SUMMARY
Rhabdomyosarcoma (RMS) is a pediatric soft tissue sarcoma that histologically resembles embryonic skeletal muscle. RMS occurs throughout the body and an exclusively myogenic origin does not account for RMS occurring in sites devoid of skeletal muscle. We previously described a RMS model activating a conditional constitutively active Smoothened mutant (SmoM2) with aP2-Cre. Using genetic fate mapping, we show SmoM2 expression in Cre expressing endothelial progenitors results in myogenic transdifferentiation and RMS. We illustrate endothelium and skeletal muscle within the head and neck arise from Kdr expressing progenitors and that hedgehog pathway activation results in aberrant expression of myogenic specification factors as a potential mechanism driving rhabdomyosarcomagenesis. These findings suggest that RMS can originate from aberrant development of non-myogenic cells.
Keywords: Rhabdomyosarcoma, skeletal muscle, endothelium, myogenesis, Tbx1, hedgehog, sarcoma
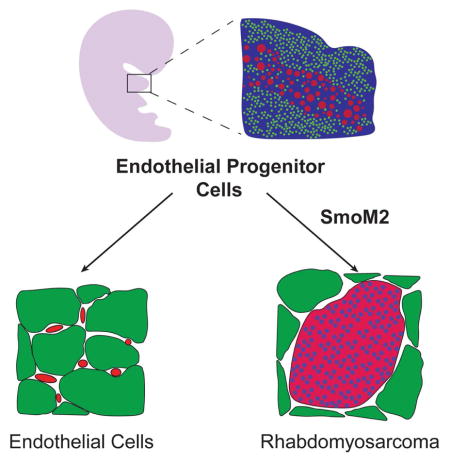
Graphical abstract
Using genetic fate mapping, Drummond et al. show that hedgehog pathway activation in endothelial progenitors results in aberrant expression of myogenic specification factors, myogenic transdifferentiation, and rhabdomyosarcoma (RMS). The finding may explain how RMS develops in sites devoid of skeletal muscle.
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma with an incidence of 4.5 cases per million children (Perez et al., 2011). Intensive clinical trials over the past three decades have not improved survival rates or treatment options for high risk patients (Oberlin et al., 2008). RMS is classified into two major histological subtypes, alveolar RMS (ARMS) or embryonal RMS (ERMS) (Parham and Barr, 2013). ARMS tumors typically (~80%) harbor chromosomal translocations resulting in the expression of a PAX3-FOXO1 or PAX7-FOXO1 fusion protein (Davis et al., 1994; Galili et al., 1993) which foretells a worse prognosis. The molecular features and clinical outcomes of ARMS patients lacking PAX3/PAX7-FOXO1 gene translocations resemble ERMS, thus classifying RMS as either fusion negative (FN-RMS) or fusion positive (FP-RMS) better reflects both the biology and clinical outcomes (Williamson et al., 2010). In addition to tumor histology, pretreatment staging and post-surgical grouping are important prognostic indicators of RMS. Tumor location is a key feature of staging and nearly 40% of all RMS occurs in the head and neck (Sultan et al., 2009). It remains unknown how the cell of origin impacts location and clinical outcome of FN-RMS.
RMS resembles developing skeletal muscle and is hence viewed as an arrested state in normal skeletal muscle development (Kashi et al., 2015). During myogenesis the temporal expression of myogenic regulatory factors (Mrfs) Myogenic Differentiation 1 (MYOD1), MYF5, MRF4 (MYF6) and Myogenin drive differentiation and a terminal cell cycle exit (Buckingham and Rigby, 2014). RMS cells express Mrfs, yet fail to execute terminal muscle differentiation. Thus, RMS is thought to originate in muscle progenitor cells. However, an exclusively myogenic origin of RMS does not account for FN-RMS occurring in sites devoid of skeletal muscle such as the salivary gland, gallbladder, prostate and bladder suggesting additional non-myogenic origins for FN-RMS.
Muscles in the head and neck are derived from the branchial arches and cranial mesoderm and have distinct embryonic origins from somite derived trunk and limb muscles (Michailovici et al., 2015). The specification of head and neck muscle progenitor cells also differs from the somite. In contrast to the trunk and limbs where PAX3 drives Mrf expression, a combination of transcription factors including TBX1, Musculin, TCF21, ISL1, LHX2, and PITX2 act upstream of Mrfs in the head and neck (Buckingham, 2017). It remains unclear how these differing developmental programs contribute to tumorigenesis in RMS.
The Sonic Hedgehog (Shh) pathway is critically involved in tissue morphogenesis including skeletal muscle but not in the muscle of the head and neck (Borycki et al., 1999; Munsterberg et al., 1995). Hedgehog signaling is maintained inactive by the transmembrane receptor Patched1 (PTCH1) binding and repressing Smoothened (SMO). Upon Shh ligand binding PTCH1, SMO is released from inhibition and activates the Gli family of transcription factors inducing downstream target gene expression (Pak and Segal, 2016). Aberrant Shh signaling drives a number of experimental FN-RMS models (Hahn et al., 1998; Hatley et al., 2012; Lee et al., 2007; Mao et al., 2006). Furthermore, active Shh signaling is observed in a high proportion of sporadic FN-RMS with 53% harboring amplification of 12q13.3 containing GLI1, 73% displaying increased GLI1 protein by immunohistochemistry (IHC), and 33% exhibiting loss of chromosome 9q22 containing PTCH1 (Bridge et al., 2000; Paulson et al., 2011; Pressey et al., 2011; Zibat et al., 2010). Hedgehog signaling controls self-renewal of FN-RMS tumor propagating cells and hedgehog pathway inhibition reduces chemotherapy resistance (Satheesha et al., 2016). Together, these studies highlight a role for Shh activation in FN-RMS pathogenesis.
Previously, we described a highly penetrant mouse model of FN-RMS, aP2-Cre;SmoM2/+, driven by conditional expression of constitutively active SmoM2 by Cre recombinase expressed from the adipocyte protein 2 (aP2) gene promoter. The histology and gene expression of aP2-Cre;SmoM2/+ tumors recapitulate both other mouse models and human FN-RMS (Hatley et al., 2012). Interestingly, tumors are anatomically restricted, occurring exclusively in the head and neck. In this study we leverage the aP2-Cre;SmoM2/+ mouse model to interrogate the cellular origins of FN-RMS.
RESULTS
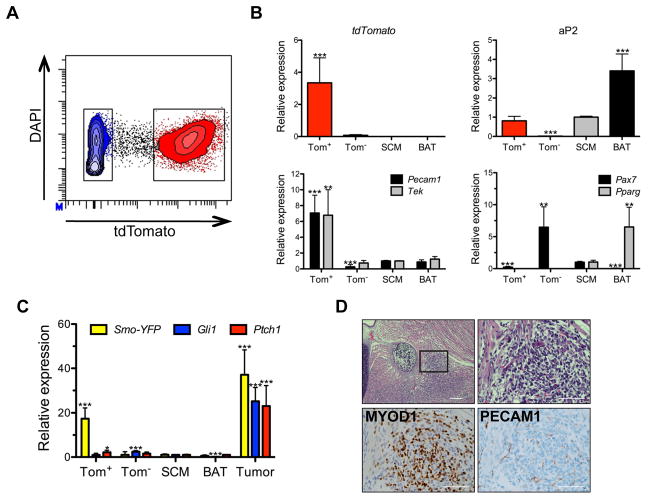
aP2-Cre labels cells within both adipose tissue and skeletal muscle
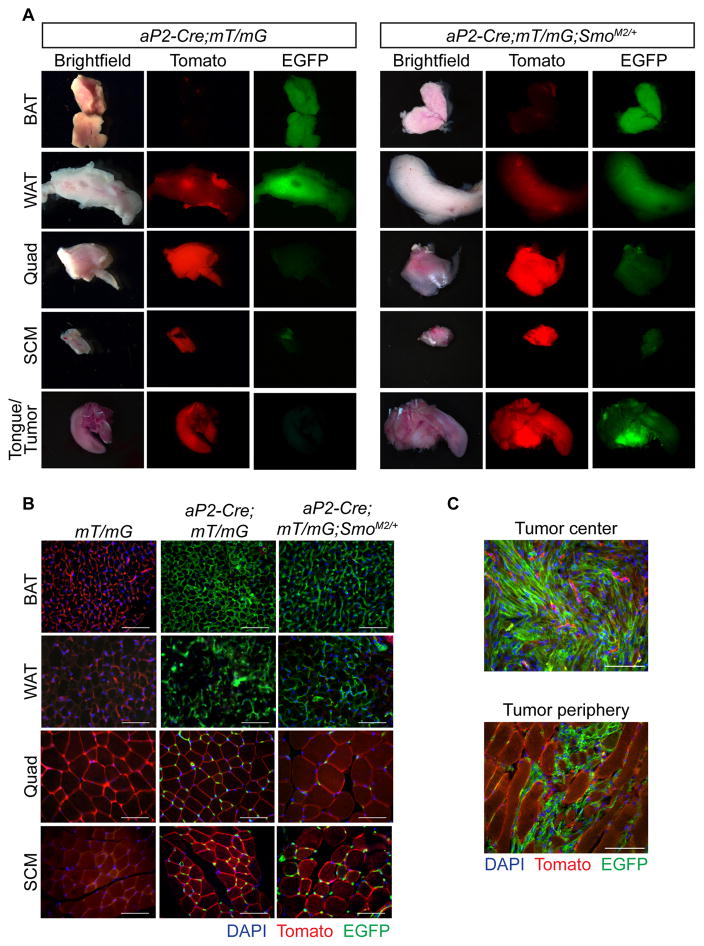
The development of FN-RMS from conditional, oncogenic Smoothened allele, SmoM2, activation by aP2-Cre was surprising. Therefore, we sought to determine the cell of origin of FN-RMS in the aP2-Cre;SmoM2/+ (AS) mouse model. Previously, aP2-Cre (also known as Fabp4-Cre) was thought to be adipose restricted, but recent reports indicate a broader tissue expression (Lee et al., 2013; Tang et al., 2008; Urs et al., 2006). First, we bred Rosa26mT/mG (mT/mG) reporter mice to aP2-Cre mice in the presence and absence of SmoM2 to localize aP2-Cre expression. The mT/mG reporter expresses membrane-targeted Tomato (mT) in all tissues in the absence of Cre recombinase (Figures S1A&B). After breeding to aP2-Cre, the mT-stop cassette is deleted in all cells expressing aP2-Cre resulting in the indelible labeling of cells and their progeny with membranous EGFP. We generated aP2-Cre;mT/mG and aP2-Cre;mT/mG;SmoM2/+ mice to explore the role of oncogenic SmoM2 in aP2-Cre expressing cells. Consistent with aP2 expression in mature adipose tissue, interscapular brown adipose tissue (BAT), inguinal white adipose tissue (WAT) and perirenal adipose were EGFP positive in both aP2-Cre;mT/mG and aP2-Cre;mT/mG;SmoM2/+ mice (Figures 1A&B and S1C). Discrete EGFP positive cells were also observed within both the kidney and the lung (Figure S1C), reflective of aP2 expression in pulmonary and renal capillary endothelial cells (Elmasri et al., 2009). EGFP expression in the developing sperm indicates aP2-Cre expression in the male germline accounting for the high rate of global Cre-mediated recombination observed in offspring from male aP2-Cre mice (Figure S1D). No EGFP was observed in the ovary and only 0.4% global reporter activation noted in offspring of female aP2-Cre mice. Therefore, we restricted our breeding strategies to exclusively use aP2-Cre female mice.
Figure 1. aP2-Cre labels cells within both adipose tissue and skeletal muscle.
(A) Representative whole mount images of BAT, WAT, quad, SCM and tongue from aP2-Cre;mT/mG and aP2-Cre;mT/mG;SmoM2/+ mice (n = 3). Scale bars, 3 mm. Arrowhead denotes tumor.
