Abstract
Temporal control of physiology requires the interplay between gene networks involved in daily timekeeping and tissue function across different organs. How the circadian clock interweaves with tissue-specific transcriptional programs is poorly understood. Here, we dissected temporal and tissue-specific regulation at multiple gene regulatory layers by examining mouse tissues with an intact or disrupted clock over time. Integrated analysis uncovered two distinct regulatory modes underlying tissue-specific rhythms: tissue-specific oscillations in transcription factor (TF) activity, which were linked to feeding-fasting cycles in liver and sodium homeostasis in kidney; and colocalized binding of clock and tissue-specific transcription factors at distal enhancers. Chromosome conformation capture (4C-seq) in liver and kidney identified liver-specific chromatin loops that recruited clock-bound enhancers to promoters to regulate liver-specific transcriptional rhythms. Furthermore, this looping was remarkably promoter-specific on the scale of less than 10 kilobases (kb). Enhancers can contact a rhythmic promoter while looping out nearby nonrhythmic alternative promoters, confining rhythmic enhancer activity to specific promoters. These findings suggest that chromatin folding enables the clock to regulate rhythmic transcription of specific promoters to output temporal transcriptional programs tailored to different tissues.
A mammalian internal timing system, known as the circadian clock, orchestrates temporal physiology in organs to anticipate daily environmental cycles (Dibner and Schibler 2015). Individual cells within organs contain a molecular oscillator that, together with rhythmic systemic signals such as hormones, temperature, and feeding behavior, collectively drive diurnal oscillations in gene expression and physiology (Lamia et al. 2008; Reinke et al. 2008; Cho et al. 2012; Vollmers et al. 2012). Remarkably, the circadian clock impinges on many gene regulatory layers, from transcriptional and post-transcriptional processes, translation efficiency, to translational and post-translational processes (Mermet et al. 2016).
Transcriptome analysis of different cell types and tissues has highlighted the breadth of tissue-specific transcriptional regulation (Merkin et al. 2012; Yue et al. 2014). However, physiological processes are dynamic at the timescale of hours and often under circadian control, such as hormone secretion, drug and xenobiotic metabolism, and glucose homeostasis (Takahashi et al. 2008). Adding the temporal dimension to tissue-specific gene regulation is needed for an integrated understanding of physiology.
Chronobiology studies have shown that tissues utilize the circadian clock to drive tissue-specific rhythmic gene expression (Storch et al. 2002; Korenčič et al. 2014; Zhang et al. 2014), presumably to schedule physiological functions to optimal times of day. Indeed, genetic ablation of the circadian clock in different tissues can lead to divergent pathologies, such as diabetes in pancreas-specific Bmal1 knockout (KO) and fasting hypoglycemia in liver-specific Bmal1 KO, suggesting that the clock interweaves with tissue-specific transcriptional programs (Bass and Lazar 2016), but how diurnal and tissue-dependent regulatory landscapes interact to generate tissue-specific rhythms is poorly understood.
Results
Contributions of tissue, daily time, and circadian clock to global variance in mRNA expression
To estimate the respective contributions of tissues, daily time, and circadian clock to global variance in gene expression, we analyzed available temporal transcriptomes across 11 tissues in WT mice (Zhang et al. 2014) and generated temporal RNA-seq data of liver and kidney from Bmal1 KO mice and WT littermates (Supplemental Tables S1, S2; Methods). The Zhang et al. data set was obtained under dark–dark (DD), ad libitum feeding, sampled every 2 h. The liver and kidney Bmal1 KO and WT data sets were obtained under light–dark (LD), night-restricted feeding conditions, sampled every 4 h.
To avoid mixing different experimental designs (e.g., temporal resolution and number of repeats) (Deckard et al. 2013; Li et al. 2015), we analyzed these data sets separately. We performed principal component analysis (PCA) on the entire set of conditions (11 tissues × 24 time points) to obtain an unbiased overview into the contributions of tissue- and time-specific variance. Most of the variance concerned differences in expression between tissues (Fig. 1A; Supplemental Fig. S1A–D). Temporal variance, in particular, 24-h periodicity, was present among a group of principal components carrying lower amounts of variance (Fig. 1A; Supplemental Fig. S1E–G). Focusing on genome-wide temporal variation within each tissue, we found that 24-h rhythms constituted the largest contribution of temporal variance, followed by 12-h rhythms, which were close to background levels for many tissues (Fig. 1B; Hughes et al. 2009). We thus focused the rest of our analysis on 24-h rhythms.
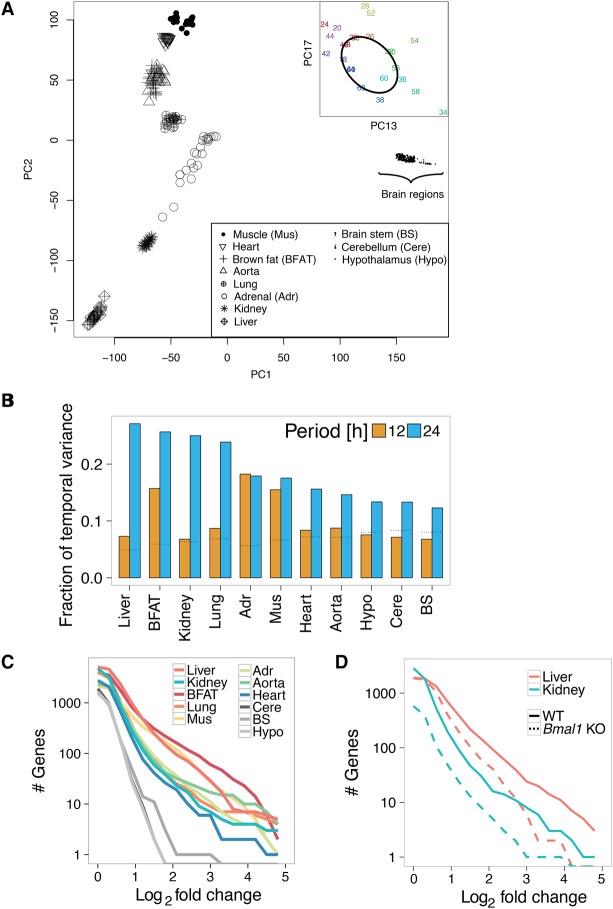
Figure 1.
