ABSTRACT
Rabbit hemorrhagic disease virus (RHDV) and European brown hare syndrome virus (EBHSV) are two lagoviruses from the family Caliciviridae that cause fatal diseases in two leporid genera, Oryctolagus and Lepus, respectively. In the last few years, several examples of host jumps of lagoviruses among leporids were recorded. In addition, a new pathogenic genotype of RHDV emerged, and many nonpathogenic strains of lagoviruses have been described. The molecular mechanisms behind host shifts and the emergence of virulence are unknown. Since RHDV uses glycans of the histo-blood group antigen type as attachment factors to initiate infection, we studied if glycan specificities of the new pathogenic RHDV genotype, nonpathogenic lagoviruses, and EBHSV potentially play a role in determining the host range and virulence of lagoviruses. We observed binding to A, B, or H antigens of the histo-blood group family for all strains known to primarily infect European rabbits (Oryctolagus cuniculus), which have recently been classified as GI strains. However, we could not explain the emergence of virulence, since similar glycan specificities were found in several pathogenic and nonpathogenic strains. In contrast, EBHSV, recently classified as GII.1, bound to terminal β-linked N-acetylglucosamine residues of O-glycans. Expression of these attachment factors in the upper respiratory and digestive tracts in three lagomorph species (Oryctolagus cuniculus, Lepus europaeus, and Sylvilagus floridanus) showed species-specific patterns regarding susceptibility to infection by these viruses, indicating that species-specific glycan expression is likely a major contributor to lagovirus host specificity and range.
IMPORTANCE Lagoviruses constitute a genus of the family Caliciviridae comprising highly pathogenic viruses, RHDV and EBHSV, that infect rabbits and hares, respectively. Recently, nonpathogenic strains were discovered and new pathogenic strains have emerged. In addition, host jumps between lagomorphs have been observed. The mechanisms responsible for the emergence of pathogenicity and host species range are unknown. Previous studies showed that RHDV strains attach to glycans expressed in the upper respiratory and digestive tracts of rabbits, the likely portals of virus entry. Here, we studied the glycan-binding properties of novel pathogenic and nonpathogenic strains looking for a link between glycan binding and virulence or between glycan specificity and host range. We found that glycan binding did not correlate with virulence. However, expression of glycan motifs in the upper respiratory and digestive tracts of lagomorphs revealed species-specific patterns associated with the host ranges of the virus strains, suggesting that glycan diversity contributes to lagovirus host ranges.
KEYWORDS: EBHSV, lagovirus, RHDV, attachment, glycan, histo-blood group antigen, host range, rabbit hemorrhagic disease
INTRODUCTION
High mutation rates, vast population sizes, and short generation times make RNA viruses capable of rapidly adapting to a large number of hosts and thus prone to cross species boundaries (1, 2). Viruses are most likely to jump between closely related species (3), and this may result in occasional and unique spillover infections or severe epidemics, depending on how successfully the virus adapts to the new host population. Host switching constitutes an important mechanism of virus evolution (4) and is important in several families of viruses, including Caliciviridae. These nonenveloped viruses have a single-stranded, positive-sense RNA genome. Caliciviruses comprise five recognized genera: Norovirus, Sapovirus, Vesivirus, Nebovirus, and Lagovirus (5). The genus Lagovirus encompasses two presently recognized species: rabbit hemorrhagic disease virus (RHDV), highly fatal to the European rabbit (Oryctolagus cuniculus), and European brown hare syndrome virus (EBHSV), which affects European brown and mountain hares (Lepus europaeus and Lepus timidus, respectively) (6).
Both RHDV and EBHSV emerged in the 1980s (7, 8), and they are similar in terms of morphology, genome organization, and the epidemiological courses of the respective diseases, rabbit hemorrhagic disease (RHD) and European brown hare syndrome (EBHS) (9, 10). For EBHSV, only a single serotype is known (11), and cases of nonpathogenic forms of the virus circulating in hare populations have just been described (12). In contrast, several serological subgroups are recognized for RHDV (13), including the antigenic variant RHDVa (14) and several nonpathogenic and moderately pathogenic strains also circulating in European rabbit populations from different parts of the world (15–19). In 2010, new pathogenic RHDV strains emerged and rapidly spread throughout Europe and Australia (20–28). Due to the large number of new lagovirus strains described, confusion in the current nomenclature, and close relationship between RHDV and EBHSV, a new classification of lagoviruses has recently been proposed, distinguishing a single species with two genogroups and several genotypes within these genogroups (29). In this new nomenclature, all classical RHDV strains are classified as GI.1 and the new pathogenic strains (called RHDV2 or RHDVb) are classified as GI.2, while the related nonpathogenic strains (RCV-E and RCV-A) are classified as GI.3 and GI.4. All pathogenic EBHSV strains fall into the GII.1 genotype (see the nomenclature concordance table in the supplemental material).
Virus attachment to the cells of any new host is an initial step for virus entry and subsequent replication and thus constitutes a crucial step for a species jump. Attachment factors can include proteins, carbohydrates, and lipids. Regarding caliciviruses, the most common ligands are carbohydrates: murine norovirus and feline caliciviruses use sialic acid (30, 31), human noroviruses recognize heparan sulfate (32), and histo-blood group antigens (HBGAs) are used by noroviruses and RHDV (33–35).
In Europe, three genera of lagomorphs exist: Oryctolagus (rabbits), Lepus (hares), and Sylvilagus (cottontails), which diverged about 12 million years ago (36). Sylvilagus is a genus native to the Americas, one species of which, the eastern cottontail (Sylvilagus floridanus), has been introduced into Europe. In Italy, eastern cottontails were successfully introduced in the Po valley in the 1960s and may have caused a decline in hare populations due to competition with the native lagomorph (37). Massive introductions of S. floridanus took place in France in the 1970s and 1980s, but the species failed to establish (38). The European rabbit (O. cuniculus) is widely distributed across Europe and may occur in sympatry with hares and locally in Italy with cottontails.
Experimental cross-infections with RHDV and EBHSV in hares and rabbits have been attempted, but the results were quite disparate, with some studies failing to induce disease (39–41) while others reported successful cross-infection (42, 43). Eastern cottontail challenges with EBHSV resulted in infection and death of one animal (44). Recently, several cases of cross-species infection occurring under natural conditions were reported: an Iberian hare (Lepus granatensis) was infected with RHDV (45); an eastern cottontail (S. floridanus) was susceptible to EBHSV (44); and Lepus capensis, Lepus corsicanus, and L. europaeus were found to be fatally susceptible to the new RHDV genotype (GI.2) (46–49).
Several studies have detected the presence of low levels of RHDV RNA in micromammals, such as mice, voles, and shrews, sharing a habitat with RHDV-infected rabbits (50, 51). While these findings, likely a result of ingestion of RHDV-contaminated materials, indicate the possibility of micromammals acting as a mechanical vector for RHDV, no conclusive evidence has been presented so far suggesting that lagoviruses can productively infect species outside the lagomorph family.
Two non-mutually exclusive hypotheses for the emergence of pathogenicity in lagoviruses are currently proposed. The first suggests the emergence of virulence from nonpathogenic circulating viruses through acquisition of key mutations that, for reasons not directly related to the host, resulted in high virulence. This hypothesis is supported by the detection of antibodies against RHDV and EBHSV in samples collected before pathogenic virus emergence (52, 53) and by the characterization of widespread nonpathogenic forms (12, 15–18, 54). The other hypothesis involves a species jump, most likely from S. floridanus (25, 55), in which viruses would likely circulate as benign forms. This is consistent with the dates of introduction of S. floridanus in Europe and subsequent emergence of RHD and EBHS (7, 8, 56, 57), although lagoviruses have not yet been reported in cottontails in their native range.
We showed previously that RHDV strains recognized fucosylated glycans of the HBGA type (58). Later, binding to blood group B, A, and H type 2 epitopes in a strain-dependent manner was observed, with slight differences in specificity for A, B, or H epitopes so that not all animals were equally recognized by a single strain. Synthesis of these carbohydrate antigens proceeds by stepwise addition of monosaccharides to precursor disaccharides, such as the Galβ4GlcNAc so-called type 2 precursor that appeared to be the main precursor in rabbits (59). Addition of a fucose in α1,2 linkage to its galactose residue generates the H type 2 epitope, which itself serves as a precursor for the A and B epitopes characterized by an additional N-acetylgalactosamine or galactose, respectively, linked in α1,3 to the galactose of the precursor (60). Following devastating outbreaks, selection of resistant animals based on their weak expression of these attachment factors could be documented, showing the role of these HBGAs as functional virus ligands and of their intraspecies polymorphism in contributing to susceptibility or resistance (35, 59, 61, 62). Considering the recent reports of lagovirus species jumps and the close phylogenetic relationship of leporids (36), together with their overlapping geographic ranges (63) and an overall conservation of glycans among vertebrates, we sought to investigate the potential role of host glycan recognition in lagovirus cross-species jumps. With this aim, the abilities of the new RHDV genotype (GI.2), EBHSV (GII.1), and nonpathogenic lagoviruses from Europe and Australia (GI.3 and GI.4) to recognize glycans were examined, in addition to that of classical RHDV strains (GI.1). Furthermore, we investigated the expression of the corresponding glycan epitopes in tissues from European rabbits (O. cuniculus), European brown hares (L. europaeus), and eastern cottontails (S. floridanus) in order to relate the expression of these glycans to the documented susceptible host species.
RESULTS
Lagovirus GI strains attach to HBGA-type glycans.
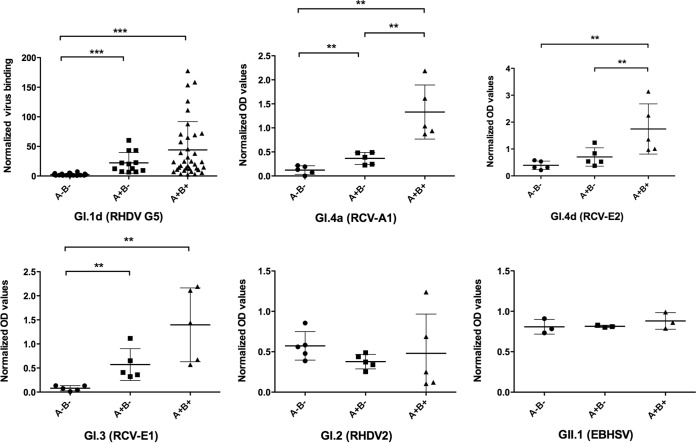
We previously showed that classical strains of RHDV (GI.1) recognized glycans of the HBGA type and that their ability to recognize individual rabbits depended on the animals' AB phenotypes. Rabbits can be classified as A+ B+, A+ B−, A− B+, and A− B−, depending on their expression of the A and B histo-blood group antigens in the gut. In order to determine if the other lagovirus strains were also influenced by the AB types of rabbits, we analyzed the binding of virus-like particles (VLPs) prepared from the new RHDV genotype (GI.2), EBHSV (GII.1), and nonpathogenic rabbit strains (GI.3 and GI.4) to duodenum scrapings of A+ B+, A+ B−, and A− B− rabbits. Due to its very low frequency, the A− B+ subgroup of rabbits was not used. A classical strain (GI.1d, or RHDV G5) was used as a control. The positions of the strains used in the lagovirus phylogenetic tree are shown in Fig. 1. As depicted in Fig. 2, binding of the classical strain (GI.1d) to tissue extracts of A+ B+ animals was significantly stronger than binding to tissue extracts of A− B− animals. Binding to A+ B− animals was also stronger than to A− B− animals. These combined observations are consistent with the previously reported preference of the strain for the B antigen over the A antigen and its weak ability to recognize the H antigen, which constitutes the precursor of both the A and B antigens. All three nonpathogenic strains tested (GI.4a, GI.4d, and GI.3) presented similar patterns of binding, showing a preferred recognition of A+ B+ animals over A+ B− animals and poor recognition of A− B− animals. In contrast, both the new pathogenic variant RHDV2 (GI.2) and EBHSV (GII.1) did not present this pattern of recognition. For these viruses, binding to rabbit tissues occurred independently of the histo-blood group AB type, suggesting either an equal recognition of the A, B, and H motifs or binding to an unrelated ligand.
