Abstract
We present here a full update of the PMut predictor, active since 2005 and with a large acceptance in the field of predicting Mendelian pathological mutations. PMut internal engine has been renewed, and converted into a fully featured standalone training and prediction engine that not only powers PMut web portal, but that can generate custom predictors with alternative training sets or validation schemas. PMut Web portal allows the user to perform pathology predictions, to access a complete repository of pre-calculated predictions, and to generate and validate new predictors. The default predictor performs with good quality scores (MCC values of 0.61 on 10-fold cross validation, and 0.42 on a blind test with SwissVar 2016 mutations). The PMut portal is freely accessible at http://mmb.irbbarcelona.org/PMut. A complete help and tutorial is available at http://mmb.irbbarcelona.org/PMut/help.
INTRODUCTION
Single nucleotide variants (SNVs) are responsible for ∼90% of human variability (1). When mapped on coding regions (single amino acid variants, SAVs) may affect the function of the transcribed proteins (2), leading to phenotype variations, and often to pathology. Last generation sequencing and genotyping techniques are reporting a large amount of human genetic variation data (3), fueling initiatives to derive links between genome alterations and pathologies. Thus, the HapMap consortium (4) is characterizing common variation and linkage disequilibrium patterns that can be related to common diseases (5). The Human Variation Project (6) collects, curates, and makes accessible information on genetic variations affecting human health. The 1000 Genomes Project (www.1000genomes.org) is expected to produce the most complete catalog of genetic variations in human population (7). Already in 2005, the Wellcome Trust Case Control Consortium genotyped ∼14 000 patients for seven common diseases performing one of the largest Genome-Wide Association Study (GWAS) (8) to date. As a result of these and other projects the dbSNP database at the NCBI (9) collects, nowadays, ∼20 million of validated human SNPs. The manually curated SwissVar database (10) reports on the pathological effect of ∼61 000 missense SNPs, the public version of the HGMD database (11) includes >78 000 missense mutations causing, or associated with human inherited diseases, plus disease-associated/functional polymorphisms and ClinVar reports over 125 000 clinically relevant variants (12). Systematic sequencing through NGS of cancer patients (projects like ICGC, www.icgc.org, and TGCA, cancergenome.nih.gov) expanded the range of mutations in the human genome. For example, the present public version of ICGC reports ∼520 000 new somatic SAVs.
Despite the amount of data available, the issue of predicting the functional consequences of SAVs is still open, and there is a continuous effort in developing more accurate and flexible predictors (13–15). There is no consensus on the type of approach used to obtained predictions. PMUT (16), one of the oldest and still widely used methods, uses neural networks, as so does for instance SNAP (17); SIFT (18), Polyphen (19), PROVEAN (20), LRT (21) and MutationAssessor (22; Hidden Markov Models are used in PANTHER (23) and FATHMM (24); Random Forests are used in PON-P2 (25), CHASM (26), CanPredict (27) and MuD (28); Support Vector Machines in CADD (29), SNPs&GO (30), SeqProfCod (31), LS-SNP (32), SNPs3D (33), MetaSVM (34) and MetaLR (34); Naïve Bayes is used in MutationTaster (35), and a gradient boosting tree is used in M-CAP (36). Also predictors giving consensus predictions like Condel (37), are available. All of them use input features that represent the change in amino acid sequence, structure and evolutionary properties resulting from the amino acid replacement.
PMut (16), first released in 2005, predicted the pathological nature of a given SAV, and also hot-spot positions on protein sequences. PMut used a neural network-based classifier trained by a manually curated dataset extracted from SwissProt (38), and used sequence conservation and predicted physico-chemical properties as main features. Here, we present a major update of the PMut predictor and web portal. While maintaining the philosophy of the original application, the backend classification engine has been completely renewed and automated, taking advantage of the increase in the amount of data available for training. The engine powering PMut is provided as a separate software package (PyMut) that allows users to prepare their own predictors for specific families of proteins. The new PMut web site also provides access to a complete data repository, including all possible SAVs on human known proteins. The web portal is available at http://mmb.irbbarcelona.org/PMut and has already received more than 900 requests between 1 January 2017 and 1 April 2017.
