Abstract
Molecular and genomic studies have shown the presence of a large number of SPX gene family members in plants, some of which have been proved to act in P signalling and homeostasis. In this study, the molecular and evolutionary characteristics of the SPX gene family in plants were comprehensively analysed, and the mechanisms underlying the function of SPX genes in P signalling and homeostasis in the model plant species Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), and in important crops, including wheat (Triticum aestivum), soya beans (Glycine max) and rapeseed (Brassica napus), were described. Emerging findings on the involvement of SPX genes in other important processes (i.e. disease resistance, iron deficiency response, low oxygen response and phytochrome-mediated light signalling) were also highlighted. The available data suggest that SPX genes are important regulators in the P signalling network, and may be valuable targets for enhancing crop tolerance to low P stress. Further studies on SPX proteins should include more diverse members, which may reveal SPX proteins as important regulatory hubs for multiple processes including P signalling and homeostasis in plants.
Keywords: plant, SPX gene, gene family, P signalling and homeostasis, functional analysis
1. Introduction
The SPX family was named after SYG1, PHO81 and Xpr1, the first three SPX gene members identified [1]. SYG1 and PHO81 encode yeast gpa1 suppressor and cyclin-dependent kinase, respectively, while Xpr1 codes for the xenotropic and polytropic retrovirus receptor 1 in humans. More and more studies have shown that SPX genes are involved in phosphorus (P) signalling and homeostasis and are prevalent in plants, and phosphate transport is impaired if the SPX domain is mutated [2]. P is an indispensable macroelement required for normal plant growth and development, and P content is quite high in plant tissues. P is not only an important component of membranes and nucleic acids, but also plays important roles in diverse physiological processes, including photosynthesis, enzyme activity regulation, respiration, signal transduction, oxidation–reduction reactions, energy metabolism and carbon metabolism [3–6]. Plants absorb P mainly from the soil through their roots [7,8]. Soil P exists primarily in the forms of calcium, iron and aluminium salts, and organic molecules, which are difficult for the roots to absorb [9]. This decreases the bioavailability of P, leading to an available P content in the soil that is far lower than that required for normal plant growth [10–12]. In plant cells, P concentration in the cytosol is approximately 60–80 µM [13], which is much higher than the concentration of available P in the soil (less than 10 µM) [14]. Thus, plants are usually under low P conditions. The phenotypic symptoms of P deficiency are mainly dark-green leaf colour, reduced elongation rate of shoot and decreased leaf size [15]. To improve crop yield in agricultural production, a large quantity of P fertilizers is often applied to solve the problem of P deficiency. However, this approach can cause not only water eutrophication but also overexploitation and consumption of phosphate ore, which is non-renewable. Solving this problem is critical to environmental protection and sustainable development. To adapt to P-deficient environments, plants undergo phenotypic changes in the root system to increase the absorption of P from the soil [16–19]. A series of P transport mechanisms at the molecular level are gradually established to overcome P starvation and to maintain P homeostasis, in which phosphate transporter is the basic effector involved in P uptake, transfer and storage [20,21]. These mechanisms are controlled by complex and sophisticated molecular regulatory networks. Recently, many genes have been identified and functionally linked to these molecular regulatory networks, including various protein-encoding genes and non-coding RNA genes [22]. Hence, the discovery of genes conferring low P tolerance has great importance.
In this study, the structural and evolutionary characteristics of the SPX gene family in plants were systematically analysed, the latest research progress on SPX gene functions was summarized and the mode of action of these genes in the P regulatory signalling network was discussed.
2. Structural characteristics of SPX genes and proteins in plants and their relationships with phosphorus metabolism
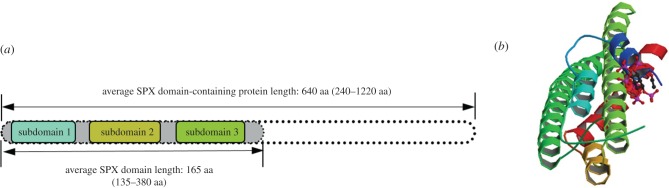
The SPX domain found in eukaryotic proteins is rather conserved and has hydrophilic properties [23]. It is often located at the N-terminus of eukaryotic proteins [23]. SPX domain has an average length of 165 amino acids and can be divided into three subdomains with 30–40 amino acids in each (figure 1). They are separated from each other by low similarity regions [24]. In plants, SPX domains can be grouped into several distinct subfamilies: SPX proteins carrying only SPX domain, SPX–EXS proteins containing SPX and an EXS (ERD1, XPR1 and SYG1) domain, SPX–MFS proteins with SPX and the major facility superfamily (MFS) domain and SPX–RING proteins containing SPX and the RING-type zinc finger domain [25]. Many studies have shown that the SPX domain is closely related to P signalling and plays an important role in maintaining P homeostasis [22,26–28].
Figure 1.