(B) Representative tissue sections from mT/mG, aP2-Cre;mT/mG and aP2-Cre;mT/mG;SmoM2/+ mice (n = 3). Scale bars, 50 μm.
(C) Representative images of tumor center and tumor periphery of aP2-Cre;mT/mG;SmoM2/+ mice (n = 3). Scale bars, 50 μm.
See also Figure S1.
Next, we explored the contribution of aP2-Cre labeled cells to skeletal muscle development in the absence or presence of SmoM2. As previously reported, aP2-Cre labeled cells surround the myofiber periphery (Lee et al., 2013). We noted this localization was unaffected by SmoM2 expression (Figure 1B). Examination of hindlimb quadriceps femoris (quad) and neck sternocleidomastoid (SCM) skeletal muscles revealed that individual muscle fibers remained aP2-Cre negative in both aP2-Cre;mT/mG and aP2-Cre;mT/mG;SmoM2/+ mice. Thus, skeletal muscle did not generally derive from aP2-Cre expressing cells irrespective of SmoM2 expression providing further evidence that FN-RMS in our AS model does not originate from myogenic precursors. Since our data is based on the lack of staining, we cannot conclude that skeletal muscle never arises from aP2-Cre expressing cells. In contrast, FN-RMS tumors were EGFP positive reflecting tumor cell autonomous Cre-mediated activation of hedgehog signaling driving tumorigenesis. These tumors are invasive with EGFP positive cells infiltrating adjacent normal muscle at the tumor periphery (Figures 1A&C).
Given BAT and skeletal muscle share a common progenitor (Seale et al., 2008) and adipocytes express aP2, we asked whether adipocytes are the origin of tumors in aP2-Cre;SmoM2/+ (AS) mice. To express SmoM2 in adipocytes we utilized adipose specific Adipoq-Cre (Eguchi et al., 2011) and brown adipose specific Ucp1-Cre (Kong et al., 2014). However, Adipoq-Cre;SmoM2/+ or Ucp1-Cre;SmoM2/+ mutant mice did not develop tumors suggesting mature adipose is not the FN-RMS origin in AS mice (Figures S1E–H). Together, these results demonstrate that oncogenic SmoM2 in aP2-Cre expressing cells does not alter adipocytes or skeletal muscle, nor result in skeletal muscle fibers derived from aP2-Cre labeled cells.
SmoM2 dependent proliferation of aP2-Cre labeled cells during embryogenesis
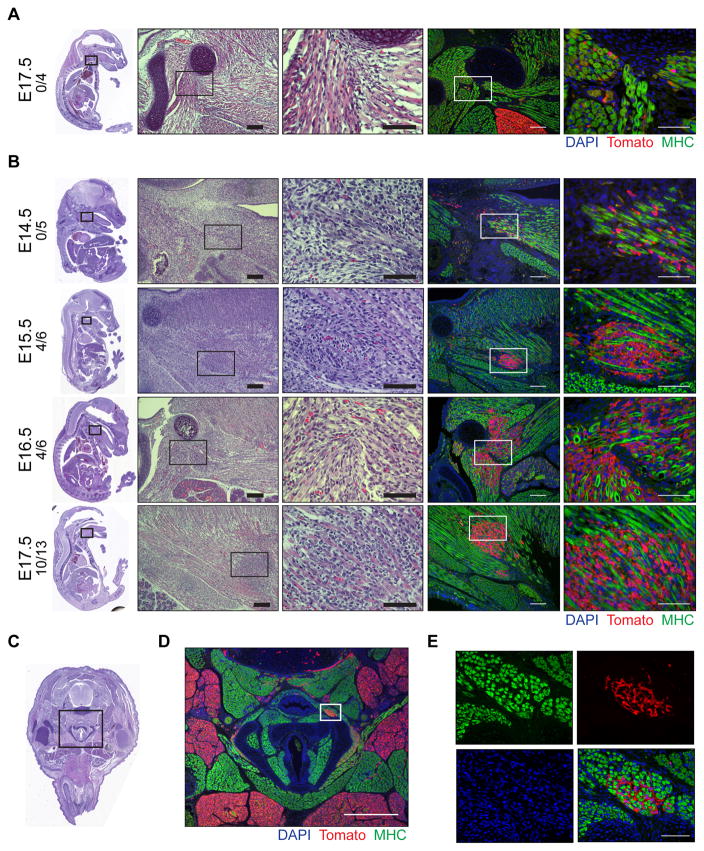
Next, we focused on determining how SmoM2 expression affects aP2-Cre labeled cells in early tumor development. Mice sacrificed as early as P7 had microscopic FN-RMS lesions on H&E staining (Figure S2A). We thus performed genetic fate mapping of aP2-Cre labeled cells in the anterior head and neck during embryonic development in AS mice. We utilized ROSA26-loxP-stop-loxP-tdTomato (R26-tdTom) reporter mice to compare aP2-Cre expressing cells indelibly labeled with the Tomato fluorescent protein in aP2-Cre;R26-tdTom (AT) and aP2-Cre;SmoM2/+;R26-tdTom (AST) mice (Figure S2B). Consistent with the mT/mG fluorescent labeling of FN-RMS (Figure 1C), Tomato and MYOD1 colocalized in tumors from AST mice (Figure S2C). At embryonic day 17.5 (E17.5), regions of BAT in the anterior neck were labeled with Tomato in AT embryos along with scattered cells within skeletal muscle that were clearly distinct from myosin heavy chain (MHC) positive myofibers (Figure 2A). Expression of oncogenic SmoM2 led to expansion of aP2-Cre labeled cells between skeletal muscle fibers in AST embryos. Starting at E15.5 the Tomato positive cells were distinctly expanding between MHC positive myofibers at sites where tumors develop in the adult (Figures 2B–E). We confirmed these findings using another independently derived aP2-Cre driver, Fabp4-Cre (He et al., 2003), to induce the expression of SmoM2. Although Fabp4-Cre;SmoM2/+ mice exhibited perinatal lethality, E16.5 embryos displayed cell expansions between muscle fibers of the head and neck identical to the AS mice (Figure S2D). These data suggest that SmoM2 expression in aP2-Cre labeled cells adjacent to the myofibers in the neck results in FN-RMS. Thus, we focused on the aP2-Cre labeled cells in the muscle interstitium as a potential origin of FN-RMS in the AS mouse model.
Figure 2. SmoM2 dependent proliferation of aP2-Cre labeled cells during embryogenesis.
(A–B) Representative H&E and immunostaining of sagittal sections from AT (A) and AST (B) embryos. Embryonic age and number of embryos with Tomato positive expansions/total embryos shown on left. Right panels show high magnification of boxed insets. Scale bars, 100 μm (left) and 50 μm (right).
(C) H&E staining of transverse section of E17.5 AST embryo.
(D) Magnification of boxed inset from (C) of AST embryo immunostained as in (A). Scale bar, 500 μm.
(E) Magnification of boxed inset (D) of AST embryo immunostained as in (A)(n = 3). Scale bar, 50 μm.
* denotes regions of adipose.
See also Figure S2.
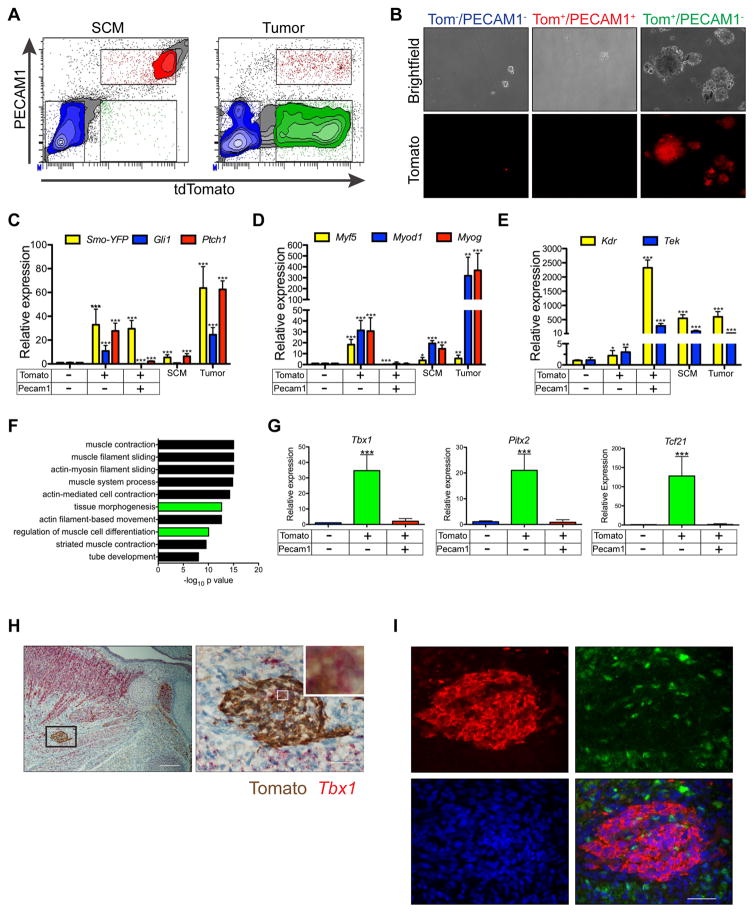
aP2-Cre is not expressed in satellite cells
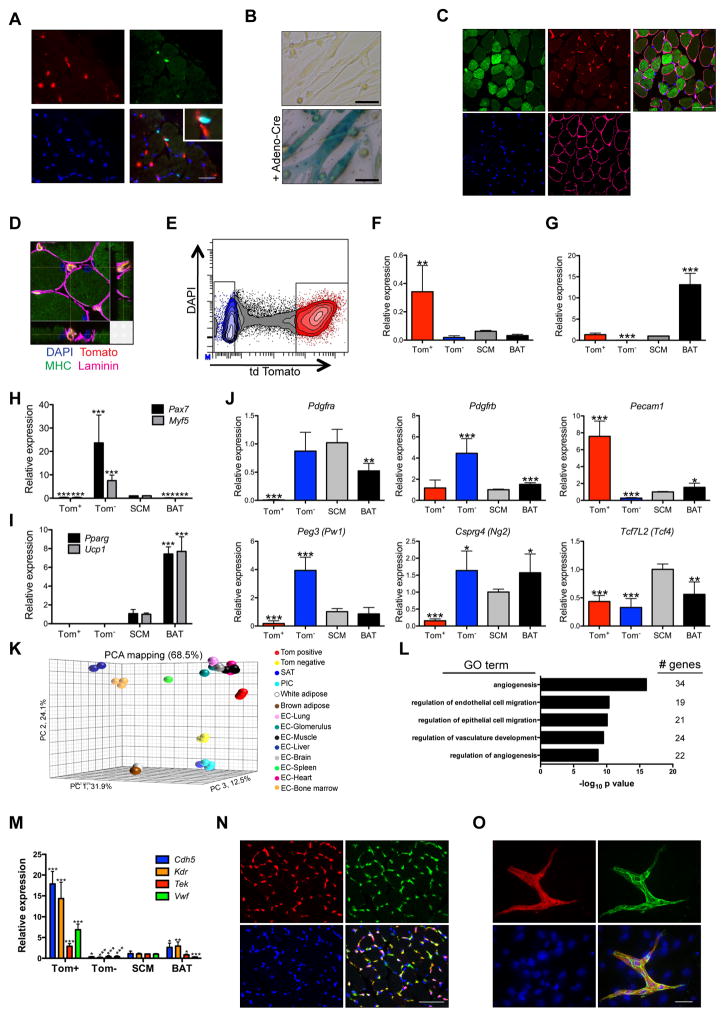
Activating oncogenes and deleting tumor suppressors in muscle stem cells (satellite cells) results in myogenic sarcomas (Blum et al., 2013; Rubin et al., 2011; Van Mater et al., 2015; Zhang et al., 2015). Although satellite cells constitute a rare population of cells in the myofiber, their localization within muscle is reminiscent of aP2-Cre labeled cells that undergo SmoM2 dependent expansion. Thus, we sought to determine whether aP2-Cre is expressed during satellite cell specification or following activation and subsequent myogenic differentiation. First, we determined aP2-Cre labeled cells within the SCM of adult AT mice were distinct from cells stained with satellite cell marker PAX7 illustrating aP2-Cre is not expressed during satellite cell specification (Figures 3A and S3A&B). To determine if aP2-Cre is expressed during satellite cell activation and differentiation, we purified satellite cells from aP2-Cre;R26-LacZ mice, differentiated cells in vitro and performed β-galactosidase staining. Myotubes from aP2-Cre;R26-LacZ mice showed no β-galactosidase staining following in vitro differentiation in contrast to intense staining observed in differentiated myoblasts transduced with adenovirus expressing Cre recombinase (Figure 3B). Furthermore, aP2-Cre labeled cells clearly localized outside the laminin sheath in the interstitium between myofibers in the SCM of AT mice (Figures 3C&D). Thus, satellite cell activation and myogenic differentiation does not induce aP2-Cre expression eliminating a satellite cell origin of FN-RMS in the AS model.