Contribution of tissue, daily time, and circadian clock to global variance in mRNA expression. (A) Principal component analysis (PCA) across 11 WT tissues sampled over 2 d. PC1 and PC2 show clustering of samples by tissues; each point represents a tissue sample (see key) at a specific time point (not labeled). (Inset) Loadings for PC13 and PC17 for the liver samples labeled with circadian time (CT), showing temporal variation along an elliptic path. Labels indicate CT time; samples that are 24 h apart are in the same color. (B) Fractions of temporal variance in each tissue explained by 24- and 12-h periods, obtained by applying spectral analysis genome-wide for each tissue. Dotted horizontal lines represent the expected background level, assuming white noise. (C,D) Cumulative number of rhythmic genes (P < 0.01, harmonic regression) with log2 fold change larger than the value on the x-axis. (C) Analysis on 11 WT tissues. (D) Analysis on four conditions: Bmal1 KO mice and WT littermates in liver and kidney.
We analyzed the peak-to-trough amplitudes (hereafter also referred to as fold change) of 24-h rhythmic transcripts. Metabolic tissues, notably liver, brown fat, and skeletal muscle exhibited more (on the order of 100 transcripts) intermediate- to high-amplitude (between two- and 10-fold) transcript rhythms. Brain tissues showed virtually no rhythmic transcripts above fourfold (Fig. 1C). In liver and kidney of Bmal1 KO mice, the number of rhythmic mRNAs was reduced by threefold compared to WT littermates. This effect increased for larger amplitudes. Few transcripts in tissues of Bmal1 KO oscillated by more than 10-fold (Fig. 1D). Thus, a functional circadian clock is required for high-amplitude transcript rhythms across diverse tissues, while systemic signals regulate lower amplitude rhythms that persist in clock-deficient liver (Hughes et al. 2012; Atger et al. 2015; Sobel et al. 2017) and kidney (Nikolaeva et al. 2012).
Combinatorics of rhythmic transcript expression across tissues and genotypes
We reasoned that identifying sets of genes with shared rhythms across subsets of tissues would allow finding underlying regulatory mechanisms. We developed a model selection (MS) algorithm extending harmonic regression (Fisher 1929) to classify genes into modules sharing rhythmic mRNA profiles across subsets of tissues (Fig. 2A; Methods). Phase (time of peak) and amplitude (log2 fold change) relationships between genes and tissues are summarized using complex-valued singular value decomposition (SVD) (Fig. 2B; Methods). We applied MS to the 11 tissues, which identified gene modules involving rhythmic mRNA accumulation in nearly all tissues (tissue-wide) (Fig. 2C), in single tissues (tissue-specific), or in several tissues (tissue-restricted) (examples shown in Fig. 2D; Supplemental Fig. S2A; Supplemental Table S3).
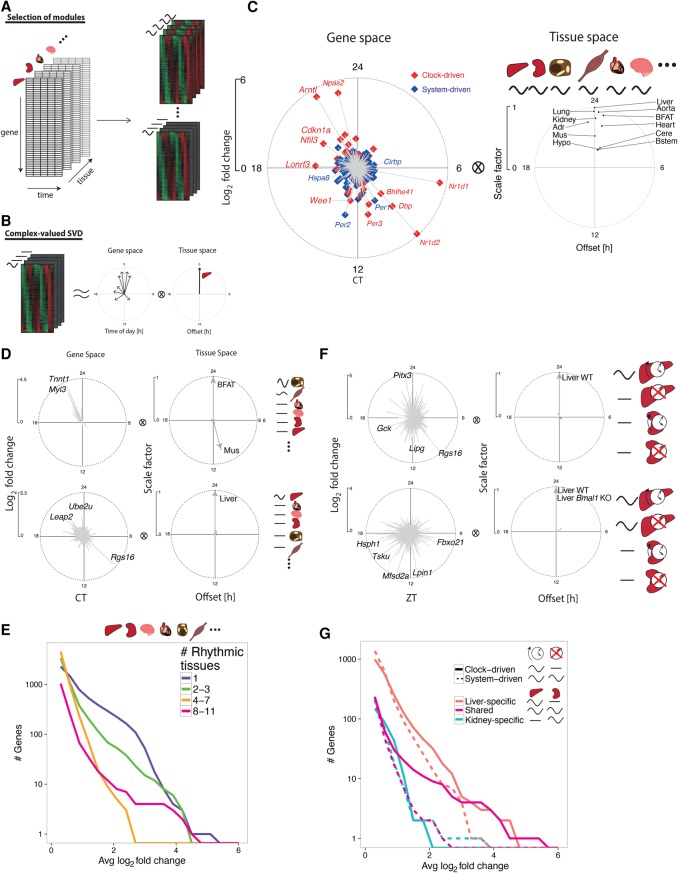
Figure 2.
Combinatorics of rhythmic transcript expression across tissues and genotypes. (A) Schema for the model selection (MS) algorithm to identify rhythmic gene expression modules across tissues. Temporal transcriptomes of different tissues represented as a three-dimensional array (left). Gene modules are probabilistically assigned among different combinations of 24-h rhythms across tissues (e.g., tissue-specific or tissue-wide rhythms schematically shown on right). (B) Gene modules are summarized by the first component of complex-valued singular value decomposition (SVD) to highlight phase (peak time shown as the clockwise angle) and amplitude (log2 fold change shown as the radial distance) relationships between genes (gene space) and between tissues (tissue space). SVD representation is scaled such that the genes show log2 fold changes, while tissue vectors are scaled such that the highest amplitude tissue has length of 1 and a phase offset of 0 h. (C–E) MS applied to 11 WT tissues. (C) SVD representation of tissue-wide mRNA rhythms from the 11 tissues. Genes are labeled as system-driven (blue) or clock-driven (red) according to the comparison of the corresponding temporal profiles in Bmal1 KO and WT littermates. (D) Examples of anti-phasic rhythms (brown fat and muscle, n = 20, first SVD component explains 81% of variance), and tissue-specific rhythms (liver, n = 846, first SVD component explains 59% of variance). Representative genes with large amplitudes are labeled. (E) Number of transcripts showing rhythms (P-value < 0.01, harmonic regression) in different numbers of tissues, in function of increasing peak to trough amplitudes on the x-axis. x-axis: average log2 fold change calculated from the identified rhythmic tissues. (F,G) MS applied to Bmal1 KO and WT littermates in liver and kidney. (F) SVD representation of clock- (top, n = 991, 83% of variance) and system-driven (bottom, n = 1395, 84% of variance) liver-specific rhythms. (G) Number of transcripts showing clock- (solid) or system-driven (dotted) rhythms (P-value < 0.01, harmonic regression) in liver (red), kidney (blue), or both (magenta).
