Abstract
Recent exponential advances in genome sequencing and engineering technologies have enabled an unprecedented level of interrogation into the impact of DNA variations (genotype) on cellular function (phenotype). Furthermore, these advances have also prompted realistic discussion of writing and radically re-writing complex genomes. In this Perspective, we detail the motivation for large-scale engineering, discuss the progress made from such projects in bacteria and yeast and describe how various genome-engineering technologies will contribute to this effort. Finally, we describe the features of an ideal platform and provide a roadmap to facilitate the efficient writing of large genomes.
Graphical Abstract
Advances in genome sequencing, editing, and synthetic biology have enhanced the feasibility of large-scale genome engineering, termed genome writing. In this Opinion article, Chari and Church discuss the strengths and limitations of diverse strategies for genome writing, including extensively modifying existing genomes versus synthesizing genomes de novo, and they provide future visions for writing large genomes.
DNA reading (sequencing) and writing (synthesis) are intimately connected: meaningful use of either requires the other. For example, the most widely used sequencing platforms read DNA during the synthesis of a complementary strand1,2,3. Targeted DNA reading, such as exome sequencing, depends on vast libraries of synthetic oligonucleotides for target capture, and the annotation and understanding of genome reads benefit greatly from genome-scale synthesis and testing. Furthermore, meaningful use of genome engineering generally requires whole-genome sequencing to design strategies for avoiding off-target writing and to verify the quality of the final genome product. Genome editing and writing has benefitted from diverse nanomachines harvested by reading the biosphere. Since 1975, reading and writing have exhibited increases in throughput of three-million-fold (for reading; see https://www.genome.gov/sequencingcostsdata/) and one-billion-fold (for writing of raw oligonucleotides)1, with a steep increase at an exponential rate since 2004. The technical progress of both reading and writing has depended not only on mere automation of human protocols, but on rethinking the whole process to permit miniaturization and multiplexing (in which millions of reactions can be performed in the space, at the same cost and with the same effort as a single reaction).
Although much of the attention has been on the progress in DNA reading, namely, advances in DNA sequencing technologies2,3, the improvements in writing, which can largely be attributed to harnessing and stimulating natural cellular processes such as homologous recombination (HR) to enable genome modification, should not be overlooked. From the initial work of describing HR at a very low level in higher-order eukaryotic cells to current work, where we can achieve reasonably high efficiencies of HR in stem cells4–7 (Figure 1), our ability to direct targeted changes in genomes has opened a large number of avenues that previously may have not seemed possible.
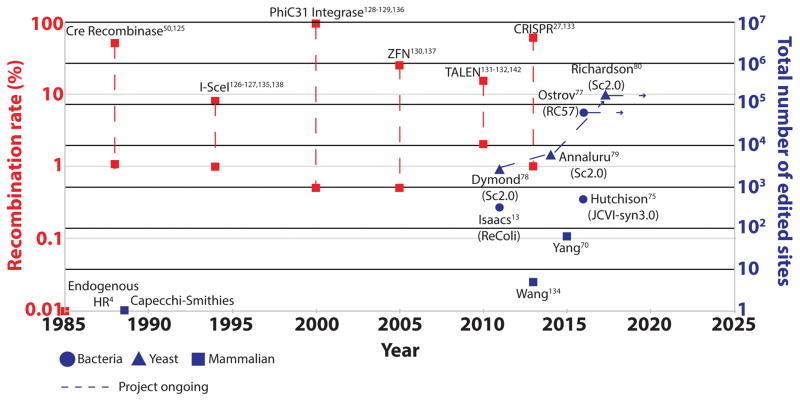
Figure 1. Timeline of gene targeting.
The timeline depicts progress in gene targeting rates through recombination (red) and the number of edited sites in a single genome (blue). Recombination rates with dashed lines denote ranges observed in the published literature4,27,28,50,58,63,64,125–147. In the mid to late 1980s, it was observed that in the presence of exogenous DNA, homologous recombination (HR) could occur at a very low frequency (0.01%). Subsequently, it was shown that by inducing a double-strand break (DSB), the rates of HR could be dramatically improved. Thus, different modalities have been discovered and employed to create DSBs at higher frequencies, from meganucleases (such as I-SceI) to zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) to the current CRISPR-associated nucleases. In addition, approaches that do not use DSB-stimulated HR such as Cre recombinase or phiC31 integrase, have also shown tremendous promise for DNA replacement, but are restricted by the limited range of sequences that can be targeted. As a corollary, in combination with the progress in DNA synthesis technologies, our improved ability to edit DNA has permitted increases in the total number of modifications being made in a single genome. In the past, 1 to 2 edits were typically made in a single genome. Ambitious projects such as RC57 and Sc2.0 have aimed to make up to four or even five orders of magnitude more edits in a single genome.
One of the main areas that can be realistically pursued is the ability to perform large-scale editing of genomes, even multiplexing libraries of billions of changes8. From an industrial perspective to a healthcare perspective and to researchers aiming for fundamental understanding of how biological systems work, the motivations to pursue ambitious genome engineering projects originate from a diverse group of stakeholders, and the results of these projects could have considerable positive impact on multiple aspects of society. Combining the technological advancements in genome engineering with those in both DNA synthesis and DNA sequencing, the key components that are necessary to approach large-scale genome engineering have fallen into place.
In this Opinion article, we describe the motivation for and current state of large-scale genome engineering and compare strategies for how it can be achieved. We provide an overview of key genome engineering technologies and how they can be utilized for large-scale editing. Finally, we detail the key features that will be necessary for a robust genome-writing platform and provide a roadmap to engineer a large genome. As seen for whole-genome reading, the cost and quality of methods for genome writing are improving by factors of millions, and rigorous, comprehensive tests for the accuracy of genome writing will shift from being a luxury to becoming essential and routine.
Why should we engineer genomes?
When we refer to genome engineering and genome editing, our use of ‘genome’ specifically means large-scale engineering or alteration of several loci across the genome. We feel that such terminology is unhelpfully misapplied when referring to single-locus applications, such as using genome engineering to refer to editing that is done directly in the genome rather than in a plasmid or transgenic setting or using the term ‘genome editing’ when the changes are neither precise editing nor genome scale. The question for this section is why should we design and generate whole (or nearly whole) genomes? An initiative known as Genome Project Write has recently advocated large-scale genome engineering9. The motivation and goals are well aligned for both small-scale and large-scale genome engineering. With respect to non-human organisms, bacteria are common chassis for various bioproduction applications, from uses in bioremediation to pharmaceuticals to bioenergy10–12 (Figure 2). From a purely functional perspective, re-engineering the genomes of particular strains could render the bacteria less susceptible to viruses and/or more efficient at producing a biomolecule of interest13,14. Similarly, yeast strains, plants and livestock could be thought of in the same manner, and re-engineering their genomes could ensure the highest and most efficient productivity. For some of these particular entities, genome engineering could address global problems such as food shortage and environmental deterioration, for example, by altering sets of genes in metabolic pathways, which would permit either increased or reduced production of specific biomolecules. Thus, an ability to perform genome engineering at a larger scale would only enhance our chances of achieving these goals.