Figure 3. aP2-Cre labels endothelial cells within skeletal muscle interstitium.
(A) Immunostaining of AT SCM cross sections illustrating Tomato (red, arrowheads), PAX7 (green, arrows), DAPI (blue) positive cells (high magnification inset shown)(n = 3). Scale bars, 25 μm.
(B) β-galactosidase staining of myoblasts isolated from aP2-Cre;R26-LacZ mice following in vitro differentiation. Cells infected with adeno-Cre as control (n = 3). Scale bars, 50 μm.
(C) Immunostaining of AT SCM cross sections showing Tomato (red), MHC (green), LAMININ (magenta) and DAPI (blue)(n = 3). Scale bar, 50 μm.
(D) Confocal microscopy of (C) illustrating 3D cross sections with Tomato labeled cells in muscle interstitium. Scale bar, 10 μm.
(E–F) Isolation of Tom+ and Tom− cells from AT SCM by FACS (E) and confirmation by real-time PCR for Tomato (F). Data shown are normalized to Actb (n = 3, mean ± SEM).
(G–J) Gene expression by real-time PCR from Tom+ and Tom− cells isolated as in (E) in comparison to mature SCM and BAT from SmoM2/M2 mice: aP2 (G), myogenic genes Myf5 and Pax7 (H), adipose genes Pparg and Ucp1 (I), and muscle interstitial cell genes (J). Data shown are normalized to Actb expression and expressed relative to SCM (n = 3, mean ± SEM).
(K) Principle component analysis of Tom+ and Tom− cells isolated as in (E), microvascular endothelial cells, satellite cells, PW1+ interstitial cells (PICs) and adipose. Coordinates describe 68.5% of the data.
(L) Gene ontology analysis of genes enriched in Tom+ vs Tom− cells isolated as in (E).
(M) Expression of endothelial genes in Tom+ and Tom− cells isolated as in (E) by real-time PCR. Data analyzed as in (G)(n = 3, mean ± SEM).
(N) Immunostaining of AT SCM cross sections showing Tomato (red), PECAM1 (green), DAPI (blue) (n = 2). Scale bar, 50 μm.
(O) Immunocytochemistry of cells isolated from AT SCM and grown to confluence showing Tomato (red), PECAM1 (green), DAPI (blue) (n = 4). Scale bar, 50 μm.
*p < 0.05, ** p < 0.01, *** p < 0.001.
See also Figure S3.
aP2-Cre labels endothelial cells in muscle interstitium
Several distinct cell types have been described within the muscle interstitium with some having myogenic potential (Malecova and Puri, 2012). To identify the cell type within the muscle interstitial populations indelibly labeled by aP2-Cre, we sorted Tomato positive (Tom+) and negative (Tom−) cells from digested SCM of AT mice (Figures 3E–G). Further confirming that aP2-Cre does not label satellite cells, myogenic markers Pax7 and Myf5 were exclusively expressed in the Tom− population (Figure 3H). Mature adipose markers Pparg and Ucp1 were not expressed in either Tom+ or Tom− cells, suggesting intramuscular adipose was not a large component of AT SCM (Figure 3I). Muscle interstitial cells display distinct gene expression patterns with fibroadipogenic progenitors expressing Pdgfra and Pdgfrb (Uezumi et al., 2010), mesangioblasts and Pw1 expressing interstitial cells (PICs) expressing Pdgfra and Peg3 (Pw1) (Pannerec et al., 2013; Tagliafico et al., 2004), pericytes expressing Pdgfrb and Cspg4 (Ng2) (Lindahl et al., 1997; Ozerdem et al., 2001), connective tissue fibroblasts expressing Tcf7l2 (Tcf4) and Cspg4 (Mathew et al., 2011) and endothelial cells expressing Pecam1 (Cd31). Of these interstitial cell markers, only Pecam1 was enriched in the Tom+ population (Figure 3J).
To further identify aP2-Cre labeled cells, we next used mRNA expression profiling to compare the gene expression profiles of isolated Tom+ and Tom− cells with that of cell types found within skeletal muscle. Endothelial specific genes Pecam1, Cdh5, Kdr, Tek, Gata2 and Sox18 were significantly enriched in Tom+ cells while the myogenic marker Myf5 and interstitial cell markers Pdgfra and Peg3 were significantly enriched in Tom− cells (Figure S3C). Principle component analysis (PCA) illustrated Tom+ cells clustered closely to microvascular endothelial cells (Nolan et al., 2013). In contrast, Tom− cells clustered more closely with satellite cells and PW1+ interstitial cells. No similarities were identified between either Tom+ or Tom− cells and brown or white adipose (Seale et al., 2007) (Figure 3K). The gene expression profile of Tom+ cells was also clearly distinct from recently described Twist2 expressing interstitial cells (Figure S3D)(Liu et al., 2017). Gene ontology analysis of 489 genes enriched in Tom+ cells compared to Tom− cells (Log ratio ≥ 2, p < 0.05) revealed genes involved in angiogenesis, endothelial cell migration and vascular development were significantly enriched in the Tom+ population (Figure 3L). Altogether, these results suggest that aP2-Cre labels endothelial cells within the muscle interstitium. Confirming these findings, Tom+ cells express endothelial genes Tek, Cdh5, Vwf and Kdr (Figure 3M). As well, aP2-Cre labeled cells co-localize with PECAM1 in both cross sections and cultured cells from the SCM of AT mice (Figures 3N&O). Furthermore, aP2-Cre labeled cells colocalize with PECAM1 in the anterior neck of E17.5 AT embryos (Figure S3E&F).
Previously, Hettmer et al. defined discrete cell types isolated by FACS from adult Cdkn2a−/− mouse skeletal muscle with capacity to form myogenic sarcomas after ectopic expression of KrasG12V. This strategy separates satellite cells (CD45−MAC1−TER119−Sca1−β1-integrin+CXCR4+) from endothelial cells and cells with both fibrogenic and adipogenic potential (CD45−MAC1−TER119−Sca1+). Myogenic tumors resembling human pleomorphic RMS developed only from the satellite cell population (Hettmer et al., 2011; Schulz et al., 2011). Using this strategy, we identified 0.08% of the Tom+ muscle interstitial cells from AT mice were within the satellite cell population, in contrast to 18% of Tom− cells. Instead, 92% of Tom+ cells were Sca1+ (Figure S3G) and 96% of these Tom+Sca1+ cells were also PECAM1+ (Figure S3H). Since fibroadipogenic progenitors are also contained within the Sca1+ population, we evaluated gene expression on cells sorted for both Tomato and Sca1 fluorescence (Figure S3I). Tom+Sca1+ cells expressed endothelial markers Pecam1 and Tek, and not the fibroadipogenic marker Pdgfra. Consistent with satellite cells being among the Tom− population, sorted Tom−Sca1− cells also expressed Pax7 and Myf5 (Figure S3J). Thus, the aP2-Cre labeled cells within the skeletal muscle are endothelial cells and previously did not give rise to myogenic sarcomas with oncogenic KrasG12V.
Oncogenic Kras drives angiosarcoma in aP2-Cre expressing cells
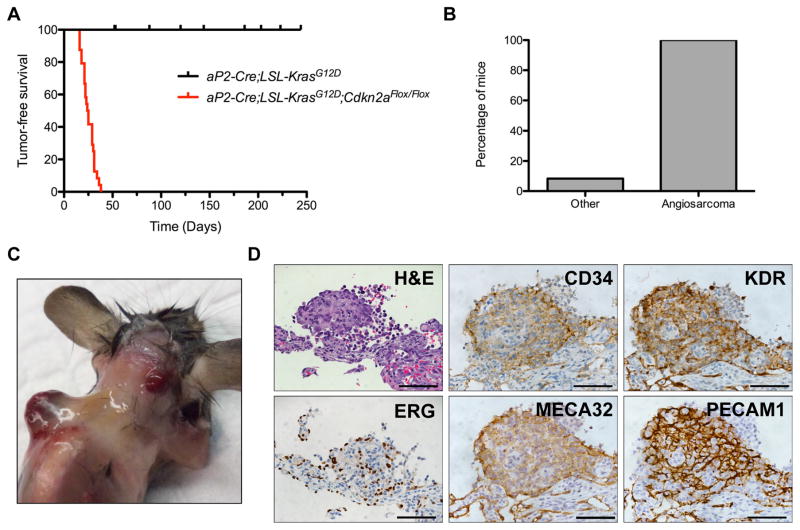
Next, we investigated whether other drivers of RMS similarly transform aP2-Cre expressing cells in vivo. Ras mutations and CDKN2A loss are common in FN-RMS (Chen et al., 2013b; Paulson et al., 2011; Shern et al., 2014). Thus, we bred aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox compound mutant mice to conditionally activate oncogenic Ras signaling and delete Cdkn2a in aP2-Cre expressing cells. These mice did not develop RMS but instead all mice developed hemorrhagic tumors in multiple organs including skeletal muscle, lung, heart and adipose with median onset of 29 days (Figures 4A–C). These tumors expressed endothelial markers PECAM1, CD34, PLVAP (Meca32), ERG and KDR, which are diagnostic for angiosarcoma (Figures 4D and S4A). In addition, 2 of the 24 mice developed solid tumors on the lower chest wall one consistent with malignant triton tumor (Figure S4B) and the other pleomorphic spindle cell carcinoma (Figure S4C). These results highlight the dependence of Shh pathway activation by illustrating oncogenic Ras does not phenocopy oncogenic SmoM2 in aP2-Cre expressing cells driving FN-RMS, but results in an endothelial tumor. Thus, illustrating that the origin of FN-RMS in the AS mice are distinct from reported Ras-driven models.
Figure 4. Oncogenic KRAS drives angiosarcoma in aP2-Cre expressing cells.
(A) Kaplan-Meier survival curve illustrating survival of aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox(n = 24) and aP2-Cre;LSL-KrasG12D(n = 10) mice. p < 0.0001.
(B) Percentage of aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox mice developing angiosarcoma or other malignancies.
(C) Angiosarcomas (arrowheads) in aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox mice.
(D) Representative H&E staining and IHC of angiosarcoma markers in aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox tumors(n = 6). Scale bars, 50 μm.
See also Figure S4.
SmoM2 promotes a myogenic fate switch during endothelial cell development
After identifying aP2-Cre labeled muscle interstitial cells are endothelial cells, we next asked whether SmoM2 expression alters their development. We isolated Tom+ and Tom− cells by FACS from the SCM of adult AST mice (Figure 5A). Tom+ cells express aP2 as well as Pecam1 and Tek (Figure 5B). In addition, aP2-Cre labeled cells co-localize with PECAM1 in cross sections and cultured cells from the SCM of AST mice (Figures S5A&B). In contrast, aP2-Cre expressing cells did not colocalize with PAX7 and Pax7 and interstitial cell markers Pdgfra, Pdgfrb, Pw1 and Ng2 were enriched in the Tom− fraction (Figures 5B and S5C–D). Neither fraction expressed the mature adipocyte marker Pparg (Figure 5B). These results demonstrate that SmoM2 expression does not block endothelial development within the muscle interstitium.
