Abstract
The signaling networks that control the immune system are coordinated by a myriad of interconnecting phosphorylation and ubiquitylation events. This review provides an overview of mutations in human genes encoding these proteins that give rise to immune diseases. Analysis of the biological effects of these mutations has revealed the true physiological roles of particular signaling networks and promises to revolutionize the treatment of these diseases.
Nearly all aspects of a cell’s life are controlled by the phosphorylation and ubiquitylation of proteins, and a substantial proportion of human genes encode the enzymes that catalyze these modifications. Over 500 protein kinases and 140 protein phosphatases mediate reversible protein phosphorylation in human cells, and many other proteins interact with them to modulate their activity, substrate specificity and subcellular location. Similarly, some 40 E2 ubiquitin-conjugating enzymes, over 600 E3 ubiquitin ligases and about 90 deubiquitylases (DUBs) mediate the reversible ubiquitylation of proteins. Proteins can undergo monoubiquitylation or any of eight types of polyubiquitylation, in which the C-terminal glycine residue of one ubiquitin forms an isopeptide bond with an ε-amino moiety of one of the seven lysine residues in another ubiquitin molecule or a peptide bond with the α-amino group of the N-terminal methionine of another ubiquitin molecule (termed ‘Met1-linked’ or ‘linear ubiquitin’). The ubiquitin code is decoded by ubiquitin-binding proteins, which interact with the different types of ubiquitin chains. Over 200 such proteins have been identified so far1, and the list is still growing.
Protein phosphorylation and ubiquitylation are frequently interlinked processes. For example, some E3 ubiquitin ligases require phosphorylation to become catalytically active2 or can only ubiquitylate their substrates when the latter are phosphorylated3. Conversely, some protein kinases form complexes with ubiquitin-binding proteins and the activation of these kinases is triggered when ubiquitin interacts with these proteins4. The interplay between phosphorylation and ubiquitylation events is a particular feature of innate immune signaling networks.
About 10% of all human genes encode proteins that control the reversible phosphorylation or ubiquitylation of proteins, so it is not surprising that mutations or polymorphisms in protein kinases and components of the ubiquitin system should underlie or be associated with many human diseases (Tables 1 and 2). Moreover, mutations in other proteins, such as ligands, receptors and adaptors, affect protein phosphorylation and ubiquitylation indirectly by suppressing or enhancing the activation of particular signaling networks. Here I focus mainly on rare mutations in kinases and components of the ubiquitin system that cause immune diseases, but also mention some polymorphisms in these proteins that appear to predispose to immune diseases. In recent years, protein kinases have become one of the most important classes of drug target5, and the ubiquitin system is also likely to furnish drug targets in the future6. A mechanistic understanding of the diseases and conditions associated with mutations in some of these proteins has already led to the improved treatment of some immune diseases and is likely to revolutionize the treatment of others in the years to come.
Table 1. Human diseases caused by mutations in ubiquitin-binding proteins (UBPs), E3 ligases and kinases discussed in this review.
| Protein | Type | Human diseases caused by mutation | Refs. |
|---|---|---|---|
| NEMO | UBP | Incontinentia pigmenti, anhidrotic ectodermal dysplasia with immuno-insufficiency, X-linked recessive Mendelian susceptibility to mycobacterial disease | 10,11 |
| Optineurin (OPTN) | UBP | Paget’s disease of bone, ALS, normal-tension glaucoma | 38,40,42 |
| Sequestosome 1 (SQSTM1) | UBP | Paget’s disease of bone, ALS | 39,41 |
| LUBAC | E3 ligase | Upon mutation of the HOIL-1 component, susceptibility to infection by pyogenic bacteria and autoinflammation | 18 |
| XIAP | E3 ligase | X-linked lymphoproliferative syndrome type 2 (XLP2) | 22–24 |
| ACT1 | E3 ligase | Chronic mucocutaneous candidiasis | 29 |
| IRAK4 | Protein kinase | Childhood susceptibility to infection by pyogenic bacteria, causing meningitis, sepsis, abcesses and osteomyelitis | 16,81,82 |
| TBK1 | Protein kinase | HSV1 encephalitis caused by loss of function. Normal-tension glaucoma caused by gene duplication | 43,106 |
| JAK2 | Protein kinase | Primary polycythemia, thrombocytopenia and myelofibrosis | 83–85 |
| JAK3 | Protein kinase | Severe combined immunodeficiency syndrome | 86,87 |
| TYK2 | Protein kinase | Increased susceptibility to viral and mycobacterial infections; P104A variant associated with rheumatoid arthritis | 89,90,92 |
| BTK | Protein kinase | X-linked agammaglobulinemia, mantle cell lymphoma and chronic lymphocytic leukemia | 93–96 |
| ZAP70 | Protein kinase | Recurrent infections due to loss of T cell function | 98–101 |
| PI(3)Kδ | Lipid kinase | Respiratory infection, airway damage and susceptibility to EBV and cytomegalovirus infection, owing to impaired development of functional memory T cells and B cells | 108,109 |
Table 2. Polymorphisms in ubiquitin-binding proteins (UBPs), E3 ligases, DUBs and protein kinases that predispose to human disease.
