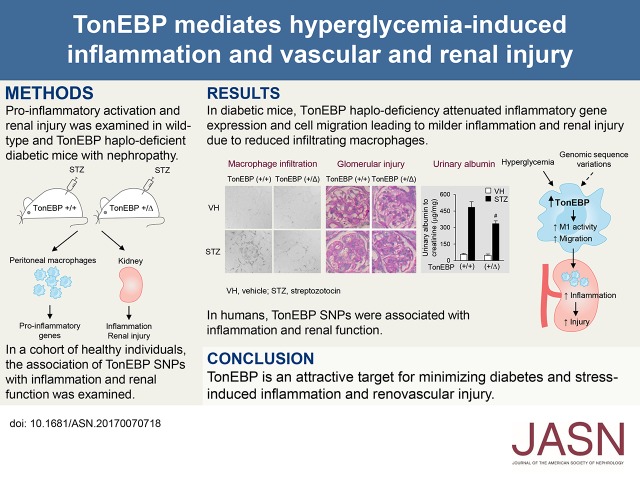
Abstract
Diabetic nephropathy (DN) has become the single leading cause of ESRD in developed nations. Bearing in mind the paucity of effective treatment for DN and progressive CKD, novel targets for treatment are sorely needed. We previously reported that increased activity of tonicity-responsive enhancer-binding protein (TonEBP) in monocytes was associated with early DN in humans. We now extend these findings by testing the hypotheses that TonEBP in macrophages promotes hyperglycemia-mediated proinflammatory activation and chronic renal inflammation leading to DN and CKD, and TonEBP genetic variability in humans is associated with inflammatory, renal, and vascular function–related phenotypes. In a mouse model of DN, compared with the wild-type phenotype, TonEBP haplodeficiency associated with reduced activation of macrophages by hyperglycemia, fewer macrophages in the kidney, lower renal expression of proinflammatory genes, and attenuated DN. Furthermore, in a cohort of healthy humans, genetic variants within TonEBP associated with renal function, BP, and systemic inflammation. One of the genetic variants associated with renal function was replicated in a large population-based cohort. These findings suggest that TonEBP is a promising target for minimizing diabetes- and stress-induced inflammation and renovascular injury.
Keywords: Diabetic nephropathy, chronic kidney disease, blood pressure, macrophages, genetic variants
Diabetic nephropathy (DN) is a complex disease with progressive decline in renal function that involves multiple pathways including inflammation, endothelial injury, and tubular injury. Only 30% of patients with type 1 diabetes and 40% of those with type 2 diabetes develop DN, indicating considerable individual variations in susceptibility.1 Genome-wide studies have uncovered many genetic variants associated with CKD; nevertheless, they only account for a small proportion of the total genetic contribution.2–4 As such, complementary approaches, such as animal models and translational candidate gene approaches, are needed to better help delineate genetic contributors and novel targets for treatment.
In macrophages, TonEBP (tonicity-responsive enhancer-binding protein) is the rate-limiting component of “NFκB enhanceosome” in which TonEBP binds activated NFκB, histone acetylatransferase p300, and RNA polymerase II on the promoters of the TNF-α and other inflammatory genes.5,6 Haplo-deficiency of TonEBP results in a reduced NFκB enhanceosome activity and blunted macrophage activation in response to inflammatory signals. Thus, TonEBP haplo-deficient animals display dramatically reduced inflammatory injury in animal models of rheumatoid arthritis7,8 and atherosclerosis.9 We previously reported that among patients with approximately 30 years of type 1 diabetes, proteinuria was associated with approximately 50% higher TonEBP activity in monocytes.10 Here, we extend these results by showing in a mouse model that TonEBP mediates the hyperglycemia-induced proinflammatory activation of macrophages leading to renal infiltration of macrophages and DN. In humans, we find TonEBP-associated single nucleotide polymorphisms (SNPs) to be associated with systemic inflammatory markers, BP, and renal function. Our animal-based findings along with consistent SNP-based association in related phenotypes in humans suggest that genetic variability in TonEBP leads to differential susceptibility to inflammatory responses, vascular injury, and CKD, in response to stressors such as hyperglycemia.
Results
Hyperglycemia Stimulates M1 Polarization and Migration of Macrophages via Upregulation of TonEBP
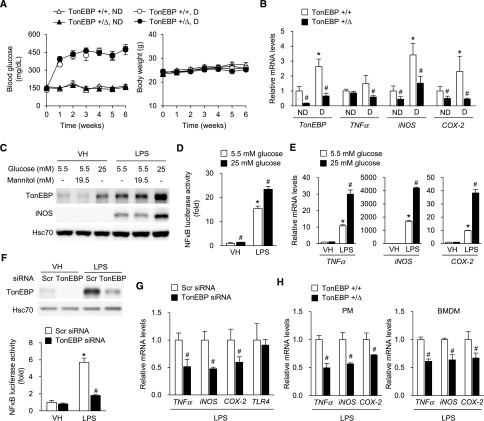
We sought to investigate the underlying molecular mechanism for our previously noted association between TonEBP activity in monocytes and DN in patients with type 1 diabetes.10 Because previous studies demonstrated the role of macrophage-mediated inflammation in the development of DN,11,12 we decided to examine macrophages in a mouse model of type 1 diabetes. In order to mimic the differences in the level of TonEBP activity, we used the TonEBP heterozygous (TonEBP+/∆) mice because they display TonEBP haplo-deficiency.7,9,13 We made the TonEBP+/∆ mice and their TonEBP wild-type (TonEBP+/+) littermates hyperglycemic by injecting streptozotocin (STZ) as shown in Figure 1A. When peritoneal macrophages (PMs) from the TonEBP+/+ animals were examined, higher mRNA expression for TonEBP and M1 polarization, as indicated by increased proinflammatory gene expression, in diabetic animals compared with nondiabetic animals was observed (Figure 1B). These changes were reproduced in Raw264.7 cells cultured in high glucose (Figure 1C): raising glucose concentration to 25 mM resulted in higher TonEBP expression in a manner synergistic with LPS, whereas addition of mannitol to the same osmolality did not. Furthermore, the high glucose–enhanced TonEBP expression was associated with elevated NFκB activity in the presence of LPS (Figure 1D). Because most of the M1 genes were NFκB target genes, expression of M1 genes was elevated as expected (Figure 1E). These observations indicate that the enhanced TonEBP expression and M1 polarization of macrophages from diabetic animals (Figure 1B) are due, at least in part, to hyperglycemia.
Figure 1.
Hyperglycemia induces TonEBP and stimulates M1 gene expression in macrophages. (A and B) TonEBP+/+ or TonEBP+/∆ mice were injected with vehicle (nondiabetic [ND], n=6) or STZ (diabetic [D], n=16). (A) Blood glucose and body weight, mean+SEM. (B) mRNA abundance for TonEBP, TNF-α, iNOS, and COX-2 in PMs relative to TonEBP+/+,ND. Mean+SEM, n=6. (C–E) High glucose induces TonEBP, and stimulates NFκB and its target genes in Raw264.7 cells. (C) Cells were treated with various combinations of high glucose, mannitol, vehicle (VH), and LPS as indicated for 24 hours. Immunoblots for TonEBP, iNOS, and Hsc70 are shown. (D) Cells transfected with an NFκB luciferase reporter were treated for 6 hours with VH or LPS in combination with 5.5 or 25 mM glucose, as indicated. Normalized luciferase activity relative to 5.5 mM glucose and VH (fold) is shown as mean+SD, n=3. (E) Cells were treated for 6 hours with various combinations of LPS and 25 mM glucose. mRNA abundance relative to VH and 5.5 mM glucose (fold) is shown as mean+SD, n=3. (F–H) TonEBP deficiency inhibits NFκB and reduces proinflammatory genes in macrophages under high-glucose conditions. (F) Raw264.7 cells were transfected with scrambled (Scr) or TonEBP siRNA followed by transfection with the NFκB luciferase reporter. After 6 hours of treatment with VH or LPS in 25 mM glucose, TonEBP and Hsc70 were immunoblotted and luciferase was measured. n=3. (G) Raw264.7 cells were transfected with siRNA followed by treatment with LPS in high glucose. mRNA abundance for TNF-α, iNOS, COX-2, and TLR4 relative to Scr siRNA, n=3. (H) PMs and BMDMs obtained from TonEBP+/+ or TonEBP+/∆ mice were treated and analyzed as above. n=6. Mean+SEM. *P<0.01 compared with corresponding ND (B) or VH (D–F). #P<0.01 compared with corresponding TonEBP+/+ (B and H), 5.5 mM glucose (D and E), and Scr siRNA (F and G).