FIG 1.
Phylogenetic relationships between lagoviruses studied for their glycan attachment properties. The maximum-likelihood (ML) phylogenetic tree was built as previously described using 1,000 bootstrap replicates; bootstrap values of >70 are shown (29). The scale bar represents the number of substitutions per site. The red asterisks indicate strains that had been previously studied (59); the blue asterisks indicate new strains studied in the present study.
FIG 2.
Histo-blood group A and B phenotype dependence of the binding of lagovirus strains to duodenum extracts of rabbits (O. cuniculus). VLPs from each strain were incubated on ∼3-μg/ml protein-coated tissue extracts from animals of the A− B−, A+ B−, and A+ B+ phenotypes, and their binding was quantified by ELISA. To account for variations in extract material concentrations, the data points correspond to normalized mean OD values for each animal. For GI.1d (RHDV G5), the values were normalized to ConA binding values as previously described (59). For the other strains, OD values were normalized to the protein concentrations of the extracts. The horizontal bars represent mean values and standard deviations (SD). Statistically significant differences between groups are indicated: **, P < 0.01; ***, P < 0.0001 (two-tailed Mann-Whitney test).
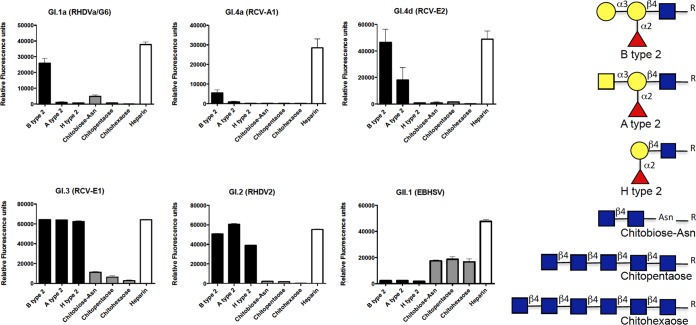
To gain further insight into the glycans potentially recognized by these lagoviruses, we assayed their binding to a printed glycan array that displayed a large number of glycan motifs (see Table S1A to F in the supplemental material). Since binding of the VLPs was detected using polyclonal antisera, with the exception of the nonpathogenic RCV-A1 strain (GI.4a), which was detected using a monoclonal antibody, natural anti-carbohydrate antibodies present in the sera gave a relatively high and uneven background. We therefore applied a stringent selection criterion for specificity by considering only the glycan motifs for which a fluorescence intensity ratio of >10 between the assay performed in the presence or in the absence of VLPs was obtained. Under these conditions, although we might have missed some weakly bound glycan motifs, the major specific ligands could be detected (Fig. 3). For all the strains, the highest signal was obtained with heparin, indicating strong recognition of this sulfated polysaccharide. Some strains also bound to sulfated oligosaccharides, as shown by the glycan microarray data (see the supplemental material). The classical RHDV strain (GI.1a), as well as the nonpathogenic strains GI.4a and GI.4d, additionally bound to HBGA motifs with a clear preference for the B type 2 epitope. For these strains, much weaker binding to the A type 2 motif was observed, consistent with their weaker ability to bind to duodenal tissue extracts from animals lacking the B epitope. The new pathogenic strain (GI.2, or RHDV2) and the nonpathogenic strain GI.3 also bound to HBGA motifs. However, they showed equally strong signals on the B type 2, A type 2, and H type 2 motifs. Importantly, in this experiment, for the GI.3 VLPs, the signal was saturating, making it impossible to determine if differential recognition of the A, B, and H type 2 epitopes could occur. We therefore tested, by enzyme-linked immunosorbent assay (ELISA), the binding of the three strains GI.2, GI.3, and GI.4d to the same oligosaccharides coupled to polyacrylamide (Fig. 4). Under these conditions, the new RHDV genotype GI.2 showed strong binding to A, B, and H type 2. This was confirmed by assaying another strain of the same genotype on a set of HBGA-related oligosaccharides immobilized on ELISA plates (the structures are provided in Table S2 in the supplemental material). Both RHDV2 (GI.2) strains showed strong binding to A, B, and H type 2. Weak binding to the Lewis Y difucosylated motif was additionally observed (Fig. 5). The much stronger binding to B type 2 over A type 2 and the very weak binding to H type 2 of the nonpathogenic GI.4d strain were confirmed (Fig. 4), while the nonpathogenic GI.3 VLPs showed strong binding to B type 2 only, indicating a similar strong preference for this motif that could not be seen in the data from the printed glycan microarray due to the signal saturation.
FIG 3.
Results from the printed glycan microarrays. Direct fluorescence measurements were obtained from the printed glycan microarrays incubated with VLPs of each virus strain. Only glycans to which specific binding by at least one of the strains was observed are shown. They are either HBGA-related structures (black bars) or GlcNAc-terminated structures (gray bars). Except for that of heparin (white bars), to which all strains bound most strongly, their structures are shown on the right (blue square, N-acetylglucosamine; yellow circles, galactose; red triangles, fucose; yellow square, N-acetylgalactosamine). The full list of arrayed oligosaccharides (n = 360) is presented in Table S1 in the supplemental material. The results are shown as mean values of six replicates; the error bars represent SD.
FIG 4.
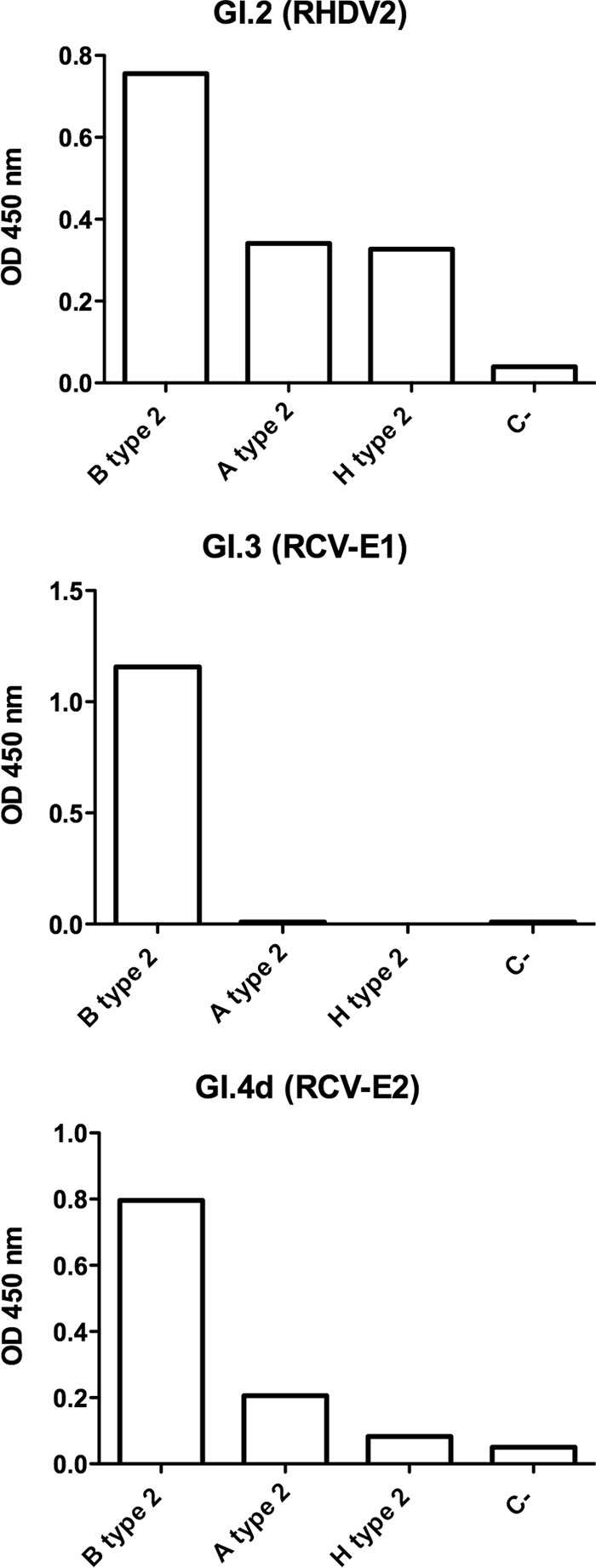
Comparison of the binding of VLPs from three lagovirus strains to A, B, and H type 2. Following incubation of VLPs to polyacrylamide-conjugated glycans on ELISA plates, binding was detected using rat polyclonal antibodies. The data are shown as mean values of triplicates; the error bars represent SD. The negative control (C-) corresponds to OD values obtained in the absence of VLPs.
FIG 5.
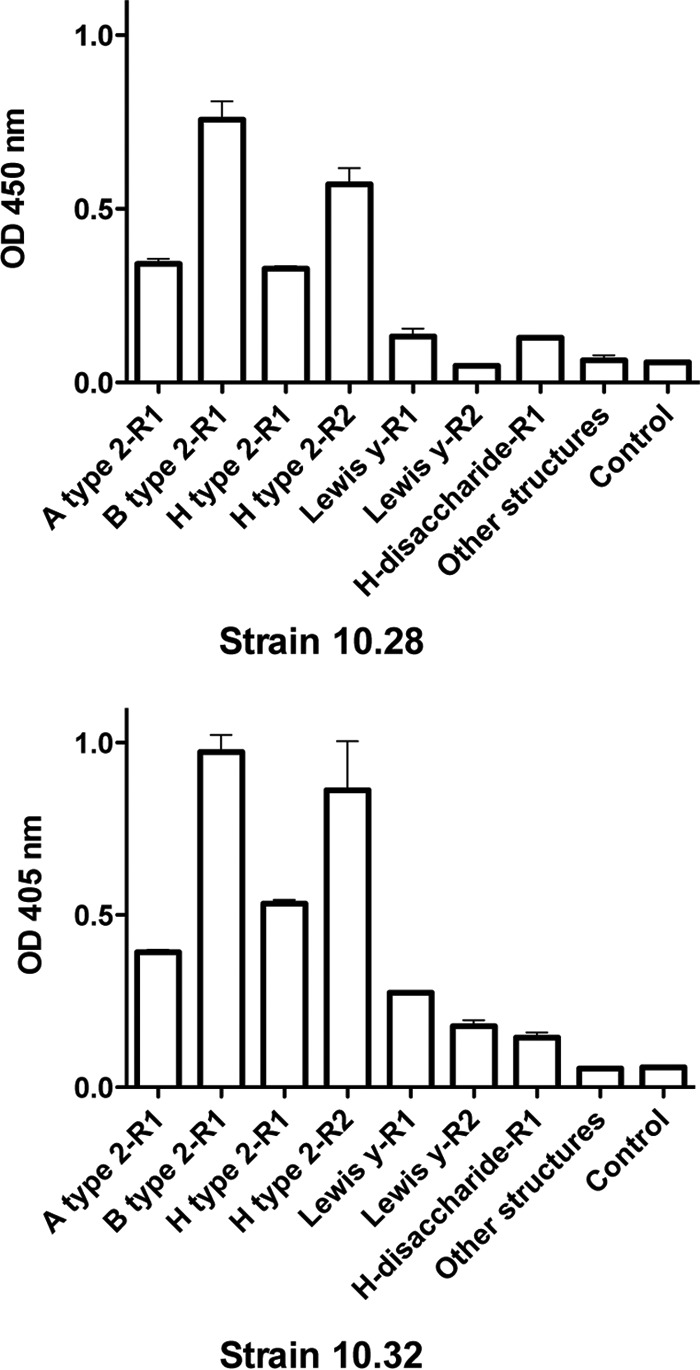
Comparison of the HBGA specificities of two strains of the new RHDV variant (RHDV2, or GI.2). Neoglycoconjugates, as polyacrylamide (R1) or human serum albumin conjugates (R2), were immobilized on ELISA plates. After incubation with VLPs from strain 10.28 or 10.32, binding was detected using a specific rat antiserum. The data are shown as mean values of triplicates; the error bars represent SD. Other structures, mean OD values of 48 other HBGA-related neoglycoconjugates to which no binding was observed (the structures are listed in Table S2 in the supplemental material).