Main structural features of SPX proteins. (a) The N-terminal SPX domain can be divided into three well-conserved subdomains separated by low similarity regions, with 30–40 amino acids in each subdomain (Secco et al. [24]). (b) Crystal structure of the SPX domain in the Vtc4 protein of Chaetomium thermophilum in complex with inositol hexakisphosphate (InsP6). Classification: inositol phosphate-binding protein. SPX helical bundles provide a positively charged ligand-binding surface (Wild et al. [2]; http://www.rcsb.org/pdb/explore.do?structureId=5IJP).
The SPX domain can indicate the phosphate status in fungal, plant and human cells. SPX domain-containing proteins are indispensable for the absorption, transport, storage and signal transduction of inorganic P in eukaryotes. Wild et al. [2] studied the ligand of the SPX domain and suggested that the domain provided a binding surface for small molecules (inositol polyphosphate signalling molecules, InsPs). In this way, the balance of P in plant cells can be regulated by the binding of different InsPs to SPX. In phosphate-deficient plant cells, InsPs bind to SPX domains, being able to interact with several other proteins involved in the regulation of P signalling in plants [2,29]. If the SPX domain is mutated, then phosphate transport capacity is impaired [2], highlighting the unique importance of the SPX domain in P metabolism.
P exists in different molecular forms in plants to serve their needs at different times [30]. One form is inorganic polyphosphate (polyP). PolyP includes hundreds of types of phosphoric anhydrides, which can be hydrolysed to meet the needs of various molecular processes [31–33]. In yeast, the vacuolar transporter chaperone (VTC) complex can synthesize polyP [34]. The VTC is a fairly large protein complex (Vtc1–5) located on the vacuole membrane [35,36]. Approximately 80% of VTC proteins contain an SPX domain, which may contribute to P homeostasis in the cells [5]. Additionally, an important role for the phosphate starvation response 1 (PHR1) protein has been identified in the P signalling network. This MYB-like transcription factor is homologous to phosphorus starvation response 1 (PSR1), which participates in the P sensing process in Chlamydomonas reinhardtii [37,38]. PHR1 regulates the expression of AtACP5, AtIPS1, PHT1.1 and RNS1 [39,40], as well as the expression of several PSR genes, including microRNA399 and the SPX genes [38], by binding to their promoters through the cis-element PHR1-binding sequence (P1BS; GNATATNC) [40–42].
3. Evolutionary analysis of SPX gene family in plants
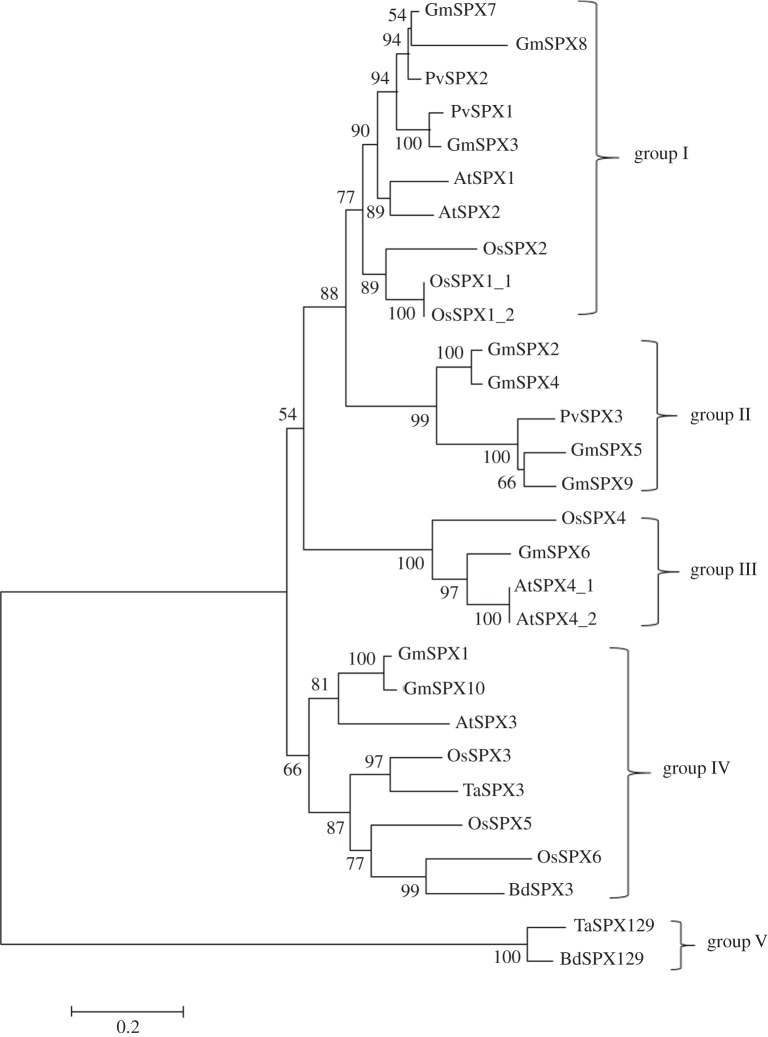
Through analysis of existing plant genomic sequences, 20 SPX gene family members have been identified in Arabidopsis, including four SPX genes whose deduced proteins contain only the SPX domain [26]. Meanwhile, 15 SPX gene family members are identified in rice, including six SPX genes whose products carry only the SPX domain [24]. Numerous SPX gene family members also exist in the genomes of legumes and other important crops [43,44]. A phylogenetic tree was constructed for some of the important SPX genes using the multi-sequence alignment generated by ClustalW and the neighbour-joining method with 1000 bootstrap replicates in MEGA software (figure 2). The results showed evolutionary divergence in SPX genes, and the compared genes were clustered into five types of sub-structures (numbered I, II, III, IV and V). Most of the SPX genes fell into types I and IV, whereas no more than five genes fell into each of the other three types. GmSPX8, GmSPX5, OsSPX4, OsSPX6 and TaSPX129 genes exhibited the most rapid evolution for each type. The paralogues of these genes included AtSPX1/AtSPX2, GmSPX2/GmSPX4, GmSPX5/GmSPX9, OsSPX1/OsSPX2, GmSPX1/GmSPX10 and OsSPX5/OsSPX6. Each gene may evolve under different evolutionary pressure and may possibly acquire new function during the course of evolution.