| Protein | Type | Polymorphisms that predispose to immune diseases in humans or mice | Refs. |
|---|---|---|---|
| ABIN1 | UBP | Polymorphisms predispose to autoimmune diseases (human). Failure to bind ubiquitin chains causes lupus nephritis (mice) | 45–50 |
| A20 | UBP and DUB | Polymorphisms predispose to autoimmune diseases (human) | 127,128 |
| TOLLIP | UBP | Polymorphisms associated with increased risk of tuberculosis (human) | 68 |
| PARKIN | E3 ligase | Polymorphisms cause susceptibility to leprosy (human) | 69 |
| ALPK1 | Protein kinase | Polymorphisms cause susceptibility to IBD and gout (human). Candidate gene for susceptibility to colitis and colon cancer (mice) | 110–113 |
| IRAK1 | Protein kinase | Polymorphisms associated with SLE (human) | 121,122 |
| SIK2 | Protein kinase | Polymorphisms associated with increased risk of primary sclerosing cholangitis (human) | 126 |
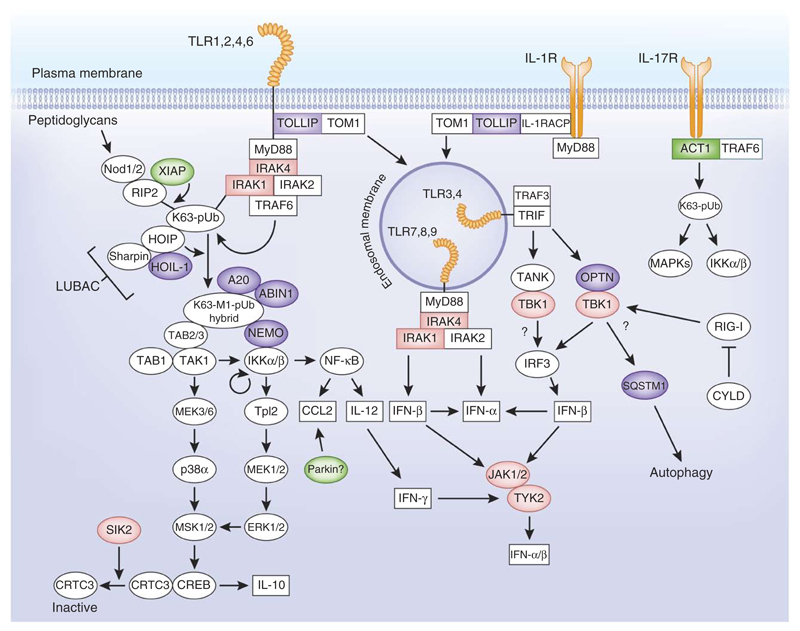
Many of the proteins discussed in this article are components of innate immune signaling networks (Fig. 1). For example, in the signaling networks that are controlled by myeloid differentiation response gene 88 (MyD88) (ref. 7) and nucleotide-binding oligomerization domain (Nod) proteins8, the activation of the canonical IκB kinase (IKK) complex is facilitated by the formation of hybrid ubiquitin molecules. These hybrid chains are formed when the linear ubiquitin assembly complex (LUBAC) uses preformed Lys63-linked ubiquitin oligomers as a substrate. The Lys63-linked chains are themselves formed by the actions of other E3 ubiquitin ligases, such as TRAF6 and XIAP (Fig. 1). The Lys63-Met1 hybrid ubiquitin chains become attached covalently to other proteins discussed in this review, such as the protein kinases IRAK1 and RIP2, while other proteins, such as ABIN1 and A20, bind non-covalently to these ubiquitin chains to prevent the hyperactivation of the network. Other E3 ligases, such as Parkin, have been proposed to control more ‘downstream’ signaling events, while ACT1 is an E3 ligase with an essential role in the interleukin (IL)-17 signaling network. TOLLIP is a ubiquitin-binding protein that recognizes ubiquitylated receptors and may target them to early endosomes for degradation. The protein kinase TBK1 forms heterodimers with several proteins, including the ubiquitin-binding protein optineurin (OPTN), and has several roles in the innate immune system9, which include the control of autophagy and the production of interferon (IFN)-β by a pathway dependent on the adaptor protein TRIF that is triggered by agonists of Toll-like receptors (TLRs) 3 or 4. During autophagy one substrate of TBK1 is the ubiquitin-binding protein sequestosome 1 (SQSTM1, also called p62). Mutations in either SQSTM1 or OPTN cause Paget’s disease of bone, probably because of their involvement in osteoclast proliferation, which is triggered by the tumor necrosis family (TNF) member RANK. This particular example serves to highlight the fact that many of the proteins discussed in this review participate not just in the immune signaling networks illustrated in this review but also in others, explaining the unexpected diseases that can sometimes arise from their mutation.
Figure 1.
Components of the ubiquitin system and protein kinases whose mutation causes or may predispose to human immune diseases. Highlighted are the polyubiquitin-binding proteins (purple), the E3 ubiquitin ligases (green) and the protein kinases (red) discussed in this review. Receptors are shown in orange. The figure indicates where these components are thought to be positioned in the signaling networks initiated by the activation of TLRs and the IL-1 and IL-17 receptors. The figure represents a generic myeloid cell. Owing to space limitations, not all the events described in the text are depicted. pUb, polyubiquitin; TAB1, TAK1-binding protein; MSK1/2, mitogen- and stress-activated kinases 1 and 2; ERK1/2, extracelullar signal–regulated kinases 1 and 2; p38α, p38α MAP kinase; MEK1/2, ERK kinases 1 and 2.
Mutations in the ubiquitin system: NEMO and LUBAC
The canonical IKK complex has many crucial roles in the immune system, including the activation of NF-κB and the protein kinase Tpl2 (ref. 9). The IKK complex, which is composed of NEMO (NF-κB essential modifier) and the catalytic subunits IKKα and IKKβ, is activated by many stimuli, including IL-1, TNF and agonists that activate TLRs. The IKBKG gene, encoding NEMO, is located on the X chromosome in humans, and its mutation or truncation causes several diseases, including incontinentia pigmenti10, anhidrotic ectodermal dysplasia with immunodeficiency, and X-linked recessive Mendelian susceptibility to mycobacterial disease11. These mutations cause recurrent infections with invasive pyogenic bacteria causing meningitis, sepsis, osteomyelitis and abscesses. Some years after these discoveries were made, the mutation of Asp311 to asparagine or glycine, which causes anhidrotic ectodermal dysplasia with immunodeficiency, and the mutation of Glu315 to alanine, which causes X-linked recessive Mendelian susceptibility to mycobacterial disease, were shown to be located in the UBAN domain (the ubiquitin binding domain found in ABINs and NEMO) and to prevent the binding of Lys63-linked12,13 ubiquitin dimers to NEMO. NEMO interacts with Met1-linked ubiquitin chains 100-fold more strongly than with Lys63-linked ubiquitin chains14,15, and the interaction of Met1-linked ubiquitin oligomers with NEMO permits phosphorylation of the canonical IKK complex by TAK1, priming it for autoactivation (J. Zhang and P. Cohen, unpublished data). Thus, the D311N and E315A mutations impair activation of the canonical IKK complex in human cells and hence the activation of NF-κB. This prevents, for example, CD40 (co-stimulatory factor found on antigen-presenting cells)-dependent IL-12 production by B cells and dendritic cells, resulting in defective interferon-γ production by T cells and an inability to clear infections11. More detailed descriptions of the pathological consequences of NEMO mutations may be found elsewhere16.
Human mutations causing a complete loss of the HOIL-1 component of LUBAC destabilize the complex, which leads to greatly reduced expression of HOIP, the catalytic subunit of LUBAC. This suggests that the LUBAC-catalyzed formation of Met1-linked ubiquitin chains17 is impaired in these patients, explaining why the TNF- or IL-1–stimulated activation of IKK and NF-κB activation is reduced in the fibroblasts and B cells of these patients18. These findings can also explain why, like those with mutations in NEMO, patients with HOIL-1 mutations are susceptible to infection by pyogenic bacteria. Notably, the patients with HOIL-1 deficiency also display an autoinflammatory phenotype, and, in accord with this, their mononuclear leukocytes and monocytes display increased levels of pro-inflammatory cytokine mRNAs (ref. 18). This inflammatory phenotype is similar to that seen in mice that do not express Sharpin19, another component of LUBAC20,21. Why the loss of the HOIL-1 and Sharpin components of LUBAC leads to hypersensitivity to IL-1 has yet to be clarified.