Overall, these data are consistent with a preferential recognition of the B epitope (presented on rabbit gut tissue) by the classical RHDV strains and the nonpathogenic strains. The new pathogenic strain GI.2 also recognizes HBGA motifs, yet its preference for the B epitope over the A and H epitopes is much less pronounced, consistent with its ability to attach to duodenum extracts regardless of the AB phenotype.
EBHSV (GII.1) attaches to a distinct glycan motif.
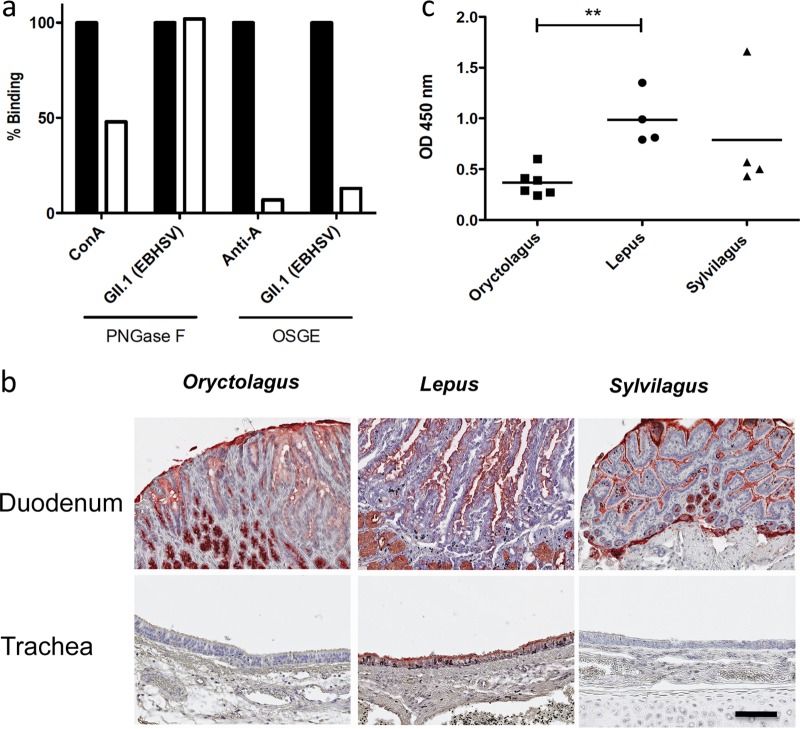
When testing by ELISA the binding of GII.1 VLPs to the set of HBGA-related glycans presented in Table S2 in the supplemental material, we failed to detect any signal (data not shown). However, on the printed glycan array, besides that of heparin, strong signals were observed for several structures composed of N-acetylglucosamines in β-anomeric linkage (Fig. 3). The printed glycan microarray also confirmed the lack of binding to HBGA-related motifs. Thus, the GII.1 strain presented a glycan specificity clearly distinct from those of the GI strains. We have previously shown that the HBGA motifs recognized by classical strains of RHDV were mainly expressed on O-glycans in the rabbit duodenum (59). To determine whether the motifs recognized by the GII.1 strain were preferentially expressed on N-linked or O-linked glycan chains, a European brown hare duodenal extract was treated with either peptide-N-glycosidase F (PNGase F) or O-sialoglycoprotein endopeptidase (OSGE) in order to selectively remove N-linked and O-linked glycans, respectively. Following treatment, the VLPs were incubated, and their binding was detected. As shown in Fig. 6a, PNGase treatment resulted in a substantial decrease (50%) in concanavalin A (ConA) lectin binding, used as a control for the efficacy of enzyme treatment. However, it had no effect on GII.1 VLP attachment. In contrast, OSGE treatment resulted in nearly complete loss of anti-A antibody binding, used as a control for enzyme efficacy, as well as in strongly decreased attachment of VLPs, indicating that GII.1 VLPs attach to O-glycans on duodenal extracts from hares, similar to the previously described attachment to O-glycans of rabbit tissue by GI strains (59).
FIG 6.
EBHSV binding to hare duodenal tissue extracts following enzyme treatments and expression of terminal GlcNAc residues in lagomorph tissues. (a) ELISA plates were coated with duodenum mucosal extracts and treated with either PNGase F or OSGE. Untreated control wells were incubated in the presence of the enzyme buffers only. Following the treatments, the tissue extracts were incubated with biotinylated ConA, an anti-A blood group MAb, or EBHSV. The data are shown as means of duplicate percentages of binding in treated wells (white bars) versus untreated wells (black bars). The ratios of the OD values of untreated wells to those of negative controls in the absence of ConA, anti-A, or EBHSV were 9.8, 11.7, and 6.4, respectively. (b) Staining of O. cuniculus, L. europaeus, and S. floridanus duodenums and tracheas was performed using biotinylated sWGA. Tissues from 6 animals of each species were analyzed. Representative images from each species corresponding to ×200 magnification are shown (scale bar = 100 μm). The specificity of sWGA attachment was assessed by coincubation with chitobiose-polyacrylamide conjugate, which completely inhibited staining, unlike a dissacharide conjugate without N-acetylglucosamine (not shown). (c) sWGA binding was quantified by ELISA on duodenum extracts coated at ∼3 μg/ml protein. The data represent mean OD values normalized for protein content variations for each individual animal. Statistically significant differences between groups are indicated: **, P < 0.01 (two-tailed Mann-Whitney test).
Since the data from printed glycan arrays indicated a specificity of the GII.1 strain for β-linked N-acetylglucosamine residues, such as GlcNAcβ4GlcNAcβ-R, that are not present on O-glycans, we hypothesized that the ligands present on hare tissues could comprise an accessible β-linked N-acetylglucosamine residue. We thus examined the expression of terminal N-acetylglucosamine residues on tissues from European hares (L. europaeus), as well as from the European rabbit (O. cuniculus) and the eastern cottontail (S. floridanus), using the lectin succinylated wheat germ agglutinin (sWGA). As shown in Fig. 6b, binding sites for this terminal N-acetylglucosamine-specific lectin were observed on surface epithelial cells of the duodenum from the three lagomorph species. Staining was also observed on the tracheal surface epithelial cells, albeit of L. europaeus only. It was undetectable on the same tissues from the other two species. Staining by sWGA of the duodenal epithelium appeared weaker on the O. cuniculus tissue sections, which was confirmed by ELISA using duodenal tissue extracts from the three lagomorph species (Fig. 6c).
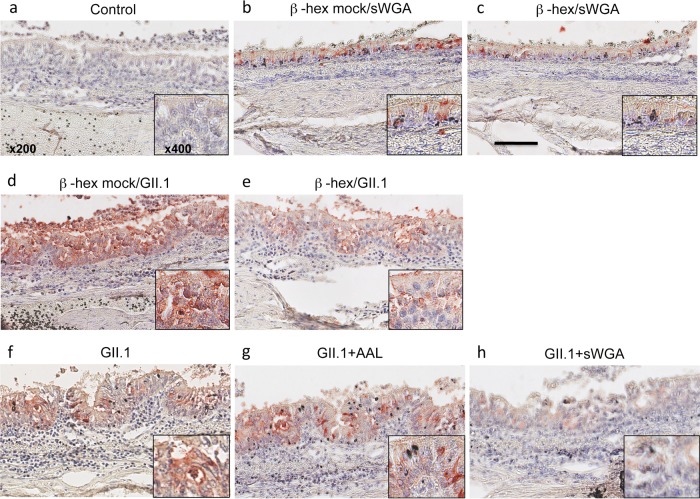
Having observed that terminal N-acetylglucosamine residues are present in a species-specific manner in the trachea and to some extent in the duodenum in leporids, we next sought to determine whether GII.1 attached to these epitopes on hare tissues. With this aim, GII.1 virus was incubated on tissue sections from hares, and the same binding pattern was observed (data not shown). Terminal N-acetylglucosamine residues were removed by pretreatment of the tissue sections using a β-hexosaminidase prior to incubation with the virus. The enzyme treatment partly removed the sWGA binding sites, ascertaining efficacy (Fig. 7). Compared with mock-treated sections (Fig. 7d), staining by GII.1 of the treated sections was also clearly diminished (Fig. 7e). Next, tracheal tissue sections were preincubated in the presence of unlabeled sWGA or a fucose-specific lectin (Aleuria aurantia lectin [AAL]) prior to incubation with the virus. Under these conditions, virus binding was clearly decreased on the sWGA-pretreated sections compared to control sections or to sections pretreated with AAL, indicating blocking of their binding sites by sWGA (Fig. 7f, g, and h). These results indicate that GII.1 (EBHSV) recognizes terminal β-linked N-acetylglucosamine residues present on O-glycans that are preferentially expressed in the trachea and in the small intestine in hares compared to the same tissues in rabbits.
FIG 7.
Blocking terminal GlcNAc residues decreases EBHSV binding. (Top row) (a) For negative-control sections, all incubation steps were followed, but the virus was omitted. (b and c) L. europaeus trachea tissue sections were either left untreated (b) or treated with β-hexosaminidase (β-hex) (c) prior to incubation with sWGA, showing a decrease of accessible terminal N-acetylglucosamine residues following treatment. (Middle row) Tissue sections were either left untreated (d) or treated with β-hexosaminidase (e). Detection of virus binding was then performed using a monoclonal anti-EBHSV antibody (5F5). (Bottom row) L. europaeus tissue sections were incubated with the virus alone (f), the virus in the presence of the unlabeled fucose-specific lectin AAL (g), or the virus in the presence of unlabeled sWGA (h). The images correspond to ×200 magnification (scale bar = 100 μm), or to ×400 magnification (insets).
Variations in histo-blood group antigen expression across lagomorph species.
The differences in expression of the N-acetylglucosamine residues recognized by the GII.1 VLPs between hares and rabbits were associated with the host species specificity of EBHSV (GII.1), which readily infects European brown hares but is not known to infect European rabbits. This prompted us to examine the expression of the A, B, and H motifs in the same lagomorph tissues in search of potential host preferences based on the abilities of the diverse virus strains to recognize these glycan motifs. The results are summarized in Fig. 8 and Table 1.
FIG 8.
