalian cells involves the RNA interference machinery and most especially the Argonaute members AGO1 and AGO2 (20–22). Of particular interest for CHARGE syndrome, both human AGO2 (but not AGO1) and CHD7 have been reported to interact with the core proteins of the SWI/SNF chromatin-remodeling complexes BRG1 and BAF155 (9, 23). Whether any of these observations is relevant for CHARGE syndrome is, however, unknown.
Via a forward genetic screen in mice, we report here the generation and detailed characterization of a mouse model for CHD7 mutation-negative CHARGE syndrome. Fam172a, the gene disrupted in this mouse model, codes for a nuclear-specific Ago2-binding protein that appears to couple transcription with alternative splicing. Analysis of Chd7 mutant mice and cells from human patients further allow us to suggest that problems with cotranscriptional alternative splicing are likely common to all cases of CHARGE syndrome and that these splicing problems can be corrected in vitro by short exposure to rapamycin.
Results
The Toupee Line Is a Model for CHARGE Syndrome.
Toupee is the fourth mouse line issued from a forward genetic screen aimed at identifying genes with key roles in NCCs (24). As with the other lines issued from this screen (25–27), Toupee was generated via random insertion of a tyrosinase (Tyr) minigene into the FVB/N genetic background (28) and was identified using incomplete rescue of pigmentation as an indicator of NCC defects. The name Toupee was chosen in recognition of the white spot present on the head of heterozygous animals, which otherwise do not show any overt phenotype (Fig. 1A).
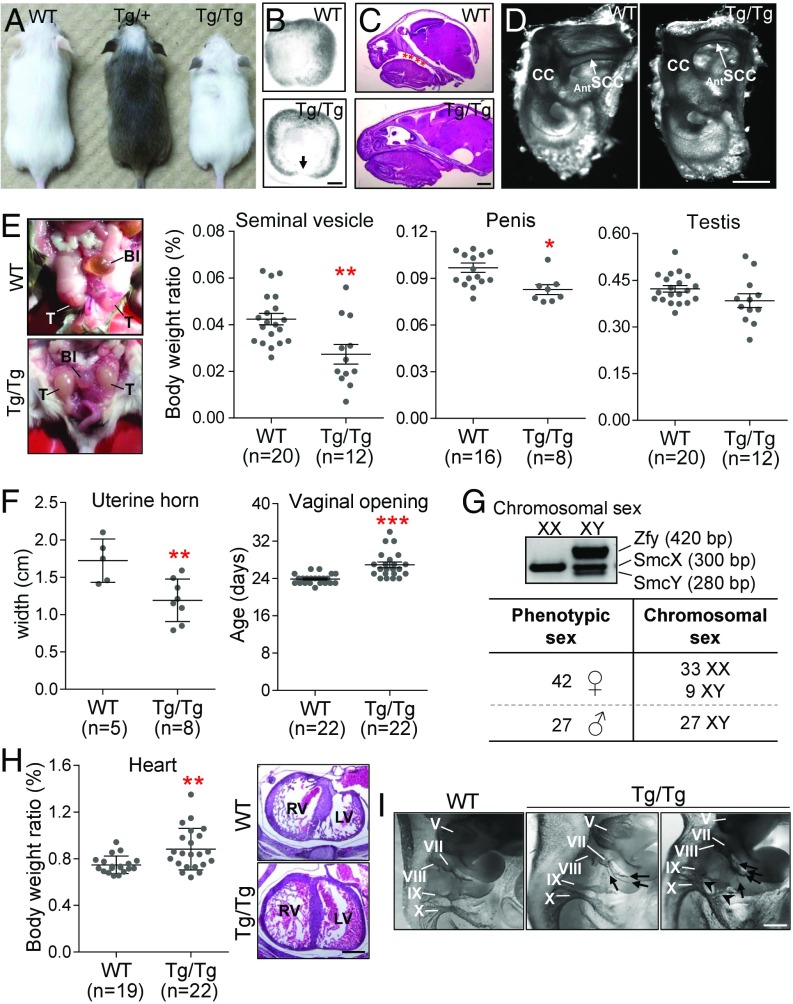
Fig. 1.
Major and minor features of CHARGE syndrome in ToupeeTg/Tg mice. (A) Comparison between WT, ToupeeTg/+, and ToupeeTg/Tg animals at P25. (B) Bright-field images of E12.5 eyes showing incomplete closure of the choroidal fissure in ToupeeTg/Tg embryos (n = 10 WT, n = 14 ToupeeTg/Tg). (C) H&E-stained sagittal sections of E18.5 heads (n = 10 WT, n = 14 ToupeeTg/Tg) with asterisks indicating cleft palate. (D) Bright-field images of Alizarin red- and Alcian blue-stained inner ears from P25 mice (n = 7 WT, n = 11 ToupeeTg/Tg). AntSCC, anterior semicircular canal; CC, common crus. (E) Overview of genital anomalies in P25 ToupeeTg/Tg males (Left, cryptorchidism; Middle, smaller androgen-sensitive seminal vesicles and penis; Right, normally sized testes). B1, bladder; T, testes. (F) ToupeeTg/Tg females present hypoplastic uterine horns at P25 (Left) and delayed opening of the vaginal cavity after P20 (Right). (G) PCR-based sexing of ToupeeTg/Tg animals revealed male-to-female sex reversal for 25% of XY animals. (H) A subset of ToupeeTg/Tg mice display heart malformation (Left, increased weight at P25; Right, hypertrophy of the left ventricle at E15.5). LV, left ventricle; RV, right ventricle. (I) Whole-mount staining of cranial nerves in E10.5 embryos using antineurofilament immunohistochemistry. ToupeeTg/Tg embryos exhibit supernumerary sprouting in the facial (VII) nerve (arrows) and extensive mingling between glossopharyngeal (IX) and vagal (X) nerves (arrowheads). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t test). [Scale bar: 50 µm (B); 1 mm (C and D); 500 µm (H and I).]
Intercrosses of Toupee heterozygotes produce almost fully depigmented homozygous animals (Fig. 1A). Born at approximate Mendelian ratios, about 20% of these homozygotes (ToupeeTg/Tg) die before postnatal day 25 (P25) (SI Appendix, Fig. S1 A and B). As summarized in SI Appendix, Table S1, ToupeeTg/Tg animals also display a complex phenotype mimicking both the major and minor features of CHARGE syndrome (2, 3). As seen in CHARGE patients, ToupeeTg/Tg mice have different combinations of such features, which also vary in their severity. Among the major features, ToupeeTg/Tg animals display retinal coloboma (Fig. 1B), cleft palate (Fig. 1C), and hypoplastic semicircular canals (Fig. 1D). As observed in other mouse models of CHARGE syndrome (29, 30), such malformations of semicircular canals are believed to be the cause of hyperactive circling behavior (Movie S1). The most frequently observed minor features (i.e., in more than 50% of animals) are retarded growth (Fig. 1A and SI Appendix, Fig. S1C), genital anomalies (Fig. 1 E–G), and malformation of the heart (Fig. 1H) and cranial nerves (Fig. 1I). Other notable but less frequently observed minor features include hypoplasia of the thymus (SI Appendix, Fig. S2A), hypoplasia of the olfactory bulbs associated with a decreased sense of smell (SI Appendix, Fig. S2 B and C), and diverse craniofacial malformations such as asymmetry of facial bones (SI Appendix, Fig. S2D), partial atresia of the oropharynx (SI Appendix, Fig. S2E), and delayed closure of the fontanelles (SI Appendix, Fig. S2F). On very rare occasions, we also found evidence of hypoplastic kidneys (SI Appendix, Fig. S2G), malformed outer ears (SI Appendix, Fig. S2H), and exencephaly/hydrocephaly (SI Appendix, Fig. S2I) in ToupeeTg/Tg animals. Finally, we noted that the gastrointestinal tract of mutant animals found dead before P25 was often filled with air bubbles (SI Appendix, Fig. S2J). Combined with the poor postnatal growth and anomalies of the oropharynx and cranial nerves described above, this strongly suggests that a subset of ToupeeTg/Tg animals have feeding difficulties—another hallmark of CHARGE syndrome (31).
CHARGE syndrome-related genital anomalies include delayed puberty in both sexes, small uterus in females, and cryptorchidism and small penis in males. ToupeeTg/Tg mice differ from previously described mouse models with Chd7 mutations (32, 33) in that both females and males appear to be affected (Fig. 1 E and F). Interestingly, we found that both male and female ToupeeTg/Tg animals are subfertile (SI Appendix, Table S2). We further noticed that the sex ratio is distorted toward females in the ToupeeTg/Tg population. This quite unexpected finding prompted us to verify the concordance between phenotypic and chromosomal sex, which revealed that 25% of ToupeeTg/Tg genetic males are phenotypic females (Fig. 1G). Although male-to-female sex reversal has not been reported in CHARGE syndrome, this phenotype appears not to be specific to the Toupee line as it was also detected in ∼12% of XY Chd7Gt/+animals (SI Appendix, Table S3)—a well-recognized mouse model of CHARGE syndrome that contains a gene-trapped allele of Chd7 (30).
To further test whether Toupee is a model for CHARGE syndrome, we asked whether the Toupee allele could genetically interact with the gene-trapped allele of Chd7 (SI Appendix, Fig. S3 and Table S3). In comparison with corresponding single heterozygotes, ToupeeTg/+;Chd7Gt/+ double heterozygotes were found to be markedly smaller at weaning age (SI Appendix, Fig. S3A) and to exhibit a higher frequency of premature postnatal death, circling behavior, and male-to-female sex reversal (SI Appendix, Table S3). Coloboma as determined by incomplete closure of the choroidal fissure in E12.5 embryos was also found to be much more severe in double heterozygous mutants (SI Appendix, Fig. S3B). Moreover, the lower-than-expected number of such mutants at birth suggests that an important subset of them dies in utero, an outcome known to occur in Chd7 homozygous mutants (30). All these observations strongly suggest that Toupee is a valid model for CHARGE syndrome.