Figure 2.
Phylogenetic tree of SPX domain-containing proteins from different plant species. The tree was constructed with the neighbour-joining method by MEGA with 1000 bootstrap replicates. At, Arabidopsis thaliana; Os, Oryza sativa; Pv, Phaseolus vulgaris; Gm, Glycine max; Ta, Triticum aestivum; Bd, Brachypodium distachyon.
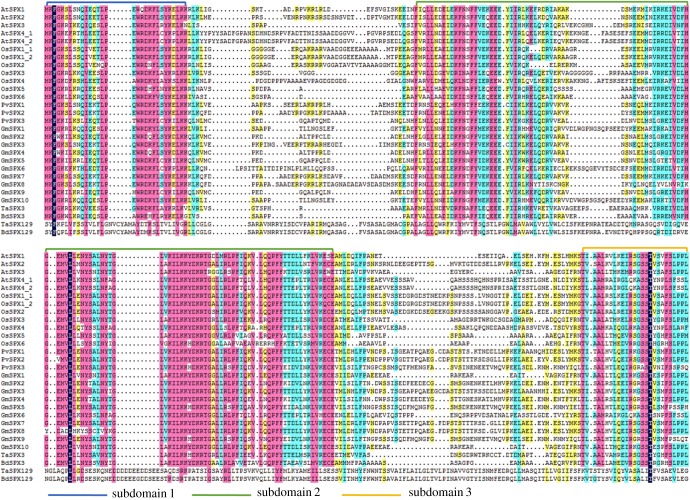
A multiple sequence alignment of the SPX domains was analysed (figure 3). The result showed a high degree of similarity in the SPX domains in Arabidopsis, rice, common beans, soya beans, wheat and Brachypodium. The SPX domains in TaSPX129 and BdSPX129 were of SPX–MFS type, and have an increased length, whereas the remaining SPX domains ranged from 200 to 400 amino acids. Amino acid point mutations are the main sources of variation, which may affect the role of SPX genes in P regulatory networks in plants. Notably, the SPX domains in TaSPX129 and BdSPX129 had many insertions (figure 3); whether this is caused by the presence of an additional MFS domain is a matter for further study.
Figure 3.
Multiple alignment of the SPX domains in different SPX proteins. The multiple alignment was generated using DNAMAN with different colours representing different homology of amino acids. The three subdomains are distinguished by coloured brackets. At, Arabidopsis thaliana; Os, Oryza sativa; Pv, Phaseolus vulgaris; Gm, Glycine max; Ta, Triticum aestivum; Bd, Brachypodium distachyon.
4. Research progress on the functional analysis of SPX genes in plants
A large number of SPX domain-containing proteins have been identified in plants [11]. Here, SPX proteins refer to the proteins that contain only the SPX domain. Owing to their presence in different subcellular structures, they may have different functions in the P signalling network. Here, we summarize the studies on the genes whose proteins contain only the SPX domain, including SPX1–4 in Arabidopsis, SPX1–6 in rice, SPX1–10 in soya beans, SPX1–3 in common bean and TaSPX129 in wheat (table 1).
Table 1.
