Abstract
Lipids are ubiquitous metabolites with diverse functions; abnormalities in lipid metabolism appear to be related to complications from multiple diseases, including type 2 diabetes. Through technological advances, the entire lipidome has been characterized and researchers now need computational approaches to better understand lipid network perturbations in different diseases. Using a mouse model of type 2 diabetes with microvascular complications, we examined lipid levels in plasma and in renal, neural, and retinal tissues to identify shared and distinct lipid abnormalities. We used correlation analysis to construct interaction networks in each tissue, to associate changes in lipids with changes in enzymes of lipid metabolism, and to identify overlap of coregulated lipid subclasses between plasma and each tissue to define subclasses of plasma lipids to use as surrogates of tissue lipid metabolism. Lipid metabolism alterations were mostly tissue specific in the kidney, nerve, and retina; no lipid changes correlated between the plasma and all three tissue types. However, alterations in diacylglycerol and in lipids containing arachidonic acid, an inflammatory mediator, were shared among the tissue types, and the highly saturated cholesterol esters were similarly coregulated between plasma and each tissue type in the diabetic mouse. Our results identified several patterns of altered lipid metabolism that may help to identify pathogenic alterations in different tissues and could be used as biomarkers in future research into diabetic microvascular tissue damage.
Keywords: diabetes, eye, kidney, mass spectrometry, nerve, systems biology
Lipids are vital metabolites that serve as structural components of cell membranes, signaling mediators, and energy depots. The advent of MS-based lipidomics has expanded our knowledge on lipid alterations in disease and has identified lipid biomarkers of disease states (1). A limitation of current comprehensive shotgun lipidomic techniques is that lipids are identified by class group along with the total number of carbon molecules and double bonds in the aliphatic side chains. This information does not allow for the definitive identification of most lipid species because structural isomers cannot be positively assigned. This limits input primarily to the lipid class level and restricts the ability to map these lipid metabolites to databases such as Kyoto Encyclopedia of Genes and Genomes. Although a typical shotgun lipidomic experiment can characterize hundreds of unique lipid identities, these get condensed to a handful (10–20) when only considering lipid class. Furthermore, individual lipids are under-represented in databases, such as Kyoto Encyclopedia of Genes and Genomes, which are frequently used in systems analysis and pathway mapping, limiting the potential of using multi-omics datasets for understanding disease processes (2). To that end, reconstruction of interaction networks among lipid species from their quantitative MS profiles would provide insights into associated biological mechanisms (3).
Both type 1 and type 2 diabetes are accompanied by dyslipidemia, which is characterized clinically by elevated plasma triglycerides and cholesterol with an enrichment of LDL cholesterol and decreased HDL cholesterol. Recently, lipidomics has revealed abnormalities in other lipid classes in diabetes along with intra-class variation (4–9). Expansion of lipid coverage has improved diabetes risk prediction (4, 6, 9) and increased research on the impact of other lipid classes on diabetes and diabetic complications (10–12). Three devastating complications of diabetes are diabetic kidney disease (DKD), diabetic peripheral neuropathy (DPN), and diabetic retinopathy (DR), and most diabetic patients develop at least one of these complications in their lifetime. Dyslipidemia is associated with the onset and progression of DKD (10, 13–15), DPN (16, 17), and potentially DR (18, 19), although how dyslipidemia correlates with tissue lipid metabolism and disease progression is poorly understood. While plasma lipid composition is used to determine biomarkers for progression of DKD, DPN, and DR, it is unknown whether these extrinsic systemic lipid alterations are reflected in the tissues themselves and can potentially be used to provide insight into specific tissue metabolism or if they are relatively nonspecific and reflective of broader changes.
DKD, DPN, and DR are classically termed diabetic microvascular complications. Although the metabolic response of these tissues to diabetes has historically been thought to be similar and downstream of altered glucose metabolism (20), we recently found that glucose and fatty acid oxidation are tissue specific and dissimilar among the three diabetic end-organ target tissues of kidney, nerve, and retina (21). Transcriptomic analysis identified several pathways involved in lipid biosynthesis that were enriched in both the diabetic mouse kidney and nerve (21), although dysregulation of gene expression in the diabetic kidney and nerve was overall discordant (22, 23). Transcriptomic data indicates altered lipid synthesis, but there is a lack of information on broader lipid changes derived from synthesis in these diabetic tissues and how any lipid changes relate to mRNA expression. In the current study, we performed lipid profiling on mouse plasma, kidney, nerve, and retina. We used the BKS db/db mouse model of type 2 diabetes because this model develops all three diabetic microvascular complications (DKD, DPN, and DR) and is a representative model of human dyslipidemia (24). Using these profiles, we compared the lipidome from plasma to each tissue and compared lipidomes for all three tissues to determine similarities among profiles. We also used correlation analysis to provide precursory information on interaction networks in each tissue and focused on the diabetic kidney for a proof of concept of a lipid-centric approach to multi-omic data integration. These analyses will help determine which classes of lipids in plasma are most useful for analysis of diabetic tissue health and provide a unique approach for linking lipid changes to enzymatic pathways.
MATERIALS AND METHODS
Reagents
All HPLC grade reagents were from Sigma-Aldrich (St. Louis, MO). The lipid internal standards were from Avanti Polar Lipids (Alabaster, AL).
Animal studies
Male BKS db/+ and db/db mice (BKS.Cg-m+/+ Leprdb/J; Jackson Laboratory, Bar Harbor, ME) were used at 24 weeks of age, corresponding to advanced DKD, DPN, and DR (25–28). Food and water were provided ad libitum. Mice (n = 10 per group) were fasted 2 h prior to euthanasia. At the time of euthanasia, plasma, liver, kidney cortex, sciatic nerve, and retina were collected, snap-frozen, and stored at −80°C until use. The University of Michigan committee on use and care of animals reviewed and approved all animal protocols for this study.
Internal standards and quality controls
Lipid standards [cholesteryl ester (CE) 17:0, ceramide d18:1/17:0, diacylglycerol (DAG) d5 19:0, lysophosphatidylcholine (LPC) 17:0/0:0, monoacylglycerol (MAG) 17:0, phosphatidic acid (PA) 17:0, phosphatidylcholine (PC) 17:0, phosphatidylethanolamine (PE) 17:0, phosphatidylglycerol (PG) 17:0, phosphatidylinositol (PI) 17:0/20:4, phosphatidylserine (PS) 17:0, SM d18:1/17:0, and triacylglycerol (TAG) 17:0] were prepared at 1 mg/ml in chloroform/methanol/water and stored at −20°C. For analysis, the final concentration of each standard was 100 pmol/μl. To monitor instrument performance, 10 μl of a dried matrix-free mixture of internal standards reconstituted in 100 μl buffer B were analyzed. To monitor the lipid extraction process, a standard pool of plasma samples and a pool comprised of each test sample were analyzed at the beginning and end of each batch, as well as after every 20 samples.
Sample preparation for MS analysis
Lipids were extracted from plasma (30 μl) or homogenized tissues in a randomized order using a modified Bligh-Dyer method (29), as previously described (10). Briefly, extraction was performed using a 2:2:2 vol ratio of water/methanol/dichloromethane at room temperature after spiking internal standards. The organic layer was collected, dried completely under nitrogen, resuspended in 100 μl buffer B (5:10:85 water:acetonitrile:isopropanol containing 10 mM ammonium acetate) and analyzed using LC/MS/MS-based lipidomics.