PMut PREDICTION ENGINE
PMut prediction engine (PyMut) is prepared as a Python 3 module. PyMut is based on the widely used libraries NumPy (www.numpy.org) and Scipy (www.scipy.org), for fast numerical computing, Pandas (data management, pandas.pydata.org), Scikit-learn (machine learning, scikit-learn.org), Matplotlib (matplotlib.org), and Seaborn (graphical representation, seaborn.pydata.org). PyMut performs all operations regarding calculation of features, selection of classifiers, validation and results analysis. It is distributed as a separate software module that can be downloaded and installed locally. Specific functions available are:
Compute protein features and plot their distribution (see Supplementary Tables S1 and S2 in the Supplementary Material for a complete list, and details of the procedures).
Select the most informative features. Selection of features is performed in an iterative way, following the improvement of MCC obtained in cross-validation. Supplementary Figure S1 shows a schema of the algorithm used, and Supplementary Figure S2 a plot of such MCC evolution.
Train classifiers, evaluate them using several cross-validation protocols, and obtain their Receiver Operating Characteristic (ROC) curves (see Supplementary Table S4, and Supplementary Figure S3 for a list of the available classifiers and its comparative performance).
Prepare and evaluate a pathology predictor.
Predict the pathology of mutations.
PyMut module covers all operations required to generate new predictors in a fully automated way (see Supplementary Table S5 for a detailed list of software functions, and Supplementary Table S6 for a list of software dependencies), allowing the user to easily explore alternative datasets, classifiers or collections of features, enabling to fine tune the predictor to cover not only pathology, but other structural or functional characteristics of the proteins. As the module can be downloaded and run locally, it allows the user to analyze private data to derive tailored predictors without uploading it to a server.
PyMut source code is available at https://github.com/inab/pymut and in the official Python package repository (https://pypi.python.org/pypi/pymut), and can be downloaded from the PMut Web portal, where a tutorial following the main functions of PyMut is also available.
PMut2017 PREDICTOR
PMut2017 default predictor was trained using the manually curated variation database SwissVar (10) (October 2016 release), which contains 27 203 disease and 38 078 neutral mutations on 12 141 proteins. Two hundred fifteen numerical features were first computed for each mutation, accounting for (i) physical property differences between wild type and mutated amino acids, (ii) protein interactome information and (iii) amino acid conservation. The conservation features are derived from local searches over UniRef100 and UniRef90 cluster databases (39), using PSI-Blast (40), and multiple sequence alignments generated using Kalign2 (41). After evaluation of the different machine learning algorithms (Supplementary Table S4 and Supplementary Figure S3), the chosen predictor is based on a Random Forest (42) classifier, trained with only 12 selected features (see Supplementary Table S3). The classifier outputs a prediction score between 0 and 1; mutations scoring from 0 to 0.5 are classified as neutral, and those scoring from 0.5 to 1 are classified as pathological. To evaluate the confidence degree of such score, we have analyzed the accuracy of the predictions based on their score (Supplementary Figure S4). It can be seen that accuracy increases with extreme score values. The analysis of these results allows us to qualify the prediction with a statistically meaningful reliability score.
PMut predictor has been validated following several approaches:
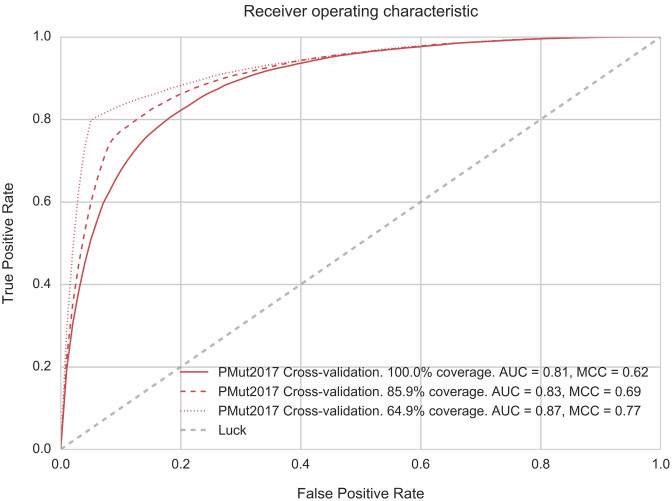
A traditional 10-fold cross-validation on protein families with 50% sequence identity exclusion. No sequence in the testing set shares >50% sequence identity with any protein in the training set. Figure 1 shows the corresponding ROC curves. Detailed performance metrics are summarized in Table 1. Restriction of the analysis to most confident predictions lead to a significant increase in the performance of the prediction. See Supplementary Figure S4 for a measured confidence of PMut scores.