Figure 5. SmoM2 promotes a myogenic fate switch during endothelial cell development.
(A) FACS isolation of Tom+ and Tom− cells from AST SCM.
(B) Gene expression (aP2, Pecam1, Tek, Pax7 and Pparg) of Tom+ and Tom− cells isolated as in (A) compared to mature SCM and BAT from SmoM2/M2 mice by real-time PCR. Data shown are normalized to Actb expression (tdTomato) or normalized to Actb expression and expressed relative to SCM (n = 3, mean ± SEM).
(C) Gene expression of SmoM2-YFP and Shh target genes by real-time PCR in isolated Tom+ and Tom− cells, mature SCM and BAT from SmoM2/M2 mice as well as whole tumor from AST mice. Data shown are normalized to Actb expression expressed relative to SCM (n = 3, mean ± SEM).
(D) Representative H&E staining and IHC of myogenic marker (MYOD1) and endothelial marker (PECAM1) in sagittal section of E17.5 AST embryo(n = 3). Scale bars, 100 μm (left), 50 μm (right). High magnification of boxed area (right) shown.
*p < 0.05, ** p < 0.01, *** p < 0.001.
See also Figure S5.
Next, we assessed the fidelity of the Shh pathway in Tom+ cells isolated from the SCM of P21 AST mice. Despite expression of SmoM2, Tom+ cells did not increase expression of Shh pathway genes suggesting mature endothelial cells are not the origin of FN-RMS in the AS model (Figure 5C). As Shh is a key developmental morphogen, we hypothesized that SmoM2 expression in aP2-Cre labeled endothelial progenitor cells induces myogenic transdifferentiation. SmoM2 dependent embryonic proliferations at E17.5 were largely MYOD1 positive while PECAM1 was limited to the vasculature (Figure 5D). These findings suggest that prior to endothelial cell terminal differentiation, aP2-Cre expressing endothelial progenitor cells undergo a SmoM2-dependent transformation resulting in FN-RMS.
Purified tumor cells express myogenic specification factors critical for head and neck muscle development
Previously we illustrated similarity in the gene expression profile of FN-RMS from aP2-Cre;SmoM2/+ and Myogenin-Cre;SmoM2/+ mice (Hatley et al., 2012). We hypothesized that differences between AS and Myogenin-Cre;SmoM2/+ tumors should reflect their differing cellular origins. 79 genes were enriched (≥ 2 fold, p < 0.05) in AS tumors with respect to Myogenin-Cre;SmoM2/+ tumors (Table S1) and a subset of these genes were involved in angiogenesis and vascular development (Figure S6A).
Given solid tumors contain heterogeneous cell types, we sought to purify tumor cells for further analysis. We digested AST tumors and separated cells with FACS by Tomato fluorescence and PECAM1 staining to isolate aP2-Cre-labeled tumor cells from aP2-Cre-labeled tumor vasculature and Tom− stroma. In contrast to the AST SCM where the majority of Tom+ cells were PECAM1+, tumors contained an additional Tom+PECAM1− population (Figure 6A). Of the cell populations isolated from tumors, only Tom+PECAM1− cells displayed sphere initiating capacity (Figure 6B). Although both Tom+PECAM1− and Tom+PECAM1+ populations expressed SmoM2, only Tom+PECAM1− cells had increased expression of Shh pathway target genes (Figure 6C). Furthermore, myogenic genes were enriched in Tom+PECAM1− cells whereas Tom+PECAM1+ cells expressed endothelial genes (Figures 6D&E).
Figure 6. Purified tumor cells retain expression of myogenic specification factors critical for head and neck muscle development.
(A) FACS isolation of PECAM1 and Tomato stained cells from AST SCM and tumors.
(B) Sphere formation of tumor cell populations sorted as in (A)(n = 4). Scale bars are 100 μm.
(C–E) Expression of SmoM2-YFP and Shh target genes (C), myogenic markers (D) and endothelial markers (E) in populations isolated as in (A), mature SCM from SmoM2/M2 mice and whole tumor from AST mice. Data shown are normalized to Actb expression and expressed relative to sorted Tom−PECAM1− cells (n = 3, mean ± SEM).
(F) Gene ontology analysis of genes upregulated in Tom+PECAM1− cells versus Tom+PECAM1− cells isolated from AST tumors.
(G) Real-time PCR of myogenic specification factors in cells isolated in (A). Data are analyzed as in (C)(n = 3, mean ± SEM).
(H) Representative dual ISH for Tbx1 and IHC for Tomato in sagittal section of E15.5 AST embryo. Right panel is enlarged boxed region on left with high magnification inset shown (n = 4). Scale bars, 100 μm (left) and 20 μm (right).
(I) Representative immunostaining for Tomato (red), MYOD1 (green), and DAPI (blue) in sagittal sections of E15.5 AST embryo shown in H. (n = 4). Scale bar, 50 μm.
*p < 0.05, ** p < 0.01, *** p < 0.001.
See also Figure S6 and Tables S1 and S2.
To further define cell populations isolated by FACS from AST tumors, we utilized gene expression arrays. Gene expression separated the cell populations into three distinct groups by PCA of Tom−PECAM1−, Tom+PECAM1− and Tom+PECAM1+ cells (Figure S6B). We first compared the genes enriched in purified Tom+PECAM1− cells compared to SCM to genes enriched in unsorted tumors from AS mice compared to SCM (Hatley et al., 2012) and found agreement in 55% of 16,957 ortholog gene pairs (Figure S6C). Further validating that Tom+PECAM1− cells represent the FN-RMS tumor cell population, genes enriched in the Tom+PECAM1− cells compared to the Tom−PECAM1− stroma cells closely resembled that of both FN- and FP-RMS cell lines (Figure S6D). To confirm that the Tom+PECAM1+ population represented the tumor endothelial cells, we compared sorted Tom+PECAM1+ cells to Tom+ endothelial cells isolated from AT SCM (Figure 3) and 97% of genes were similarly expressed (Figure S6E). Genes with increased expression in Tom+PECAM1+ cells compared to Tom−PECAM1− cells were involved in angiogenesis and vasculature development (Figure S6F). Thus, we were able to separate tumor cells (Tom+PECAM1−) and tumor vascular cells (Tom+PECAM1+) from heterogeneous tumor stroma (Tom−PECAM1−) in tumors from AST mice using differential Tomato and PECAM1 labeling.
To explore mechanisms responsible for FN-RMS formation, we further examined gene expression of Tom+PECAM1− tumor cells. First, we identified 385 genes enriched in Tom+PECAM1− tumor cells with respect to Tom−PECAM1− stroma cells (≥ 2 fold increase, p < 0.05) including genes encoding a number of transcription factors that regulate development, including the forkhead box transcription factors FOXD1 and FOXF1, Sox family protein SOX9, T-box transcription factor TBX3 and homeobox protein PROX1. Key components of Notch (HEY1, HEYL), Bmp (BMP4) and Wnt (LEF1, LGR4) signaling pathways were also upregulated as were stem cell markers ALDH1A3 and PROX1 (Figure S6G). Gene ontology analysis of enriched genes identified muscle related processes and muscle cell differentiation among the most significant GO terms (Figure 6F). Muscle cell differentiation genes including the Mrfs Myod1, Myf5, Myf6 and Myog were enriched in Tom+PECAM1− cells (Figure 6D & Table S2).
Among genes significantly enriched in Tom+PECAM1− tumor cells with respect to both Tom+PECAM1+ endothelial and Tom−PECAM1− stromal cells were Tbx1, Pitx2, Tcf21 and Msc (Figure 6G and S6G) which are transcription factors with critical roles in head and neck muscle specification acting upstream of Mrfs (Buckingham, 2017). Pax3, whose expression is required for specification of limb and trunk muscles (Buckingham and Relaix, 2007), was not enriched in Tom+PECAM1− cells (Figure S6H). Since Tbx1 is a known Shh target gene (Garg et al., 2001), we hypothesize that SmoM2 expression in aP2-Cre expressing endothelial progenitors induces TBX1 that in turn activates Mrf expression driving rhabdomyosarcomagenesis. Consistent with this, 34% of aP2-Cre labeled cells in E15.5 AST embryos expressed Tbx1 but only 2% express MYOD1 (Figures 6H&I). At E17.5, the aP2-Cre labeled cells in AST are MYOD1 positive (Figure 5D) indicating that Tbx1 expression precedes Myod1 expression. As only a fraction of the aP2-Cre labeled cells co-localized with Tbx1, TBX1 is not likely the lone acting factor downstream of SmoM2 and likely there is redundancy with PITX2, TCF21 and MSC.
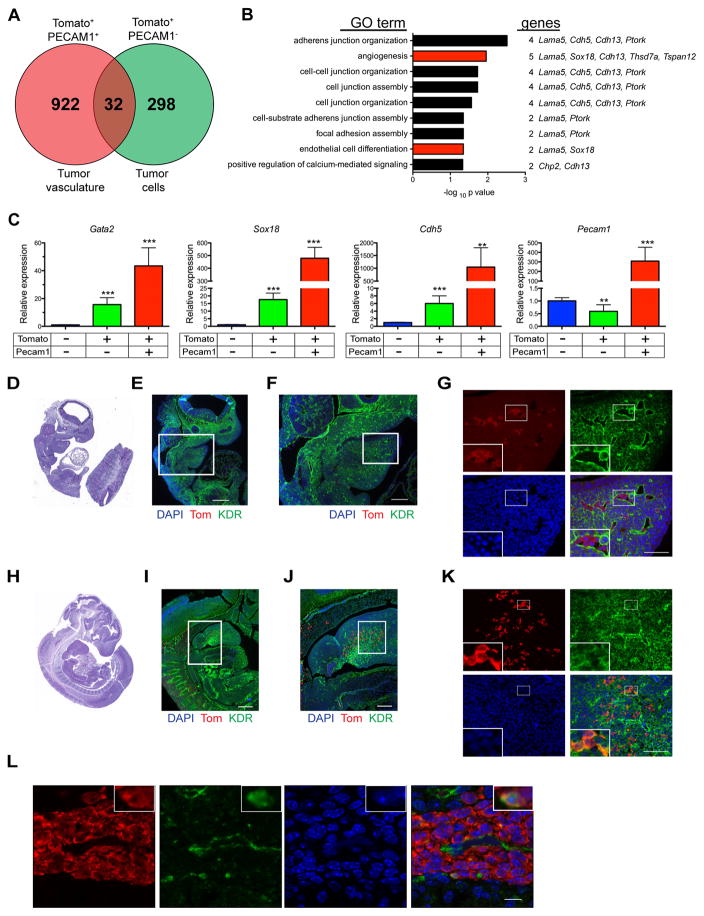
aP2-Cre;SmoM2/+ FN-RMS tumor cells retain evidence of endothelial origin
Genes involved in tube development were also enriched in Tom+PECAM1− tumor cells with respect to Tom−PECAM1− cells (Figures 6F, S6G and Table S2). To explore whether Tom+PECAM1− cells retain evidence of their cellular origins we compared the gene profile of Tom+PECAM1− cells with Tom+PECAM1+ tumor vasculature. 32 genes were enriched in both Tom+PECAM1− and Tom+PECAM1+ cells compared to Tom−PECAM1− tumor stroma including genes involved in angiogenesis and endothelial cell differentiation (Figures 7A&B and Table S3). In addition, genes encoding endothelial cell transcription factors GATA2 and SOX18 and vascular endothelial cadherin CDH5 were among the 32 overlapping genes (Figure 7C). This was most likely not due to contaminating tumor vasculature as Pecam1 expression was solely observed in Tom+PECAM1+ cells (Figure 7C).
Figure 7. Purified tumor cells and tumor vasculature express common endothelial markers.