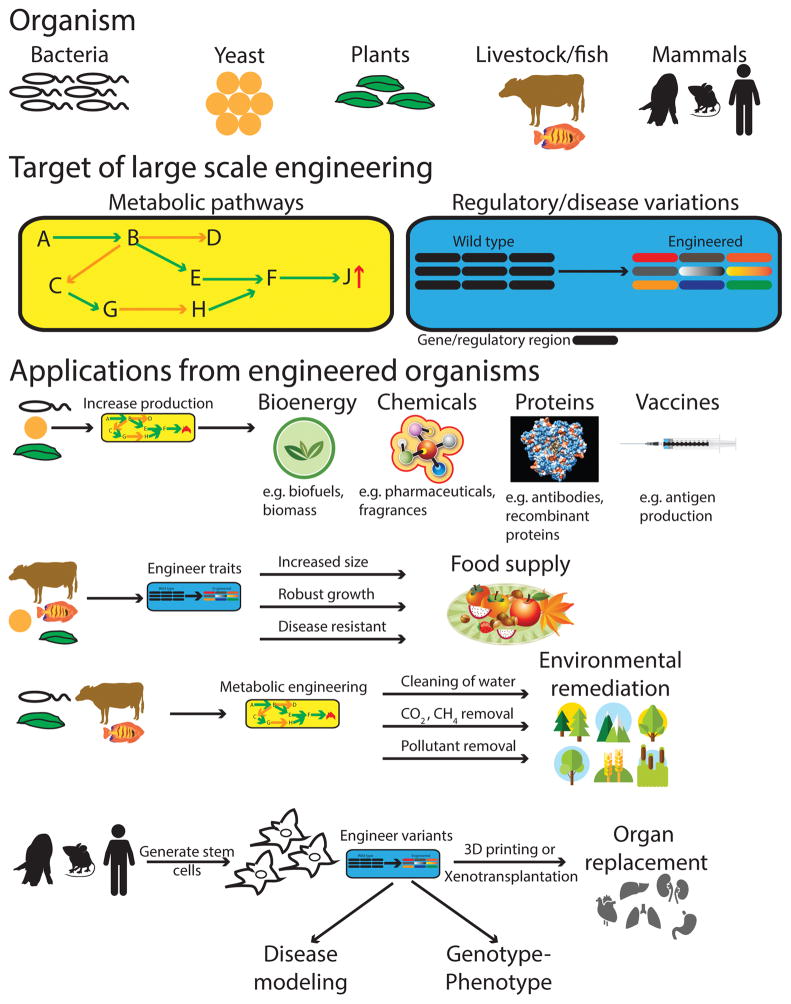
Figure 2. Potential applications of large-scale genome engineering of different organisms.
For applications such as bioenergy, chemicals, proteins, and vaccines, the goal is to increase production of these entities by engineering metabolic pathways in particular organisms. Similarly, engineering of the metabolic pathways in these organisms could also be used for environmental remediation, from maintaining clean water supplies to removing pollutants and CO2 from the air. While for applications such as food supply, in addition to making plants and livestock larger, large-scale engineering of regulatory variants would also aim to make these sources grow more robustly (that is, tolerant to different environmental conditions) as well as less susceptible to disease to minimize loss. Finally, for mammalian genome engineering, some of the main key applications would be for deciphering complex genotype–phenotype relationships, modelling disease, and producing cells of interest. Subsequently, these engineered cells could be built into tissues and organs ex vivo using approaches such as 3D printing or, if the engineering was performed in pigs, organs could be harvested for xenotransplantation.
Genome-wide association studies (GWAS) along with large-scale high-throughput sequencing of exomes and genomes at scale have uncovered hundreds, if not thousands, of small and large DNA sequence variations associated with a given phenotype, including medically important human traits15. For example, in numerous genetic disorders, studies are advancing from correlation to cause and effect (for instance, via genome engineering in animals or organoids) to prove that particular mutations are the likely causes and drivers of specific diseases16–22. Subsequently, from a therapeutic perspective, repair of such defects would probably necessitate small-scale genome editing only, whereby a limited number of genomic targets would need to be altered in order to restore normal function. Whether the correction becomes a lasting cure for the disease will largely depend on the proportion of target cells in which editing is achieved and the persistence of the edited cells in their biological niche. The proportions and persistence of edited and unedited cells are particularly important in diseases such as cancer, where residual unedited, defective cells may have acquired a selective growth advantage over edited cells that could confer them with the ability, over time, to eliminate edited cells from the population.
It is also clear that many normal and disease phenotypes are governed by concurrent genetic variants across multiple alleles throughout the genome. Large-scale genome editing would enable, in the immediate term, the understanding of complex genotype–phenotype associations. From a more ambitious perspective, engineering human cells to be less susceptible to disease and generating tissues to eventually produce organs of high value are additional applications that would require large-scale genome editing. Thus, it is likely that the most immediate impact from editing human cells will emanate from small-scale engineering, whereas large-scale approaches will be crucial for obtaining a better understanding of phenotypes with complex genetics.
Tactically, if the number of desired changes is large and the changes span different areas of the genome, there is a compelling reason to approach this problem in a completely synthetic manner. From a practical perspective, this approach is currently limited to genomes that are megabases in size (at most), such as those in unicellular organisms. Here, rather than modifying an existing genomic template, designing and building the genome of interest completely from chemically synthesized DNA would not only provide the most efficient way to incorporate many changes in a desired DNA target but also enable complete control of the final sequence. The most compelling cases so far have included extensive changes in the genetic code (for biocontainment, efficient use of non-standard amino acids and multi-virus resistance) and ‘refactoring’ to reveal the absence or presence of unknown regulatory motifs23–25.
Genome editing systems — not just Cas9
The excitement and enthusiasm surrounding the ability to edit large regions and genomes can be mainly attributed to the recent developments in genome engineering technologies. Much of the recent spotlight has been on the CRISPR–Cas9 system from Streptococcus pyogenes that has been repurposed and adapted for genome engineering in human cells26–28. However, we have access to a large number of genome editing nanomachines, most of which have different mechanisms of action. Broadly, these tools can be classified into how each system recognizes its target site, that is, using DNA, protein, or RNA (Figure 3; Table 1) and whether they make double-strand breaks (DSBs).
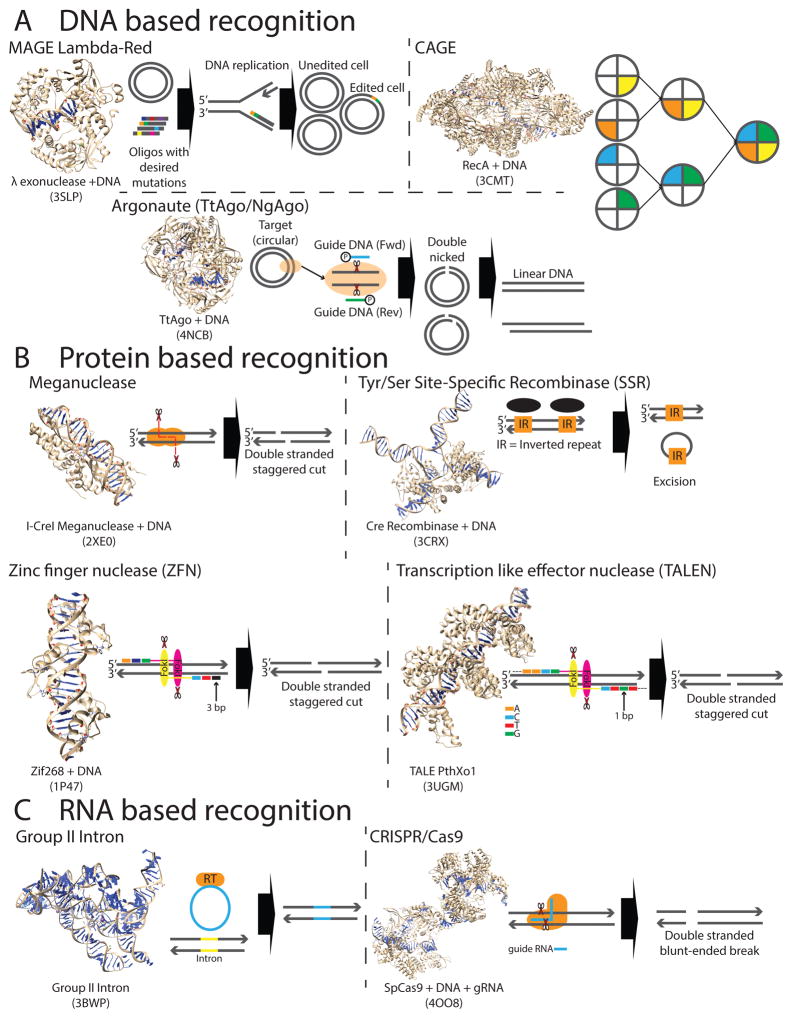
Figure 3. DNA-editing nanomachines.