NCC Development Is Globally Affected in ToupeeTg/Tg Embryos.
Based on previous studies showing that NCCs are a major cell population impaired in CHARGE syndrome (9, 11, 12, 34), we undertook a detailed analysis of this cell lineage in ToupeeTg/Tg embryos. The aim of this analysis was to determine which of the main basic cellular processes (i.e., proliferation, survival, migration, and/or differentiation) might be affected. We chose to focus on the E10.5 developmental stage since it allows analysis of virtually all key steps of NCC development in different anteroposterior regions of a single embryo, with the cranial region having the “oldest” and the elongating posterior region having the “youngest” NCCs.
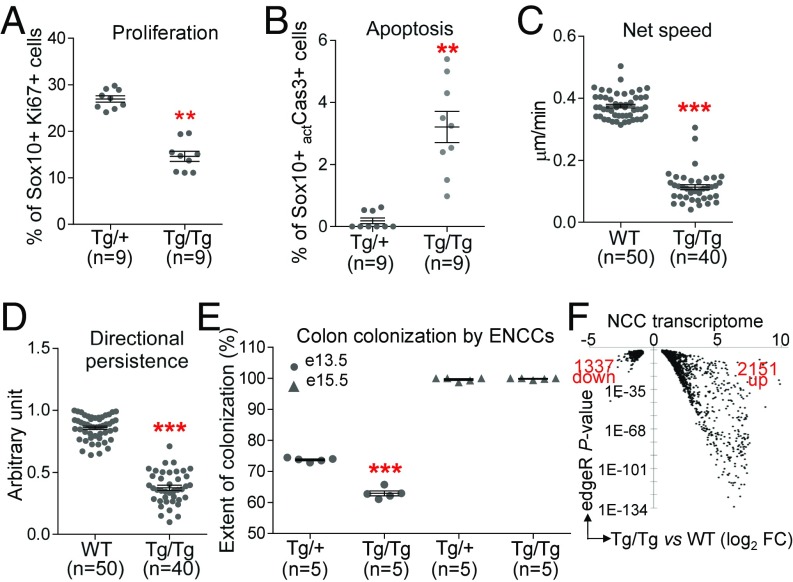
We first monitored the number of NCCs (Sox10+) undergoing apoptosis (actCaspase3+) as well as those actively proliferating (Ki67+) via immunofluorescence analyses of trunk cross-sections. We detected significant variations in both processes, with proliferation being decreased (Fig. 2A) and apoptosis being increased (Fig. 2B) in ToupeeTg/Tg embryos. Of note, these defects were found not to be exclusive to NCCs (SI Appendix, Fig. S4 A and B). Interestingly, we further noticed that mutant NCCs were closer to the dorsal neural tube than normal, suggesting that trunk NCC migration was also affected in ToupeeTg/Tg embryos. To directly verify this possibility, we transferred the Toupee allele onto a Gata4p[5kb]-red fluorescent protein (RFP) (G4-RFP) transgenic background and followed movements of recently induced trunk NCCs via time-lapse imaging. In accordance with the previously described expression pattern of the G4-RFP transgene (35), an anterior-to-posterior wave of transgene activation was noted in control NCCs delaminating from the dorsal neural tube and ventrally migrating through the somites (Movie S2). In contrast, ToupeeTg/Tg NCCs were found to accumulate in the vicinity of the dorsal neural tube, to migrate more slowly, and to oscillate rather than persist in their ventrally oriented migration (Movie S3). Quantification of NCC speed and directionality at the leading edge of migration streams revealed that both parameters are significantly impaired in ToupeeTg/Tg;G4-RFP explants (Fig. 2 C and D). However, these defects appeared stronger than expected from the relatively mild phenotypic presentation of postnatal ToupeeTg/Tg animals, suggesting that mutant NCCs might still be able to reach some of their final destinations. Accordingly, in similar fashion to what was previously reported for Tcof1+/− embryos (36), we found that hindgut colonization by RFP-labeled ToupeeTg/Tg enteric NCCs of vagal origin is delayed at E13.5 but not at E15.5 [i.e., a day before and a day after the end of the normal period of colonization (37)] (Fig. 2E and SI Appendix, Fig. S4 C and D). These observations indicate that some of the NCC migration defects detected in ToupeeTg/Tg embryos at early stages can be compensated for at later stages.
Fig. 2.
Global impairment of NCC development in ToupeeTg/Tg embryos. (A and B) Quantification of Ki67+ proliferating (A) and actCasp3+ apoptotic (B) NCCs (expressed in percentage of Sox10+ NCCs) in 30-μm transverse sections of E10.5 embryos at hindlimb level (ToupeeTg/+ vs. ToupeeTg/Tg). (C and D) Quantification of NCC migration speed (C) and movement persistence (D) in E10.5 embryos (WT;G4-RFP vs. ToupeeTg/Tg;G4-RFP). (E) Quantification of the extent of colon colonization by enteric NCCs (expressed in percentage of colon length from cecum to anus) at E13.5 and E15.5 (ToupeeTg/+;G4-RFP vs. ToupeeTg/Tg;G4-RFP). (F) Volcano plot summarizing a RNA-seq–based analysis of differential gene expression levels in E10.5 NCCs (WT;G4-RFP vs. ToupeeTg/Tg;G4-RFP). Only genes modulated at least 1.5-fold with a P value below 0.01 are displayed. **P ≤ 0.01 and ***P ≤ 0.001 (Student’s t test).
To complement our cellular analyses, we next evaluated the impact of the Toupee mutation on the NCC transcriptional profile using RNA sequencing (RNA-seq). To this end, we again took advantage of the G4-RFP transgene to specifically recover NCCs from whole E10.5 embryos (control G4-RFP vs. ToupeeTg/Tg;G4-RFP) by FACS before deep sequencing of total RNA depleted of ribosomal RNA. Differential analysis of gene expression levels identified several thousand genes that are dysregulated at least 1.5-fold in ToupeeTg/Tg NCCs (3,488 genes at a DESeq P-value cutoff of 0.01), with a bias toward up-regulated genes (61.7% up-regulated vs. 38.3% down-regulated) (Fig. 2F and Dataset S1). Of note, this bias appears much stronger when considering only those genes dysregulated at least fourfold (622 up-regulated genes vs. 19 down-regulated genes). Gene Ontology (GO) analysis of the 3,488 genes list identified 132 enriched terms (with an ontology level ≥5 and P < 0.05) that can be classified into seven main categories (from most to less significant): metabolic processes; cell differentiation and morphogenesis; cell signaling; cell motility and transport; control of cell number; gene expression; and nervous system development (SI Appendix, Fig. S5). Interestingly, restricting our analysis to genes that form the constantly expanding NCC gene regulatory network (38–41) revealed that 90.6% (96 of 106 identified) of those found to be affected are down-regulated in ToupeeTg/Tg embryos (SI Appendix, Table S4). Essentially every aspect of NCC development is represented in this list of down-regulated genes—from induction/specification to anteroposterior patterning, delamination/migration, and formation of specific cell types/structures (peripheral neurons and glia, melanocytes, craniofacial skeleton, and enteric nervous system).
Toupee Is a Hypomorphic Allele of Fam172a.
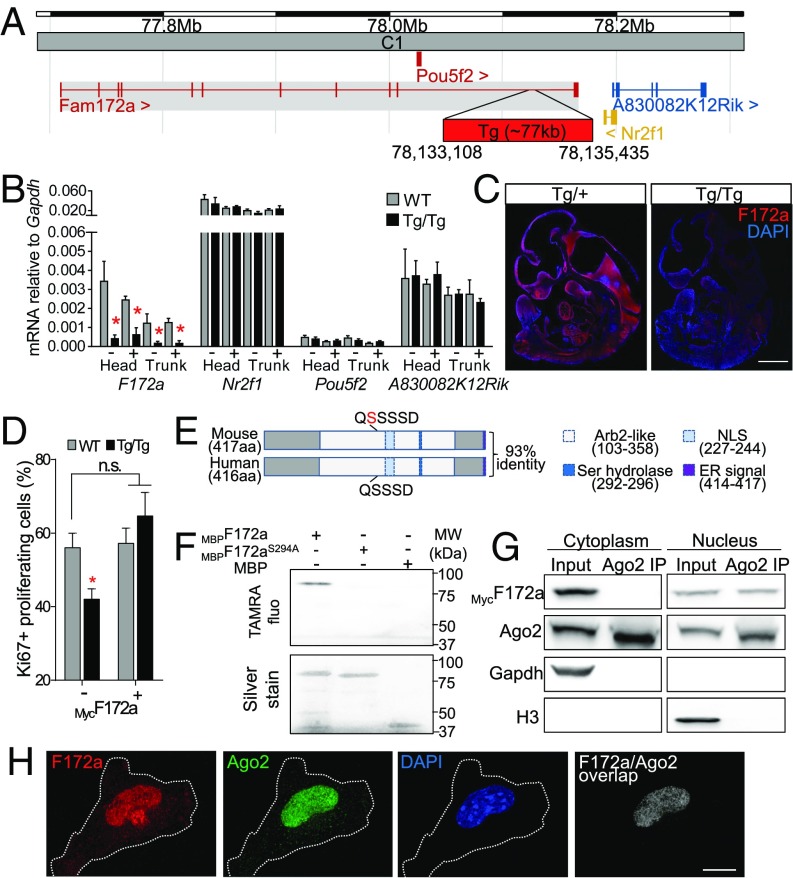
Using whole-genome sequencing, we localized the transgene insertion site of the Toupee line in the last intron of Fam172a (family with sequence similarity 172, member A), a poorly characterized but highly conserved gene (93% identity with its human ortholog on Chr.5q15) (Fig. 3A). Expression analyses using RT-qPCR and immunofluorescence at multiple embryonic stages revealed that Fam172a is normally widely expressed during development—including prominent expression in neural tissues—and robustly down-regulated (down to ∼15% on average) in ToupeeTg/Tg embryos (Fig. 3 B and C and SI Appendix, Figs. S6 and S7). This effect appears highly specific as no significant change in gene expression was detected for the other genes flanking the insertion (Pou5f2, Nr2f1, and A830082K12Rik) regardless of the stage (E10.5, E13.5 and E15.5), region (cranial and trunk), or cell population (NCCs and non-NCCs) analyzed (Fig. 3 B and C and SI Appendix, Fig. S6). Importantly, ex vivo transfection of a Myc-tagged Fam172a expression vector in primary cultures of dissociated E10.5 ToupeeTg/Tg embryos fully rescued the global proliferation defect previously identified (SI Appendix, Fig. S4A), thereby confirming causality of the Fam172a mutation (Fig. 3D).