Data acquisition through LC/MS/MS analysis
Chromatographic separation was performed on a Shimadzu CTO-20A Nexera X2 UHPLC system (Shimadzu, Kyoto, Japan) with a 1.8 μm particle 50 × 2.1 mm internal diameter Waters Acquity HSS T3 column (Waters, Milford, MA), as previously described (10). The injection volume was 5 μl for all analyses. The data acquisition of each sample was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a Turbo VTM ion source (AB Sciex, Concord, Canada). The mass range of both modes was m/z 50–1,200. Acquisition of MS/MS spectra was controlled by data-dependent acquisition function of the Analyst TF software (AB Sciex) with the application of the following parameters: dynamic background subtraction, charge monitoring to exclude multiply charged ions and isotopes, and dynamic exclusion of former target ions for 9 s. A collision energy spread of 20 V was set whereby the software calculated the collision energy value to be applied as a function of m/z. Mass accuracy was maintained by the use of an automated calibrant delivery system (AB Sciex) interfaced to the second inlet of the DuoSpray source. Calibrations were performed at the start of a workday and whenever ionization polarity was changed. MS data files were processed using MultiQuant 1.1.0.26 (Applied Biosystems/MDS Analytical Technologies, Foster City, CA) (30). Identified lipids were normalized to plasma volume or tissue weight. Quality control samples were used to monitor the overall quality of the lipid extraction and MS analyses (31). The quality control samples were mainly used to remove technical outliers and lipid species that were detected below the lipid class-based lower limit of quantification.
Data processing
After data acquisition, the identified lipids underwent a compound-by-compound review to combine different adducts of the same lipid feature. Missing values were imputed using the k-nearest neighbor method (32). Data were log transformed followed by normalization using the cross-contribution compensating multiple internal standard normalization method (33). Data from positive and negative modes were combined and scaled by the standard deviation.
Statistical analysis
Differentials at individual lipid level.
For any tissue and all lipids that were detected for this tissue, we used t-tests to test differentials between groups (control vs. diabetic) followed by Benjamini-Hochberg false discovery rate (FDR) correction (34) to control the family-wise type I error. Specifically, for each tissue, the t-statistic for lipid i is calculated as
where is the pooled standard deviation of the two groups under consideration, and and are the unbiased estimator of the variance for the two groups, respectively. For any specific lipid, the -value is calculated as
where is the Student’s t-distribution with being the degree of freedom; note that for this data set, . Finally, we performed FDR correction on all s for any specific tissue/plasma to get the adjusted p-values, . The null hypothesis that the lipid does not differ at the mean level for the two groups was rejected if .
Correlation analysis at the individual lipid level.
To obtain the correlations for all lipids that are common across all three tissues and plasma, we calculated the Pearson correlation for lipid i and lipid j as
where is the th sample of lipid and is the sample mean for lipid i. Further, we only retained the correlations that were statistically significantly different than 0. To do so, we first calculated Fisher’s z-score for each , given by
which under the null hypothesis, , is normally distributed with mean zero and standard deviation , where denotes the sample size. The corresponding -value is given by
Moreover, to control the family-wise type I error rate, we adjusted all pairs of the resulting -values using the Benjamini-Hochberg FDR correction, and denote the adjusted -values by . Finally, the statistically significant correlations, , are given by
where is the family-wise type I error rate under control. In this study, we set .
Correlation analysis at the lipid set level.
To test whether any prespecified sets of lipids are differentially coexpressed between plasma and any of the tissues under study (e.g., diabetic plasma vs. diabetic retina), we propose the following procedure, which provides adequate power in detecting the differentially coexpressed sets, while simultaneously keeping the FDR controlled at a prespecified level (e.g., 0.1). The proposed test is in the spirit of pathway enrichment tests, such as Gene Set Enrichment Analysis (35), and its variants that look for persistent activity in the quantities under study (e.g., correlation differentials in our setting). Specifically, the testing procedure is carried out in three steps:
Step 1.
Test for individual pairs of lipids between tissues. For each pair of lipids indexed by , we test whether they are differentially coexpressed between plasma and the tissue under consideration; that is, the null hypothesis is given by versus the alternative , where represents the correlation between lipid and lipid for tissue . The test statistic is given by
where and respectively denote the sample size for tissues and . The -value is given by
where is distributed as a standard normal random variable. Based on a thresholding level , which controls the type I error of this individual test, the decision of the test is given by
that is, we reject if the -value is smaller than the threshold. In this study, we set .
Step 2.
Calculate -values for each lipid set. For any prespecified set , we test whether at the set level, it is differentially coexpressed between tissues and . The test statistic is defined as
which counts the total number of pairs of lipids for which the null hypothesis is rejected based on the individual test in Step 1. Under the null hypothesis that set is commonly coexpressed between tissues and , follows a binomial distribution with size and success probability , that is, , where is the total number of correlations and is the number of lipids in set . The binomial distribution can be approximated by a normal distribution when is sufficiently large. The corresponding -value is then given by
Step 3.
Control FDR across lipid sets. Next, for all lipid sets under consideration, we calculate their -values based on Step 1 and Step 2. We perform FDR correction on these -values and get the adjusted -values. Finally, lipid sets whose adjusted p-values are smaller than the thresholding level (e.g., 0.1) are declared as differentially coexpressed sets.
Data and software availability
The lipidomic data have been deposited in the Metabolomics Workbench Data Repository (www.metabolomicsworkbench.org). Raw and processed microarray data are available in the National Center for Biotechnology Information Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo), accession number GSE86300 (21).
RESULTS
Diabetes alters the lipidome in a tissue-specific manner
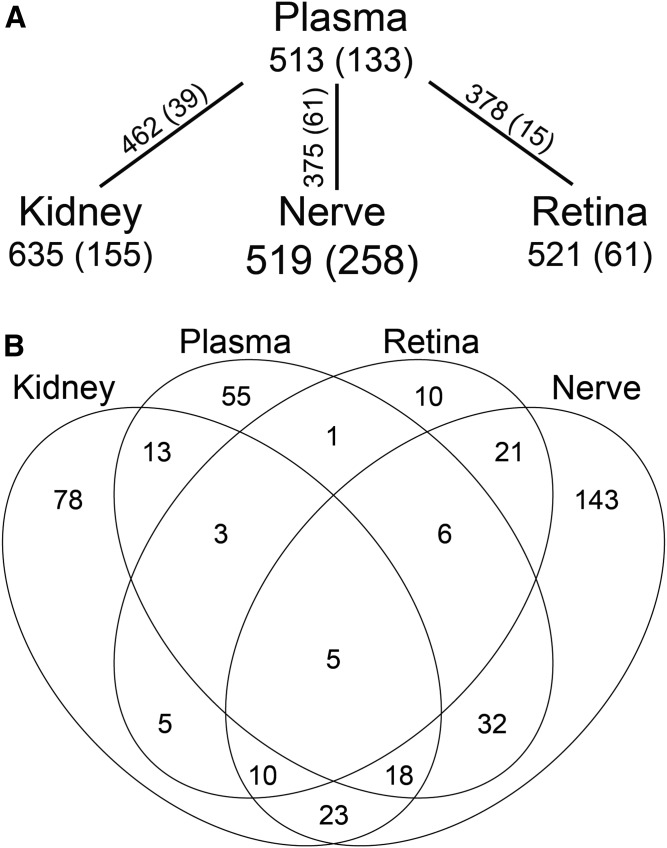
Shotgun lipidomics was used to determine differences in complex lipids present in plasma, kidney cortex, sciatic nerve, and retina from db/db type 2 diabetic mice and db/+ normoglycemic littermate controls. Over 500 unique lipid features were detected in each sample matrix, with 364 lipid features present in plasma and all three tissues (Fig. 1A). The number of lipids that were significantly altered (P < 0.01) between control and diabetic mice in each sample type varied, with 61 (11.7%) in retina, 155 (24.4%) in kidney, 133 (25.9%) in plasma, and 258 (49.7%) in nerve (Fig. 1B, supplemental Tables S1–S4). Of the 364 unique lipid features present in every sample matrix, only 15 were significantly different between control and diabetic mice in all three complication-prone tissues and only five were significantly changed in plasma, kidney, nerve, and retina (Fig. 1B, Table 1). Even in these shared features, however, the direction of the change with diabetes was often inconsistent. Only plasma and nerve showed the same direction of change in diabetes among all five shared features (Table 1).