List of the plant SPX genes whose function has been analysed to some extent. N, cell nucleus; M, cell membrane; C, cytoplasm; +, increase; +(*), increase (except seeds); +(**), increase (except flowers and seeds); =, no difference; −, decrease; Pr, positive regulation; Pr*, positive regulation (except PvPDR2-like); Nr, negative regulation.
| species | gene | protein location | expression after Pi starvation | regulation of PSI gene | main functional characteristics | source |
|---|---|---|---|---|---|---|
| Oryza sativa | OsSPX1 | N | + | Nr | OsSPX1 can interact with OsPHR2 and acts as a negative regulator of OsPHR2. OsSPX1 regulates OsSPX2, 3 and 5 at the transcriptional level, and the repression of OsSPX1 results in excessive P accumulation in the shoot | Wang et al. [27,28] |
| OsSPX2 | N | + | OsSPX2 can interact with OsPHR2 and acts as a negative regulator of OsPHR2. PHR2, SPX1 and SPX2 constitute a regulatory feedback loop in P signalling | Wang et al. [28,45] | ||
| OsSPX3 | N/C | + | Nr | OsSPX3 plays an important role in OsIPS1/miR399-mediated long distance regulation on OsPHO2 and acts as a negative regulator of OsPHR2. OsSPX3 negatively regulates the root-to-shoot transportation of P. Overexpression of OsSPX3 inhibits plant growth, which is more severe under P-deficient conditions | Wang et al. [28] Shi et al. [46] |
|
| OsSPX4 | N/C | = | OsSPX4 can interact with OsPHR2 in the cytoplasm and inhibits translocation of PHR2 into the nucleus. OsSPX4 functions as a negative regulator of PHR2 and can affect the activity of OsPHR2, sequentially regulating downstream gene expression | Lv et al. [47] | ||
| OsSPX5 | N/C | + | Nr | OsSPX5 and OsSPX3 are paralogous genes. SPX3/5 proteins act as repressors of PHR2. Overexpression of SPX3 and SPX5 completely rescues the excessive shoot of P accumulation. SPX3/5 negatively regulates P transport from roots to leaves with redundant function | Shi et al. [46] Zhang et al. [43] |
|
| OsSPX6 | + | OsSPX6, as a paralogue of SPX3/5, may play a compensatory role | Shi et al. [46] | |||
| Arabidopsis thaliana | AtSPX1 | N | + | Pr | AtSPX1 can interact with AtPHR1 and may act as a negative regulator of AtPHR1 in P concentration | Duan et al. [26] Qi et al. [48] |
| AtSPX2 | N | + | AtSPX2 can interact with AtPHR1 in the cell nucleus. AtSPX1 and AtSPX2 have functional redundancy with one another | Puga et al. [49] | ||
| AtSPX3 | M/C | + | Nr | Partial repression of AtSPX3 can exacerbate phosphate-deficiency symptoms, alter P allocation and enhance the expression of a subset of phosphate starvation responsive genes including AtSPX1 | Duan et al. [26] | |
| AtSPX4 | M | − | AtSPX4 can interact with AtPHR1 in the cytoplasm | Duan et al. [26] | ||
| Glycine max | GmSPX1 | N/C | +(*) | Nr | GmSPX1 interacts with a newly identified P starvation-induced transcription factor GmMYB48, and this interaction may represent a potential suppressor of P signalling network in soya bean | Zhang et al. [43] |
| GmSPX2, 4, 6, 9 and 10 | N/C | + | — | Yao et al. [50] | ||
| GmSPX3, 7 and 8 | N | + | GmSPX3 overexpression results in increased P concentration in both leaf and root tissues under high P conditions, which correlates with elevated transcript levels of several PSI genes in the root hairs | Yao et al. [50] | ||
| GmSPX5 | N/C | +(**) | — | Yao et al. [50] | ||
| Phaseolus vulgaris | PvSPX1 | N | + | Pr* | Overexpression of PvSPX1 results in increased P concentration in the roots, morphological change in root hairs, inhibition of main root growth, more numerous lateral roots and upregulated transcription of 10 PSR genes | Yao et al. [44] |
| PvSPX2 | N | + | Pr* | PvSPX2 participates in P signalling pathway in both shoot and root tissues. Overexpression of PvSPX2 results in increased transcription of several genes downstream from PvSPX1, suggesting that PvSPX2 might have a similar regulatory role as PvSPX1 | Yao et al. [44] | |
| PvSPX3 | N/C | + | = | PvSPX2 participates in P signalling pathway in both shoot and root tissues. PvSPX3 expression is less sensitive to P deficiency compared with that of PvSPX1 and PvSPX2 | Yao et al. [44] | |
| Triticum aestivum | TaSPX129 | + | — | Fang et al. [51] | ||
| TaSPX | TaSPX participates in high temperature-induced resistance to wheat stripe rust | Wei et al. [52] |
4.1. Functional analysis of SPX genes in Arabidopsis
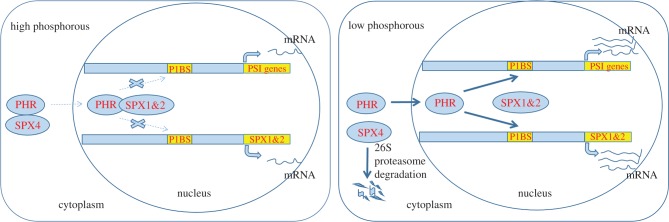
Twenty SPX domain-containing proteins were found in Arabidopsis, among which four proteins contained only the SPX domain (AtSPX1–AtSPX4) [26]. AtSPX1, localized in the nucleus, is a P-dependent suppressor of AtPHR1 in Arabidopsis [53]. AtPHR1 overexpression results in an increase in P concentration in the shoot and induces the expression of a series of Pi starvation-induced (PSI) genes that encode phosphate transporter, phosphatase or RNase [54,55]. Co-immunoprecipitation experiment showed that the AtSPX1/AtPHR1 interaction was strongly dependent on P level. AtSPX1 is a competitive suppressor that binds AtPHR1 through its recognition sequence. The working model in figure 4 depicts the interactions of AtSPX with AtPHR1 in response to cellular Pi concentration for PSI transcription. Under high P conditions, AtSPX1 has a high binding affinity for AtPHR1, and thus the process by which AtPHR1 regulates PSI genes through the P1BS is inhibited, resulting in a decrease in PSR gene expression. Under P deficiency conditions, the AtSPX1/AtPHR1 interaction weakens, thus facilitating the binding of AtPHR1 to the P1BS to regulate PSR gene expression [49].