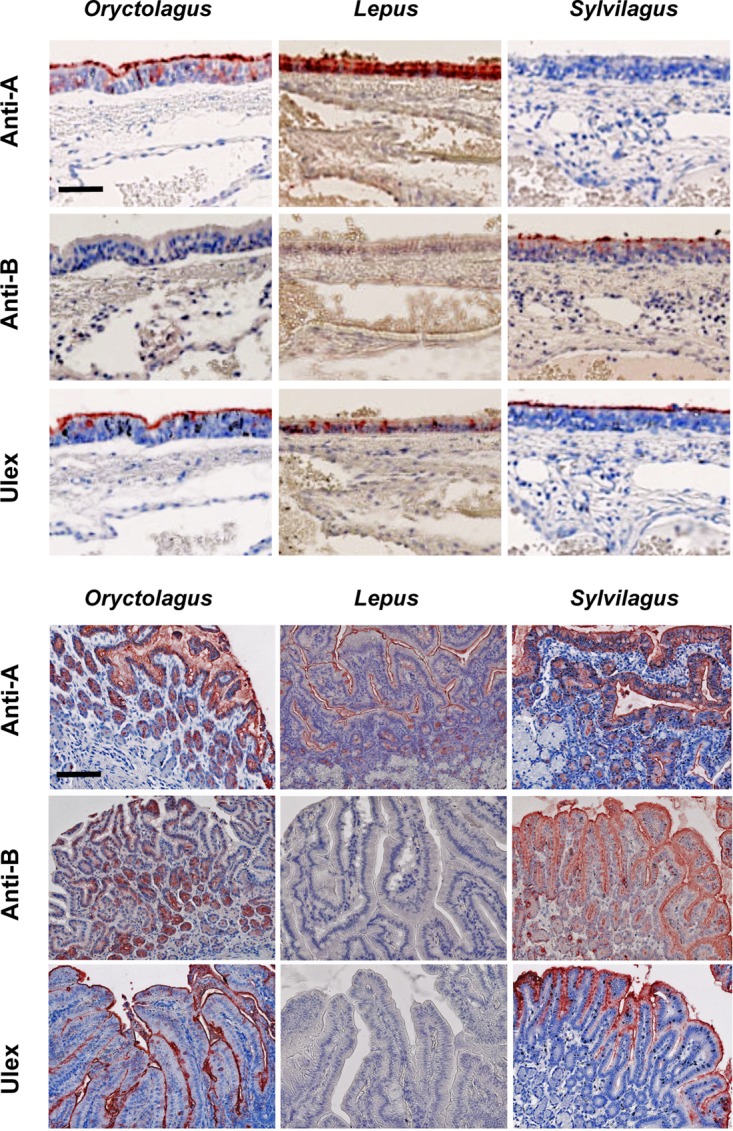
Expression of A, B, and H antigens in lagomorph tissues. Staining of sections of tracheas (top) and duodenums (bottom) from 3 species of lagomorphs was performed using specific monoclonal antibodies for A and B antigens (anti-A and anti-B) and UEA-I for H antigen. Tissues from 21 O. cuniculus, 6 L. europaeus, and 6 S. floridanus individuals were analyzed. Representative images corresponding to ×400 magnification (top; scale bar = 50 μm) or ×200 magnification (bottom; scale bar = 100 μm) for each species are shown. No detectable staining was visible on negative controls performed in the presence of irrelevant primary antibodies (not shown).
TABLE 1.
Expression of ABH antigens in tissues of lagomorphs
| Antigen | Expressiona |
|||||
|---|---|---|---|---|---|---|
| Tracheab |
Small intestinec |
|||||
| Oryctolagus | Lepus | Sylvilagus | Oryctolagus | Lepus | Sylvilagus | |
| Anti-A | 5/21d | 6/6 | 0/6 | 5/21d | 6/6 | 4/6 |
| Anti-B | 0/21 | 0/6 | 6/6 | 16/21d | 0/6 | 6/6 |
| UEA-I | 20/21 | 0/6e | 6/6 | 19/21 | 0/6f | 6/6 |
Number of animals with positive staining/total number tested.
Labeling of the epithelium.
Labeling of villus and crypt epithelial cells.
A and B antigen expression is polymorphic, with variable frequencies between populations (59).
Weak labeling of glandular cells of the submucosa only.
Weak, irregular labeling only.
Major differences in A, B, or H antigen expression were observed across the three species. Indeed, A antigen expression was clearly confirmed in the trachea and the small intestine in O. cuniculus and L. europaeus, although not all O. cuniculus individuals expressed the antigen due to the genetic polymorphism of A antigen expression. Accordingly, the animals that expressed the A antigen did so in both the trachea and the small intestine. In contrast, none of the six S. floridanus animals tested expressed the A antigen in the trachea, despite clear expression in the gut by four of them. The B antigen was not detected in the trachea in O. cuniculus, although it was strongly expressed in the small intestine of 16 out of 21 animals tested. The remaining 5 animals were classified as B−, as previously described (59). The same antigen was completely absent from all six L. europaeus animals tested, but it was detected in both the trachea and the small intestine in all S. floridanus individuals. Interestingly, as previously described (59), expression of the B antigen in the O. cuniculus small intestine was patchy and heterogeneous, with areas of strong expression among negative areas. In contrast, in S. floridanus, staining by anti-B was always strong and homogeneous. Finally, the H type 2 antigen, detected by the Ulex europaeus I (UEA-I) lectin, was found to be strongly expressed in the trachea and the small intestine in both O. cuniculus and S. floridanus, but not in the corresponding tissues of L. europaeus, where it was either completely undetectable or present at very low levels, but not at the apical surfaces of cells. These data indicate that, besides the intraspecific genetic polymorphism of A and/or B antigen expression, there are overarching species-specific features. Strikingly, hares lack both the B and H antigens in the trachea and the small intestine. O. cuniculus and S. floridanus can express both the A and B antigens in the intestine, but in the trachea, only the A antigen is present in O. cuniculus, and conversely, only the B antigen is present in S. floridanus. The trachea and small intestine were chosen, as they are easy to sample in wild animals captured in naturo. Expression of the same glycans in the noses of two hares and two European rabbits was also assessed to confirm that their expression was similar to that in the trachea. The results indicated that tracheal glycan expression corresponds to that in the nasal epithelium, a likely portal of entry for lagoviruses in a natural setting (Fig. 9).
FIG 9.
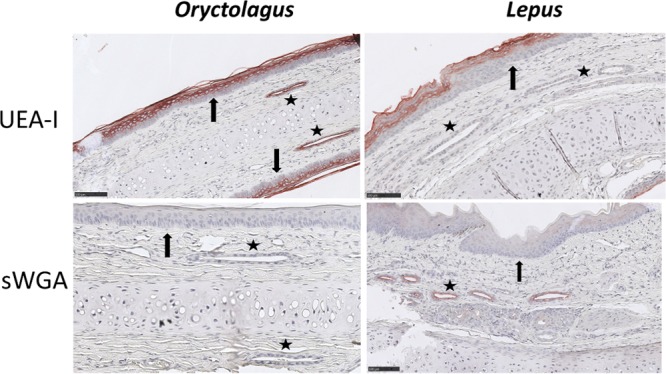
Glycan expression in the trachea matches that in the nose. Shown is staining of the nose epithelium of European rabbit (O. cuniculus) and European brown hare (L. europaeus) by the UEA-I and sWGA lectins illustrating binding patterns compared to the trachea. The stars indicate glandular ducts, strongly stained by UEA-I in O. cuniculus only and inversely by sWGA in L. europaeus only. The arrows indicate the dying and dead cells of the stratum granulosum and stratum corneum layers of the nose stratified epithelium that were labeled by the UEA-I lectin in both species. Scale bars = 100 μm.
DISCUSSION
RNA viruses have commonly crossed species barriers, probably because of their high mutation rates, short generation times, and large population sizes, which enable them to quickly adapt to new hosts (1). Phylogenetically related species are more likely to experience species jumps, as they have more similarity in cell receptors and other components critical to viral replication (64). Caliciviruses appear to be good models to study the role of molecular factors involved in species jumps, as they are quickly evolving single-stranded RNA viruses and as several instances of likely host species jumps have been reported within the virus family, in particular, the vesiviruses and noroviruses (65–67). Within the genus Lagovirus, cross-species infections involving closely related host species of the family Leporidae (order Lagomorpha) have recently been reported, including the classical RHDV (GI.1) in L. granatensis, the new RHDV genotype (GI.2) infecting several hare species, and EBHSV (GII.1) infecting S. floridanus (44–49). Previous studies conducted on RHDV showed involvement of glycans of the HBGA type in attachment of the virus to epithelia of the upper respiratory tract or of the gut, which constitute the most likely common portals of entry of the virus (35, 58, 59, 61, 62). HBGA structures are highly conserved among vertebrates, and this conservation might facilitate cross-species infections. However, species differences also exist in terms of HBGA expression. They include the absence of motifs based on type 1 precursors in many species; loss of the alpha-Gal motif in apes; and differences in cellular distribution, such as the lack of ABH antigens on the erythrocytes of most mammals (68–70). Nevertheless, comparative analysis of the expression of these glycan epitopes in lagomorph species had never been performed. Here, we analyzed the expression of these glycans in the trachea, small intestine, and nose.
Overall, our observations are consistent with a role of the glycan attachment factors in determining lagovirus host specificity (or lack thereof) in three species of leporids.
Indeed, we observed that all GI strains (pathogenic or not) could attach to O. cuniculus epithelial cells through attachment to either the A or the H antigen in the trachea and to the B antigen in the small intestine. We had previously observed that European rabbits' survival of outbreaks of RHD was associated with the absence or low expression of these antigens, clearly establishing their role in the infection process in vivo (35, 59, 61, 62).
The lack of infection of hares by many GI strains might reflect the lack of expression of the B antigen and the lack of accessibility of the H antigen, since in hares, among HBGAs, only the A antigen appears to be available at cell or tissue surfaces. Early strains of RHDV (GI.1c) that emerged during the second half of the 1980s did not recognize the A antigen at all (59), which might explain why these strains did not infect hares even under experimental conditions (39–41). However, the virus evolved to progressively acquire recognition of the A antigen, diminishing in parallel its ability to recognize the H antigen (59). This led to the circulation of strains, such as those that we used in the present study, that can bind to A+ B− animals but hardly recognize A− B− animals. This newly evolved ability of RHDV (GI.1) strains to bind to the A antigen might explain the recent report of natural infection of hares in the Iberian Peninsula (45).
The broad HBGA specificity of the new RHDV genotype (GI.2) that recognizes A, B, and H type 2 epitopes almost equally well is associated with its ability to infect both European rabbits and hares, the latter species being recognized through the presence of the A antigen in the trachea and the small intestine. Interestingly, evidence for the development of genetic resistance to classical RHDV (GI.1) has been obtained in Australia (71), and selection of genetically resistant rabbits involving the B− phenotype was observed in Australia and France (35, 59), suggesting that the broad HBGA specificity of the GI.2 strains might allow infection of animals resistant to GI.1 strains. This could help to explain why the GI.2 virus has been spreading so successfully and supplanting GI.1 strains. In contrast, the exclusive expression of terminal N-acetylglucosamine residues on the trachea in hares and their lower expression in the gut in European rabbits correlate with the host species specificity of EBHSV (GII.1), which does not infect European rabbits. In terms of glycan specificity, EBHSV thus appears quite distinct from the GI lagoviruses. Nonetheless, both types of strains attach to O-glycans expressed at the surface of the upper respiratory tract and of the small intestine, indicating a common mechanism of infection. Unlike RHDV, EBHSV did not show any agglutination of human erythrocytes regardless of their ABO phenotype (data not shown) or binding to the polymorphic HBGAs. Instead, it attached to terminal β-linked N-acetylglucosamine residues, which are present in glycans of all species in all cell types. These motifs are generally masked by addition of other monosaccharides. The patterns of binding to tissues observed using the lectin sWGA and VLPs from EBHSV indicate restriction in their availability as ligands. In a previous study, the occurrence of O-glycans from the rabbit small intestine presenting terminal β-GlcNAc residues was observed by mass spectrometry (59). However, these structures had relatively low abundance. It would be interesting to perform the same type of analysis on hare tissues in order to determine the precise O-glycan oligosaccharides that harbor these terminal β-GlcNAc residues and their abundance. It would also be interesting to determine if an intraspecific polymorphism of their expression exists that might contribute to generating different individual susceptibilities to the virus, similar to what was observed between HBGAs and susceptibility to RHDV. Figure 10 summarizes the expression of carbohydrate epitopes in the tracheas of the three lagomorph species studied (O. cuniculus, S. floridanus, and L. europaeus) and their recognition by the known lagovirus genotypes.