A blind validation using new SwissVar entries. To perform this analysis, PMut has been trained using the same protocol, but limited to the data available at the SwissVar December 2015 release, and tested with SwissVar 2016 entries (3166 new mutations on 762 proteins. 1656 mutations were tagged as pathological and 1510 as neutral). For comparative purposes, we have also performed a complete series of analyses of the same test-set using other prediction methods. Table 2 shows the good performance of PMut when applied to the entire test set. The new engine used in PMut2017 represents a large improvement over the original PMut predictor (MCC 0.03 in the blind validation, data not shown). If predictions are limited to those cases where more reliable scores are obtained (PMut 85%), MCC value raises to 0.53, what puts PMut as the most accurate predictor within the test performed, covering yet >80% of the mutations. This can be further improved taking only >90% confidence scores (MCC 0.62), but in this case predictions can be obtained only in half of the cases. The availability of PyMut module allows for a seamless update of PMut predictor when new releases of SwissVar become available.
A blind validation using ClinVar entries not included in the training set. ClinVar (12) is an alternative well known source for disease related variants. ClinVar includes a much larger set of variants, although only 34 024 can be directly mapped to the protein sequences used in PMut. From those, 13 716 were already present in the SwissVar training set. To further analyze the performance of the PMut2017 predictor we have analyzed the variants reported in ClinVar that were not used in the training set (20 308 variants). Results are shown in Table 2. MCC value (0.49) is slightly better but comparable with those obtained in the SwissVar 2016 delta release. This behavior further confirms the prediction power of PMut2017.
Comparative test on selected genes. Global validation schemas provide an averaged estimation of the performance of a prediction method. Applying the predictor to specific protein families may result in a degraded performance due to the individual features of such families. We have selected a number of genes to evaluate PMut2017 performance in comparison with some other methods. To avoid a statistical bias, genes have been selected in a way that the number of neutral and pathological mutations were equilibrated and reasonably large. Results are reported in Table 3. As expected, MCC values obtained are specific to the analyzed gene and range widely around the average value obtained in the global test. Although some of them show a clearly poorer result, it can be seen that the behavior is consistent with the other methods assayed, and shows a good predicting power. These differences additionally support the need to develop specific predictors for protein families showing non-standard behavior.
Figure 1.
ROC Curves corresponding to PMut2017 10-fold cross-validation based on protein families. No sequence in the validation set has more than 50% identity with sequences in the training set. Additional curves correspond to the subsets of prediction with 85% and 90% confidence. AUC: area under the curve, MCC: Matthews Correlation Coefficient.
Table 1. Summary of performance metrics for PMut2017 predictor.
| Confidence | Coverage | Accuracy | Sensitivity | Specificity | AUC | MCC |
|---|---|---|---|---|---|---|
| All | 100 | 0.82 | 0.76 | 0.86 | 0.81 | 0.62 |
| >85% a | 85.9 | 0.85 | 0.75 | 0.92 | 0.83 | 0.69 |
| >90% a | 64.9 | 0.90 | 0.80 | 0.95 | 0.87 | 0.77 |
aOnly predictions with scores corresponding to higher confidence levels are considered (see Supplementary Figure S4).