We asked whether the elevated expression of TonEBP in response to hyperglycemia was responsible for the elevated NFκB activity and M1 polarization. When TonEBP was knocked down, the LPS-induced activation of NFκB (Figure 1F) and M1 gene expression was reduced without changes in TLR4 mRNA expression (Figure 1G) in high-glucose conditions. Likewise, the induction of M1 genes by LPS was reduced in PMs and bone marrow–derived macrophages (BMDMs) obtained from the TonEBP+/∆ mice compared with those obtained from their TonEBP+/+ littermates (Figure 1H). These data demonstrate that in macrophages TonEBP is induced by hyperglycemia leading to activation of NFκB and elevation of M1 gene expression. In the TonEBP haplo-deficient animals, the M1 gene induction in response to diabetes was blunted (Figure 1B), as expected.
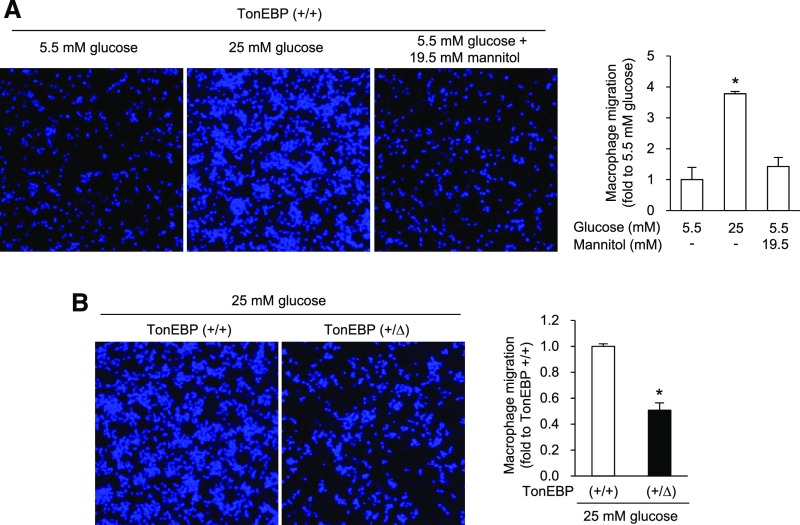
We showed previously that haplo-deficiency of TonEBP in bone marrow cells resulted in approximately 80% reduction in the size of atherosclerotic plaques in a mouse model of atherosclerosis.9 The reduced atherosclerotic lesion was associated with reduced cell migration of BMDMs. We asked whether hyperglycemia affected migration of macrophages. We found that BMDMs from wild-type animals were stimulated by an increase in glucose concentration to 25 mM, but not by addition of mannitol to the same osmolality (Figure 2A). This cell migration was dramatically reduced in BMDMs obtained from the TonEBP haplo-deficient animals (Figure 2B). Taken together, the data in Figures 1 and 2 demonstrate that hyperglycemia enhances TonEBP expression in macrophages, leading to M1 polarization and increased migration of macrophages. A modest, approximately 50% reduction in TonEBP expression resulted in a dramatic decrease in the M1 polarization and migration.
Figure 2.
High glucose–induced macrophage migration is defective in TonEBP deficiency. (A) BMDMs isolated from wild-type (TonEBP+/+) animals were cultured in normal glucose (5.5 mM), high glucose (25 mM), or 5.5 mM glucose +19.5 mM mannitol (osmotic control for high glucose). The cells were then plated on membranes with 5-μm pores and cell migration for 16 hours through the pores in response to MCP-1 was visualized by DAPI staining. Cells migrated were counted and expressed relative to 5.5 mM glucose condition (fold). (B) BMDMs isolated from TonEBP+/+ or TonEBP+/∆ mice were cultured in high glucose and analyzed as above. Mean+SEM, n=3. *P<0.01 compared with 5.5, − (A) or (+/+) (B). +/+, wild-type; +/Δ, heterozygous.
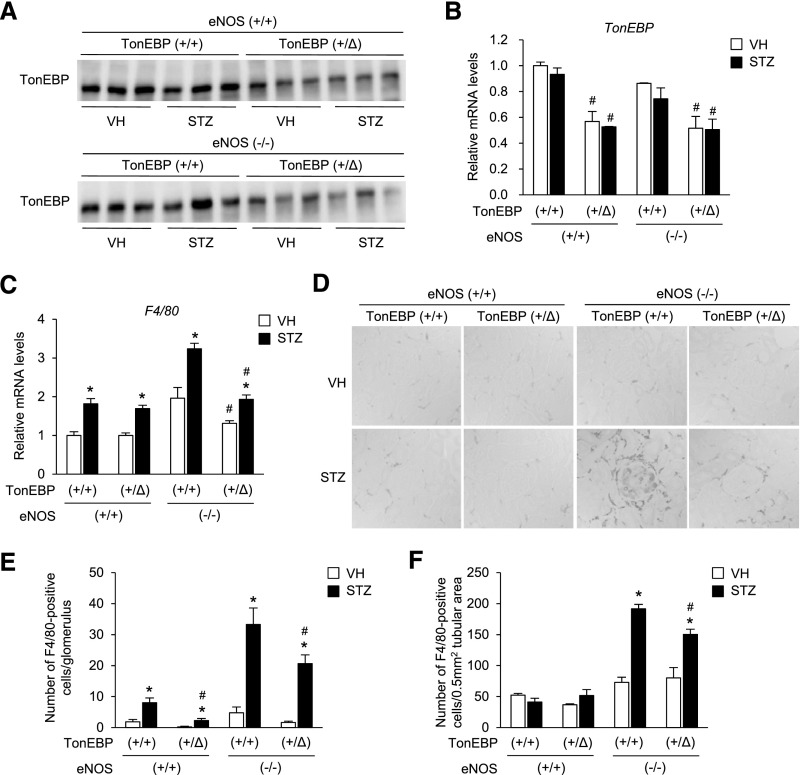
TonEBP Haplo-Deficiency Displays Reduced Number of Renal Macrophages and Renal Inflammation in Mouse Model of DN
Given the reduced activation and migration of macrophages in response to hyperglycemia in the TonEBP haplo-deficient animals (see above), we asked whether reduced renal macrophages were observed in these animals in a mouse model of DN: deficiency of endothelial nitric oxide synthase (eNOS), i.e., TonEBP+/∆, eNOS−/− versus TonEBP+/+, eNOS−/− (Figure 3, A and B; see also Supplemental Material for details). Renal macrophage numbers assessed by F4/80 mRNA expression (Figure 3C) and immunohistochemical analyses of F4/80 (Figure 3D) were higher in the TonEBP+/+ animals on the eNOS−/− background compared with those on the eNOS+/+ background. In those animals on the eNOS−/− background, but not those on the eNOS+/+ background, both the mRNA abundance and the number of F4/80-positive cells were lower in the TonEBP+/∆ mice compared with their TonEBP+/+ littermates. The decrease in the F4/80-positive cells was observed both in the glomerular (Figure 3E) and the tubular regions (Figure 3F). Thus, the reduced cell migration of the TonEBP haplo-deficient macrophages (Figure 2) was translated into reduced renal macrophage infiltration on the eNOS−/− animals. This pattern of reduced renal macrophage numbers in TonEBP haplo-deficiency on the background of endothelial deficiency was maintained in the renal expression of NFκB-driven, proinflammatory genes—IL-6, MCP-1, IP-10, IL-8, TNF-α, IL-1β, RANTES, IL-18, and INF-γ (Figure 4, A–I). All of these genes have been implicated in DN both in patients14–16 and animals.15,17 In correlation with the reduced IL-6 mRNA expression, IL-6 signaling measured by phosphorylation of STAT3 was reduced (Figure 4J), suggesting that the lower gene expression in TonEBP haplo-deficiency led to reduced inflammation in the kidney.
Figure 3.
TonEBP haplo-deficiency reduces renal macrophages in a mouse model of DN. Mice were bred to generate littermates of TonEBP+/+ or TonEBP+/∆ animals on eNOS+/+ or eNOS−/− background as indicated. Animals were injected with vehicle (VH) or STZ to induce diabetes as described in Figure 1. Seven weeks later, kidneys were analyzed. (A and B) Immunoblot images and quantification of TonEBP. (C) F4/80 mRNA was measured using qRT-PCR. (D) Representative images of immunohistochemical staining for F4/80. (E and F) Number of F4/80-positive cells per glomerulus (E) or 0.5 mm2 of tubular area (F) was counted. Mean+SEM, n=8. *P<0.05 compared with corresponding VH. #P<0.05 compared with corresponding TonEBP+/+. +/+, wild-type; +/Δ, heterozygous; −/−, knock out.
Figure 4.