The tissue-wide module contained a set of both clock- and system-driven rhythmic mRNAs, as determined by comparing Bmal1 KO data in liver and kidney (Fig. 2C, left). Moreover, these transcripts oscillated in synchrony across all tissues and peaked at fixed times of day, although their amplitudes varied between tissues, with brain regions showing the smallest amplitudes (Fig. 2C, right). The clock drove synchronized oscillations at high amplitudes, notably clock genes (e.g., Arntl, Npas2, Nr1d1,2; note that Arntl and Nr1d1,2 are also named Bmal1 and Rev-erba,b, respectively), clock output genes (e.g., Dbp, Nfil3), and cell cycle regulators (Cdkn1a and Wee1) (Matsuo et al. 2003; Gréchez-Cassiau et al. 2008). Interestingly, clock genes Per1,2 continued to oscillate in Bmal1 KO in multiple tissues, extending previous studies in liver (Kornmann et al. 2007). Other clock-independent oscillations included mRNAs of heat- and cold-induced genes, such as Hspa8 and Cirbp (Morf et al. 2012; Gotic et al. 2016), that peaked 12 h apart near CT18 and CT6 (CT: circadian time; CT0 corresponds to subjective dawn and start of the resting phase; CT12 corresponds to subjective dusk and start of the activity phase), concomitantly with highs and lows in body temperature rhythms (Refinetti and Menaker 1992).
Tissue-restricted modules contained rhythmic transcripts that peaked in synchrony, such as in liver and kidney, or with fixed offsets, such as the nearly 12-h shifted rhythms in brown fat and skeletal muscle (Supplemental Fig. S3A). Overall, transcripts with large amplitudes (FC > 8) oscillated in either a few tissues (three or less) or tissue-wide (eight or more) (Fig. 2E).
To distinguish clock- and system-driven mRNA rhythms, we applied the MS algorithm to the liver and kidney transcriptomes in WT and Bmal1 KO mice (Fig. 2F; Supplemental Fig. S3B; Supplemental Table S4). This separation identified clock- and system-driven modules that oscillated in liver but were flat in kidney (Fig. 2F), as exemplified by mRNAs of Lipg and Lpin1 (Supplemental Fig. S2B). Indeed, both transcripts oscillated in WT liver with robust amplitudes, peaking near ZT11, but were flat in kidney (ZT: Zeitgeber time; ZT0 corresponds to onset of lights-on; ZT12 corresponds to onset of lights-off). However, in Bmal1 KO, Lpin1 continued to oscillate, while Lipg was flat.
Summarizing, we found that shared clock-driven mRNA rhythms, which contained core clock and clock-controlled genes, oscillated with significantly larger amplitudes than system-driven genes (Fig. 2G, magenta solid versus dotted). Similarly, clock-driven liver-specific mRNA rhythms also oscillated at higher amplitudes compared with system-driven mRNA rhythms (Fig. 2G, red solid versus dotted). On the other hand, kidney-specific clock- and system-driven transcripts oscillated with comparable amplitudes (Fig. 2G, blue solid versus dotted) and were less numerous overall, which could reflect the distinct cell types constituting the kidney (Lee et al. 2015). The uncovered diversity of clock- and system-driven mRNA rhythms involving distinct combinations of tissues hints at complex transcriptional or post-transcriptional regulation.
Oscillatory TF activity in one tissue but not others can drive tissue-specific mRNA rhythms
We focused on WT and Bmal1 KO liver and kidney to identify rhythmic TF activities underlying clock- and system-driven tissue-specific mRNA rhythms. We first analyzed liver-rhythmic genes driven by systemic signals (n = 1395, MS) (Fig. 3A), which were associated with feeding and fasting rhythms (Gene Ontology analysis around the clock) (Methods). Indeed, ribosome biogenesis was up-regulated most strongly during the first 6 h of the feeding phase (from ZT12 to ZT18) (Jouffe et al. 2013; Chauvin et al. 2014), while insulin signaling was down-regulated during first 6 h of the fasting phase (from ZT0 to ZT6) (Ravnskjaer et al. 2013), consistent with daily responses to nutrient fluctuations in liver (Sinturel et al. 2017).
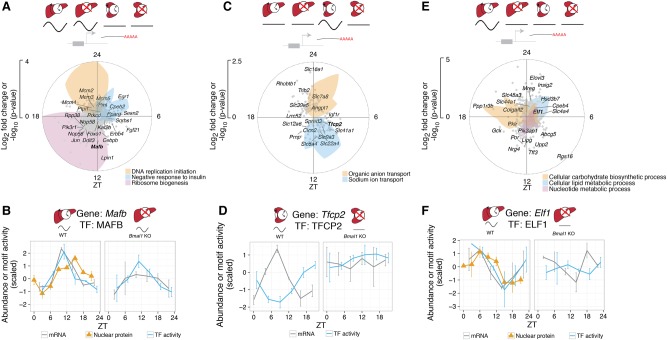
Figure 3.
Oscillatory TF activity in one tissue but not others can drive tissue-specific rhythms. (A) Module describing system-driven liver-specific rhythms (n = 1395, first SVD component explains 84% of variance). Radial coordinate of the colored polygons represents enrichment of the indicated GO terms at each time point, obtained by comparing the genes falling in a sliding window of ±3 h to the background set of all 1395 genes assigned to the module (P-value computed from Fisher's exact test). (B) MAFB is a candidate TF for the module in A. Predicted MAFB activity (blue), nuclear protein abundance (orange triangles), and mRNA accumulation (gray) oscillate in WT and Bmal1 KO, with peak mRNA preceding peak nuclear protein and TF activity. Error bars in nuclear protein, mRNA, and TF activity show SEM (n = 2). (C) Clock-driven kidney-specific module (n = 156, first SVD component explains 80% of variance). Colored polygons as in A. (D) TFCP2 is a candidate TF for the module in C. The temporal profile of predicted TFCP2 activity (blue) is anti-phasic with Tfcp2 mRNA accumulation (gray) in WT, and both are flat in Bmal1 KO. Error bars in mRNA and TF activity show SEM (n = 2). (E) Clock-driven liver-specific module (n = 991, first SVD explains 83% of variance). (F) ELF is a candidate TF for the module in E. The temporal profile of predicted ELF activity (blue) in WT matches that of nuclear protein abundance in liver (orange triangles), and both are delayed compared to Elf1 mRNA accumulation (gray). In Bmal1 KO, ELF activity and Elf1 mRNA are nonrhythmic. Error bars in nuclear protein, mRNA, and TF activity show SEM (n = 2).