Mutations in the ubiquitin system: the E3 ligase XIAP
Mutations in XIAP, the human gene encoding X-linked inhibitor of apoptosis (XIAP), cause X-linked lymphoproliferative syndrome type 2 (XLP2), an immunodeficiency disease associated with susceptibility to infection by Epstein-Barr virus (EBV), chronic colitis, hepatitis or persistent splenomegaly22. XIAP, together with other members of the IAP family of E3 ligases (cIAP1, cIAP2), is essential in the signaling network triggered by the interaction of Nod1 and Nod2 with fragments of bacterial peptidoglycans. Mutations in the RING domain of XIAP2 that carries the E3 ligase function23 impair Nod1- and Nod2-dependent immune signaling, whereas missense mutations in the BIR2 domain abolish interaction with receptor-interaction protein kinase 2 (RIPK2), resulting in impaired RIPK2 ubiquitylation and failure to recruit LUBAC to the Nod2 complex in primary cells from XLP2 patients (Fig. 1)24. NOD2 was the first gene shown to be associated with Crohn’s disease, and mutations that impair the activation of the Nod2-RIPK2 signaling network confer a considerable risk of developing Crohn’s disease25. Thus, mutations in XIAP could underlie the chronic ulcerative colitis seen patients with XLP2.
Mutations in the ubiquitin system: the E3 ligase ACT1
IL-17 family members are produced by CD4+ T helper cells and are important in coordinating local tissue inflammation by enhancing the production of many pro-inflammatory cytokines, chemokines and matrix metalloproteinases. The interaction of IL-17 with its receptor leads to the activation of the canonical IKK complex and MAP kinases, and hence to the production of inflammatory mediators via a signaling network in which the E3 ubiquitin ligases, TNF receptor–associated factor 6 (TRAF6)26 and NF-κB–activating protein (ACT1, also called TRAF3IP2 and CIKS) are essential27. However, in contrast to the IL-1 and TLR signaling networks, where TRAF6 is also an essential protein, it is ACT1, and not TRAF6, that is thought to generate the Lys63-linked ubiquitin chains needed to activate the pathway, with TRAF6 functioning as a coupling factor in this process28.
ACT1 is recruited to IL-17 receptors (IL-17Rs) in response to IL-17, its interaction with IL-17Rs requiring the SEFIR (similar expression to fibroblast growth factor genes and IL-17R) domain of ACT1. The mutation of Thr536 in the SEFIR domain of ACT1 to alanine disrupts the interaction between ACT1 and IL-17 receptors, and causes chronic mucocutaneous candidiasis in humans, a disease characterized by persistent infections of the skin, nails, and oral and genital mucosae with the fungus Candida albicans29. A similar phenotype is seen in patients with deficiencies in IL-17 or IL-17 receptors. Conversely, IL-17 contributes to the development and pathogenesis of human inflammatory conditions, such as rheumatoid arthritis, lung infections, psoriasis and multiple sclerosis30–32. Thus, components of the IL-17 signaling network merit attention as potential targets for the development of improved drugs to treat these diseases.
Mutations in the ubiquitin-binding proteins OPTN and SQSTM1
OPTN is the protein that most closely resembles NEMO in structure. This includes conservation of the ubiquitin-binding domain, and, like NEMO, OPTN interacts with both Met1-linked and Lys63-linked ubiquitin chains33,34. Interestingly, OPTN interacts with the IKK-related kinase TBK1 (TANK-binding kinase 1)35, and nearly all the OPTN in macrophages is present as a complex with TBK1 (ref. 33), indicating that many of the physiological functions of OPTN, such as its involvement in the lipopolysaccharide (LPS)-stimulated production of IFN-β33,36 might be mediated via TBK1 (Fig. 1).
TBK1 phosphorylates OPTN at Ser177 in vitro33 and in cells34, and phosphorylation of OPTN is reported to strengthen its interaction with the ATG8 family of proteins, which are essential for autophagy and facilitate the autophagic clearance of the bacterium Salmonella enterica34. Other studies also identified TBK1 as a key regulator of immunological autophagy that promotes the maturation of autophagosomes into the bactericidal organelles required for the elimination of mycobacteria by macrophages37. TBK1 appears to phosphorylate the autophagic adaptor protein SQSTM1 at Ser403, which is located in the ubiquitin-binding UBA domain and is essential for autophagic clearance34. It would therefore be interesting to investigate whether it is the OPTN-TBK1 complex, rather than other forms of TBK1, that phosphorylate SQSTM1 at Ser403 in vivo (Fig. 1).
Notably, human mutations in OPTN and SQSTM1 both cause Paget’s disease of bone38,39, as well as amyotrophic lateral sclerosis (ALS), the most common form of motor neuron disease40,41. Both diseases have been linked to defective autophagy, leading to a failure to degrade protein aggregates. Furthermore, the mutations in SQSTM1 that cause Paget’s disease are predominantly located in the UBA domain, although only some of these mutations affect the ability of SQSTM1 to interact with ubiquitin oligomers. The mutation of Glu478 in OPTN to glycine, which causes ALS40, prevents OPTN from binding to ubiquitin oligomers33.
In contrast to the above mutations, the mutation of Glu50 of OPTN to lysine causes a normal-tension form of glaucoma, in which the optic nerve heads are thought to become hypersensitive to inflammation-induced damage, leading to the destruction of the optic nerves and blindness42. The Glu50-to-lysine mutation enhances the interaction of OPTN with TBK1 in overexpression experiments35, suggesting that it may be a gain-of-function mutation that enhances the activity of TBK1 in the OPTN-TBK1 complex. Consistent with this hypothesis, duplication of the gene encoding TBK1 has been identified in other patients with the same type of glaucoma43.
Ubiquitin system polymorphisms and disease predisposition
A20-binding inhibitor of NF-κB 1 (ABIN1) contains a ubiquitin-binding domain similar to that found in NEMO and interacts with both Met1-linked and Lys63-linked ubiquitin chains44. Knock-in mice in which wild-type ABIN1 is replaced by the polyubiquitin-binding-defective D485N mutant progressively develop the classical symptoms of systemic lupus erythematosus (SLE)44. The myeloid and B cells of these mice show hyperactivation of the MyD88 signaling network and overproduce pro-inflammatory cytokines. Importantly, autoimmunity is prevented by crossing the ABIN1 mutant mice to MyD88-deficient mice, establishing that hyper-activation of this pathway causes the disease. How the binding of ubiquitin oligomers to ABIN1 restricts activation of the MyD88 signaling network is not yet understood, but one possibility is that ABIN1 competes with NEMO for binding to Met1-linked ubiquitin chains and that this competition is lost in cells carrying the D485N mutant of ABIN1 (Fig. 1).
Notably, polymorphisms in the TNIP1 gene (which encodes the ABIN1 protein) predispose humans to a variety of autoimmune disorders, including SLE, psoriasis, psoriatic arthritis, myasthenia gravis and vasculitis in at least five different human populations45–49. Certain human polymorphisms predispose to types III and IV SLE (lupus nephritis), and mice with the D485N knock-in mutation of ABIN1 develop this type of SLE50.