TonEBP haplo-deficiency reduces renal expression of proinflammatory genes and STAT3 activation in a mouse model of DN. mRNA for IL-6 (A), MCP-1 (B), IP-10 (C), IL-8 (D), TNF-α (E), IL-1β (F), RANTES (G), IL-18 (H), and IFN-γ (I) was measured from the kidney samples described in Figure 3 using qRT-PCR. Animals injected with VH are shown in open bars, and those injected with STZ in filled bars. Mean+SEM, n=8. *P<0.05 compared with corresponding VH. #P<0.05 compared with corresponding TonEBP+/+. (J) Representative images of immunohistochemical staining for phosphorylated STAT3. Arrow heads denote intense signals in tubulointerstitial areas. VH, vehicle; +/+, wild-type; +/Δ, heterozygous; −/−, knock out.
TonEBP Haplo-Deficiency Displays Reduced Renal Injury in Mouse Model of DN
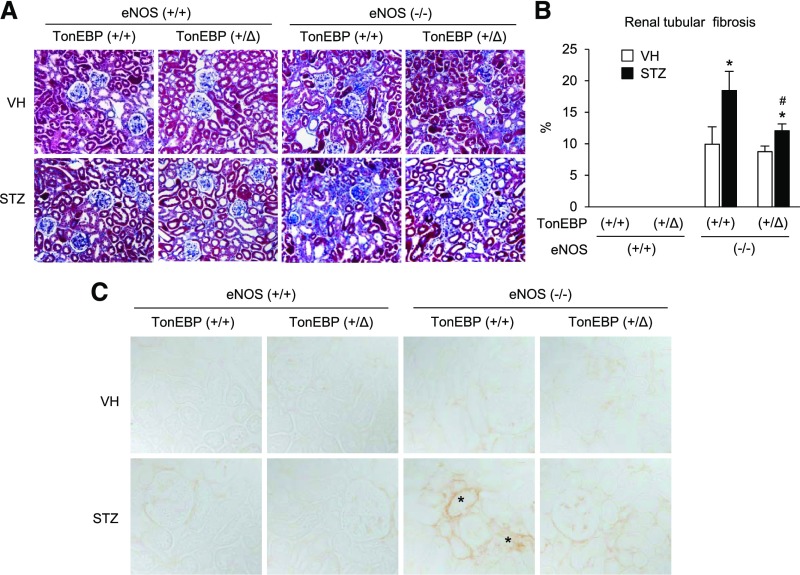
Compared with animals with normal endothelial function, eNOS-deficient animals displayed significantly more mesangial expansion and glomerulosclerosis in basal conditions, as reported previously (Figure 5, A and B).18,19 The glomerular injury worsened dramatically in diabetic animals as previously reported18,20: mesangiolysis, mesangial expansion, microaneurysm, and diffuse and nodular sclerotic lesions (Figure 5A). The glomerular injury was significantly milder in the TonEBP haplo-deficient animals on the basis of visual impression (Figure 5A) and the percentage of injured glomeruli (Figure 5B). Albuminuria displayed the same pattern of changes (Figure 5C). Tubulointerstitial fibrosis was absent in animals with normal endothelial function but dramatically induced with endothelial dysfunction (Figure 6, A and B). Diabetes exacerbated the fibrosis, which was ameliorated in the TonEBP-deficient animals. Interestingly, immunohistochemical signals of TGF-β (Figure 6C) mirrored the changes in fibrosis, suggesting that changes in TGF-β expression were responsible for the variations in fibrosis. Thus, diabetic renal injuries were dramatically tempered in the TonEBP haplo-deficient animals in association with reduced macrophage infiltration and renal inflammation.
Figure 5.
TonEBP haplo-deficiency reduces glomerular injury and albuminuria in a mouse model of DN. (A) Representative images of periodic acid–Schiff staining of kidney sections. (B) Percentage of glomeruli with injury: mesangial matrix expansion, mesangiolysis, and glomerulosclerosis. (C) Albumin and creatinine were measured from urine samples, and micrograms albumin per milligram of creatinine was calculated. Mean+SEM, n=8. *P<0.05 compared with corresponding VH. #P< 0.05 compared with corresponding TonEBP+/+. VH, vehicle; +/+, wild-type; +/Δ, heterozygous; −/−, knock out.
Figure 6.
TonEBP haplo-deficiency reduces tubulointerstitial fibrosis and TGF-β expression. (A) Representative images of Masson’s trichrome staining of kidney sections. (B) Blue areas representing collagen deposition were measured and expressed as percentage. Mean+SEM, n=8. *P<0.05 compared with corresponding VH. #P<0.05 compared with corresponding TonEBP+/+. (C) Representative images of immunohistochemical staining for TGF-β. Asterisks denote intense signals in tubulointerstitial areas. VH, vehicle; +/+, wild-type; +/Δ, heterozygous; −/−, knock out.
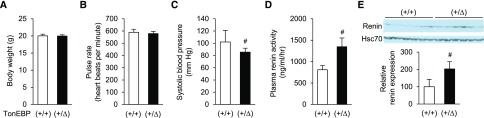
Hypotension and Hyperreninemia in TonEBP Haplo-Deficiency
A recent report shows that the SNP rs33063 in the TonEBP gene is associated with pulse pressure.21 Because the functional significance of the associated variant was not established, we tested whether reduced TonEBP levels were associated with BP using the TonEBP haplo-deficient mice. Consistent with the human association studies, these animals displayed reduced systolic BP (SBP). We also noted elevated renal renin expression and circulating renin levels (Figure 7), presumably as a compensatory reaction to the reduced BP.
Figure 7.
TonEBP haplo-deficiency is associated with reduced SBP, hyperreninemia, and elevated renal expression of renin. Male TonEBP+/∆ mice or their TonEBP+/+ littermates were analyzed for body weight (A), pulse rate (B), SBP (C), plasma renin activity (D), and renal renin immunoblot (E). Mean+SEM, n=8. #P<0.05 compared with corresponding TonEBP+/+. VH, vehicle; +/+, wild-type; +/Δ, heterozygous; −/−, knock out.
Association of TonEBP Polymorphisms with Inflammatory, Vascular, and Renal Function Markers in Humans
Data in the previous sections demonstrate the relevance of TonEBP to glycemic stress–induced inflammation, renal function, and BP. Moreover, previously published associations of TonEBP expression with inflammation,6,22 rheumatoid arthritis,7,8 atherosclerosis,9 and DN in humans10 raised the possibility that variations in TonEBP might affect similar phenotypes in humans. Accordingly, we performed a look-up of TonEBP variant association in a highly homogeneous cohort of healthy individuals with minimal confounders (see Methods and Supplemental Material for details) with measures of inflammation, renal function, and BP.
We identified a total of 320 SNPs on the basis of full sequencing of the TonEBP gene region, from which we identified 16 haplotype blocks23 and selected 16 single haplotype tagging SNPs (Supplemental Figure 1). We then looked at the association between the 16 identified SNPs and our phenotypes. On the basis of Bonferroni correction for the number of SNPs, a P value <0.05/16=0.003 was considered statistically significant. We also identified SNP-based associations with P values <0.05 but >0.003 as suggestively associated. On the basis of this, we identified five significant and eight suggestive associations, as noted in Table 1. For our inflammatory phenotypes, we found significant associations between rs72783114 and serum matrix metalloprotease-1 (MMP-1) (P<0.001) and suggestive association with serum white blood cell count (P=0.04). We also found rs564919090 to be significantly associated with serum MMP-1 (P=0.001), independently of rs72783114. Given our previous findings of higher TonEBP expression in monocytes from individuals with DN,10 we also looked at the association with absolute monocyte values, which were available in a different group of 473 healthy Amish, and found a significant association between rs118095741 and absolute monocyte count (P=0.002). We also found rs74956396 to be suggestively associated with serum IL-1β (P<0.01) and homocysteine (P<0.01), whereas rs244416 was independently also suggestively associated with IL-1β (P<0.01). For our BP phenotypes, we found rs2287970 to be significantly associated with diastolic BP (DBP) (β=1.4, P=0.003) and suggestively with SBP (β=1.65, P=0.04). Lastly, for our renal phenotypes, we found a significant association between rs17297179 and eGFR (β=6.3, P=0.003) and suggestive association between rs17232663 and albuminuria (β=0.36, P<0.01). Further details are provided in the Supplemental Material. Secondary adjustments for eGFR and BP, as appropriate, did not modify our findings (see Supplemental Material). Functional annotations for our top identified SNPs are also summarized in Supplemental Table 3.
Table 1.