Fig. 3.
Functional characterization of Fam172a, the Toupee causative gene. (A) Schematic representation of the Toupee transgene insertion site in cytoband C1 of chromosome 13 (adapted from the Ensembl website), where ∼10 copies of a tyrosinase minigene are inserted in a 2,327-bp deletion in the last intron of Fam172a (red box). (B) RT-qPCR analysis of gene expression in E10.5 embryos (WT;G4-RFP vs. ToupeeTg/Tg;G4-RFP). Transcript levels of genes around the transgene insertion site were monitored in FACS-recovered NCCs (+) and non-NCCs (−) from the head and the trunk (n = 3 per condition). (C) Immunofluorescence labeling of the Fam172a protein (red) in sagittal sections of E10.5 mouse embryos (ToupeeTg/+ vs. ToupeeTg/Tg, n = 3 per genotype). DAPI (blue) was used to counterstain nuclei. (D) Quantification of proliferation in cultures of dissociated E10.5 embryos (WT vs. ToupeeTg/Tg; n = 3 per condition) after transfection with a MycFam172a-expressing vector. (E) Schematic representation of the functional domains of mouse Fam172a protein compared with its human ortholog. The serine in position 215 (highlighted in red) corresponds to the supernumerary amino acid between mouse (417 aa) and human (416 aa) sequences. Arb2-like, domain homologous to yeast Arb2 (Argonaute-binding protein 2); ER, endoplasmic reticulum retention signal; NLS, nuclear localization signal; Ser hydrolase, esterase-like serine hydrolase motif. (F) Hydrolase activity is demonstrated by covalent binding of a TAMRA-labeled fluorophosphonate probe on MBPFam172a protein (Upper). No binding is detected on MBPFam172a bearing a S294A mutation nor on the MBP tag alone. Lower panel shows that silver-stained MBP-tagged Fam172a proteins (∼90 kDa; slightly higher in the presence of fluorophosphonate probe) and MBP alone (42 kDa) were all present at the expected size in the same gel. (G) Co-IP assays using cytoplasmic (Gapdh+) and nuclear (H3+) fractions of Neuro2a cells transfected with a MycFam172a-expressing vector (n = 3). Inputs correspond to 10% of protein extracts used for IP. (H) Double immunofluorescence labeling of Fam172a and Ago2 in dissociated cells obtained from WT E10.5 embryos (n = 7) and counterstained with DAPI. (Right) The overlap of Fam172a and Ago2 signals (Pearson’s correlation coefficient of 0.82). *P ≤ 0.05 (Student’s t test). [Scale bar: 500 μm (C); 25 μm (H).]
Bioinformatics-based analysis of Fam172a protein sequences (using the meta sites MyHits, MOTIF, and MetaDBsite) notably predicted an esterase-like serine hydrolase motif (G-X-S-X-G) and a bipartite Lys/Arg-rich nuclear localization signal, both overlapping with a large domain originally described in Schizosaccharomyces pombe Arb2 [Argonaute binding protein-2 (42)] (Fig. 3E). In vitro labeling of WT and mutant maltose-binding protein (MBP)-tagged Fam172a with a serine hydrolase-specific fluorescent probe [tetramethylrhodamine (TAMRA)-Fluorophosphonate] confirmed serine hydrolase activity and identified the serine 294 as the nucleophilic residue (Fig. 3F), while double-immunofluorescence and coimmunoprecipitation (co-IP) in multiple cell types/tissues showed that Fam172a is found in the vicinity of, and physically interacts with, the nuclear fraction of the Argonaute member Ago2 (Fig. 3 G and H and SI Appendix, Fig. S8 A and B). Of note, Fam172a is apparently not a general Argonaute binding protein as no interaction was detected with Ago1 (SI Appendix, Fig. S8 D–F). Moreover, in accordance with the nuclear specificity of the Fam172a-Ago2 interaction, transfection of the psi-CHECK2-let-7 × 8 luciferase reporter in primary cultures of dissociated E10.5 embryos revealed that the Toupee mutation has no impact on cytoplasmic posttranscriptional gene silencing (SI Appendix, Fig. S8C). However, Fam172a is present in the cytoplasm where, in accordance with the prediction of a relevant C-terminal retention signal (HEEL), high amounts are frequently detected in the endoplasmic reticulum (SI Appendix, Fig. S7C).
Characterization of Fam172a Function Suggests a Role for Alternative Splicing in the Pathogenesis of CHARGE Syndrome.
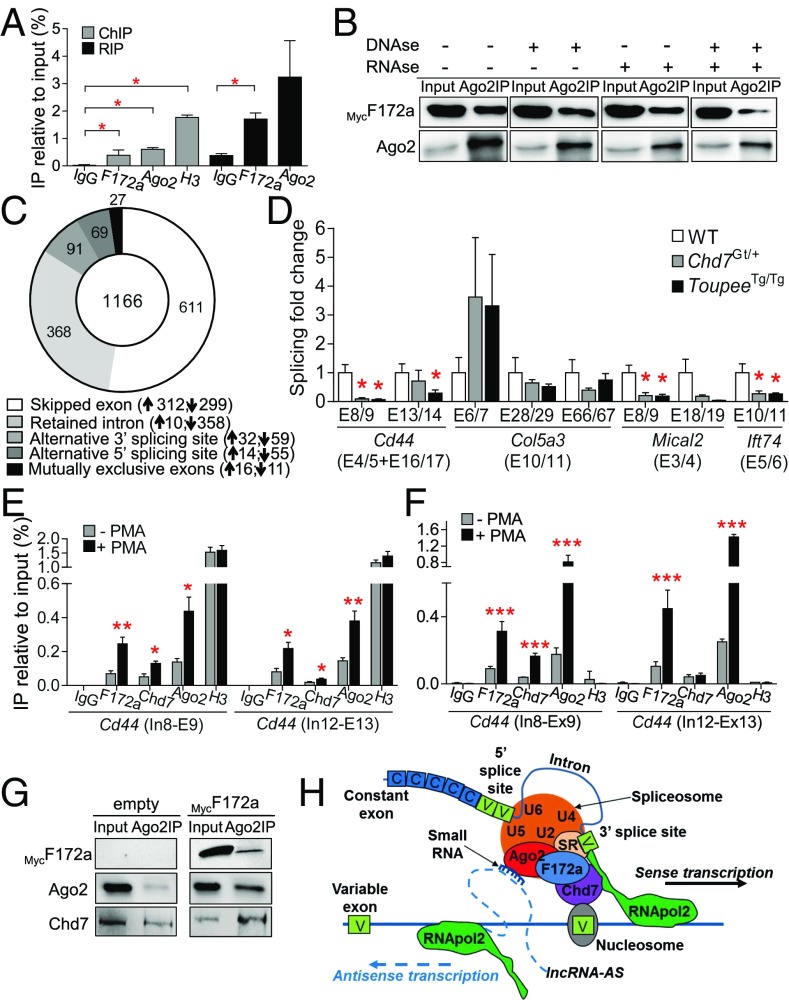
To follow up on our discovery of a nuclear-specific Fam172a-Ago2 interaction, we first evaluated the ability of Fam172a to interact with chromatin and/or RNA via untargeted chromatin immunoprecipitation (ChIP) and RNA immunoprecipitation (RIP) assays. Using Neuro2a cells to model the NCC lineage, we found that Fam172a, like Ago2, can bind both chromatin and RNA (Fig. 4A). Interestingly, co-IP assays in the presence of DNase and/or RNase further revealed that efficient formation of the Fam172a-Ago2 complex requires at least one type of nucleic acid (Fig. 4B and SI Appendix, Fig. S8G).
Fig. 4.
Both Fam172a and Chd7 play a role in alternative splicing. (A) Untargeted ChIP and RIP assays in Neuro2A cells transfected with a Fam172a-expressing plasmid (n = 3). (B) Co-IP assays using RNase- and/or DNase-treated whole-cell extracts of Neuro2A cells transfected with a MycFam172a expression vector. Inputs correspond to 10% of protein extracts used for IP (n = 3 per condition). Impact of each treatment on the integrity of proteins, DNA, and RNA is shown in SI Appendix, Fig. S8G. (C) Donut chart showing the distribution of the 1,166 differentially modulated alternative splicing events (P < 0.01; variation in inclusion level ≥0.1) in ToupeeTg/Tg E10.5 NCCs. Upward- and downward-pointing arrows indicate splicing events that are over- and under-represented in ToupeeTg/Tg E10.5 NCCs, respectively. (D) RT-qPCR analysis of splicing events for Cd44, Col5a3, Mical2, and Ift74 in G4-RFP (WT), ToupeeTg/Tg;G4-RFP (ToupeeTg/Tg), and Chd7Gt/+ heads of E12.5 embryos (n = 5 per genotype). Expression levels of variable regions are normalized with levels of corresponding constant regions (indicated between parentheses). (E and F) ChIP (E) and RNA-ChIP (F) assays of the PMA-inducible Cd44 gene in Neuro2a cells. (G) Co-IP assays in Neuro2A cells transfected with empty or MycFam172a-expressing vector. Inputs correspond to 10% of protein extracts used for IP (n = 3 per condition). (H) Potential mode of action of Fam172a and Chd7 in Ago2-mediated alternative splicing (adapted from ref. 21). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 (Student’s t test).
