Significance
We report that huntingtin mutation affects specific aspects of human neurodevelopment at the level of neuronal progenitor specification and its early commitment, leading to an abnormal cell organization and acquisition of mature neuronal identities in cerebral organoids. We also show that down-regulation of mutant huntingtin or pharmacological inhibition of one of its effectors, ADAM10, successfully rescues neuronal differentiation, suggesting that an early intervention may revert neurodegeneration later in life.
Keywords: Huntington’s disease, neurodevelopment, striatal differentiation, organoids, human iPS lines
Abstract
Increasing evidence suggests that early neurodevelopmental defects in Huntington’s disease (HD) patients could contribute to the later adult neurodegenerative phenotype. Here, by using HD-derived induced pluripotent stem cell lines, we report that early telencephalic induction and late neural identity are affected in cortical and striatal populations. We show that a large CAG expansion causes complete failure of the neuro-ectodermal acquisition, while cells carrying shorter CAGs repeats show gross abnormalities in neural rosette formation as well as disrupted cytoarchitecture in cortical organoids. Gene-expression analysis showed that control organoid overlapped with mature human fetal cortical areas, while HD organoids correlated with the immature ventricular zone/subventricular zone. We also report that defects in neuroectoderm and rosette formation could be rescued by molecular and pharmacological approaches leading to a recovery of striatal identity. These results show that mutant huntingtin precludes normal neuronal fate acquisition and highlights a possible connection between mutant huntingtin and abnormal neural development in HD.
Huntington’s disease (HD) is an inherited neurodegenerative disease caused by an expansion of CAG repeats in the huntingtin gene (HTT). While normal huntingtin (HTT) has a ubiquitous cellular localization and pleiotropic functions in the adult brain, mutant HTT (muHTT) appears to have a more selective pathogenic effect, as symptoms mostly relate to the loss of neurons within the striatum (1) and cortex (2). HTT also plays an essential role during early development in the organization of embryonic tissues, and complete loss-of-function of HTT leads to early embryonic death (3, 4). The fact that mutant HTT can, however, rescue lethality in HTT-KO embryos, indicates that the extraembryonic function of huntingtin does not depend on the length of the CAG tract (5), so that muHTT can rescue for loss of the wild-type form. Thus, different parts of the protein are involved in different functions during early development and late in life. Abnormal brain development has also been linked to the loss of HTT (6) and increasing evidence suggests a neurodevelopmental component in HD (7, 8). However, what remains poorly understood is whether the increased striatal and cortical neuronal vulnerability caused by muHTT is due to specific effects in the adult brain or if it is caused by abnormal development that leads to neurodegeneration in late life.
So far, the main evidence suggesting a neurodevelopmental component in HD is threefold. First, HTT interacts with a number of developmental factors (9, 10) and is implicated in several aspects of neural development, such as epiblast formation (11, 12), neural rosette and neural tube formation (13, 14), as well as neuronal survival and cortical maturation and migration (12, 15, 16). Second, there are predictions that the pathological process develops linearly from birth (17) and, accordingly, measurement of the intracranial volume as a representation of the maximal brain growth obtained during development has shown consistent changes in HD subjects before symptoms arise (18–20). Third, muHTT knockin mice show delayed acquisition of early striatal cytoarchitecture in the developing brain, and display high numbers of cells positive for the SOX2 immature marker (21). Moreover, conditional expression of muHTT from conception and until postnatal day 21 produces the same profiles of neurodegeneration observed in mice expressing muHTT throughout adult life (21). Furthermore, omics studies on neuronal cultures derived from HD human induced pluripotent stem cells (HD-hiPSC) documented developmental alterations in gene expression and highlighted the down-regulation of the key neurogenic factor NeuroD1 (8, 22).
However, these studies did not address which precise steps of development are affected by the mutation, or whether reversal of an early abnormal phenotype has any consequences on more advanced stages of maturation of HD neurons. To test this hypothesis, we evaluated the ability of human iPSCs to generate dorsal cortical and ventral striatal telencephalic identities in the presence of muHTT. We report that muHTT carrying different CAG repeat expansions leads to impaired down-regulation of the pluripotency marker OCT4, combined with lower expression of the neuroectodermal fate determinant PAX6, and defective neural rosette formation. Moreover, muHTT led to defects in cortical and striatal progenitor and neuronal specification, cytoarchitecture, and terminal neuronal maturation, with the HD lines showing an overall immature profile compared with control cultures and human embryonic brain tissue. Importantly, both zinc finger protein (ZFP) repressor directed against muHTT and a pharmacological approach by small molecule efficiently rescued early and late phenotypes in HD lines.
These data show that muHTT affects neuronal differentiation from early neuroectodermal fate acquisition to late striatal and cortical identity. Our data highlight a role for muHTT in neurodevelopment that could lead to the pathological phenotypes seen in HD patients in later life.
Materials and Methods
Human iPSC Cultures.
Nonintegrating HD and control (CTR) iPSC lines were obtained from Mattis et al. (23). Cell lines were generated by reprogramming using the Amaxa Human Dermal Fibroblast Nucleofector Kit to express episomal plasmids, as previously reported (23). The lines/clones used in this study were regularly tested and maintained mycoplasma-free. Cultures tested in this work are reported in Table S1. Karyotype for each cell line/clone was regularly monitored every 3 mo during the passages by Q-banding analyses (ISENET). Cells were maintained in mTeSR1 medium (Voden) and plated on Matrigel (Becton Dickinson). At 80% of confluence, iPSC colonies were mechanically isolated and transferred onto new plates.
Striatal Differentiation.
Human HD and CTR iPSC lines were exposed to striatal differentiation protocol published in Delli Carri et al. (24). All differentiation experiments performed in this study have been conducted using cells from passage 20 to passage 50 (Table S1). Briefly, cells were plated at a density of 0.6 × 105 cells/cm−2 on Matrigel-coated dishes in Matrigel with 10 μM ROCK inhibitor (Y-27632; Sigma). Cultures were expanded for 2 d until 70% confluent. The starting differentiation medium included DMEM/F12 (Thermo Fisher) supplemented with N2 and B27 (Life Technologies), 10 μM SB431542 and 500 nM of LDN (provided by Evotec). Medium was replaced every day. Starting from day in vitro (DIV) 5, 200 ng/mL SHHC-25II (Tocris) and 100 ng/mL DKK1 (Peprotech) were added and maintained for 3 wk. At DIV15 the cell population was detached by Accutase (Millipore) and replated on Matrigel at a density of 2.5 × 104 cells/cm−2. Finally, the cells were differentiated in terminal differentiation medium by adding 30 ng/mL BDNF together with N2 and B27.
Cortical Differentiation.
Cortical neurons were generated exposing cells into cortical differentiation using a three-stage protocol that recapitulates cortical development (25). The in vitro cortical protocol generates cortical progenitor cells from iPSCs within 15 d of neural induction by neural induction medium supplemented with 10 μM SB431542 and 500 nM of LDN (provided by Evotec). Next, a 1:1 mixture of N2 and B27 media was used within 3 wk.
Three-Dimensional Cerebral Organoids.
To generate 3D cortical organoids, adapted Lancaster’s protocols were applied. Confluent cells were detached by Accutase and plated 10 × 103 cells per well of a 96-well plate (Perfecta3D hanging drop HDP1096; 3D Biomatrix) in DMEM/F12 with 20% KSR (KSR medium). At DIV3, spheroids were removed and collected in a 15-mL canonical tube. After being resuspended in KSR medium, spheroids were placed in dish plate on an orbital shaker at 70 rpm. When the embryoid bodies were about 500–600 μm in diameter, we changed the KSR medium with Neural Induction Medium [same as in Lancaster’s protocol (26)]. After 2–7 d the embryoid bodies become brighter on the outside, indicating neuroectodermal differentiation. Next, aggregates were transferred into Matrigel droplets. First the dish are placed into the 37 °C incubator and incubated for about 20 min to allow the Matrigel to polymerize. When Matrigel is solidified, 6 mL of Neuronal Differentiation Medium (same as Lancaster’s) is added in the dish. Half of the neuronal differentiation medium was changed three times a week, keeping the dish on the orbital shaker.
Electrophysiology.
Human iPSC-derived neurons were recordered by whole-cell patch-clamp at DIV30 of differentiation. Whole-cell patch-clamp recordings have been performed at room temperature (23 °C) in voltage- and current-clamp mode by using patch-pipettes (3–5 MΩ) made from borosilicate glass. Acquisition and data analysis were done using Multiclamp 700B amplifier, digitized with Digidata 1322, and analyzed with PCLAMP (Molecular Devices) and Origin (Microcal) software. The series resistance compensation was applied to obtain the maximal response time constants. Current density was calculated by dividing the amplitude of the current peak by membrane capacitance. Action potential discharges and total ionic current have been evoked in bath solution containing: 155 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 3 mM KCl, and 10 mM Hepes/NaOH (pH 7.4). The pipettes were filled with a solution composed of: 128 mM KCl, 10 mM NaCl, 11 mM EGTA, and 10 mM Hepes/KOH (pH 7.4). The patch pipette for the study of the biophysical properties of voltage-gated Na+ currents was filled with 120 mM CsCl, 10 mM NaCl, 20 mM TEA-Cl, 10 mM EGTA, 2 mM MgCl2, 4 mM Mg-ATP, and 10 mM Hepes/CsOH (pH 7.4). The extracellular solution contained: 135 mM NaCl, 1 mM CaCl2, 2 mM MgSO4, 10 mM glucose, 5 mM tetraethylammonium-Cl, and 10 mM Hepes/NaOH (pH 7.4).