To infer rhythmic TF activities that may underlie these mRNA rhythms, we applied a penalized regression model (MARA) (Balwierz et al. 2014) that integrates TF binding site predictions near promoters with mRNA accumulation. TF analysis of this module notably identified TFs related to insulin biosynthesis and gluconeogenesis, such as MAFB (Matsuoka et al. 2003) and EGR1 (Matsuoka et al. 2003; Shen et al. 2015), whose activities peaked at ZT11 and ZT3, respectively (Fig. 3B; Supplemental Fig. S4A). Integrating temporal activities of candidate TFs with RNA-seq and our previously described temporal nuclear protein data set (Wang et al. 2017), we found that rhythmic activity of MAFB and EGR1 was supported by rhythmic mRNA abundance, followed by rhythmic nuclear protein abundance (Fig. 3B; Supplemental Fig. S4B), likely reflecting the delayed protein abundance after mRNA accumulation (Mermet et al. 2016).
Next, we analyzed clock-driven transcripts oscillating specifically in the kidney (n = 156, MS) (Fig. 3C), among which sodium ion and organic anion transporters peaked near ZT12 and ZT0, respectively. The up-regulation of sodium ion transporters in kidney during the behaviorally active phase may underlie clock-dependent increase of sodium excretion (Nikolaeva et al. 2012). Similarly, the up-regulation of organic anion transporters during the resting phase may explain increased transport activity for precursors of gluconeogenesis, such as pyruvate and lactate, during fasting (Stumvoll et al. 1998; Ekberg et al. 1999). TF analysis predicted TFCP2 to regulate mRNAs that peaked during the resting phase (Fig. 3D; Supplemental Fig. S4C). The predicted TFCP2 activity was anti-phasic with Tfcp2 mRNA abundance, suggesting a repressive activity, consistent with the ability of TFCP2 to recruit histone deacetylase HDAC1 (Kim et al. 2016).
Finally, liver-specific clock-driven rhythmic transcripts (n = 991, MS) were comprised of genes associated with glucose metabolism (enriched at ZT18), such as Gck and Ppp1r3b (Kelsall et al. 2009; Oosterveer and Schoonjans 2014), as well as lipid, cholesterol, and bile acid metabolism genes (enriched at ZT2), such as Elovl3, Insig2, Hsd3b7, and Cyp8b1 (Fig. 3E; Shea et al. 2007; Le Martelot et al. 2009; Guillou et al. 2010; Sayin et al. 2013). Predicted activity of ELF oscillated and peaked near ZT3 in WT liver but was flat in Bmal1 KO (Fig. 3F; Supplemental Fig. S4D; Fang et al. 2014). Interestingly, mRNA abundance of Elf1, as well as its nuclear protein abundance, also oscillated in WT, supporting Elf1 as a potential regulator of oscillating transcriptions peaking near ZT6.
Colocalized binding of clock- and liver-specific TFs drives liver-specific mRNA rhythms
To further dissect liver-specific clock-driven rhythms, we reasoned that accessible chromatin regions specific to the liver could harbor regulatory sites for clock TFs, which could then regulate mRNA rhythms liver-specifically. Comparing DNase I hypersensitive sites (DHSs) in liver and kidney (DNase-seq data from ENCODE) (Yue et al. 2014), we found that liver-specific clock-driven genes were enriched with liver-specific DHSs (within 40 kb of promoters), compared to system-driven as well as nonrhythmic genes (Fig. 4A). Using TF binding site predictions underlying these liver-specific DHSs, we applied MARA to predict rhythmic TF activities that explain gene expression of this module (Supplemental Fig. S5A). In WT liver, the predicted activity of RORE oscillated with robust amplitudes and peaked near ZT21. RORE activity became high and flat in Bmal1 KO liver, consistent with loss of REV-ERB expression and consequently de-repression of REV-ERB target genes (Fig. 4B, top; Bugge et al. 2012). Activity of E-box in WT liver peaked at ZT7, consistent with BMAL1:CLOCK activity (Rey et al. 2011), albeit with weaker amplitudes compared to RORE activity, likely reflecting fewer E-box target genes compared to RORE in this module. In Bmal1 KO mice, E-box activity was low and flat in liver, as expected.
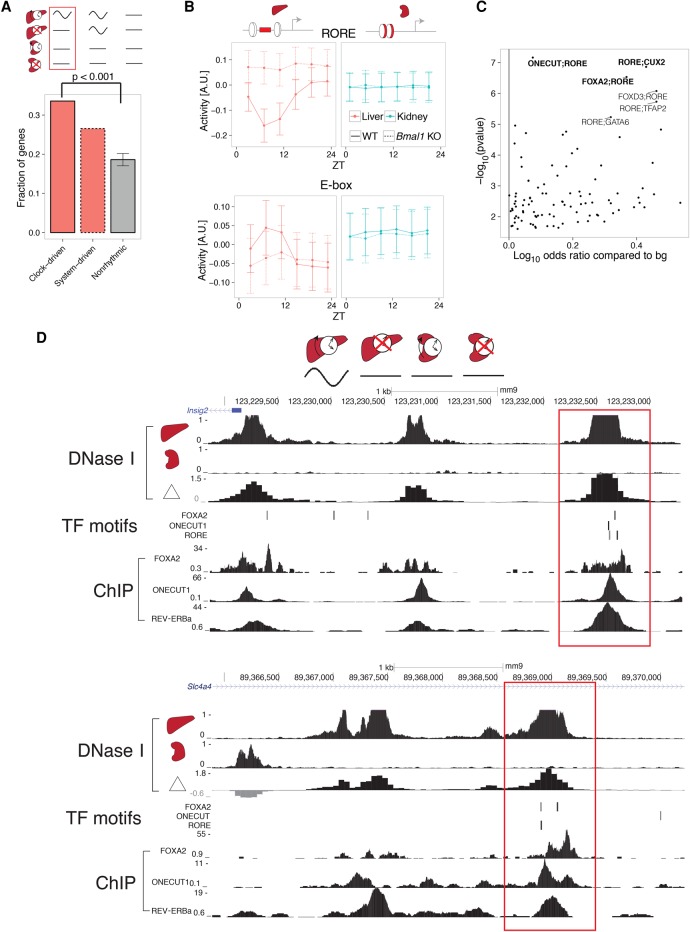
Figure 4.