Table 2. Comparative performance of PMut2017 predictor.
| Method | Coverage (%) | Accuracy | Specificity | Sensitivity | AUC | MCC |
|---|---|---|---|---|---|---|
| SIFT (18) | 89.6 | 0.61 | 0.33 | 0.88 | 0.60 | 0.25 |
| Polyphen2 (19) | 92.1 | 0.64 | 0.35 | 0.91 | 0.63 | 0.32 |
| PROVEAN (20) | 91.5 | 0.64 | 0.41 | 0.87 | 0.64 | 0.31 |
| FATHMM (24) | 90.5 | 0.55 | 0.45 | 0.64 | 0.55 | 0.09 |
| PON-P2 (25) | 42.4 | 0.72 | 0.52 | 0.9 | 0.71 | 0.45 |
| CADD (29) | 95.0 | 0.65 | 0.33 | 0.94 | 0.64 | 0.35 |
| M-CAP (36) | 91.5 | 0.60 | 0.19 | 0.95 | 0.57 | 0.22 |
| Condel (37) | 91.0 | 0.63 | 0.40 | 0.84 | 0.62 | 0.26 |
| LRT (21)a | 95.1 | 0.73 | 0.58 | 0.87 | 0.73 | 0.47 |
| MutationAssessor (22) a | 95.1 | 0.63 | 0.46 | 0.78 | 0.62 | 0.26 |
| MetaSVM (34) a | 95.1 | 0.63 | 0.51 | 0.74 | 0.62 | 0.26 |
| MetaLR (34) a | 95.1 | 0.6 | 0.46 | 0.73 | 0.60 | 0.20 |
| MutationTaster (35) a | 95.1 | 0.65 | 0.31 | 0.96 | 0.64 | 0.36 |
| PMut | 100.0 | 0.71 | 0.65 | 0.76 | 0.71 | 0.42 |
| PMut (85%)b | 81.0 | 0.76 | 0.76 | 0.77 | 0.76 | 0.53 |
| PMut (90%)b | 51.2 | 0.81 | 0.78 | 0.84 | 0.81 | 0.62 |
| PMut (ClinVar)c | 100.0 | 0.73 | 0.88 | 0.85 | 0.75 | 0.49 |
Blind validation based on new variants added to SwissVar during 2016 (3166 variants), CADD predictor has been evaluated using a threshold of 20. AUC: area under the ROC curve, MCC: Matthews correlation coefficient.
aAnalysis performed from ANNOVAR data (42).
bAnalysis restricted to most reliable PMut predictions (reliability level in parentheses).
cBlind validation based on variants reported on ClinVar (43), not present in the SwissVar dataset (20,308 variants). Indicated coverage is calculated on ClinVar dataset.
Table 3. Comparative performance of the PMut2017 predictor on selected genes.
| Gene | Disease | #D | #N | PMut | SIFT | Polyphen | LRT | Mut. Taster | Mut. Assessor | PROVEAN |
|---|---|---|---|---|---|---|---|---|---|---|
| MECP2 | Rett syndrome | 46 | 22 | 0.86 | 0.66 | 0.85 | 0.69 | 0.64 | 0.41 | 0.53 |
| COL1A2 | Osteogenesis Imperfecta | 78 | 20 | 0.77 | 0.74 | 0.62 | 0.55 | 0.55 | 0.74 | 0.74 |
| SLC4A1 | Distal Renal Tubular Acidosis | 38 | 36 | 0.69 | 0.65 | 0.65 | 0.54 | 0.55 | 0.68 | 0.60 |
| ADAMTS13 | Upshaw-Schulman syndrome | 43 | 17 | 0.62 | 0.76 | 0.46 | 0.00 | 0.71 | 0.54 | 0.62 |
| ATM | Hereditary cancer-predisposing syndrome | 46 | 54 | 0.60 | 0.53 | 0.57 | 0.32 | 0.42 | 0.48 | 0.55 |
| ATP7B | Wilson disease | 195 | 25 | 0.48 | 0.34 | 0.49 | 0.37 | 0.43 | 0.29 | 0.52 |
| MLH1+MSH2+MSH6+PMS2 | Lynch syndrome | 159 | 78 | 0.48 | 0.32 | 0.31 | 0.23 | 0.16 | 0.43 | 0.32 |
| MYOC | Primary open angle glaucoma | 57 | 24 | 0.47 | 0.37 | 0.45 | 0.38 | 0.50 | 0.47 | 0.49 |