Figure 4.
Interaction of SPX1, SPX2 and SPX4 with PHR under high or low P conditions. Under high P conditions, SPX1 and SPX2 in the nucleus and SPX4 in the cytoplasm bind PHR with high affinity, thus inhibiting PHR binding to the P1BS motif in the promoters of PSI genes and leading to repression of the transcription of PSI genes, including SPX1 and SPX2. Under low P conditions, SPX4 in the cytoplasm is degraded via the 26S proteasome pathway, which promotes the targeting of PHR to the nucleus, thereby releasing PHR to activate downstream PSI gene expression. Meanwhile, SPX1 and SPX2 in the nucleus interact with PHR at a low affinity, which facilitates PHR to bind to the P1BS motif in the promoters of PSI genes, further enhancing the expression of PSI genes, including SPX1 and SPX2. The thick arrow denotes enhancement. The dotted lines represent reduced effects. This working model is drawn based on the studies of SPX1, SPX2 and SPX4 genes in Arabidopsis and rice (Lv et al. [47]; Wang et al. [27,28]; Zhou et al. [53]; Puga et al. [49]).
In Arabidopsis, no significant phenotypic differences have been found among the single-gene knockout mutants of Atspx1, Atspx2 and Atspx4 under P-sufficient or -starvation conditions. However, in plants with AtSPX1 overexpression, the expression levels of some PSI genes (i.e. ACP5, PAP2 and RNS1) are significantly increased regardless of P concentration, thus suggesting that AtSPX1 may function in the transcriptional regulation of P starvation. Additionally, inhibition of AtSPX3 through RNAi can change the phenotypes and gene expression levels under P-starvation conditions, rendering an increase in P concentration in the shoot tissues and a reduced P concentration in the roots [26,43]. The expression levels of AtPHT1–4, AtPHT1–5, AtACP5, AtRNS and AtAT4 in spx3 deletion mutants are increased irrespective of P concentration [43], indicating that AtSPX3 is a negative regulator of the signalling process of P starvation. Collectively, these results indicate that SPX proteins have functional redundancy with one another and can serve as an important role in regulating Pi signalling and homeostasis in plants.
4.2. Functional analysis of SPX genes in rice
P is an important nutrient element that limits the yield of rice. Studies of P relevant genes in rice, especially SPX genes, can potentially aid rice yield improvement. A total of 15 SPX domain-containing proteins have been identified in rice, of which only six are SPX proteins (OsSPX1–OsSPX6) [5]. OsSPX1 inhibits P uptake and P-starvation signalling through negative feedback regulation [27,55]. OsSPX1 is induced by P starvation in the roots, and inhibition of OsSPX1 by RNAi leads to an excessive accumulation of P and thus induces severe toxicity. This phenotype is similar to that observed in the plants overexpressing OsPHR2 and the pho2 mutant. OsPHR2 overexpression leads to increased PSI gene expression, including IPS1 and PT2, which promotes excessive P absorption and accumulation and results in leaf necrosis. Quantitative polymerase chain reaction (qRT-PCR) assay showed that OsSPX1 expression was strongly induced in the plants with OsPHR2 overexpression and the pho2 mutant, suggesting that OsSPX1 may function downstream from PHO2 and OsPHR2. Wang et al. [27] analysed the expression levels of 10 genes involved in the rice P-starvation signalling pathway. OsPT2 and OsPT8 were significantly induced in OsSPX1 RNAi plants, pointing to increased P transport and accumulation. By contrast, OsSPX1 overexpression inhibited the expression of 10 phosphate starvation-mediated genes, including IPS1, IPS2, OsPAP1, OsSQD2 (sulfo quinovosyl diacylglycerol 2), miR399d, miR399j, OsPT2, OsPT3, OsPT6 and OsPT8. However, in the double mutant plants with overexpression of OsPHR2 and OsSPX1, P concentration and PSR gene expression levels were basically the same as those in wild-type plants, which indicated that OsSPX1 was a negative regulator of OsPHR2-mediated signal transduction. Previous studies found that overexpression of OsSPX1, OsSPX2, OsSPX3, OsSPX4 and OsSPX5 attenuated the phenotype of OsPHR2 overexpression [45–47,53]. Thus, OsSPX1, OsSPX2 and OsSPX4 may interact with OsPHR2 and inhibit its binding to the P1BS cis-acting element (figure 4). The interaction between SPX proteins and PHR1/2 was strongly dependent on P concentration [45,47]. Knockout of OsSPX1, OsSPX2 and OsSPX4 results in P accumulation in the shoot and significant leaf tip necrosis [45,47]. P accumulation and leaf necrosis also occurred in the Osspx3 and Osspx5 double mutant, and the expression levels of PSI genes, including IPS1, miR399, PT2, miR827, PAP10 and SQD2, were significantly upregulated [46]. This observation indicated that OsSPX3 and OsSPX5 were homologous and that they responded to P stress at the transcriptional and post-transcriptional levels. Collectively, the data gathered so far support the function of the studied OsSPX genes in rice tolerance to P deficiency by regulating P acquisition and its transport from roots to leaves.