a | DNA-based recognition. Multiplex automated genome engineering (MAGE) utilizes machinery from the phage λ, namely the single stranded binding protein beta and an exonuclease (Exo). Designer oligonucleotides, along with these two proteins, can enable the incorporation of desired changes at multiple genomic locations8. Conjugative assembly genome engineering (CAGE) enables the hierarchical assembly of parallel edited genomes. For every pair of edited genomes (strains), one strain is designated a donor strain and the other a recipient strain such that the direction of DNA transfer is engineered to be in a single direction13. This cycle can be repeated until the result is a single strain with all desired changes. The Argonaute (Ago) system utilizes a 24 nucleotide guide DNA sequence along with the Ago protein to direct a double-strand break (DSB) in a site-specific manner. b | Protein-based recognition., Zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and meganucleases utilize staggered-end forming DSBs to facilitate DNA editing. For each site that is being targeted, a new custom protein is required. Tyrosine (Tyr) and serine (Ser) site-specific recombinases (SSRs) mediate editing with (Ser) or without (Tyr) using double stranded breaks. A pair of short inverted repeats (IRs), flanking both the target site and donor DNA molecule, are required for DNA to be exchanged. Currently, the repertoire of different recombinase recognition sites is quite limited. c | RNA-based recognition. Group II introns encode both a self-catalyzing RNA moiety along with a reverse transcriptase, which enable site-specific disruption within a sequence of interest. The CRISPR–Cas9 system has demonstrated the most promise amongst current genome engineering technologies. A single-guide RNA (sgRNA) specifies the location in which a blunt-end-forming DSB is made by the Cas9 protein and due to this simplicity, enables simultaneous targeting of multiple sites by including multiple sgRNAs. Protein crystal structures were generated using Chimera148 and Protein Data Bank (PDB) accession numbers are provided underneath each structure149–157.
Table 1.
DNA editing nanomachines
| Name | Target scanning | Use in human cells | Target diversity | Double strand break | Maximum number of simultaneously altered sites* | Maximum observed HDR (%)‡ | Maximum observed NHEJ (%)‡ | Refs |
|---|---|---|---|---|---|---|---|---|
| Unstimulated Endogenous HR | DNA | Yes | High | No | 2 | 10−6 | NA | 158 |
|
|
||||||||
| MAGE (λ-Red) | DNA | Yes | High | No | 2¶ | NA | NA | 32,159 |
|
|
||||||||
| CAGE | DNA | No | High | No | NA | NA | NA | 13 |
|
|
||||||||
| SSOs only | DNA | Yes | High | No | 2 | 10−4 | NA | 33,35 |
|
|
||||||||
| ZFNs | Protein | Yes | Moderate | Yes§ | 6 | 43 | 65 | 160–162 |
|
|
||||||||
| TALENs | Protein | Yes | High | Yes§ | 2 | 10 | 50 | 6,163 |
|
|
||||||||
| Meganucleases | Protein | Yes | Low | Yes | 2 | 1 | 5 | 164,165 |
|
|
||||||||
| Tyr/Ser-SSRs | Protein | Yes | Low | No|| | 6 | 10−5 | NA | 166 |
|
|
||||||||
| Group II Introns | RNA | Yes | High | Yes | NA | NA | NA | 58,59 |
|
|
||||||||
| CRISPR–Cas9 | RNA | Yes | High | Yes§ | 62 | 33 | 79 | 7,70,167 |
In mammalian cells (per individual genome). Homozygous changes counted as two altered sites.
Observed in mammalian stem cells without antibiotic selection.
Can be programmed as nickases (single-strand nucleases).
Although Ser-SSRs utilize double stranded breaks mechanistically, these breaks are probably not exposed to the NHEJ machinery in the manner of the various designer nucleases.
In bacteria, 12 sites have been simultaneously edited.
CAGE, conjugative assembly genome engineering; HDR, homology-directed repair; HR, homologous recombination; MAGE, multiplex automated genome engineering; NA, data not available or not applicable; NHEJ, non-homologous end-joining; SSO, single-stranded oligonucleotides; SSRs, site-specific recombinases; TALENs, transcription activator-like effector nucleases; ZFNs, zinc finger nucleases.
DNA-based target recognition
For DNA-based target recognition systems (Fig 3a), the two major approaches, multiplex automated genome engineering (MAGE) using λ-red recombination8 and conjugative assembly genome engineering (CAGE)13 have been widely used in prokaryotes and demonstrated a high degree of multiplexability. Briefly, the λ-red system, similar to recET29, originates from phage and when imported into bacterial cells, can stimulate high levels of recombination in the presence of homologous DNA30. Although the machinery used in these approaches has not been fully optimized yet for eukaryotic cells, some conjugal transfer has been demonstrated31 and use of an optimized version of Redβ in human cells has yielded limited success32. Moreover, these technologies can be used to edit and propagate large pieces of DNA in bacteria for eventual use as donor DNA molecules for HR in human cells.
Transfection of single-stranded oligonucleotides (SSOs) alone can also be used to create small, site-specific insertions, deletions and substitutions. Typically, these oligonucleotides range between 20 and 200 bases in length, may contain modified bases to increase stability, and have a high degree of homology to the target site. Mechanistically, during DNA replication, in a very small percentage of instances, SSOs will be used as a template and the change encoded by the SSO will be incorporated in the host genome. SSOs alone have demonstrated efficiencies ranging from 10−5 to 2% in mammalian cells (Table 1)33–35. However, the most prevalent utility of SSOs are in conjunction with a designer nuclease to generate much higher rates of modification5,36.
The Argonaute protein (Ago) from the bacterium Thermus thermophilus (TtAgo) had been shown to provide DNA-based DNA interference, where a single stranded DNA guide could direct Argonaute-based cleavage of a plasmid DNA target, albeit only at temperatures ≥65°C37. Editing activity in human cells with TtAgo has not yet been published. Although a different Ago from Natronobacterium gregoryi, NgAgo, was initially reported to edit DNA in eukaryotic cells at physiological temperatures38, these results were questioned39–41 and the original paper was subsequently retracted. If an Ago-based system can be shown to have reliable cleavage efficacy at DNA target sites in eukaryotic cells, a key potential advantage is that, unlike CRISPR–Cas9, there is no requirement of a protospacer adjacent motif (PAM) which would, in theory, make any DNA targetable for editing.
Protein-based target recognition
The majority of systems use protein-based target recognition, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), meganucleases and Tyrosine/Serine site-specific recombinases (Tyr/Ser SSRs) (Fig 3b). ZFNs and TALENs recognize DNA target sites, ranging from 25 to 40 bp in size, in a sequence-specific manner through their DNA-binding domains and generate staggered DSBs through the action of FokI nuclease domains on opposite DNA strands. These systems have previously been discussed in great detail and, thus, will not be elaborated further here42,43. Meganucleases, also known as homing endonucleases, recognize a specific DNA sequence between 14 to 40 bp upon which they cut and induce a DSB44. The efficiencies of meganucleases are reasonably high and they only require a single custom biopolymer for each target site. However, the natural repertoire of targeting sites is quite limited. To address this key limitation, work has been done to systematically increase the diversity of targeting sites using mutagenesis approaches and by deriving fusions to programmable DNA-binding domains45–48.
Tyr/Ser SSRs, which typically recognize target sequences between 30 to 40 bp in length, were one of the earliest genome engineering tools to enable homology-directed repair (HDR) in mammalian genomes49,50. Briefly, the target site is comprised of three parts, a short DNA sequence flanked by two inverted repeats, and recombination can occur between a pair of target sites, where the DNA sequence between the target sites could be deleted, inverted or replaced. Notably, whereas Tyr SSRs utilize a mechanism of strand exchange without creating DSBs, Ser SSRs do create DSBs, but unlike simpler designer double-strand nucleases, SSR requires concerted cleavage and re-ligation with the donor-DNA present51. Although this mechanism of genome engineering is highly efficient and works across both prokaryotes and eukaryotes, the requirement of two naturally occurring copies of a very specific target site in relatively close proximity is limiting and there are very few Tyr/Ser SSRs that recognize different target sites. However, the potential for recombinases in the scheme of large-scale genome engineering is vast; thus, work to increase the diversity of DNA sequences that SSRs can target by engineering new specificities, which has already started and is continuing52–57, will be highly valuable.