Immunocytochemistry.
Cell cultures were fixed in 4% (vol/vol) paraformaldehyde for 15 min at room temperature at different time points analyzed. Subsequently, cells were blocked in 5% normal goat serum (NGS; Vector), 0.5% Triton X-100 diluited in PBS at room temperature. Cells were then incubated overnight at 4 °C with primary antibodies diluted in solution containing 2.5% NGS and 0.25% Triton X-100. Images were acquired with a Leica DMI 6000B microscope (10× and 20× objectives) and analyzed with LAS-AF imaging software and then processed using ImageJ, only to adjust contrast for optimal RGB. Additional images were acquired with confocal microscope (40× objectives) and then counted using Cell Profiler. The following primary antibodies were used: OCT4 (mouse, 1:100; Santa Cruz), PAX6 (rabbit, 1:300; Covance), SOX2 (rabbit 1:200; Millipore), PAX6 (mouse, 1:1,000; DHSB), FLAG (rabbit, 1:500; Sigma), N-cadherin (1:800; Becton Dickinson), PALS1 (rabbit, 1:500; Santa Cruz), FOXG1 (rabbit, 1:500; StemCulture), GSX2 (rabbit, 1:500; Millipore), ASCL1 (mouse, 1:500; Becton Dickinson), ISLT1/2 (mouse, 1:1,000; Hybridoma Bank), NKX2.1/TTF1 (rabbit, 1:200; Abcam), βIII-tubulin (rabbit, 1:1,000; Promega), MAP2a/B (mouse, 1:500; Becton Dickinson), CTIP2 (rat, 1:500; Abcam), DARPP32 (rabbit, 1:200; Abcam), TBR1 (rabbit, 1:1,000; Abcam), TBR2 (rabbit, 1:100; Abcam). Secondary antibodies conjugated to Alexa fluorophores 488 or 568 (Molecular Probe, Life Technologies) were used at a 1:1,000 diluition in PBS at room temperature for 1 h. Nuclei were visualized with Hoechst 33258 (5 μg mL−1; Life Technologies).
Immunohystochemistry.
Organoids were fixed in 4% (vol/vol) paraformaldehyde overnight at 4 °C. They were washed in PBS and transferred in a 30% sucrose solution at 4 °C for 48–72 h. Next, organoids were transferred into Tissue-Tek OCT compound, frozen immediately on dry ice and stored at −80 °C. Next, 15-μm-thick sections were generated using a cryostat. Cryosections were washed with PBS and blocked 10% NGS, 0.3% Triton X-100 in PBS at room temperature. Primary antibodies used were diluited in solution containing 5% NGS and 0.1% Triton X-100: NESTIN (mouse, 1:300; Millipore), Ki67 (rabbit, 1:500; Abcam), BLBP (rabbit, 1:500; Millipore), p-VIMENTIN (mouse, 1:100; Hybridoma Bank), SOX2 (rabbit, 1:200; Millipore), MAP2a/B (mouse, 1:500; Becton Dickinson), TBR1 (rabbit, 1:1,000; Abcam), PAX6 (mouse, 1:1,000; DHSB), TBR2 (rabbit, 1:100; Abcam), CTIP2 (rat, 1:500; Abcam). Secondary antibodies conjugated to Alexa fluorophores 488 or 568 (Molecular Probe, Life Technologies) were used 1:500 diluition in PBS at room temperature mixed with Hoechst 33258 (5 μg mL−1; Life Technologies) to visualize nuclei.
ZFPs Cloning Strategy.
The coding sequence for ZFP-A repressor designed to repress muHTT was cut from the corresponding pVAX plasmid using EcoRI-XbaI. Instead, the ZFPΔDBD sequence was obtained by EcoRI-HindIII digestion from the plasmid pAAV-6P-SEWB-ZFPΔDBD (Evotec) and blunted at the HindIII site. The sequences were then subcloned into the tetracycling-regulated lentiviral plasmid FUW-tetO-MCS (Addgene #84008). Constructs were designed as bicistronic vectors carrying a puromycin resistance gene located downstream of the 3′ region of the ZFP coding sequence separated by a “self-cleaving” T2A peptide (SI Materials and Methods), allowing for enrichment of ZFP-expressing cells upon induction with doxycycline and selection with puromycin. For the cloning of the ZFPs-T2A-puroR sequences into constitutive plasmid under the control of chicken β-actin promoter, both ZFPs-T2A-puroR were cut from the lentiviral constructs using EcoRI-AfeI and ligated into the pCAG-Cre (Addgene #26647) recipient plasmid upon excision of Cre by EcoRI-NotI digestion and 5′ blunting at the NotI site. Additional details are reported in SI Materials and Methods.
Real-Time qPCR.
MRNA was isolated using TRIzol reagent according to the manufacture’s instruction (Life Technologies). Potential contaminating DNA was removed by DNA-free kit (Ambion). Template cDNA was prepared by reverse transcription of 500 μg RNA using iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed using the CFX96 real-Time System (Bio-Rad). All reactions were performed in 15 μL containing 50 ng cDNA and SsoFast EVAGreen Supermix (Bio-Rad). Primer pairs used were reported in SI Materials and Methods.
Western Blot Analysis.
Cell cultures were homogenized in RIPA buffer (Tris⋅HCl pH8 50 mM, NaCl 150 mM, SDS 0.1%, NP401%, Sodium deoxycholate 0.5%) supplemented with PMSF 1 mM, DTT 0.5 mM and protease inhibitor mixture (Sigma). Total protein was quantified using the BCA Kit (Thermo Scientific). Twenty micrograms of protein was loaded per track onto a 7.5% SDS/PAGE gels for OCT4 and NANOG detection. Fifty micrograms of protein was loaded for N-cadherin per track onto a 7.5% SDS/PAGE gel. Fifty micrograms of protein was loaded for MAP2a/b and DARPP32 per track onto a 6% and 10% SDS/PAGE gels, respectively. Protein were transferred onto a nitrocellulose membrane and blocked in TBS-T with 5% nonfat dry milk (Bio-Rad). Nitrocellulose membranes were immunoprobed with OCT3/4 (mouse, 1:1,000; Santa Cruz), OCT4 1–14 aa (rabbit, 1:1,000; Abcam), OCT4 C-end (rabbit, 1:1,000; Abcam); NANOG (rabbit, 1:500; Abcam); MAP2a/B (mouse, 1:500; Becton Dickinson); DARPP32 (rabbit, 1:200; Abcam), N-CAD (mouse, 1:500; Becton Dickinson) at 4 °C overnight. Secondary antibody probing and detection was by use of the ECL Kit (Bio-Rad). α-Tubulin (1:10,000, mouse; Sigma) and GAPDH (1:3,000, mouse; Abcam) were used to normalize. The acquisitions were performed by Chemidoc MP (Bio-Rad) and densitometric analysis was performed by Image Lab software.
Microarray Analysis.
To perform the microarray analysis, the Illumina HumanHT-12 v4 Expression BeadChip platform was used to perform differential expression analysis on DIV45 and DIV105 of differentiation of iPSC Q21n1 and Q109n1. A starting amount of 300 ng of high-quality total RNA was used to generate cDNA and cRNA with the Illumina TotalPrep RNA Amplification Kit (Illumina). The procedure consisted of RT with an oligo(dT) primer bearing a T7 promoter using Array-Script. The obtained cDNA became a template for in vitro transcription with T7 RNA polymerase and biotin UTP, which generated multiple copies of biotinylated cRNA. The purity and concentration of the cRNA were determined using a NanoDrop ND-1000 Spectrometer. Quality cRNA was subsequently hybridized using a direct hybridization array kit (Illumina). Each cRNA sample (1.5 μg) was hybridized overnight using the HumanHT-12v4 BeadChip array in a multiple-step procedure; the chips were washed, dried, and scanned on the BeadArray Reader (Illumina).
Data Processing and Pathway Analysis.
Raw microarray data were generated using BeadStudio v3.0 (Illumina) and background-corrected for P < 0.01. Absolute expression values were log2-transformed and normalized using quantile normalization. Principal component analysis (PCA) was performed using the R package ade4 (27). Transcriptional profiles of CTR and HD organoids at DIV45 and DIV105 were compared with those of the developing human fetal brain using a machine-learning algorithm called CoNTExT that is trained on 1,340 primary tissue samples (28). In particular, we employed the laminar expression data dissected via laser-capture microdissection from the fetal human brain. To identify differentially expressed genes at DIV45, a t test was applied and only genes with P < 0.01 were considered significant. Gene Ontology (GO) analysis of genes with a higher expression in CTR organoids was performed using the ClueGo plugin of Cytoscape. The GO “Biological component” was used to query these genes against the background of all genes expressed in the organoids. Only GO with a P < 0.01 were considered (κ-score threshold = 0.4). The rest of the settings were left as defaults. To calculate the semantic similarity score between GO terms the R package GOSemSim was used (29). The resulting matrix was then subjected to hierarchical clustering to find the most represented GO terms. We confirmed the presence of these GO term clusters with the REVIGO webtool (30). Functional annotation of the genes differentially expressed in both our organoids at DIV45 and in the study performed by Ring et al. (22), was performed using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). Microarray data RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/) under accession no. E-MTAB-5964. More detail is available in SI Materials and Methods.