Colocalized binding of clock- and liver-specific TFs underlies liver-specific mRNA rhythms. (A) The fraction of genes containing liver-specific DNase I hypersensitive sites (DHSs) in the clock-driven liver-specific module is higher compared with both nonrhythmic and system-driven liver-specific modules. Error bars and P-values calculated from 10,000 bootstrap iterations. (B) Predicted temporal activities of RORE (top) and E-box (bottom) TF motifs located within liver-specific DHSs. Error bars show standard deviation of the estimated activities. (C) Co-occurrence of RORE with all other TFs in the SwissRegulon database (Pachkov et al. 2007) (189 TF motifs). Positive log10 odds ratios (ORs) represent pairs of motifs enriched in the clock-driven liver-specific module compared to the flat module. P-values for the motif pairs were calculated from χ2 tests applied to three-way contingency tables (Myšičková et al. 2012). Selected pairs are in bold. (D) DNase I hypersensitivity in liver, kidney, and the corresponding differential signal (in log2 fold change) near two representative genes (top: Insig2; bottom: Slc4a4). RORE, ONECUT1, and FOXA2 TF binding motifs (posterior probability > 0.5, MotEvo) co-occur at liver-specific DHSs (red boxes). Predicted TF binding sites correspond to experimentally observed TF binding in publicly available ChIP-exo data sets for REV-ERBa, ONECUT1, and FOXA2 (bottom).
We hypothesized that cooperativity of liver-specific and clock TFs at liver-specific DHSs can regulate liver-specific mRNA rhythms. Pairwise analysis of TF binding sites at liver-specific DHSs found enrichment of co-occurrence between RORE and liver-specific TF motifs, FOXA2, ONECUT, and CUX2 (Fig. 4C). Enrichment of both CUX2 and ONECUT (also named HNF6) is consistent with ONECUT1 binding to both ONECUT and CUX2 motifs (Conforto et al. 2015). mRNAs of genes with co-occurrence of RORE and liver-specific TF motifs peaked near ZT1, consistent with peak RORE activity (near ZT21) preceding peak mRNA abundance of REV-ERB targets (Supplemental Fig. S5B). Analysis of ChIP-exo data sets targeting FOXA2, ONECUT1, and REV-ERBa in liver (Wang et al. 2014; Zhang et al. 2015; Iwafuchi-Doi et al. 2016) confirmed colocalized TF binding at liver-specific DHSs distal from clock-driven liver mRNAs such as Insig2 and Slc4a4 (Fig. 4D).
Liver-specific chromatin loops regulate liver-specific mRNA rhythms
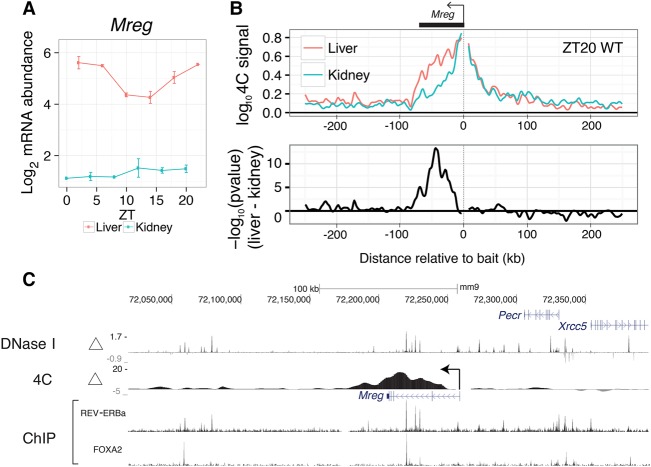
To test whether distally located liver-specific DHSs can contact promoters of clock-driven liver-rhythmic genes, we selected the promoters of Mreg, Pik3ap1, and Slc44a1 as baits for 4C-seq experiments in liver and kidney harvested at the time of peak mRNA accumulation for the selected genes (Methods; Fig. 5A; Supplemental Figs. S6A, S7A). Upstream of Mreg, the 4C-seq signal, which measures frequency of promoter-enhancer contacts (van de Werken et al. 2012), decayed rapidly to background level in both liver and kidney (Fig. 5B, top). Downstream from Mreg, however, the 4C-seq signal showed a tissue-dependent pattern, decaying slowly in the liver but more rapidly in the kidney. This difference in decay suggests increased frequency of promoter-enhancer contacts in the liver compared to the kidney. Indeed, differential analysis identified liver-specific chromatin contacts 40 kb downstream from the promoter (Fig. 5B, bottom). Overlaying the contact data with DNase-seq, we found that liver-specific chromatin contacts downstream from Mreg connected liver-specific DHSs with the Mreg promoter (Fig. 5C). Furthermore, ChIP-exo showed colocalization of REV-ERBa and FOXA2 binding at liver-specific DHSs contacting the promoters (Fig. 5C). In contrast, accessible regions upstream of the Mreg promoter did not show liver-specific chromatin contacts. The 4C-seq data thus suggest that liver-specific chromatin loops can recruit clock-bound distal elements to promoters to regulate liver-specific transcriptional rhythms. Other liver-specific rhythmic transcripts, Pik3ap1 and Slc44a1, also displayed liver-specific chromatin loops between promoter and liver-specific open chromatin regions (Supplemental Figs. S6, S7). In sum, tissue-specific chromatin looping can drive tissue-specific mRNA rhythms.
Figure 5.
Liver-specific chromatin loops regulate liver-specific mRNA rhythms. (A) Temporal mRNA profile for Mreg, a clock-driven liver-rhythmic gene. Error bars are SEM (n = 2). (B) 4C-seq profiles (summary from two replicates, each pooling two different mice) using the Mreg promoter as a bait in liver and kidney at ZT20. Data are shown in a window of ±250 kb from the bait (top). Profiles of differential contacts between liver and kidney (bottom) represented as signed log P-values (regularized t-test, positive values denote liver-enriched 4C contacts). (C) Tracks of differential 4C contacts (signed log P-values), log2 fold change of DNase I hypersensitivity between liver and kidney, and ChIP-exo of REV-ERBa and FOXA2. Regions of significant differential 4C contacts correspond to liver-specific DNase I hypersensitive regions and REV-ERBa binding sites.