The ABIN1-binding protein A20 also interacts with Lys63-linked and Met1-linked ubiquitin chains via NZF domains near its C terminus51–53, and the tissue-specific deletion of A20 causes a variety of inflammatory and autoimmune diseases in mice54 (Fig. 1). Moreover, human polymorphisms in the TNFAIP3 gene, encoding A20, are reported to predispose to autoimmune diseases (Table 2). A20 is an immediate-early gene that is induced within an hour of TLR ligation. It is therefore possible that competition between NEMO and ABIN1 for binding to ubiquitin oligomers is enhanced when ABIN1 and A20 form a complex (Fig. 1). Autoimmune diseases are frequently associated with an increased risk of lymphoma. Consistent with this notion, mutations in TNFAIP3 that impair the expression and/or function of A20 are a frequent cause of some lymphomas55,56. Somatic mutations in TNFAIP3 are found in patients with diffuse large B cell lymphoma (19%) more commonly than is the case for TNIP1 (4%) (ref. 57). In addition to its C-terminal ubiquitin-binding NZF motifs, the N-terminal region of A20 has DUB activity. Although it was originally thought to deubiquitylate Lys63-linked ubiquitin chains58, subsequent studies have shown that it hydrolyzes Lys48-linked ubiquitin chains specifically in vitro59, but physiological substrates have yet to be identified. Knock-in mice that express mutants of A20 lacking either DUB activity or the ability to interact with ubiquitin chains do not display the drastic phenotype associated with the complete loss of the A20 protein. However, they are more sensitive to dextran sodium sulfate–induced ulcerative colitis than wild-type control littermates60.
Other polyubiquitin-binding proteins that have been associated with human disease are Toll-interacting protein (TOLLIP) and its interaction partner target of Myb1 (TOM1). TOLLIP and TOM1 target ubiquitylated proteins to early endosomes, where they are either sorted for recycling to the cell surface or degraded after endosomal-lysosomal fusion61,62. TOLLIP interacts with the IL-1 accessory protein (IL-1RACP), IRAK1 (ref. 63) and TLRs64, and is thought to exert anti-inflammatory effects by targeting ubiquitylated IL-1 receptor (IL-1R) and TLRs to endosomes for degradation (Fig. 1)65–67. Notably, two single-nucleotide polymorphisms (SNPs) have been identified in the gene encoding human TOLLIP that are associated with increased risk of tuberculosis68. An important outstanding question in this area concerns the identity of the E3 ubiquitin ligase(s) that ubiquitylates TLRs and the IL-1R to allow their recognition by the TOLLIP-TOM1 complex.
Mutations in the PARK2 gene that inactivate the E3 ligase function of its protein product Parkin cause Parkinson’s disease in humans. Subsequently, PARK2 was identified as a leprosy-susceptibility gene69, and the short interfering RNA knockdown of Parkin in human monocytes reduces the production of IL-6 and the chemokine CCL2 (MCP-1) upon infection by Mycobacterium leprae or other mycobacteria70. The monocytes of leprosy patients have a reduced ability to produce CCL2 upon infection by M. leprae71, and there is genetic evidence implicating CCL2 in susceptibility to mycobacterial infection and tuberculosis. These observations suggest that mutations causing a decrease in the expression or E3 ligase activity of Parkin may increase susceptibility to infection by decreasing the production of CCL2, and perhaps other inflammatory mediators. However, the substrate(s) of Parkin, whose ubiquitylation leads to an enhanced production of CCL2 has yet to be identified.
Two DUBs that cleave Met1-linked ubiquitin chains have been identified, termed Otulin and Cylindromatosis (CYLD). Otulin hydrolyzes Met1-linked ubiquitin chains exclusively72, whereas CYLD also hydrolyzes Lys63-linked ubiquitin chains in vitro 59. There is evidence that Otulin hydrolyzes the Met1-linked ubiquitin chains formed in response to TNF or viral double-stranded RNA72, while studies with CYLD knockout mice have implicated this DUB in hydrolyzing the ubiquitin chains required for the activation of JNK and NF-κB in lymphocytes, but not in myeloid cells73–75. CYLD also prevents the activation of one or more forms of TBK1 and the production of IFN-β during viral infection, probably by hydrolyzing ubiquitin chains attached to the cytosolic RNA receptor RIG-I (Fig. 1)76. However, inactivating mutations in the human CYLD gene do not result in any immune disease, but instead cause cylindromatosis, familial trichoepithelioma and Brooke-Spiegler syndrome, which are conditions involving skin tumors thought to originate from the hair follicles. These tumors are generally benign but can occasionally become malignant. However, humans with CYLD mutations are more susceptible to other cancers77. Similarly, mutations in Otulin that impair its DUB activity also do not cause immune disease, but instead cause impaired angiogenic, craniofacial and neural development in Gumby mice, probably by affecting the Wnt signaling pathway78. In summary, several DUBs implicated in regulating immune signaling networks in mice have been identified, but their mutation in humans or mice gives rise to different diseases and phenotypes, due to the participation of these DUBs in the regulation of many other cellular processes.
Immune diseases and protein kinase mutations: IRAK4
IL-1R–associated kinase 4 (IRAK4) is essential in the signaling network triggered by TLR ligands and the receptors for IL-1, IL-18 and IL-33. The activation of these receptors induces the recruitment of MyD88, which is followed by the binding of IRAK4 to MyD88 and then the recruitment of the other members of the IRAK family, such as IRAKs 1 and 2, to form a structure termed the ‘myddosome’79,80 (Fig. 1). Since the original report81, about 50 human patients with IRAK4 deficiency from over 30 families worldwide have been identified and followed for many years. IRAK4 deficiency is an autosomal recessive disease, heterozygotes being asymptomatic, and affected individuals either have homozygous or compound heterozygous mutations in the IRAK4 gene that mostly result in the complete loss of expression of the protein. The cells from IRAK4-deficient patients produce very little IL-6 or IL-8 in response to IL-1 or TLR agonists.
IRAK4-deficient patients are highly susceptible to pyogenic bacteria, especially infection by Streptococcus pneumoniae, Staphylococcus aureus and Pseudomonas aeruginosa, causing meningitis, sepsis, arthritis, osteomyelitis, and deep inner organ and tissue abscesses. Even when broad-spectrum antibiotics (crimoxazole and penicillin 5) are administered daily from a very early age, nearly half the patients die before the age of eight, mostly before the age of two. However, no patients die from bacterial infections after the age of eight, and invasive bacterial infections gradually decline and are rarely seen after affected individuals reach the age of 20. Surprisingly, IRAK4-deficient patients have normal resistance to common fungi, parasites, viruses and many other types of bacteria. An almost identical phenotype is seen in patients with MyD88 deficiency, suggesting that the main role of the MyD88-IRAK4 signaling network in humans might be to protect young children against infection by a few bacterial species16,82.
Mutation of the JAK family of protein kinases
Several of the classical immune cytokine receptors associate with the JAK family of protein tyrosine kinases, which comprise JAKs 1, 2 and 3 and TYK2. These protein kinases form heterodimers that become activated in response to many cytokines and phosphorylate the STAT (signal transducers and activators of transcription) family of transcription factors, STATs 1–5 (Fig. 1).