| TTC21B | Jeune thoracic dystrophy | 16 | 28 | 0.42 | 0.20 | 0.22 | 0.18 | 0.16 | 0.28 | 0.26 |
| SCN5A | Brugada syndrome | 154 | 46 | 0.40 | 0.32 | 0.26 | 0.43 | 0.31 | 0.34 | 0.34 |
| KCNH2+SCN5A | Congenital long QT syndrome | 270 | 54 | 0.38 | 0.32 | 0.28 | 0.36 | 0.32 | 0.30 | 0.38 |
| ABCA1 | Tangier disease | 32 | 31 | 0.37 | 0.43 | 0.31 | 0.32 | 0.47 | 0.43 | 0.47 |
| PKHD1+PKD1 | Polycystic kidney disease | 197 | 96 | 0.37 | 0.43 | 0.37 | 0.30 | 0.41 | 0.36 | 0.45 |
| FBN1 | Marfan syndrome | 385 | 20 | 0.35 | 0.31 | 0.25 | 0.21 | 0.33 | 0.32 | 0.30 |
| RYR1 | Central core disease | 147 | 25 | 0.34 | 0.27 | 0.31 | 0.00 | 0.36 | 0.28 | 0.34 |
| LDLR | Familial hypercholesterolemia | 103 | 23 | 0.32 | 0.29 | 0.08 | 0.17 | 0.09 | 0.26 | 0.25 |
| DYSF | Limb-Girdle Muscular Dystrophy | 48 | 16 | 0.31 | 0.35 | 0.27 | 0.15 | 0.21 | 0.41 | 0.39 |
| BRCA2 | Breast-ovarian cancer, familial 2 | 43 | 61 | 0.31 | 0.10 | 0.18 | 0.18 | 0.14 | 0.19 | 0.01 |
| BRCA1 | Breast-ovarian cancer, familial 1 | 27 | 36 | 0.31 | 0.24 | 0.20 | 0.38 | 0.29 | 0.30 | 0.17 |
| WFS1 | WFS1-Related Spectrum Disorders | 40 | 17 | 0.30 | 0.25 | 0.35 | 0.20 | 0.18 | 0.16 | 0.26 |
| PINK1 | Parkinson Disease | 23 | 39 | 0.25 | 0.33 | 0.48 | 0.40 | 0.41 | 0.44 | 0.30 |
| LRRK2 | Parkinson Disease | 21 | 24 | 0.19 | 0.06 | 0.14 | 0.01 | 0.13 | 0.09 | 0.14 |
| CFTR | Cystic fibrosis | 146 | 32 | 0.15 | 0.06 | 0.20 | 0.21 | 0.12 | 0.20 | 0.27 |
| PROC | Thrombophilia | 36 | 28 | 0.12 | -0.15 | -0.08 | 0.07 | 0.14 | 0.08 | -0.01 |
MCC values obtained restraining the analysis to variants on the indicated genes. Analysis for non-PMut methods performed from ANNOVAR data (42). #N Neutral mutations, #D Disease causing mutations.
PMut WEB PORTAL
The access to PMut is possible through a Web portal (http://mmb.irbbarcelona.org/PMut/) which is implemented in Python using the Django Web framework (www.djangoproject.com). The variants’ features and predictions are stored in a MongoDB database (www.mongodb.com). All calculations are performed under the control of a SGE queuing system configured to deploy additional backend workers on peaks of demand. Id mapping and keyword searches are performed using the appropriate services at EBI (www.ebi.ac.uk). Sequences, features, variants and 3D structures are obtained from MMB-IRB data repository (mmb.irbbarcelona.org/api). Most functionalities of the portal are available anonymously, but the users may register to keep records of the activity in the server. After registering, a private workspace is created with links to the prediction requests, and to their customly trained predictors.