To gain more insight into Fam172a function, we next sought to identify its interactors in an unbiased manner via pull-down assays coupled to mass spectrometry. Using stringent binding conditions and three different fractions of Neuro2a cells (chromatin, nucleoplasm, and cytoplasm), this analysis notably revealed a marked enrichment for chromatin proteins and RNA splicing factors among the interacting partners of MBP-tagged Fam172a (SI Appendix, Tables S5–S7). Taken together with our ChIP, RIP, and co-IP data described above, this finding strongly suggested that Fam172a might bridge the chromatin with the alternative splicing machinery as previously suggested for both Chd7 (14) and Ago2 (14, 21), which were not identified in our proteomic screen most likely because the Fam172a-Ago2 interaction is impaired in the high-salt–binding conditions used (SI Appendix, Fig. S8H). This intriguing possibility prompted us to reanalyze our transcriptome data for the presence of aberrant splicing events in ToupeeTg/Tg NCCs. Using the rMATS computational tool (43) to compare WT and mutant RNA-seq data, we discovered that 1,166 transcripts are aberrantly spliced in ToupeeTg/Tg NCCs (using variation in inclusion level >0.1 and P < 0.01 as cutoff values) (Fig. 4C and Dataset S2). Among the different rMATS categories, the vast majority of these affected splicing events were found to fall into either the Skipped exon (52.4%) or the Retained intron (31.5%) categories. GO analysis of the 1,166 abnormally spliced transcripts yielded 75 enriched terms (with ontology level ≥5 and P < 0.05) that can be classified into six main categories fully overlapping with those of the differential expression level analysis (from most to less significant): metabolic processes; cell motility and transport; cell signaling; gene expression; control of cell number; and cell differentiation and morphogenesis (SI Appendix, Fig. S9). Cross-comparison of both RNA-seq analyses revealed that 30% of all aberrantly spliced transcripts (350 transcript isoforms of 1,166) correspond to genes also affected at the transcriptional level, this latter group representing 7.4% of all differentially expressed genes (259 single genes of 3,488).
To determine whether dysregulation of alternative splicing might represent a common signature for CHARGE syndrome, we then analyzed the splicing pattern of several previously reported Ago2 target genes (21) in both the ToupeeTg/Tg and the Chd7Gt/+ mouse models in comparison with WT. Because the selected genes (Cd44, Col5a3, Mical2, and Ift74) are all expressed in the developing brain (www.brain-map.org), this analysis was performed using whole heads from E12.5 embryos. Remarkably, our RT-qPCR data revealed that variable exons of Cd44, Mical2, and Ift74 are specifically underrepresented in both ToupeeTg/Tg and Chd7Gt/+ mutants (Fig. 4D and SI Appendix, Fig. S10A). Among the four tested genes, only Col5a3 did not appear to be significantly affected at the alternative splicing level. However, in contrast to Cd44, Mical2, and Ift74, global transcription of Col5a3 as determined by the level of constant exons was found to be robustly down-regulated in both mutant lines (SI Appendix, Fig. S10A). As suggested by previous work (14), we further found that Chd7 can regulate alternative splicing of a large number of genes in different spatiotemporal contexts. Indeed, rMATS-based reanalysis of recently published RNA-seq data from P7 granule neuron progenitors (18) revealed dysregulated alternative splicing of 227 and 252 transcripts upon heterozygous or homozygous loss of Chd7, respectively (using variation in inclusion level >0.1 and P < 0.05 as cutoff values) (Datasets S3 and S4 and SI Appendix, Fig. S10B). These aberrant splicing events were found to affect 208 and 226 genes, respectively, 60 of them being detected in both datasets. Given the limited impact on gene expression levels previously reported in the original analysis (only 151 dysregulated genes, most likely because of low sequencing depth) (18), the presence of over 200 splicing defects appears highly significant in this case. It is also interesting to note that these splicing anomalies are distributed into the different rMATS categories in a way very similar to what we observed with ToupeeTg/Tg NCCs (Fig. 4C), with the Skipped exon and Retained intron categories again being overrepresented (SI Appendix, Fig. S10B).
Focusing on the PMA-inducible Cd44 gene model, ChIP assays in Neuro2a cells then showed that both Fam172a and Chd7 are, like Ago2 (21), normally present on transcribed chromatin regions containing alternative splice sites (Fig. 4E). Importantly, using RNA immunoprecipitation of chromatin (RNA-ChIP), a similar pattern of interaction was also observed on corresponding regions of the chromatin-associated pre-mRNA (Fig. 4F). Moreover, co-IP data showed that Chd7 can physically interact with both Fam172a and Ago2 in low-stringency conditions (Fig. 4G and SI Appendix, Fig. S8I). Interestingly, this analysis further showed that the presence of exogenous MycFam172a seems to promote the Chd7-Ago2 interaction (Fig. 4G). Together, these results strongly suggest that CHARGE syndrome-related malformations are caused not only by defective transcription (as generally thought) but also by dysregulated alternative splicing of genes that are commonly targeted by Fam172a, Ago2, and Chd7. Although the exact relationship between these three proteins is currently unknown, we propose a model in which Fam172a appears to be required for stabilizing protein–protein interactions at the chromatin–spliceosome interface (Fig. 4H).
Alternative Splicing Defects Are Common in CHARGE Syndrome Patients, and These Defects Can Be Corrected with Acute Rapamycin Treatment.
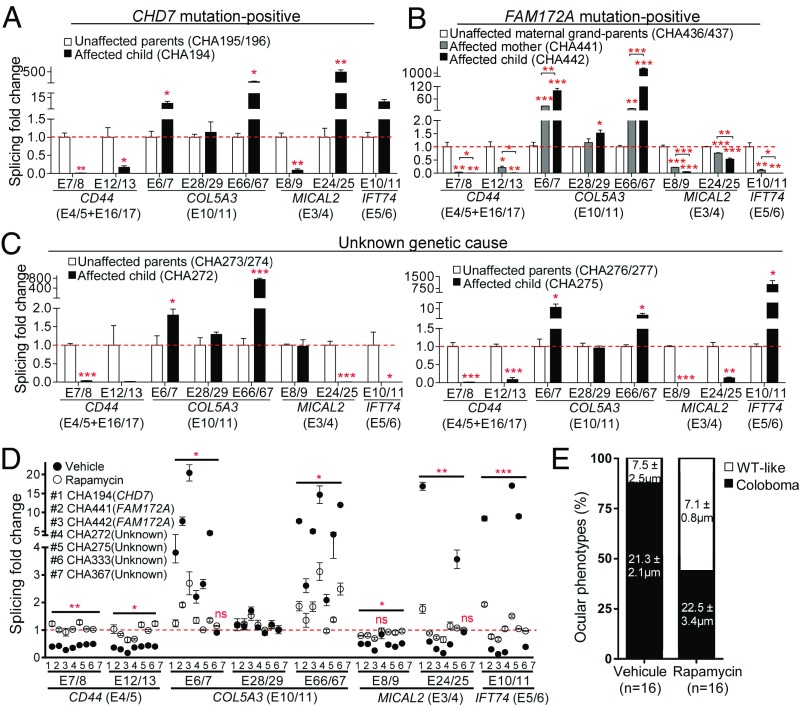
To test our findings in humans, we generated lymphoblastoid cell lines (LCLs) from genetically and phenotypically heterogeneous CHARGE syndrome patients (SI Appendix, Table S8) and their unaffected parents. All patients were enrolled based on CHD7 mutation-negative status according to clinical genetic testing, and all individuals were characterized by exome sequencing. Careful analysis of CHD7 sequences revealed a likely deleterious variant (c.5050+1G > T) in one patient while the other six patients were confirmed as CHD7 mutation-negative, including a mother–child pair. As previously reported for other familial cases with or without the CHD7 mutation (4–6), the mother (ear malformations and choanal atresia) was found to be less affected than her child (ear malformations, choanal atresia, retarded growth, genital hypoplasia, and a small kidney). Very interestingly, exome sequencing data revealed the presence of two rare FAM172A variants in this mother–child pair. The variant c.682G > C (p.Glu228Gln; frequency of 1.7e-05 in ExAC browser) was detected in both the mother and the child as well as in the unaffected maternal grandmother, while the second variant, c.916C > T (p.Arg306*, not found in ExAC browser), was detected only in the child. No paternal sample was available for this analysis. Of note, the co-occurrence of two rare and likely deleterious variants in single patients has also been reported for CHD7 (44, 45), including at least one confirmed case of compound heterozygosity (46). To experimentally evaluate the pathogenicity of the identified FAM172A variants, we introduced the same mutations in the corresponding conserved residues of murine Fam172a. In accordance with their location in the Arb2 domain, both variants were found to prevent the formation of a Fam172a-Ago2 complex in co-IP assays, and the nonsense mutation was further found to affect Fam172a protein levels (SI Appendix, Fig. S11A). Both variants also appeared to impact the subcellular distribution of Fam172a (SI Appendix, Fig. S11B). Taken together with the finding of the c.682G > C variant in another mother–child pair of CHARGE patients from a different cohort (SI Appendix, Table S9), these results support the hypothesis that FAM172A deficiency may play a role in the pathogenesis of a subset of CHD7 mutation-negative cases of CHARGE syndrome. Intriguingly, obvious signs of coloboma were not observed in any of these four FAM172A-affected individuals, suggesting that they might even form a distinct group of people with CHARGE (or CHARGE-like) syndrome. No convincing candidate gene could be identified in the other four CHD7 mutation-negative patients.