A notable feature of the JAKs is the presence of two kinase domains. The N-terminal kinase domain lacks several amino acid residues essential for catalysis by classical protein kinases and is thought to be an inactive pseudokinase. The mutation of Val617 in the pseudokinase domain of JAK2 to phenylalanine is a principal cause of human myeloproliferative diseases83,84, which are hematological neoplasias characterized by excessive proliferation of one or more myeloid lineages. The V617F mutation underlies 95% of all cases of primary polycythemia and about 50% of primary thrombocythemia and primary myelofibrosis, and generates a form of JAK2 in which the C-terminal catalytic domain is constitutively active in the absence of cytokine stimulation. This suggests that one role of the pseudokinase domain is to prevent activation of the C-terminal kinase domain until receptor activation by the appropriate cytokine. The V617F mutation may also prevent the inhibition of JAK2 by SOCS3 (suppressor of cytokine synthesis 3) (reviewed in ref. 85). The JAK inhibitor ruxolitinib, which inhibits the C-terminal kinase domain of JAK2, has been approved for the treatment of these myeloproliferative disorders. In contrast to JAK1, JAK2, and TYK2, which interact with many cytokine receptors, JAK3 only associates with one—namely, the IL-2 receptor γ-chain (IL-2RG). Mutations in this receptor or the loss of JAK3 in humans cause most cases of severe combined immunodeficiency syndrome, which is characterized by the absence of T cells or NK cells in humans86,87. This life-threatening disorder can now be treated by hemopoietic stem-cell transplantation with a survival rate of up to 95% (ref. 88). TYK2 deficiency in humans is associated with increased susceptibility to infections by viruses and other pathogens, such as mycobacteria, probably as a result of compromised activation of the IFN-α/β and IFN-γ signaling networks. A patient with autosomal recessive hyperimmunoglobulinemia E was found to have mutations in TYK2 and to suffer from opportunistic infections. Signaling by many cytokines was found to be impaired in this patient89,90. More recently, a human variant was described in which Ile684 in the N-terminal pseudokinase domain is mutated to serine, and another patient was identified in which Pro1104 in the C-terminal kinase domain is mutated to alanine. Although both mutations were found to impair the catalytic activity of TYK2, the mutated TYK2s are as competent as wild-type TYK2 in rescuing IFN-α-induced STAT signaling in TYK2-deficient cells. Because TYK2 forms heterodimers with JAK1 and JAK2 (ref. 91), these observations are probably explained by the formation of heterodimers between TYK2 and other JAK family members, the loss of TYK2 activity being compensated for by the JAK-binding partner in vivo (Fig. 1). These findings have identified a key non-catalytic function for TYK2 in inducing catalytic competence in the other JAKs with which it interacts. The TYK2 P1104A mutation has also been associated with rheumatoid arthritis92.
Tofacitinib is a potent inhibitor of JAK3, and it also inhibits JAK1 and JAK2, albeit less strongly. Approved for the treatment of rheumatoid arthritis in the United States and Russia in 2012, tofacitinib exerts its beneficial effects by suppressing the production of inflammatory cytokines in both T cells and myeloid cells. It will be interesting to see whether tofacitinib and ruxolitinib are also efficacious for the treatment of several types of leukemia that are associated with mutations in JAK1 and JAK3, and more frequently with the fusion of JAK2 to transcription factors85.
Bruton’s tyrosine kinase (BTK)
The protein kinase BTK is critical to the maturation of B cells and the activation of mast cells. It is switched on in vivo by the binding of phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3) to its pleckstrin homology domain and phosphorylates and activates phospholipase C, the enzyme that hydrolyzes phosphatidylinositol-4,5-bisphosphate to inositol-1,4,5-trisphosphate and diacylglycerol. These second messengers mobilize calcium ions and activate protein kinase C, respectively, thereby regulating many cellular functions.
The gene encoding BTK is located on the X chromosome, and mutations in this gene underlie nearly all cases of X-linked agamma-globulinemia in humans. Over 400 different mutations in the BTK gene have been identified93–96. The BTK inhibitor ibrutinib (now marketed as Imbruvica) was approved by the US Food and Drug Administration in 2013 for the treatment of mantle cell lymphoma and in 2014 for the treatment of chronic lymphocytic leukemia. Approval for the treatment of other lymphomas is expected in the near future.
Zeta chain associated protein kinase (ZAP70)
The protein tyrosine kinase ZAP70 is critical to T cell function. By phosphorylating the essential adaptor proteins LAT and LCP2, it leads to the recruitment of a number of proteins that together trigger the activation of T cell signaling networks97. Several families with severe T cell deficiency have been identified with mutations in ZAP70. These patients present in the first few months after birth with recurrent infections, similar to those observed in combined immunodeficiency diseases. Mutations are mostly, but not exclusively, in the kinase domain and lead to loss of catalytic activity because the mutated protein is absent, presumably as a result of destabilization and subsequent degradation. The T cells of these patients fail to produce IL-2 and do not proliferate in response to T cell antigen receptor stimulation with mitogens or antigens98,99. A mutation causing low-level expression of ZAP70 has also been identified and leads to reduced but detectable T cell signaling. The patient with this mutation had a later onset of combined immunodeficiency, with low but detectable numbers of CD4+ and CD8+ T cells in the periphery and less severe clinical consequences than patients with a complete absence of the ZAP70 protein100. Conversely, a mutation in the C-terminal Src-homology 2 (SH2) domain of ZAP70 that causes altered signal transduction from the T cell antigen receptor and changes the threshold of T cells to thymic selection leads to the positive selection of otherwise negatively selected autoimmune T cells, explaining why it causes autoimmune arthritis in humans101. A mutation in the ZAP70 substrate LAT also causes autoimmunity102,103.
The protein kinase TBK1
Herpes simplex virus 1 (HSV1) encephalitis (HSE) is a rare disease of the CNS that was fatal before the introduction of the antiviral agent acyclovir in the early 1980s. Nevertheless, lifelong neurological consequences are frequently observed even in survivors of the disease. HSE is caused by mutations in genes that encode proteins that participate in the TLR3 signaling network. In this pathway, TLR3 signals from within endosomes, to which it is targeted by interaction with UNC93B, a protein that also targets TLR7, TLR8 and TLR9 to endosomal membranes. TLR3 is activated by double-stranded RNA formed during the replication of some viruses, which is followed by the recruitment of the adaptor protein TRIF and its interaction partner TRAF3 and the activation of one or more forms of TBK1, which phosphorylate interferon regulatory factor 3 (IRF3; Fig. 1). IRF3 dimerizes and translocates to the nucleus, where it stimulates IFN-β gene transcription. IFN-β then drives a gene transcription program required to mount an antiviral state in the cell.
Some patients with HSE have TLR3, UNC93B, TRAF3 or TRIF deficiency104,105, but two children were identified carrying different heterozygous mutations in the TBK1 gene, which result in the mutation of Asp50 or Gly159 to Ala. These are both loss-of-function mutations, one (D50A) caused by the instability of the protein and one (G159A) by the loss of its kinase activity. The fibroblasts of these patients display high rates of HSV1 replication and high rates of cell death that can be rescued by IFNα2b (ref. 106). Together, the genetic analysis of this system has established that the TLR3 signaling network leading to IFN production is essential for protective immunity to HSV1 infection of the CNS during childhood. As discussed earlier in the section on OPTN, duplication of TBK1 has been identified in patients with a normal-tension form of glaucoma, suggesting that enhanced TBK1 activity is one of the causes of this type of glaucoma43.