Top SNP associations with selected traits
| SNP | Allele | MAF (%) | Trait | n | Effect Size | P Value |
|---|---|---|---|---|---|---|
| rs564919090 | G | 5 | MMP-1 | 710 | 0.28 | 0.001a |
| rs72783114 | G | >5 | MMP-1 | >710 | >−0.31 | <0.001a |
| WBC | 771 | 0.06 | 0.04 | |||
| Monocyte | 473 | 56 | 0.01 | |||
| rs118095741 | G | 12 | Monocyte | 473 | 47.3 | 0.002a |
| rs74956396 | C | 3 | Homocysteine | 766 | −0.61 | <0.01 |
| IL-1β | 710 | 0.52 | <0.01 | |||
| rs244416 | C | 18 | IL-1β | 710 | −0.23 | <0.01 |
| DBP | 776 | −1.2 | 0.04 | |||
| rs2287970 | G | 37 | DBP | 776 | 1.4 | 0.003a |
| SBP | 776 | 1.65 | 0.04 | |||
| rs17232663 | G | 3 | Albuminuria | 668 | 0.36 | <0.01 |
| rs17297179 | A | 5 | eGFR | 776 | 6.34 | 0.003a |
For IL-1β, WBC, c-IMT, and MMP-1, log transformations were used to calculate effect size—IL-1β in pg/ml, MMP-1 in ng/ml. SNP, the SNP nucleotide associated with the outcome; allele, reference allele; MAF, minor allele frequency; n, sample size; effect size, effect size for SNP for selected traits, adjusted for age, sex, and family structure; MMP-1, MMP-1 (interstitial collagenase); WBC, white blood cell count; monocyte, absolute monocyte cell count; DBP, DBP (mmHg); albuminuria, log-transformed urinary albumin-to-creatinine ratio (mg/gm); eGFR, eGFR (ml/min per 1.73 m2).
Statistical significance on the basis of P<0.003 (0.05/16 haplotypes=0.003).
We also attempted to replicate our renal function associated SNP within the open source CKDGen consortium meta-GWAS results for eGFR24 (n=67,093) and albuminuria.25 Neither our albuminuria sequence based variant rs17232663 nor any SNPs in strong linkage disequilibrium (LD) with it were identified in the more limited HapMap based CKDGen database and hence could not be tested for replication. Our eGFR variant rs17297179 or other variants in strong LD were similarly not available. However, we were able to identify rs1064825, our second-best association with eGFR, which is in moderate LD with our top SNP (rs17297179), to be associated with eGFR in CKDGen (β=0.006, P<0.003), hence validating our finding.
In summary, in a highly homogeneous cohort of healthy individuals with minimal confounders, gene variants in TonEBP are associated with inflammation, renal function, and BP, all in accordance with our mouse TonEBP haplo-deficiency findings described here and previously. The association with renal function is replicated in the CKDGen cohort, suggesting its relevance to renal function.
Discussion
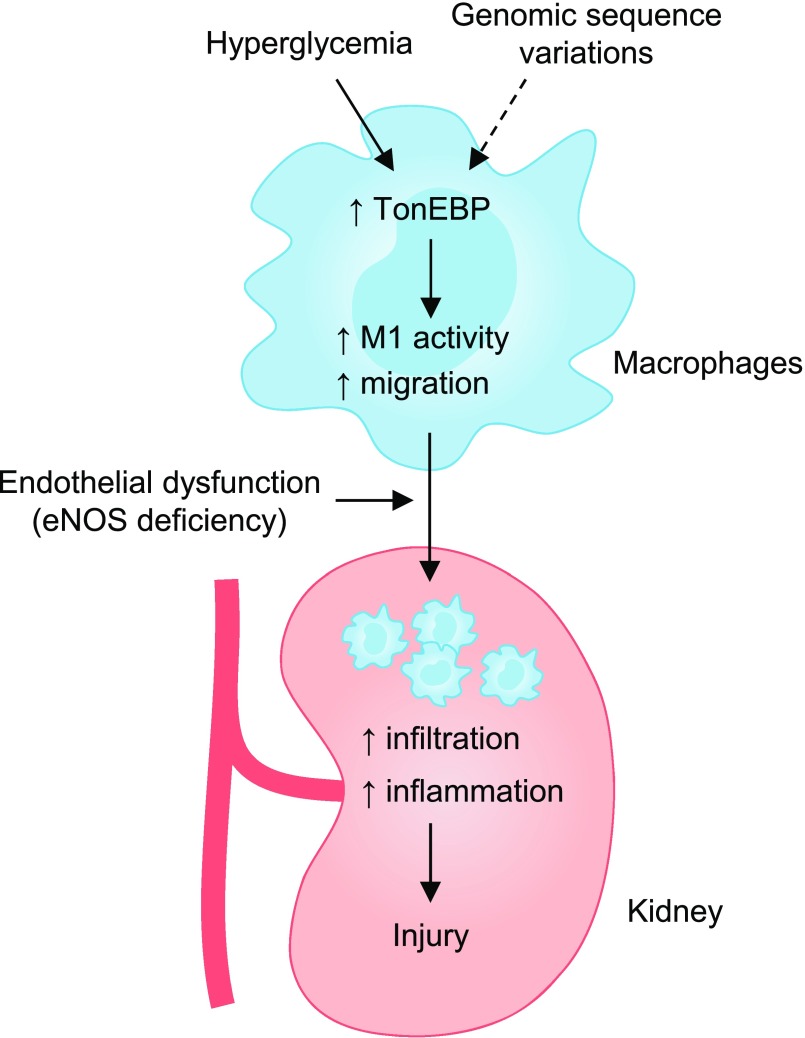
Numerous underlying molecular mechanisms involved in glucose toxicity have been defined, including oxidative stress and advanced glycan end products (for a review, see 26). Here, we have demonstrated that hyperglycemia is a proinflammatory signal for macrophages (Figure 8). The effects of hyperglycemia are similar to those of LPS6: like LPS, hyperglycemia enhances TonEBP expression which in turn drives the expression of proinflammatory genes via stimulation of NFκB. In addition, hyperglycemia stimulates the migration of macrophages, another key proinflammatory phenotype, which is also TonEBP-dependent. Even more interesting is the finding that TonEBP haplo-deficiency is associated with dramatically reduced expression of the proinflammatory genes as well as migration in macrophages. The reduced renal injury in response to STZ-induced diabetes in the TonEBP haplo-deficient animals can be readily explained by the reduced macrophage numbers in the kidney in combination with reduced expression of proinflammatory genes by the macrophages. These data provide a mechanistic basis for the association of TonEBP activity in monocytes and DN in patients with type 1diabetes.10 In line with this, we previously reported that TonEBP haplo-deficiency in bone marrow cells led to an 80% reduction in the size of atherosclerotic lesions.9 The same pathway in monocytes can also explain the dramatically reduced rheumatoid arthritis7,8 in the TonEBP haplo-deficient animals.
Figure 8.
TonEBP mediates hypergycemia-induced DN. In a mouse, TonEBP promotes inflammatory gene expression and cell migration leading to higher inflammation and renal injury due to enhanced infiltrating macrophages. TonEBP SNPs are associated with inflammation and renal function in humans. ↑, increase; M1, proinflammatory response.
Other investigators have reported that monocytes isolated from humans with both type 1 and type 2 diabetes display an inflammatory phenotype because they secrete higher levels of proinflammatory cytokines.27–29 The data presented here reveal that hyperglycemia stimulates NFκB via induction of TonEBP: TonEBP is a transcriptional cofactor of NFκB by way of recruiting histone acetyl transferase p300 leading to increased expression of proinflammatory cytokines and COX-26. Thus, TonEBP is a critical mediator of diabetes-induced inflammation in macrophages.
Given our animal-based findings, we examined whether TonEBP gene variants in humans were associated with phenotypes that are affected by TonEBP expression levels in our animal model and noted multiple consistent findings. This included significant associations with serum MMP-1 and absolute monocyte counts. This corroborates our previous finding of increases in TonEBP expression in monocytes of individuals with DN.10 Because IL-1β is a direct transcriptional target of TonEBP,5,6 the suggestive association of TonEBP SNPs with circulating levels of IL-1β and homocysteine observed in this study supports the notion that TonEBP genetic variability may possibly affect IL-1β expression in humans. The previously noted association between TonEBP variants and pulse pressure21 also confirms our association with BP, whereas the associations with eGFR and albuminuria are novel. Interestingly, in a cohort which included >700 individuals with DN, Kavanagh et al.30 found rs17297207 in TonEBP to be weakly associated with DN (OR, 0.66; P=0.04). The rs17297207 SNP from the Kavanagh et al. study is in perfect LD with rs118095741, which we found to be significantly associated with serum monocyte count (P<0.002) and MMP-1 measures in our population. This is also in agreement with our previously noted association of a near 50% increase in TonEBP expression in monocytes of type 1 diabetic individuals with proteinuria.10
Although several variants were each associated with multiple related phenotypes, as in our animal model, we also noted a significant amount of distinct associations between several variants and phenotypes. A plausible explanation for this is that these variants are associated with differential expression of distinct TonEBP isoforms. Indeed, given that this gene can regulate hundreds of genes,5,7 both as enhancer and suppressor, a complex regulatory process is arguably required. For example, it has been shown to stimulate genes as a DNA binding transcription factor31 or as a transcriptional cofactor for NFkB and other DNA binding proteins,6 while suppressing other genes by recruiting histone methylase32 or by recruiting DNA methylase to promoter regions (unpublished observations by H.H.L.). It has also been noted to have seven splice isoforms (Ensembl, GRCh38.p10), which in addition to tissue-specific enhancer and repressor can provide additional mechanisms for differential regulation of various downstream pathways. Consistent with our findings, three distinct TonEBP signals have been associated with various phenotypes, such as: (1) rs7193778 with uric acid and CRP,33,34 rs33063 with pulse pressure,21,35 and rs9980 with plasma osmolality,35 which are all in strong LD with each other and rs244416 from our analysis (associated with IL-1β and DBP); (2) rs12599391 with age at menarche36 and rs6499244 with allogeneic hematopoietic stem cell transplantation outcomes,37 which are in LD with each other but not with any of our identified SNP’s; and (3) rs17297207 with DN,30 which is in LD with our monocyte count–associated SNP (rs118095741). Lastly, in an interesting human case of TonEBP haplo-insufficiency, disturbances in both innate and adaptive immunity leading to intestinal autoimmunity38 were noted, again supporting our findings.