Although our cohort was relatively small, it allowed us to directly test the hypothesis that dysregulation of alternative splicing is common to all cases of CHARGE syndrome irrespective of their CHD7 or FAM172A mutation status. To this end, we used the same approach as described above for mice and evaluated alternative splicing of CD44, COL5A3, MICAL2, and IFT74 in our collection of LCLs via RT-qPCR. Strikingly, the splicing patterns of all four genes were found to be severely dysregulated in all seven patients compared with their unaffected parents (Fig. 5 A–C and SI Appendix, Fig. S12). For most of the tested splicing events, the same trend could be noted regarding the inclusion/exclusion of a given exon. However, no global rule could be observed. Indeed, tested variable exons appeared preferentially excluded for CD44 and preferentially included for COL5A3, while either inclusion or exclusion was promoted in the case of MICAL2 and IFT74. All these results thus support the idea that impairment of cotranscriptional alternative splicing is a unifying pathogenic mechanism for all cases of CHARGE syndrome. Interestingly, our RT-qPCR analyses also revealed that FAM172A expression is decreased in a majority of patients within this cohort, thereby further supporting its role in the pathogenesis of CHARGE syndrome (SI Appendix, Fig. S13A).
Fig. 5.
Rapamycin-correctable dysregulation of alternative splicing in CHARGE syndrome patients. (A–D) RT-qPCR analysis of splicing events for CD44, COL5A3, MICAL2, and IFT74 in lymphoblastoid cell lines. Expression levels of variable regions were normalized with levels of corresponding constant regions (indicated between parentheses). Results for unaffected parents were combined and used as reference value for calculation of splicing fold change (red dashed line). In A–C, each graph depicts the results obtained for a given family (for each individual, n = 9 from three independent experiments). Results for other families can be found in SI Appendix, Fig. S12, while detailed information about each patient can be found in SI Appendix, Table S8. D depicts the results obtained after a 30-min treatment with rapamycin (10 µM) or vehicle only (ethanol), each vertically aligned pair of dots corresponding to a single CHARGE patient (for each individual, n = 6 from two independent experiments). In D, statistic tests refer to the difference between rapamycin and vehicle treatments. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ns, not significant (Student’s t test). (E) Occurrence of coloboma in E12.5 ToupeeTg/Tg embryos (for each condition, n = 8 embryos/16 eyes) following a 3-d in utero exposition to rapamycin (1 mg/kg) or vehicle (20% ethanol). For each phenotypic group (coloboma and WT-like), the average width of choroidal fissure is indicated in their corresponding bar subdivision.
We then reasoned that our set of LCLs could serve in addition as a tool to explore the possibility of developing a small-molecule–based therapy for CHARGE syndrome. As a proof of concept, we evaluated rapamycin, a well-known TOR inhibitor for which analogs (i.e., rapalogs) have been approved by the Food and Drug Administration for multiple clinical conditions (47). One of the best known roles of the nutrient-sensing TOR pathway is to stimulate ribosome biogenesis by activating the transcription of ribosomal protein genes (48). Accordingly, one of the main effects of acute rapamycin treatments is the down-regulation of ribosomal protein gene expression, which in turn promotes splicing of other pre-mRNAs due to relief of competition for a likely limiting pool of splicing factors (49). Since rapamycin-mediated repression of ribosomal protein gene expression was reported to occur within a few minutes in yeast (49), we treated LCLs with a moderate dose of rapamycin (10 µM) during 30 min and then analyzed alternative splicing of CD44, COL5A3, MICAL2, and IFT74 as described above (Fig. 5D). Of note, CD44 is a known mTOR target gene (50), meaning that absolute levels of CD44 constant exons also allowed us to confirm efficacy of the rapamycin treatment (SI Appendix, Fig. S13B). Remarkably, such acute rapamycin treatment was sufficient for positively impacting the splicing defects detected in all patients of our cohort (Fig. 5D). Both previously down-regulated and up-regulated splicing events were found to be corrected, as indicated by the splicing fold change levels that became closer to the levels observed in their respective unaffected parents (i.e., splicing fold change of 1). Analysis of FAM172A expression in this context revealed a very modest increase, if any, upon rapamycin treatment (SI Appendix, Fig. S13B).
These most encouraging results finally prompted us to directly evaluate the in vivo potential of rapamycin as a therapeutic agent for CHARGE syndrome. To this end, we administered a moderate dose of rapamycin (1 mg/kg) via i.p. injection to pregnant females from ToupeeTg/Tg intercrosses twice a day between E9.5 and E11.5 (i.e., during the peak of NCC migration) and then analyzed E12.5 embryos for the presence of coloboma—the phenotype with highest penetrance in ToupeeTg/Tg animals (SI Appendix, Table S1). Very interestingly, this approach allowed us to decrease the incidence of coloboma by 50% (Fig. 5E). It should, however, be noted that, although the rapamycin dose that we used was lower than the previously described teratogenic dose (3 mg/kg), most treated embryos did not look healthy, showing signs of growth retardation and/or resorption (SI Appendix, Fig. S13C). We thus conclude that rapamycin is a promising drug for the treatment of CHARGE syndrome but that more work will be required to balance therapeutic benefits with adverse effects.
Discussion
The Toupee insertional mutant line is a viable mouse model for CHD7 mutation-negative cases of CHARGE syndrome. Extensive characterization of this mouse line notably allowed us to unveil a previously overlooked partially penetrant male-to-female sex reversal phenotype, which we found to be present in a subset of Chd7 mutants as well. We further discovered that Fam172a, the Toupee mutated gene, codes for a nuclear-specific binding partner of Ago2 that appears to couple chromatin structure with alternative splicing. Finally, by analyzing a genetically heterogeneous cohort of CHARGE patients, we found that dysregulation of cotranscriptional alternative splicing could well be the pathogenic mechanism underlying all cases of CHARGE syndrome. This finding is expected to help in guiding gene discovery for CHD7 mutation-negative cases and, most importantly, opens up the possibility of eventually developing small-molecule–based therapeutic interventions for this devastating pediatric disease.
Fam172a, a Highly Conserved Protein with Enigmatic Serine Hydrolase Activity.
Using different experimental approaches, we confirmed the presence of four functional domains/motifs in murine Fam172a: an Arb2-like domain, a bipartite nuclear localization signal, an esterase-like serine hydrolase motif, and an endoplasmic reticulum (ER) retention signal. With the exception of the ER retention signal, all these domains/motifs are also present in Fam172a orthologs from evolutionarily distant species such as humans (417 aa; 93% overall identity with mouse ortholog), zebrafish (415 aa; 71% overall identity with mouse ortholog), lamprey (311 aa; 47% overall identity with mouse ortholog), and Caenorhabditis elegans (313 aa; 33% overall identity with mouse ortholog). The C-terminal ER retention signal (HEEL) appears as a “recent” evolutionary gain, being present in mouse and human orthologs but not in those from zebrafish, lamprey, and C. elegans. From an evolutionary point of view, this suggests that Fam172a is thus especially important in the nucleus, where our data collectively reveal that it can connect the alternative splicing machinery to specific transcribed chromatin regions (Fig. 4H).
Since Fam172a is devoid of any predicted DNA- or RNA-binding domain, its ability to target specific transcribed regions is most likely mediated by some of the chromatin- and spliceosome-associated proteins that compose most of the Fam172a interactome (SI Appendix, Fig. S5 and Table S5). In fact, all of our observations point to the chromatin–spliceosome interface as the preferred site of Fam172a’s action in the nucleus, a notion further supported by our findings that the Fam172a-Ago2 interaction is destabilized by DNase/RNase treatments (Fig. 4B and SI Appendix, Fig. S8G). Of note, given the particular enrichment in relevant gene-expression–associated GO terms in our NCC RNA-seq data (SI Appendix, Figs. S5 and S9), Fam172a also appears to regulate transcription and/or splicing of several genes through indirect means (i.e., through regulation of genes encoding other regulatory proteins). For example, the expression levels or splicing of many genes encoding splicing accessory factors of the hnRNP family (Hnrnpa2b1, Hnrnph2, Hnrnph3, Hnrnpk, and Hnrnpul1) are dysregulated in ToupeeTg/Tg NCCs (Datasets S1 and S2). Although we cannot currently exclude the possibility that Fam172a might also regulate transcription and splicing independently of each other, the existence of indirect splicing targets represents a very plausible reason as to why gene expression levels and splicing patterns only partially overlap in our NCC RNA-seq data.
More work will definitely be required to determine the nature of the interactions within the Fam172a interactome (i.e., direct protein–protein or indirect as part of larger complexes) and the role played by the Arb2-like domain in this regard. Determining which part of Fam172a function relies on its intriguing esterase-like serine hydrolase activity as well as identifying its physiological substrate(s) will represent other exciting, and most likely challenging, lines of future research. Like other “metabolic” serine hydrolases, esterases form a structurally heterogeneous group of enzymes for which very few endogenous substrates have been reported (51). To the best of our knowledge, CIB [CCG1/TAFII250-interacting factor B] (52), also named ABHD14B [α/β-hydrolase domain protein 14B] (53), is the only other known example of a nuclear protein with esterase-like serine hydrolase activity (54). Although both CIB (54) and Fam172a (this work) are similarly involved in transcription-related processes, their respective physiological substrates are expected to be different. Indeed, the sequences flanking the nucleophilic serine in CIB (S-X-S-X-S) differ substantially from the consensus observed in Fam172a (G-X-S-X-G). Moreover, we failed to detect in vitro cleavage of p-nitrophenyl butyrate by purified MBP-tagged Fam172a (not shown) whereas this general hydrolase substrate was reported cleavable by CIB (54).
Is CHARGE Syndrome a Spliceosomopathy?