(A) Venn diagram of genes upregulated in Tom+PECAM1− and Tom+PECAM1+ cells compared to Tom−PECAM1− cells isolated from AST tumors.
(B) Gene ontology analysis of 32 overlapping genes from (A).
(C) Gene expression of endothelial genes identified in (A) by real-time PCR. Data shown are normalized to Actb expression and expressed relative to sorted Tom−PECAM1− cells (n = 3, mean ± SEM).
(D, E) Representative H&E staining (D) and immunostaining (E) for Tomato (red), KDR (green) and DAPI (blue) on sagittal section of E9.5 AT embryo (n = 2). Scale bars, 250 μm.
(F) Magnification of boxed inset in (E). Scale bars, 100 μm.
(G) Magnification of boxed inset in (F) with high magnification inset in lower left. Scale bars, 50 μm.
(H, I) Representative H&E staining (H) and immunostaining (I) for Tomato (red), KDR (green) and DAPI (blue) on sagittal section of E10.5 AT embryo (n = 4). Scale bars, 250 μm.
(J) Magnification of boxed inset in (I). Scale bars, 100 μm.
(K) Magnification of boxed inset in (J) with high magnification inset in lower left. Scale bars, 50 μm.
(L) Representative immunostaining of Tomato positive proliferations in sagittal sections of E15.5 AST embryo (Figure 6H–I). Tomato (red), KDR (green), DAPI (blue)(n = 4). High magnification insets are shown in upper right. Scale bar, 10 μm.
*p < 0.05, ** p < 0.01, *** p < 0.001.
See also Table S3.
Vascular endothelial growth factor (Vegf) signaling mediated by KDR (VEGFR2) regulates aP2 expression in endothelial cells (Elmasri et al., 2009). Consistent with this, KDR expression at E9.5 precedes and then co-localizes with aP2-Cre expression at E10.5 in the branchial arches with 31% of Tom+ cells colocalized with KDR (Figures 7D–K). Next, we asked whether the cell expansions observed in AST embryos similarly express KDR. At E15.5, 8% of aP2-Cre expressing cells colocalize with KDR (Figure 7L). These findings provide further evidence that FN-RMS originate from endothelial progenitor cells in AST mice.
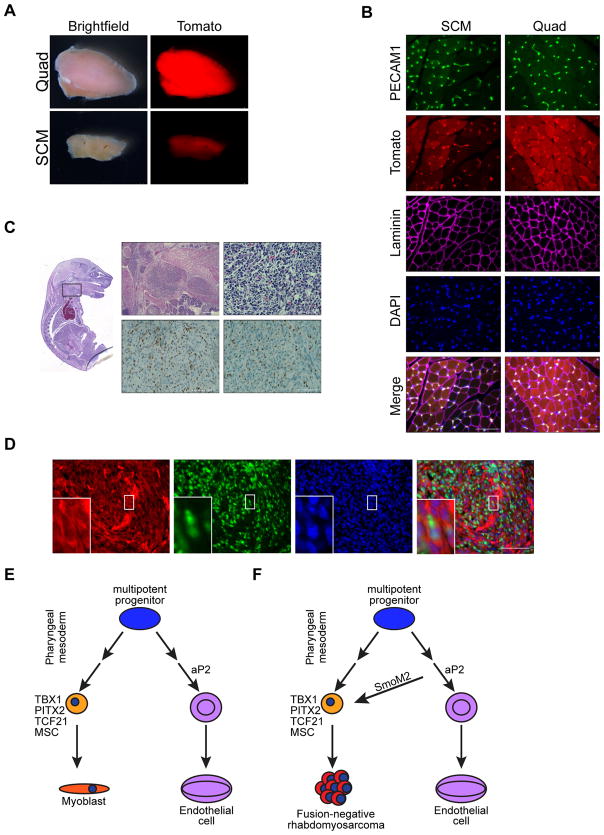
FN-RMS in Kdr::Cre;SmoM2/+ mice
In the trunk and limbs, endothelial cells and skeletal muscle originate from a common KDR and PAX3 expressing progenitor in the somite (Kardon et al., 2002; Motoike et al., 2003). To determine whether head and neck muscles are similarly derived from KDR+ cells, we bred Kdr::Cre;R26-tdTom mice and assessed tomato expression in head and neck muscle and endothelial cells. At P14, both the quad and SCM exhibited tomato fluorescence grossly (Figure 8A). In contrast to AT mice, both PECAM1+ endothelial cells and myofibers in both the SCM and quad of Kdr::Cre;R26-tdTom mice display native Tomato fluorescence histologically (Figure 8B). Thus, muscle and endothelial cells within the head and neck arise from common Kdr::Cre expressing progenitors. Together these data suggest that KDR acts upstream of aP2 in a multipotent progenitor of endothelium and head and neck muscles. As well, aP2-Cre expression is restricted to progenitors committed to an endothelial fate.
Figure 8. FN-RMS in Kdr::Cre;SmoM2/+ mice.
(A) Representative whole mount images of Quad and SCM muscle from Kdr::Cre;R26-tdTom mice (n = 3). Scale bar, 1mm.
(B) Representative immunostaining of PECAM1 (green), Tomato (red), LAMININ (magenta) and DAPI (blue) in SCM and quad of Kdr::Cre;R26-tdTom mice (n = 3). Scale bars, 50 μm.
(C) Representative sagittal sections of E17.5 Kdr::Cre;SmoM2/+ embryo with H&E staining (top) and IHC for MYOD1 and MYOG (bottom) (n = 4). High magnification of boxed inset shown on top right. Scale bars, 250 μm (top left) and 50 μm (top right and bottom IHC panels).
(D) Representative immunostaining of Tomato (red), MYOD1 (green), DAPI (blue) in tumor from Kdr::Cre;SmoM2/+;R26-tdTom mice (n = 3). High magnification inset at bottom left of boxed area in center. Scale bar, 50 μm.
(E) Model highlighting KDR expressing multipotent muscle and endothelial cell progenitors in the pharnygeal mesoderm.
(F) Model highlighting rhabdomyosarcomagenesis in aP2-Cre expressing cells.
To determine whether activation of the Shh pathway in KDR expressing cells would result in RMS, we utilized Kdr::Cre to drive SmoM2 expression. No Kdr::Cre;SmoM2/+ mice were born alive and those delivered dead had malformed hindlimbs and large umbilical protrusions. Interestingly, E18.5 Kdr::Cre;SmoM2/+ embryos had MYOD1 and MYOGENIN positive small round blue cell proliferations in the same location as the developing FN-RMS in the AS mice (Figure 8C). The MYOD1 positive tumor cells colocalized with Tomato in Kdr::Cre;SmoM2/+;R26-tdTom mice (Figure 8D). These results highlight that activating Shh signaling in Kdr expressing cells leads to similar myogenic lesions and FN-RMS as in AS mice.
DISCUSSION
The use of lineage specific cis-regulatory regions to express Cre recombinase to activate the expression of oncogenes and to delete tumor suppressor genes illustrates that the respective lineage is permissive to transformation. It is not surprising that activation of oncogenic RAS in the context of either Trp53 or Cdkn2a loss using myogenic Cre-drivers results in transformation and myogenic tumors such as FN-RMS (Kashi et al., 2015). However, the serendipitous finding of FN-RMS in aP2-Cre;SmoM2/+ compound mutant mice offers insight into the development of RMS that could not have been predicted a priori. The most surprising aspect of our FN-RMS model is that it results from SmoM2 activation in non-muscle aP2-Cre expressing cells and it only occurs exclusively in the head and neck. This allowed us the opportunity to understand the origins of RMS that occurs in tissues devoid of skeletal muscle and the development of RMS in the most common location for RMS, the head and neck.
Through genetic fate mapping we find that aP2-Cre is not expressed in skeletal muscle but in endothelial cells within the muscle interstitium. Activation of oncogenic SmoM2 with aP2-Cre reprogrammed aP2-Cre labeled interstitial endothelial progenitor cells leading to FN-RMS formation only in the head and neck. This suggests that expression of SmoM2 in endothelial progenitor cells within the muscle interstitium results in a myogenic fate switch and FN-RMS formation. Consistent with these results, isolated tumor cells are enriched in transcription factors involved in both myogenic specification as well as endothelial cell regulation. Interestingly, recent work using chromatin conformation capture to map topographical associating domains and characterization of the Foxo1 enhancer speculates a vascular origin in PAX3-FOXO1 driven fusion–positive RMS (Vicente-Garcia et al., 2017).
Although our characterization focused on skeletal muscle, the expression of aP2-Cre in endothelial cells including the adipose vasculature (Jeffery et al., 2014) would account for the occurrence of FN-RMS in adipose in both AS mice (Hatley et al., 2012) as well as Fabp4-Cre;SmoM2/+ embryos (Nosavanh et al., 2015). Shh pathway responsiveness is reduced in postnatal vasculature, (Passman et al., 2008) and we similarly observed that mature endothelial cells within skeletal muscle do not respond to hedgehog signaling despite the expression of constitutively active Smoothened. These results suggest that SmoM2 promotes myogenic transdifferentitation during a permissive window during endothelial cell development. Supporting this developmental competence model, Fabp4-Cre;Ptch1flox/− embryos, in which Shh pathway activation was achieved by conditional deletion of Ptch1, similarly develop myogenic lesions within the ventral neck (Nosavanh et al., 2015) whereas aP2-Cre;LSL-KrasG12D;Cdkn2aFlox/Flox mice develop angiosarcoma and not FN-RMS.
Previously, KDR+ cells were shown to contribute to limb skeletal muscle and cardiac muscles development as well as that of endothelial and hematopoietic lineages (Kardon et al., 2002; Motoike et al., 2003). β-galactosidase staining is evident in skeletal muscle from Kdr::Cre;R26-LacZ compound mutant mice but not Kdr::LacZ knock-in mice indicating that skeletal muscle is derived from a KDR+ cell that no longer expresses KDR (Motoike et al., 2003). Our results with Kdr::Cre;R26-tdTom mice show that endothelial cells and skeletal muscle within the head and neck are similarly derived from KDR+ cells. Lineage tracing and clonal analysis demonstrate that both branchiometric neck skeletal muscle and the second heart field within the pharyngeal mesoderm are derived from a common progenitor (Harel et al., 2009; Lescroart et al., 2015). Within the second heart field in the pharyngeal mesoderm, KDR along with ISL1 and NKX2.5 label a progenitor cell capable of giving rise to cardiac muscle, smooth muscle and endothelial cells (Moretti et al., 2006). Thus, the KDR+ multipotent progenitor upstream of aP2-Cre in the branchial arches are potentially derived from the second heart field (Diogo et al., 2015; Lescroart et al., 2015). Although KDR signaling regulates aP2 expression in endothelial cells (Elmasri et al., 2009) and aP2-Cre is expressed during embryonic development (Urs et al., 2006), our results clearly demonstrate that aP2-Cre expressing cells do not contribute to the myogenic lineage and therefore aP2-Cre is not expressed in this KDR+ early multipotent progenitor population. However, our findings that KDR expression precedes aP2-Cre expression in KDR expressing cells within the branchial arch instead suggests aP2-Cre is expressed in KDR+ committed endothelial progenitors (Figure 8E).
In KDR+ bipotent progenitors of the developing trunk and limb, an equilibrium between the transcriptional activity of FOXC1/C2 and PAX3 dictates endothelial or skeletal muscle fate respectively, thus illustrating cellular pliancy of the KDR+ progenitors (Lagha et al., 2009). Our results suggest that aberrant Shh pathway activation mediated by SmoM2 leads to transdifferentation of aP2-Cre expressing committed endothelial progenitor cells in the head and neck into myoblast-like cells in a manner analogous to increased PAX3 expression in trunk and limb progenitors (Figure 8F).