4.3. Functional analysis of SPX genes in legumes
Phylogenetic analysis demonstrated that GmSPXs 1–10 can be divided into three groups [43]. Quantitative PCR assay showed that the expression of these genes was significantly increased under low P conditions and decreased rapidly one day after P supplementation [43]. The expression of these genes was highly sensitive to low P conditions. Overexpression of GmSPX3 led to increased P concentration in both leaf and root tissues, and increased transcriptional levels of seven PSI genes in the root hairs, under high P conditions [44]. Analysis of GmSPX1 overexpression in Arabidopsis spx3 mutant showed that GmSPX1 negatively regulated many PSR genes, including AtPHT1–4, AtPHT1–5, AtACP5, AtRNS and AtAT4, in a P level-dependent manner [43]. Furthermore, GmSPX1 interacted with a newly identified P starvation-induced transcription factor GmMYB48, and this interaction may represent a potential suppressor module of the P signalling network in soya bean [43]. Three SPX proteins (PvSPX1–PvSPX3) have been found in common bean (Phaseolus vulgaris), and their expression levels were significantly increased in roots and leaves under P-starvation conditions [44]. PvSPX1 is localized in the nucleus and exhibits a more sensitive and rapid response to P starvation. Overexpression of PvSPX1 resulted in an increase in P concentration in root tissues, a configuration change of root hairs, growth inhibition of the main root and an increase in the number of lateral roots, accompanied by upregulated transcription of 10 PSR genes [44]. Further studies showed that PvPHR1 overexpression increased PvSPX1 transcription level, and thus PvSPX1 may act downstream from PvPHR1 [44,50].
4.4. Functional analysis of SPX genes in wheat
P deficiency is also a primary factor constraining the yield of wheat [15]. Therefore, identification of P-regulated genes and breeding of low P-tolerant cultivars are of prime importance to increasing global wheat productivity without excessive use of P fertilizers. Owing to the possession of a complex hexaploid genome, functional analysis of P-regulated genes (including SPX members) in common wheat (2n = 6x = 42) is lagging behind that in Arabidopsis and rice. Shang et al. [56] found that TaSPX3 was strongly induced by low P stress, but became significantly downregulated when P supply was restored; the expression profile of TaSPX3 differed among cultivars, indicating that the mechanism of low P stress response may vary among different wheat genotypes. Shukla et al. [57] demonstrated that the relative transcriptional level of TaSPX1 was higher in the aleurone than in the endosperm in developing wheat grains, which paralleled the accumulation of more P in aleurone tissues.
4.5. Functional analysis of SPX genes in rapeseed
Du et al. [11] analysed 69 SPX gene family members in rapeseed (Brassica napus) and found that the expression levels of different BnaSPX genes differed under P-starvation conditions. The expression levels of nine genes in the SPX subfamilies were significantly induced by P starvation and rapidly declined upon P supplementation. Analysis of two BnaSPX1 genes (i.e. BnaA2.SPX1 and BnaC3.SPX1) in transgenic Arabidopsis revealed functional difference between them: the transgenic lines of BnaA2.SPX1, but not those of BnaC3.SPX1, showed retarded growth and higher sensitivity to P deficiency when compared with wild-type control [11]. In two other studies, BnSPX3;1 and BnSPX3;2 were found specifically induced by P deficiency and that the induction was rapid and reversible [58,59]. Unlike P deficiency, the deprivation of other nutrients (N, K, S or Fe) did not affect the transcription of BnSPX3;1 and BnSPX3;2, and thus the two genes may be used as markers for assessing P-starvation status in plants [58,59].