While the term “neurocristopathy” is appropriate to highlight NCCs as the main cell population affected in CHARGE syndrome, the term “spliceosomopathy” should also be considered to highlight this potential underlying pathogenic mechanism. Defined by the presence of germline mutation of spliceosome-associated proteins, spliceosomopathies recently emerged as a subgroup of rare diseases that notably include neurodegenerative conditions such as retinitis pigmentosa, spinal muscle atrophy, and amyotrophic lateral sclerosis as well as multiple craniofacial disorders such as Nager syndrome, cerebrocostomandibular syndrome, Richieri–Costa–Pereira syndrome, Burn–McKeown syndrome, and the Guion–Almeida type of mandibulofacial dysostosis (55, 56). As observed in CHARGE syndrome, most spliceosomopathies are believed to be caused by de novo dominant mutations although familial cases as well as examples with autosomal recessive inheritance have also been reported (55). Moreover, in accordance with all being considered neurocristopathies as well (57, 58), the phenotypic presentation of the craniofacial disorders mentioned above can be very similar to CHARGE syndrome. One especially striking example is the Guion–Almeida type of mandibulofacial dysostosis, which is caused by mutation of the spliceosomal GTPase-encoding gene EFTUD2 (59). Like CHARGE syndrome, clinical diagnosis of the Guion–Almeida type of mandibulofacial dysostosis may be difficult to establish due to variable phenotypic presentation characterized by different combinations of both major (external and inner ear anomalies, choanal atresia/cleft palate) and minor (facial asymmetry, micrognathia, intellectual disability, growth retardation, heart defects, esophageal atresia, genitourinary defects) features of CHARGE syndrome (55). In line with such an extensive overlap between CHARGE syndrome and the Guion–Almeida type of mandibulofacial dysostosis, EFTUD2 mutations have been reported in seven patients initially referred for possible CHARGE syndrome (60). Of note, the wide range of possible differential diagnosis for craniofacial spliceosomopathies is also highlighted by the fact that EFTUD2 mutations have been reported in patients initially diagnosed for Nager (61) and Feingold (60) syndromes.
Dysregulation of alternative splicing as a common pathogenic mechanism for CHARGE syndrome is in agreement with previous findings regarding the contribution of activated p53. Indeed, while p53 expression and activity have been shown to be increased upon CHD7 deficiency in both mouse and human cells, p53 heterozygosity only partially rescues the malformations found in Chd7-null mouse embryos (13). Moreover, p53 knockdown completely failed to rescue chd7 loss in zebrafish (62) while p53 protein levels, localization and activity—as deduced from the unaffected expression levels of its CHARGE-associated downstream targets Noxa, Perp, and Dr5 (13)—do not appear to be affected in ToupeeTg/Tg embryos (Dataset S1 and SI Appendix, Fig. S14). Considering that p53 may be activated upon splicing impairment (63), these observations thus suggest that p53 activation might contribute partially to the pathogenic cascade downstream of dysregulated splicing and only in a context-dependent manner (i.e., in some CHD7 mutation-positive cases). Importantly, our findings are also in agreement with previous studies showing that knockdown of kdm2b can partially rescue chd7 loss in zebrafish (62). Indeed, given the prominent role for H3K36 methylation in promoting alternative splicing (15, 64, 65), it is reasonable to think that deficiency of a H3K36 demethylase such as Kdm2b could help compensate for reduced splicing efficiency. Along these lines, it is further noteworthy that the Drosophila ortholog of Chd7 (Kismet) has been reported to promote H3K36 methylation by Ash1 (66), the Drosophila ortholog of Ash1l that we found to be part of the Fam172a interactome (SI Appendix, Table S5).
Like other genes coding for spliceosome-associated proteins, Fam172a appears almost ubiquitously expressed in the developing embryo (Fig. 3C and SI Appendix, Fig. S7 A and B). Alternative splicing is a very pervasive process (67), raising the question as to why mutation of spliceosome genes results in tissue-specific malformations. One possibility would be that some cell types are more vulnerable to splicing defects than others, as suggested for retinal neurons in the context of retinitis pigmentosa (68). An explanation for such differential vulnerability could be that, although spliceosome-associated proteins are ubiquitous, their relative proportion may fluctuate as a function of tissue-specific variations in expression levels. These differences can even be functionally amplified through competition between pre-mRNAs that have different affinities for limiting splicing factors (49). In this context, the phenotypic outcome would also depend on compensatory and/or antagonistic activity of available splicing regulatory proteins on their cognate cis-acting RNA elements (69). Another important aspect to consider is that many genes are believed to exert tissue-specific functions through tissue-specific transcript isoforms, which may also depend on the tissue-specific activity of alternative enhancers, silencers, and promoters (67). Considering all of the above, it is not surprising that deficiency of a given splicing regulatory protein can lead to tissue-specific perturbations of a large repertoire of transcripts, as reported for SMN mutation-dependent cases of spinal muscular atrophy (70). In conclusion, regardless of the exact mechanism involved, NCCs appear as vulnerable as retinal and motor neurons to splicing defects.
Toward the Development of Small-Molecule–Based Treatment Strategies for CHARGE Syndrome.
In the current study, we demonstrated that an acute rapamycin treatment is sufficient to correct alternative splicing defects in LCLs from CHARGE patients and partially rescue coloboma in ToupeeTg/Tg embryos, thereby providing hope that small-molecule–based strategies can be envisaged for CHARGE syndrome and other spliceosomopathies. While we recognize that the specific therapeutic value of rapamycin may depend on the relative weight of transcription versus splicing defects in the pathogenesis of CHARGE syndrome (which is currently impossible to determine), it is important to bear in mind that splicing defects are sufficient by themselves to cause neurocristopathy-related malformations (55). Therefore, the correction of splicing defects in the context of CHARGE syndrome can only be advantageous. As highlighted by our first in utero attempt (Fig. 5E and SI Appendix, Fig. S13C), additional work will clearly be required to determine to what extent rapamycin treatments can be effective and safe at the organismal level. In theory, both in utero and early postnatal treatments are possible (71, 72) if care is taken to not administer teratogenic doses (73). Feasibility of in utero treatments is notably supported by ex vivo studies with chicken embryos, which revealed that a 48-h exposure to a low dose of rapamycin (200 nM) is well-tolerated by WT NCCs (72). Since prenatal diagnosis of CHARGE syndrome is particularly difficult to establish (74), it is also very interesting to note that an early postnatal treatment with rapamycin can correct NCC-related craniofacial bone defects in mice (71). Apart from issues related to dosage, frequency, and duration of rapamycin treatments, other outstanding questions include: What is the global impact of rapamycin treatment on the NCC transcriptome? Do acute treatments have long-lasting effects? Can all or only a subset of CHARGE syndrome-related malformations be prevented/corrected? Is it possible to correct some these malformations/dysfunctions postnatally? Both Toupee and Chd7 mutant mouse lines will be especially useful in answering these very important questions.
Materials and Methods
Animals.
Experiments involving mice were performed following the biomedical research guidelines of the Canadian Council of Animal Care. Further details about animal ethics approval could be found in SI Appendix, SI Methods. The Toupee transgenic mouse line was generated by standard pronuclear injection of a previously described tyrosinase minigene in FVB/N zygotes (26), while the Chd7Gt/+ transgenic mouse line has been described previously (30). Details about epistasis studies can be found in SI Appendix, SI Methods. For some studies in embryos, the Toupee allele was backcrossed on the Gata4p[5kb]-RFP transgenic background (FVB/N) in which the DsRed2 fluorescent marker is expressed by NCCs (35). Embryos were all generated by natural mating and staged by considering noon of the day of vaginal plug detection as E0.5. Except for studies involving time-lapse imaging or FACS, control and mutant embryos to be compared were obtained from the same litter and processed in parallel. Olfaction tests were performed in accordance with a previously described protocol (32) on a total of 20 adult mice (10 males and 10 females; aged between 8 and 12 wk) per genotype (control FVB/N vs. ToupeeTg/Tg). Further details about olfaction tests can be found in SI Appendix, SI Methods.
Analysis of Prenatal and Postnatal Tissues.
H&E staining of 10-µm paraffin-embedded tissue sections, immunofluorescence staining of 30-µm cryosections or dissociated cells, neurofilament immunohistochemistry of whole embryos, alizarin red-alcian blue staining of skeletons, and microcomputed tomography analysis of cranial bones were performed as previously described (25, 26, 75, 76). Details about all antibodies used in this study, as well as image acquisition, can be found in SI Appendix, Table S10, and SI Methods, respectively.
Ex Vivo Time-Lapse Imaging of NCCs.
Live imaging of RFP-labeled NCCs was adapted from a previously described suspension culture technique (27). Migration speed and persistence were calculated for 10 individual cells from at least four embryos per genotype. See SI Appendix, SI Methods for further details.
FACS and RNA Extraction.
FACS-mediated recovery of RFP-labeled NCCs was adapted from previously described protocols (27) as described in SI Appendix, SI Methods. RNA extraction from FACS-recovered cells or whole embryonic tissues was performed using the RNeasy Plus Purification Kit (Qiagen) in accordance with the manufacturer’s protocol.
Genotyping PCR and RT-qPCR.
The Toupee allele of Fam172a and the gene-trapped allele of Chd7 were genotyped by PCR using standard Taq DNA polymerase (Feldan) and primers flanking the respective insertion/deletion. Chromosomal sexing was performed by PCR amplification of the male-specific Zfy (423 bp) and the Smcx/Smcy paralogous gene pair, the latter generating a single 300-bp amplicon in XX genomes that is combined with a second 280-bp amplicon in XY genomes due to different intronic lengths. RT-qPCR analyses were performed on 50 ng of total RNA using the Ssofast EvaGreen Supermix and C1000 Touch thermal cycler (BioRad) in accordance with the manufacturer’s protocol. The Gapdh gene was used for normalization of absolute expression levels while constant exons of selected genes were used for normalization of splicing events. Details about the primers used for RT-qPCR can be found in SI Appendix, Table S11, while all other primers are listed in SI Appendix, Table S12.