Fig. 1.
Overlap of differential lipid changes in diabetic mouse plasma, kidney, nerve, and retina. A: Shotgun lipidomics identified over 500 lipids in each plasma, kidney cortex, sciatic nerve, and retina from 24-week-old diabetic versus control mice. Shown are the number of lipids identified in each tissue and the number (in parentheses) that differed between control and diabetic conditions. The numbers of unique features shared between plasma and each tissue are shown on the connecting line, followed by those (in parentheses) that were significantly different between control and diabetic conditions in both plasma and each tissue. B: Venn diagram of significantly altered lipid features between control and diabetic mice in plasma, kidney cortex, sciatic nerve, and retina. P < 0.01 based on a two-sample t-test with FDR correction, n = 10 per group.
TABLE 1.
Significantly different lipid features in diabetic plasma, kidney, nerve and retina
| Lipid Name | Plasma | Kidney | Nerve | Retina | ||||
| P | Mean Change (db/db) | P | Mean Change (db/db) | P | Mean Change (db/db) | P | Mean Change (db/db) | |
| Significant in plasma, kidney, nerve, and retina | ||||||||
| DAG 34:2 | 0.0084 | 1.2625 | 0.0070 | 1.2870 | <0.0001 | 1.9351 | 0.0065 | −1.3539 |
| DAG 38:5 | 0.0089 | 1.2540 | 0.0003 | 1.5501 | <0.0001 | 1.6358 | 0.0005 | −1.5475 |
| LPC 18:1 | 0.0092 | −1.2484 | <0.0001 | 1.6997 | <0.0001 | −1.6510 | 0.0001 | −1.6771 |
| PC 36:4 | 0.0001 | 1.5937 | 0.0009 | −1.4768 | <0.0001 | 1.8168 | 0.0004 | 1.5771 |
| SM 36:2 | <0.0001 | 1.7578 | <0.0001 | 1.7121 | <0.0001 | 1.6427 | 0.0001 | 1.6602 |
| Significant in kidney, nerve and retina | ||||||||
| DAG 36:1 | 0.0334 | 1.0850 | 0.0022 | 1.4065 | <0.0001 | 1.8305 | 0.0002 | −1.6232 |
| DAG 40:5 | 0.0537 | 1.0044 | 0.0029 | 1.3812 | 0.0002 | 1.4952 | 0.0031 | −1.4129 |
| DAG 40:7 | 0.0133 | 1.2075 | 0.0020 | 1.4147 | <0.0001 | 1.7180 | 0.0022 | −1.4473 |
| DAG 44:12 | ND | ND | 0.0034 | 1.3650 | <0.0001 | 1.7500 | 0.0005 | −1.5430 |
| LPC 18:2 | 0.2907 | 0.6101 | <0.0001 | 1.6473 | 0.0003 | 1.4885 | 0.0065 | −1.3511 |
| PC 18:0 | 0.1757 | −0.7546 | 0.0001 | −1.5931 | 0.0045 | 1.2603 | 0.0005 | −1.5423 |
| PE 34:2 | 0.0569 | 0.9926 | 0.0033 | 1.3680 | <0.0001 | 1.7749 | 0.0005 | −1.5451 |
| PE 40:9 | ND | ND | 0.0001 | 1.6079 | <0.0001 | 1.5950 | <0.0001 | 1.7529 |
| Plasmenyl-PE 36:4 | 0.4021 | −0.4952 | <0.0001 | 1.8557 | <0.0001 | 1.9067 | 0.0002 | 1.6183 |
| PS 36:4 | ND | ND | <0.0001 | 1.6834 | <0.0001 | 1.5901 | 0.0024 | 1.4383 |
Values are given as mean difference in 24-week-old db/db versus db/+. P < 0.01 based on a two-sample t-test with FDR correction, n = 10 per group. ND, not detected.
The differentially altered lipids in plasma, kidney, nerve, and retina were widely distributed across the 18 lipid classes (supplemental Fig. S1). The direction of change in lipid levels within most classes varied by tissue type (Table 2, supplemental Table S5). The overall trend was that the majority of altered lipid features in the diabetic nerve were increased, while those in the diabetic retina were mostly decreased. There were a few subtleties. For example, the direction of change in LPCs in the diabetic nerve and PEs in the diabetic retina were dependent on acyl-chain length and, apart from PI, glycerophospholipids with 36 carbons and four double bonds (36:4) either trended toward an increase or were significantly increased in the diabetic retina. In the diabetic kidney, the class-level direction of change was similar to the diabetic nerve except for the glycerophospholipids, where there was a greater degree of intra-class variance that was largely dependent upon acyl-chain length (Table 2, supplemental Table S5). The short- and medium-chain cardiolipins (CLs) were mostly increased, including the immature CL species (<72:8), while the long-chain CLs were decreased. This pattern of increased short-chain and decreased long-chain species was also seen within the LPC, lysoPE, and PA classes. However, the pattern was reversed within the PC and PE classes, where the short acyl-chain lipid features were all decreased and several long-chain lipids were increased.
TABLE 2.
Lipid changes by class in diabetic plasma, kidney, nerve, and retina
| Lipid Class | Plasma | Kidney | Nerve | Retina | ||||
| Increase | Decrease | Increase | Decrease | Increase | Decrease | Increase | Decrease | |
| CE | 7.1% (1) | 7.1% (1) | 9.1% (1) | 0% (0) | 0% (0) | 0% (0) | 0% (0) | 0% (0) |
| CerP | 0% (0) | 16.7% (1) | 0% (0) | 0% (0) | 0% (0) | 0% (0) | 0% (0) | 0% (0) |
| CL | 12.7% (7) | 3.6% (2) | 17.2% (15) | 11.5% (10) | 10.7% (3) | 0% (0) | 0% (0) | 7.1% (2) |
| DAG | 20.0% (7) | 0% (0) | 25.0% (14) | 1.8% (1) | 66.7% (32) | 0% (0) | 0% (0) | 29.2% (14) |
| FFA | ND | ND | ND | ND | 56.3% (9) | 6.3% (1) | 0% (0) | 37.5% (6) |
| LPC | 4.3% (1) | 60.9% (14) | 13.0% (3) | 13.0% (3) | 33.3% (8) | 20.8% (5) | 0% (0) | 39.1% (9) |
| LysoPE | 11.8% (2) | 11.8% (2) | 5.9% (1) | 11.8% (2) | 0% (0) | 30.0% (6) | 0% (0) | 5.0% (1) |
| MAG | 18.2% (2) | 0% (0) | 20.0% (2) | 0% (0) | 0% (0) | 0% (0) | 25.0% (1) | 0% (0) |
| PA | 0% (0) | 6.3% (1) | 23.8% (5) | 9.5% (2) | 20.0% (4) | 0% (0) | 0% (0) | 0% (0) |
| PC | 6.2% (5) | 34.6% (28) | 7.0% (7) | 24.0% (24) | 46.2% (36) | 1.3% (1) | 2.6% (2) | 2.6% (2) |
| PE | 16.3% (7) | 7.0% (3) | 12.3% (8) | 15.4% (10) | 66.7% (34) | 0% (0) | 5.9% (3) | 7.8% (4) |
| PG | 0% (0) | 0% (0) | 2.9% (1) | 22.9% (8) | 29.6% (8) | 3.7% (1) | 0% (0) | 10.0% (3) |
| PI | 6.7% (1) | 20.0% (3) | 6.7% (1) | 20.0% (3) | 20.0% (3) | 6.7% (1) | 0% (0) | 0% (0) |
| Plasmenyl-PC | 11.1% (2) | 33.3% (6) | 22.2% (2) | 0% (0) | 0% (0) | 0% (0) | 0% (0) | 0% (0) |
| Plasmenyl-PE | 28.0% (7) | 24.0% (6) | 25.9% (7) | 11.1% (3) | 44.8% (13) | 6.9% (2) | 3.2% (1) | 19.4% (6) |
| PS | 0% (0) | 0% (0) | 26.3% (5) | 0% (0) | 51.4% (19) | 0% (0) | 2.9% (1) | 8.8% (3) |
| SM | 36.0% (9) | 12.0% (3) | 34.6% (9) | 3.8% (1) | 47.8% (11) | 0% (0) | 9.1% (2) | 4.5% (1) |
| TAG | 5.6% (6) | 5.6% (6) | 6.7% (7) | 0% (0) | 71.1% (59) | 2.4% (2) | 0% (0) | 0% (0) |
Percent and total number (in parentheses) of members of lipid classes that were significantly different in 24-week-old db/db compared with db/+ mice. P < 0.01 based on a two-sample t-test with FDR correction, n = 10 per group. CerP, ceramide-1-phosphate; ND, not determined. Trends are summarized in supplemental Table S5.