FIG 10.
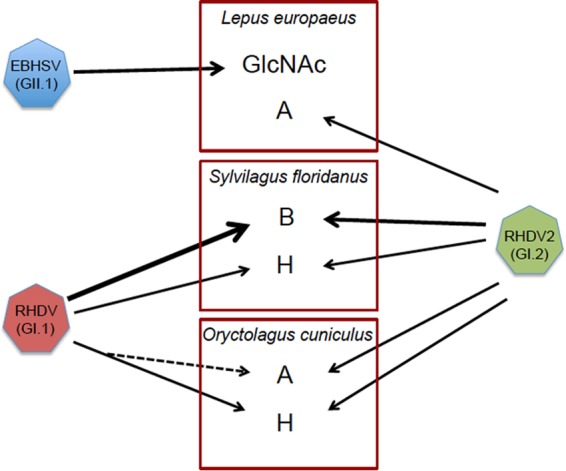
Relationships between species-specific glycan expression and glycan recognition by lagoviruses. Expression of glycans in the trachea is shown for each host species (A, B, H, and GlcNAc). The arrows indicate glycan recognition of each virus, with thicker arrows showing preferred recognition. The dashed arrow indicates acquisition of A antigen recognition by GI.1 through coevolution with O. cuniculus (nonpathogenic GI viruses are similar to late GI.1 that binds A antigen). The broad HBGA specificity of GI.2 may contribute to explaining its wider host range and ability to replace GI.1 strains, while conversely, the unique restricted specificity of GII.1 for N-acetylglucosamine (GlcNAc) is associated with its limited host range.
The glycan specificity of lagoviruses nevertheless fails to explain the lack of infection of S. floridanus by classical RHDV (GI.1) both experimentally and in naturo in northern Italy, where European rabbits and free-living cottontail rabbits exist in sympatry (44), despite strong binding to B antigen expressed in the trachea and the small intestine of that potential host species. It is also unclear why infection of S. floridanus by EBHSV occurs (44) despite the absence of expression of terminal β-GlcNAc residues in the trachea in these animals. In that case, the role of glycan recognition cannot be excluded, since the virus strain that we used was older than the strain for which records of cross-infection have been obtained, and differential glycan specificity has evolved since then (35). Alternatively, an intraspecific genetic polymorphism of β-GlcNAc expression might also exist, but too few animals were studied to investigate this possibility, which warrants further study. Regardless, it is most likely that the absence of cross-infection involves factors related, not to the initial attachment step, but rather, to subsequent steps within the infection cycle, such as entry receptor incompatibility, replication mechanism incompatibilities, or the presence of species-specific antiviral factors.
Printed glycan microarrays revealed that a common feature of all the virus strains that we examined was strong binding to heparin. Binding to heparin cannot be compared to that on the other glycans that were printed on the microarray, since it is a polysaccharide constituted of a large number of repeating units. It is structurally similar to heparan sulfate, which is expressed by all animal tissues. Heparan sulfate is a complex polysaccharide composed of repeating variably sulfated disaccharide units that can display remarkable structural diversity (72). Interactions of heparan sulfate with proteins are established mainly through electrostatic interactions of its negatively charged sulfates with basic amino acids (73). Heparan sulfate is a primary receptor or coreceptor for viruses from various families, including Parvoviridae, Retroviridae, Herpetoviridae, and Togaviridae, as well as Filoviridae (74–76). Within the family Caliciviridae, binding of GII noroviruses to heparan sulfate has been reported (32). However, its exact role in the infection process remains unknown. Our observation of the binding of diverse lagoviruses to heparin suggests a property shared between lagoviruses and other noroviruses, but its functional importance remains to be examined.
Highly virulent lagoviruses have emerged independently on at least three separate occasions, first in the early 1980s with EBHSV (GII.1) and RHDV (GI.1), and then in 2010 with the new RHDV genotype (GI.2). The causes of the emergence of these highly pathogenic strains in a short interval are not known at present. Clearly, the ability to recognize HBGA glycan motifs shared by pathogenic and nonpathogenic strains indicates that it does not constitute a virulence factor and therefore cannot explain the acquisition of virulence by some strains. Cocrystallization of the P domain from a GI.2 strain with the H type 2 trisaccharide allowed characterization of the carbohydrate-binding site (77). Examination of presently available protein sequences indicated that the main amino acids involved in glycan binding are conserved across all lagoviruses, including GI and GII. Nonetheless, amino acid differences exist within the binding site itself or in its vicinity (data not shown). They may contribute to explaining the differences in glycan specificity of the various strains. Evolution of the specificity for HBGAs of GI.1 strains has already been documented and was associated with changes in the frequencies of HBGA polymorphisms within wild rabbit populations, strongly suggesting a phenomenon of coevolution between the virus and its host (35, 59). Thus, conservation of the carbohydrate-binding site, albeit with some variation, underscores its major importance in adaptation to the host species. A recent study pointed to the presence of several amino acid- coding positions of the GI virus genome that distinguished pathogenic strains from nonpathogenic strains (78). None of the positions concerned the carbohydrate-binding site, consistent with the notion that that part of the capsid protein does not directly contributes to virulence. It has been proposed that the emergence of virulence might have involved a species jump, with S. floridanus being a possible candidate species of origin because of the coincidence between its repeated introductions in Europe from the 1970s and the emergence of highly pathogenic lagoviruses (55). Here, we observed that the B histo-blood group antigen, which is the preferred ligand of all GI strains, is strongly and homogeneously expressed in the trachea and small intestine in S. floridanus, which is compatible with this hypothesis (Fig. 10). Regardless of the answer to this difficult question, the data presented here strongly indicate that species-specific glycan expression represents an important element of the host species specificity and range of lagoviruses.
MATERIALS AND METHODS
VLPs and virus preparations.
VLPs from the first nonpathogenic lagovirus strain described in Australia were prepared and described previously (79). The strain was originally called RCV-A1 (GenBank accession number EU871528) and is now called GI.4a. VLPs of seven other strains of lagoviruses were generated using a previously described method (80). Recombinant baculoviruses containing the VP1 sequence from the following viruses were generated: two classical RHDV strains from France, a GI.1d strain and a GI.1a strain (previously G5 and G6, or RHDVa), GenBank accession numbers AM085133 and AJ969628, respectively; two strains of the new pathogenic genotype from France (GI.2; previously RHDV2, or RHDVb), strains 10.28 (GenBank accession number HE800531) and 10.32 (GenBank accession number HE800532); two nonpathogenic strains from France, i.e., a GI.3 strain previously called RCV-E1, strain 06-11 (GenBank accession number AM268419) and a GI.4d strain previously called RCV-E2, strain B09/08-117 (GenBank accession number LT708121); and one strain of EBHSV (GII.1), strain B/EBHS/6, from France (GenBank accession number KY801206). Briefly, recombinant baculoviruses expressing the VLPs were used to infect Sf9 cells. Five days postinfection, cellular debris and baculovirus were removed by centrifugation (12,000 × g; 30 min), and freeze-thaw cycles released VLPs from the cells. The supernatant was once again centrifuged at 100,000 × g for 3 h, and the pellets were resuspended in 200 μl phosphate-buffered saline (PBS). A cesium chloride solution at a 1.3-g/ml density was added to the preparation and ultracentrifuged for 18 h at 180,000 × g. Fractions (500 μl) were collected by puncture, and their refractive index was determined on a Nanodrop 2000 spectrophotometer (ThermoFisher Scientific). Fractions with refractive indexes between 1.362 and 1.363 were kept and dialyzed against PBS. Cesium chloride was eliminated through serial washes on Vivaspin columns (30,000 MWCO PES; Sartorius, Göttingen, Germany). The integrity and quality of the VLPs were checked by Coomassie blue staining of SDS-PAGE gels and Western blotting. Protein amounts were quantified using a Nanodrop 2000 spectrophotometer.
In some experiments, virus samples were used. They were obtained from liver homogenates (10% [wt/vol] in PBS) from dead animals prepared as previously described (59).
Antibodies to VLPs.
A previously prepared high-titer rabbit serum, Lp4, was used for GI.1d and GI.1a detection (59). A hyperimmune serum that recognizes the new RHDV genotype (GI.2) and the nonpathogenic lagoviruses RCV-E1 and RCV-E2 (GI.3 and GI.4d) was produced. For this, two rats were immunized through four subcutaneous injections of 40 μg VLPs from the 10-28 strain (GI.2) at 2-week intervals. The anti-EBHSV polyclonal antibody was generated by serial inoculation of two rats with VLPs from GII.1 (EBHSV). Recognition of target VLPs using the antibodies generated was confirmed using ELISA. Rat inoculations were performed at the animal experimentation core facility of the University of Nantes (IRT-UN facility agreement number 4478) and were approved by the national ethics review board of the French Ministry of Enseignement Supérieur et de la Recherche (project license number CEEA.2012.83). The animal care and use protocol adhered to European Directive number 2010/063 and to the French national regulation (Décret no. 2013-118 du 1er Février 2013 Relatif à la Protection des Animaux Utilisés à des Fins Scientifiques). A previously described mouse monoclonal antibody (MAb 11F12) that specifically recognizes the Australian rabbit calicivirus RCV-A1 (GI.4a) was also used for detection of RCV-A1 VLPs (79). Isolation of the first nonpathogenic Australian lagovirus strain (RCV-A1) that allowed preparation of VLPs and their antibodies was approved by the CSIRO Sustainable Ecosystems Animal Ethics Committee (SEAEC 06-31) and performed using the guidelines of the Australian code of practice for the care and use of animals for scientific purposes.
Tissue sampling.
Tracheal and duodenal tissue samples from European rabbits, hares, and cottontails were used for histochemistry following fixation in formalin for 48 to 96 h and embedding in paraffin. Duodenum mucosal extracts were prepared as follows. The first 5 cm posterior to the gastroduodenal junction was removed after clearing the section of intestinal contents, and the sample was vigorously rinsed in PBS and stored in RNAlater (Ambion, Life Technologies, Paisley, United Kingdom) at −20°C. One-centimeter sections of the duodenum were then rinsed in PBS, opened, and scraped into RLT lysis buffer (Qiagen, Hilden, Germany) containing β-mercaptoethanol. The tissue scrapings were homogenized and boiled for 10 min. After clearing, the protein contents were determined using a Nanodrop 2000 apparatus and kept at −20°C prior to being used for ELISA.