5. Discussion and prospects
The available studies clearly suggest that SPX proteins occupy a very important position in the P signalling network, which is tightly related to P uptake, transport, storage and homeostasis. Most of the studied SPX genes are low P inducible and can influence the transcription of downstream PSI (PSR) genes by regulating PHR activity, likely via controlling the movement of PHR from the cytoplasm to the nucleus and by decreasing the binding of PHR to the P1BS cis-element [45,47,49]. Not surprisingly, current understandings on SPX genes are largely based on the data from model plants (i.e. Arabidopsis and rice). The results from complex crop plants (e.g. legumes, common wheat and rapeseed) are much less. Nevertheless, they complemented and expanded the insights obtained from model species, and yielded potential clues and targets for enhancing crop tolerance to low P stress. Considering the urgent need in developing P-efficient cultivars [60], more efforts should be devoted to studying SPX gene functions in crop plants. The accumulation of ever more genomic resources [61], as well as the rapid development of gene-editing technologies for diverse plant species [62], will facilitate such efforts.
Past investigations have mainly concerned relatively simple SPX proteins, i.e. those with only one SPX domain. Future studies should cover more complex SPX proteins that carry extra domains in addition to SPX. The presence of extra domains may confer multiple functions to SPX proteins. This possibility may be illustrated by the analysis of Arabidopsis PHO1 (AtPHO1), which carries the EXS domain in addition to SPX. Although originally found required for xylem loading of inorganic phosphate [63,64], recent investigations have indicated the likely involvement of AtPHO1 in the cross talk among P, sucrose and phytohormone signalling pathways [65].
Future studies will also shed new light on the involvement of SPX proteins in other vital plant processes. There is emerging evidence for the participation of SPX domain-containing proteins in disease resistance [52], iron deficiency response [66], low oxygen response [67] and phytochrome-mediated light signalling [68]. Considering the diverse and fundamental roles of P in cellular organisms, it may not be surprising to find that SPX proteins act as important regulatory hubs for multiple processes including the fine tuning of P signalling and homeostasis in plants.
Data accessibility
This article has no additional data.
Author contributions
All authors contributed to the production of this article.
Competing interests
We declare we have no competing interests.
Funding
This study was supported by the Distinguished Scholar Program of Henan Province (154200510024), the Key Project of Henan Province (161100110400) and the Innovation Fund of Henan Agricultural University (KJCX2017A13).
References
- 1.Wang Y, Ribot C, Rezzonico E, Poirier Y. 2004. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. (doi:10.1104/pp.103.037945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild R, et al. 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. (doi:10.1126/science.aad9858) [DOI] [PubMed] [Google Scholar]
- 3.Chen JY. 2008. QTL mapping of phosphorus efficiency and P-related traits in maize (Zea mays L.). Doctoral dissertation, Southwest University, Chongqing, People's Republic of China: In Chinese. [Google Scholar]
- 4.Fang ZY, Shao C, Meng YJ, Wu P, Chen M. 2009. Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci. 176, 170–180. (doi:10.1016/j.plantsci.2008.09.007) [Google Scholar]
- 5.Secco D, Wang C, Shou H, Whelan J. 2012. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 586, 289–295. (doi:10.1016/j.febslet.2012.01.036) [DOI] [PubMed] [Google Scholar]
- 6.Marschner H. 1995. Mineral nutrition of higher plants, 2nd edn London, UK: Academic Press. [Google Scholar]
- 7.Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. 2011. Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2, 83 (doi:10.3389/fpls.2011.00083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu FP, Zhang LJ, Gu JT, Li XJ, Guo CJ, Lu WJ, Xiao K. 2012. Cloning and molecular characterization analysis of TaPT4, a phosphate transporter gene in wheat (Triticum aestivum L.). J. Agric. Univ. Hebei 35, 1–7 (in Chinese). (doi:10.3969/j.issn.1000-1573.2012.03.001) [Google Scholar]
- 9.Raghothama KG, Karthikeyan AS. 2005. Phosphate acquisition. Plant Soil 274, 37–49. (doi:10.1007/s11104-004-2005-6) [Google Scholar]
- 10.Hinsinger P. 2001. Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237, 173–195. (doi:10.1023/A:1013351617532) [Google Scholar]