Phosphatidylinositol 3-OH kinase-δ (PI(3)Kδ)
This lipid kinase is expressed only in immune cells and is activated by the interaction of the SH2 domains in its p85 regulatory subunit with phosphotyrosine-containing pYXXM motifs (where pY is phosphotyrosine and X indicates any amino acid) present in cell-surface receptors. This interaction relieves the inhibition of the catalytic p110δ component exerted by the p85 subunit, enabling p110δ to catalyze the formation of PtdIns(3,4,5)P3 (ref. 107).
Recently patients were identified with mutations in the p110δ catalytic subunit that enhance both the basal and agonist-stimulated catalytic activity of PI(3)Kδ, leading to enhanced amounts of cellular PtdIns(3,4,5)P3 and the hyperactivation of the PtdIns(3,4,5)P3-dependent protein kinases Akt and mTOR108,109. These patients have a combined immunodeficiency due to defects in both B cell and T cell function but also show lymphoproliferation. More detailed analysis revealed that their B cells have impaired secretion of class-switched immunoglobulin isotypes, and T cell memory responses are also defective. These findings can explain the susceptibility of these patients to respiratory infection and airway damage, even after vaccination against S. pneumoniae and Hemophilus influenza type B, and why they are unable to control infection by EBV or cytomegalovirus. Indeed, several of the patients had an EBV-driven form of Hodgkin’s lymphoma.
These studies have identified critical roles for the PI(3)Kδ-dependent, PtdIns(3,4,5)P3-stimulated activation of the Akt-mTOR network in controlling the balance between the formation of short-lived effector T cells that respond to the acute phase of infection and the long-lived memory cells that ensure a rapid response if the same antigen is re-encountered. The hyperactivation of the PtdIns(3,4,5)P3-dependent signaling network severely impairs the development of functional memory T cells and B cells. Treatment of one severely affected patient with the mTOR inhibitor rapamycin restored CD8+ T cell numbers to normal, increased the frequency of naive T cells and restored IL-2 secretion and proliferative responses108. Drugs that inhibit PI(3)Kδ specifically may prove even more effective for the treatment of this genetic disease.
Kinase polymorphisms that predispose to immune diseases
The cytokine deficiency–induced colitis susceptibility locus Cdcs1 is a major modifier of mouse inflammatory bowel disease (IBD). A gene in this region identified as an attractive candidate for regulating susceptibility to IBD encodes α-protein kinase 1 (ALPK1), a member of the atypical α-kinase family, whose catalytic domains bear no sequence similarity to other protein kinases110. The mouse Alpk1 gene is also present in the genetic interval termed ‘Hiccs’ that regulates Helicobacter hepaticus–induced colitis and associated sensitivity to colon cancer in mice. The Hiccs locus controls the induction of the innate inflammatory response by regulating cytokine expression and granulocyte recruitment by Thy1+ innate lymphoid cells. Alpk1 mRNA was expressed at higher levels in bone marrow–derived macrophages from mice that were susceptible to colitis compared to protected mice, and its expression increased in response to LPS or after infection by H. hepaticus. Of the 17 non-synonymous nucleotide variations in the Alpk1 gene found between the susceptible and protected strains, two were located in the kinase domain111. The ALPK1 gene is also located within a human susceptibility region for IBD on chromosome 4 (refs. 112,113).
The substrates of ALPK1 and how its kinase activity may be regulated are unknown, but it is present in polarized epithelial cell lines, where it may be required for the transport of lipid rafts carrying vesicles from the Golgi apparatus to the plasma membrane, perhaps by phosphorylating myosin Ia (ref. 114). As lipid rafts are thought to have important functions in TLR signaling, including the secretion of TNF, antigen presentation and phagocytosis in macrophages or B cell and T cell antigen receptor signaling in lymphocytes, ALPK1 could well be important for responses to bacterial stimuli and mucosal homeostasis115,116. ALPK1 has also been identified as a gout-susceptibility factor in humans and shown to help increase the production of pro-inflammatory cytokines in monocytes upon challenge with urate crystals117.
The catalytic activity of IRAK1 is critical for the production of type 1 IFNs by plasmacytoid dendritic cells but is not required for cytokine production by macrophages in mice118,119. Autoimmune diseases have been linked to the overproduction of type 1 IFNs by plasmacytoid dendritic cells120, and polymorphisms in IRAK1 have been associated with SLE in humans121,122. These observations suggest that IRAK1 inhibitors merit investigation as a potential therapy for SLE and other autoimmune diseases.
Salt-inducible kinase 2 (SIK2) restricts the production of the anti-inflammatory cytokine IL-10 by TLR agonists in macrophages. It exerts this effect by phosphorylating CREB-regulated transcriptional coactivator 3 (CRTC3), a coactivator of the cyclic AMP response element–binding protein (CREB). Phosphorylation of CRTC3 induces its dissociation from CREB and its exit from the nucleus, where it is sequestered in the cytosol by 14-3-3 proteins (Fig. 1)123. SIK2 activity can be suppressed by incubation of macrophages with ligands that elevate the intracellular concentration of cyclic AMP, such as prostaglandin E2 (ref. 124). Prostaglandin E2 or pharmacological inhibition of SIK family members greatly enhances TLR-driven IL-10 production and induces the conversion of macrophages from a pro-inflammatory (M1) to the anti-inflammatory (M2b/c) phenotype thought to be important for the resolution of inflammation123,124. It is therefore of interest that, in humans, CRTC3 has been identified as a gene that predisposes to IBD125, while SIK2 has been identified as a risk factor for primary sclerosing cholangitis (PSC)126. PSC is a severe liver disease that leads to fibrotic destruction of the bile ducts and the need for liver transplantation. Notably, 60–80% of PSC patients suffer from IBD, while 25% have autoimmune disease. How changes in the expression or activity of SIK2 and its substrate CRTC3 impinge on these diseases will require further research. However, it will clearly be of considerable interest to know whether the inhibition of SIK2 has a beneficial effect in patients with these conditions123–126.