For analysis of the classical RHDV G5 strain (GI.1d), 103 wild rabbits (O. cuniculus) harvested by hunting in Southern France were used. For the other analyses, rabbit samples were collected from 12 domestic animals and 9 wild animals from western France that had been freshly killed by hunters. Four samples of European brown hares (L. europaeus) from Spain reared in captivity were collected. Samples from two additional L. europaeus animals hunted in western France were also used, and six eastern cottontails (S. floridanus) reared in captivity were bought from a French farm.
The use of domestic European rabbits was carried out in a group V animal facility (agreement no. 44267) and approved under specific agreement no. 006933 by the National Committee of Ethics on Animal Experiments of the French Ministry of Higher Education and Research. The breeders from whom animals were obtained approved the study. Animal care and handling were performed in strict accordance with the recommendations of the French National Guide for the Ethics of Animal Experiments, and euthanasia was performed using xylazine and ketamine anesthesia. Tissues from wild European rabbits and hares were taken from animals killed by hunters during rabbit and hare hunting seasons in the Aveyron area of France. No wild animal was killed specifically for the purpose of this study, and the hunters approved the study. Therefore, no animal ethics permit was required.
Phenotyping ELISA.
Duodenum scrapings were phenotyped using ELISA. Briefly, the duodenum scrapings were diluted in duplicate in 11 2-fold dilutions, with final dilutions ranging from 1/100 to 1/102,400 in 0.1 M sodium carbonate buffer, pH 9.5, on a Nunc MaxiSorp plate (ThermoFisher Scientific, Waltham, MA). Antibody-dependent assays were blocked with 5% nonfat dry milk (Régilait, Saint-Martin-Belle-Roche, France) diluted in PBS, while lectin assays were blocked with Synblock (AbD Serotec, Oxford, United Kingdom). The A antigen was detected using mouse monoclonal anti-A antibody (2A21), and the B antigen was detected using a specific mouse monoclonal antibody (B49) (81). H type 2 expression was determined using the horseradish peroxidase (HRP)-conjugated lectin UEA-I at 2 μg/ml (Sigma-Aldrich, St. Louis, MO). Expression of terminal β-linked N-acetylglucosamine residues was determined using sWGA at 10 μg/ml (Vector Laboratories, Burlingame, CA). Secondary HRP-conjugated anti-mouse (Uptima/Interchim, Montlucon, France) was used for detection of anti-A and anti-B primary antibodies. HRP-avidin D (Vector Laboratories) was used for detection of UEA-I and sWGA. TMB (BD Bioscience, San Jose CA) was used as a substrate for all assays, and optical density values were measured at 450 nm (OD450).
VLPs and virus binding assays.
VLP binding to animal duodenum scrapings or synthetic sugars was analyzed as previously described (59, 82). Briefly, plates were coated overnight at 4°C with duodenum scrapings, normalized for protein concentrations, and diluted at a range of dilutions starting from 200 μg/ml in 0.1 M sodium carbonate buffer, or with 1 μg of synthetic sugars in the same buffer. The plates were blocked with 5% nonfat dry milk diluted in PBS or distilled water. VLPs at 8 μg/ml were then added to the plates. Binding of VLPs was detected using primary antibodies against the strains described above. Secondary anti-rabbit, anti-rat, or anti-mouse antibodies conjugated with HRP were then used, depending on the primary antibodies. TMB (BD Bioscience, San Jose, CA) was used as a substrate for all the assays, and OD450 values were measured.
In the case of EBHSV (GII.1), additional assays were performed in order to test the effect of enzyme treatments on binding, as follows. Nunc MaxiSorp plates were coated with hare duodenum tissue extracts at 200 μg/ml at 4°C overnight. Prior to the blocking step, the plates were incubated for 6 h at 37°C with 1,000 U of PNGase F (New England BioLabs, Evry, France). After incubation with the enzyme, ELISA steps were performed as described above for binding to tissue scrapings. For treatment with OSGE (Cedarlane, Burlington, Canada), a slightly modified protocol was adopted. Hare duodenum samples were incubated overnight at 37°C with 15 μl (36 μg) of the enzyme, and then Nunc MaxiSorp plates were coated with the samples at 4°C overnight. After blocking the plate, binding was carried out as described above.
Printed glycan microarray assay.
Printed glycan array slides were manufactured and profiled as described previously (83). Briefly, six replicates of 353 mono- and oligosaccharides (see Table S1 in the supplemental material) (50 μM as ω-aminopropyl glycosides of 95 to 98% purity) were diluted in 300 mM PBS-0.001% Tween 20 (pH 8.5) and printed by robotic-pin deposition on N-hydroxysuccinimide-activated glass slides (Schott Nexteron); the array also contained 150 bacterial polysaccharides (not shown in Table S1). Free N-hydroxysuccinimide-activated groups were blocked with 25 mM ethanolamine in 100 mM boric acid with 0.2% Tween 20 at a final pH of 8.5. The slides were then rinsed with MilliQ grade water, dried, and stored at 4°C in a desiccator.
Each VLP, diluted in PBS-Tween bovine serum albumin (BSA) (Sigma, St. Louis, MO; 0.1% [vol/vol] Tween 20 and 1% [wt/vol] BSA) was incubated on slides in a humid chamber overnight at 4°C with gentle shaking. Monoclonal antibodies or hyperimmune sera, also diluted in PBS-Tween BSA, were incubated at 37°C for 60 min with gentle shaking. A final incubation with Cy5-labeled secondary antibodies diluted in PBS-Tween (0.01% [vol/vol] Tween 20) was performed at room temperature for 60 min at 37°C. Between incubations, the slides were washed with a series of 0.1% and 0.01% Tween 20 in PBS. Fluorescence signals were measured with an Agilent scanner (G2565CA), using the same settings for all assays, and analyzed using ImaGene analysis software version 7.5 (BioDiscovery, El Segundo, CA, USA).
Immunohistochemistry.
Tissue sections were used individually, or tissue blocks were used to prepare a tissue microarray that contained duplicate tissue samples from tracheas and duodenums from 10 rabbits, 8 hares, and 6 cottontail rabbits. The sections were deparaffinated through baths of LMR-SOL (1-bromopropane, 2-methylpropane-2-ol, and acetonitrile), followed by rehydration with successive baths of 100, 90, 70, and 50% ethanol. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide in PBS. Nonspecific binding was blocked with 3% BSA in PBS. HRP-conjugated UEA-I (Sigma-Aldrich, St. Louis, MO) at 0.8 μg/ml, HRP-conjugated sWGA (EY Laboratories, Burlingame, CA) at 2 μg/ml, anti-A monoclonal antibody 2A21, and anti-B monoclonal antibody B49 were used for binding to H antigen, A antigen, and B antigen, respectively. Lectins and antibodies were incubated with the tissue sections in 1% BSA in PBS at 4°C (UEA-I), at room temperature (sWGA), or at 37°C (antibodies) overnight. After three washes in PBS, a biotinylated anti-mouse antibody (Vector Laboratories, Burlingame, CA) diluted in 1% BSA in PBS was added to the assay mixtures with primary mouse antibodies. The sections were washed three times in PBS prior to addition of HRP-conjugated avidin D (Vector Laboratories, Burlingame, CA), also diluted in 1% BSA in PBS. Substrate (3-amino-ethyl-carbazole) was added to the slides (AEC kit; Vector Laboratories, Burlingame, CA), followed by Mayer's hematoxylin solution (Merck, Whitehouse Station, NJ) for contrast staining.
In order to confirm the role of terminal β-linked N-acetylglucosamine residues in EBHSV attachment, deparaffinated and hydrogen peroxide-blocked tissue sections were treated with 25 U β-N-hexosaminidasef (New England BioLabs, Ipswich, MA) for 2 h at 37°C. Fresh enzyme was then added, and the sections were further incubated overnight at 37°C. Control sections were made in parallel with the corresponding enzyme buffers (sodium citrate, pH 4.5). After overnight incubation, the sections were washed twice in PBS and blocked with PBS-5% BSA for 1 h at room temperature. A competition assay was also performed by preincubating the sections with unlabeled sWGA (Vector Laboratories) or with the fucose-specific lectin AAL (Vector Laboratories) as a negative control. Both lectins were used at 10 μg/ml in PBS-1% BSA and incubated for 1 h at room temperature prior to incubation with EBHSV. B/EBHS/6-infected liver homogenate diluted 1/5 in PBS-1% BSA was then added and incubated at 4°C overnight. After 3 washes with PBS, monoclonal anti-EBHSV antibody (5F5; a kind gift from L. Capucci, IZSLER, Brescia, Italy) was added at 1/100 dilution for 2 h at 37°C, followed by 3 washes with PBS and incubation with biotinylated anti-mouse antibody (Vector Laboratories) at 1/1,000 dilution for 2 h at 37°C. The rest of the protocol was as described above.
Accession number(s).
The sequence for the capsid protein gene of isolate B-EBHS-6 is available in GenBank under accession number KY801206.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Anne Pleney and Jean-Claude Ricci (IMPCF) for providing hunt-harvested rabbit samples from Aveyron and to Lorenzo Capucci for monoclonal anti-EBHSV. We also thank Nadezhda Shilova for discussions of PGA data.
This work was supported in part by a grant from the Agence Nationale de la Recherche (France), CALILAGO, and by a grant from the Région des Pays de la Loire (France) ARMINA to J.L.P. It was performed within the framework of the ECALEP project selected during the 2nd joint call of the Animal Health and Welfare ERA-Net (Anihwa) initiative, a coordination action funded under the European Commission's ERA-Net scheme within the Seventh Framework Programme (contract no. 291815). The ECALEP project is funded by the Agence Nationale de la Recherche (France); the Ministry of Health, Department for Veterinary Public Health, Nutrition and Food Safety (Italy); and the Research Council FORMAS (Sweden). FCT-Foundation for Science and Technology, Portugal, supported the FCT Investigator grant of J.A. (reference no. IF/01396/2013) and the postdoctoral grant of A.M.L. (SFRH/BPD/115211/2016). N.V.B. was supported by Russian Science Foundation grant 14-50-00131. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01759-17.