RNA-based target recognition
The class of RNA-based target recognition includes group II introns and CRISPR–Cas9 (Fig 3c). For group II introns, in a process known as retrohoming, DNA is first transcribed into a catalytically active RNA as well as an mRNA encoding a reverse transcriptase. These two components form a complex and invade DNA with sequence similarity to a part of the intronic RNA. Subsequently, reverse transcription of the RNA molecule into cDNA permits insertion in a targeted manner to disrupt gene function, or when homologous DNA is introduced, promote HDR58,59. At this current juncture, this technology is not likely to be a main contributor to large-scale genome engineering owing to its low activity in mammalian cells.
Finally the CRISPR–Cas9 system has been shown to be one of the most robust gene-editing platforms currently available26–28. Review articles have covered the development of the CRISPR–Cas9 system in terms of its mechanism of action, ability to be tethered to different effector domains, and improvements in terms of higher specificity and multiplex editing potential42,60–62. Furthermore, specific variants of Cas9 have been made to enable gene editing without creating DSBs. A single mutation to the Cas9 protein can generate a version that only creates a single strand DNA cut (nick)63,64. Fusion of a nuclease-dead Cas9 (dCas9) to a cytidine deaminase enables the site-specific conversion of cytidine to uracil65–67. With respect to large-scale genome editing, the key relevant aspect of CRISPR is the ease of designing and multiplexing guide RNAs for targeting diverse genomic loci, which could enable excision and/or replacement of DNA segments of various sizes. This aspect has been demonstrated in the epigenetic context of studying enhancer function22,68,69, multi-kilobase gene replacements6, and multiplexed disruption of 62 different porcine endogenous retrovirus (PERV) sites in the pig genome70.
Genome editing versus de novo synthesis
Various strategies have been outlined to perform large-scale genome engineering, from completely synthesizing genomes de novo, to massively editing an existing genome scaffold, or a combination of both71. In a fully synthetic approach, genomes of interest are designed computationally, and then a rational, hierarchical strategy is devised to assemble long oligonucleotides into larger pieces (Figure 4a). Alternatively, an existing genome scaffold can be substantially edited, whereby the scaffold is delineated into a manageable number of segments of roughly equal size and then each segment is edited independently using approaches such as MAGE or CRISPR (Figure 4b). Subsequently, these segments can be re-assembled into the final desired sequence. In this section, we describe successful applications of large-scale engineering in bacteria and yeast and how these approaches could be translated for higher-order eukaryotes.
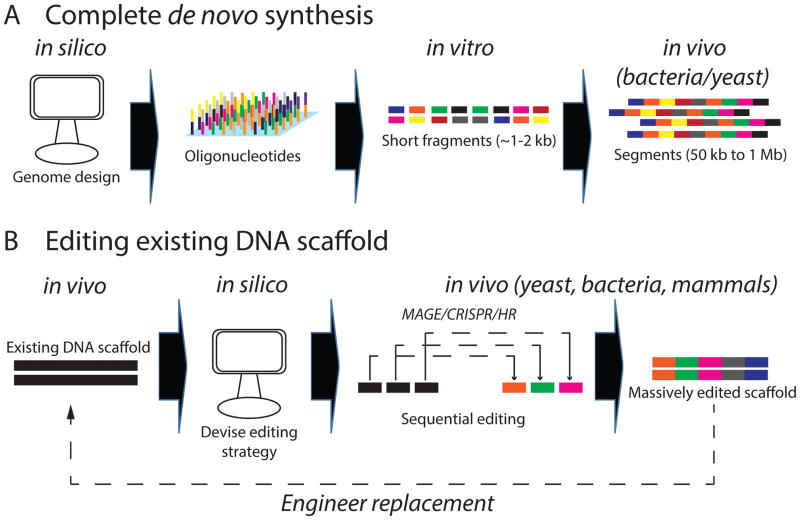
Figure 4. Two main approaches to large-scale genome engineering in bacteria and yeast.
a | For complete de novo synthesis, one must first computationally design oligonucleotide sequences that comprise the desired product while ensuring highest compatibility with the experimental strategy used for assembly. For example, thermodynamic properties of the sequence such as GC% and secondary structure formation are important parameters to consider. Subsequently, using in vitro PCR-based approaches, oligonucleotides (~150–200 bases in length) can then be assembled into fragments 1–2 kbp in size. Finally, using either bacteria or yeast as a chassis, these fragments can be assembled into segments in the order of Megabase pairs (Mbp). b | Extensive editing of an existing genome scaffold. Depending on the size of the DNA segment being edited, a segment is computationally divided into pieces with approximately similar level of editing. Each piece can then be edited independently in bacteria or yeast and upon completion, can be re-assembled into a single final segment and then placed into the final recipient cell.
Previously, our laboratory engineered an Escherichia coli strain in which all known instances of the UAG stop codon were replaced with UAA, with the motivation of freeing the UAG codon to encode for novel amino acids13,14. In this work, the E. coli genome was separated into 32 different segments, such that each segment contained approximately 10 UAG codons, then these segments were edited in parallel using MAGE. Briefly, MAGE utilizes the phage λ-red recombination machinery along with short oligonucleotides carrying desired mutations to enable cumulative recombination in a cyclical process that can be repeated multiple times. Once each segment obtained all the necessary UAG changes, the segments were hierarchically assembled into a single strain using CAGE. Notably, this final strain exhibited decreased phage infectivity and the UAG codon was successfully re-assigned to different non-standard amino acids14,24,25, demonstrating that this approach can be used to create potentially industrially robust strains.
By contrast, genomes have also been constructed completely de novo from synthesized DNA. Work from the J. Craig Venter Institute (JCVI) has resulted in the synthesis of numerous small genomes72–75. Here, the eventual goal was to generate a minimal genome (JCVI-syn3.0) using a completely synthetic genome (JCVI-syn1.0) as a starting point. Briefly, for JCVI-syn1.0, the approach taken was to hierarchically assemble 1 kb DNA fragments into larger and larger segments by using homologous recombination in yeast and to subsequently transfer the constructed genome (~1 Mbp in size) into a new recipient strain76. Similarly. our group recently used synthesized DNA fragments for an entire E. coli genome (~4 Mbp in size) along with yeast HR for assembly to re-design the E. coli genome to use 57 out of 64 codons, enabling re-assignment of the seven unused codons to additional, non-standard amino acids77.
In yeast, the major project that is well underway is the synthetic yeast genome project (Sc2.0)78,79. Sc2.0 utilizes a full synthetic approach whereby chromosomes (or chromosome arms) are made entirely from chemically synthesized DNA. The two chromosome arms (synIXR and semi-synVIL) were synthesized de novo as a bacterial artificial chromosome and transformed into yeast; subsequently, the native wild-type versions of these arms were inactivated. By contrast, for synthesis of chromosome III, a hierarchical approach was taken: starting from thousands of short nucleotides (70 bases), ~400 750 bp building blocks were assembled using PCR-based protocols and eventually, 83 overlapping segments (2–4 kbp) were made in yeast by homologous recombination. For the replacement of the native chromosome, synthetic segments were iteratively replaced over multiple cycles until the entire chromosome was replaced. Most recently, in addition to chromosome III, synthetic versions of chromosomes II, V, VI, X, and XII have been made, marking the completion of more than one-third of the chromosomes in the Sc2.0 project80–84.
Technical challenges
It is clear that both large-scale editing of existing scaffolds as well as de novo synthesis of DNA have yielded successful end products, each with its own advantages and disadvantages. Furthermore, when scaling these approaches to larger genomes (such as human), additional considerations would also need to be addressed. Here, we discuss a number of key technical challenges, many of which are shared by both approaches, and potential solutions.
