Abstract
The purpose of this study is to identify microRNA (miRNA) related polymorphism, including single nucleotide variants (SNVs) in mature miRNA-encoding sequences or in miRNA target sites, and their association with cardiovascular disease (CVD) risk factors in African-American population. To achieve our objective, we examined 1,900 African-Americans from the Atherosclerosis Risk in Communities (ARIC) study using SNVs identified from whole-genome sequencing (WGS) data. A total of 971 SNVs found in 726 different mature miRNA-encoding sequences and 16,057 SNVs found in the three prime untranslated region (3’UTR) of 3,647 protein coding genes were identified and interrogated their associations with 17 CVD risk factors. Using single-variant-based approach, we found 5 SNVs in miRNA-encoding sequences to be associated with serum Lipoprotein(a) (Lp(a)), high-density lipoprotein (HDL) or triglycerides, and 2 SNVs in miRNA target sites to be associated with Lp(a) and HDL, all with false discovery rates of 5%. Using a gene-based approach, we identified 3 pairs of associations between gene NSD1 and platelet count, gene HSPA4L and cardiac troponin T, and gene AHSA2 and magnesium. We successfully validated the association between a variant specific to African-American population, NR_039880.1:n.18A>C, in mature hsa-miR-4727-5p encoding sequence and serum HDL level in an independent sample of 2,135 African-Americans. Our study provided candidate miRNAs and their targets for further investigation of their potential contribution to ethnic disparities in CVD risk factors.
Keywords: microRNA, whole genome sequencing, 3’ untranslated regions, single nucleotide variant, African American, cardiovascular disease
Introduction
Cardiovascular disease (CVD) is a leading cause of morbidity and mortality in the U.S. (Benjamin et al. 2017). In 2013, 796,494 deaths were attributable to CVD, especially in people age 45 and up (777,474 deaths, 97.6%) (Xu et al. 2016). Among all ethnic groups in the U.S., African Americans (AAs) are disproportionately affected, suffering from the highest age-adjusted death rate caused by CVD (Benjamin et al. 2017). To better understand CVD pathogenesis, independent risk factors have been reported by multiple studies, such as type 2 diabetes (Kannel and McGee 1979), hypertension (Vasan et al. 2001), low high-density lipoprotein (HDL) cholesterol and high low-density lipoprotein (LDL) cholesterol levels (Barter et al. 2007). While European Americans (EAs) and AAs share similar risk factors, studies have shown that AAs suffer from higher morbidity of several of these CVD risk factors which leads to elevated incidence and increased severity of adverse cardiovascular outcomes compared to those in EAs (Graham 2015; Kurian and Cardarelli 2007; Magnani et al. 2016).
It is known that many of the CVD risk factors have an underlying genetic component (Do et al. 2013; Meyer et al. 2010; Weissglas-Volkov and Pajukanta 2010). For example, HDL, LDL and blood pressure have estimated heritability of up to 60% (Austin et al. 1987; Mitchell et al. 1996; Shih and O'Connor 2008). Only handful of studies have been conducted to investigate the difference between AAs and EAs in terms of the heritability of CVD risk factors, however, some differences have been noted (Malhotra et al. 2005). Thus, understanding genetics of these risk factors is one fundamental step in explaining inter-individual and inter-population variation in CVD risk. Although the genetic components of these risk factors have been widely investigated, e.g. through whole-genome association studies (GWAS), most of the associated variants that have been identified thus far lack clear functional interpretation. Furthermore, a large proportion of the heritability has not yet been explained, indicating that more genetic variants remain to be characterized that can contribute to the variation in CVD risk factors (Middelberg et al. 2011; Whitfield 2014).
Research has shown that microRNA (miRNA), as an important class of regulatory RNA, is involved in CVD pathogenesis (Huang et al. 2015; Wang et al. 2014). MiRNAs are ~22 nucleotide long non-coding RNAs, and they mostly bind to the three prime untranslated region (3’UTR) of protein coding genes to post-transcriptionally repress expression of target transcript(s). Same as other regulatory RNAs, the functionality of miRNA is subject to the effect of genetic variants. MiRNA-related genetic variants can impact miRNA efficacy through two different mechanisms. First, genetic variants in miRNA-encoding sequences can affect the processing of primary miRNAs (Duan et al. 2007) and precursor miRNA (pre-miRNA) (Ding et al. 2013) hairpin structures, thus influencing miRNA maturation. Second, genetic variants in miRNA target site or miRNA recognition element (MRE), the region of the target transcript with which the miRNA pairs, can disrupt the interaction between miRNA and its target and/or create new MRE for other miRNAs (Bracken et al. 2016). Moreover, population specific genetic variants could be one important source causing miRNA expression difference between AAs and EAs (Huang et al. 2011). And this miRNA level difference could lead to downstream transcriptome and proteome change, contributing to CVD disparity between populations. Therefore, miRNA related variants could be exciting new targets to help us not only understand the ‘missing heritability’ of CVD risk factors, but also explain the observed disparity in CVD risk at population level.
Unfortunately, large-scale studies that systematically investigate variants in the miRNA-related network and their relationship with CVD risk factors in human populations are still lacking. In this study, we evaluated the DNA variants in the miRNA genes and MREs in 1,900 whole genomes of AAs, a relatively under-examined population which is at higher risk of CVD, from the Atherosclerosis Risk in Communities (ARIC) study and their association with 17 CVD risk factors. We identified several novel single nucleotide variants (SNVs) that are significantly associated with some of those risk factors in AA population and validated our findings using an additional 2,135 independent AA samples.