Distinct transcription factors including TBX1, PITX2, TCF21, and MSC drive specification of head and neck skeletal muscle and regulate Mrf expression (Buckingham, 2017; Kelly et al., 2004; Lu et al., 2002; Shih et al., 2007). Interestingly, Tbx1, Pitx2, Tcf21 and Msc are enriched in FN-RMS cells isolated from AST tumors. The Shh pathway activates Tbx1 expression (Garg et al., 2001) and our findings that TBX1 is expressed prior to MYOD1 in embryonic expansions of aP2-Cre expressing cells are consistent with constitutive Hh signaling driving transdifferentiation of aP2-Cre expressing endothelial progenitor cells by increasing TBX1 and consequently, Mrf expression (Figure 8F). In addition, TBX1 negatively regulates Kdr expression (Lania et al., 2015), and thus may further suppress endothelial cell fate. However, Shh activation occurs after endothelial specification in our AS model of FN-RMS resulting in these cells lacking developmental priming necessary for terminal skeletal muscle differentiation and thus resemble an arrested state of skeletal muscle development.
This work offers a more complete understanding of the intersection between normal development and RMS. As we continue to interrogate the genomics of RMS, it is becoming clear that FN-RMS is likely driven by many distinct genetic events resulting in similar histology. As we strive for more for personalized medicine, including agents to differentiate pediatric embryonal tumors, increased understanding of the developmental underpinnings for these tumors becomes even more critical. For example, FN-RMS in the head and neck derived from endothelial progenitors may not respond to a “differentiation therapy” identified in a Ras-driven tumor of satellite cell origin. Further work investigating likely multiple cellular origins of RMS will compliment extensive genomic analysis to provide therapeutic insights to improve clinical outcomes.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Mark Hatley (mark.hatley@stjude.org).
Experimental model and subject details
Mouse strains
All mouse strains used have been previously reported: aP2-Cre (Tang et al., 2008), Adipoq-Cre (#10803, Jackson Laboratories) (Eguchi et al., 2011), Ucp1-Cre (#24670, Jackson Laboratories) (Kong et al., 2014), Fabp4-Cre (#5069, Jackson Laboratories) (He et al., 2003), Flk1::Cre (Kdr::Cre) (#018976, Jackson Laboratories) (Motoike et al., 2003), R26-mTmG (#7676, Jackson Laboratories) (Muzumdar et al., 2007), R26-tdTomato (#7914, Jackson Laboratories) (Madisen et al., 2010), R26-LacZ (#3474, Jackson Laboratories) (Soriano, 1999), SmoM2 (#5130, Jackson Laboratories) (Mao et al., 2006), LSL-KrasG12D (#8179, Jackson Laboratories) (Jackson et al., 2001) and Cdkn2AFlox (Aguirre et al., 2003). For Kaplan-Meier tumor free survival analysis, Adipoq-Cre;SmoM2/+ and Ucp1-Cre;SmoM2/+ animals were observed from birth and confirmed to be tumor free at necropsy. For Kaplan-Meier survival analysis, aP2-Cre; LSL-KrasG12D and aP2-Cre; LSL-KrasG12D;Cdkn2aFlox/Flox animals were observed from birth and sacrificed when showing signs of obvious tumor burden or other distress. Full necropsies were performed. All experimental procedures involving animals were reviewed and approved by SJCRH Institutional Animal Care and Use Committee.
Method Details
Histology and immunostaining
Dissected tissues were submerged in PBS and whole mount images acquired on Nikon SMZ 1500 and Leica M165FC fluorescent stereomicroscopes using Nikon Elements and Leica Application Suite X software. Frozen sections were prepared by fixing tissues overnight in 4% paraformaldehyde (PFA) and cryoprotecting in 30% Sucrose, 2 mM MgCl2. Soft tissues were mounted in tissue freezing medium (#TFM-5, General Data) and muscle/tumors snap frozen in 2-methyl-butane cooled in liquid nitrogen prior to sectioning using a conventional cryostat. Formalin fixed Paraffin-embedded (FFPE) sections were prepared by fixation in 10% neutral-buffered formaldehyde (NBF) before embedding in paraffin. Embryos and animals for head and neck sectioning were initially fixed in 4% PFA by transcardial perfusion, and head and neck sections decalcified for 48 hours in CAL-RITE (#5501 Richard Allan Scientific) prior to embedding. Hematoxylin and eosin (H&E) and immunostaining were performed using standard procedures. Dual Tbx1 RNAscope (#481919, Advanced Cell Diagnostics) and IHC were carried out according to manufacturer’s instructions. Antibody concentrations and antigen retrieval conditions are listed in Table S4. Images were captured on Nikon Eclipse 80i upright and Nikon C2 confocal microscopes using Nikon Elements software.
Fluorescent Activated Cell Sorting
Mononuclear cells were isolated from the SCM of 3–4 week old AT or AST animals and tumors from AST animals. For each experiment, SCM was pooled from two aged matched litters. Adjacent adipose was removed from SCM and normal muscle from tumors prior to dissociation. Tissues were manually dissociated and then digested for 1 hour at 37°C in 2 U/mL Collagenase B (#11088831001, Roche)/Dispase II (#04942078001, Roche), 50mM HEPES/KOH pH 7.4; 150 mM NaCl. Following the addition of 2X volume 10% fetal bovine serum (FBS) (#SH30910.03, GE Hyclone) in phosphate buffered saline (PBS) to inactive digestion enzymes, samples were sequentially filtered through 70 μm (#22363548, Fisher) and 40 μm (#22363547, Fisher) filters to yield single cell suspensions. Single cells were blocked in 5% FBS in PBS and then stained for 30 minutes on ice prior to sorting on a FACS Aria Cellsorter (BD Biosciences). Data was acquired and analyzed using FACSDiva software. Antibody concentrations are listed in Table S5 and DAPI used as a live/dead cell marker.
Cell culture and differentiation assays
Primary mouse myoblasts were isolated from hindlimb muscles of 3–4 week old aP2-Cre;R26-LacZ mice by manual disassociation and subsequent digestion for 90 minutes in 0.15 U/mL Liberase DL (#0540116000, Roche) at 37°C. Digestion was stopped by the addition of 1X volume 20% FBS in PBS and then samples were sequentially filtered through 100 μm (#352360, BD Falcon) and 70 μm (#22363548, Fisher) filters to yield a single cell suspension. Myoblasts were enriched from cell suspensions after pre-plating for 2 hours. Unadherred cells were cultured in Hams F10 medium (#10070CV, Corning) supplemented with 20% Cosmic Calf serum (#SH30087.03, GE Hyclone), 1% penicillin, streptomycin and amphotericin (PSA) (#A5955, Sigma Aldrich) and 4ng/mL fibroblast growth factor (FGF) (#233-FB-025, R&D systems) on 1% gelatin coated plates at 37°C in 5% CO2. For differentiation assays, myoblasts were grown until 90% confluence and transferred to DMEM (#SH30243, GE Hyclone) containing 2% horse serum. Following differentiation, cells were fixed for 15 minutes in 4% PFA, 0.1 M phosphate buffer (pH 7.4), 0.01% sodium deoxycholate, and 0.02% Nonidet P-40 and stained for β-galactosidase activity overnight in 1 mg/mL X-gal (5-bromo-4-chloro-3-indoyl-β-D-galactoside), 5 mM potassium ferricyanide, and 5 mM potassium ferrocyanide, 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% Nonidet P-40. Cells transduced with Ad5CMVeGFP (VVC-U of Iowa-4, University of Iowa, Iowa City, IA, USA) prior to differentiation were utilized as a positive control for β-galactosidase staining. Mononuclear cells were isolated from SCM of 3–4 week old AT and AST animals by digesting disassociated tissue in 2 U/mL Collagenase B (#11088831001, Roche)/Dispase II (#04942078001, Roche) and then culturing until confluent in DMEM (#SH30243, GE Hyclone) supplemented with 10% FBS (#SH30910.03 GE Hyclone) and 1% PSA (#A5955, Sigma Aldrich). Immunofluorescence was performed according to standard techniques using antibodies listed in Table S4. Tumor spheres were grown on ultra low attachment plates (#3471, Corning) in neurobasal medium (#12348 Gibco) supplemented 10 ng/mL epidermal growth factor (#23626, R and D systems), 20 ng/mL FGF (#233-FB-025, R and D systems), 2x B27 supplement (#17504-44, Gibco), 1x Glutamax (#35050, Gibco) and 1% PSA (#A5955, Sigma Aldrich) with from cell populations isolated from AST tumors by FACS. Images were captured on a Nikon Eclipse Ti-S inverted microscope using Nikon Elements Software.