Perspective
The last quarter of the twentieth century was undoubtedly a golden era for the study of cell regulation, when many components of the cell signaling networks were identified and their molecular actions elucidated, at least in outline. In contrast, the twenty-first century is proving to be a golden era for human genetics, with technical advances in genome-wide analysis now permitting the identification of proteins that cause or may predispose to human diseases with unprecedented speed. These studies have suggested many polymorphisms that are risk factors for disease, but the function and regulation of the proteins they encode are frequently unknown. These observations are now stimulating a renaissance of the field of cell signaling as researchers try to understand how these proteins impinge on signaling networks to increase or decrease susceptibility to disease. The genes whose mutation or polymorphism cause or predispose to human disease include the E3 ubiquitin ligases, deubiquitylases, ubiquitin-binding proteins and protein kinases discussed in this article (Tables 1 and 2). However, many other proteins mutated in immune disease that have not been discussed here alter ubiquitylation and phosphorylation events indirectly by suppressing or enhancing particular signaling networks. Such proteins include receptors, adaptors and the substrates of protein kinases. The next two decades should reveal a vast amount of new knowledge about the physiological roles of these proteins and the pathological consequences of their variation. This will gradually change the focus of the pharmaceutical industry from developing drugs to treat diseases to developing drugs that prevent diseases before they happen. This will require the formulation of new models for their development and approval, but should be the long-term goal of medicine over the next century.
Acknowledgments
I thank S. Nanda, C. Emmerich and K. Clark for suggestions and A. Nicoll for assistance in preparing the manuscript. Supported by a Wellcome Trust Senior Investigator award (WT100294), the UK Medical Research Council (MRC_MR/K000985/1), AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Janssen Pharmaceuticals, Merck-Serono and Pfizer.
Footnotes
Competing Financial Interests
The author declares no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 2.Smith H, et al. Identification of the phosphorylation sites on the E3 ubiquitin ligase Pellino that are critical for activation by IRAK1 and IRAK4. Proc Natl Acad Sci USA. 2009;106:4584–4590. doi: 10.1073/pnas.0900774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strack P, et al. SCFβ-TRCP and phosphorylation dependent ubiquitination of IκBα catalyzed by Ubc3 and Ubc4. Oncogene. 2000;19:3529–3536. doi: 10.1038/sj.onc.1203647. [DOI] [PubMed] [Google Scholar]
- 4.Xia ZP, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P, Alessi DR. Kinase drug discovery–what’s next in the field? ACS Chem Biol. 2013;8:96–104. doi: 10.1021/cb300610s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Emmerich CH, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc Natl Acad Sci USA. 2013;110:15247–15252. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiil BK, et al. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol Cell. 2013;50:818–830. doi: 10.1016/j.molcel.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark K, Nanda S, Cohen P. Molecular control of the NEMO family of ubiquitin-binding proteins. Nat Rev Mol Cell Biol. 2013;14:673–685. doi: 10.1038/nrm3644. [DOI] [PubMed] [Google Scholar]
- 10.Courtois G, Israel A. IKK regulation and human genetics. Curr Top Microbiol Immunol. 2011;349:73–95. doi: 10.1007/82_2010_98. [DOI] [PubMed] [Google Scholar]
- 11.Döffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 12.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Wu C-J, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 14.Kensche T, et al. Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J Biol Chem. 2012;287:23626–23634. doi: 10.1074/jbc.M112.347195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo YC, et al. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33:602–615. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IκBα deficiency. Clin Microbiol Rev. 2011;24:490–497. doi: 10.1128/CMR.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boisson B, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–1186. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.HogenEsch H, Janke S, Boggess D, Sundberg JP. Absence of Peyer’s patches and abnormal lymphoid architecture in chronic proliferative dermatitis (cpdm/cpdm) mice. J Immunol. 1999;162:3890–3896. [PubMed] [Google Scholar]
- 20.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaud S, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–114. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 23.Damgaard RB, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell. 2012;46:746–758. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Damgaard RB, et al. Disease-causing mutations in the XIAP BIR2 domain impair NOD2-dependent immune signalling. EMBO Mol Med. 2013;5:1278–1295. doi: 10.1002/emmm.201303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K, Girardin SE. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol. 2014;14:9–23. doi: 10.1038/nri3565. [DOI] [PubMed] [Google Scholar]
- 26.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med. 2000;191:1233–1240. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, et al. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian Y, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 29.Boisson B, et al. An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39:676–686. doi: 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conti HR, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. J Clin Invest. 2006;116:1218–1222. doi: 10.1172/JCI28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gleason CE, Ordureau A, Gourlay R, Arthur JS, Cohen P. Polyubiquitin binding to optineurin is required for optimal activation of TANK-binding kinase 1 and production of interferon β. J Biol Chem. 2011;286:35663–35674. doi: 10.1074/jbc.M111.267567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morton S, Hesson L, Peggie M, Cohen P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 2008;582:997–1002. doi: 10.1016/j.febslet.2008.02.047. [DOI] [PubMed] [Google Scholar]
- 36.Munitic I, et al. Optineurin insufficiency impairs IRF3 but not NF-κB activation in immune cells. J Immunol. 2013;191:6231–6240. doi: 10.4049/jimmunol.1301696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilli M, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albagha OM, et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat Genet. 2010;42:520–524. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am J Hum Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 41.Rubino E, et al. SQSTM1 mutations in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Neurology. 2012;79:1556–1562. doi: 10.1212/WNL.0b013e31826e25df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezaie T, et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science. 2002;295:1077–1079. doi: 10.1126/science.1066901. [DOI] [PubMed] [Google Scholar]
- 43.Kawase K, et al. Confirmation of TBK1 duplication in normal tension glaucoma. Exp Eye Res. 2012;96:178–180. doi: 10.1016/j.exer.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregersen PK, et al. Risk for myasthenia gravis maps to a (151) Pro→Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012;72:927–935. doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 47.He CF, et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19:1181–1186. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
- 48.Nair RP, et al. Psoriasis bench to bedside: genetics meets immunology. Arch Dermatol. 2009;145:462–464. doi: 10.1001/archdermatol.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Q, et al. Investigation of 20 non-HLA (human leucocyte antigen) psoriasis susceptibility loci in Chinese patients with psoriatic arthritis and psoriasis vulgaris. Br J Dermatol. 2013;168:1060–1065. doi: 10.1111/bjd.12142. [DOI] [PubMed] [Google Scholar]
- 50.Caster DJ, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. J Am Soc Nephrol. 2013;24:1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matmati M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 52.Skaug B, et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhelst K, et al. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ngo VN, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nocturne G, et al. Germline and somatic genetic variations of TNFAIP3 in lymphoma complicating primary Sjogren’s syndrome. Blood. 2013;122:4068–4076. doi: 10.1182/blood-2013-05-503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong G, et al. A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:1440–1451. doi: 10.1158/1078-0432.CCR-10-1859. [DOI] [PubMed] [Google Scholar]