Results
We used a systematic approach to investigate the association between miRNA-related variants and the 17 CVD risk factors measured in fasting serum samples of 1,900 AA individuals from the ARIC study (see Materials and Methods for details). In our discovery analysis, both single-variant-based and region-based association analyses were conducted. Our discovery analysis included 971 mature miRNA SNVs found in 726 mature miRNA-encoding sequences and 16,057 MRE SNVs found in the 3’UTR of 3,647 protein coding genes. Of these SNVs, we found 343 in mature miRNA-encoding sequences to be novel (Supplementary Table S1), and 5,946 in MREs to be novel (Supplementary Table S2). A total of 76.6% of all studied SNVs were rare and had a minor allele frequency (MAF) < 0.01.
Single-variant-based association analysis
As our preliminary analysis, we assumed an additive model for all SNVs being tested. We used linear regression to estimate the association between miRNA-related SNVs and CVD risk factors of interest. False discovery rate (FDR) corrected P-value of 0.05 was used to identify candidate significant results. For those candidate significant variants with at least one minor allele homozygote, we tested other models, i.e. dominant, recessive and genotypic models (Supplementary Table S3). For each CVD risk factor, only variants with a minor allele count (MAC) of at least 3 were kept in subsequent analyses to minimize the false positive rate.
SNVs in mature miRNA-encoding sequences
After filtering for the MAC, we had approximately 500 variants in the mature miRNA-encoding sequences for each CVD risk factor. The five SNVs that passed the multiple-testing corrected FDR significance threshold (Benjamini and Hochberg 1995) are listed in Table 1, and all nominally significant results are reported in Supplementary Table S4.
Table 1.
Significant associations between the mature miRNA sequence variants and CVD risk factors reached FDR-corrected P-values (n = 1,900)
| Host miRNA |
Variant | SNPs(rs#) | MAF | Trait | Beta(SE) | P-value | ΔΔG (kcal/mol) |
|---|---|---|---|---|---|---|---|
| hsa-miR-524-5p | NR_030200.1: n.20A>G* | rs374426690 | 0.00085 | Triglycerides | 1.13(0.26) | 9.67×10−6 | 0.3 |
| hsa-miR-4704-3p | NR_039853.1: n.58T>C | rs76595065 | 0.00621 | Triglycerides | 0.39(0.10) | 4.27×10−5 | −1.6 |
| hsa-miR-4782-3p | NR_039943.1: n.66T>C* | - | 0.00084 | HDL | 49.77(9.82) | 4.43×10−7 | −3.0 |
| hsa-miR-4727-5p | NR_039880.1: n.18A>C* | rs73295187 | 0.09725 | HDL | 4.31(0.97) | 9.45×10−6 | −0.2 |
| hsa-miR-449b-5p | NR_030387.1: n.27A>G | rs10061133 | 0.04637 | Lp(a) | −0.30(0.07) | 2.74×10−5 | 0.2 |
Variants that were selected to validate.
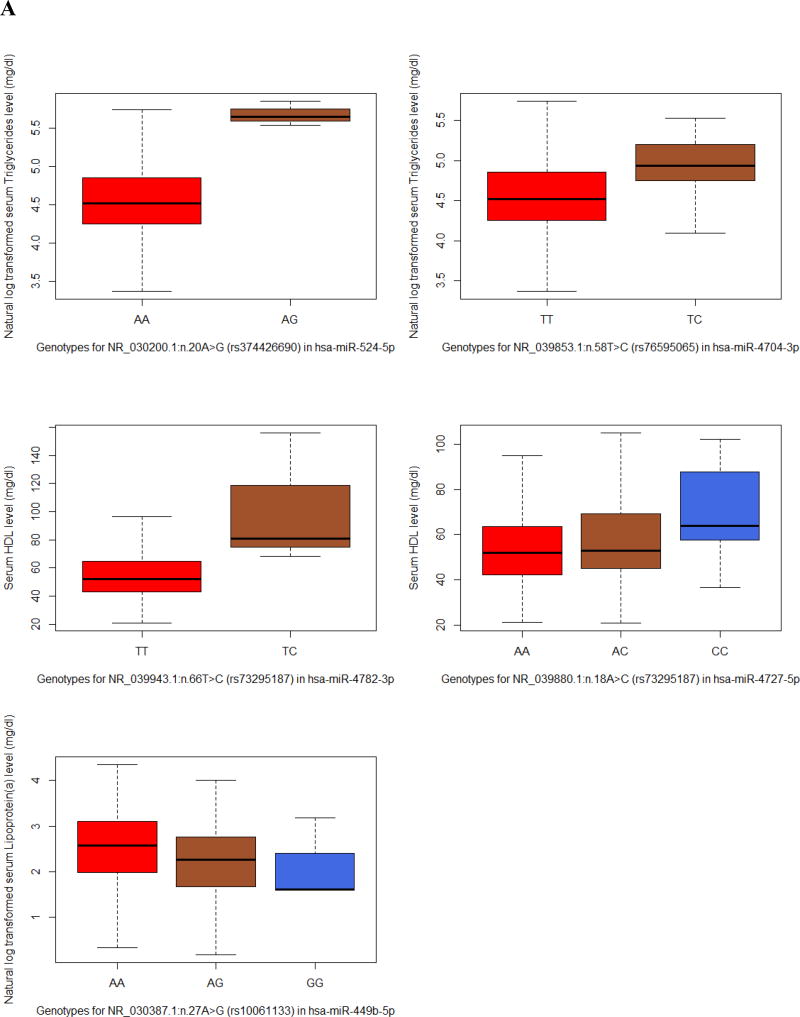
We found that the top two SNVs in our list were associated with HDL, with the minor alleles associated with an elevated level of HDL (Fig. 1A). The most significant SNV was NR_039943.1(MIR4782): n.66T>C (Beta(SE) = 49.77(9.82), Praw = 4.43×10−7) which resided in the mature hsa-miR-4782-3p encoding sequence. It also showed the highest predicted free energy change of the pre-miRNA hairpin structure (ΔΔG = −3.0 kcal/mol) by introducing the variant, indicating the increase in stability and expression of the mature miRNA. The second most significant SNV was NR_039880.1(MIR4727): n.18A>C (rs73295187, Beta(SE) = 4.31(0.97), Praw = 9.45×10−6) in mature hsa-miR-4727-5p encoding sequence.