RNA isolation and Gene Expression analysis
Total RNA was isolated from whole tissues using a miRNEasy mini kit (#217004, Qiagen, Valencia, CA, USA) and from sorted cell populations using a miRNEasy micro kit (#217084, Qiagen) according to manufacturer’s instructions. Reverse transcription was performed using Superscript III First Strand Synthesis using random hexamer primers. Real-time PCR was performed utilizing SYBR primers and Taqman probes listed in Table S6 or Key Resources Table and normalized to Actb expression. Relative quantity of tomato expression was determined using the ΔCT method and of other genes of interest using the ΔΔCT method.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-CD34 (Clone RAM 34) | BD Bioscience | Cat# 553731; RRID:AB_395015 |
| Rabbit polyclonal anti-DESMIN | ThermoFisher | Cat # RB-9014; RRID:AB_149771 |
| Rabbit monoclonal anti-ERG | Abcam | Cat # ab133264; RRID:AB_11156852 |
| Mouse monoclonal anti-CK-Oscar (Clone OSCAR) | Covance | Cat #465-01; RRID:AB_2565083 |
| Mouse monoclonal anti-CK-HMV (Clone 34βE12) | DAKO | Cat # M06302 |
| Goat polyclonal anti-KDR | R&D Systems | Cat #AF644; RRID:AB_355500 |
| Rabbit polyclonal anti-Laminin | Sigma | Cat # L9393; RRID:AB_477163 |
| Rat monoclonal anti-Meca32 (Clone Meca 32) | BD Bioscience | Cat #553849; RRID:AB_395086 |
| Mouse monoclonal anti-MHC (Clone MF20) | DSHB | Cat #MF20; RRID:AB_2147781 |
| Mouse monoclonal anti-MYOD1 (Clone 5.8A) | DAKO | Cat #M3512 RRID:AB_2148874 |
| Rabbit monoclonal anti-MYOD1 (Clone EP 212) | Cell Marque | Cat #386R-18 |
| Mouse monoclonal anti-MYOGENIN (Clone F5D) | DAKO | Cat #M3559; RRID:AB_2250893 |
| Mouse monoclonal anti-PAX7 | DSHB | Cat #Pax7; RRID:AB_2299243 |
| Rat monoclonal anti-PECAM1 (Clone Mec 13.3) | BD Bioscience | Cat #550274; RRID:AB_393571 |
| Rat monoclonal anti-PECAM1 (Clone SZ31) | Dianova | Cat #Dia 310; RRID:AB_2631039 |
| Rabbit polyclonal anti-S100 | DAKO | Cat #Z031129; RRID:AB_2315306 |
| Rabbit polyclonal anti-RFP | Rockland | Cat #600-401-379; RRID:AB_2209751 |
| Goat polyclonal anti-tDTomato | LSBio | Cat #LS-C340696 |
| Rabbit monoclonal anti-VIMENTIN | Novus | Cat #NBP1-40730; RRID:AB_10004971 |
| Hamster monoclonal anti-β1-INTEGRIN (Clone Ha2/5) – FITC conjugated | BD Bioscience | Cat #561796 RRID:AB_10894590 |
| Rat monoclonal anti-CD11b (Clone M1/70) APC conjugate | BD Bioscience | Cat # 553312 RRID:AB_398535 |
| Rat monoclonal anti-CD45 (Clone 30 F11) APC conjugate | BD Bioscience | Cat #561018 RRID:AB_10584326 |
| Rat monoclonal anti-CXCR4 (Clone 2B11/CXCR4) Biotin conjugate | BD Bioscience | Cat #551968 RRID:AB_394307 |
| Rat monoclonal anti-PECAM1 (Clone Mec 13.3) APC conjugate | BD Bioscience | Cat #551262 RRID:AB_398497 |
| Rat monoclonal anti-PECAM1 (Clone Mec 13.3) FITC conjugate | BD Bioscience | Cat #553372 RRID:AB_394818 |
| Rat monoclonal anti-Sca1 (Clone D7) PerCPCy5.5 conjugate | eBioscience | Cat #45-5981-82 RRID:AB_914372 |
| Rat monoclonal anti-TER-119 (Clone TER 119) APC conjugate | BD Bioscience | Cat #557909 RRID:AB_398635 |
| Goat anti-Rat IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Fluor 488 conjugate | ThermoFisher | Cat #A11006 RRID:AB_2534074 |
| Goat anti-Mouse IgG1 (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 conjugate | ThermoFisher | Cat #A21121 RRID:AB_141514 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Fluor 488 conjugate | ThermoFisher | Cat #A11029 RRID:AB_138404 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Fluor 568 conjugate | ThermoFisher | Cat #A11036 RRID:AB_10563566 |
| Donkey anti-Rabbit IgG (H+L) Highly Cross-Absorbed Secondary Antibody, Alexa Fluor 568 conjugate | ThermoFisher | Cat# A10042 RRID:AB_2534017 |
| Goat anti-Mouse Affinity Purified IgG (H+L) Secondary HRP conjugate | BioRad | Cat# 170-6516 RRID:AB_11125547 |
| Goat anti-Rat IgG Antibody, HRP conjugate | Millipore | Cat# AP136P RRID:AB_11214444 |
| Donkey anti-Goat IgG (H+L) Highly Cross-Absorbed Secondary Antibody, HRP conjugate | ThermoFisher | Cat# A16005 RRID:AB_2534679 |
| Bacterial and Virus Strains | ||
| Ad5CMV-Cre-eGFP | University of Iowa | VVC-U of Iowa-4 |
| Biological Samples | ||
| Sorted cells from aP2-Cre;R26-tdTom mouse sternocleidomastoid | This paper | |
| Sorted cells from aP2-Cre;SmoM2/+;R26-tdTom mouse sternocleidomastoid | This paper | |
| Sorted tumor cells from aP2-Cre;SmoM2/+;R26-tdTom mouse rhabdomyosarcoma | This paper | |
| Mononuclear cells from aP2-Cre;SmoM2/+;R26-tdTom mouse sternocleidomastoid | This paper | |
| Mononuclear cells from aP2-Cre;SmoM2/+;R26-tdTom mouse sternocleidomastoid | This paper | |
| aP2-Cre;R26-LacZ hindlimb muscle myoblast | This paper | |
| SmoM2/M2 mouse sternocleidomastoid | This paper | |
| SmoM2/M2 mouse brown adipose tissue | This paper | |
| Rhabdomyosarcoma from aP2-Cre;SmoM2/+;R26-tdTom mice | This paper | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase B | Roche | Cat # 11088831001 |
| Dispase II | Roche | Cat # 4942078001 |
| Liberase DL | Roche | Cat # 05401160001 |
| bFGF | R&D Systems | Cat # 233-FB-025 |
| EGF | R&D Systems | Cat # 23626 |
| B27 Supplement | Gibco | Cat # 17504-44 |
| Critical Commercial Assays | ||
| TSA plus Fluorescein System | Perkin-Elmer | Cat# 741E001KT |
| Vectashield Hardset mounting medium with DAPI | Vector Laboratories | Cat# H1500 |
| miRNeasy micro kit | Qiagen | Cat# 217084 |
| miRNeasy mini kit | Qiagen | Cat# 217004 |
| Superscript Reverse Transcriptase | ThermoFisher | Cat# 18080051 |
| Fast SYBR Green Master Mix | ThermoFisher | Cat# 4385610 |
| TaqMan Fast Advanced Master Mix | ThermoFisher | Cat# 4444964 |
| Ovation Pico WTA system V2 | Nugen | Cat# 3302 |
| Mouse gene 2.0 ST microarray | Affymetrix | Cat#902118 |
| RNAscope 2.5 VS-probe-Mm-Tbx1 | ACD | Cat#481919 |
| Deposited Data | ||
| Microarray data | This Paper | GSE 98059 |
| Microarray data – aP2-Cre;SmoM2 RMS, MyoG-Cre;SmoM2 RMS | (Hatley et al., 2012) | GSE 40359 |
| Microarray data – C57/Bl6 sternocleidomastoid muscle | (Hanna et al., 2017) | GSE 85834 |
| Microarray data – microvascular endothelial cells | (Nolan et al., 2013) | GSE 47067 |
| Microarray data – brown and white adipose | (Seale et al., 2007) | GSE 8044 |
| Microarray data – satellite cells and PWI expressing interstitial cells | (Pannerec et al., 2013) | GSE 40523 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.SJL-Tg(aP2-Cre)Jmg | ||
| Mouse: B6;FVB-Tg(Adipoq-Cre)1Evdr/J | Jackson Laboratories (Eguchi et al., 2011) | Cat # 10803 RRID:IMSR_JAX:010803 |
| Mouse: B6.FVB-Tg(Ucp1-Cre)1Evdr/J | Jackson Laboratories (Kong et al., 2014) | Cat # 24670 RRID:IMSR_JAX:024670 |
| Mouse: B6.Cg-Tg(Fabp4-Cre)1Rev/J | Jackson Laboratories (He et al., 2003) | Cat # 5069 RRID:IMSR_JAX:005069 |
| Mouse: Kdrtm1(Cre)Sato/J | Jackson Laboratories (Motoike et al., 2003) | Cat # 018976 RRID:IMSR_JAX:018976 |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories (Muzumdar et al., 2007) | Cat # 7676 RRID:IMSR_JAX:007676 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratories (Madisen et al., 2010) | Cat # 7914 RRID:IMSR_JAX:007914 |
| Mouse: B6.129S4-Gt(ROSA)26Sortm1Sor/J | Jackson Laboratories (Soriano, 1999) | Cat # 3474 RRID:IMSR_JAX:003474 |
| Mouse: Gt(ROSA)26Sortm1(Smo/EYFP)Amc/J | Jackson Laboratories (Mao et al., 2006) | Cat # 5130 RRID:IMSR_JAX:005130 |
| Mouse: B6.129S4-Krastm4Tyj/J | Jackson Laboratories (Jackson et al., 2001) | Cat # 8179 RID:IMSR_JAX:008179 |
| Mouse: Cdkn2aflox/flox | (Aguirre et al., 2003) | |
| Oligonucleotides | ||
| SYBR primers for real-time PCR – see Table S6 | ||
| Taqman FAM Probe – Gli1 | ThermoFisher | Cat # Mm00494654_m1 |
| Taqman FAM Probe – MyoG | ThermoFisher | Cat # Mm00446195_g1 |
| Taqman FAM Probe – Msc | ThermoFisher | Cat # Mm00447887_m1 |
| Taqman FAM Probe – Pax3 | ThermoFisher | Cat # Mm00435491_m1 |
| Taqman FAM Probe – Pitx2 | ThermoFisher | Cat # Mm01316994_m1 |
| Taqman FAM Probe – Tbx1 | ThermoFisher | Cat # Mm00448949_m1 |
| Taqman FAM Probe – Lgr4 | ThermoFisher | Cat # Mm00554385_m1 |
| Software and Algorithms | ||
| Nikon Elements Basic Research v4.13 | Nikon Instruments Inc | |
| Leica Application Suite X | Leica Microsystems | |
| FACSdiva v8.0.1 | BD Biosystems | |
| Graphpad Prism 6 | Graphpad software | |
| Image J v1.50 | NIH | |
| STATA/MP 14.2 | StataCorp | |
| Partek Genome Suite 6.6 | Partek | |
| Enrichr gene enrichment analysis tool | (Chen et al., 2013b; Kuleshov et al., 2016) | |
| Adobe Photoshop CS5.1 | Adobe Software | |
| Adobe Illustrator CS5.1 | Adobe Software | |
| Other | ||
For gene expression profiling, amplified cDNA was prepared using the Ovation Pico WTA System V2 (#3302, Nugen, San Carlos, CA, USA) from RNA isolated from sorted cell populations. Gene expression analysis was performed on Mouse gene 2.0 ST microarrays (#902118, Affymetrix, Santa Clara, CA, USA). Data were imported into Partek Genome Suite 6.6, visualized by principal component analysis to check for consistency of samples and class variance. Data were then batch corrected and statistically tested using an unequal variance t test. After filtering unannotated transcripts, transcripts were imported into STATA/MP 14.2 and p-values adjusted for multiple comparisons by the Benjamini-Hochberg FDR method (Benjamini and Hochberg, 1995). Volcano plots were produced in STATA/MP 14.2 by plotting the Log10 transformed p value from the unequal variance t test against the Log ratio of expression for datasets being compared. Array data is deposited in GEO database (GSE98059). The gene expression profile of Tom+ and Tom− cells isolated from AT SCM were compared to microvascular endothelial cells (GSE47067) (Nolan et al., 2013), satellite cells and PW1+ interstitial cells (GSE40523) (Pannerec et al., 2013) as well as brown and white adipose (GSE8044) (Seale et al., 2007). The gene expression profile of Tom+PECAM1− cells isolated from AST tumors were expressed relative to normal SCM from C57BL/6 animals (GSE85834) (Hanna et al., 2017) and compared to that of whole aP2-Cre;SmoM2/+ tumors normalized to SCM (GSE40359) (Hatley et al., 2012). Comparisons between the gene expression profile of Tom+PECAM1+ cells isolated from AST tumors and Tom+ cells isolated from AT SCM (this study) were also made. Gene Ontology analysis was performed using Enrichr gene enrichment analysis tool (Chen et al., 2013a; Kuleshov et al., 2016). Reported p values are adjusted using the Benjamini-Hochberg method.
Quantification and Statistical Analysis
Sample size and replicates for each experiment are listed in the figure legends. Embryonic expansions of aP2-Cre expressing cells (Figure 2) were defined as tomato positive cell proliferations that displaced and engulfed the MHC+ myofibres. Real-time PCR error bars indicate SEM of three independent biological experiments performed in technical replicate using two tailed unpaired student t tests for pairwise comparisons (Graphpad Prism6). Kaplan-Meier survival analysis was performed by Log Rank (Mantel-Cox) test. p values less than 0.05 were considered significant.
Data Availability
The microarray data has been deposited in the Gene Expression Omnibus database under number GSE98059.
Supplementary Material
SIGNIFICANCE.
RMS is the most common pediatric soft tissue sarcoma and occurs anywhere in the body including tissues devoid of skeletal muscle. The cell of origin for RMS remains unknown. Identification of the cell lineages that contribute to RMS formation provides insight into the mechanisms of tumorigenesis. We demonstrate using aP2-Cre to activate the hedgehog pathway that fusion negative RMS originates from endothelial progenitor cells specifically in the head and neck, the most common location of RMS. Our findings illustrate how normal muscle developmental programs are perturbed to drive location specific tumor formation through transdifferentiation induced by hedgehog activation in endothelial progenitors. These studies implicate cell of origin as a major determinate of RMS location and therefore survival.
HIGHLIGHTS.