Statistical Analysis.
One-way ANOVA, Tukey posttest was used for all biological experiments performed on more HD and CTR lines (*P < 0.05, **P < 0.01, ***P < 0.001).
Student t test was used to compare only two groups (#P < 0.05) (Fig. 6 A and B and Figs. S4 B, C, L, and M and S5 C and D).
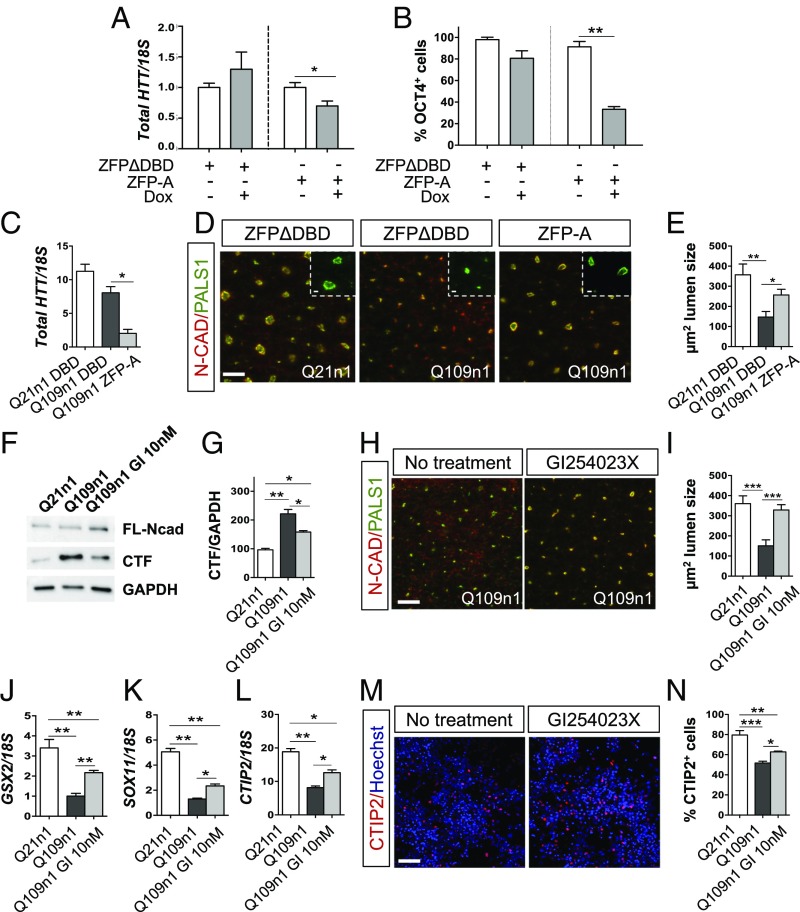
Fig. 6.
HTT down-regulation by ZFP rescues defects in neural induction. (A) qPCR for total HTT mRNA in Q109n5 line after doxy treatment (+Dox) and relative control (−Dox) at DIV5 of differentiation. (A.U., *P < 0.05 Student t test. n = 3 biological experiments, data are represented as mean ± SEM.) (B) Counts of OCT4+ cells by cell profile in ZFP-A and ZFPΔDBD-infected cells in the absence or presence of Dox. (**P < 0.01 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (C) qPCR for total HTT mRNA in ZFP-A and ZFPΔDBD constitutive Q109n1 line at DIV15 of differentiation. (A.U., *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (D) Immunostaining for PALS1/N-cadherin in ZFP-A and ZFPΔDBD constitutive Q109n1 lines and ZFPΔDBD Q21n1 line at DIV15 of differentiation. [Scale bar, 50 μm; Insets (crops of the PALS1 of the same images), 50 μm.] (E) Counts of lumen size in rosettes in ZFP-A and ZFPΔDBD constitutive Q109n1 and Q21n1 (μm2). (*P < 0.05, **P < 0.01 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (F) Western blot for N-CAD in Q109n1 cells treated with 10 nM ADAM10 inhibitor GI254023X with respect to untreated cells and Q21n1 at DIV15 of differentiation. MuHTT leads to increase N-CAD cleavage and affects rosettes formation, a phenotype that is partially rescued by GI254023X treatment. (G) Densitometric analysis of CTF level in Q109n1 cells in the presence or absence of GI254023X and relative to Q21n1 control line. (*P < 0.05, **P < 0.01 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (H) Immunostaining for PALS1/N-CAD in Q109n1 treated with GI254023X relative to untreated control at DIV15 of differentiation. (Scale bar, 50 μm.) (I) Counts of area lumen size in the presence or absence of GI254023X (μm2) (***P < 0.001 Student t test). (J) qPCR for GSX2, (K) SOX11, and (L) CTIP2 transcripts at DIV30 of differentiation in Q109n1 treated with GI254023X relative to untreated control. (*P < 0.05, **P < 0.01 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (M) Confocal images of CTIP2 in Q109n1 without and with GI254023X at DIV30 of differentiation. (Confocal images, scale bar, 50 μm.) (N) Counts of percentage of CTIP2+ cells with cell profile pipeline in Q109n1 without and with GI254023X. (***P < 0.001, *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.)
Results
Large CAG Repeats in Huntingtin Gene Lead to Neuroectodermal Acquisition Defects.
Integration-free HD and CTR iPSC lines were previously generated from fibroblasts of subjects carrying Q21, Q28, Q33, Q60, Q109, and Q180 (respectively with 21, 28, 33, 60, 109, and 180 CAG repeats) (Table S1) (23). Total HTT mRNA was similar among all iPSC lines and clones (Fig. S1A).
With the aim of investigating the role of muHTT in the early phases of neural development, we exposed cells to a stepwise protocol that mimics human ventral telencephalon development (24, 31) (Fig. 1B).
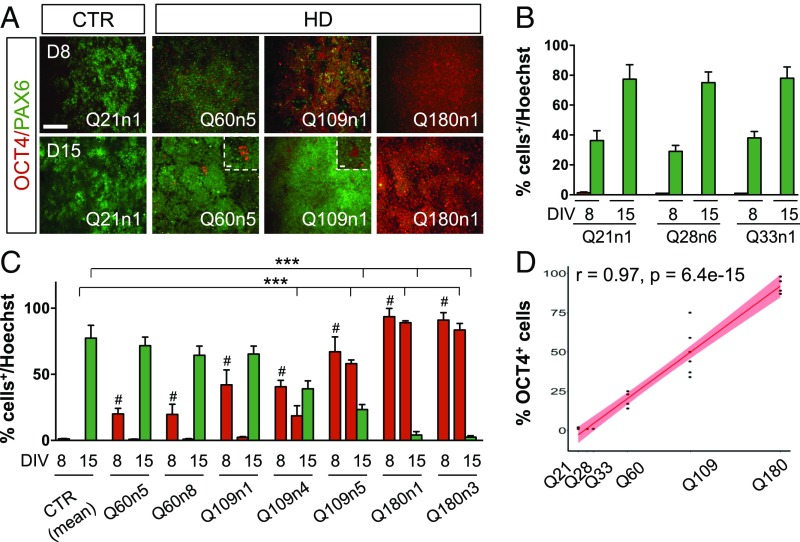
Fig. 1.
Neural induction analysis in HD and CTR iPSCs following striatal differentiation. (A) Immunocytochemistry for OCT4 and PAX6 at DIV8 and 15 of differentiation in Q21n1 and three HD lines (Q60n5, Q109n1, Q180n1). (Scale bar, 100 μm; Inset, 50 μm.) (B) Counts of OCT4+ (red column) and PAX6+ (green column) cells by the Automatic Nuclei Counter plug (ITCN) ImageJ plugin both at DIV8 and 15. (C) Counts of OCT4+ (red column) cells at DIV 8 and 15 and PAX6+ (green column) cells at DIV 15 by ITCN in CTR and HD lines. (One-way ANOVA, OCT4: #P < 0.01 between HD and CTRs at DIV8, one-way ANOVA; OCT4 and PAX6: ***P < 0.001 between HD and CTRs at DIV15, one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (D) Graph of OCT4 and CAG length correlation for all seven HD lines/clones (Q60n5, Q60n8, Q109n1, Q109n4, Q109n5, Q180n1, and Q180n3) (r = 0.97, P = 6.4e-15 calculated using Pearson correlation).