The portal is divided in four sections:
Data repository, which allows the user to access the set of pre-calculated PMut2017 predictions covering all human protein sequences in UniRef100. Search options available include protein name and id (UniprotKB (44) or PDB (45)), gene id (Ensembl (46)), dbSNP id (9) and free keywords. Data in the repository can be accessed programmatically through a REST API. The repository is continuously updated with new information appearing in UniRef100 (39). Detailed statistics of the repository can be found in Table 4.
Pathology prediction, which allows the user to evaluate the pathological profile of SAVs, input options include protein id(s), or uploaded sequences. PMut Data Repository is used to speed up analysis in the case of known protein sequences. The default predictor is PMut2017, but Custom Predictors can also be used. In the case of mutations mapping on known sequences all possible single SAVs are precomputed. The output includes a variety of graphical and numerical results which are presented in different formats. In all cases, the output combines the information with known variants and sequence features of the protein, giving a comprehensive context of the mutations. When 3D structure is available, the mutations are also mapped onto the structure, using a JsMol visualizer (www.jmol.com). Several public databases (UniprotKB (44), PDB (45), PFam (47), InterPro (48) and Interactome3D (49)) are linked to the results card. All intermediate data including alignments and calculated features, are available for download in the appropriate formats. See representative screenshots at Help pages on PMut portal, http://mmb.irbbarcelona.org/PMut/help).
Batch predictions, users requesting larger series of predictions can send them in a single batch. The options available, and output are equivalent to single requests. The user is informed when the work is finished and results are stored in his/her private workspace.
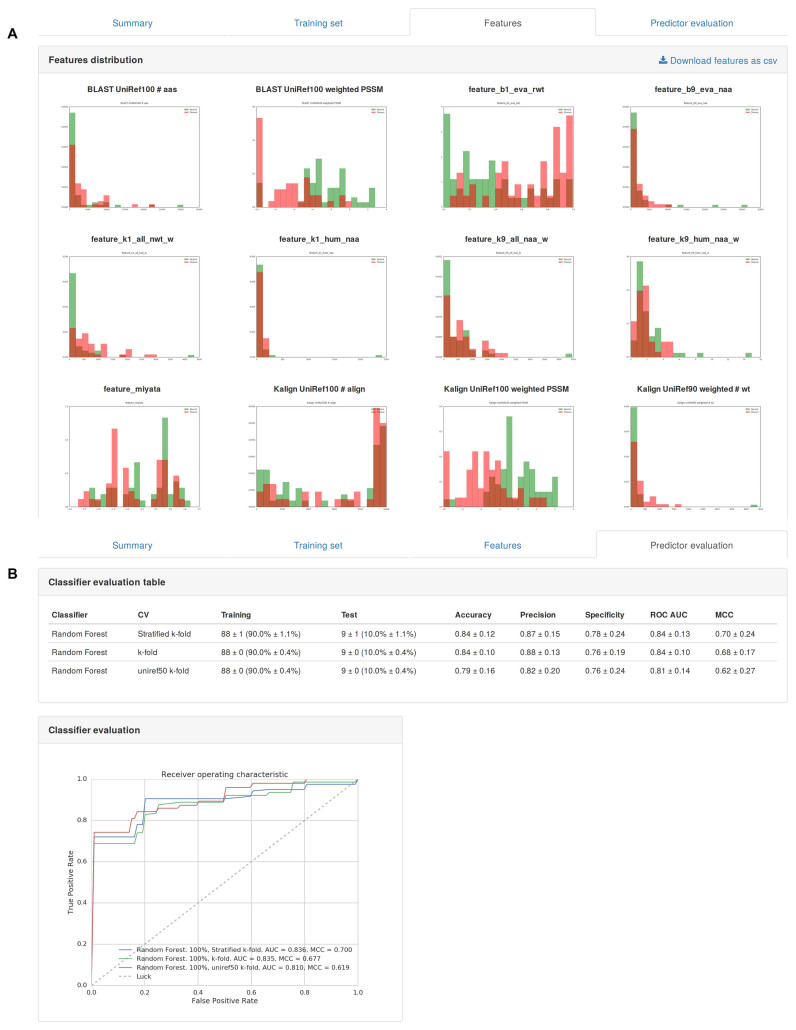
Train you own predictor, this section provides a frontend to the PyMut engine. Users can specify a training set, select a classifier and a validation procedure. Figure 2 shows some screenshots of the output. Available information includes the original training set, a graphical view of the calculated features (Figure 2A), and a summary of the evaluation results (Figure 2B). The newly trained predictor becomes automatically available in the prediction section of the portal. Please note that the use of trained predictors requires to log in the personal workspace and is restricted to the user developing it.