REFERENCES
- 1.Woolhouse MEJ, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jump. Trends Ecol Evol 20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geoghegan JL, Duchene S, Holmes EC. 2017. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog 13:e1006215. doi: 10.1371/journal.ppat.1006215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM. 2014. The evolution and genetics of virus host shifts. PLoS Pathog 10:e1004395. doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longdon B, Hadfield JD, Webster CL, Obbard DJ, Jiggins FM. 2011. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog 7:e1002260. doi: 10.1371/journal.ppat.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke IN, Estes MK, Green KY, Hansman GS, Knowles NJ, Koopmans MK, Matson DO, Meyers G, Neill JD, Radford A, Smith AW, Studdert MJ, Thiel H-J, Vinjé J. 2012. Family Caliciviridae, p 977–986. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 6.Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. 2012. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43:12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu SJ, Xue HP, Pu BQ, Quian NH. 1984. A new viral disease in rabbit. Anim Husb Vet Med 16:235–255. [Google Scholar]
- 8.Gavier-Widén D, Mörner T. 1991. Epidemiology and diagnosis of the European brown hare syndrome in Scandinavian countries: a review. Rev Sci Tech 10:453–458. doi: 10.20506/rst.10.2.555. [DOI] [PubMed] [Google Scholar]
- 9.Capucci L, Scicluna MT, Lavazza A. 1991. Diagnosis of viral haemorrhagic disease of rabbits and the European brown hare syndrome. Rev Sci Tech 10:347–370. doi: 10.20506/rst.10.2.561. [DOI] [PubMed] [Google Scholar]
- 10.Laurent S, Vautherot JF, Le Gall G, Madelaine MF, Rasschaert D. 1997. Structural, antigenic and immunogenic relationships between European brown hare syndrome virus and rabbit haemorrhagic disease virus. J Gen Virol 78:2803–2811. doi: 10.1099/0022-1317-78-11-2803. [DOI] [PubMed] [Google Scholar]
- 11.Lopes AM, Capucci L, Gavier-Widén D, Le Gall-Reculé G, Brocchi E, Barbieri I, Quemener A, Le Pendu J, Geoghegan J, Holmes EC, Esteves PJ, Abrantes J. 2014. Molecular evolution and antigenic variation of European brown hare syndrome virus (EBHSV). Virology 468-470:104–112. doi: 10.1016/j.virol.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Cavadini P, Molinari S, Pezzoni G, Chiari M, Brocchi E, Lavazza A, Capucci L. 2016. Identification of a new non-pathogenic lagovirus in brown hares (Lepus europeaus), p 81 In Kelly P, Phillips S, Smith A, Browning C (ed), 5th World Lagomorph Conference. California State University, Stanislaus, Turlock, CA, USA. [Google Scholar]
- 13.Le Gall-Reculé G, Zwingelstein F, Laurent S, De Boisseron C, Portejoie Y, Rasschaert D. 2003. Phylogenetic analysis of rabbit haemorrhagic disease virus in France between 1993 and 2000 and the characterization of RHDV antigenic variants. Arch Virol 148:65–81. doi: 10.1007/s00705-002-0908-1. [DOI] [PubMed] [Google Scholar]
- 14.Capucci L, Fallacara F, Grazioli S, Lavazza A, Pacciarini ML, Brocchi E. 1998. A further step in the evolution of rabbit hemorrhagic disease virus: the appearance of the first consistent antigenic variant. Virus Res 58:115–126. doi: 10.1016/S0168-1702(98)00106-3. [DOI] [PubMed] [Google Scholar]
- 15.Bergin IL, Wise AG, Bolin SR, Mullaney TP, Kiupel M, Maes RK. 2009. Novel calicivirus identified in rabbits, Michigan, USA. Emerg Infect Dis 15:1955–1962. doi: 10.3201/eid1512.090839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capucci L, Fusi P, Lavazza A, Pacciarini M, Rossi LC. 1996. Detection and preliminary characterization of a new rabbit calicivirus related to Rabbit Haemorrhagic Disease Virus but nonpathogenic. J Virol 70:8614–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Gall-Reculé G, Zwingelstein F, Fages MP, Bertagnoli S, Gelfi J, Aubineau J, Roobrouck A, Botti G, Lavazza A, Marchandeau S. 2011. Characterisation of a non-pathogenic and non-protective infectious rabbit lagovirus related to RHDV. Virology 410:395–402. doi: 10.1016/j.virol.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Strive T, Wright JD, Robinson AJ. 2009. Identification and partial characterisation of a new lagovirus in Australian wild rabbits. Virology 384:97–105. doi: 10.1016/j.virol.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Nicholson LJ, Mahar JE, Strive T, Zheng T, Holmes EC, Ward VK, Duckworth JA. 2017. Discovery and characterization of a benign rabbit calicivirus in New Zealand. Appl Environ Microbiol 83:e00090-17. doi: 10.1128/AEM.00090-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Gall-Reculé G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, Decors A, Bertagnoli S, Guérin JL, Marchandeau S. 2011. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet Rec 168:137–138. doi: 10.1136/vr.d697. [DOI] [PubMed] [Google Scholar]
- 21.Le Gall-Reculé G, Lavazza A, Marchandeau S, Bertagnoli S, Zwingelstein F, Cavadini P, Martinelli N, Lombardi G, Guérin JL, Lemaitre E, Decors A, Boucher S, Le Normand B, Capucci L. 2013. Emergence of a new lagovirus related to rabbit hemorrhagic disease virus. Vet Res 44:81. doi: 10.1186/1297-9716-44-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalton KP, Nicieza I, Balseiro A, Muguerza MA, Rosell JM, Casais R, Alvarez AL, Parra F. 2012. Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerg Infect Dis 18:2009–2012. doi: 10.3201/eid1812.120341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrantes J, Lopes AM, Dalton KP, Melo P, Correia JJ, Ramada M, Alves PC, Parra F, Esteves PJ. 2013. New variant of rabbit hemorrhagic disease virus, Portugal, 2012-2013. Emerg Infect Dis 19:1900–1902. doi: 10.3201/eid1911.130908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westcott DG, Frossard JP, Everest D, Dastjerdi A, Duff JP, Steinbach F, Choudhury B. 2014. Incursion of RHDV2-like variant in Great Britain. Vet Rec 174:333. doi: 10.1136/vr.g2345. [DOI] [PubMed] [Google Scholar]
- 25.Almeida T, Lopes AM, Magalhaes MJ, Neves F, Pinheiro A, Goncalves D, Leitao M, Esteves PJ, Abrantes J. 2015. Tracking the evolution of the G1/RHDVb recombinant strains introduced from the Iberian Peninsula to the Azores islands, Portugal. Infect Genet Evol 34:307–313. doi: 10.1016/j.meegid.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Duarte M, Henriques M, Barros SC, Fagulha T, Ramos F, Luis T, Fevereiro M, Benevides S, Flor L, Barros SV, Bernardo S. 2015. Detection of RHDV variant 2 in the Azores. Vet Rec 176:130. doi: 10.1136/vr.h497. [DOI] [PubMed] [Google Scholar]
- 27.Neimanis AS, Ahola H, Zohari S, Larsson Pettersson U, Brojer C, Capucci L, Gavier-Widen D. 13 April 2017. Arrival of rabbit haemorrhagic disease virus 2 to northern Europe: emergence and outbreaks in wild and domestic rabbits (Oryctolagus cuniculus) in Sweden. Transbound Emerg Dis. doi: 10.1111/tbed.12650. [DOI] [PubMed] [Google Scholar]
- 28.Hall RN, Mahar JE, Haboury S, Stevens V, Holmes EC, Strive T. 2015. Emerging rabbit hemorrhagic disease virus 2 (RHDVb), Australia. Emerg Infect Dis 21:2276–2278. doi: 10.3201/eid2112.151210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Pendu J, Abrantes J, Bertagnoli S, Guiton JS, Le Gall-Recule G, Lopes AM, Marchandeau S, Alda F, Almeida T, Alves PC, Barcena J, Burmakina G, Blanco E, Calvete C, Cavadini P, Cooke B, Dalton KP, Delibes MM, Deptula W, Eden JS, Fang W, Ferreira CC, Fereira P, Foronda P, Gonçalves D, Gavier-Widén D, Hall R, Hukowska-Szematowicz B, Kerr P, Kovaliski J, Lavazza A, Mahar JE, Malogolovkin A, Marques R, Marques S, Martin-Alonso A, Monterroso P, Moreno S, Mutze G, Neimanis AS, Niedzwiedzka-Rystwej P, Peacock DE, Parra F, Rocchi M, Rouco C, Ruvöen-Clouet N, Silva E, Silvério D, Strive T, Thompson G, Tokarz-Deptula B, Esteves P. 2017. Proposal for a unified classification system and nomenclature of lagoviruses. J Gen Virol 98:1658–1666. doi: 10.1099/jgv.0.000840. [DOI] [PubMed] [Google Scholar]
- 30.Stuart AD, Brown TD. 2007. Alpha2,6-linked sialic acid acts as a receptor for feline calicivirus. J Gen Virol 88:177–186. doi: 10.1099/vir.0.82158-0. [DOI] [PubMed] [Google Scholar]
- 31.Taube S, Perry JW, Yetming K, Patel SP, Auble H, Shu L, Nawar HF, Lee CH, Connell TD, Shayman JA, Wobus CE. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol 83:4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. 2004. Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J Virol 78:3817–3826. doi: 10.1128/JVI.78.8.3817-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruvoën-Clouet N, Belliot G, Le Pendu J. 2013. Noroviruses and histo-blood groups: the impact of common host genetic polymorphisms on virus transmission and evolution. Rev Med Virol 23:355–366. doi: 10.1002/rmv.1757. [DOI] [PubMed] [Google Scholar]
- 34.Tan M, Jiang X. 2014. Histo-blood group antigens: a common niche for norovirus and rotavirus. Expert Rev Mol Med 16:e5. doi: 10.1017/erm.2014.2. [DOI] [PubMed] [Google Scholar]
- 35.Le Pendu J, Nystrom K, Ruvoen-Clouet N. 2014. Host-pathogen co-evolution and glycan interactions. Curr Opin Virol 7:88–94. doi: 10.1016/j.coviro.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Matthee CA, van Vuuren BJ, Bell D, Robinson TJ. 2004. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst Biol 53:433–447. doi: 10.1080/10635150490445715. [DOI] [PubMed] [Google Scholar]
- 37.Rosin A, Gilio N, Merrigi A. 2008. Introduced lagomorphs as a threat to “native” lagomorphs: the case of the eastern cottontail (Sylvilagus floridanus) in northern Italy, p 153–164. In Alves P, Ferrand N, Hackl̈ander K (ed), Lagomorph biology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 38.Sasse C. 1983. Ecological consequences of the introduction of the American cottontail, Sylvilagus floridanus, into Europe, p 33. Report of the Council of Europe SN-US, vol 83 Research Institute for Nature Management, Leersum, The Netherlands. [Google Scholar]
- 39.Lavazza A, Scicluna MT, Capucci L. 1996. Susceptibility of hares and rabbits to the European brown hare syndrome virus (EBHSV) and rabbit haemorrhagic disease virus (RHDV) under experimental conditions. Zentralbl Veterinarmed B 43:401–410. [DOI] [PubMed] [Google Scholar]
- 40.Chasey D, Lucas M, Westcott D, Williams M. 1992. European brown hare syndrome in the U.K.; a calicivirus related to but distinct from that of viral haemorrhagic disease in rabbits. Arch Virol 124:363–370. doi: 10.1007/BF01309816. [DOI] [PubMed] [Google Scholar]
- 41.Nauwynck H, Callebaut P, Peeters J, Ducatelle R, Uyttebroek E. 1993. Susceptibility of hares and rabbits to a Belgian isolate of European brown hare syndrome virus. J Wildl Dis 29:203–208. doi: 10.7589/0090-3558-29.2.203. [DOI] [PubMed] [Google Scholar]
- 42.di Modigno G, Nasti R. 1990. La malattia emorragica virale in Puglia. Contributo sperimentale. Riv Conigliol 27:25–32. [Google Scholar]
- 43.Morisse J-P, Picault J-P, Boilletot E, Morin M. 1990. Relations étiologiques entre le syndrome du lièvre brun européen (EBHS) et la maladie hémorragique virale du lapin (VHD). Rev Med Vet 141:463–467. [Google Scholar]