Limitations of our findings include the lack of information on the putative functional SNPs driving our associations, as well as their effect on TonEBP expression. Accordingly, we cannot prove that the noted associations between the TonEBP SNPs in our and previous studies are due to differential expression of TonEBP. Moreover, our set of statistically suggestive findings should not be over-interpreted without further validation with more data in independent cohorts. However, there are factors that suggest these suggestive findings, in addition to our significantly associated findings, are worth further study. Most notably, we have already established, via our animal-based experiments, mechanistic pathways by which TonEBP deficiency can affect our selected outcomes. Second, many of our findings are at least partially corroborated by related previous publications. Last, our findings are highly consistent across our mouse model and identified human phenotypes. These findings provide further impetus for the targeting of TonEBP as a potential novel treatment for inhibiting inflammation-based renal injury. Lastly, we previously showed that cerulenin could suppress the TonEBP-mediated proinflammatory activation of NFκB and downstream inflammation,16 demonstrating that the targeting of TonEBP may be a viable therapeutic option.
Concise Methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee at Ulsan National Institute of Science and Technology. The TonEBP+/∆ mice on C57BL/6 background were obtained from Dr. S.N. Ho.13 They were crossed back to the C57BL/6 line (The Jackson Laboratory, Bar Harbor, ME) to produce TonEBP+/∆ animals and their TonEBP+/+ littermates. Where indicated, the animals were bred to the eNOS-deficient (eNOS−/−) line on C75BL/6 (The Jackson Laboratory) to produce TonEBP+/∆, eNOS−/− mice and their littermates—TonEBP+/+, eNOS−/−. Mice were kept on a 12-hour light/dark cycle with free access to standard chow and water. Males were selected and made diabetic by daily intraperitoneal injections of freshly prepared STZ (50 mg/kg body weight; Sigma-Aldrich, St. Louis, MO) in 0.1 M citrate buffer (pH 4.5) for 4 days. Animals displaying fasting blood glucose levels >250 mg/dl after 2 weeks of STZ injections were considered diabetic. Control, nondiabetic animals were injected with the buffer. Six or 7 weeks post the STZ injections the animals were analyzed for macrophages (see below) and nephropathy. Body weight and blood glucose levels were monitored weakly. After spot urine was collected, animals were euthanized with intraperitoneal injection of Zoletil 50 (10 mg/kg; Virbac Laboratories, Carros, France) and Rompun (15 mg/kg; Bayer, Leuverkusen, Germany) to collect blood samples and tissue specimens. Fractional excretion of sodium (FENA), urine osmolality, urine creatinine, urine albumin, and BUN were measured as described previously with slight modification.39,40 Tissue sections were stained with periodic acid–Schiff and Trichrome to assess glomerular injury and renal interstitial fibrosis, respectively. Randomly selected fields were analyzed to quantify injuries and fibrosis. For BP and renin analyses, 5-week-old animals were used.
Raw264.7 Cells, PMs, and BMDMs
Raw264.7 cells were obtained from the American Type Culture Collection (Manassas, VA) and were grown in DMEM (Hyclone, Logan, UT) supplemented with 10% (v/v) FBS (Gibco BRL, Grand Island, NY). Where indicated, cells were switched to 25 mM D-glucose or 5.5 mM D-glucose plus 19.5 mM mannitol (osmotic control for high glucose). Growth media were changed every 12 hours to prevent glucose depletion. PMs were isolated from nondiabetic or diabetic mice as described.41 In short, 1 ml thioglycollate (30 mg/ml) was injected intraperitoneally and the macrophages were collected 4 days later. The cells were adhesion-purified for 1 hour followed by a wash with PBS to remove nonadherent cells. Bone marrow cells obtained from femurs were differentiated for 7 days using 20% L929-conditioned medium, as a source of M-CSF, to obtain BMDMs. Where indicated, cells were stimulated with 100 ng/ml LPS for 6 hours (RNA analysis) or 24 hours (for protein analysis).
Transfection and Luciferase Reporter Assay
Dicer-substrate small interfering RNA (siRNA) targeting TonEBP (5′-CCAGUUCCUACAAUGAUAACACUga-3′) and nontargeting negative control (scrambled) siRNA (5′-CGUUAAUCGCGUAUAAUACGCGUA-3′) were purchased from Integrated DNA Technologies (Coralville, IA). Raw264.7 cells were transfected for 1 day with 2 nM siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Transfected cells were cultured for another day in fresh culture medium before LPS treatment. For NF-κB luciferase assays, siRNA-transfected cells were transfected with the NF-κB–dependent luciferase reporter plasmid (pGL4.32[luc2P/NF-KB-RE/Hygro]; Promega, Madison, WI) using Lipofectamine 2000 reagent (Invitrogen). The phRL-TK vector was cotransfected to normalize for transfection efficiency. After 24 hours, the cells were treated with LPS for 8 hours. The cell lysates were prepared with a passive lysis buffer and used to measure the luciferase activity according to the manufacturer’s instructions for the luciferase reporter assay system (Promega). The luciferase assays were carried out using a GloMax 96 Microplate Luminometer (Promega).
RNA Isolation and Real-Time PCR
Total RNA was isolated using the TRIzol reagent according to the manufacturer’s instructions (Invitrogen). After reverse transcription, quantitative PCR was performed using SYBR Green I Master and LightCycler 480 II (Roche Applied Sciences, Indianapolis, IN) and primers described in Supplemental Table 4. Resulting cycle threshold (Ct) values were normalized with cyclophilin A and the ΔΔCt method was then used to express values as fold over control samples.
Western Blotting
Protein extraction from tissues was performed as previously described.39 Equal amounts of protein from samples were separated by SDS-PAGE and immunoblotted using specific antibodies. The antigen-antibody binding was detected by enhanced chemiluminescence western blotting detection reagents (GE Healthcare, Little Chalfont, UK). Primary antibodies used were anti-TonEBP antibody,31 anti-iNOS antibody (BD Biosciences, Franklin Lakes, NJ), and anti-Hsc70 (Rockland, Gilbertsville, PA).
Macrophage Migration Assay
Macrophage migration was measured using a modified Boyden chamber (NeuroProbe, Gaithersburg, MD). BMDMs were plated in the upper chamber on a 5-μm porous membrane and cultured in DMEM containing normal glucose (5.5 mM), high glucose (25 mM), or 5.5 mM glucose +19.5 mM mannitol. MCP-1 (10 ng/ml) was added to the lower chamber as a chemoattractant. After 16 hours, cells were removed from the upper side of membranes and nuclei of migratory cells on the lower side of the membrane were stained with DAPI. The number of migratory cells was visualized by fluorescence microscopy and quantified using ImageJ software.
Immunohistochemistry
Immunohistochemistry was performed as described previously.39 The F4/80, STAT3, and TGF-β1 were detected in 4-µm tissue sections by incubating tissue slides for 12 hours with specific antibodies against F4/80 (Serotec, Oxford, UK), STAT3 (Cell signaling, Boston, MA), and TGF-β1 (Proteintech, Chicago, IL) at 4°C.
Histologic Analyses
Kidneys were fixed with 2% paraformaldehyde-lysine-periodate and embedded in paraffin. Sections were stained with periodic acid–Schiff reaction plus hematoxylin counterstain. For quantitative assessment of glomerular injury, >50 glomeruli were examined from each animal. Percentage of glomeruli displaying mesangiolysis, mesangial expansion, microaneurysm, nodular lesions, and sclerosis was assessed in a blinded manner.