Although computationally designing and subsequently producing chemically synthesized DNA offers much greater control than starting from existing genomes, the cost to synthesize an entire human genome is probably more expensive than editing an existing scaffold. It should be noted that this cost comprises the raw materials (synthetic DNA) and the process and labor of assembling the raw material into the final product. Thus, a further reduction in the cost will be necessary. However, this difference in cost is much smaller than it once was, as we have observed a million-fold reduction in cost in a decade for genome sequencing85 and solid-phase DNA synthesis86 to the point where the cost for sequencing is about US$1,000 whereas synthesis runs about US$6,0009 per human genome. Furthermore, genomic regions with low sequence complexity (i.e. those which are highly repetitive) can also pose difficulties with current DNA synthesis platforms. Relatedly, the genome sequences of many higher-order eukaryotes are incomplete, largely due to difficulties reading low-complexity genomic regions. For example, further development and application of newer sequencing technologies that can generate long reads such as those obtained from Pacific Biosciences (PacBio) single-molecule real-time (SMRT) sequencing87 and Oxford Nanopore Technologies (ONT) nanopore sequencing88–91 or high-resolution visualization approaches could be used to complete our map of these genomes92.
Massively editing an existing scaffold also has limitations and consequences. First, the multiple rounds of editing required to achieve the desired genome sequence can be quite laborious and time consuming to perform. If the efficiency for multiple targeting is not sufficiently high, cell populations would need be screened for the correct sequence over each iteration. Second, depending on the genome editing technology used, the sequence of the genome itself can present challenges in targeting. For example, regions with high percent of cytosines could present challenges for site-specific deaminases, or regions that are A/T rich-would have problems for CRISPR–Cas9. The discovery of the CRISPR system Cpf1 nuclease (as an alternative to Cas9) could address that as the PAM is composed of a thymidine homopolymer93.
A notable consequence of genome editing is the targeting of unintended sites. Targeting particular single nucleotide variants (SNVs) or point mutations is difficult because all the genome editing platforms can tolerate some level of mispairing. For unintended mutagenesis, the use of a non-DSB inducing technology could alleviate this problem, but in general, the efficiencies of these approaches are lower than the efficiencies of DSB-inducing technologies. Thus, if a DSB-inducing approach were used, any off-target sites that are unintentionally altered could be identified in a high-throughput manner61,94–97, and even if there were DSBs, these still could be tolerated if the DSBs were not introduced in important regions in the genome such as oncogenes or tumor suppressor genes. Similar to the above suggestion, an iterative approach could be used whereby after each iteration, the target DNA is sequenced to verify the absence of off-target mutations. However, as the number of iterations required increases, the costs associated with sequencing would also increase.
For both de novo genome synthesis and substantial editing of existing genomes, designing and introducing many changes at once relative to the normal genome sequence may result in a non-viable phenotype as a consequence of the editing process itself (in the case of radically modifying an existing scaffold) or as a result of the DNA sequence being incompatible with normal function. Although we have gained a better understanding of how to design prokaryotic77 and lower-order eukaryotic genomes80, this level of understanding does not exist for more complex mammalian genomes. Thus, it may be imperative that an upper limit of the number of modifications that can be made in a single iteration be identified for each cell type, and if the number of changes exceeds this limit, the modifications would be incorporated in multiple iterations.
As each approach provides distinct advantages and challenges, it may very well be a hybrid of these two approaches that will ultimately be most successful. One can envision a scenario in which an initial library of DNA building blocks that spans an entire genome will be synthesized (or generated through PCR of genomic DNA) and subsequent sequence variations to particular sets of building blocks will be generated using genome engineering technologies. For example, starting with a bacterial artificial chromosome (BAC) library comprising of overlapping 100–300 kbp segments of the human genome98, one can select an individual BAC encompassing a desired genomic location, edit this DNA segment in E. coli using MAGE and subsequently use HR to incorporate this new DNA segment in the destination cell population.
Desired genome-writing properties
We have detailed two different approaches to performing large-scale genome engineering. Although building of large genomes de novo has many advantages, as we stated earlier, we believe that a combination of de novo DNA synthesis and genome editing technologies will probably be utilized moving forward. Thus, it will be important to understand what properties we desire in a truly robust, high-throughput genome-writing platform (Table 2).
Table 2.
Ideal features for genome writing or editing platforms
| Feature | Description |
|---|---|
| Simple | Only one custom biopolymer for each HDR or HR event |
| Multiplex | 100% targeting at multiple sites, not merely ‘high efficiency’ at a single site |
| Robust | High efficiency in post-mitotic cells (e.g. neurons), broad sequence compatibility |
| Large regions | High efficiency of 10 kbp to 10 Mbp scale replacements, alternative strategies possible |
| Low toxicity | Low apoptosis, high specificity, genome integrity intact |
| Germline editing | Specific safeguards might include restriction to gametes and/or clonal analysis to reduce mosaicism |
| Reading and testing | Rapid feedback on many synthetic genomes via reporters, organoids and sequencing |
HDR, homology-directed repair; HR, homologous recombination.
Of the different genome engineering technologies described, although it has not been shown to work in human cells, the λ-red (and the similar recET) recombination systems have the ideal characteristic that only one custom biopolymer (the donor DNA molecule carrying desired changes) is necessary to make site-specific changes, whereas the other technologies would require at least two. For example, for CRISPR–Cas9 or TALENs and ZFNs, each target site would require a new guide RNA or new designer nuclease, respectively, in addition to the donor DNA molecule specifying the new sequence.
Next, the majority of editing experiments have been performed in cells that are continually undergoing mitosis, so another ideal feature would be a system that can also operate in post-mitotic cells in a highly efficient manner. Although the level of editing desired in post-mitotic cells may not be as large in scale as the level desired in other cells, this feature would at least increase the repertoire of cell types that could be edited. To date, Cre recombinase and CRISPR–Cas9 have been shown to be effective in cardiomyocytes and neurons respectively99–101, with application in other cell types yet to be demonstrated.
In addition to increasing the diversity of cell types, it is also important to be able to have broad target sequence specificity. As mentioned earlier, if Ago systems could be optimized in the future for efficient DNA target cleavage in eukaryotic cells, one of their more compelling features is the lack of a requirement for a PAM sequence. Although this system would provide a theoretically unlimited range of targetable sequences, it may also increase the likelihood of unintended mutagenesis via off-target activity. In the same vein, when improving CRISPR–Cas9 specificity, depending on the approach taken, a reduction in off-target activity may be accompanied by a reduction in on-target activity as well61. Thus, it is going to be a fine balance of having a system that has a broad range of targeting, high editing efficiency and high specificity.
The ability to achieve 100% gene disruption at a single locus has become feasible for some loci with many of the technologies discussed. However, the ideal feature with respect to efficiency is obtaining a high frequency of multiplexed editing where we have the ability to make hundreds of changes per cell simultaneously. Highly efficient multiplexed editing would need to be done in conjunction with an approach that minimizes toxicity to the cell. If the method of choice utilizes DSBs, creating hundreds of DSBs could be problematic in that it could either induce cellular apoptosis or chromosomal rearrangements. As highlighted earlier, disruption of 62 PERV genes in a single genome with minimal impact on genome architecture was probably a unique case as there were no apparent rules for multiplex targeting that could be discerned moving forward, and only a single guide RNA was required to target all 62 sites70. Thus, a more sophisticated strategy, such as manipulating DNA repair machinery or utilizing non-DSB creating entities, would be necessary if this many genes were simultaneously targeted in other applications.
Nonetheless, this level of disruption will be crucial for large-scale editing. Although efforts have been made to identify approaches to improve HR and HDR, the efficiencies are still typically one order of magnitude lower than that of an alternative mechanism of DSB repair, non-homologous end-joining (NHEJ), in which the DNA ends are re-ligated, typically without incorporating a donor DNA sequence102–107. Intriguingly, attempts at utilizing the NHEJ repair pathway to incorporate specific DNA sequences have also been made with some degree of success108,109. Thus, work will need to continue to identify innovative ways to achieve high editing frequency simultaneously across multiple loci.