Fig. 1.
Single-variant-based association analysis that reached FDR-corrected P-values. Red box represents major allele homozygotes; sienna box represents heterozygotes; blue box represents minor allele homozygotes. (A) Significant associations between the mature miRNA sequence variants and CVD risk factors. (B) Significant associations between the MRE variants and CVD risk factors.
Variant NR_030200.1(MIR524): n.20A>G (rs374426690) in hsa-miR-524-5p encoding sequence and NR_039853.1(MIR4704): n.58T>C (rs76595065) in hsa-miR-4704-3p encoding sequence were associated with elevated serum triglycerides (Beta(SE) = 1.13(0.26), Praw = 9.67×10−6 and Beta(SE) = 0.39(0.10), Praw = 4.27×10−5, respectively). It is worth mentioning that variant rs374426690 is in the seed region of mature hsa-miR-524-5p encoding sequence. The seed region refers to nucleotide 2–7 of a mature miRNA, which plays crucial role in miRNA target recognition and miRNA efficacy. Therefore, the observed 3-fold increase in triglyceride level of the variant carriers may be partially explained by the potential change in hsa-miR-524-5p’s targeting specificity (Fig. 1A).
One variant NR_030387.1(MIR449B): n.27A>G in hsa-miR-449b-5p encoding sequence was associated with Lipoprotein(a) (Lp(a)) (rs10061133, Beta(SE) = −0.30(0.07), Praw = 2.74×10−5).
SNVs in 3’UTR MRE
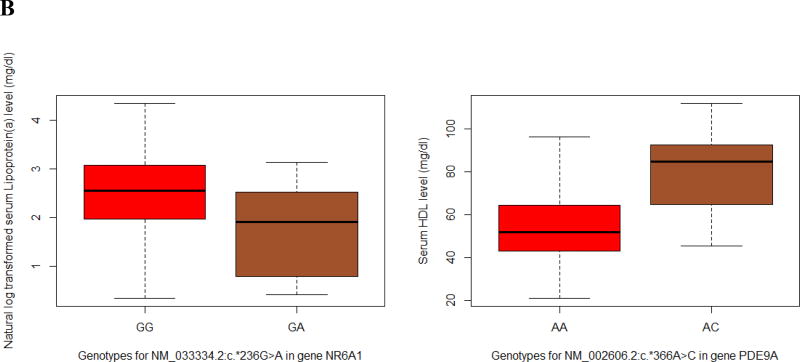
After filtering for the MAC, there were approximately 8,000 eligible variants for each CVD risk factor. Among these variants, we found NM_033334.2(NR6A1): c.*236G>A in the 3’UTR of gene NR6A1 and NM_002606.2(PDE9A): c.*366A>C in the 3’UTR of gene PDE9A that passed the FDR significance threshold to be associated with Lp(a) (P = 3.18×10−7) and HDL (P = 2.66×10−7), respectively (Table 2; Fig. 1B). Other nominally significant results are reported in Supplementary Table S5.
Table 2.
Significant associations between the MRE variants and CVD risk factors reached FDR-corrected P-values (n = 1,900)
| Gene name | Trait | Variant | MAF | Beta(SE) | P-value |
|---|---|---|---|---|---|
| NR6A1 | Lp(a) | NM_033334.2:c.*236G>A | 0.00315 | −1.34(0.26) | 3.18×10−7 |
| PDE9A | HDL | NM_002606.2:c.*366A>C* | 0.00252 | 29.23(5.66) | 2.66×10−7 |
Variants that were selected to validate.
Region-based association analysis
To increase power for analyzing rare variants, MRE variants in the same transcript were collapsed together to estimate their cumulative effect on CVD risk factors. Variants in the MRE often disrupt the binding of one miRNA and, at the same time, create new MRE for other miRNAs, which can lead to either upregulated or downregulated target expression. Therefore, sequence kernel association tests (SKAT) with two fixed thresholds (MAF ≤ 1%; MAF ≤ 5%) were implemented to account for the possible bidirectional effect of MRE variants. While the same effect of the variants could be observed in miRNA-encoding sequences, there lacked enough variants in our study for each miRNA gene to yield noticeable gain in power. Therefore, the SKAT test was only performed for the 16,057 MRE variants in the 3’UTR of 4,636 different protein coding genes. And the Bonferroni correction was used to control for the false positive rate since no independent validation was performed for these region-based results.