Precise promoter-enhancer contacts underlie liver-specific mRNA rhythms
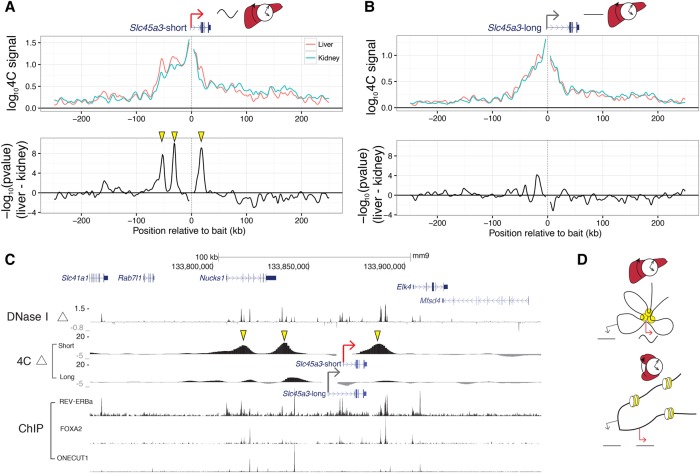
To test whether distinct chromatin loops would form at alternative nearby gene promoters with distinct temporal mRNA profiles, we searched for candidate genes where one promoter was rhythmically transcribed while the alternative one was nonrhythmic (Supplemental Fig. S8). Slc45a3 has two alternative transcripts using different promoters 8 kb apart. The shorter pre-mRNA oscillated in the liver (rhythmic promoter, Slc45a3-short), while the longer did not (flat promoter, Slc45a3-long). In kidney, neither Slc45a3-short nor Slc45a3-long showed robust transcript rhythms (Supplemental Fig. S9). Targeting the Slc45a3-short promoter with 4C-seq in liver and kidney showed liver-specific chromatin loops at three distal regions (two upstream, one downstream) (Fig. 6A). These same regions did not form liver-specific chromatin loops with the Slc45a3-long promoter (Fig. 6B), suggesting that promoters 8 kb apart can contact distinct enhancers. Overlaying 4C-seq with DNase-seq showed that these chromatin loops link liver-specific DHSs specifically to the Slc45a3-short promoter (Fig. 6C). These liver-specific DHSs were bound by liver-specific TFs, FOXA2 and ONECUT1, and clock TF, REV-ERBa, as shown in ChIP-seq. The 4C experiments suggest that enhancers can contact a rhythmic promoter while looping out nearby nonrhythmic alternative promoters, confining rhythmic enhancer activity to specific promoters (Fig. 6D). Furthermore, rhythmically active enhancers can contact promoters in a tissue-specific manner. Thus, chromatin folding not only regulates tissue-specific rhythms but also differentiates between closely spaced promoters to control rhythmic transcription with spatial precision.
Figure 6.
Precise promoter-enhancer contacts underlie liver-specific mRNA rhythms. (A,B) 4C-seq profiles for the (A) Slc45a3-short and (B) Slc45a3-long isoforms within ±250 kb around baits targeting the two TSSs (top). Signed log P-values for differential contacts between liver and kidney (bottom) as in Figure 5B. TSSs for Slc45a3-short and Slc45a3-long are 8 kb apart. (C) Differential 4C contacts (signed log P-values), log2 fold change of DNase I hypersensitivity between liver and kidney, and ChIP-exo signal of REV-ERBa, FOXA2, and ONECUT1. Regions of significant differential contacts in Slc45a3-short correspond to liver-specific DNase I hypersensitive regions. Yellow arrowheads in A and C show liver-specific distal contacts recruited to the Slc45a3-short TSS. These contacts are absent for Slc45a3-long TSS (B). (D) Schematic model illustrating enhancer-promoter interactions in liver and kidney that may generate liver-specific rhythms. Yellow circles illustrate liver-active enhancers contacting the rhythmic promoter (red arrow) but not the alternative nonrhythmic promoter (gray arrow). In kidney, the enhancer is not accessible, and both promoters are nonrhythmic.
Discussion
The mammalian genome encodes transcriptional programs that allow the molecular clock to robustly oscillate across diverse tissue transcriptomes while maintaining flexibility to regulate distinct clock outputs in different combinations of tissues. Here, we identified two regulatory modes underlying tissue-specific transcript rhythms: Regulatory sequences can recruit individual TFs bearing rhythmic activity; coordinated binding of clock- and tissue-specific TFs can generate tissue-specific rhythms. Moreover, we found that clock- and tissue-specific TFs bound at distal enhancers can be recruited to promoters through precise chromatin loops.
Several of our predictions of transcription regulators and regulated genes (e.g., Egr1, Por, Upp2) corroborated with previous analyses of independent data sets (Yan et al. 2008; Bozek et al. 2009; Bhargava et al. 2015). Further analysis incorporating outputs of enhancer activity, such as eRNAs (Fang et al. 2014), across multiple tissues may uncover additional rhythmically active regulators.
Colocalized binding of clock- and tissue-specific TFs at enhancers provides a putative mechanism for the clock to regulate clock output genes in a tissue-specific manner. In mouse liver, clock TFs can colocalize with liver-specific TFs, such as FOXA2 and ONECUT1, consistent with multiple TFs associating with liver-specific DHSs (Iwafuchi-Doi et al. 2016). Our findings are currently based on sequence-specific DNA binding of TFs, comparison of tissues, and ChIP-seq data sets. Further mechanistic basis for the functional significance of colocalization could be gained, for example, by using inducible knockout models for tissue-specific regulators. Moreover, the observed colocalization does not exclude other cooperative modes, such as tethering of REV-ERBa to ONECUT1 through protein-protein interactions (Zhang et al. 2015).
Our 4C analysis showed that chromatin looping might mediate interaction between clock- and tissue-specific transcriptional programs by recruiting clock-bound distal elements to promoters in a tissue-specific manner. Such loops can surgically discriminate between nearby promoters as close as 8 kb apart, suggesting a way to separate temporal regulation of neighboring promoters. A previous 4C study on a core clock gene enhancer proposed that cohesion-mediated promoter-enhancer looping can compartmentalize rhythmic gene expression within genomic regions spanning 150 kb (Xu et al. 2016). Here, chromatin interactions that differed between tissues were localized to a small genomic region (<10 kb) near promoters (<100 kb). Future studies integrating temporal data across tissues with large-scale promoter-enhancer networks may reveal regulatory sequences that encode promoter-enhancer compatibility and elucidate whether this compatibility is tissue-specific (Li and Noll 1994; Merli et al. 1996; Zabidi et al. 2014; Nguyen et al. 2016).