Relatedly, another important feature will be to increase the efficiency of larger changes. There is an inverse relationship observed between the efficiency of alteration and the size of the alteration, where smaller changes are much easier to incorporate than larger ones6,110. Two potential strategies could be pursued to address the efficiency. The first strategy would be to utilize recombinases, and the second would be to tether homologous DNA directly to genome-editing reagents. Recombinase-mediated cassette exchange (RMCE) has been routinely used in mammalian cells, with its most prevalent use in creating conditional mouse strains111–114. This approach requires two separate gene editing events — one event to place recombination sites flanking the area to exchange and then the exchange event itself — and thus could be quite laborious. Intriguingly, RMCE has been combined with CRISPR–Cas9 to make this approach more efficient115. As stated previously, the main challenge with recombinases is the limited diversity of targetable sites. In terms of tethering or pairing, placing the donor template in close proximity to the site of cleavage should increase the frequency at which replacements would occur. For example, when using CRISPR–Cas9, this could be accomplished by covalently attaching the donor DNA to the guide RNA.
Last, but not least, is germline editing. The ability to edit the germline poses the usual ethical issues of risks, benefits, safety and efficacy (adequately handled by organizations such as the US Food and Drug Administration (FDA), the European Medicines Agency (EMA) and the China Food and Drug Administration (CFDA))116–119. One of the most difficult challenges, from a logistical perspective, will be understanding the long-term, potentially trans-generational, societal impact of germline editing applications, which may otherwise be deemed ‘safe’ in a conventional medical sense. Ironically, because reproductive technologies, such as spermatogonial stem cell engineering, can use clonal sibling cells, which can be checked for genomic goals, including zero undesired on-target mutations (caused by NHEJ rather than HDR), off-target errors or epigenetic issues, high-fidelity editing may be more critical for somatic gene therapy. Additionally, editing of sperm or egg precursor cells could result in considerable reduction in embryo loss or spontaneous or induced abortion relative to in vitro fertilization (IVF) and hence might be attractive to recessive carrier parents with concerns about risks to embryos. At this juncture, it seems difficult to draw sharp red lines for what may be permitted versus forbidden; for example, designating areas of the genome, such as the mitochondrial genome, where editing would be permitted. It should also be noted that mitochondrial replacement therapy has been utilized in cases of IVF120. A key component will be engaging various communities to imagine unexpected scenarios, including respect for diverse cultures and faiths.
A roadmap to writing large genomes
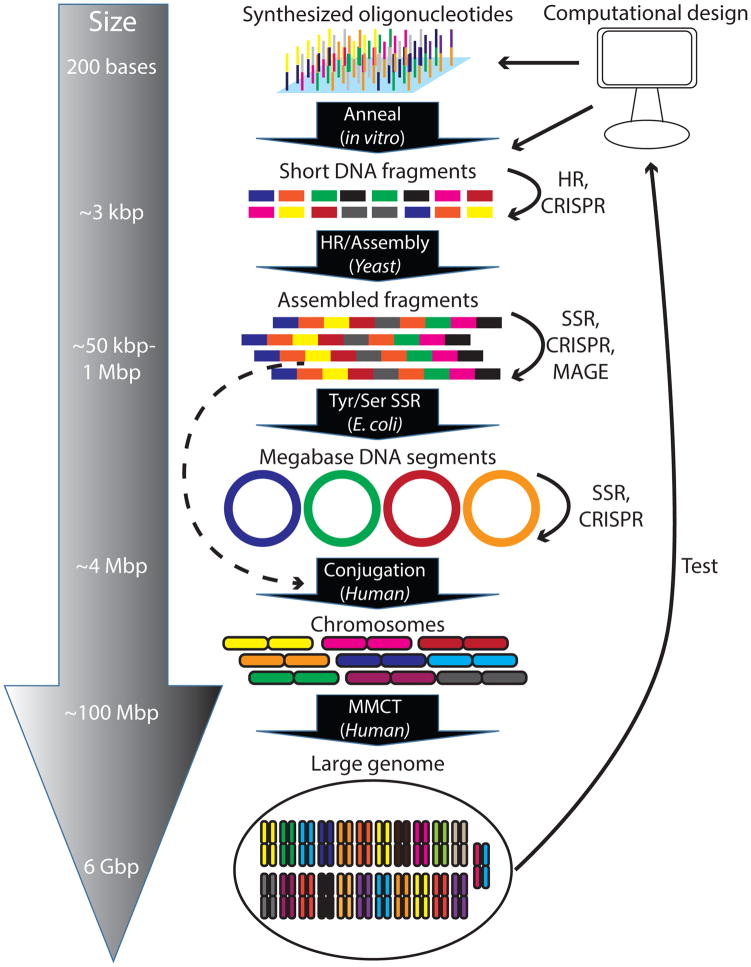
After these desired features of a large-scale genome-writing platform are achieved, the final step is implementation. Here, we consider a roadmap for how the implementation could be accomplished (Figure 5). Starting with libraries of DNA oligonucleotides, small DNA fragments in the 3-kbp range can be constructed in vitro using a combination of DNA annealing, ligation and amplification. By taking advantage of the high efficiency of HR in yeast, these smaller fragments could then be assembled into larger pieces of approximately 50 kbp in size (or possibly into the megabase scale). Next, these 50-kbp segments can be built into larger fragments (~ 4 Mbp) using an SSR system in E. coli. Multiple bacterial or yeast strains carrying different 4 Mbp fragments could be delivered via conjugation (or spheroplast fusion) to plant or animal cells to build chromosome-sized elements31,121. Finally, these large (40–250 Mbp) segments could be hierarchically assembled (usually with selectable markers) into single cells by microcell-mediated chromosomal transfer (MMCT)122. As the efficiency of HR increases, we can miniaturize and multiplex the HR reactions and reduce the need for selection. High-throughput and multiplex testing of large numbers of combinatorially created genomes greatly increases the chances of rapid success.
Figure 5. A roadmap to building large genomes using a combination of de novo synthesis and genome editing.
Here, a hierarchical strategy is employed starting from short oligonucleotides (150 to 200 bases in length), which can be assembled into short DNA fragments ~3 kbp in size. Short DNA fragments would then be assembled into larger segments between 50 kbp and 1 Mbp in size. Segments ~4 Mbp in size can be built in bacteria or yeast and subsequently transferred into human cells using conjugation. These can then be built into segments up to ~100 Mbp in size and be transferred into recipient cells by microcell-mediated chromosomal transfer (MMCT). At each stage where a segment is built, genome-editing tools such as multiplex automated genome engineering (MAGE), CRISPR, homologous recombination (HR) and site specific recombinases (SSRs) can be utilized to make further edits from what was specified in the initial set of oligonucleotides.
It should also be noted that at various points of this process, DNA segments of different sizes could be modified using smaller-scale genome-editing technologies, contingent on the ability to retrieve and replace individual segments efficiently (such as using a multiplex barcoded library). Additional editing is advantageous for various reasons. First, if a particular segment was deemed incompatible for viability, the segment could be edited until it no longer created problems. Second, if additional sequence diversity was desired, it may be more efficient to edit the DNA segment as opposed to re-synthesizing it entirely. For example, if the editing was to be performed in yeast, native HR could be used, which already is highly efficient, or a CRISPR-based approach could be used for replacement123,124. Similarly, if the segments were in E. coli, use of MAGE and/or CAGE could be employed.
Taken together, achieving this collection of reading, editing, writing and testing goals may move us closer to a safe, robust, inexpensive, egalitarian and effective genome editing that can change our environment and ourselves, provided that such strategies are constantly under technical and ethical scrutiny and feedback.
Acknowledgments
This work was supported by NIH grant RM1HG008525 “Center for Genomically Engineered Organs”.
Glossary
- Reading
The use of massively parallel sequencing technologies to decipher nucleotide composition.
- Writing
The use of either genome editing tools and/or DNA synthesis to make desired changes in DNA sequence.
- Homologous recombination
(HR). Exchange of sequence between two highly similar DNA molecules.
- Protospacer adjacent motif
(PAM). A short, characteristic sequence (typically between 3–8 nucleotides in length) that the CRISPR-associated nuclease (such as Cas9 or Cpf1) recognizes prior to making its DNA break. Depending on the CRISPR system used, this sequence must either flank the 3′ end or 5′ end of the sequence of interest.
- Homology-directed repair
(HDR). The process by which double stranded breaks (DSBs) in DNA are repaired when in the presence of an additional DNA moiety that has homology to the DNA sequence surrounding the DSB.