Plasma may reflect changes in specific subclasses of lipids
To reduce dimensionality due to the high number of lipids identified versus the low sample size, we grouped the individual lipid features present in plasma and all three tissues by either saturation or acyl-chain length per class and examined these biologically relevant subclasses across plasma and each kidney, nerve, or retina in both control and diabetic animals. As liver lipid composition is arguably the most likely to reflect plasma composition, we also included analysis of lipids in liver tissue to look for broader trends and insight into whether similarities may be more systemic. For saturation, each class was split into saturated/monounsaturated (low) or polyunsaturated (high), while for chain length we split each lipid class into thirds (supplemental Table S6). We performed correlation analysis at the set level to determine commonly coexpressed lipid sets between plasma and tissues. We declared sets in plasma and tissues to be commonly coregulated if the proposed test described in the Materials and Methods failed to reject the null hypothesis at a FDR of 0.1 (supplemental Fig. S2). Not surprisingly, liver tissue most closely reflected plasma lipid composition (Table 3). Interestingly, this was much more pronounced in nondiabetic control mice than in mice with diabetes. Furthermore, the majority of lipid subclasses that were similarly coregulated between diabetic plasma and diabetic liver tissue were also similar between control plasma and control liver tissue. This association was not as pronounced between plasma and the other tissues (kidney, nerve, and retina), where there were fewer similarities across the control and diabetic conditions for each plasma/tissue comparison. This was particularly true between plasma and kidney, as only one chain-length lipid subclass and two saturation lipid subclasses were commonly regulated in both control and diabetic conditions (Table 3; supplemental Tables S7, S8). Although a slightly greater number of lipid subclasses overall were commonly coregulated in the control tissues than the diabetic tissues, the saturated/monounsaturated CEs were the only commonly coregulated subclass between diabetic plasma and each diabetic kidney, nerve, and retina, but not between control plasma and control tissues.
TABLE 3.
Commonly coregulated lipid subclasses between plasma and tissues
| Tissue | Saturation (Plasma vs. Tissue) | Acyl-Chain Length (Plasma vs. Tissue) | ||
| Control | Diabetic | Control | Diabetic | |
| Kidney | DAG low, LPE low, PA low, PA high, PC low | CE low, LPE low, PA low, PG low, SM high | LPC bottom, LPE mid, PA mid, PA top, PI top | LPE top, PA bottom, PI top, PlPE bottom, SM bottom, SM mid |
| Nerve | CL high, DAG low, LPE low, LPE high, PA high | CE low, LPC low, LPE low, PA low, PA high, PG low, PlPE high | CL mid, LPC bottom, LPC mid, LPE bottom, LPE mid, LPE top, PA mid, PA top, PE bottom, PG mid, SM bottom | LPC bottom, LPE bottom, LPE mid, LPE top, PA bottom, PA mid, PA top, PI top |
| Retina | CE high, PA high, PG low, PG high, PlPE low | CE low, LPE high, PA high, PG low | CE mid, LPE top, PA bottom, PA mid, PA top, PG bottom, PG mid, SM bottom, SM mid | LPC bottom, LPE bottom, LPE mid, LPE top, PA mid, PA top |
| Liver | CE low, CE high, DAG low, LPC low, LPE low, LPE high, PG low, PG high, PlPE low, SM low | CE low, LPE low, PA low, PA high, PG low | CE mid, DAG bottom, DAG top, LPC bottom, LPE bottom, LPE mid, LPE top, PA mid, PA top, PG bottom, PG mid, PI top, PlPE bottom | LPC bottom, PA top, PI top |
Commonly coregulated lipid subclasses between plasma and each tissue in 24-week-old db/+ and db/db mice. For saturation, each class was separated into low (unsaturated/monounsaturated) and high (polyunsaturated). For acyl-chain length, each class was divided into thirds (bottom, mid, top). P < 0.1 using correlation analysis at the set level, n = 10 per group. PlPE, plasmenyl-phosphatidylethanolamine. Data are also presented in supplemental Tables S7 and S8.
Organization of dysregulated lipid metabolism is tissue specific
To examine how lipid levels were associated within and across classes, we performed correlation analyses in control and diabetic plasma, kidney, nerve, and retina using the 364 lipid features identified in all samples (P < 0.1; supplemental Fig. S3). Of the 66,066 possible combinations, no lipid-to-lipid correlations were significantly different in each diabetic sample matrix (plasma, kidney, nerve, and retina) that were not present in the control samples, revealing that there was not an underlying lipid-to-lipid signature in the diabetic milieu. When considering the three tissues without plasma, there were two intra-class correlations between lipid species that were significantly different in diabetic tissues, but not in control tissues (supplemental Table S9).
Lipids in plasma and kidney displayed much more correlation than did nerve and retina. Plasma had the highest degree of significant lipid correlations from the control mice (supplemental Fig. S3A). While there were similarities between control and diabetic plasma, there were also large intra- and inter-class differences, such as within the CL and the TAG classes (supplemental Fig. S4). Of the tissues examined, the diabetic kidney had the largest number of significantly correlated lipids (supplemental Fig. S3B). The pattern was vastly different than in the control kidney, with many intra- and inter-class correlations, and there was a relatively equal mix of lipids that were positively and negatively correlated in the diabetic kidney. There were few significant correlations seen in the nerve (supplemental Fig. S3C), even though the nerve had the largest number of differentially expressed lipid features (49.7%). The primary discernable pattern in the diabetic nerve was an intra-class association within TAGs, particularly the short-chain to short-chain and the medium-chain to medium-chain lengths (supplemental Fig. S5). The diabetic retina had several intra- and inter-class significant lipid correlations and the vast majority of these were positively correlated, suggesting an overall change in lipogenesis or tissue uptake (supplemental Fig. S3D). Regardless, in plasma, kidney, nerve, and retina, more lipids were significantly correlated in the diabetic tissues than in the nondiabetic tissues. This was particularly pronounced in the diabetic kidney, where there was much more organization across classes.