While our work focused on transcriptional mechanisms, studying other mechanisms such as post-transcriptional, translational, and post-translational processes using PRO-seq, Ribo-seq, and proteomics data may provide additional insights. Expanding our 24-h analysis to 12-h or other harmonics would broaden the view of tissue-specific temporal gene expression but may require experimental designs of higher temporal resolution (Hughes et al. 2009; Krishnaiah et al. 2017). In sum, integrating the temporal axis into tissue-specific gene regulation offers an integrated understanding of how tissue physiology resonates with daily cycles in the environment.
Methods
Animal experiments
Eight- to 14-wk-old C57Bl/6 mice have been purchased from Charles River Laboratory. Bmal1 KO mice have been previously described (Jouffe et al. 2013). Without further indications, mice are kept under a 12-h light/12-h dark regimen and ad libitum feeding. All animal care and handling was performed according to the Canton de Vaud (Fred Gachon, authorization no. VD 2720) laws for animal protection.
RNA-seq experiments
To complement the mouse liver WT and Bmal1 KO RNA-seq data (GSE73554) (Atger et al. 2015), transcriptomes of kidneys from Bmal1 KO and WT littermates (12-h light/12-h regimen; night-restricted feeding) were measured from poly(A)-selected mRNA using single-end reads of length 100. mRNA levels were quantified using kallisto version 0.42.4 (mm10) (Bray et al. 2015).
Global temporal variance
For each tissue, we estimated the contribution of temporal variance for each gene, broken down by its Fourier components. We calculated the background level assuming temporally unstructured data (white noise), whose magnitude (strength of the white noise) was estimated from the mean of squared magnitudes of Fourier coefficients that were not submultiples of 24 h (i.e., the mean of 48-, 16-, 9.6-, 6.9-, 5.3-, 4.4-h components).
Model selection
We fitted harmonic regression models that integrated temporal gene expression across different combinations of rhythms in different conditions (Atger et al. 2015). We used a g-prior for the rhythmic parameters as a penalization scheme (Liang et al. 2008). We set g = 1000, which we found to maximize temporal variations captured in the shared rhythms model while minimizing temporal variations captured in the flat model.
Complex singular value decomposition representation of gene and tissue module
We first transformed the time domain to the frequency domain corresponding to 24-h rhythms. The resulting matrix was decomposed using SVD; the first left-singular and first right-singular values were visualized in separate polar plots (Supplemental Methods).
Functional analysis by GO terms
We used Fisher's exact test to assess statistical significance of gene enrichment for each GO term. Foreground genes were genes with phases within a 6-h window. Background genes were genes assigned to a model. For each GO term, we slid the 6-h window with a step size of 1 h and calculated the P-value enrichment. GO terms were chosen by visualizing significant GO terms in the tree and choosing GO terms that were comparably deep in the tree.
Chromatin conformation experiments and analysis
C57Bl/6 mice were sacrificed at ZT08 and ZT20 to extract liver and kidneys. Liver and kidney nuclei were prepared as previously described (Ripperger and Schibler 2006) with some minor changes. 4C-seq assays were performed as in Gheldof et al. (2012). Raw read counts for each sample were normalized by library size by the sum of the read counts on the cis-chromosome (excluding 10 fragments around the bait). We used a weighted linear model to fit the log10 signal around each fragment f. A Gaussian window of standard deviation = 2.5 kb centered on f was used to incorporate signal from neighboring fragments (Supplemental Methods). Differential contacts were estimated using t-statistics.
Data access
Raw and processed RNA-seq and 4C-seq data generated from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE100457.
Competing interest statement
C.J., J. Marquis, A.C., and F.G. are employees of Nestlé Institute of Health Sciences SA.
Supplementary Material
Acknowledgments
We thank Eric Paquet for critical reading and Saeed Omidi for help and discussions in bioinformatics. This work was supported by Swiss National Science Foundation Grant 31003A-153340, European Research Council Grant ERC-2010-StG-260667, and the Ecole Polytechnique de Lausanne. J.Y. benefits from the Natural Sciences and Engineering Research Council of Canada Postgraduate Studies Doctoral scholarship.
Author contributions: Conceptualization, J.Y., J. Mermet, and F.N; formal analysis, J.Y. and F.N.; investigation, J. Mermet, C.J., J. Marquis, and A.C.; writing, review, and editing, J.Y., J. Mermet, F.G., and F.N.; supervision, F.N. and F.G.; funding acquisition, F.N. and F.G.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222430.117.
Freely available online through the Genome Research Open Access option.