Blood Collection, Renin Activity, and Renin Immunoblot
Blood was taken from conscious mice by tail vein puncture and collected into a 75-μl hematocrit tube that contained 1 μl of 125 mM EDTA at its tip. Plasma was collected by centrifugation and stored frozen. Renin concentration was measured by Gammacoat plasma renin activity radioimmunoassay kit (DiaSorin, Stillwater, MN). Renal renin expression was evaluated using immunoblot analysis. Renin was detected by incubating for 12 hours with specific antibody, goat polyclonal renin antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Relative OD of the band in each lane was normalized relative to the density of the Hsc70 band from the same gel.
BP and Heart Rate
SBP and heart rate of TonEBP heterozygotes and their wild-type littermates were determined by noninvasive tail-cuff BP system (Hatteras Instruments SC-1000, Cary, NC). Animals were conditioned by placing them into the holding device on three consecutive days before the first measurement. BP was determined for 3 days in a row, and values were calculated as averages of these three measurements for each individual mouse.
Human Population, Genotyping, and Phenotype Analyses
Our primary population consisted of 868 participants from the HAPI Heart study.42 These participants were healthy white individuals from the Old order closed Amish founder population in Lancaster, PA. Subjects recruited for this study were ≥20 years old and were excluded if they had severe hypertension (BP>180/105 mmHg); malignancy; or kidney, liver, or untreated thyroid disease. Additionally, all participants were not on any active medications at the time of the study.42 This unique genetically and environmentally homogenous population was selected to minimize potential confounders and enhance the ability to detect genetic contributions to a variety of cardiovascular phenotypes, including selected markers of systemic inflammation. Resting protocol–based SBP and DBP measures were also obtained while participants were off any antihypertensive medications. Baseline measures of inflammatory markers: serum IL-1β an inflammatory cytokine, homocysteine, MMP-1, matrix metalloprotease-9 (MMP-9), C-reactive protein (CRP), monocyte count, and white blood cell count as markers of systemic inflammation.43,44 Albuminuria was on the basis of spot morning collected urine albumin-to-creatinine ratio. GFR was estimated on the basis of serum creatinine using the Modification of Diet in Renal Disease formula.45 Details regarding methods for selection of individuals and phenotype measures have been previously published.42,43,46,47
Genotyping by whole-genome sequencing was done under the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine program (http://www.nhlbi.nih.gov/research/resources/nhlbi-precision-medicine-initiative/topmed). We selected all SNPs with MAF≥3% within a 2-kb window of the TonEBP gene. We then used Haploview to identify the number of haplotype blocks and tagging SNPs.23 We then looked at the association between the identified tagging SNPs and our selected phenotypes. Once we identified our top candidate SNPs for each phenotype, we used the HaploReg software package V4.1 to extract the functional annotation for each SNP of interest.36 For each variant, we looked for its predicted chromatin segmentation, including histone markers, focusing on enhancers as well as DNase I hypersensitive sites across a variety of tissue and cell types. Expression quantitative trait loci annotation was on the basis of the Genotype-Tissue Expression Project, the Geuvadis RNA-sequencing project, and the latest NHLBI-supported GRASP databases.
All SNP to phenotype associations were adjusted for age, sex, and family relatedness using the Mixed Models Analysis for Populations and Pedigrees (MMAP) program (http://edn.som.umaryland.edu/mmap/index.php). All baseline inflammatory marker and albuminuria values were natural logarithm transformed to remove skewness.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation grants (NRF-2011-0020163, 2016R1D1A1B03932335) and Health Technology Research and Development Project grant (HI16C1837) of Korea, and Ulsan National Institute of Science and Technology Intramural Support (1.160078.01). This work was also supported in part from National Institutes of Health (NIH) grants U01 HL072515 and P30 DK072488. The whole-genome sequencing in Trans-Omics for Precision Medicine (TOPMed) was performed at the Broad Institute of Massachusetts Institute of Technology and Harvard (HHSN268201500014C). Centralized joint genotype calling, mapping, and harmonization, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1, Principle Investigator Abecasis). Dr. Salimi is supported by NIH training grant T32 AG00262.
S.Y.C., S.W.L., A.P., and H.M.K. conceived and designed the experiments. S.Y.C., S.W.L., S.S., E.J.Y., W.L-K., H.H.L., J.H.L., B.D.M., and S.S. performed the experiments and analyzed data. H.M.K. and A.P. supervised the research and wrote the manuscript with input and editing from all of the authors.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017070718/-/DCSupplemental.
References
- 1.US Renal Data System: 2015 Annual Data Report: Chapter 2. End-stage Renal Disease in the US, 2015. Available at: www.usrds.org.
- 2.Wuttke M, Köttgen A: Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol 12: 549–562, 2016 [DOI] [PubMed] [Google Scholar]
- 3.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, Li Y, Tayo B, Tin A, Feitosa MF, Aspelund T, Attia J, Biffar R, Bochud M, Boerwinkle E, Borecki I, Bottinger EP, Chen MH, Chouraki V, Ciullo M, Coresh J, Cornelis MC, Curhan GC, d’Adamo AP, Dehghan A, Dengler L, Ding J, Eiriksdottir G, Endlich K, Enroth S, Esko T, Franco OH, Gasparini P, Gieger C, Girotto G, Gottesman O, Gudnason V, Gyllensten U, Hancock SJ, Harris TB, Helmer C, Höllerer S, Hofer E, Hofman A, Holliday EG, Homuth G, Hu FB, Huth C, Hutri-Kähönen N, Hwang SJ, Imboden M, Johansson Å, Kähönen M, König W, Kramer H, Krämer BK, Kumar A, Kutalik Z, Lambert JC, Launer LJ, Lehtimäki T, de Borst M, Navis G, Swertz M, Liu Y, Lohman K, Loos RJF, Lu Y, Lyytikäinen LP, McEvoy MA, Meisinger C, Meitinger T, Metspalu A, Metzger M, Mihailov E, Mitchell P, Nauck M, Oldehinkel AJ, Olden M, Wjh Penninx B, Pistis G, Pramstaller PP, Probst-Hensch N, Raitakari OT, Rettig R, Ridker PM, Rivadeneira F, Robino A, Rosas SE, Ruderfer D, Ruggiero D, Saba Y, Sala C, Schmidt H, Schmidt R, Scott RJ, Sedaghat S, Smith AV, Sorice R, Stengel B, Stracke S, Strauch K, Toniolo D, Uitterlinden AG, Ulivi S, Viikari JS, Völker U, Vollenweider P, Völzke H, Vuckovic D, Waldenberger M, Jin Wang J, Yang Q, Chasman DI, Tromp G, Snieder H, Heid IM, Fox CS, Köttgen A, Pattaro C, Böger CA, Fuchsberger C: 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7: 45040, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsa A, Kanetsky PA, Xiao R, Gupta J, Mitra N, Limou S, Xie D, Xu H, Anderson AH, Ojo A, Kusek JW, Lora CM, Hamm LL, He J, Sandholm N, Jeff J, Raj DE, Böger CA, Bottinger E, Salimi S, Parekh RS, Adler SG, Langefeld CD, Bowden DW, Groop PH, Forsblom C, Freedman BI, Lipkowitz M, Fox CS, Winkler CA, Feldman HI; FIND Consortium; and the Chronic Renal Insufficiency Cohort (CRIC) Study Investigators : Genome-wide association of CKD progression: The chronic renal insufficiency cohort study. J Am Soc Nephrol 28: 923–934, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buxadé M, Lunazzi G, Minguillón J, Iborra S, Berga-Bolaños R, Del Val M, Aramburu J, López-Rodríguez C: Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med 209: 379–393, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee HH, Sanada S, An SM, Ye BJ, Lee JH, Seo YK, Lee C, Lee-Kwon W, Küper C, Neuhofer W, Choi SY, Kwon HM: LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding. Sci Rep 6: 24921, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon HJ, You S, Yoo SA, Kim NH, Kwon HM, Yoon