Integration of lipid correlation data with transcriptomic data
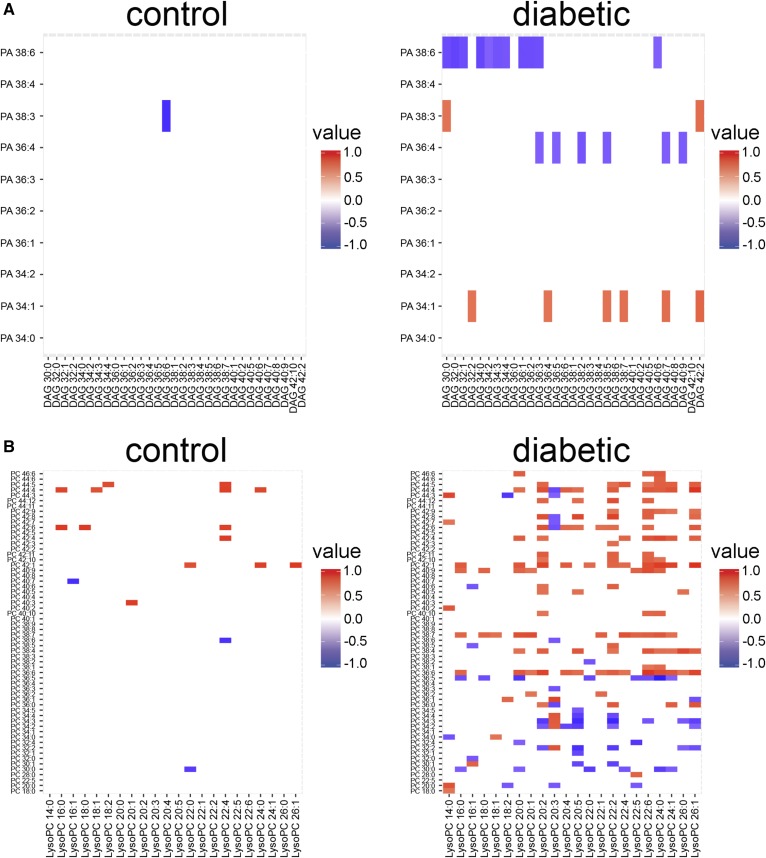
One dilemma faced when trying to integrate lipidomic data with other -omics datasets is that lipids are not direct products of single enzymes that are subject to control by gene expression regulation. Lipids can be synthesized from acetyl-CoA in the cytoplasm, followed by elongation and desaturation to generate chain diversity. Further enzymatic modifications generate class diversity, with lipids ultimately being synthesized de novo or through interconversion between classes. To examine the feasibility of using the correlation matrices as an integration point with transcriptomic data, we focused on the diabetic kidney because there were significantly correlated lipid features in the diabetic kidney and because high complexity transcriptomic data have been generated from kidney tissue from the same diabetic model. We examined the kidney correlation matrix (supplemental Fig. S3B) in parallel with a focused lipid-centric list of 283 transcripts from our microarray data (GSE86300) (21) to validate patterns of lipid levels. Correlation analysis of the kidney lipidomic data identified an inverse relationship between the PA and the DAG classes, with apparent specificity for the PA, as several DAG species were negatively correlated with PA 38:6 or PA 36:4 (Fig. 2A). Transcriptomic analysis supported this, as mRNA levels of four DAG kinase (Dgk) isoforms were decreased and expression of two PA phosphatase (Ppap2) isoforms were increased (Table 4). This expression pattern would suggest a conversion of PA to DAG. Another pattern identified in the correlation analysis was a strongly correlated network between PC and LPC levels in the diabetic kidney (Fig. 2B). PC is a major membrane lipid that serves as a precursor for lipid mediators, such as LPC following hydrolysis by phospholipase A2 (PLA2) (36). In concert with PLA2, lysophospholipid acyltransferases facilitate remodeling of the glycerophospholipid acyl-chains (37). Transcriptomic analysis revealed that several PLA2 and lysophospholipid acyltransferase mRNAs were significantly altered in the diabetic kidney, confirming mRNA expression level changes that affect glycerophospholipid remodeling (Table 4).
Fig. 2.
Class-specific correlation analysis in the control and diabetic mouse kidneys for integration with transcriptomic data. Zoom-in views of correlations between PA and DAG lipid classes (A) and PC and LPC lipid classes (B) in kidney cortex from 24-week-old control and diabetic mice. Significant correlations are shown as positive (red) or negative (blue). P < 0.1 using Pearson’s correlation with Fisher’s transformation and FDR correction, n = 10 per group. Corresponds to supplemental Fig. S3B.
TABLE 4.
Mouse kidney cortex mRNA expression of genes involved in lipid metabolism
| Enzyme Class | Enzyme Subclass | Gene Identification | Log2 Fold Change | Q Value |
| DAG kinase (DAG to PA) | N/A | Dgka | 0.06 | NS |
| Dgkb | −0.48 | <0.01 | ||
| Dgkd | N/A | |||
| Dgke | −0.06 | NS | ||
| Dgkg | −0.22 | NS | ||
| Dgkh | −0.79 | 0.02 | ||
| Dgki | −0.18 | NS | ||
| Dgkk | N/A | |||
| Dgkq | −0.42 | 0.03 | ||
| Dgkz | −0.31 | <0.01 | ||
| PA phosphatase (PA to DAG) | N/A | Ppap2a | 0.25 | <0.01 |
| Ppap2b | 0.19 | 0.03 | ||
| Ppap2c | 0.13 | NS | ||
| PLA2 | cPLA2 | Pla2g4a | 0.81 | 0.04 |
| Pla2g4b | 0.2 | NS | ||
| Pla2g4c | −0.34 | 0.01 | ||
| Pla2g4d | N/A | |||
| Pla2g4e | N/A | |||
| Pla2g4f | N/A | |||
| sPLA2 | Pla2g1b | −0.04 | NS | |
| Pla2g10 | N/A | |||
| Pla2g12a | 0.16 | NS | ||
| Pla2g12b | −0.32 | NS | ||
| Pla2g2a | N/A | |||
| Pla2g2d | N/A | |||
| Pla2g2e | 0.29 | 0.03 | ||
| Pla2g2f | 0.05 | NS | ||
| Pla2g3 | N/A | |||
| Pla2g5 | 0.59 | <0.01 | ||
| iPLA2 | Pla2g6 | −0.58 | <0.01 | |
| PAFAH | Pla2g7 | 2.80 | 0.03 | |
| Pafah1b1 | −0.13 | NS | ||
| Pafah1b2 | −0.03 | NS | ||
| Pafah1b3 | 0.50 | 0.03 | ||
| Lysophospholipid acyltransferases | AGPAT | Gpat1 | −0.29 | NS |
| Gpat2 | −0.25 | NS | ||
| Gpat3 | 0.66 | <0.01 | ||
| Gpat4 (Agpat 6) | −0.20 | NS | ||
| Lpaat1 | −0.27 | 0.03 | ||
| Lpaat2 (Agpat2) | N/A | |||
| Lpgat1 | −0.61 | <0.01 | ||
| Lclat1 | 0.03 | NS | ||
| Lpcat2 | 1.75 | NS | ||
| Lpeat2 | 0.23 | NS | ||
| MBOAT | Lpcat3 (Mboat5) | −0.15 | <0.01 | |
| Lpcat4 (Mboat2) | 0.53 | 0.10 | ||
| Lpeat1 (Mboat1) | 1.06 | 0.02 | ||
| Lpiat1 (Mboat7) | −0.16 | 0.05 |
Log2 fold change is in 24-week-old db/db versus db/+ mice. Q < 0.05 (bold) based on a two-sample t-test, n = 5 per group. AGPAT = 1-acylglycerol-3-phosphate O-acyltransferase, MBOAT = membrane-bound O-acyltransferase, PAFAH = platelet-activating factor acetylhydrolase, cPLA2 = cytosolic PLA2, iPLA2 = calcium-independent PLA2, sPLA2 = secreted PLA2, N/A = not on the array or below signal threshold.