After Bonferroni correction, three significant genes with a minimum of 3 cumulative MAC were identified (Table 3). The most significant association was between the gene HSPA4L and cardiac troponin T (cTnT) level (P = 1.67×10−9), followed by gene AHSA2 and magnesium (Mg) (P = 1.32×10−8) and gene NSD1 and platelet count (P = 2.85×10−7).
Table 3.
Significant SKAT test results with cumulative MAC ≥ 3
| Gene name | P-value* | Qmeta | CMAF | nSNPs | Trait |
|---|---|---|---|---|---|
| NSD1 | 2.85×10−7 | 5.13 | 0.00084 | 3 | platelet count |
| HSPA4L | 1.67×10−9 | 85844.73 | 0.00142 | 3 | cTnT |
| AHSA2 | 1.32×10−8 | 830962.44 | 0.00083 | 3 | Mg |
All three associations reached genome-level significance after Bonferroni correction.
Validation with independent AA participants
Four candidate SNVs passed FDR significance threshold that were associated with CVD risk factors in our single-variant-based analysis were selected for further validation using an independent set of ARIC AA participants. These included three miRNA gene SNVs: the variant in hsa-miR-4782-3p and hsa-miR-4727-5p encoding sequences were selected because of their small P-values; the variant in hsa-miR-524-5p encoding sequence was selected because it was located in the seed region of the mature miRNA and the carriers of the minor allele showed substantial increase in triglycerides (Fig. 1A). One SNV within the MRE in the 3’UTR of PDE9A with the smallest P-value was also selected for validation.
Of these four SNVs, the variant in hsa-miR-4727-5p encoding sequence had a MAC of 2, so it was removed from further analysis. As the variant hsa-miR-4727-5p/rs73295187 has minor allele homozygotes, we examined its genotype-phenotype association under the additive, dominant, recessive and genotypic model (Table 4). The result showed significant association under the genotypic (P = 0.00135) and recessive (Beta(SE) = 13.25(3.655), P = 0.00030) models. The variant in the 3’UTR of PDE9A showed the same direction of association with HDL as our discovery result, but it was not statistically significant (P = 0.849). The variant hsa-miR-524-5p/rs374426690 showed neither a significant association nor the same direction of association as our discovery study (P = 0.915).
Table 4.
Association between variant hsa-miR-4727-5p/rs73295187 and HDL under different modes of inheritance
| Genetic model | Discovery result | Validation result | ||
|---|---|---|---|---|
|
|
||||
| Beta | P-value | Beta | P-value | |
| Additive | 4.312(0.9707) | 9.45×10−6 | 1.224(0.9018) | 0.1748 |
| Recessive | 13.47(4.572) | 0.003265 | 13.25(3.655) | 0.00030* |
| Dominant | 4.198(1.035) | 5.19×10−5 | 0.5029(0.9846) | 0.6096 |
| Genotypic | - | 2.07×10−5 | - | 0.00135* |
The mode of inheritance showed significant association with HDL after Bonferroni correction.
Discussion
Based on both discovery and validation results, the strongest evidence of association in our single-variant-based analysis was the variant hsa-miR-4727-5p/rs73295187, which showed a significant association with serum HDL level under various models. The variant carriers, especially those homozygous rare allele carriers, presented significantly elevated HDL level (Fig. 1A; Supplementary Fig. S1; Supplementary Table S7). In our discovery study, the additive (Beta(SE) = 4.312(0.9707), P = 9.45×10−6), dominant (Beta(SE) = 4.198(1.035), P = 5.19×10−5) and genotypic models (P = 2.07×10−5) were the most supportive, whereas in our validation study, the recessive (Beta(SE) = 13.25(3.655), P = 0.00030) and genotypic models (P = 0.00135) were significant (Table 4). While we do not have a clear picture of the mode of inheritance of this variant, the results are a promising starting point for further analyses. First, we used the Mfold web server (Zuker 2003) to evaluate the free energy change of the miRNA hairpin structure caused by introducing the variant. We observed a −0.2 kcal/mol free energy change which would likely increase the stability of mir-4727 hairpin structure, suggesting an elevated expression level of mature hsa-miR-4727-5p. In addition, we investigated the known HDL-related genes collected by the Global Lipids Genetics Consortium (Willer et al. 2013). To check whether this miRNA is more involved in HDL metabolism, we retrieved the summary of all miRNA family-target gene pairs from the TargetScan website, which included 7,733,952 entries of miRNA family-target gene pairs. We found that hsa-miR-4727-5p regulates 24 HDL-related genes (85th percentile), whereas the rest of the 2,484 miRNA families regulate an average of 14.98 HDL-related genes. Compared to other miRNAs (including those that showed known associations with HDL), hsa-miR-4727-5p exhibited an enriched signal for the HDL regulatory pathway. This percentile ranking for hsa-miR-4727-5p was 88th among the miRNAs investigated in our study. This indicates that the discovered regulatory effect of hsa-miR-4727-5p on HDL could be partially explained by the dysregulation of these known HDL-related genes. Moreover, using the mirPath web service (Vlachos et al. 2015), we found that two of hsa-miR-4727-5p’s targets, FUT4 and B3GNT3, were involved in glycosphingolipid biosynthesis. Animal models have shown that an inhibitor of glycosphingolipid synthesis diet has led to increased serum HDL levels (Chatterjee et al. 2014). Since the variant was not discovered in EAs in ARIC study and has a high MAF in AA population (MAF = 0.097), gene FUT4 and B3GNT3 might suggest a novel mechanism of hsa-miR-4727-5p influencing serum HDL levels specific to AA population. For future studies, multi-ethnic participants combined with whole-transcriptome analysis could help elucidate the function of hsa-miR-4727-5p and its contribution to the disparity in serum HDL levels between AAs and EAs.