We first compared the neurogenic potential of all CTR and HD-iPSC lines and clones by measuring their transition from pluripotency to neuroectoderm formation to telencephalic specification, as judged by total numbers of OCT4+, PAX6+, and FOXG1+ cells at early stages of differentiation. All iPSC lines were pluripotent, as attested to by OCT4 and SOX2 staining performed at DIV0 (Fig. S1C). Following dual SMAD-inhibition, CTR lines Q21n1, Q28n6, and Q33n1 showed the expected PAX6+ ectodermal acquisition and OCT4 down-regulation at DIV15 (Fig. 1 A and B and Fig. S1D). In contrast, HD lines Q60n5, Q60n8, Q109n1, Q109n4, Q109n5, Q180n1, and Q180n3 showed that OCT4+ cells were still present at the same time point (Fig. 1 A–C and Fig. S1D). Remarkably, at day 8 there was a clear correlation (r = 0.97, P = 6.4e-15) between the number of OCT4+ cells and the number of CAG repeats (Fig. 1D). To further check the validity of the OCT4 immunocytochemistry data, we performed Western blot and qPCR analyses on two or three, respectively, control lines and three HD lines (Q60n5, Q109n5, Q180n1), confirming that HD lines showed defects in OCT4 down-regulation following differentiation (Fig. S1 E and F). Interestingly, and contrary to this finding, NANOG was completely down-regulated upon differentiation in all CTR and HD lines (Fig. S1 G and H). Increased PAX6+ cells and mRNA level were found during neuroectodermal differentiation in all three CTR and in Q60n5 lines, whereas Q109n1 and Q180n1 lines showed only slowest or modest increases during differentiation (Fig. 1 A–C and Fig. S1D). In agreement with this high expression of OCT4 and lower expression of PAX6, Q109n5 and Q180n1 lines, but not the lines with fewer CAG repeats, showed expression of mesodermal markers like α-smooth muscle actin (α-SMA) and α-fetoprotein (AFP) at a later stage in culture (32, 33) (Fig. S1 J–L).
These data indicate that CAG expansion in HTT gene specifically interferes with the down-regulation of the OCT4 pluripotency gene, and with the proper acquisition of a PAX6+ neuroectodermal fate.
Defective Ventral Telencephalic and Striatal Identity Acquisition in HD-Derived Neural Progenitors.
Next, we tested the effect of muHTT at subsequent stages of the developmental program (i.e., in the emergence of neural rosettes and in the steps that precede neuronal specification). We focused on HD lines Q60n5 and Q109n1, because they show loss of pluripotency at day 15 and are therefore expected to progress properly toward differentiation (Fig. 1D). Fig. 2A shows that at DIV15 cultures of Q21n1 CTR were composed of typical N-CADHERIN+ (N-CAD+) large rosettes made of polarized NESTIN+ (NES+) neuroepithelial stem cells, radially aligned around a central lumen, whose mean size is of 216.4 ± 80.7 μm2. In contrast, a significant reduction in lumen size was found in Q60n5 and Q109n1 lines (area of lumen size in square micrometers Q60n5: 80.13 ± 15.8, Q109n1: 87.1 ± 31.8) (Fig. 2 A and B).
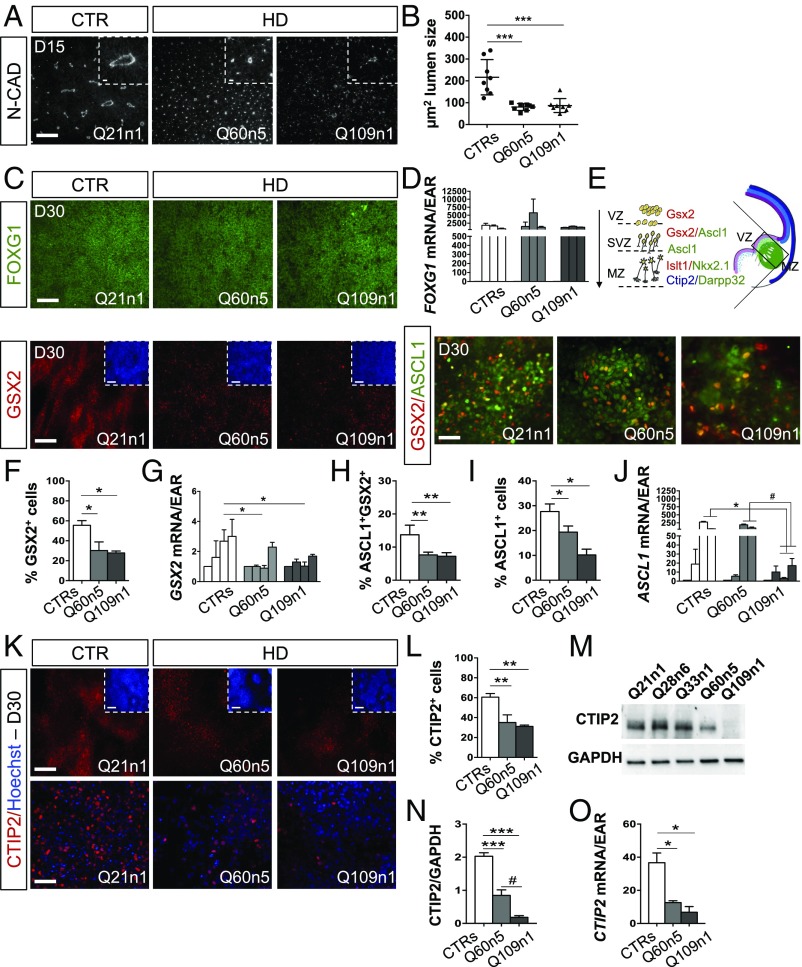
Fig. 2.
HD lines show defects in ventral telencephalic identity acquisition. (A) Immunocytochemistry for N-CAD in Q21n1 and in Q60n5 and Q109n1 HD lines at DIV15 of differentiation. [Scale bar, 100 μm; Inset (the N-CAD staining for each image), 50 μm.] (B) Counts of lumen size’s area (μm2) in rosettes from HD and CTRs lines performed by Cell Profiler. (***P < 0.001, one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (C) Immunocytochemistry for FOXG1, GSX2, and GSX2/ASCL1 in Q21n1, Q60n5, and Q109n1 after neural induction (DIV30). [FOXG1, GSX2 scale bar: 100 μm; Inset (Hoechst staining for each image), 100 μm; GSX2/ASCL1, 50 μm.] (D) qPCR for FOXG1 in CTRs and Q60n5 and Q109n1 lines at DIV0, 8, 15, and 30 (A.U.). (E) Scheme of cells transitioning during development in the striatal anlage. GSX2+ cells found in the VZ migrate in the SVZ while acquiring identity of progenitors GSX2+/ASCL1+. Their complete maturation in striatal neurons is confirmed by colocalization of CTIP2 and DARPP32. (F) Quantification of GSX2+ cells by the ITCN ImageJ plugin. (CTRs = 55.55 ± 11.36%; Q60n5 = 30.22 ± 15.1%; Q109n1 = 27.8 ± 3.52%; *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (G) qPCR GSX2 in CTRs, Q60n5, and Q109n1 lines at DIV0, 8, 15, and 30. (A.U., GSX2, CTRs = 2.69 ± 0.76-fold increase at DIV30 vs. DIV0 vs. 0.9 ± 0.17 and 1.03 ± 0.28 in Q60n5 and Q109n1 fold-increase, respectively; *P < 0.05, one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (H and I) Quantification of ASCL1+/GSX2+ (H) and ASCL1+ (I) cells by the ITCN ImageJ plugin. (ASCL1+/GSX2+: CTRs = 13.7% ± 2.9; Q60n5 = 7.63 ± 0.85; Q109n1 = 7.23 ± 1.08; **P < 0.01 one-way ANOVA; ASCL1+: CTRs = 27.67 ± 3.05%; Q60n5 = 19.3 ± 2.52%; Q109n1 = 9.5 ± 1.80%; *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (J) qPCR for ASCL1 in CTRs and Q60n5 and Q109n1 lines at DIV0, 8, 15, and 30. (A.U., ASCL1 DIV 15: CTRs = 275 ± 32.53-fold induction vs. 186.5 ± 37.47 and 2.93 ± 1.58 in Q60n5 and Q109n1 fold-increase, respectively; DIV30 CTRs = 60.5 ± 4.95 vs. 68.4 ± 4.4 and 17.24 ± 10.98 in Q60n5 and Q109n1 fold-increase, respectively; *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM; #P < 0.05 one way ANOVA.) (K) Immunocytochemistry for CTIP2 at DIV30 of differentiation. [Scale bar, 100 μm; Inset (Hoechst staining for each image), 100 μm.] (Lower) High magnification for CTIP2 staining. (Confocal images, scale bar, 50 μm.) (L) Counts of CTIP2+ postmitotic progenitors at DIV30 of differentiation by ITCN ImageJ plugin. (CTIP2+ cells decreased of 42 ± 5.3% and 45 ± 2.6%, respectively, in Q60 and Q109 lines compared with CTRs, **P < 0.01, one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (M) Western blot for CTIP2 at DIV30 of differentiation. CTIP2 protein level was normalized on GAPDH. (N) Graph represents densitometric analysis performed on Western blot results from three biological differentiation experiments. (CTRs = 2.02 ± 0.32; Q60n5 = 0.85 ± 0.29; Q109n1 = 0.18 ± 0.09, HD vs. CTRs ***P < 0.001 one-way ANOVA; Q60n5 vs. Q109n1 #P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (O) qPCR for CTIP2 in CTRs and Q60n5 and Q109n1 lines at DIV30. (A.U., CTRs = 31.5 ± 11.4; Q60n5 = 12.6 ± 1.08; Q109n1 = 6.75 ± 3.46; *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.)
We then analyzed whether the early defects in neuroectodermal acquisition and rosette formation influence the ability of HD lines to acquire a ventral telencephalic fate, as judged by FOXG1 expression, a transcription factor (TF) required for the development of the mammalian ventral telencephalon (31, 34). At DIV30 of differentiation, no differences in FOXG1 expression were found between CTR and HD lines, regardless of their genotype, indicating that the HD mutation does not interfere with the acquisition of proper telencephalic identity (Fig. 2 C and D).