DISCUSSION
Most lipid profiling studies describe overall changes in individual lipid species levels without providing a deeper understanding of the biological pathways resulting in these changes. Although acetyl-CoA is the primary building block for all lipids, lipids undergo fatty acyl-chain remodeling and multi-step transformations between major lipid classes (1, 38). While simulations have been used with MS data for specific lipid classes to better understand lipid metabolic pathways (1, 39), a relatively quick and comprehensible method is needed to provide insight into the complex changes in lipid metabolism. Here, we describe an approach using correlation analysis to combine transcriptomic and lipidomic data. Lipid levels are more closely related to the biological state of a tissue, so we used a lipid-centric approach by examining lipid-lipid interactions through correlation analysis. We then limited our transcriptomic input to buffer unrelated inputs and biological noise (40, 41). This approach has the ability to identify underlying mechanisms resulting in the altered lipid profiles in disease states such as DKD. Combining this approach with the traditional method of analyzing individual lipid levels independently of one another revealed that, similar to glucose and fatty acid oxidation (21), complex lipid metabolism is differentially regulated in the diabetic kidney, nerve, and retina.
There was an overall directional change in lipid levels in the diabetic nerve and retina, with an increase in the diabetic nerve and a decrease in the diabetic retina. In the diabetic nerve, we previously found decreased levels of acylcarnitines and acyl-CoAs (21), possibly contributing to the increase in FFAs. Furthermore, metabolic flux analysis identified lipid oxidation to be shunted away from the TCA cycle after citrate in the diabetic nerve (21). Citrate can be exported to the cytosol and metabolized into acetyl-CoA for lipogenesis. Therefore, it is likely that oxidized FFAs in the diabetic nerve are ultimately used for lipid synthesis via citrate. As a neural tissue, the retina is thought to be obligated to glucose metabolism, but the expression of proteins involved in lipid transport and β-oxidation have been identified in human and rodent retinas (42–45) and the retina has recently been shown to be capable of oxidizing palmitate (21, 46). Thus, lowering of retinal lipids in diabetes could be due to either decreased transport or increased oxidation. In VLDL-knockout mice, limiting FFA uptake into the retina resulted in stabilization of hypoxia-inducible factor (HIF-1α) and overexpression of vascular endothelial growth factor, resulting in angiogenesis (46). Furthermore, diminished lipid levels in the diabetic retina have been linked to retinal neuronal cell apoptosis (47). Collectively, these studies suggest a decrease in retinal tissue lipid levels or FFA uptake could contribute to the development of proliferative DR and should be an area of future study.
In the diabetic kidney, glycerolipids and plasmalogens were mostly increased, while levels of most glycerophospholipid features varied depending on chain length, suggesting that glycerophospholipid remodeling may be an important feature of early DKD. Of particular interest was the increase of immature CLs in diabetic kidneys. CLs are primarily found in the inner mitochondrial membrane and the presence of immature CLs has been linked with impaired mitochondrial function and bioenergetics (48, 49), connecting mitochondrial structural abnormalities to respiratory defects and the Warburg effect in brain tumors (50). In our previous report, we identified the association between mitochondrial electron transport chain defects with increased glycolysis, lactate formation, and pyruvate oxidation in diabetic kidneys (21). Further studies are needed to determine whether there is a functional link between immature CL species and decreased electron transport chain function and altered glucose metabolism.
Although complex lipid metabolism in the diabetic kidney, nerve, and retina was tissue-specific, there were some common characteristics. Altered levels of DAG lipid features were relatively enriched in each tissue, and DAGs comprised 6 of the 15 shared lipid features significantly altered in each of the tissues. The DAG-protein kinase C pathway has been implicated in diabetic complications, as activation of protein kinase C is hypothesized to be a key modulator through which a variety of effectors enhance complications (51, 52). Another shared feature between the tissues is the potential importance of the inflammatory mediator, arachidonic acid (AA). While our LC/MS analysis did not positively identify the individual acyl-chain composition in each lipid feature, lipids of the 36:4 composition were frequently increased in each of the tissues, including 3 of the 15 shared altered lipid features. These 36:4 lipids most likely include linoleic acid (18:2 + 18:2) and/or palmitic acid (16:0) and AA (20:4). AA can be converted from linoleic acid by Fads1, Elovl2 or Elovl5, and Fads2, hydrolyzed from a glycerophospholipid by PLA2 or hydrolyzed from phospholipid-derived DAG by phospholipase C and DAG lipase (53). In db/db kidney cortex (GSE86300) (21) and sciatic nerve (GSE27382) (54), mRNA levels of several secreted PLA2 enzymes were increased along with several phospholipase C isoforms. Also, mRNA levels of enzymes involved in the conversion from linoleic acid were also significantly altered in both kidney and nerve, albeit in a bi-directional manner. Interestingly, the mRNA levels of Ptgs1, Ptgs2, and Alox5, responsible for metabolism of AA to prostaglandins and leukotrienes, were increased in the diabetic kidney, while Alox15 and Alox8, for metabolism of AA to 15-, 12-, or (15S)-hydroperoxyeicosatetraenoic acid, were significantly decreased.
Several studies have found DKD, DPN, and potentially DR onset and progression to correlate with plasma lipid levels (13, 14, 16–19). Our findings suggest that certain subclasses of lipids in diabetic plasma may be useful as biomarkers for diabetic tissue lipid metabolism. Although plasma reflects systemic lipid metabolism, there were several lipid subclasses similarly regulated (co-“expressed”) between plasma and kidney, nerve, and/or retina that were not similarly regulated between plasma and liver tissue. As there was a lower degree of shared commonly coregulated lipid subclasses between plasma and each kidney, nerve, and retina, respectively, between the control and diabetic states, it suggests that diabetes uniquely alters the plasma/tissue lipid relationship for the kidney, nerve, and retina. Exactly how plasma lipid levels relate to tissue lipid metabolism needs to be further defined.
Our approach had several limitations including a relatively small sample size and confounding tissue heterogeneity, which could mask important changes in specific cell types. Additionally, in a recent study, James and colleagues (55) reported targeted metabolomics of 165 metabolites (organic acids, amino acids, and lipids including acylcarnitines, acyl-CoAs, and ceramides) on skeletal muscle from three different inbred mouse strains that were fed two different diets. Using random forest, the authors determined a metabolic signature of insulin resistance, regardless of strain or diet. This work highlighted that there is a significant amount of metabolic diversity between the strains, diets, and individual animals. While our study included a pathophysiologically relevant model of diabetic complications, it was restricted to only one mouse strain and the lipid metabolic response to diabetes could be different between strains. Finally, no MS platform is capable of detecting every lipid metabolite, limiting analysis to only those detected and excluding analysis of others that may play an important role, such as sphingosine-1-phosphate. Although we do not compare correlation networks with transcriptomic data for all three tissues, we illustrate the potential of this approach using data from diabetic and control kidney cortex as a proof of principle. Correlation analysis is not a new technique, but the simplicity of employing a widely-used and understood method to allow for network analysis is the primary strength of the study. Using this approach, we observed PA specificity in the conversion of PA to DAG, generating a future testable hypothesis regarding enzyme isoforms on lipid acyl-chain preference. Correlation analysis is already built in to freely available analytical pipelines for lipidomic data analysis, such as MetaboAnalyst (www.metaboanalyst.ca) (56, 57), which also provides an integrated pathway mapping feature. Therefore, this approach could provide a user-friendly method to integrate lipidomic data with other -omics datasets. Although LC/MS analysis cannot positively identify individual acyl-chain composition, combining this analysis with other approaches can help impute such changes computationally. This combinatorial approach may identify mechanistic and potentially pathogenic mechanisms to explore, such as the enrichment of AA (20:4) or lipokines such as palmitoleate (16:1) (58). These approaches will enhance our understanding of the pathophysiology of lipid metabolic pathways in disease.