Compared to variants in mature miRNA sequences, those in miRNA binding sites in general presented a weaker influence on associated phenotypes, so it would be more difficult for us to obtain sufficient power to detect such associations. For these variants, a region-based or gene-based approach would be more robust, such as the SKAT test we adopted. The three significant SKAT results we obtained were not discovered in single-variant-based association analysis, indicating that an increased power could be achieved by limiting and grouping functional variants based on the predicted MRE. All these 3 pairs of associations were novel. The association between the AHSA2 gene and serum magnesium could be explained by their shared association with inflammatory bowel disease (Jostins et al. 2012) and celiac disease (Dubois et al. 2010), where both of these diseases were reported to be associated with AHSA2 gene and paired with magnesium deficiency. Elucidating the explanations for the other two pairs of associations, namely, NSD1 and platelet count and HSPA4L and cTnT level, would require further investigation.
Our study had strengths as well as limitations. To our knowledge, our study is the only genome-level analysis of miRNA-related variants in the AA population to date, which could aid in identifying population-specific variants and potential novel therapeutic targets for CVD. Our study benefited from the large and well-designed cohort, which enabled us to discover rare variants with adequate power. Another strength was the application of the SKAT test collapsing on putative MREs, which increased our power to detect rare variants with a moderate effect size. Some limitations of our study are also worth noting. First, the MRE variants were based on in silico predictions; however, we believe, this would most likely increase our false positive rate, but not the false negative rate because: 1) TargetScan has been considered one of the best miRNA target prediction tools, and their predictions of miRNA efficacy were comparable to most of the experimental methods (Agarwal et al. 2015); and 2) TargetScan favors sensitivity over specificity, so our chance of missing variants located in the 3’UTR was relatively low. To minimize false positive results, we adopted a strict P-value threshold and conducted a validation study using independent data sets. Second, the expression profiles of miRNAs and their targets were not considered in our study. Because both the miRNA and its target genes should be abundantly expressed in the same tissue in order for them to interact, additional data (e.g. RNA sequencing data) would be needed to further demonstrate the robustness of our findings. Additionally, by taking into consideration the expression profiles, future studies could reduce the number of comparisons even further by including only those miRNA-target pairs that are sufficiently expressed in the same tissue. Third, a common drawback that our validation study and other similar studies share would be the difficulty and lack of power to validate extremely rare variants (MAF < 0.1%). It is sometimes difficult to observe the variant in validation population. Moreover, the low number of variants is vulnerable to the random effects of sampling error. Some potential solutions have been proposed by previous studies. For a single-variant-based association, one possible solution is to design the study to only sample individuals with extreme phenotypes to increase statistical power (Li et al. 2011). For a gene-based/functional annotation-based association, future study would benefit from the sequencing of the entire miRNA gene/entire 3’UTR. The possibility of identifying novel variants could increase the power to replicate the gene-phenotype association (Liu and Leal 2010). Furthermore, it might be interesting to look at the entire miRNA primary transcript instead of just mature miRNA sequences to further gain power by increasing the number of variants in gene-based analysis. This increase in power can be especially important for non-coding rare variants where their effect is considered weaker than coding variants.
In summary, we conducted a genome-wide association study of variants in miRNA and their targets with 17 CVD risk factors using 1,900 AA study participants. We identified 5 SNVs in mature miRNA sequences, 2 SNVs in predicted MREs, and 3 genes that are significantly associated with CVD risk factors. We conducted a validation study using independent AA samples and confirmed the association of an AA specific variant, hsa-miR-4727-5P/rs73295187, with HDL. Our study provided candidate miRNAs and their targets for further investigation of their potential contribution to ethnic disparities in CVD risk factors.
Materials and Methods
Study participants and phenotype measurements
All samples were taken from the ARIC study. Our discovery study population was comprised of 1,900 AAs with available whole-genome sequencing (WGS) data. The 17 CVD risk factors were obtained from fasting serum samples, including: c reactive protein (CRP), Lp(a), small dense LDL (sdLDL), pro-brain natriuretic peptide (proBNP), cTnT, hemoglobin (Hb), neutrophil count, platelet count, phosphorus, Mg, sodium, potassium, HDL, LDL, triglycerides, systolic blood pressure (SBP), and diastolic blood pressure (DBP). The detailed method for WGS and phenotype measurements of the ARIC study have been described previously (Morrison et al. 2013; Yu et al. 2016). Study participants with available WGS data were used as our discovery population (n = 1,900, median age = 52 with an interquartile range = 9, proportion of females = 0.64). A total of 2,331 AAs from the ARIC cohort with no WGS data were genotyped on candidate loci and 2,135 study participants (median age = 54 with an interquartile range = 10, proportion of females = 0.60) with at least one non-missing genotype were used as our validation population.
Identification of miRNA-related SNVs
All miRNA-related variants were annotated according to reference genome GRCh37. All mature miRNA data were downloaded from the miRBase ftp site (ftp://mirbase.org/pub/mirbase/21/genomes/). The chromosomal locations of all 2,588 mature miRNAs (excluding those in chromosome X and Y) were obtained. MiRNA-encoding variants were defined as variants located in any of these 2,588 mature miRNA sequences. The miRNA seed region was defined as position 2–7 in the mature miRNA sequence.