We then looked at TF expression patterns to distinguish progenitor, immature, and mature neuronal populations coexisting within the human lateral ganglionic eminence (Fig. 2E). To begin with, we analyzed GSX2 expression, a key regional marker that identifies early striatal progenitors (31, 35, 36). Strikingly, we found that the percentage of GSX2+ cells and GSX2 mRNA levels were significantly reduced in HD lines compared with CTR lines (Q60n5: 30.22% ± 15%; Q109n1 27.8% ± 3%, vs. CTR 55.55 ± 11%) (Fig. 2 C, F, and G).
In vivo studies of human fetal development (31) demonstrate that the GSX2+ cells from the ventricular zone (VZ) migrate toward the subventricular zone (SVZ) while acquiring a new positional identity characterized, first, by a double-positive GSX2+/ASCL1+ state, followed by a GSX2−/ASCL1+ state, before entering into the marginal zone (MZ) (Fig. 2E). We detected a 47% and 53% reduction in GSX2+/ASCL1+ cells and a 32% and 64.3% reduction in ASCL1+ cells, respectively, in Q60n5 and in Q109n1 lines compared with CTR (Fig. 2 C, H, and I), as well as lower ASCL1 expression in the Q109n1 line (Fig. 2J).
In the developing human striatum, the postmitotic progenitors migrate into the MZ and acquire CTIP2 expression (Fig. 2E) (31, 37). Interestingly, at DIV30 of differentiation the HD lines showed a marked reduction in the percentage of CTIP2 immunopositive cells (Fig. 2K). Compared with CTRs, Q60n5 and Q109n1 lines had a 25 ± 5.3% and 30 ± 2.6% drop in the percentage of CTIP2+ cells (Fig. 2L). Western blot and qPCR analysis confirmed the significant reduction in CTIP2 protein and mRNA expression in HD lines (Fig. 2 M–O). In addition, immunostaining and qPCR analysis revealed reduced levels of ISLT1 and NKX2.1, two TF involved in ventralization and expressed both in the SVZ and MZ, together with CTIP2 (31) (Fig. S2).
We next evaluated the capacity of the HD cells to complete neuronal maturation and generate striatal neurons. As expected, CTR lines were very efficient in their neuronal differentiation capacity, giving rise to dense clusters of βIII-tubulin+ (TUBB3+) and MAP2+ neurons. Conversely, a decrease in TUBB3+ and MAP2+ immunoreactive neurons was found in HD lines (Fig. 3A). Western blot analysis revealed that the decrease in MAP2a/b isoform levels in HD lines is inversely correlated to the CAGs expansion (Fig. 3 B and C). The presence of the 70-KDa MAP2c in the Q109n1 line, an isoform specifically found early in development, further confirmed the incomplete or delayed maturation of these cells (Fig. 3B). Finally, we analyzed the expression of markers for terminal striatal differentiation, like CTIP2 and DARPP32. At DIV50, coexpression of these two markers was observed both in CTR and, to a minor extent, in HD lines (Fig. 3D). Western blot and qPCR analysis for DARPP32 showed a clear reduction in protein and mRNA levels confirming a defect of the HD lines when undergoing terminal maturation into medium spiny neurons (MSNs) (Fig. 3 D–F and Fig. S3A).
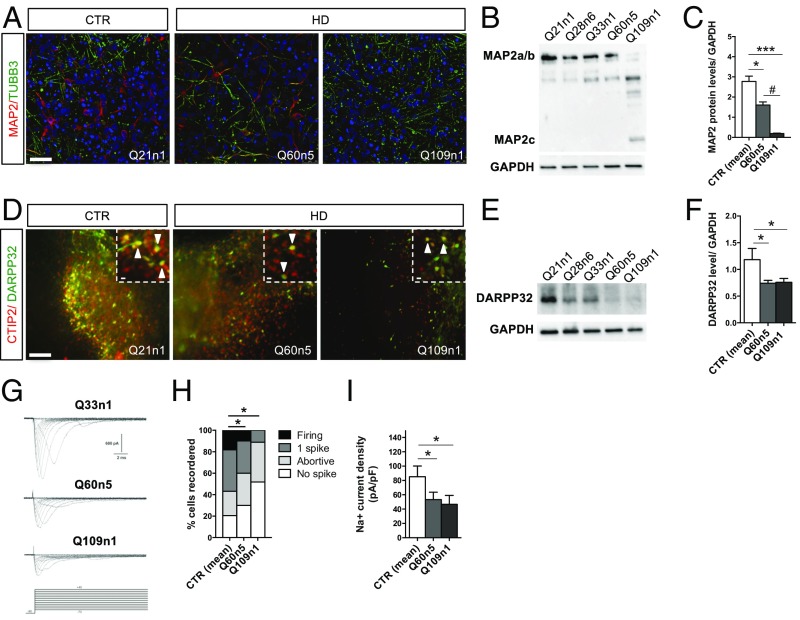
Fig. 3.
Defective striatal terminal differentiation and maturation in HD lines. (A) Immunostaining for TUBB3/MAP2 at DIV30 of differentiation of Q60n5 and Q109n1 and CTR Q21n1. (Confocal images, scale bar, 50 μm.) (B) Representative Western blot for MAP2a/b at DIV30 with corresponding GAPDH levels. (C) Densitometric analysis on Western blot analysis normalized with GAPDH. (CTRs = 2.77 ± 0.74; Q60n5 = 1.61 ± 0.27; Q109n1 = 0.19 ± 0.04; *P < 0.05, ***P < 0.001, #P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (D) Double immunostaining for CTIP2 and DARPP32 at DIV 50 of differentiation. Arrows indicate striatal neurons double positive for CTIP2 (red) and DARPP32 (green). [Scale bar, 100 μm; Insets (crops of the same images), 50 μm.] (E) Representative Western blot for DARPP32 at DIV50 with corresponding GAPDH levels. (F) Densitometric analysis on Western blot analysis. (CTRs = 1.19 ± 0.21; Q60n5 = 0.85 ± 0.29; Q109n1 = 0.18 ± 0.09; *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (G) Families of Na+ current traces evoked by the protocol (Lower traces) from iPSC-derived striatal neurons differentiated in vitro for 30 d. (H) The graph represents the percentage of cells able to generate 1 spike or firing at DIV30 of differentiation. 62% of CTRs, 53% of Q60n5, and only 8% of Q109 when activated displayed single spike or repetitive firing. (*P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.) (I) The Na+ current density recorded at −20 mV from the different iPSC-derived neurons. In the graph, each column represents the average of Na+ current density in recorded cells. (Q21n1 plus Q33n1 n = 78; Q60 n = 27, Q109 n = 17, *P < 0.05 one-way ANOVA; n = 3 biological experiments, data are represented as mean ± SEM.)
Next, we profiled neurons derived from CTR and HD lines for their electrophysiological properties by patch-clamp analysis at DIV30. A total number of 328 CTR cells, 180 Q60n5 cells, and 170 Q109n1 cells were registered for the different electrophysiological parameters. Voltage-gated Na+ inward currents evoked at different membrane potentials showed that 64% (n = 140) of CTR neurons, 51% (n = 73) of Q60n5 and 20% (n = 41) of Q109n1 displayed Na+ current peak larger than 500 pA, an arbitrary threshold defined to select in vitro mature stem cell-derived neurons (24) (Fig. 3G). In parallel, we found that 58% of CTRs, 41% of Q60n5, but only 8% of Q109n1 neurons displayed single spike or repetitive firing upon stimulation (Fig. 3H). Accordingly, the analysis of the Na+ current revealed that its density peak measured at −20 mV is higher in CTRs (89 ± 7 pA/pF; n = 78) than in Q60n5 (61 ± 10; n = 27) and in Q109n1 (51 ± 6; n = 17) (Fig. 3I). Interestingly, qPCR analysis for the levels of sodium channel transcripts SCN1B, SCN2B, SCN3B, and SCN4B, demonstrated a significant decrease in their expression in HD compared with CTRs (Fig. S3 B–E).
Overall, our data suggest that HD lines are defective in the acquisition of a GSX2+, ASCL1+, ISLT1+, NKX2.1+ ventral telencephalic progenitor identity and in striatal fate determination and terminal maturation.
MuHTT Expression Precludes Cortical Specification and Cell Compartmentalization.
To investigate whether muHTT affects also human cortical neuronal specification, we differentiated one control (Q21n1) and one HD line (Q109n1) to cortical projection neuronal cultures (25). Immunostaining and quantification for cortical-specific markers revealed a significant reduction in the percentage of TBR2+ and TBR1+ cells in Q109n1-derived cortical cultures compared with Q21n1 line (Fig. S4 A and B). Moreover, qPCR analysis also showed a significant reduction in PAX6, TBR1, and TBR2 transcript levels in Q109n1 cells (Fig. S4C).