Supplementary Material
Footnotes
Abbreviations:
- AA
- arachidonic acid
- CE
- cholesteryl ester
- CL
- cardiolipin
- DAG
- diacylglycerol
- Dgk
- diacylglycerol kinase
- DKD
- diabetic kidney disease
- DPN
- diabetic peripheral neuropathy
- DR
- diabetic retinopathy
- FDR
- false discovery rate
- LPC
- lysophosphatidylcholine
- MAG
- monoacylglycerol
- PA
- phosphatidic acid
- Ppap2
- phosphatidic acid phosphatase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PKC
- protein kinase C
- PLA2
- phospholipase A2
- PS
- phosphatidylserine
- QC
- quality control
- TAG
- triacylglycerol
This work was supported, in part, by National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Grants DK094292, DK089503, DK082841, DK081943, and DK097153 (S.P.) and National Eye Institute Grant EY20582 (T.W.G.; S.F.A.); and Michigan Institute for Clinical and Health Research CTSA Award 2UL1TR000433 (K.M.S.) from the National Center for Advancing Translational Sciences. Additional support was provided by the American Diabetes Association, the Program for Neurology Research and Discovery, Novo Nordisk Grant NNF14SA0006 (E.L.F.), and the Juvenile Diabetes Research Foundation (postdoctoral fellowship to L.M.H.). F.C.B., E.L.F., and T.W.G. are supported by the Taubman Institute at the University of Michigan. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Han X. 2016. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 12: 668–679. [DOI] [PubMed] [Google Scholar]
- 2.Yetukuri L., Ekroos K., Vidal-Puig A., and Oresic M.. 2008. Informatics and computational strategies for the study of lipids. Mol. Biosyst. 4: 121–127. [DOI] [PubMed] [Google Scholar]
- 3.Creixell P., Reimand J., Haider S., Wu G., Shibata T., Vazquez M., Mustonen V., Gonzalez-Perez A., Pearson J., Sander C., et al. ; Mutation Consequences and Pathway Analysis Working Group of the International Cancer Genome Consortium. 2015. Pathway and network analysis of cancer genomes. Nat. Methods. 12: 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floegel A., Stefan N., Yu Z., Muhlenbruch K., Drogan D., Joost H. G., Fritsche A., Haring H. U., Hrabe de Angelis M., Peters A., et al. 2013. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 62: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haus J. M., Kashyap S. R., Kasumov T., Zhang R., Kelly K. R., Defronzo R. A., and Kirwan J. P.. 2009. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 58: 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rhee E. P., Cheng S., Larson M. G., Walford G. A., Lewis G. D., McCabe E., Yang E., Farrell L., Fox C. S., O’Donnell C. J., et al. 2011. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J. Clin. Invest. 121: 1402–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ståhlman M., Fagerberg B., Adiels M., Ekroos K., Chapman J. M., Kontush A., and Borén J.. 2013. Dyslipidemia, but not hyperglycemia and insulin resistance, is associated with marked alterations in the HDL lipidome in type 2 diabetic subjects in the DIWA cohort: impact on small HDL particles. Biochim. Biophys. Acta. 1831: 1609–1617. [DOI] [PubMed] [Google Scholar]
- 8.Ståhlman M., Pham H. T., Adiels M., Mitchell T. W., Blanksby S. J., Fagerberg B., Ekroos K., and Borén J.. 2012. Clinical dyslipidaemia is associated with changes in the lipid composition and inflammatory properties of apolipoprotein-B-containing lipoproteins from women with type 2 diabetes. Diabetologia. 55: 1156–1166. [DOI] [PubMed] [Google Scholar]
- 9.Wang-Sattler R., Yu Z., Herder C., Messias A. C., Floegel A., He Y., Heim K., Campillos M., Holzapfel C., Thorand B., et al. 2012. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 8: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afshinnia F., Rajendiran T. M., Karnovsky A., Soni T., Wang X., Xie D., Yang W., Shafi T., Weir M. R., He J., et al. 2016. Lipidomic signature of progression of chronic kidney disease in the chronic renal insufficiency cohort. Kidney Int. Rep. 1: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaurasia B., and Summers S. A.. 2015. Ceramides - lipotoxic inducers of metabolic disorders. Trends Endocrinol. Metab. 26: 538–550. [DOI] [PubMed] [Google Scholar]
- 12.García-Fontana B., Morales-Santana S., Díaz Navarro C., Rozas-Moreno P., Genilloud O., Vicente Pérez F., Perez Del Palacio J., and Muñoz-Torres M.. 2016. Metabolomic profile related to cardiovascular disease in patients with type 2 diabetes mellitus: a pilot study. Talanta. 148: 135–143. [DOI] [PubMed] [Google Scholar]
- 13.Chen S. C., and Tseng C. H.. 2013. Dyslipidemia, kidney disease, and cardiovascular disease in diabetic patients. Rev. Diabet. Stud. 10: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herman-Edelstein M., Scherzer P., Tobar A., Levi M., and Gafter U.. 2014. Altered renal lipid metabolism and renal lipid accumulation in human diabetic nephropathy. J. Lipid Res. 55: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadler K., Goldberg I. J., and Susztak K.. 2015. The evolving understanding of the contribution of lipid metabolism to diabetic kidney disease. Curr. Diab. Rep. 15: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hur J., Sullivan K. A., Callaghan B. C., Pop-Busui R., and Feldman E. L.. 2013. Identification of factors associated with sural nerve regeneration and degeneration in diabetic neuropathy. Diabetes Care. 36: 4043–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiggin T. D., Sullivan K. A., Pop-Busui R., Amato A., Sima A. A., and Feldman E. L.. 2009. Elevated triglycerides correlate with progression of diabetic neuropathy. Diabetes. 58: 1634–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chew E. Y., Klein M. L., Ferris F. L. 3rd, Remaley N. A., Murphy R. P., Chantry K., Hoogwerf B. J., and Miller D.. 1996. Association of elevated serum lipid levels with retinal hard exudate in diabetic retinopathy. Early Treatment Diabetic Retinopathy Study (ETDRS) Report 22. Arch. Ophthalmol. 114: 1079–1084. [DOI] [PubMed] [Google Scholar]
- 19.Ioannidou E., Tseriotis V. S., and Tziomalos K.. 2017. Role of lipid-lowering agents in the management of diabetic retinopathy. World J. Diabetes. 8: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giacco F., and Brownlee M.. 2010. Oxidative stress and diabetic complications. Circ. Res. 107: 1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sas K. M., Kayampilly P., Byun J., Nair V., Hinder L. M., Hur J., Zhang H., Lin C., Qi N. R., Michailidis G., et al. 2016. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 1: e86976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinder L. M., Park M., Rumora A. E., Hur J., Eichinger F., Pennathur S., Kretzler M., Brosius F. C. 3rd, and Feldman E. L.. 2017. Comparative RNA-Seq transcriptome analyses reveal distinct metabolic pathways in diabetic nerve and kidney disease. J. Cell. Mol. Med. 21: 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hur J., O’Brien P. D., Nair V., Hinder L. M., McGregor B. A., Jagadish H. V., Kretzler M., Brosius F. C. 3rd, and Feldman E. L.. 2016. Transcriptional networks of murine diabetic peripheral neuropathy and nephropathy: common and distinct gene expression patterns. Diabetologia. 59: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin W., Carballo-Jane E., McLaren D. G., Mendoza V. H., Gagen K., Geoghagen N. S., McNamara L. A., Gorski J. N., Eiermann G. J., Petrov A., et al. 2012. Plasma lipid profiling across species for the identification of optimal animal models of human dyslipidemia. J. Lipid Res. 53: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brosius F. C. III, Alpers C. E., Bottinger E. P., Breyer M. D., Coffman T. M., Gurley S. B., Harris R. C., Kakoki M., Kretzler M., Leiter E. H., et al. ; Animal Models of Diabetic Complications Consortium. 2009. Mouse models of diabetic nephropathy. J. Am. Soc. Nephrol. 20: 2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H. T., Dauch J. R., Hayes J. M., Hong Y., and Feldman E. L.. 2009. Nerve growth factor mediates mechanical allodynia in a mouse model of type 2 diabetes. J. Neuropathol. Exp. Neurol. 68: 1229–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma K., McCue P., and Dunn S. R.. 2003. Diabetic kidney disease in the db/db mouse. Am. J. Physiol. Renal Physiol. 284: F1138–F1144. [DOI] [PubMed] [Google Scholar]