MiRNA target prediction data were retrieved from TargetScan version 7.0 (http://www.targetscan.org/vert_70/). The search for potential MREs was limited to the 3’UTR of protein coding genes for this is the most active and well-studied region of miRNA regulation. For MRE variants, we focused our study on variants located in the region of MRE that pairs with the seed of the targeting miRNA, because this region was most decisive in miRNA target recognition. MRE variants were defined as variants in the focused MRE regions predicted by TargetScan v7.0 all predictions (http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_70). TargetScan utilizes 14 different site- and UTR-level features to model the interaction between miRNA and its target. It requires a minimum 6-nucleotide pairing between miRNA seed and target mRNA, which has been shown to effectively predict the regulatory outcome of miRNAs (Agarwal et al. 2015).
Statistical analysis
Single-variant-based analyses were conducted using the PLINK v1.07 (Purcell et al. 2007). SKAT test was conducted using the R (R Development Core Team 2011) package seqMeta (Voorman et al. 2017). Since most of the SNVs were rare, with MAF < 0.01, we required a minor allele count (MAC) of ≥ 3 for our single variant based analysis and cumulative MAC ≥ 3 for the SKAT test.
In our discovery study, each of the 17 risk factors was analyzed separately using multiple linear regressions to examine the additive genetic model for a single-variant effect. Risk factors CRP, SBP and DBP were controlled for age, sex, body mass index and the first three principal components to account for population stratification. Risk factor neutrophil count was controlled for age, sex, three principal components and current smoking status. All other risk factors were controlled for age, sex and three principal components (Supplementary Table S6). Genotypic, dominant and recessive models were assessed for significant variants with minor allele homozygotes. Benjamini & Hochberg’s (1995) step-up FDR corrected P-values were used to identified significant results for the single-variant-based association analysis, and raw P-values were reported. While the Bonferroni correction could be too conservative for these regulatory variants and might lead to high false negatives, we used FDR as the first step screening methods, which was proven to be successful (Nelson et al. 2017). To minimize false positives, we restricted the results to have minor allele count of at least 3 and conducted validation study with a larger sample size.
The SKAT test was performed to evaluate the association between aggregated MRE variants and the 17 CVD risk factors, where MRE variants in the 3’UTR of the same gene were grouped together. R package seqMeta was exclusively used for SKAT T1 (MAF range = [0, 0.01]) and T5 (MAF range = [0, 0.05]) analysis. Since most of our variants had MAF < 0.01, the T5 test did not gain much additional power, and all of the significant SKAT test results that were reported were T1 results. Statistical significance for the SKAT test was defined as P < 1.05×10−6 (Bonferroni correction of 47,600 tests: 1300×17 T1 tests + 1500×17 T5 tests).
Four of the reported results were selected to validate our study findings. A total of 2,331 AAs were genotyped, and 2,135 had at least one non-missing genotype for these four loci. Linear regression was used to examine these four loci, controlling for age, sex and study center. Statistical significance for the validation study was defined as P < 0.0125 (Bonferroni correction of 4 tests). Detailed model and covariate information for each tested phenotype are presented in Supplementary Table S6.
DNA isolation and validation genotyping
Whole blood samples were collected from each participant upon enrollment in EDTA lavender top vacutainers which were subsequently centrifuged for collection of buffy coat aliquots that were stored at −80°C. Genomic DNA was isolated from the stored buffy coats using the Gentra Puregene Blood Kit (Qiagen N.V., Venlo, Netherlands) in accordance with the manufacturer’s instructions. Isolated genomic DNA was quantified using the Quant-iT™ PicoGreen™ dsDNA Assay Kit (ThermoFisher Scientific; Waltham, Massachusetts, USA).
Four variants were validated using TaqMan® Custom Genotyping Assays (ThermoFisher Scientific, formerly Applied Biosystems; Waltham, MA, USA) in accordance with the manufacturer’s protocol (https://tools.thermofisher.com/content/sfs/manuals/TaqMan_SNP_Genotyping_Assays_man.pdf). Alleles were detected and genotypes were called using Life Technologies’ ABI 7900HT and Sequence Detection System software. All genotype calls were visually verified and a list of SNPs genotyped, including dbSNP ID (if available), chromosome, position, functional prediction, and primer sequences are detailed in Supplementary Table S8.
Supplementary Material
Acknowledgments
The Atherosclerosis Risk in Communities study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700003I, HHSN268201700005I, HHSN268201700004I, and HHSN2682017000021). The authors thank the staff and participants of the ARIC study for their important contributions. Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Sequencing was carried out at the Baylor College of Medicine Human Genome Sequencing Center and supported by the National Human Genome Research Institute grants U54 HG003273 and UM1 HG008898.