Table 4. Statistics of PMut repository (January 2017).
| Proteins available (from human UniRef) | 106 407 |
|---|---|
| Analysed variants | 725 596 928 |
| Analysed variants (>85% prediction reliability) | 586 383 428 (80%) |
| Analysed variants (>90% prediction reliability) | 370 444 279 (51%) |
Figure 2.
Partial screenshots of output of Predictor's training section. (A) Comparative plot of the selected protein features. (B) ROCs curves of performance evaluation.
CONCLUSIONS
The 2017 new release of the PMut portal constitutes a novel approach that largely improves our previous 2005 PMut server. The new portal offers not only a generally trained predictor that performs in a competitive manner with current available methods, but allows the user to access an automatic procedure to train new predictors with specific datasets or features. The possibility of enriching the analysis with alternative predictors, or training predictors with specific information of a single protein family, largely increases the scope of usability of the portal. Overall, the 2017 release of PMut is a powerful tool to approach the issue of predicting functional consequences of protein sequence variants, and will surely contribute to improve the quality of the annotation of pathological variants. The server and platform are already available and accessible without restrictions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Patrick Aloy for providing protein interactome data. We thank Jose A. Alcantara, and Dr. Adam Hospital, for technical assistance.
Footnotes
Present address: Andrea Gazzo, Interuniversity Institute for Bioinformatics, ULB-VUB, Brussels, 1050 Brussels, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science [BIO2015-64802-R-32868, SEV-2015-0493, TIN2012-34557, TEC2015-67774-C2-2-R, SAF2016-80255-R]; Catalan Government [2014-SGR-134, 2014-SGR-1051]; Instituto de Salud Carlos III-Instituto Nacional de Bioinformática [INB; PT13/0001/0019, PT13/0001/0028]; European Union, H2020 programme [Elixir-Excelerate: 676559; BioExcel: 674728 and MuG: 676566]; La Caixa Foundation fellowship [to V.L.F.]. Funding for open access charge: European Union and Spanish Ministry of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1. Collins F.S., Brooks L.D., Chakravarti A.. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998; 8:1229–1231. [DOI] [PubMed] [Google Scholar]
- 2. Cargill M., Altshuler D., Ireland J., Sklar P., Ardlie K., Patil N., Shaw N., Lane C.R., Lim E.P., Kalyanaraman N. et al. . Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999; 22:231–238. [DOI] [PubMed] [Google Scholar]
- 3. Fernald G.H., Capriotti E., Daneshjou R., Karczewski K.J., Altman R.B.. Bioinformatics challenges for personalized medicine. Bioinformatics (Oxford, England). 2011; 27:1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M. et al. . A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007; 449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D.G., Fan J.B., Siao C.J., Berno A., Young P., Sapolsky R., Ghandour G., Perkins N., Winchester E., Spencer J. et al. . Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science (New York, N.Y.). 1998; 280:1077–1082. [DOI] [PubMed] [Google Scholar]
- 6. Cotton R.G., Auerbach A.D., Axton M., Barash C.I., Berkovic S.F., Brookes A.J., Burn J., Cutting G., den Dunnen J.T., Flicek P. et al. . GENETICS. The human variome project. Science (New York, N.Y.). 2008; 322:861–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A.. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007; 447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yip Y.L., Famiglietti M., Gos A., Duek P.D., David F.P., Gateau A., Bairoch A.. Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase. Hum. Mut. 2008; 29:361–366. [DOI] [PubMed] [Google Scholar]
- 11. Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N.. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014; 133:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al. . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karchin R. Next generation tools for the annotation of human SNPs. Brief. Bioinform. 2009; 10:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief. Bioinform. 2005; 6:44–56. [DOI] [PubMed] [Google Scholar]
- 15. Tavtigian S.V., Greenblatt M.S., Lesueur F., Byrnes G.B.. In silico analysis of missense substitutions using sequence-alignment based methods. Hum. Mut. 2008; 29:1327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrer-Costa C., Gelpi J.L., Zamakola L., Parraga I., de la Cruz X., Orozco M.. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics (Oxford, England). 2005; 21:3176–3178. [DOI] [PubMed] [Google Scholar]