Committed endothelial progenitors can give rise to rhabdomyosarcoma (RMS)
Aberrant activation of muscle development programs in non-myogenic cells drive RMS
SmoM2 transdifferentiates endothelial progenitors resulting in RMS
Cell of origin emerges as major determinant of RMS location
Acknowledgments
We thank Shannon McKinney-Freeman for mouse generosity. We also wish to thank St. Jude Children’s Research Hospital Cancer Center shared resources including Cell and Tissue Imaging (Victoria Frohlich, Jennifer Peters), Flow Cytometry and Cell Sorting (Richard Ashmun), Veterinary Pathology (Peter Vogel, Dorothy Bush, Sean Savage) and the Hartwell Center and Functional Genomics (Melanie Lloyd) as well as Jonathan Go for technical assistance. Work in M.E.H.’s laboratory is supported by the V Foundation for Cancer Research, NIH (NCI-R01CA216344 and K08CA151649), St. Jude Cancer Center Support Grant (P30 CA021765), and American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, C.J.D., J.A.H, D.J.D and M.E.H.; Methodology, C.J.D., J.A.H., M.R.G., D.J.D. and M.E.H.; Formal Analysis, C.J.D., J.A.H., J.E.R. and M.E.H.; Investigation, C.J.D., J.A.H., M.R.G., D.J.D., and A.J.H.; Data Curation, D.F.; Writing-Original Draft, C.J.D and M.E.H.; Writing – Reviewing and Editing, C.J.D. J.A.H., M.R.G, D.J.D, A.J.H, J.E.R, D.F. and M.E.H.; Visualization, C.J.D., J.A.H. and M.E.H.; Supervision M.E.H.; Funding Acquisition M.E.H.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Blum JM, Ano L, Li Z, Van Mater D, Bennett BD, Sachdeva M, Lagutina I, Zhang M, Mito JK, Dodd LG, et al. Distinct and overlapping sarcoma subtypes initiated from muscle stem and progenitor cells. Cell reports. 2013;5:933–940. doi: 10.1016/j.celrep.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP., Jr Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126:4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Liu J, Weibolt V, Baker KS, Perry D, Kruger R, Qualman S, Barr F, Sorensen P, Triche T, Suijkerbuijk R. Novel genomic imbalances in embryonal rhabdomyosarcoma revealed by comparative genomic hybridization and fluorescence in situ hybridization: an intergroup rhabdomyosarcoma study. Genes, chromosomes & cancer. 2000;27:337–344. doi: 10.1002/(sici)1098-2264(200004)27:4<337::aid-gcc1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Buckingham M. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1610605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annual review of cell and developmental biology. 2007;23:645–673. doi: 10.1146/annurev.cellbio.23.090506.123438. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Chen EY, Dobrinski KP, Brown KH, Clagg R, Edelman E, Ignatius MS, Chen JY, Brockmann J, Nielsen GP, Ramaswamy S, et al. Cross-species array comparative genomic hybridization identifies novel oncogenic events in zebrafish and human embryonal rhabdomyosarcoma. PLoS Genet. 2013a;9:e1003727. doi: 10.1371/journal.pgen.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer cell. 2013b;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, D’Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Diogo R, Kelly RG, Christiaen L, Levine M, Ziermann JM, Molnar JL, Noden DM, Tzahor E. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature. 2015;520:466–473. doi: 10.1038/nature14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmasri H, Karaaslan C, Teper Y, Ghelfi E, Weng M, Ince TA, Kozakewich H, Bischoff J, Cataltepe S. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J. 2009;23:3865–3873. doi: 10.1096/fj.09-134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, 3rd, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nature genetics. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Garg V, Yamagishi C, Hu T, Kathiriya IS, Yamagishi H, Srivastava D. Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol. 2001;235:62–73. doi: 10.1006/dbio.2001.0283. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nat Med. 1998;4:619–622. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- Hanna JA, Drummond CJ, Garcia MR, Go JC, Finkelstein D, Rehg JE, Hatley ME. Biallelic Dicer1 loss mediated by aP2-Cre drives angiosarcoma. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel I, Nathan E, Tirosh-Finkel L, Zigdon H, Guimaraes-Camboa N, Evans SM, Tzahor E. Distinct origins and genetic programs of head muscle satellite cells. Dev Cell. 2009;16:822–832. doi: 10.1016/j.devcel.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, Graff J, Galindo RL, Olson EN. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer cell. 2012;22:536–546. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Barak Y, Hevener A, Olson P, Liao D, Le J, Nelson M, Ong E, Olefsky JM, Evans RM. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettmer S, Liu J, Miller CM, Lindsay MC, Sparks CA, Guertin DA, Bronson RT, Langenau DM, Wagers AJ. Sarcomas induced in discrete subsets of prospectively isolated skeletal muscle cells. Proc Natl Acad Sci U S A. 2011;108:20002–20007. doi: 10.1073/pnas.1111733108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Berry R, Church CD, Yu S, Shook BA, Horsley V, Rosen ED, Rodeheffer MS. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte. 2014;3:206–211. doi: 10.4161/adip.29674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardon G, Campbell JK, Tabin CJ. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev Cell. 2002;3:533–545. doi: 10.1016/s1534-5807(02)00291-5. [DOI] [PubMed] [Google Scholar]
- Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer. 2015;15:426–439. doi: 10.1038/nrc3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RG, Jerome-Majewska LA, Papaioannou VE. The del22q11.2 candidate gene Tbx1 regulates branchiomeric myogenesis. Hum Mol Genet. 2004;13:2829–2840. doi: 10.1093/hmg/ddh304. [DOI] [PubMed] [Google Scholar]
- Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham ME. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell. 2009;17:892–899. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Lania G, Ferrentino R, Baldini A. TBX1 Represses Vegfr2 Gene Expression and Enhances the Cardiac Fate of VEGFR2+ Cells. PLoS One. 2015;10:e0138525. doi: 10.1371/journal.pone.0138525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, McKinnon PJ. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- Lescroart F, Hamou W, Francou A, Theveniau-Ruissy M, Kelly RG, Buckingham M. Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc Natl Acad Sci U S A. 2015;112:1446–1451. doi: 10.1073/pnas.1424538112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- Liu N, Garry GA, Li S, Bezprozvannaya S, Sanchez-Ortiz E, Chen B, Shelton JM, Jaichander P, Bassel-Duby R, Olson EN. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol. 2017;19:202–213. doi: 10.1038/ncb3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JR, Bassel-Duby R, Hawkins A, Chang P, Valdez R, Wu H, Gan L, Shelton JM, Richardson JA, Olson EN. Control of facial muscle development by MyoR and capsulin. Science. 2002;298:2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecova B, Puri PL. “Mix of Mics”-Phenotypic and Biological Heterogeneity of “Multipotent” Muscle Interstitial Cells (MICs) J Stem Cell Res Ther. 2012 [PMC free article] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailovici I, Eigler T, Tzahor E. Craniofacial Muscle Development. Curr Top Dev Biol. 2015;115:3–30. doi: 10.1016/bs.ctdb.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Motoike T, Markham DW, Rossant J, Sato TN. Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis. 2003;35:153–159. doi: 10.1002/gene.10175. [DOI] [PubMed] [Google Scholar]
- Munsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9:2911–2922. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, Ding BS, Schachterle W, Liu Y, Rosenwaks Z, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosavanh L, Yu DH, Jaehnig EJ, Tong Q, Shen L, Chen MH. Cell-autonomous activation of Hedgehog signaling inhibits brown adipose tissue development. Proc Natl Acad Sci U S A. 2015;112:5069–5074. doi: 10.1073/pnas.1420978112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin O, Rey A, Lyden E, Bisogno G, Stevens MC, Meyer WH, Carli M, Anderson JR. Prognostic factors in metastatic rhabdomyosarcomas: results of a pooled analysis from United States and European cooperative groups. J Clin Oncol. 2008;26:2384–2389. doi: 10.1200/JCO.2007.14.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozerdem U, Grako KA, Dahlin-Huppe K, Monosov E, Stallcup WB. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev Dyn. 2001;222:218–227. doi: 10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- Pak E, Segal RA. Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell. 2016;38:333–344. doi: 10.1016/j.devcel.2016.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannerec A, Formicola L, Besson V, Marazzi G, Sassoon DA. Defining skeletal muscle resident progenitors and their cell fate potentials. Development. 2013;140:2879–2891. doi: 10.1242/dev.089326. [DOI] [PubMed] [Google Scholar]
- Parham DM, Barr FG. Classification of rhabdomyosarcoma and its molecular basis. Advances in anatomic pathology. 2013;20:387–397. doi: 10.1097/PAP.0b013e3182a92d0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passman JN, Dong XR, Wu SP, Maguire CT, Hogan KA, Bautch VL, Majesky MW. A sonic hedgehog signaling domain in the arterial adventitia supports resident Sca1+ smooth muscle progenitor cells. Proc Natl Acad Sci U S A. 2008;105:9349–9354. doi: 10.1073/pnas.0711382105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, Cameron S. High-resolution array CGH identifies common mechanisms that drive embryonal rhabdomyosarcoma pathogenesis. Genes, chromosomes & cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- Perez EA, Kassira N, Cheung MC, Koniaris LG, Neville HL, Sola JE. Rhabdomyosarcoma in children: a SEER population based study. The Journal of surgical research. 2011;170:e243–251. doi: 10.1016/j.jss.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Pressey JG, Anderson JR, Crossman DK, Lynch JC, Barr FG. Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57:930–938. doi: 10.1002/pbc.23174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin BP, Nishijo K, Chen HI, Yi X, Schuetze DP, Pal R, Prajapati SI, Abraham J, Arenkiel BR, Chen QR, et al. Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer cell. 2011;19:177–191. doi: 10.1016/j.ccr.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satheesha S, Manzella G, Bovay A, Casanova EA, Bode PK, Belle R, Feuchtgruber S, Jaaks P, Dogan N, Koscielniak E, Schafer BW. Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene. 2016;35:2020–2030. doi: 10.1038/onc.2015.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer discovery. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Gross MK, Kioussi C. Cranial muscle defects of Pitx2 mutants result from specification defects in the first branchial arch. Proc Natl Acad Sci U S A. 2007;104:5907–5912. doi: 10.1073/pnas.0701122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sultan I, Qaddoumi I, Yaser S, Rodriguez-Galindo C, Ferrari A. Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: an analysis of 2,600 patients. J Clin Oncol. 2009;27:3391–3397. doi: 10.1200/JCO.2008.19.7483. [DOI] [PubMed] [Google Scholar]
- Tagliafico E, Brunelli S, Bergamaschi A, De Angelis L, Scardigli R, Galli D, Battini R, Bianco P, Ferrari S, Cossu G, Ferrari S. TGFbeta/BMP activate the smooth muscle/bone differentiation programs in mesoangioblasts. J Cell Sci. 2004;117:4377–4388. doi: 10.1242/jcs.01291. [DOI] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Urs S, Harrington A, Liaw L, Small D. Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic research. 2006;15:647–653. doi: 10.1007/s11248-006-9000-z. [DOI] [PubMed] [Google Scholar]
- Van Mater D, Ano L, Blum JM, Webster MT, Huang W, Williams N, Ma Y, Cardona DM, Fan CM, Kirsch DG. Acute tissue injury activates satellite cells and promotes sarcoma formation via the HGF/c-MET signaling pathway. Cancer Res. 2015;75:605–614. doi: 10.1158/0008-5472.CAN-14-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Garcia C, Villarejo-Balcells B, Irastorza-Azcarate I, Naranjo S, Acemel RD, Tena JJ, Rigby PWJ, Devos DP, Gomez-Skarmeta JL, Carvajal JJ. Regulatory landscape fusion in rhabdomyosarcoma through interactions between the PAX3 promoter and FOXO1 regulatory elements. Genome Biol. 2017;18:106. doi: 10.1186/s13059-017-1225-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, Thway K, Orbach D, Lae M, Freneaux P, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–2158. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- Zhang M, Qiu Q, Li Z, Sachdeva M, Min H, Cardona DM, DeLaney TF, Han T, Ma Y, Luo L, et al. HIF-1 Alpha Regulates the Response of Primary Sarcomas to Radiation Therapy through a Cell Autonomous Mechanism. Radiation research. 2015;183:594–609. doi: 10.1667/RR14016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibat A, Missiaglia E, Rosenberger A, Pritchard-Jones K, Shipley J, Hahn H, Fulda S. Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. 2010;29:6323–6330. doi: 10.1038/onc.2010.368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data has been deposited in the Gene Expression Omnibus database under number GSE98059.