Footnotes
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:1–38. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MA, King MC, Bawol RD, Hulley SB, Friedman GD. Risk factors for coronary heart disease in adult female twins. Genetic heritability and shared environmental influences. Am J Epidemiol. 1987;125:308–18. doi: 10.1093/oxfordjournals.aje.a114531. [DOI] [PubMed] [Google Scholar]
- Barter P, Gotto AM, LaRosa JC, Maroni J, Szarek M, Grundy SM, Kastelein JJ, Bittner V, Fruchart J-C. HDL cholesterol, very low levels of LDL cholesterol, and cardiovascular events. New England Journal of Medicine. 2007;357:1301–1310. doi: 10.1056/NEJMoa064278. [DOI] [PubMed] [Google Scholar]
- Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jimenez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu SM, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JHY, Alger HM, Wong SS, Muntner P, Amer Heart Assoc Stat C, Stroke Stat S Heart Disease and Stroke Statistics-2017 Update A Report From the American Heart Association. Circulation. 2017;135:E146–E603. doi: 10.1161/cir.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nature reviews. Genetics. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Bedja D, Mishra S, Amuzie C, Avolio A, Kass DA, Berkowitz D, Renehan M. Inhibition of glycosphingolipid synthesis ameliorates atherosclerosis and arterial stiffness in apolipoprotein E−/−Mice and rabbits fed a high-fat and -cholesterol diet. Circulation. 2014;129:2403–2413. doi: 10.1161/CIRCULATIONAHA.113.007559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Wang JX, Jiao JQ, Tu X, Wang Q, Liu F, Li Q, Gao J, Zhou QY, Gu DF, Li PF. A Pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the puma protein in apoptosis. Journal of Biological Chemistry. 2013;288:26865–26877. doi: 10.1074/jbc.M112.440453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang HY, Demirkan A, Den Hertog HM, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkila K, Hypponen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan J, Lyytikainen LP, Magnusson PK, Mangino M, Mihailov E, Montasser ME, Muller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CD, Perola M, Petersen AK, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney AS, Doring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen AL, Hayward C, Hernandez D, Hicks AA, Holm H, Hung YJ, Illig T, Jones MR, Kaleebu P, Kastelein JJ, Khaw KT, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Pak CH, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Human Molecular Genetics. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Dubois PCA, Trynka G, Franke L, Hunt KA, Romanos J, Curtotti A, Zhernakova A, Heap GAR, Adány R, Aromaa A, Bardella MT, van den Berg LH, Bockett NA, de la Concha EG, Dema B, Fehrmann RSN, Fernández-Arquero M, Fiatal S, Grandone E, Green PM, Groen HJM, Gwilliam R, Houwen RHJ, Hunt SE, Kaukinen K, Kelleher D, Korponay-Szabo I, Kurppa K, MacMathuna P, Mäki M, Mazzilli MC, McCann OT, Mearin ML, Mein CA, Mirza MM, Mistry V, Mora B, Morley KI, Mulder CJ, Murray JA, Núñez C, Oosterom E, Ophoff RA, Polanco I, Peltonen L, Platteel M, Rybak A, Salomaa V, Schweizer JJ, Sperandeo MP, Tack GJ, Turner G, Veldink JH, Verbeek WHM, Weersma RK, Wolters VM, Urcelay E, Cukrowska B, Greco L, Neuhausen SL, McManus R, Barisani D, Deloukas P, Barrett JC, Saavalainen P, Wijmenga C, van Heel DA. Multiple common variants for celiac disease influencing immune gene expression. Nature genetics. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham G. Disparities in cardiovascular disease risk in the United States. Curr Cardiol Rev. 2015;11:238–45. doi: 10.2174/1573403X11666141122220003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Gamazon ER, Ziliak D, Wen Y, Im HK, Zhang W, Wing C, Duan S, Bleibel WK, Cox NJ, Dolan ME. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8:692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou S, Zhang Y, Lv Z, Li S, Xie C, Ke Y, Deng P, Geng Y, Zhang Q, Chu X, Yi Z, Zhang Y, Wu T, Cheng J. Association of the genetic polymorphisms in pre-microRNAs with risk of ischemic stroke in a Chinese population. PLoS One. 2015;10:e0117007. doi: 10.1371/journal.pone.0117007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar J-P, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel WB, McGee DL. Diabetes and cardiovascular disease: the Framingham study. Jama. 1979;241:2035–2038. doi: 10.1001/jama.241.19.2035. [DOI] [PubMed] [Google Scholar]
- Kurian AK, Cardarelli KM. Racial and ethnic differences in cardiovascular disease risk factors: a systematic review. Ethn Dis. 2007;17:143–52. [PubMed] [Google Scholar]
- Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genetic Epidemiology. 2011;35:790–799. doi: 10.1002/gepi.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM. Replication strategies for rare variant complex trait association studies via next-generation sequencing. American Journal of Human Genetics. 2010;87:790–801. doi: 10.1016/j.ajhg.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani JW, Norby FL, Agarwal SK, Soliman EZ, Chen LY, Loehr LR, Alonso A. Racial Differences in Atrial Fibrillation-Related Cardiovascular Disease and Mortality: The Atherosclerosis Risk in Communities (ARIC) Study. JAMA Cardiol. 2016;1:433–41. doi: 10.1001/jamacardio.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Wolford JK, American Diabetes Association GSG Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 2005;54:3007–14. doi: 10.2337/diabetes.54.10.3007. [DOI] [PubMed] [Google Scholar]
- Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, Ehret GB, Boerwinkle E, Felix JF, Leak TS, Harris TB, Yang Q, Dehghan A, Aspelund T, Katz R, Homuth G, Kocher T, Rettig R, Ried JS, Gieger C, Prucha H, Pfeufer A, Meitinger T, Coresh J, Hofman A, Sarnak MJ, Chen YD, Uitterlinden AG, Chakravarti A, Psaty BM, van Duijn CM, Kao WH, Witteman JC, Gudnason V, Siscovick DS, Fox CS, Kottgen A, Genetic Factors for Osteoporosis C, Meta Analysis of G, Insulin Related Traits C Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelberg RPS, Ferreira MAR, Henders AK, Heath AC, Madden PAF, Montgomery GW, Martin NG, Whitfield JB. Genetic variants in LPL, OASL and TOMM40/APOE-C1-C2-C4 genes are associated with multiple cardiovascular-related traits. BMC medical genetics. 2011;12:123. doi: 10.1186/1471-2350-12-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–70. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Morrison AC, Voorman A, Johnson AD, Liu X, Yu J, Li A, Muzny D, Yu F, Rice K, Zhu C, Bis J, Heiss G, O'Donnell CJ, Psaty BM, Cupples LA, Gibbs R, Boerwinkle E. Whole-genome sequence–based analysis of high-density lipoprotein cholesterol. Nature Genetics. 2013;45:899–901. doi: 10.1038/ng.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, Giannakopoulou O, Jiang T, Hamby SE, Di Angelantonio E, Assimes TL, Bottinger EP, Chambers JC, Clarke R, Palmer CNA, Cubbon RM, Ellinor P, Ermel R, Evangelou E, Franks PW, Grace C, Gu D, Hingorani AD, Howson JMM, Ingelsson E, Kastrati A, Kessler T, Kyriakou T, Lehtimaki T, Lu X, Lu Y, Marz W, McPherson R, Metspalu A, Pujades-Rodriguez M, Ruusalepp A, Schadt EE, Schmidt AF, Sweeting MJ, Zalloua PA, AlGhalayini K, Keavney BD, Kooner JS, Loos RJF, Patel RS, Rutter MK, Tomaszewski M, Tzoulaki I, Zeggini E, Erdmann J, Dedoussis G, Bjorkegren JLM, Consortium E-C, CardioGramplusC4D, group UKBCCCw, Schunkert H, Farrall M, Danesh J, Samani NJ, Watkins H, Deloukas P. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team R. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2011;1:409. doi: 10.1007/978-3-540-74686-7. [DOI] [Google Scholar]
- Shih PA, O'Connor DT. Hereditary determinants of human hypertension: strategies in the setting of genetic complexity. Hypertension. 2008;51:1456–64. doi: 10.1161/HYPERTENSIONAHA.107.090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan RS, Larson MG, Leip EP, Evans JC, O'Donnell CJ, Kannel WB, Levy D. Impact of high-normal blood pressure on the risk of cardiovascular disease. N Engl J Med. 2001;345:1291–7. doi: 10.1056/NEJMoa003417. [DOI] [PubMed] [Google Scholar]
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Research. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorman AA, Brody J, Chen H, Lumley T, Davis B, Briandavisgmailcom MBD. Package ‘seqMeta’ 2017 [Google Scholar]
- Wang L, Zhi H, Li Y, Ma G, Ye X, Yu X, Yang T, Jin H, Lu Z, Wei P. Polymorphism in miRNA-1 target site and circulating miRNA-1 phenotype are associated with the decreased risk and prognosis of coronary artery disease. Int J Clin Exp Pathol. 2014;7:5093–102. [PMC free article] [PubMed] [Google Scholar]
- Weissglas-Volkov D, Pajukanta P. Genetic causes of high and low serum HDL-cholesterol. J Lipid Res. 2010;51:2032–57. doi: 10.1194/jlr.R004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB. Genetic insights into cardiometabolic risk factors. Clinical Biochemist Reviews. 2014;35:15–36. [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, Beckmann JS, Bragg-Gresham JL, Chang H-Y, Demirkan A, Den Hertog HM, Do R, Donnelly LA, Ehret GB, Esko T, Feitosa MF, Ferreira T, Fischer K, Fontanillas P, Fraser RM, Freitag DF, Gurdasani D, Heikkilä K, Hyppönen E, Isaacs A, Jackson AU, Johansson A, Johnson T, Kaakinen M, Kettunen J, Kleber ME, Li X, Luan Ja, Lyytikäinen L-P, Magnusson PKE, Mangino M, Mihailov E, Montasser ME, Müller-Nurasyid M, Nolte IM, O'Connell JR, Palmer CDCNA, Perola M, Petersen A-K, Sanna S, Saxena R, Service SK, Shah S, Shungin D, Sidore C, Song C, Strawbridge RJ, Surakka I, Tanaka T, Teslovich TM, Thorleifsson G, Van den Herik EG, Voight BF, Volcik KA, Waite LL, Wong A, Wu Y, Zhang W, Absher D, Asiki G, Barroso I, Been LF, Bolton JL, Bonnycastle LL, Brambilla P, Burnett MS, Cesana G, Dimitriou M, Doney ASF, Döring A, Elliott P, Epstein SE, Eyjolfsson GI, Gigante B, Goodarzi MO, Grallert H, Gravito ML, Groves CJ, Hallmans G, Hartikainen A-L, Hayward C, Hernandez D, Hicks AA, Holm H, Hung Y-J, Illig T, Jones MR, Kaleebu P, Kastelein JJP, Khaw K-T, Kim E, et al. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Murphy SL, Kochanek KD, Bastian BA. National Vital Statistics Reports Deaths : Final Data for 2013. National Center for Health Statistics. 2016;64:1–118. doi: May 8, 2013. [PubMed] [Google Scholar]
- Yu B, de Vries PS, Metcalf GA, Wang Z, Feofanova EV, Liu X, Muzny DM, Wagenknecht LE, Gibbs RA, Morrison AC, Boerwinkle E. Whole genome sequence analysis of serum amino acid levels. Genome Biology. 2016;17:237. doi: 10.1186/s13059-016-1106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.