CH, Cho CS, Hwang D, Kim WU: NF-AT5 is a critical regulator of inflammatory arthritis. Arthritis Rheum 63: 1843–1852, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi S, You S, Kim D, Choi SY, Kwon HM, Kim HS, Hwang D, Park YJ, Cho CS, Kim WU: Transcription factor NFAT5 promotes macrophage survival in rheumatoid arthritis. J Clin Invest 127: 954–969, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halterman JA, Kwon HM, Leitinger N, Wamhoff BR: NFAT5 expression in bone marrow-derived cells enhances atherosclerosis and drives macrophage migration. Front Physiol 3: 313, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, Hodgkinson AD, Oates PJ, Kwon HM, Millward BA, Demaine AG: Elevated activity of transcription factor nuclear factor of activated T-cells 5 (NFAT5) and diabetic nephropathy. Diabetes 55: 1450–1455, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Chow F, Ozols E, Nikolic-Paterson DJ, Atkins RC, Tesch GH: Macrophages in mouse type 2 diabetic nephropathy: Correlation with diabetic state and progressive renal injury. Kidney Int 65: 116–128, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Nguyen D, Ping F, Mu W, Hill P, Atkins RC, Chadban SJ: Macrophage accumulation in human progressive diabetic nephropathy. Nephrology (Carlton) 11: 226–231, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Go WY, Liu X, Roti MA, Liu F, Ho SN: NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc Natl Acad Sci U S A 101: 10673–10678, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro-González JF, Mora-Fernández C: The role of inflammatory cytokines in diabetic nephropathy. J Am Soc Nephrol 19: 433–442, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Wolkow PP, Niewczas MA, Perkins B, Ficociello LH, Lipinski B, Warram JH, Krolewski AS: Association of urinary inflammatory markers and renal decline in microalbuminuric type 1 diabetics. J Am Soc Nephrol 19: 789–797, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M; European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Lim AK, Tesch GH: Inflammation in diabetic nephropathy. Mediators Inflamm 2012: 146154, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa T, Sato W, Glushakova O, Heinig M, Clarke T, Campbell-Thompson M, Yuzawa Y, Atkinson MA, Johnson RJ, Croker B: Diabetic endothelial nitric oxide synthase knockout mice develop advanced diabetic nephropathy. J Am Soc Nephrol 18: 539–550, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Li F, Wang CH, Wang JG, Thai T, Boysen G, Xu L, Turner AL, Wolberg AS, Mackman N, Maeda N, Takahashi N: Elevated tissue factor expression contributes to exacerbated diabetic nephropathy in mice lacking eNOS fed a high fat diet. J Thromb Haemost 8: 2122–2132, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan S, Reddick RL, Musi N, Horn DA, Yan B, Prihoda TJ, Natarajan M, Abboud-Werner SL: Diabetic eNOS knockout mice develop distinct macro- and microvascular complications. Lab Invest 88: 515–528, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Tragante V, Barnes MR, Ganesh SK, Lanktree MB, Guo W, Franceschini N, Smith EN, Johnson T, Holmes MV, Padmanabhan S, Karczewski KJ, Almoguera B, Barnard J, Baumert J, Chang YP, Elbers CC, Farrall M, Fischer ME, Gaunt TR, Gho JM, Gieger C, Goel A, Gong Y, Isaacs A, Kleber ME, Mateo Leach I, McDonough CW, Meijs MF, Melander O, Nelson CP, Nolte IM, Pankratz N, Price TS, Shaffer J, Shah S, Tomaszewski M, van der Most PJ, Van Iperen EP, Vonk JM, Witkowska K, Wong CO, Zhang L, Beitelshees AL, Berenson GS, Bhatt DL, Brown M, Burt A, Cooper-DeHoff RM, Connell JM, Cruickshanks KJ, Curtis SP, Davey-Smith G, Delles C, Gansevoort RT, Guo X, Haiqing S, Hastie CE, Hofker MH, Hovingh GK, Kim DS, Kirkland SA, Klein BE, Klein R, Li YR, Maiwald S, Newton-Cheh C, O’Brien ET, Onland-Moret NC, Palmas W, Parsa A, Penninx BW, Pettinger M, Vasan RS, Ranchalis JE, M Ridker P, Rose LM, Sever P, Shimbo D, Steele L, Stolk RP, Thorand B, Trip MD, van Duijn CM, Verschuren WM, Wijmenga C, Wyatt S, Young JH, Zwinderman AH, Bezzina CR, Boerwinkle E, Casas JP, Caulfield MJ, Chakravarti A, Chasman DI, Davidson KW, Doevendans PA, Dominiczak AF, FitzGerald GA, Gums JG, Fornage M, Hakonarson H, Halder I, Hillege HL, Illig T, Jarvik GP, Johnson JA, Kastelein JJ, Koenig W, Kumari M, März W, Murray SS, O’Connell JR Jr, Oldehinkel AJ, Pankow JS, Rader DJ, Redline S, Reilly MP, Schadt EE, Kottke-Marchant K, Snieder H, Snyder M, Stanton AV, Tobin MD, Uitterlinden AG, van der Harst P, van der Schouw YT, Samani NJ, Watkins H, Johnson AD, Reiner AP, Zhu X, de Bakker PI, Levy D, Asselbergs FW, Munroe PB, Keating BJ: Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet 94: 349–360, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi SY, Lee HH, Lee JH, Ye BJ, Yoo EJ, Kang HJ, Jung GW, An SM, Lee-Kwon W, Chiong M, Lavandero S, Kwon HM: TonEBP suppresses IL-10-mediated immunomodulation. Sci Rep 6: 25726, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ: Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Böger CA, Chen MH, Tin A, Olden M, Köttgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tönjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarød K, Skorpen F, Syvänen AC, Illig T, Baumert J, Koenig W, Krämer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Völzke H, Stumvoll M, Mägi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlöv J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH; CKDGen Consortium : CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes JM, Cooper ME: Mechanisms of diabetic complications. Physiol Rev 93: 137–188, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Devaraj S, Glaser N, Griffen S, Wang-Polagruto J, Miguelino E, Jialal I: Increased monocytic activity and biomarkers of inflammation in patients with type 1 diabetes. Diabetes 55: 774–779, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Bradshaw EM, Raddassi K, Elyaman W, Orban T, Gottlieb PA, Kent SC, Hafler DA: Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol 183: 4432–4439, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jagannathan-Bogdan M, McDonnell ME, Shin H, Rehman Q, Hasturk H, Apovian CM, Nikolajczyk BS: Elevated proinflammatory cytokine production by a skewed T cell compartment requires monocytes and promotes inflammation in type 2 diabetes. J Immunol 186: 1162–1172, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavanagh DH, Savage DA, Patterson CC, McKnight AJ, Crean JK, Maxwell AP, McKay GJ; Warren 3/UK GoKinD Study Group : Haplotype association analysis of genes within the WNT signalling pathways in diabetic nephropathy. BMC Nephrol 14: 126, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM: Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci U S A 96: 2538–2542, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Lee HH, Ye BJ, Lee-Kwon W, Choi SY, Kwon HM: TonEBP suppresses adipogenesis and insulin sensitivity by blocking epigenetic transition of PPARγ2. Sci Rep 5: 10973, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köttgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Pistis G, Ruggiero D, O’Seaghdha CM, Haller T, Yang Q, Tanaka T, Johnson AD, Kutalik Z, Smith AV, Shi J, Struchalin M, Middelberg RP, Brown MJ, Gaffo AL, Pirastu N, Li G, Hayward C, Zemunik T, Huffman J, Yengo L, Zhao JH, Demirkan A, Feitosa MF, Liu X, Malerba G, Lopez LM, van der Harst P, Li X, Kleber ME, Hicks AA, Nolte IM, Johansson A, Murgia F, Wild SH, Bakker SJ, Peden JF, Dehghan A, Steri M, Tenesa A, Lagou V, Salo P, Mangino M, Rose LM, Lehtimäki T, Woodward OM, Okada Y, Tin A, Müller C, Oldmeadow C, Putku M, Czamara D, Kraft P, Frogheri L, Thun GA, Grotevendt A, Gislason GK, Harris TB, Launer LJ, McArdle P, Shuldiner AR, Boerwinkle E, Coresh J, Schmidt H, Schallert M, Martin NG, Montgomery GW, Kubo M, Nakamura Y, Tanaka T, Munroe PB, Samani NJ, Jacobs DR Jr, Liu K, D’Adamo P, Ulivi S, Rotter JI, Psaty BM, Vollenweider P, Waeber G, Campbell S, Devuyst O, Navarro