High-Throughput Genome and Transcriptome Sequencing.
Whole-genome and transcriptome sequencing was adapted from previously described protocols (27) as described in SI Appendix, SI Methods. Differential analysis of transcript levels was performed using the DESeq and edgeR packages whereas differential analysis of alternative splicing was performed using rMATS. Previously published RNA-seq data of P7 granule neuron progenitors [WT, Chd7-het and Chd7-null; single-end 50-bp reads, about 20 million reads per sample (18)] were retrieved from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with accession nos. GSM1857543–GSM1857549. P values were corrected via the Benjamini–Hochberg method. The GO analyses were performed using GOToolBox (genome.crg.es/GOToolBox/) and REVIGO (revigo.irb.hr/).
Plasmid Constructs and Mutagenesis.
The complete 1,251-bp Fam172a ORF (Ensembl transcript ID: ENSMUST00000163257.7) was amplified by RT-PCR from an E12.5 FVB/N embryo head using the SuperScript II Reverse Transcriptase (ThermoFisher Scientific) and the Platinum Taq DNA polymerase (ThermoFisher Scientific) in accordance with the manufacturer’s instructions. Following cloning in the pGEM-T vector (Promega) and validation by Sanger sequencing, the Fam172a ORF was subcloned into the pIRES2-EGFP mammalian expression vector (Clontech; native or modified in house to include an N-term Myc tag) as well as into the pMAL-c5X vector (New England Biolabs). Site-directed mutagenesis of the Arb2 domain (p.Glu229Gln and p.Arg307*) and the serine hydrolase motif (p.Ser294Ala) was performed in relevant Fam172a expression vectors using a previously described PCR-based approach (77). Details about the primers used for cloning and mutagenesis of Fam172a can be found in SI Appendix, Table S12. The psi-CHECK2-let-7 × 8 vector was as previously described (78).
Cell Culture and Transfection.
Propagation of Neuro2a (N2a) and COS7 cell lines as well as cell transfection using Genejuice reagents (Novagen) were performed as previously described (79). For primary culture of E10.5 embryonic cells, details can be found in SI Appendix, SI Methods. For colocalization studies, dissociated cells from FVB/N embryos were analyzed by immunofluorescence after 16 h of culture. For rescue of the Toupee proliferation defect, dissociated cells from ToupeeTg/Tg embryos were transfected with 1 µg of MycFam172a-IRES-EGFP or empty IRES-EGFP expression vector 2 h after plating and analyzed via immunofluorescence after another 48 h of culture.
Co-IP, RIP, ChIP, and RNA-ChIP.
Each IP-based assay in Neuro2a or COS7 cells was performed using a confluent 100-mm plate previously transfected with either Fam172a- or MycFam172a-IRES-EGFP expression vector. For co-IP assays in cell lines and mouse tissues, the preparation of cell extracts and Western blotting were performed using previously described protocols (80), as detailed in SI Appendix, SI Methods. The contribution of nucleic acids was verified by pretreatment of protein extracts with 100 µg/mL DNase I (Sigma) and/or 100 µg/mL RNase A (Sigma) during 2 h at 4 °C. Untargeted ChIP and RIP experiments were both adapted from previously described protocols (81, 82), as described in SI Appendix, SI Methods. Targeted ChiP and RNA-ChIP experiments of Cd44 variable regions were performed on 3 × 106 Neuro2A cells using a previously described protocol (21), as described in SI Appendix, SI Methods.
MBP Fusion Proteins and Serine Hydrolase Assay.
Details about MBP, MBPFam172a, and MBPFam172aS294A protein production as well as protein purification can be found in SI Appendix, SI Methods. Serine hydrolase activity was evaluated using the ActivX TAMRA-FP serine hydrolase probe (ThermoFisher Scientific) in accordance with the manufacturer’s instructions (SI Appendix, SI Methods). TAMRA fluorescence was directly detected in the gel using the NightOWL LB983 imaging system (Berthold), while loaded proteins were detected via standard silver nitrate staining.
Affinity Purification Coupled to Tandem Mass Spectrometry Analysis.
Neuro2a cell extracts were fractionated as previously described (83). Samples were air-dried and sent to the Proteomics Discovery Platform of the Institut de Recherches Cliniques de Montréal. Details about protein inclusion criteria can be found in SI Appendix, SI Methods. Peptides were identified using the Mascot 2.5.1 search engine (Matrix Science) and the UniProt_Mus_Musculus_txid10090 database. Results were analyzed using the Scaffold 4 software (Proteome Software).
Human Studies.
Families provided informed consent on studies approved by the respective institutional review board of the Baylor College of Medicine (experimental cohort for this study) and the University of Michigan Medical School (replication cohort for this study). Exome sequencing and analysis was done as previously described (84). Chromosomic sex was determined from sequencing results according to the presence of the SRY gene. Lymphoblastoid cell lines were established (by Epstein–Barr virus infection) and maintained as described previously (85). For each RT-qPCR experiment, about 3 × 105 cells were used per cell line (details about primers used can be found in SI Appendix, Table S11). For alternative splicing rescue experiments, cells were treated with either 10 μM rapamycin (Sigma) or vehicle only (ethanol) for 30 min just before being processed for RT-qPCR.
Statistics.
Where applicable, data are presented as the mean ± SEM with the number of independent biological replicates (n) indicated in the figure and/or legend. GraphPad Prism software version 6.0 was used to determine the significance of differences via the two-tailed Student’s t test. Differences were considered statistically significant when P values were less than 0.05.
Supplementary Material
Acknowledgments
We thank D. Raiwet (University of Montreal) for the phenotypic screening of the Toupee line; O. Souchkova [University of Quebec at Montreal (UQAM)] for help with mouse husbandry and breeding; D. Flipo (UQAM) for assistance with confocal imaging and FACS analyses; R. Moreau’s laboratory (UQAM) for help with microcomputed tomography; Dr. M. Simard for kindly providing the psi-CHECK2-let-7x8 vector; Dr. J. W. Belmont and P. Hernandez (Baylor College of Medicine) for generously providing the lymphoblastoid cell lines; the Proteomics Discovery Platform of the Institut de Recherches Cliniques de Montréal for the mass spectrometry analyses; and the Massively Parallel Sequencing Platform and the Bioinformatics Platform of McGill University and Génome Québec Innovation Center for the deep sequencing and the analyses of sequencing results, respectively. This work was funded by grants from the CHARGE Syndrome Foundation and the Canadian Institutes of Health Research Grant 376482 (to N.P.). C.B. is also supported by a doctoral scholarship from the Fondation du Grand Défi Pierre Lavoie. N.P. is a Senior Research Scholar of the Fonds de la Recherche du Québec–Santé as well as the recipient of the UQAM Research Chair on Rare Genetic Diseases. This work was also funded by NIH Grants R01NS097862 (to S.B.), R01 DC009410 (to D.M.M.), and R01 DC014456 (to D.M.M.); Donita B. Sullivan, MD Research Professorship in Pediatrics and Communicable Diseases (to D.M.M.); and NIH Training Grant Michigan Predoctoral Training in Genetics (T32GM007544 to A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715378115/-/DCSupplemental.