Biographies
Raj Chari:
Raj Chari is currently a scientist at Juno Therapeutics, moving to the US National Cancer Institute (NCI) later in 2017. He received his Bachelor’s of Science in Biochemistry and Computer Science from the University of British Columbia, Vancouver, Canada, where he then proceeded to do his PhD. He did this postdoctoral training in the department of genetics at Harvard Medical School, Boston, Massachusetts, USA, and later worked for AbVitro Inc, a small biotechnology start-up company in the single-cell sequencing space. His research interests moving forward will primarily be developing and applying genome-editing technologies in the context of cancer development and therapies.
George Church:
George Church, professor at Harvard & MIT, co-author of 425 papers, 95 patent publications & the book Regenesis, developed methods used for the first genome sequence (1994) & million-fold cost reductions since (via next-generation sequencing and nanopores), plus barcoding, DNA assembly from chips, genome editing, writing & recoding. He co-initiated the BRAIN Initiative (2011) & Genome Projects (1984, 2005) to provide & interpret the world’s only open-access personal precision medicine datasets. George Church’s homepage: http://arep.med.harvard.edu/
Footnotes
WEBLINKS:
Church lab CRISPR resources: http://crispr.med.harvard.edu
Addgene CRISPR–Cas9 Plasmids and Resources: http://www.addgene.org/crispr/
Genome Project-write: http://engineeringbiologycenter.org/
Competing interests statement
G.M.C. is a co-founder of Editas Medicine and eGenesis Bio and serves advisory roles in several companies involved in genome editing and engineering. A detailed listing of G.M.C.’s Tech Transfer, Advisory Roles, and Funding Sources can be obtained from http://arep.med.harvard.edu/gmc/tech.html. R.C. declares no competing interests.
References
- 1.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58:586–597. doi: 10.1016/j.molcel.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 5.DeWitt MA, et al. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. doi: 10.1126/scitranslmed.aaf9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne SM, Ortiz L, Mali P, Aach J, Church GM. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43:e21. doi: 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dever DP, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeke JD, et al. GENOME ENGINEERING. The Genome Project-Write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- 10.Robertson DE, et al. A new dawn for industrial photosynthesis. Photosynth Res. 2011;107:269–277. doi: 10.1007/s11120-011-9631-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salhi A, et al. DESM: portal for microbial knowledge exploration systems. Nucleic Acids Res. 2016;44:D624–633. doi: 10.1093/nar/gkv1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Isaacs FJ, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lajoie MJ, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freedman BS, et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 19.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 20.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry. 2016;6:e703. doi: 10.1038/tp.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soldner F, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan LY, Kosuri S, Endy D. Refactoring bacteriophage T7. Mol Syst Biol. 2005;1:2005.0018. doi: 10.1038/msb4100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rovner AJ, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 30.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix B, Citovsky V. Transfer of DNA from Bacteria to Eukaryotes. mBio. 2016;7 doi: 10.1128/mBio.00863-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu K, Stewart AF, Porter ACG. Stimulation of oligonucleotide-directed gene correction by Redβ expression and MSH2 depletion in human HT1080 cells. Mol Cells. 2015;38:33–39. doi: 10.14348/molcells.2015.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rios X, et al. Stable gene targeting in human cells using single-strand oligonucleotides with modified bases. PloS One. 2012;7:e36697. doi: 10.1371/journal.pone.0036697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igoucheva O, Alexeev V, Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 35.Aarts M, te Riele H. Subtle gene modification in mouse ES cells: evidence for incorporation of unmodified oligonucleotides without induction of DNA damage. Nucleic Acids Res. 2010;38:6956–6967. doi: 10.1093/nar/gkq589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quadros RM, et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017;18 doi: 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swarts DC, et al. DNA-guided DNA interference by a prokaryotic Argonaute. Nature. 2014;507:258–261. doi: 10.1038/nature12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao F, Shen XZ, Jiang F, Wu Y, Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016;34:768–773. doi: 10.1038/nbt.3547. [DOI] [PubMed] [Google Scholar]
- 39.Lee SH, et al. Failure to detect DNA-guided genome editing using Natronobacterium gregoryi Argonaute. Nat Biotechnol. 2016 doi: 10.1038/nbt.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burgess S, et al. Questions about NgAgo. Protein Cell. 2016;7:913–915. doi: 10.1007/s13238-016-0343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Javidi-Parsijani P, et al. No evidence of genome editing activity from Natronobacterium gregoryi Argonaute (NgAgo) in human cells. PLOS ONE. 2017;12:e0177444. doi: 10.1371/journal.pone.0177444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 43.Chandrasegaran S, Carroll D. Origins of Programmable Nucleases for Genome Engineering. J Mol Biol. 2016;428:963–989. doi: 10.1016/j.jmb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chevalier BS, et al. Design, activity, and structure of a highly specific artificial endonuclease. Mol Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 47.Boissel S, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42:2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfs JM, et al. MegaTevs: single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 2014;42:8816–8829. doi: 10.1093/nar/gku573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birling MC, Gofflot F, Warot X. Site-specific recombinases for manipulation of the mouse genome. Methods Mol Biol Clifton NJ. 2009;561:245–263. doi: 10.1007/978-1-60327-019-9_16. [DOI] [PubMed] [Google Scholar]
- 50.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turan S, Bode J. Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. FASEB J Off Publ Fed Am Soc Exp Biol. 2011;25:4088–4107. doi: 10.1096/fj.11-186940. [DOI] [PubMed] [Google Scholar]
- 52.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 53.Eroshenko N, Church GM. Mutants of Cre recombinase with improved accuracy. Nat Commun. 2013;4:2509. doi: 10.1038/ncomms3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci U S A. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saraf-Levy T, et al. Site-specific recombination of asymmetric lox sites mediated by a heterotetrameric Cre recombinase complex. Bioorg Med Chem. 2006;14:3081–3089. doi: 10.1016/j.bmc.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 56.Gaj T, et al. Enhancing the specificity of recombinase-mediated genome engineering through dimer interface redesign. J Am Chem Soc. 2014;136:5047–5056. doi: 10.1021/ja4130059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wallen MC, Gaj T, Barbas CF. Redesigning Recombinase Specificity for Safe Harbor Sites in the Human Genome. PloS One. 2015;10:e0139123. doi: 10.1371/journal.pone.0139123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastroianni M, et al. Group II intron-based gene targeting reactions in eukaryotes. PloS One. 2008;3:e3121. doi: 10.1371/journal.pone.0003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo H, et al. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- 60.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet. 2016;17:300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell. 2016 doi: 10.1016/j.cell.2016.10.044. [DOI] [PubMed] [Google Scholar]
- 63.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nishida K, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016 doi: 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- 67.Yang L, et al. Engineering and optimising deaminase fusions for genome editing. Nat Commun. 2016;7:13330. doi: 10.1038/ncomms13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fulco CP, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773. doi: 10.1126/science.aag2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang L, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 71.Haimovich AD, Muir P, Isaacs FJ. Genomes by design. Nat Rev Genet. 2015;16:501–516. doi: 10.1038/nrg3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith HO, Hutchison CA, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci U S A. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 74.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 75.Hutchison CA, et al. Design and synthesis of a minimal bacterial genome. Science. 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 76.Lartigue C, et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. doi: 10.1126/science.1173759. [DOI] [PubMed] [Google Scholar]
- 77.Ostrov N, et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016;353:819–822. doi: 10.1126/science.aaf3639. [DOI] [PubMed] [Google Scholar]