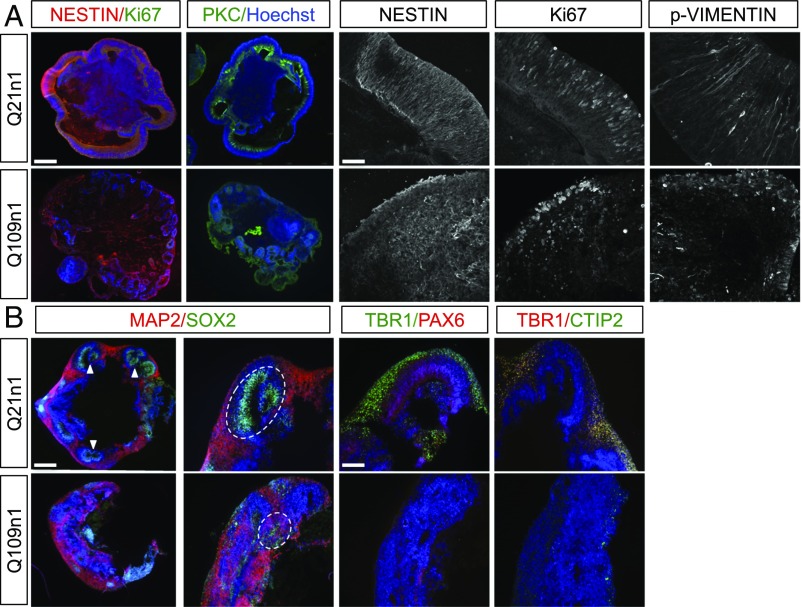
In addition, we exposed two CTR (Q21n1 and Q28n6) and two HD (Q60n5 and Q109n1) lines to a 3D differentiation protocol to obtain human cerebral organoids that more closely resemble in vivo cortical cytoarchitecture (Fig. S4D) (26, 38). We observed that following neuroectodermal differentiation, CTR cells presented a radial NES+ and p-VIM+ neuroepithelium surrounding a fluid-filled cavity reminiscent of an embryonic ventricle, with polarized distribution of Ki67+ cells and αPKC+ staining at DIV60 (Fig. 4A). Strikingly, these structures were completely absent in HD-derived corticoids. Moreover, NES and p-VIM staining showed an altered columnar organization with absence of Ki67+ cell polarity (Fig. 4A). Accordingly, SOX2+ progenitor and MAP2+ neuronal populations clearly localized into two different population compartments in the CTR lines but failed to segregate in the HD lines at DIV85 (Fig. 4B and Fig. S4E). In addition, PAX6, TBR1, and CTIP2 expression and compartmentalization were also disrupted in HD samples (Fig. 4B and Fig. S4E).
Fig. 4.
HD lines show defects in cortical differentiation and cytoarchitecture. (A) Immunocytochemistry for αPKC, NESTIN/Ki67 on organoids derived from Q21n1 and Q109n1. (Scale bar, 100 μm.) These images are representative of five organoids stained for each line generated in two independent experiments. Representative confocal images of organoids stained with NESTIN, Ki67, and p-VIMENTIN. (Scale bar, 25 μm.) (B) Organoids derived from Q21n1 and Q109n1, double-stained for SOX2/MAP2. (Scale bar, 100 μm.) Specific cortical markers PAX6/TBR1 and CTIP2/TBR1. Arrows indicate the structure mimicking the embryonic ventricle in Q21n1 organoids. Dotted ovals show SOX2 and MAP2 organization in Q21n1 and Q109n1. (Scale bar, 50 μm.)
Collectively, these results reveal that muHTT severely affects cortical fate differentiation and proper cell organization in 3D models in vitro.
HD Cerebral Organoids Show an Immature Transcriptional Blueprint.
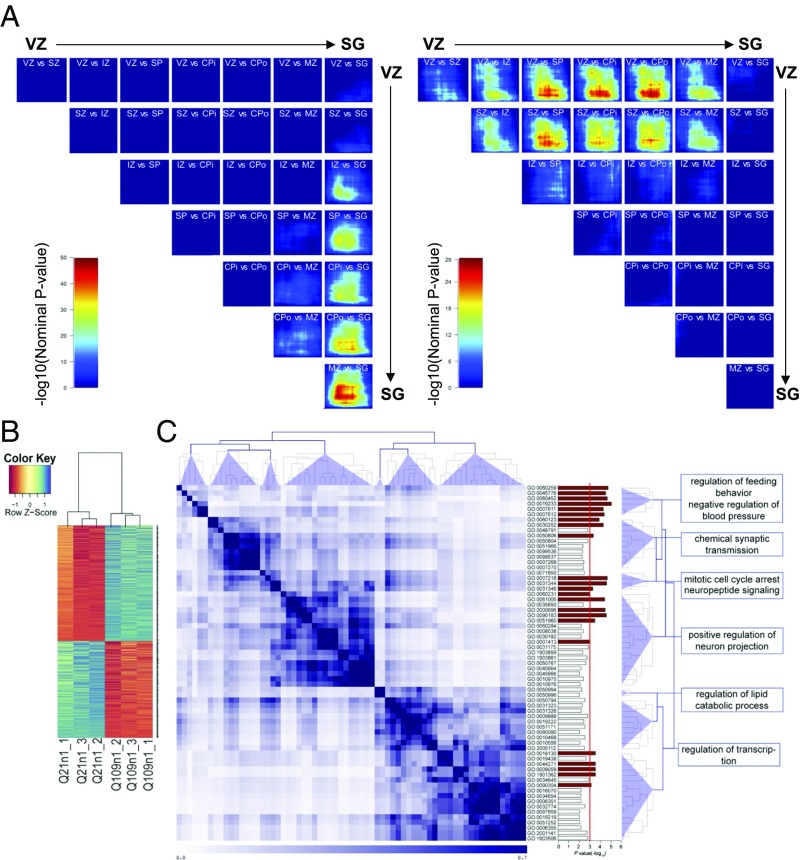
To explain in detail the impairment observed both in cortical specification and in cell organization in HD cultures, we evaluated global transcriptional differences between CTR and HD organoids at DIV45 (n = 3 for each line) and DIV105 DIV (n = 4 for CTR, n = 5 for HD). PCA showed a clear separation of the HD and CTR iPSC lines in the first principal component (PC1) at DIV45 and DIV105 (Fig. S4F). To assess the maturity reached by the CTR and HD organoids at these two time points, we compared their transcriptional profiles to those of the transitioning in vivo laminae of the developing cortex using the machine-learning algorithm called CoNTExT (28). We found that Q21n1 DIV45 and DIV105 samples had the strongest overlap [max-log10(P value) > 50] with the mature suplate zone (SP)/inner cortical plate (CPi)/outer cortical plate (CPo) and with the subpial granular layer (SG), respectively (Fig. 5A, Left). In contrast, Q109n1 samples had the strongest overlap [max-log10(P value) > 29] with the VZ/SVZ at DIV45 and with the SP at DIV105 (Fig. 5A, Right). This result suggests that HD organoids appear to remain “trapped” in the VZ/SP transitions, while CTR organoids mature to more mature entities. In line, immunocytochemistry of the organoids show that HD lines have a lower expression of TBR1 and CTIP2 that are present in the CTR line (Fig. 4B and Fig. S4E). This confirms the differences in maturity reached by the CTR line also at the protein level.
Fig. 5.
Transcriptional dynamics in CTR and HD organoids. (A) Rank-rank hypergeometric overlap maps comparing the transitions between in vivo laminae in the developing cortex to differentiation of CTR (Left) and HD (Right) organoids from DIV45 and 105. Each box shows the color associated with the −log10(P value) multiplied by the sign of either up-regulated (Bottom Left of the box) or down-regulated genes (Top Right of the box) when comparing the differentially expressed genes (DEGs) between each layer of the cortex (e.g., VZ vs. SZ) and DEGs between DIV45 and DIV105. The stronger the intensity of red, the stronger the overlap between in vivo transitions and organoid differentiation. IZ, intermediate zone. (B) Heat map diagram showing the expression levels (z-score) of the differentially expressed genes from the comparison between CTR and HD organoids (t test, P < 0.01, n = 3 different biological replicates in each experimental condition, for each line). (C) Semantic similarity matrix of down-regulated genes in HD organoids. The semantic similarity scores of all GO-term pairs were grouped by hierarchical clustering and representative GO terms are shown on the right and were identified by REVIGO. (GO terms with P < 0.05 are shown. Bar plots in red have P < 0.01.)
Subsequently, we proceeded in analyzing the differences in expression at DIV45 to determine early-stage transcriptional divergences and identified 1,452 differentially expressed genes (P < 0.01, t test) between the two groups: 685 genes had a higher expression in Q21n1 organoids and 767 had a higher expression in the Q109n1 line (Fig. 5B and Dataset S1). To further understand the biological relevance of the genes that are down-regulated in HD and that may explain the phenotype observed, we carried out a GO analysis using the clueGO plugin of Cytoscape (39). We then calculated the semantic similarity scores of all GO-term pairs (P < 0.01) and found a high enrichment for positive regulation of neuron projection development and chemical synaptic transmission (Fig. 5C), suggesting an immature transcriptional signature in HD organoids. We then searched for genes whose expression could be strictly related to muHTT effects. To do so, and to avoid noise due to the different background of the HD-iPSC lines, we compared the genes differentially expressed at DIV45 with the ones determined in another study comparing transcriptional differences between HD isogenic lines at the neural progenitor stage (22). We identified 381 genes differentially expressed between CTR and HD lines in common for both studies (Fig. S4G and Dataset S2) and submitted them to an IPA. Genes with a higher expression in HD organoids were significantly associated (P = 2.17E-07) with the main upstream regulator TGF-β, as shown by Ring et al. (22), whereas low expressed genes were significantly (P < 7.18E-04) associated with nervous system development, especially morphology of the cerebral cortex (Fig. S4H, Left, network with the highest-scored functional pathways; Right, heatmap showing genes enriched for these pathways). SOX11, GAP43, and CELSR3, all transcripts involved in neural migration and differentiation, were also down-regulated in HD cells (Fig. S4 I–K), and in E16.5 cortex from the HD mouse model R6/2 compared with control littermates (Fig. S4 L–N).