References
- 1.Hsu P, et al. CHARGE syndrome: A review. J Paediatr Child Health. 2014;50:504–511. doi: 10.1111/jpc.12497. [DOI] [PubMed] [Google Scholar]
- 2.Hale CL, Niederriter AN, Green GE, Martin DM. Response to correspondence to Hale et al. atypical phenotypes associated with pathogenic CHD7 variants and a proposal for broadening CHARGE syndrome clinical diagnostic criteria. Am J Med Genet A. 2016;170:3367–3368. doi: 10.1002/ajmg.a.37629. [DOI] [PubMed] [Google Scholar]
- 3.van Ravenswaaij-Arts CM, Blake K, Hoefsloot L, Verloes A. Clinical utility gene card for: CHARGE syndrome—Update 2015. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2011.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delahaye A, et al. Familial CHARGE syndrome because of CHD7 mutation: Clinical intra- and interfamilial variability. Clin Genet. 2007;72:112–121. doi: 10.1111/j.1399-0004.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- 5.Jongmans MC, et al. Familial CHARGE syndrome and the CHD7 gene: A recurrent missense mutation, intrafamilial recurrence and variability. Am J Med Genet A. 2008;146A:43–50. doi: 10.1002/ajmg.a.31921. [DOI] [PubMed] [Google Scholar]
- 6.Lalani SR, et al. Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zentner GE, Layman WS, Martin DM, Scacheri PC. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A. 2010;152A:674–686. doi: 10.1002/ajmg.a.33323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 9.Bajpai R, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouazoune K, Kingston RE. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci USA. 2012;109:19238–19243. doi: 10.1073/pnas.1213825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita K, Ogawa R, Ito K. CHD7, Oct3/4, Sox2, and Nanog control FoxD3 expression during mouse neural crest-derived stem cell formation. FEBS J. 2016;283:3791–3806. doi: 10.1111/febs.13843. [DOI] [PubMed] [Google Scholar]
- 12.Schulz Y, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet. 2014;133:997–1009. doi: 10.1007/s00439-014-1444-2. [DOI] [PubMed] [Google Scholar]
- 13.Van Nostrand JL, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014;514:228–232. doi: 10.1038/nature13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allemand E, et al. A broad set of chromatin factors influences splicing. PLoS Genet. 2016;12:e1006318. doi: 10.1371/journal.pgen.1006318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saint-André V, Batsché E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 17.Sims RJ, III, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng W, et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun. 2017;8:14758. doi: 10.1038/ncomms14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matveeva E, et al. Involvement of PARP1 in the regulation of alternative splicing. Cell Discov. 2016;2:15046. doi: 10.1038/celldisc.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alló M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 21.Ameyar-Zazoua M, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 22.Kalantari R, Chiang CM, Corey DR. Regulation of mammalian transcription and splicing by nuclear RNAi. Nucleic Acids Res. 2016;44:524–537. doi: 10.1093/nar/gkv1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carissimi C, et al. ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human transcription start sites. Nucleic Acids Res. 2015;43:1498–1512. doi: 10.1093/nar/gku1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilon N. Pigmentation-based insertional mutagenesis is a simple and potent screening approach for identifying neurocristopathy-associated genes in mice. Rare Dis. 2016;4:e1156287. doi: 10.1080/21675511.2016.1156287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergeron KF, et al. Male-biased aganglionic megacolon in the TashT mouse line due to perturbation of silencer elements in a large gene desert of chromosome 10. PLoS Genet. 2015;11:e1005093. doi: 10.1371/journal.pgen.1005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergeron KF, et al. Upregulation of the Nr2f1-A830082K12Rik gene pair in murine neural crest cells results in a complex phenotype reminiscent of Waardenburg syndrome type 4. Dis Model Mech. 2016;9:1283–1293. doi: 10.1242/dmm.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soret R, et al. Ente-Hirsch Study Group A collagen VI-dependent pathogenic mechanism for Hirschsprung’s disease. J Clin Invest. 2015;125:4483–4496. doi: 10.1172/JCI83178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Methot D, Reudelhuber TL, Silversides DW. Evaluation of tyrosinase minigene co-injection as a marker for genetic manipulations in transgenic mice. Nucleic Acids Res. 1995;23:4551–4556. doi: 10.1093/nar/23.22.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosman EA, et al. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 30.Hurd EA, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 31.Bergman JE, et al. Death in CHARGE syndrome after the neonatal period. Clin Genet. 2010;77:232–240. doi: 10.1111/j.1399-0004.2009.01334.x. [DOI] [PubMed] [Google Scholar]
- 32.Bergman JE, Bosman EA, van Ravenswaaij-Arts CM, Steel KP. Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur J Hum Genet. 2010;18:171–177. doi: 10.1038/ejhg.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet. 2011;20:3138–3150. doi: 10.1093/hmg/ddr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asad Z, et al. Rescue of neural crest-derived phenotypes in a zebrafish CHARGE model by Sox10 downregulation. Hum Mol Genet. 2016;25:3539–3554. doi: 10.1093/hmg/ddw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pilon N, Raiwet D, Viger RS, Silversides DW. Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev Dyn. 2008;237:1133–1143. doi: 10.1002/dvdy.21496. [DOI] [PubMed] [Google Scholar]
- 36.Barlow AJ, Dixon J, Dixon MJ, Trainor PA. Balancing neural crest cell intrinsic processes with those of the microenvironment in Tcof1 haploinsufficient mice enables complete enteric nervous system formation. Hum Mol Genet. 2012;21:1782–1793. doi: 10.1093/hmg/ddr611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergeron KF, Silversides DW, Pilon N. The developmental genetics of Hirschsprung’s disease. Clin Genet. 2013;83:15–22. doi: 10.1111/cge.12032. [DOI] [PubMed] [Google Scholar]
- 38.Lake JI, Heuckeroth RO. Enteric nervous system development: Migration, differentiation, and disease. Am J Physiol Gastrointest Liver Physiol. 2013;305:G1–G24. doi: 10.1152/ajpgi.00452.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simões-Costa M, Bronner ME. Establishing neural crest identity: A gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelms BL, Labosky PA. Transcriptional Control of Neural Crest Development. Morgan and Claypool Publishers; San Rafael, CA: 2010. [PubMed] [Google Scholar]
- 41.Theveneau E, Mayor R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 42.Buker SM, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- 43.Shen S, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balasubramanian R, et al. Functionally compromised CHD7 alleles in patients with isolated GnRH deficiency. Proc Natl Acad Sci USA. 2014;111:17953–17958. doi: 10.1073/pnas.1417438111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartels CF, Scacheri C, White L, Scacheri PC, Bale S. Mutations in the CHD7 gene: The experience of a commercial laboratory. Genet Test Mol Biomarkers. 2010;14:881–891. doi: 10.1089/gtmb.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu C, et al. 2016. Compound heterozygote CHD7 mutations in a female with Kallmann syndrome and CHARGE features. Endocrine Society’s 98th Annual Meeting and Expo (Endocrine Society, Washington, DC), Abstract OR21-25.
- 47.Li J, Kim SG, Blenis J. Rapamycin: One drug, many effects. Cell Metab. 2014;19:373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 49.Munding EM, Shiue L, Katzman S, Donohue JP, Ares M., Jr Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell. 2013;51:338–348. doi: 10.1016/j.molcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bachovchin DA, Cravatt BF. The pharmacological landscape and therapeutic potential of serine hydrolases. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Padmanabhan B, Kuzuhara T, Mizuno H, Horikoshi M. Purification, crystallization and preliminary X-ray crystallographic analysis of human CCG1-interacting factor B. Acta Crystallogr D Biol Crystallogr. 2000;56:1479–1481. doi: 10.1107/s0907444900010957. [DOI] [PubMed] [Google Scholar]
- 53.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padmanabhan B, Kuzuhara T, Adachi N, Horikoshi M. The crystal structure of CCG1/TAF(II)250-interacting factor B (CIB) J Biol Chem. 2004;279:9615–9624. doi: 10.1074/jbc.M312165200. [DOI] [PubMed] [Google Scholar]
- 55.Lehalle D, et al. A review of craniofacial disorders caused by spliceosomal defects. Clin Genet. 2015;88:405–415. doi: 10.1111/cge.12596. [DOI] [PubMed] [Google Scholar]
- 56.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snider TN, Mishina Y. Cranial neural crest cell contribution to craniofacial formation, pathology, and future directions in tissue engineering. Birth Defects Res C Embryo Today. 2014;102:324–332. doi: 10.1002/bdrc.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trainor PA, Andrews BT. Facial dysostoses: Etiology, pathogenesis and management. Am J Med Genet C Semin Med Genet. 2013;163C:283–294. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lines MA, et al. FORGE Canada Consortium Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon CT, et al. EFTUD2 haploinsufficiency leads to syndromic oesophageal atresia. J Med Genet. 2012;49:737–746. doi: 10.1136/jmedgenet-2012-101173. [DOI] [PubMed] [Google Scholar]
- 61.Bernier FP, et al. FORGE Canada Consortium Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am J Hum Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balow SA, et al. Knockdown of fbxl10/kdm2bb rescues chd7 morphant phenotype in a zebrafish model of CHARGE syndrome. Dev Biol. 2013;382:57–69. doi: 10.1016/j.ydbio.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allende-Vega N, et al. p53 is activated in response to disruption of the pre-mRNA splicing machinery. Oncogene. 2013;32:1–14. doi: 10.1038/onc.2012.38. [DOI] [PubMed] [Google Scholar]
- 64.Sorenson MR, et al. Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol. 2016;13:412–426. doi: 10.1080/15476286.2016.1144009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu K, et al. SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 2017;45:92–105. doi: 10.1093/nar/gkw814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dorighi KM, Tamkun JW. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development. 2013;140:4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanackovic G, et al. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang F, et al. Neural crest-specific TSC1 deletion in mice leads to sclerotic craniofacial bone lesion. J Bone Miner Res. 2015;30:1195–1205. doi: 10.1002/jbmr.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang XY, et al. High glucose environment inhibits cranial neural crest survival by activating excessive autophagy in the chick embryo. Sci Rep. 2015;5:18321. doi: 10.1038/srep18321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hentges KE, et al. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc Natl Acad Sci USA. 2001;98:13796–13801. doi: 10.1073/pnas.241184198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colin E, et al. Prenatal diagnosis of CHARGE syndrome by identification of a novel CHD7 mutation in a previously unaffected family. Prenat Diagn. 2012;32:692–694. doi: 10.1002/pd.3876. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez-Ferras O, et al. A direct role for murine Cdx proteins in the trunk neural crest gene regulatory network. Development. 2016;143:1363–1374. doi: 10.1242/dev.132159. [DOI] [PubMed] [Google Scholar]
- 76.Sanchez-Ferras O, Bernas G, Laberge-Perrault E, Pilon N. Induction and dorsal restriction of Paired-box 3 (Pax3) gene expression in the caudal neuroectoderm is mediated by integration of multiple pathways on a short neural crest enhancer. Biochim Biophys Acta. 2014;1839:546–558. doi: 10.1016/j.bbagrm.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 77.Pilon N, et al. Porcine SRY promoter is a target for steroidogenic factor 1. Biol Reprod. 2003;68:1098–1106. doi: 10.1095/biolreprod.102.010884. [DOI] [PubMed] [Google Scholar]
- 78.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanchez-Ferras O, et al. Caudal-related homeobox (Cdx) protein-dependent integration of canonical Wnt signaling on paired-box 3 (Pax3) neural crest enhancer. J Biol Chem. 2012;287:16623–16635. doi: 10.1074/jbc.M112.356394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Béland M, et al. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol Cell Biol. 2004;24:5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bérubé-Simard FA, Prudhomme C, Jeannotte L. YY1 acts as a transcriptional activator of Hoxa5 gene expression in mouse organogenesis. PLoS One. 2014;9:e93989. doi: 10.1371/journal.pone.0093989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 83.Lavallée-Adam M, et al. Discovery of cell compartment specific protein-protein interactions using affinity purification combined with tandem mass spectrometry. J Proteome Res. 2013;12:272–281. doi: 10.1021/pr300778b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campeau PM, et al. The genetic basis of DOORS syndrome: An exome-sequencing study. Lancet Neurol. 2014;13:44–58. doi: 10.1016/S1474-4422(13)70265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ling PD, Huls HM. Isolation and immortalization of lymphocytes. Curr Protoc Mol Biol. 2005;Chapter 28:Unit 28.22. doi: 10.1002/0471142727.mb2802s70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.