- 44.Lavazza A, Cavadini P, Barbieri I, Tizzani P, Pinheiro A, Abrantes J, Esteves PJ, Grilli G, Gioia E, Zanoni M, Meneguz P, Guiton JS, Marchandeau S, Chiari M, Capucci L. 2015. Field and experimental data indicate that the eastern cottontail (Sylvilagus floridanus) is susceptible to infection with European brown hare syndrome (EBHS) virus and not with rabbit hemorrhagic disease (RHD) virus. Vet Res 46:13. doi: 10.1186/s13567-015-0149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopes AM, Marques S, Silva E, Magalhaes MJ, Pinheiro A, Alves PC, Le Pendu J, Esteves PJ, Thompson G, Abrantes J. 2014. Detection of RHDV strains in the Iberian hare (Lepus granatensis): earliest evidence of rabbit lagovirus cross-species infection. Vet Res 45:94. doi: 10.1186/s13567-014-0094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puggioni G, Cavadini P, Maestrale C, Scivoli R, Botti G, Ligios C, Le Gall-Reculé G, Lavazza A, Capucci L. 2013. The new French 2010 rabbit hemorrhagic disease virus causes an RHD-like disease in the Sardinian Cape hare (Lepus capensis mediterraneus). Vet Res 44:96. doi: 10.1186/1297-9716-44-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camarda A, Pugliese N, Cavadini P, Circella E, Capucci L, Caroli A, Legretto M, Mallia E, Lavazza A. 2014. Detection of the new emerging rabbit haemorrhagic disease type 2 virus (RHDV2) in Sicily from rabbit (Oryctolagus cuniculus) and Italian hare (Lepus corsicanus). Res Vet Sci 97:642–645. doi: 10.1016/j.rvsc.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Hall RN, Peacock DE, Kovaliski J, Mahar JE, Mourant R, Piper M, Strive T. 2017. Detection of RHDV2 in European brown hares (Lepus europaeus) in Australia. Vet Rec 180:121. doi: 10.1136/vr.104034. [DOI] [PubMed] [Google Scholar]
- 49.Velarde R, Cavadini P, Neimanis AS, Cabezon O, Chiari M, Gaffuri A, Lavin S, Grilli G, Gavier-Widén D, Lavazza A, Capucci L. 2017. Spillover events of infection of brown hares (Lepus europaeus) with rabbit haemorrhagic disease type 2 virus (RHDV2) caused sporadic cases of an European brown hare syndrome-like disease in Italy and Spain. Transbound Emerg Dis 64:1750–1761. doi: 10.1111/tbed.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merchan T, Rocha G, Alda F, Silva E, Thompson G, de Trucios SH, Pages A. 2011. Detection of rabbit haemorrhagic disease virus (RHDV) in nonspecific vertebrate hosts sympatric to the European wild rabbit (Oryctolagus cuniculus). Infect Genet Evol 11:1469–1474. doi: 10.1016/j.meegid.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Rocha G, Alda F, Pagés A, Merchán T. 2017. Experimental transmission of rabbit haemorrhagic disease virus (RHDVa) from rabbit to wild mice (Mus spretus and Apodemus sylvaticus) under laboratory conditions. Infect Genet Evol 47:94–98. doi: 10.1016/j.meegid.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 52.Rodak L, Smid B, Valicek L, Vesely T, Stepanek J, Hampl J, Jurak E. 1990. Enzyme-linked immunosorbent assay of antibodies to rabbit haemorrhagic disease virus and determination of its major structural proteins. J Gen Virol 71:1075–1080. doi: 10.1099/0022-1317-71-5-1075. [DOI] [PubMed] [Google Scholar]
- 53.Robinson AJ, Kirkland PD, Forrester RI, Capucci L, Cooke BD, Philbey AW. 2002. Serological evidence for the presence of a calicivirus in Australian wild rabbits, Oryctolagus cuniculus, before the introduction of rabbit haemorrhagic disease virus (RHDV): its potential influence on the specificity of a competitive ELISA for RHDV. Wildlife Res 29:655–662. doi: 10.1071/WR00096. [DOI] [Google Scholar]
- 54.Forrester NL, Trout RC, Gould EA. 2007. Benign circulation of rabbit haemorrhagic disease virus on Lambay Island, Eire. Virology 358:18–22. doi: 10.1016/j.virol.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 55.Esteves PJ, Abrantes J, Bertagnoli S, Cavadini P, Gavier-Widén D, Guitton JS, Lavazza A, Lemaitre E, Letty J, Lopes AM, Neimanis AS, Ruvoen-Clouet N, Le Pendu J, Marchandeau S, Le Gall-Recule G. 2015. Emergence of pathogenicity in lagoviruses: evolution from pre-existing nonpathogenic strains or through a species jump? PLoS Pathog 11:e1005087. doi: 10.1371/journal.ppat.1005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arthur CP, Chapuis JL. 1983. L'introduction de Sylvilagus floridanus en France: historique, dangers et expérimentation en cours. CR Soc Biogeograph 59:333–356. [Google Scholar]
- 57.Mussa PP. 1996. Il silvilago in provincia di Torino. Habitat 61:5–11. [Google Scholar]
- 58.Ruvoën-Clouet N, Ganière JP, André-Fontaine G, Blanchard D, Le Pendu J. 2000. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol 74:11950–11954. doi: 10.1128/JVI.74.24.11950-11954.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nyström K, Le Gall-Reculé G, Grassi P, Abrantes J, Ruvoën-Clouet N, Le Moullac-Vaidye B, Lopes AM, Esteves PJ, Strive T, Marchandeau S, Dell A, Haslam SM, Le Pendu J. 2011. Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog 7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marionneau S, Cailleau-Thomas A, Rocher J, Le Moullac-Vaidye B, Ruvoën-Clouet N, Clément M, Le Pendu J. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565–573. doi: 10.1016/S0300-9084(01)01321-9. [DOI] [PubMed] [Google Scholar]
- 61.Guillon P, Ruvöen-Clouet N, Le Moullac-Vaidye B, Marchandeau S, Le Pendu J. 2009. Association between expression of the H histo-blood group antigen, α1,2 fucosyltransferases polymorphism of wild rabbits, and sensitivity to rabbit hemorrhagic disease virus. Glycobiology 19:21–28. doi: 10.1093/glycob/cwn098. [DOI] [PubMed] [Google Scholar]
- 62.Nystrom K, Abrantes J, Lopes AM, Le Moullac-Vaidye B, Marchandeau S, Rocher J, Ruvoen-Clouet N, Esteves PJ, Le Pendu J. 2015. Neofunctionalization of the Sec1 alpha1,2fucosyltransferase paralogue in leporids contributes to glycan polymorphism and resistance to rabbit hemorrhagic disease virus. PLoS Pathog 11:e1004759. doi: 10.1371/journal.ppat.1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alves PC, Hackländer K. 2008. Lagomorph species: geographical distribution and conservation status, p 395–405. In Alves PC, Ferrand N, Hackl̈ander K (ed), Lagomorph biology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 64.Holmes EC. 2013. What can we predict about viral evolution and emergence? Curr Opin Virol 3:180–184. doi: 10.1016/j.coviro.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith AW, Skilling DE, Cherry N, Mead JH, Matson DO. 1998. Caliciviruses emergence from ocean reservoir: zoonotic and interspecies movements. Emerg Infect Dis 4:13–20. doi: 10.3201/eid0401.980103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Martino B, Di Rocco C, Ceci C, Marsilio F. 2009. Characterization of a strain of feline calicivirus isolated from a dog faecal sample. Vet Microbiol 139:52–57. doi: 10.1016/j.vetmic.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caddy SL, de Rougemont A, Emmott E, El-Attar L, Mitchell JA, Hollinshead M, Belliot G, Brownlie J, Le Pendu J, Goodfellow I. 2015. Evidence for human norovirus infection of dogs in the United Kingdom. J Clin Microbiol 53:1873–1883. doi: 10.1128/JCM.02778-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oriol R, Candelier JJ, Taniguchi S, Balanzino L, Peters L, Niekrasz M, Hammer C, Cooper DK. 1999. Major carbohydrate epitopes in tissues of domestic and African wild animals of potential interest for xenotransplantation research. Xenotransplantation 6:79–89. doi: 10.1034/j.1399-3089.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 69.Oriol R, Mollicone R, Couillin P, Dalix AM, Candelier JJ. 1992. Genetic regulation of the expression of ABH and Lewis antigens in tissues. APMIS Suppl 27:28–38. [PubMed] [Google Scholar]
- 70.Macher BA, Galili U. 2008. The Galα1,3Gal1,4GlcNAc-R (α-Gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim Biophys Acta 1780:75–88. doi: 10.1016/j.bbagen.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elsworth PG, Kovaliski J, Cooke BD. 2012. Rabbit haemorrhagic disease: are Australian rabbits (Oryctolagus cuniculus) evolving resistance to infection with Czech CAPM 351 RHDV? Epidemiol Infect 140:1972–1981. doi: 10.1017/S0950268811002743. [DOI] [PubMed] [Google Scholar]
- 72.Esko JD, Lindahl U. 2001. Molecular diversity of hepan sulfate. J Clin Invest 108:169–173. doi: 10.1172/JCI200113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meneghetti MC, Hughes AJ, Rudd TR, Nader HB, Powell AK, Yates EA, Lima MA. 2015. Heparan sulfate and heparin interactions with proteins. J R Soc Interface 12:0589. doi: 10.1098/rsif.2015.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olofsson S, Bergström T. 2005. Glycoconjugate glycans as viral receptors. Ann Med 37:154–172. doi: 10.1080/07853890510007340. [DOI] [PubMed] [Google Scholar]
- 75.Gardner CL, Choi-Nurvitadhi J, Sun C, Bayer A, Hritz J, Ryman KD, Klimstra WB. 2013. Natural variation in the heparan sulfate binding domain of the eastern equine encephalitis virus E2 glycoprotein alters interactions with cell surfaces and virulence in mice. J Virol 87:8582–8590. doi: 10.1128/JVI.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salvador B, Sexton NR, Carrion R Jr, Nunneley J, Patterson JL, Steffen I, Lu K, Muench MO, Lembo D, Simmons G. 2013. Filoviruses utilize glycosaminoglycans for their attachment to target cells. J Virol 87:3295–1304. doi: 10.1128/JVI.01621-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leuthold MM, Dalton KP, Hansman GS. 2015. Structural analysis of a rabbit hemorrhagic disease virus binding to histo-blood group antigens. J Virol 89:2378–2387. doi: 10.1128/JVI.02832-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahar JE, Nicholson L, Eden JS, Duchêne S, Kerr PJ, Duckworth J, Ward VK, Holmes EC, Strive T. 2016. Benign rabbit caliciviruses exhibit evolutionary dynamics similar to those of their virulent relatives. J Virol 90:9317–9329. doi: 10.1128/JVI.01212-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J, Kerr PJ, Wright JD, Strive T. 2012. Serological assays to discriminate rabbit haemorrhagic disease virus from Australian non-pathogenic rabbit calicivirus. Vet Microbiol 157:345–354. doi: 10.1016/j.vetmic.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 80.Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J Virol 66:6527–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Pendu J, Le Cabellec M, Bara J. 1997. Immunohistological analysis of antibodies against ABH and other glycoconjugates in normal human pyloric and duodenal mucosae. Transfus Clin Biol 4:41–46. doi: 10.1016/S1246-7820(97)80009-2. [DOI] [PubMed] [Google Scholar]
- 82.Galanina OE, Chinarev AA, Sholova NV, Sablina MA, Bovin NV. 2012. Immobilization of polyacrylamide-based glycoconjugates on solid phase in immunosorbent assays. Methods Mol Biol 808:167–182. doi: 10.1007/978-1-61779-373-8_12. [DOI] [PubMed] [Google Scholar]
- 83.Huflejt ME, Vuskovic M, Vasiliu D, Xu H, Obukhova P, Shilova N, Tuzikov A, Galanina O, Arun B, Lu K, Bovin N. 2009. Anti-carbohydrate antibodies of normal sera: findings, surprises and challenges. Mol Immunol 46:3037–3049. doi: 10.1016/j.molimm.2009.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.