Overall, these data indicate that muHTT affects striatal and cortical cell specification and further neuronal maturation.
Allele-Specific Down-Regulation of muHTT Rescues Neural Induction Defects in HD Lines.
We reasoned that specific down-regulation of muHTT at early stages of differentiation might reduce or reverse its adverse effect on neural induction. To confirm this hypothesis, we used a synthetic ZFP (provided by Sangamo) shown to efficiently work as selective repressor of muHTT. To this aim, a doxycycline-inducible ZFP type A (FLAG-tagged) protein was stably expressed in the Q109n5 line, to obtain time-controlled expression of the muHTT repressor. In this assay we used Q109n5 cell lines due to the persistent presence of OCT4+ cells during neural induction (Fig. 1 A–C). As a control, an inducible Q109n5 stable cell line expressing a DNA binding-domain mutant inactive version of the ZFP was also generated (ZFPΔDBD).
ZFP-A and ZFPΔDBD expressing Q109n5 cell lines were exposed to neural induction and analyzed for OCT4 expression at DIV5 (Fig. S5 A and B). Robust induction of ZFP-A and ZFPΔDBD expression upon 5 d of doxycycline treatment was confirmed by qPCR (Fig. S5C). As expected, ZFP-A expression induced a down-regulation of total HTT mRNA that was not observed for control ZFPΔDBD (Fig. 6A). Interestingly, ZFP-A expression as judged by anti-FLAG staining, showed a significant reversion of the incomplete neuroectodermal transition observed for this line, with a 55% drop in the percentage of OCT4+ cells (Fig. 6B and Fig. S5B).
These data show that the defects observed during the pluripotency-to-neuroectodermal transition are directly dependent on expression of muHTT, and that selective down-regulation of muHTT effectively restores normal frequency of OCT4+ cells in vitro.
We then decided to investigate whether the early and continuous down-regulation of muHTT could have an impact at later maturation stages, on neural rosette formation. To achieve a robust and continuous silencing of muHTT, we decided to move the ZFPs into a constitutive plasmid under the control of the chicken β-actin promoter. Q21n1 and Q109n1 stable cell lines with constitutive expression of the ZFPs showed ZFP expression and HTT down-regulation at DIV8 (Fig. 6C and Fig. S5D). Rosette lumen size was evaluated by immunocytochemistry for PALS1 and N-cadherin in Q109n1 and control cultures expressing ZFPΔDBD and ZFP-A (Fig. 6D). Quantification of rosettes stained for PALS1 showed a significant rescue of 52.5% in lumen size in Q109n1 ZFP-A compared with cells expressing control ZFPΔDBD (Fig. 6E).
Altogether, these data demonstrate that transcriptional silencing of muHTT can have a significant impact in early developmental HD phenotypes.
Modulation of muHTT Levels During Early Patterning Rescues Later Striatal Specification Defects.
In parallel with ZFP strategy, we decided to adopt a pharmacological approach by small molecule to inhibit muHTT deleterious effect from neural rosette stage (Fig. 2A) to later stages in culture (DIV30). Our previous studies showed defects in rosette formation in HTT KO mouse ES cells, due to hyperactive metalloprotease ADAM10 and increased N-CAD cleavage, as revealed by an elevated N-CAD C-terminal fragment (CTF) by Western blot (14). We show here that the presence of muHTT in the Q109n1 line at DIV15 of differentiation leads to a similar increase in the CTF fragment (Fig. 6 F and G). The GI254023X compound is a selective inhibitor of ADAM10 activity that is able to restore neural tube defects and N-CAD cleavage in httMO-Zebrafish embryos (14). Importantly, exposure to 10 nM GI254023X from DIV5 to DIV8 of differentiation caused a significant reduction in the level of the CTF fragment, leading to a significant recovery of 54.2% in lumen size in Q109n1 cells (Fig. 6 F–I). To understand if the rescue in this early developmental phenotype influences late maturation of the HD cells at DIV30, we evaluated mRNA levels of general and striatal specific progenitor markers, such as SOX11, GSX2, and CTIP2. qPCR for these markers performed at DIV30 of differentiation showed a 50% increase of SOX11 mRNA and 30% increase in GSX2 and CTIP2 transcript levels upon GI254023X treatment (Fig. 6 J–L). Accordingly, the percentage of CTIP2+ cells was partially recovered in GI254023X treated Q109n1, compared with untreated cells (Fig. 6 M and N), and this is accompanied by an overall better neuronal differentiation as judged by Map2/TUBB3 staining (Fig. S5E).
Taken together, these results demonstrate that muHTT negatively affects striatal and cortical specification, differentiation, and maturation in vitro. This impairment is evident in both 2D monolayer neuronal cultures and 3D organoids showing an abnormal neuroectodermal transition associated with altered cortico/striatal specification, together with disrupted cellular organization. Strikingly, defects in 2D neuronal induction and striatal differentiation are successfully rescued by down-regulation of muHTT or pharmacological inhibition of one of its effectors, ADAM10, from the earliest step of the differentiation.
Discussion
HTT protein contributes to several cellular pathways during embryonic development and adulthood. Wild-type HTT is needed for normal development and its depletion in mouse causes embryonic lethality at E8.5, with severe abnormalities in neuronal development (40, 41). Mounting evidence suggests that muHTT could tamper with neuronal development with repercussions during adulthood that contribute to the HD pathology (42). However, there is no information as to how an early developmental defect may affect neuronal function in adulthood or whether early therapeutic intervention could have a significant disease modifying effect.
By using nonintegrating human HD-iPSC lines, we demonstrated that the pathologic expansion of the CAG tract affects neural cell positional identity, 3D self-organizing capacity, and terminal maturation. Problems in reaching a mature transcriptional profile by HD lines was also shown by the HD iPSC Consortium that revealed that the omics signatures of HD iPSC lines correspond to an earlier stage of mouse development compared with controls (8). In our study, HD lines show reduced efficiency in ventral specification with reduced GSX2, CTIP2, and MAP2 expression compared with controls. Moreover, HD lines manifest a delayed exit from pluripotency (Q60 and Q109 lines) that is further exacerbated in highly expanded CAG repeats (Q180 lines), where cells remained OCT4+ and committed to mesodermal differentiation even after neural induction. This is not surprising, considering that overexpression of PAX6 does not induce meso-endodermal fate while PAX6 knockdown does not inhibit meso-endodermal differentiation (43).
These data also call for a more detailed study on HTT subcellular localization during differentiation and the use of iPSC lines to reveal potential aberrant interactions between specific transcription factors and the mutant protein.
In addition to the striatal component, clear alterations in cortical fate were detected, with reduction of TBR2 and TBR1 levels in HD progenitors compared with controls. Accordingly, we reveal that polyQ-expanded HTT has a role in cell-to-cell interaction and tissue organization during cortical development because defects in neuronal positioning and cell organization were found in 3D organoids generated from HD iPSCs. These results, together with those from Barnat et al. and others showing that HTT removal in postmitotic projection neurons leads to defects in neuronal migration in the mouse neocortex, provide further evidence that HTT loss-of-function mechanisms associated with the pathological expansion can operate during neural development (40–42, 44). Our HD organoids also revealed a transcriptional profile characterized by reduced expression of genes with key roles in neuronal migration and differentiation. Among them, SOX11, GAP43, and CELSR3 were down-regulated also during 2D MSN differentiation, thus suggesting a common effect of muHTT during cortical and striatal differentiation, which contribute to explaining the selective vulnerability observed in the striatum and cortex of HD patients. This hypothesis was further sustained by the down-regulation of these transcripts in the brain of R6/2 embryos.
Importantly, studies of prodromal HD individuals revealed brain changes decades before the onset of the disease, including smaller head circumference (45), caudate and putamen atrophy (46, 47), and striatal and cortical white matter abnormalities (48), including cortical thinning (47).
Our results suggest that both the striatal and cortical signatures are flawed during development, with regards to the response to differentiation cues and ability to self-organize in complex 3D structures. We also show that the progression through subsequent steps of the developmental program is affected in a CAG-dependent manner, with the highest expansion limiting exit from pluripotency, Q109 and Q60, causing a delay in neuroectoderm formation, followed by the inability (greater in the Q109 line) to undergo full commitment to striatal specification.
Furthermore, our data attest that the defects in the transition from pluripotency to neuroectoderm can be reverted by downregulating muHTT, and that interventions aimed at restoring defects in neural rosette formation are able to significantly revert later ventral specification abnormalities.