- 28.Bogdanov P., Corraliza L., Villena J. A., Carvalho A. R., Garcia-Arumi J., Ramos D., Ruberte J., Simo R., and Hernandez C.. 2014. The db/db mouse: a useful model for the study of diabetic retinal neurodegeneration. PLoS One. 9: e97302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 30.Ejsing C. S., Duchoslav E., Sampaio J., Simons K., Bonner R., Thiele C., Ekroos K., and Shevchenko A.. 2006. Automated identification and quantification of glycerophospholipid molecular species by multiple precursor ion scanning. Anal. Chem. 78: 6202–6214. [DOI] [PubMed] [Google Scholar]
- 31.Jung H. R., Sylvanne T., Koistinen K. M., Tarasov K., Kauhanen D., and Ekroos K.. 2011. High throughput quantitative molecular lipidomics. Biochim. Biophys. Acta. 1811: 925–934. [DOI] [PubMed] [Google Scholar]
- 32.Altman N. S. 1992. An introduction to kernel and nearest neighbor nonparametric regression. Am. Stat. 46: 175–185. [Google Scholar]
- 33.Redestig H., Fukushima A., Stenlund H., Moritz T., Arita M., Saito K., and Kusano M.. 2009. Compensation for systematic cross-contribution improves normalization of mass spectrometry based metabolomics data. Anal. Chem. 81: 7974–7980. [DOI] [PubMed] [Google Scholar]
- 34.Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- 35.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu T., Ohto T., and Kita Y.. 2006. Cytosolic phospholipase A2: biochemical properties and physiological roles. IUBMB Life. 58: 328–333. [DOI] [PubMed] [Google Scholar]
- 37.Shindou H., and Shimizu T.. 2009. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5. [DOI] [PubMed] [Google Scholar]
- 38.Quehenberger O., and Dennis E. A.. 2011. The human plasma lipidome. N. Engl. J. Med. 365: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han R. H., Wang M., Fang X., and Han X.. 2013. Simulation of triacylglycerol ion profiles: bioinformatics for interpretation of triacylglycerol biosynthesis. J. Lipid Res. 54: 1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imielinski M., Cha S., Rejtar T., Richardson E. A., Karger B. L., and Sgroi D. C.. 2012. Integrated proteomic, transcriptomic, and biological network analysis of breast carcinoma reveals molecular features of tumorigenesis and clinical relapse. Mol. Cell Proteomics. 11: M111:014910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaushik A. K., Shojaie A., Panzitt K., Sonavane R., Venghatakrishnan H., Manikkam M., Zaslavsky A., Putluri V., Vasu V. T., Zhang Y., et al. 2016. Inhibition of the hexosamine biosynthetic pathway promotes castration-resistant prostate cancer. Nat. Commun. 7: 11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atsuzawa K., Nakazawa A., Mizutani K., Fukasawa M., Yamamoto N., Hashimoto T., and Usuda N.. 2010. Immunohistochemical localization of mitochondrial fatty acid beta-oxidation enzymes in Muller cells of the retina. Histochem. Cell Biol. 134: 565–579. [DOI] [PubMed] [Google Scholar]
- 43.Tserentsoodol N., Gordiyenko N. V., Pascual I., Lee J. W., Fliesler S. J., and Rodriguez I. R.. 2006. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol. Vis. 12: 1319–1333. [PubMed] [Google Scholar]
- 44.Tyni T., Paetau A., Strauss A. W., Middleton B., and Kivela T.. 2004. Mitochondrial fatty acid beta-oxidation in the human eye and brain: implications for the retinopathy of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Pediatr. Res. 56: 744–750. [DOI] [PubMed] [Google Scholar]
- 45.Vancura P., Wolloscheck T., Baba K., Tosini G., Iuvone P. M., and Spessert R.. 2016. Circadian and dopaminergic regulation of fatty acid oxidation pathway genes in retina and photoreceptor cells. PLoS One. 11: e0164665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joyal J. S., Sun Y., Gantner M. L., Shao Z., Evans L. P., Saba N., Fredrick T., Burnim S., Kim J. S., Patel G., et al. 2016. Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat. Med. 22: 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox T. E., Young M. M., Pedersen M. M., Han X., Gardner T. W., and Kester M.. 2012. Diabetes diminishes phosphatidic acid in the retina: a putative mediator for reduced mTOR signaling and increased neuronal cell death. Invest. Ophthalmol. Vis. Sci. 53: 7257–7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chicco A. J., and Sparagna G. C.. 2007. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am. J. Physiol. Cell Physiol. 292: C33–C44. [DOI] [PubMed] [Google Scholar]
- 49.Claypool S. M., and Koehler C. M.. 2012. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiebish M. A., Han X., Cheng H., Chuang J. H., and Seyfried T. N.. 2008. Cardiolipin and electron transport chain abnormalities in mouse brain tumor mitochondria: lipidomic evidence supporting the Warburg theory of cancer. J. Lipid Res. 49: 2545–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geraldes P., and King G. L.. 2010. Activation of protein kinase C isoforms and its impact on diabetic complications. Circ. Res. 106: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quagliaro L., Piconi L., Assaloni R., Martinelli L., Motz E., and Ceriello A.. 2003. Intermittent high glucose enhances apoptosis related to oxidative stress in human umbilical vein endothelial cells: the role of protein kinase C and NAD(P)H-oxidase activation. Diabetes. 52: 2795–2804. [DOI] [PubMed] [Google Scholar]
- 53.Szefel J., Kruszewski W. J., and Sobczak E.. 2015. Factors influencing the eicosanoids synthesis in vivo. BioMed Res. Int. 2015: 690692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pande M., Hur J., Hong Y., Backus C., Hayes J. M., Oh S. S., Kretzler M., and Feldman E. L.. 2011. Transcriptional profiling of diabetic neuropathy in the BKS db/db mouse: a model of type 2 diabetes. Diabetes. 60: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stöckli J., Fisher-Wellman K. H., Chaudhuri R., Zeng X. Y., Fazakerley D. J., Meoli C. C., Thomas K. C., Hoffman N. J., Mangiafico S. P., Xirouchaki C. E., et al. 2017. Metabolomic analysis of insulin resistance across different mouse strains and diets. J. Biol. Chem. 292: 19135–19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia J., Sinelnikov I. V., Han B., and Wishart D. S.. 2015. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 43: W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia J., and Wishart D. S.. 2011. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 6: 743–760. [DOI] [PubMed] [Google Scholar]
- 58.Yilmaz M., Claiborn K. C., and Hotamisligil G. S.. 2016. De novo lipogenesis products and endogenous lipokines. Diabetes. 65: 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.