References
- Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci 112: E6579–E6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. 2014. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res 24: 869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Lazar MA. 2016. Circadian time signatures of fitness and disease. Science 354: 994–999. [DOI] [PubMed] [Google Scholar]
- Bhargava A, Herzel H, Ananthasubramaniam B. 2015. Mining for novel candidate clock genes in the circadian regulatory network. BMC Syst Biol 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Relógio A, Kielbasa SM, Heine M, Dame C, Kramer A, Herzel H. 2009. Regulation of clock-controlled genes in mammals. PLoS One 4: e4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N, Pimentel H, Melsted P, Pachter L. 2015. Near-optimal RNA-Seq quantification. arXiv:1505.02710 [q-bio.QM]. [DOI] [PubMed]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, et al. 2014. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 33: 474–483. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485: 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto TL, Steinhardt GF, Waxman DJ. 2015. Cross talk between GH-regulated transcription factors HNF6 and CUX2 in adult mouse liver. Mol Endocrinol 29: 1286–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckard A, Anafi RC, Hogenesch JB, Haase SB, Harer J. 2013. Design and analysis of large-scale biological rhythm studies: a comparison of algorithms for detecting periodic signals in biological data. Bioinformatics 29: 3174–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U. 2015. Circadian timing of metabolism in animal models and humans. J Intern Med 277: 513–527. [DOI] [PubMed] [Google Scholar]
- Ekberg K, Landau BR, Wajngot A, Chandramouli V, Efendic S, Brunengraber H, Wahren J. 1999. Contributions by kidney and liver to glucose production in the postabsorptive state and after 60 h of fasting. Diabetes 48: 292–298. [DOI] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. 2014. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159: 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1929. Tests of significance in harmonic analysis. Proc R Soc Lond Ser A 125: 54–59. [Google Scholar]
- Gheldof N, Leleu M, Noordermeer D, Rougemont J, Reymond A. 2012. Detecting long-range chromatin interactions using the chromosome conformation capture sequencing (4C-seq) method. Methods Mol Biol 786: 211–225. [DOI] [PubMed] [Google Scholar]
- Gotic I, Omidi S, Fleury-Olela F, Molina N, Naef F, Schibler U. 2016. Temperature regulates splicing efficiency of the cold-inducible RNA-binding protein gene Cirbp. Genes Dev 30: 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. 2008. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem 283: 4535–4542. [DOI] [PubMed] [Google Scholar]
- Guillou H, Zadravec D, Martin PG, Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res 49: 186–199. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. 2009. Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. 2012. Brain-specific rescue of clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet 8: e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. 2013. The circadian clock coordinates ribosome biogenesis. PLoS Biol 11: e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Rosenzweig D, Cohen PTW. 2009. Disruption of the allosteric phosphorylase a regulation of the hepatic glycogen-targeted protein phosphatase 1 improves glucose tolerance in vivo. Cell Signal 21: 1123–1134. [DOI] [PubMed] [Google Scholar]
- Kim JS, Chae JH, Cheon YP, Kim CG. 2016. Reciprocal localization of transcription factors YY1 and CP2c in spermatogonial stem cells and their putative roles during spermatogenesis. Acta Histochem 118: 685–692. [DOI] [PubMed] [Google Scholar]
- Korenčič A, Košir R, Bordyugov G, Lehmann R, Rozman D, Herzel H. 2014. Timing of circadian genes in mammalian tissues. Sci Rep 4: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, et al. 2017. Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab 25: 961–974.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. 2008. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci 105: 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. 2009. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7: e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Chou CL, Knepper MA. 2015. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Noll M. 1994. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J 13: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Grant GR, Hogenesch JB, Hughes ME. 2015. Considerations for RNA-seq analysis of circadian rhythms. Methods Enzymol 551: 349–367. [DOI] [PubMed] [Google Scholar]
- Liang F, Paulo R, Molina G, Clyde MA, Berger JO. 2008. Mixtures of g priors for Bayesian variable selection. J Am Stat Assoc 103: 410–423. [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. 2003. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259. [DOI] [PubMed] [Google Scholar]
- Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. 2003. Members of the large Maf transcription family regulate insulin gene transcription in islet β cells. Mol Cell Biol 23: 6049–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. 1996. Promoter specificity mediates the independent regulation of neighboring genes. Genes Dev 10: 1260–1270. [DOI] [PubMed] [Google Scholar]
- Mermet J, Yeung J, Naef F. 2016. Systems chronobiology: global analysis of gene regulation in a 24-hour periodic world. Cold Spring Harb Perspect Biol 9: a028720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U. 2012. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338: 379–383. [DOI] [PubMed] [Google Scholar]
- Myšičková A, Vingron M. 2012. Detection of interacting transcription factors in human tissues using predicted DNA binding affinity. BMC Genomics 13(Suppl 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Jones RD, Snavely AR, Pfenning AR, Kirchner R, Hemberg M, Gray JM. 2016. High-throughput functional comparison of promoter and enhancer activities. Genome Res 26: 1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva S, Pradervand S, Centeno G, Zavadova V, Tokonami N, Maillard M, Bonny O, Firsov D. 2012. The circadian clock modulates renal sodium handling. J Am Soc Nephrol 23: 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveer MH, Schoonjans K. 2014. Hepatic glucose sensing and integrative pathways in the liver. Cell Mol Life Sci 71: 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachkov M, Erb I, Molina N, van Nimwegen E. 2007. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res 35(Database issue): D127–D131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnskjaer K, Hogan MF, Lackey D, Tora L, Dent SY, Olefsky J, Montminy M. 2013. Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. J Clin Invest 123: 4318–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R, Menaker M. 1992. The circadian rhythm of body temperature. Physiol Behav 51: 613–637. [DOI] [PubMed] [Google Scholar]
- Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U. 2008. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev 22: 331–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38: 369–374. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-β-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17: 225–235. [DOI] [PubMed] [Google Scholar]
- Shea HC, Head DD, Setchell KDR, Russell DW. 2007. Analysis of HSD3B7 knockout mice reveals that a 3α-hydroxyl stereochemistry is required for bile acid function. Proc Natl Acad Sci 104: 11526–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen N, Jiang S, Lu JM, Yu X, Lai SS, Zhang JZ, Zhang JL, Tao WW, Wang XX, Xu N, et al. 2015. The constitutive activation of Egr-1/C/EBPa mediates the development of type 2 diabetes mellitus by enhancing hepatic gluconeogenesis. Am J Pathol 185: 513–523. [DOI] [PubMed] [Google Scholar]
- Sinturel F, Gerber A, Mauvoisin D, Wang J, Gatfield D, Stubblefield JJ, Green CB, Gachon F, Schibler U. 2017. Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell 169: 651–663.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JA, Krier I, Andersin T, Raghav S, Canella D, Gilardi F, Kalantzi AS, Rey G, Weger B, Gachon F, et al. 2017. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol 15: e2001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. 2002. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83. [DOI] [PubMed] [Google Scholar]
- Stumvoll M, Meyer C, Perriello G, Kreider M, Welle S, Gerich J. 1998. Human kidney and liver gluconeogenesis: evidence for organ substrate selectivity. Am J Physiol 274: E817–E826. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. 2008. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9: 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, et al. 2012. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods 9: 969–972. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S. 2012. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab 16: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen J, Wang C, Uusküla-Reimand L, Chen K, Medina-Rivera A, Young EJ, Zimmermann MT, Yan H, Sun Z, et al. 2014. MACE: model based analysis of ChIP-exo. Nucleic Acids Res 42: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mauvoisin D, Martin E, Atger F, Galindo AN, Dayon L, Sizzano F, Palini A, Kussmann M, Waridel P, et al. 2017. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab 25: 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Guo W, Li P, Zhang Y, Zhao M, Fan Z, Zhao Z, Yan J. 2016. Long-range chromosome interactions mediated by cohesin shape circadian gene expression. PLoS Genet 12: e1005992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. 2008. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol 4: 647–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. 2014. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518: 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. 2015. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348: 1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.