- 58.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 59.Komander D, et al. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu TT, et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125:265–275. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 62.Katoh Y, et al. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J Biol Chem. 2004;279:24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 63.Burns K, et al. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat Cell Biol. 2000;2:346–351. doi: 10.1038/35014038. [DOI] [PubMed] [Google Scholar]
- 64.Zhang G, Ghosh S. Negative regulation of toll-like receptor-mediated signaling by Tollip. J Biol Chem. 2002;277:7059–7065. doi: 10.1074/jbc.M109537200. [DOI] [PubMed] [Google Scholar]
- 65.Brissoni B, et al. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr Biol. 2006;16:2265–2270. doi: 10.1016/j.cub.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 66.Bulut Y, Faure E, Thomas L, Equils O, Arditi M. Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J Immunol. 2001;167:987–994. doi: 10.4049/jimmunol.167.2.987. [DOI] [PubMed] [Google Scholar]
- 67.Didierlaurent A, et al. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol Cell Biol. 2006;26:735–742. doi: 10.1128/MCB.26.3.735-742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shah JA, et al. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Immunol. 2012;189:1737–1746. doi: 10.4049/jimmunol.1103541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mira MT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 70.de Leseleuc L, et al. PARK2 mediates interleukin 6 and monocyte chemoattractant protein 1 production by human macrophages. PLoS Negl Trop Dis. 2013;7:e2015. doi: 10.1371/journal.pntd.0002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hasan Z, et al. Elevated serum CCL2 concomitant with a reduced mycobacterium-induced response leads to disease dissemination in leprosy. Scand J Immunol. 2006;63:241–247. doi: 10.1111/j.1365-3083.2006.01733.x. [DOI] [PubMed] [Google Scholar]
- 72.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin W, et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118:1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin W, et al. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J Biol Chem. 2007;282:15884–15893. doi: 10.1074/jbc.M609952200. [DOI] [PubMed] [Google Scholar]
- 75.Reiley WW, et al. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat Immunol. 2006;7:411–417. doi: 10.1038/ni1315. [DOI] [PubMed] [Google Scholar]
- 76.Zhang M, et al. Regulation of IκB kinase-related kinases and antiviral responses by tumor suppressor CYLD. J Biol Chem. 2008;283:18621–18626. doi: 10.1074/jbc.M801451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rivkin E, et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498:318–324. doi: 10.1038/nature12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Motshwene PG, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 82.Picard C, et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 2010;89:403–425. doi: 10.1097/MD.0b013e3181fd8ec3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 84.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 85.Laurence A, Pesu M, Silvennoinen O, O’Shea J. JAK kinases in health and disease: an update. Open Rheumatol J. 2012;6:232–244. doi: 10.2174/1874312901206010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Macchi P, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 87.Russell SM, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 88.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by γc family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Minegishi Y, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 90.Watford WT, O’Shea JJ. Human tyk2 kinase deficiency: another primary immunodeficiency syndrome. Immunity. 2006;25:695–697. doi: 10.1016/j.immuni.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 91.Li Z, et al. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J Immunol. 2013;190:2335–2344. doi: 10.4049/jimmunol.1203118. [DOI] [PubMed] [Google Scholar]
- 92.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Conley ME, Mathias D, Treadaway J, Minegishi Y, Rohrer J. Mutations in Btk in patients with presumed X-linked agammaglobulinemia. Am J Hum Genet. 1998;62:1034–1043. doi: 10.1086/301828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Holinski-Feder E, et al. Mutation screening of the BTK gene in 56 families with X-linked agammaglobulinemia (XLA): 47 unique mutations without correlation to clinical course. Pediatrics. 1998;101:276–284. doi: 10.1542/peds.101.2.276. [DOI] [PubMed] [Google Scholar]
- 95.Tsukada S, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 96.Vetrie D, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 97.Fischer A, et al. ZAP70: a master regulator of adaptive immunity. Semin Immunopathol. 2010;32:107–116. doi: 10.1007/s00281-010-0196-x. [DOI] [PubMed] [Google Scholar]
- 98.Arpaia E, Shahar M, Dadi H, Cohen A, Roifman CM. Defective T cell receptor signaling and CD8+ thymic selection in humans lacking zap-70 kinase. Cell. 1994;76:947–958. doi: 10.1016/0092-8674(94)90368-9. [DOI] [PubMed] [Google Scholar]
- 99.Chan AC, et al. ZAP-70 deficiency in an autosomal recessive form of severe combined immunodeficiency. Science. 1994;264:1599–1601. doi: 10.1126/science.8202713. [DOI] [PubMed] [Google Scholar]
- 100.Picard C, et al. Hypomorphic mutation of ZAP70 in human results in a late onset immunodeficiency and no autoimmunity. Eur J Immunol. 2009;39:1966–1976. doi: 10.1002/eji.200939385. [DOI] [PubMed] [Google Scholar]
- 101.Sakaguchi N, et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003;426:454–460. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- 102.Aguado E, et al. Induction of T helper type 2 immunity by a point mutation in the LAT adaptor. Science. 2002;296:2036–2040. doi: 10.1126/science.1069057. [DOI] [PubMed] [Google Scholar]
- 103.Sommers CL, et al. A LAT mutation that inhibits T cell development yet induces lymphoproliferation. Science. 2002;296:2040–2043. doi: 10.1126/science.1069066. [DOI] [PubMed] [Google Scholar]
- 104.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 106.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209:1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 108.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat Immunol. 2014;15:88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Angulo I, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bleich A, et al. Cdcs1 a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity. Inflamm Bowel Dis. 2010;16:765–775. doi: 10.1002/ibd.21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Boulard O, Kirchberger S, Royston DJ, Maloy KJ, Powrie FM. Identification of a genetic locus controlling bacteria-driven colitis and associated cancer through effects on innate inflammation. J Exp Med. 2012;209:1309–1324. doi: 10.1084/jem.20120239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 113.Cho JH, et al. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci USA. 1998;95:7502–7507. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Heine M, et al. α-kinase 1, a new component in apical protein transport. J Biol Chem. 2005;280:25637–25643. doi: 10.1074/jbc.M502265200. [DOI] [PubMed] [Google Scholar]
- 115.Kay JG, Murray RZ, Pagan JK, Stow JL. Cytokine secretion via cholesterol-rich lipid raft-associated SNAREs at the phagocytic cup. J Biol Chem. 2006;281:11949–11954. doi: 10.1074/jbc.M600857200. [DOI] [PubMed] [Google Scholar]
- 116.Luo C, Wang K, Liu D, Li Y, Zhao QS. The functional roles of lipid rafts in T cell activation, immune diseases and HIV infection and prevention. Cell Mol Immunol. 2008;5:1–7. doi: 10.1038/cmi.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang SJ, et al. Lymphocyte α-kinase is a gout-susceptible gene involved in monosodium urate monohydrate-induced inflammatory responses. J Mol Med (Berl) 2011;89:1241–1251. doi: 10.1007/s00109-011-0796-5. [DOI] [PubMed] [Google Scholar]
- 118.Pauls E, et al. Two phases of inflammatory mediator production defined by the study of IRAK2 and IRAK1 knock-in mice. J Immunol. 2013;191:2717–2730. doi: 10.4049/jimmunol.1203268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Uematsu S, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-α induction. J Exp Med. 2005;201:915–923. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barrat FJ, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jacob CO, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007;56:4164–4173. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 122.Jacob CO, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci USA. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Clark K, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci USA. 2012;109:16986–16991. doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.MacKenzie KF, et al. PGE2 induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A–SIK–CRTC3 pathway. J Immunol. 2013;190:565–577. doi: 10.4049/jimmunol.1202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu JZ, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–675. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Graham RR, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus (SLE) Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Musone SL, et al. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–1064. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]