- 78.Dymond JS, et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature. 2011;477:471–476. doi: 10.1038/nature10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 81.Shen Y, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355 doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Z-X, et al. ‘Perfect’ designer chromosome V and behavior of a ring derivative. Science. 2017;355 doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 83.Zhang W, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355 doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 84.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355 doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 85.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 86.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 87.Gordon D, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loose M, Malla S, Stout M. Real-time selective sequencing using nanopore technology. Nat Methods. 2016 doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szalay T, Golovchenko JA. De novo sequencing and variant calling with nanopores using PoreSeq. Nat Biotechnol. 2015;33:1087–1091. doi: 10.1038/nbt.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goodwin S, et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res. 2015;25:1750–1756. doi: 10.1101/gr.191395.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fuller CW, et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc Natl Acad Sci U S A. 2016;113:5233–5238. doi: 10.1073/pnas.1601782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beliveau BJ, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 1 p following 243. [DOI] [PubMed] [Google Scholar]
- 97.Lensing SV, et al. DSBCapture: in situ capture and sequencing of DNA breaks. Nat Methods. 2016 doi: 10.1038/nmeth.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osoegawa K, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PloS One. 2014;9:e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Agah R, et al. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suzuki K, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pinder J, Salsman J, Dellaire G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015;43:9379–9392. doi: 10.1093/nar/gkv993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 104.Maruyama T, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Agudelo D, et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat Methods. 2017 doi: 10.1038/nmeth.4265. [DOI] [PubMed] [Google Scholar]
- 107.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He X, et al. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maresca M, Lin VG, Guo N, Yang Y. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013;23:539–546. doi: 10.1101/gr.145441.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Y, Seed B. Site-specific gene targeting in mouse embryonic stem cells with intact bacterial artificial chromosomes. Nat Biotechnol. 2003;21:447–451. doi: 10.1038/nbt803. [DOI] [PubMed] [Google Scholar]
- 111.Bradley A, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome Off J Int Mamm Genome Soc. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ordovás L, et al. Efficient Recombinase-Mediated Cassette Exchange in hPSCs to Study the Hepatocyte Lineage Reveals AAVS1 Locus-Mediated Transgene Inhibition. Stem Cell Rep. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Voziyanova E, et al. Efficient Flp-Int HK022 dual RMCE in mammalian cells. Nucleic Acids Res. 2013;41:e125. doi: 10.1093/nar/gkt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Osterwalder M, et al. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7:893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang X, Koolhaas WH, Schnorrer F. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 Bethesda Md. 2014;4:2409–2418. doi: 10.1534/g3.114.013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bosley KS, et al. CRISPR germline engineering--the community speaks. Nat Biotechnol. 2015;33:478–486. doi: 10.1038/nbt.3227. [DOI] [PubMed] [Google Scholar]
- 117.Baltimore D, et al. Biotechnology. A prudent path forward for genomic engineering and germline gene modification. Science. 2015;348:36–38. doi: 10.1126/science.aab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Church G. Perspective: Encourage the innovators. Nature. 2015;528:S7. doi: 10.1038/528S7a. [DOI] [PubMed] [Google Scholar]
- 119.Doudna J. Perspective: Embryo editing needs scrutiny. Nature. 2015;528:S6. doi: 10.1038/528S6a. [DOI] [PubMed] [Google Scholar]
- 120.Schandera J, Mackey TK. Mitochondrial Replacement Techniques: Divergence in Global Policy. Trends Genet TIG. 2016;32:385–390. doi: 10.1016/j.tig.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 121.Li L, Blankenstein T. Generation of transgenic mice with megabase-sized human yeast artificial chromosomes by yeast spheroplast-embryonic stem cell fusion. Nat Protoc. 2013;8:1567–1582. doi: 10.1038/nprot.2013.093. [DOI] [PubMed] [Google Scholar]
- 122.Tomizuka K, et al. Double trans-chromosomic mice: maintenance of two individual human chromosome fragments containing Ig heavy and kappa loci and expression of fully human antibodies. Proc Natl Acad Sci U S A. 2000;97:722–727. doi: 10.1073/pnas.97.2.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.EauClaire SF, Zhang J, Rivera CG, Huang LL. Combinatorial metabolic pathway assembly in the yeast genome with RNA-guided Cas9. J Ind Microbiol Biotechnol. 2016;43:1001–1015. doi: 10.1007/s10295-016-1776-0. [DOI] [PubMed] [Google Scholar]
- 125.Wang SZ, Liu BH, Tao HW, Xia K, Zhang LI. A genetic strategy for stochastic gene activation with regulated sparseness (STARS) PloS One. 2009;4:e4200. doi: 10.1371/journal.pone.0004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gouble A, et al. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J Gene Med. 2006;8:616–622. doi: 10.1002/jgm.879. [DOI] [PubMed] [Google Scholar]
- 127.Gellhaus K, Cornu TI, Heilbronn R, Cathomen T. Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells. Hum Gene Ther. 2010;21:543–553. doi: 10.1089/hum.2009.167. [DOI] [PubMed] [Google Scholar]
- 128.Robert MA, et al. Efficacy and site-specificity of adenoviral vector integration mediated by the phage φC31 integrase. Hum Gene Ther Methods. 2012;23:393–407. doi: 10.1089/hgtb.2012.122. [DOI] [PubMed] [Google Scholar]
- 129.Ou H, et al. A highly efficient site-specific integration strategy using combination of homologous recombination and the ΦC31 integrase. J Biotechnol. 2013;167:427–432. doi: 10.1016/j.jbiotec.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 130.Coluccio A, et al. Targeted gene addition in human epithelial stem cells by zinc-finger nuclease-mediated homologous recombination. Mol Ther J Am Soc Gene Ther. 2013;21:1695–1704. doi: 10.1038/mt.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beaton BP, et al. Inclusion of homologous DNA in nuclease-mediated gene targeting facilitates a higher incidence of bi-allelically modified cells. Xenotransplantation. 2015;22:379–390. doi: 10.1111/xen.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nakade S, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun. 2014;5:5560. doi: 10.1038/ncomms6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 134.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21:5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bibikova M, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Testa G, et al. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat Biotechnol. 2003;21:443–447. doi: 10.1038/nbt804. [DOI] [PubMed] [Google Scholar]
- 141.Epinat JC, et al. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mercer AC, Gaj T, Fuller RP, Barbas CF. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 2012;40:11163–11172. doi: 10.1093/nar/gks875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gaj T, Mercer AC, Sirk SJ, Smith HL, Barbas CF. A comprehensive approach to zinc-finger recombinase customization enables genomic targeting in human cells. Nucleic Acids Res. 2013;41:3937–3946. doi: 10.1093/nar/gkt071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Owens JB, et al. Transcription activator like effector (TALE)-directed piggyBac transposition in human cells. Nucleic Acids Res. 2013;41:9197–9207. doi: 10.1093/nar/gkt677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 149.Zhang J, McCabe KA, Bell CE. Crystal structures of lambda exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc Natl Acad Sci U S A. 2011;108:11872–11877. doi: 10.1073/pnas.1103467108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 151.Sheng G, et al. Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc Natl Acad Sci U S A. 2014;111:652–657. doi: 10.1073/pnas.1321032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muñoz IG, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011;39:729–743. doi: 10.1093/nar/gkq801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Peisach E, Pabo CO. Constraints for zinc finger linker design as inferred from X-ray crystal structure of tandem Zif268-DNA complexes. J Mol Biol. 2003;330:1–7. doi: 10.1016/s0022-2836(03)00572-2. [DOI] [PubMed] [Google Scholar]
- 155.Mak ANS, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hatada S, Nikkuni K, Bentley SA, Kirby S, Smithies O. Gene correction in hematopoietic progenitor cells by homologous recombination. Proc Natl Acad Sci. 2000;97:13807–13811. doi: 10.1073/pnas.240462897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang HH, et al. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu P-Q, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010 doi: 10.1002/bit.22654. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 161.Wang J, et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Karakikes I, et al. A Comprehensive TALEN-Based Knockout Library for Generating Human-Induced Pluripotent Stem Cell–Based Models for Cardiovascular DiseasesNovelty and Significance. Circ Res. 2017;120:1561–1571. doi: 10.1161/CIRCRESAHA.116.309948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lin J, et al. Creating a monomeric endonuclease TALE-I-SceI with high specificity and low genotoxicity in human cells. Nucleic Acids Res. 2015;43:1112–1122. doi: 10.1093/nar/gku1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cai D, Cohen KB, Luo T, Lichtman JW, Sanes JR. Improved tools for the Brainbow toolbox. Nat Methods. 2013;10:540–547. [PubMed] [Google Scholar]
- 167.Ding Q, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]