One major question that arises from these results is whether these developmental defects may be relevant to the in vivo pathology. Smaller intracranial volumes in presymptomatic HD patients have been observed in one study (44); however, this was not replicated in the TRACK-HD cohort (49). Whether the iPSCs models used here correctly mimic what happens in HD still need to be investigated. In general, iPSCs have been widely used to model genetically encoded pathologies in vitro. In some cases, these cells have shown remarkably good recapitulation of disease-specific features. This was the case for Ebert et al. (50), where SMA patient-derived iPSCs differentiated to motor neurons showed survival and drug responses also found in patients. Also, Koch et al. (51) successfully used iPSCs to study Ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. However, in some cases, iPSCs revealed subtle cellular characteristics that were not recapitulated (or not detected) in the patient. For example, long QT syndrome (LQT2) hiPSC-derived cardiomyocytes generated from a mutation carrier with normal QT, showed normal QT interval that was 170% longer than that of controls (52). This phenotype was only observed in the iPS-derived cells, while the patient was asymptomatic. This leads us to conclude that in vitro cell modeling could in some cases exacerbate disease-related conditions that are masked in vivo by unknown compensatory mechanisms. Thus, later symptomatic changes in the HD brain could arise from subtle changes during neurodevelopment that are in some way “camouflaged” or compensated for, in a more complex in vivo environment. However, if these small alterations were present, they may change the regular neuronal homeostasis and in the long run lead to the symptoms observed in HD. To clear how relevant iPSCs are for HD modeling needs further examination.
In conclusion, these results advocate for the theory of HD as a pathology with a disguised neurodevelopmental component, where the polyQ expansion alters the canonical developmental path of striatal and cortical neurons that are therefore more unstable and susceptible to muHTT during adulthood. Given the clinical implication of such findings, conclusive in vivo evidence in humans remains mandatory.
Supplementary Material
Acknowledgments
We thank Sangamo and Evotec for their zinc finger protein vectors. This work was supported by CHDI Foundation Grant A7333; partially by Programmi di Ricerca Scientifica di rilevanza Nazionale (Ministries of Education Universities and Research) Grant 2008JKSHKN_001; and European Commission H2020 projects Neuromics Grant 305121 and NeurostemcellRepair Grant 602278 (to E. Cattaneo).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI, https://www.ebi.ac.uk/arrayexpress/ (accession no. E-MTAB-5964).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715865115/-/DCSupplemental.
References
- 1.Reiner A, et al. Differential loss of striatal projection neurons in Huntington disease. Proc Natl Acad Sci USA. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cudkowicz M, Kowall NW. Degeneration of pyramidal projection neurons in Huntington’s disease cortex. Ann Neurol. 1990;27:200–204. doi: 10.1002/ana.410270217. [DOI] [PubMed] [Google Scholar]
- 3.Duyao MP, et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 4.Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 5.Leavitt BR, et al. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am J Hum Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auerbach W, et al. The HD mutation causes progressive lethal neurological disease in mice expressing reduced levels of huntingtin. Hum Mol Genet. 2001;10:2515–2523. doi: 10.1093/hmg/10.22.2515. [DOI] [PubMed] [Google Scholar]
- 7.Wiatr K, Szlachcic WJ, Trzeciak M, Figlerowicz M, Figiel M. Huntington disease as a neurodevelopmental disorder and early signs of the disease in stem cells. Mol Neurobiol. May 11, 2017 doi: 10.1007/s12035-017-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.HD iPSC Consortium Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci. 2017;20:648–660. doi: 10.1038/nn.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington’s disease. Neurobiol Dis. 2008;31:298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed I, et al. Huntington’s disease: Neural dysfunction linked to inositol polyphosphate multikinase. Proc Natl Acad Sci USA. 2015;112:9751–9756. doi: 10.1073/pnas.1511810112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woda JM, et al. Inactivation of the Huntington’s disease gene (Hdh) impairs anterior streak formation and early patterning of the mouse embryo. BMC Dev Biol. 2005;5:17. doi: 10.1186/1471-213X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen GD, Molero AE, Gokhan S, Mehler MF. Functions of huntingtin in germ layer specification and organogenesis. PLoS One. 2013;8:e72698. doi: 10.1371/journal.pone.0072698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White JK, et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nat Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 14.Lo Sardo V, et al. An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-Ncadherin. Nat Neurosci. 2012;15:713–721. doi: 10.1038/nn.3080. [DOI] [PubMed] [Google Scholar]
- 15.Dragatsis I, Efstratiadis A, Zeitlin S. Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development. 1998;125:1529–1539. doi: 10.1242/dev.125.8.1529. [DOI] [PubMed] [Google Scholar]
- 16.Tong Y, et al. Spatial and temporal requirements for huntingtin (Htt) in neuronal migration and survival during brain development. J Neurosci. 2011;31:14794–14799. doi: 10.1523/JNEUROSCI.2774-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penney JB, Jr, Vonsattel JP, MacDonald ME, Gusella JF, Myers RH. CAG repeat number governs the development rate of pathology in Huntington’s disease. Ann Neurol. 1997;41:689–692. doi: 10.1002/ana.410410521. [DOI] [PubMed] [Google Scholar]
- 18.Aylward EH, et al. PREDICT-HD Investigators and Coordinators of the Huntington Study Group Regional atrophy associated with cognitive and motor function in prodromal Huntington disease. J Huntingtons Dis. 2013;2:477–489. doi: 10.3233/JHD-130076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niccolini F, Politis M. Neuroimaging in Huntington’s disease. World J Radiol. 2014;6:301–312. doi: 10.4329/wjr.v6.i6.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scahill RI, et al. TRACK-HD investigators Clinical impairment in premanifest and early Huntington’s disease is associated with regionally specific atrophy. Hum Brain Mapp. 2013;34:519–529. doi: 10.1002/hbm.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molero AE, et al. Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc Natl Acad Sci USA. 2016;113:5736–5741. doi: 10.1073/pnas.1603871113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ring KL, et al. Genomic analysis reveals disruption of striatal neuronal development and therapeutic targets in human Huntington’s disease neural stem cells. Stem Cell Rep. 2015;5:1023–1038. doi: 10.1016/j.stemcr.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattis VB, et al. HD iPSC-derived neural progenitors accumulate in culture and are susceptible to BDNF withdrawal due to glutamate toxicity. Hum Mol Genet. 2015;24:3257–3271. doi: 10.1093/hmg/ddv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delli Carri A, et al. Developmentally coordinated extrinsic signals drive human pluripotent stem cell differentiation toward authentic DARPP-32+ medium-sized spiny neurons. Development. 2013;140:301–312. doi: 10.1242/dev.084608. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chessel D, Dufour AB, Thioulouse J. The ade4 package - I: One-table methods. R News. 2004;4:5–10. [Google Scholar]
- 28.Stein JL, et al. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 30.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onorati M, et al. Molecular and functional definition of the developing human striatum. Nat Neurosci. 2014;17:1804–1815. doi: 10.1038/nn.3860. [DOI] [PubMed] [Google Scholar]
- 32.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 33.Kashyap V, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manuel M, et al. The transcription factor Foxg1 regulates the competence of telencephalic cells to adopt subpallial fates in mice. Development. 2010;137:487–497. doi: 10.1242/dev.039800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nat R, Dechant G. Milestones of directed differentiation of mouse and human embryonic stem cells into telencephalic neurons based on neural development in vivo. Stem Cells Dev. 2011;20:947–958. doi: 10.1089/scd.2010.0417. [DOI] [PubMed] [Google Scholar]
- 36.Méndez-Gómez HR, Vicario-Abejón C. The homeobox gene Gsx2 regulates the self-renewal and differentiation of neural stem cells and the cell fate of postnatal progenitors. PLoS One. 2012;7:e29799. doi: 10.1371/journal.pone.0029799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bindea G, et al. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuccato C, Cattaneo E. The normal function of Huntingtin. In: Bates G, Tabrizi S, Jones L, editors. Huntington’s Disease. Oxford Univ Press; Oxford, UK: 2014. pp. 243–273. [Google Scholar]
- 41.Saudou F, Humbert S. The biology of huntingtin. Neuron. 2016;89:910–926. doi: 10.1016/j.neuron.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Mehler MF, Gokhan S. Mechanisms underlying neural cell death in neurodegenerative diseases: Alterations of a developmentally-mediated cellular rheostat. Trends Neurosci. 2000;23:599–605. doi: 10.1016/s0166-2236(00)01705-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barnat M, Le Friec J, Benstaali C, Humbert S. Huntingtin-mediated multipolar-bipolar transition of newborn cortical neurons is critical for their postnatal neuronal morphology. Neuron. 2017;93:99–114. doi: 10.1016/j.neuron.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 45.Lee JK, et al. Measures of growth in children at risk for Huntington disease. Neurology. 2012;79:668–674. doi: 10.1212/WNL.0b013e3182648b65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nopoulos PC, et al. PREDICT-HD Investigators and Coordinators of the Huntington Study Group Smaller intracranial volume in prodromal Huntington’s disease: Evidence for abnormal neurodevelopment. Brain. 2011;134:137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nopoulos PC, et al. PREDICT-HD Investigators Coordinators of Huntington Study Group (HSG) Cerebral cortex structure in prodromal Huntington disease. Neurobiol Dis. 2010;40:544–554. doi: 10.1016/j.nbd.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gómez-Tortosa E, et al. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann Neurol. 2001;49:29–34. [PubMed] [Google Scholar]
- 49.Tabrizi SJ, et al. TRACK-HD investigators Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: Cross-sectional analysis of baseline data. Lancet Neurol. 2009;8:791–801. doi: 10.1016/S1474-4422(09)70170-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch P, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 52.Lahti AL, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dull T, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–71. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.