- 17. Bromberg Y., Yachdav G., Rost B.. SNAP predicts effect of mutations on protein function. Bioinformatics (Oxford, England). 2008; 24:2397–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ng P.C., Henikoff S.. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31:3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R.. A method and server for predicting damaging missense mutations. Nat. Methods. 2010; 7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi Y., Chan A.P.. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics (Oxford, England). 2015; 31:2745–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chun S., Fay J.C.. Identification of deleterious mutations within three human genomes. Genome Res. 2009; 19:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reva B., Antipin Y., Sander C.. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011; 39:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas P.D., Kejariwal A.. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15398–15403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shihab H.A., Gough J., Cooper D.N., Stenson P.D., Barker G.L., Edwards K.J., Day I.N., Gaunt T.R.. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mut. 2013; 34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niroula A., Urolagin S., Vihinen M.. PON-P2: prediction method for fast and reliable identification of harmful variants. PLoS One. 2015; 10:e0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carter H., Chen S., Isik L., Tyekucheva S., Velculescu V.E., Kinzler K.W., Vogelstein B., Karchin R.. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009; 69:6660–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaminker J.S., Zhang Y., Watanabe C., Zhang Z.. CanPredict: a computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 2007; 35:W595–W598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wainreb G., Ashkenazy H., Bromberg Y., Starovolsky-Shitrit A., Haliloglu T., Ruppin E., Avraham K.B., Rost B., Ben-Tal N.. MuD: an interactive web server for the prediction of non-neutral substitutions using protein structural data. Nucleic Acids Res. 2010; 38:W523–W528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calabrese R., Capriotti E., Fariselli P., Martelli P.L., Casadio R.. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mut. 2009; 30:1237–1244. [DOI] [PubMed] [Google Scholar]
- 31. Capriotti E., Arbiza L., Casadio R., Dopazo J., Dopazo H., Marti-Renom M.A.. Use of estimated evolutionary strength at the codon level improves the prediction of disease-related protein mutations in humans. Hum. Mut. 2008; 29:198–204. [DOI] [PubMed] [Google Scholar]
- 32. Karchin R., Diekhans M., Kelly L., Thomas D.J., Pieper U., Eswar N., Haussler D., Sali A.. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics (Oxford, England). 2005; 21:2814–2820. [DOI] [PubMed] [Google Scholar]
- 33. Yue P., Melamud E., Moult J.. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006; 7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X.. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015; 24:2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwarz J.M., Rodelsperger C., Schuelke M., Seelow D.. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010; 7:575–576. [DOI] [PubMed] [Google Scholar]
- 36. Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G.. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016; 48:1581–1586. [DOI] [PubMed] [Google Scholar]
- 37. Gonzalez-Perez A., Lopez-Bigas N.. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011; 88:440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O’Donovan C., Phan I. et al. . The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003; 31:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H.. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics (Oxford, England). 2015; 31:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lassmann T., Sonnhammer E.L.. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005; 6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Breiman L. Random Forests. Mach. Learn. 2001; 45:5–32. [Google Scholar]
- 43. Wang K., Li M., Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uniprot UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berman H., Henrick K., Nakamura H., Markley J.L.. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007; 35:D301–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aken B.L., Achuthan P., Akanni W., Amode M.R., Bernsdorff F., Bhai J., Billis K., Carvalho-Silva D., Cummins C., Clapham P. et al. . Ensembl 2017. Nucleic Acids Res. 2017; 45:D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. . The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S. et al. . The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015; 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mosca R., Ceol A., Aloy P.. Interactome3D: adding structural details to protein networks. Nat. Methods. 2013; 10:47–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.