P, Kolcic I, Hastie N, Balkau B, Froguel P, Esko T, Salumets A, Khaw KT, Langenberg C, Wareham NJ, Isaacs A, Kraja A, Zhang Q, Wild PS, Scott RJ, Holliday EG, Org E, Viigimaa M, Bandinelli S, Metter JE, Lupo A, Trabetti E, Sorice R, Döring A, Lattka E, Strauch K, Theis F, Waldenberger M, Wichmann HE, Davies G, Gow AJ, Bruinenberg M, Stolk RP, Kooner JS, Zhang W, Winkelmann BR, Boehm BO, Lucae S, Penninx BW, Smit JH, Curhan G, Mudgal P, Plenge RM, Portas L, Persico I, Kirin M, Wilson JF, Mateo Leach I, van Gilst WH, Goel A, Ongen H, Hofman A, Rivadeneira F, Uitterlinden AG, Imboden M, von Eckardstein A, Cucca F, Nagaraja R, Piras MG, Nauck M, Schurmann C, Budde K, Ernst F, Farrington SM, Theodoratou E, Prokopenko I, Stumvoll M, Jula A, Perola M, Salomaa V, Shin SY, Spector TD, Sala C, Ridker PM, Kähönen M, Viikari J, Hengstenberg C, Nelson CP, Meschia JF, Nalls MA, Sharma P, Singleton AB, Kamatani N, Zeller T, Burnier M, Attia J, Laan M, Klopp N, Hillege HL, Kloiber S, Choi H, Pirastu M, Tore S, Probst-Hensch NM, Völzke H, Gudnason V, Parsa A, Schmidt R, Whitfield JB, Fornage M, Gasparini P, Siscovick DS, Polašek O, Campbell H, Rudan I, Bouatia-Naji N, Metspalu A, Loos RJ, van Duijn CM, Borecki IB, Ferrucci L, Gambaro G, Deary IJ, Wolffenbuttel BH, Chambers JC, März W, Pramstaller PP, Snieder H, Gyllensten U, Wright AF, Navis G, Watkins H, Witteman JC, Sanna S, Schipf S, Dunlop MG, Tönjes A, Ripatti S, Soranzo N, Toniolo D, Chasman DI, Raitakari O, Kao WH, Ciullo M, Fox CS, Caulfield M, Bochud M, Gieger C; LifeLines Cohort Study; CARDIoGRAM Consortium; DIAGRAM Consortium; ICBP Consortium; MAGIC Consortium : Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 45: 145–154, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsa A, Brown E, Weir MR, Fink JC, Shuldiner AR, Mitchell BD, McArdle PF: Genotype-based changes in serum uric acid affect blood pressure. Kidney Int 81: 502–507, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Böger CA, Gorski M, McMahon GM, Xu H, Chang YC, van der Most PJ, Navis G, Nolte IM, de Borst MH, Zhang W, Lehne B, Loh M, Tan ST, Boerwinkle E, Grams ME, Sekula P, Li M, Wilmot B, Moon JG, Scheet P, Cucca F, Xiao X, Lyytikäinen LP, Delgado G, Grammer TB, Kleber ME, Sedaghat S, Rivadeneira F, Corre T, Kutalik Z, Bergmann S, Nielson CM, Srikanth P, Teumer A, Müller-Nurasyid M, Brockhaus AC, Pfeufer A, Rathmann W, Peters A, Matsumoto M, de Andrade M, Atkinson EJ, Robinson-Cohen C, de Boer IH, Hwang SJ, Heid IM, Gögele M, Concas MP, Tanaka T, Bandinelli S, Nalls MA, Singleton A, Tajuddin SM, Adeyemo A, Zhou J, Doumatey A, McWeeney S, Murabito J, Franceschini N, Flessner M, Shlipak M, Wilson JG, Chen G, Rotimi CN, Zonderman AB, Evans MK, Ferrucci L, Devuyst O, Pirastu M, Shuldiner A, Hicks AA, Pramstaller PP, Kestenbaum B, Kardia SL, Turner ST, Study LC, Briske TE, Gieger C, Strauch K, Meisinger C, Meitinger T, Völker U, Nauck M, Völzke H, Vollenweider P, Bochud M, Waeber G, Kähönen M, Lehtimäki T, März W, Dehghan A, Franco OH, Uitterlinden AG, Hofman A, Taylor HA, Chambers JC, Kooner JS, Fox CS, Hitzemann R, Orwoll ES, Pattaro C, Schlessinger D, Köttgen A, Snieder H, Parsa A, Cohen DM: NFAT5 and SLC4A10 loci associate with plasma osmolality. J Am Soc Nephrol 28: 2311–2321, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CT, Fernández-Rhodes L, Brzyski RG, Carlson CS, Chen Z, Heiss G, North KE, Woods NF, Rajkovic A, Kooperberg C, Franceschini N: Replication of loci influencing ages at menarche and menopause in Hispanic women: The Women’s Health Initiative SHARe study. Hum Mol Genet 21: 1419–1432, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín-Antonio B, Álvarez-Laderas I, Cardesa R, Márquez-Malaver F, Baez A, Carmona M, Falantes J, Suarez-Lledo M, Fernández-Avilés F, Martínez C, Rovira M, Espigado I, Urbano-Ispizua Á: A constitutional variant in the transcription factor EP300 strongly influences the clinical outcome of patients submitted to allo-SCT. Bone Marrow Transplant 47: 1206–1211, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Boland BS, Widjaja CE, Banno A, Zhang B, Kim SH, Stoven S, Peterson MR, Jones MC, Su HI, Crowe SE, Bui JD, Ho SB, Okugawa Y, Goel A, Marietta EV, Khosroheidari M, Jepsen K, Aramburu J, López-Rodríguez C, Sandborn WJ, Murray JA, Harismendy O, Chang JT: Immunodeficiency and autoimmune enterocolopathy linked to NFAT5 haploinsufficiency. J Immunol 194: 2551–2560, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheen MR, Kim JA, Lim SW, Jung JY, Han KH, Jeon US, Park SH, Kim J, Kwon HM: Interstitial tonicity controls TonEBP expression in the renal medulla. Kidney Int 75: 518–525, 2009 [DOI] [PubMed] [Google Scholar]
- 40.Lim SW, Doh KC, Jin L, Jin J, Piao SG, Heo SB, Chung BH, Yang CW: Ginseng treatment attenuates autophagic cell death in chronic cyclosporine nephropathy. Nephrology (Carlton) 19: 490–499, 2014 [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Goncalves R, Mosser DM: The isolation and characterization of murine macrophages. Curr Protoc Immunol 83:14.1: 14.1.1–14.1.14, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell BD, McArdle PF, Shen H, Rampersaud E, Pollin TI, Bielak LF, Jaquish C, Douglas JA, Roy-Gagnon MH, Sack P, Naglieri R, Hines S, Horenstein RB, Chang YP, Post W, Ryan KA, Brereton NH, Pakyz RE, Sorkin J, Damcott CM, O’Connell JR, Mangano C, Corretti M, Vogel R, Herzog W, Weir MR, Peyser PA, Shuldiner AR: The genetic response to short-term interventions affecting cardiovascular function: Rationale and design of the Heredity and Phenotype Intervention (HAPI) Heart study. Am Heart J 155: 823–828, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng YC, Kao WH, Mitchell BD, Sharrett AR, Ryan KA, Vogel RA, Shuldiner AR, Pollin TI: Genetic effects on postprandial variations of inflammatory markers in healthy individuals. Obesity (Silver Spring) 18: 1417–1422, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keyszer G, Lambiri I, Nagel R, Keysser C, Keysser M, Gromnica-Ihle E, Franz J, Burmester GR, Jung K: Circulating levels of matrix metalloproteinases MMP-3 and MMP-1, tissue inhibitor of metalloproteinases 1 (TIMP-1), and MMP-1/TIMP-1 complex in rheumatic disease. Correlation with clinical activity of rheumatoid arthritis versus other surrogate markers. J Rheumatol 26: 251–258, 1999 [PubMed] [Google Scholar]
- 45.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D: A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of diet in renal disease study group. Ann Intern Med 130: 461–470, 1999 [DOI] [PubMed] [Google Scholar]
- 46.Post W, Bielak LF, Ryan KA, Cheng YC, Shen H, Rumberger JA, Sheedy PF 2nd, Shuldiner AR, Peyser PA, Mitchell BD: Determinants of coronary artery and aortic calcification in the old order Amish. Circulation 115: 717–724, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell GF, Verwoert GC, Tarasov KV, Isaacs A, Smith AV, Yasmin, Rietzschel ER, Tanaka T, Liu Y, Parsa A, Najjar SS, O’Shaughnessy KM, Sigurdsson S, De Buyzere ML, Larson MG, Sie MP, Andrews JS, Post WS, Mattace-Raso FU, McEniery CM, Eiriksdottir G, Segers P, Vasan RS, van Rijn MJ, Howard TD, McArdle PF, Dehghan A, Jewell ES, Newhouse SJ, Bekaert S, Hamburg NM, Newman AB, Hofman A, Scuteri A, De Bacquer D, Ikram MA, Psaty BM, Fuchsberger C, Olden M, Wain LV, Elliott P, Smith NL, Felix JF, Erdmann J, Vita JA, Sutton-Tyrrell K, Sijbrands EJ, Sanna S, Launer LJ, De Meyer T, Johnson AD, Schut AF, Herrington DM, Rivadeneira F, Uda M, Wilkinson IB, Aspelund T, Gillebert TC, Van Bortel L, Benjamin EJ, Oostra BA, Ding J, Gibson Q, Uitterlinden AG, Abecasis GR, Cockcroft JR, Gudnason V, De Backer GG, Ferrucci L, Harris TB, Shuldiner AR, van Duijn CM, Levy D, Lakatta EG, Witteman JC: Common genetic variation in the 3′-BCL11B gene desert is associated with carotid-femoral pulse wave velocity and excess cardiovascular disease risk: The AortaGen Consortium. Circ Cardiovasc Genet 5: 81–90, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.