- 11.Du H, Yang C, Din G, Shi L, Xu F.. 2017. Genome-wide identification and characterization of SPX domain-containing members and their responses to phosphate deficiency in Brassica napus. Front. Plant. Sci. 8, 35 (doi:10.3389/fpls.2017.00035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schachtman DP, Reid RJ, Ayling SM. 1998. Phosphorus uptake by plants: from soil to cell. Plant Physiol. 116, 447–453. (doi:10.1104/pp.116.2.447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratt J, Boisson AM, Gout E, Bligny R, Douce R, Aubert S. 2009. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol. 151, 1646–1657. (doi:10.1104/pp.109.144626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaxton WC, Tran HT. 2011. Metabolic adaptation of phosphate-starved plants. Plant Physiol. 156, 1006–1015. (doi:10.1104/pp.111.175281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oono Y, Kobayashi F, Kawahara Y, Yazawa T, Handa H, Itoh T, Matsumoto T.. 2013. Characterisation of the wheat (Triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: gene expression in Pi-stressed wheat. BMC Genomics 14, 77 (doi:10.1186/1471-2164-14-77) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. 2001. Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol. 126, 875–882. (doi:10.1104/pp.126.2.875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L. 2002. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129, 244–256. (doi:10.1104/pp.010934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi T, Zhao D, Li D, Wang N, Meng J, Xu F, Shi L. 2012. Brassica napus root mutants insensitive to exogenous cytokinin show phosphorus efficiency. Plant Soil 358, 61–74. (doi:10.1007/s11104-012-1219-2) [Google Scholar]
- 19.Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. (doi:10.1046/j.1469-8137.2003.00695.x) [DOI] [PubMed] [Google Scholar]
- 20.Gu M, Chen A, Sun S, Xu G. 2016. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 9, 396–416. (doi:10.1016/j.molp.2015.12.012) [DOI] [PubMed] [Google Scholar]
- 21.Luan M, Tang RJ, Tang Y, Tian W, Hou C, Zhao F, Lan W, Luan S. 2016. Transport and homeostasis of potassium and phosphate: limiting factors for sustainable crop production. J. Exp. Bot. 68, 3091–3105. (doi:10.1093/jxb/erw444) [DOI] [PubMed] [Google Scholar]
- 22.Gu M, Chen AQ, Xu GH. 2012. Signaling network in phosphate starvation response and arbuscular mycorrhiza symbiosis in plants. J. Nanjing Agric. Univ. 35, 133–146 (In Chinese). [Google Scholar]
- 23.Stefanovic A, Arpat AB, Bligny R, Gout E, Vidoudez C, Bensimon M, Poirier Y. 2011. Over-expression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 66, 689–699. (doi:10.1111/j.1365-313X.2011.04532.x) [DOI] [PubMed] [Google Scholar]
- 24.Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. 2012. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. (doi:10.1111/j.1469-8137.2011.04002.x) [DOI] [PubMed] [Google Scholar]
- 25.Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. (doi:10.1146/annurev-arplant-042110-103849) [DOI] [PubMed] [Google Scholar]
- 26.Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. 2008. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. (doi:10.1111/j.1365-313X.2008.03460.x) [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Ying S, Huang H, Li K, Wu P, Shou H. 2009. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57, 895–904. (doi:10.1111/j.1365-313X.2008.03734.x) [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. 2009. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 51, 663–674. (doi:10.1111/j.1744-7909.2009.00834.x) [DOI] [PubMed] [Google Scholar]
- 29.Jung JY, Ried MK, Hothorn M, Poirier Y. 2017. Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotech. 49, 156–162. (doi:10.1016/j.copbio.2017.08.012) [DOI] [PubMed] [Google Scholar]
- 30.Elser JJ. 2012. Phosphorus: a limiting nutrient for humanity. Curr. Opin. Biotech. 23, 833–838. (doi:10.1016/j.copbio.2012.03.001) [DOI] [PubMed] [Google Scholar]
- 31.Nocek B, et al. 2008. Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria. Proc. Natl Acad. Sci. USA 105, 17 730–17 735. (doi:10.1073/pnas.0807563105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livermore TM, Chubb JR, Saiardi A. 2016. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl Acad. Sci. USA 113, 996–1001. (doi:10.1073/pnas.1519440113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerasimaite R, Pavlovic I, Capolicchio S, Hofer A, Schmidt A, Jessen HJ, Mayer A. 2017. Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem. Biol. 12, 648–653. (doi:10.1021/acschembio.7b00026) [DOI] [PubMed] [Google Scholar]
- 34.Hothorn M, et al. 2009. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516. (doi:10.1126/science.1168120) [DOI] [PubMed] [Google Scholar]
- 35.Desfougères Y, Gerasimaite R, Jessen HJ, Mayer A. 2016. Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 291, 22 262–22 275. (doi:10.1074/jbc.M116.746784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller O, Neumann H, Bayer MJ, Mayer A. 2003. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 116, 1107–1115. (doi:10.1242/jcs.00328) [DOI] [PubMed] [Google Scholar]
- 37.Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl Acad. Sci. USA 96, 15 336–15 341. (doi:10.1073/pnas.96.26.15336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bari R, Pant BD, Stitt M, Scheible WR. 2006. PHO2, MicroRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. (doi:10.1104/pp.106.079707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Pena A, Leyva A, Paz-Ares J. 2000. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 24, 559–567. (doi:10.1046/j.1365-313x.2000.00893.x) [DOI] [PubMed] [Google Scholar]
- 40.Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. 2001. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. (doi:10.1101/gad.204401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J. 2004. The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 55, 285–293. (doi:10.1093/jxb/erh009) [DOI] [PubMed] [Google Scholar]
- 42.Hammond JP, Broadley MR, White PJ. 2004. Genetic responses to phosphorus deficiency. Ann. Bot. 94, 323–332. (doi:10.1093/aob/mch156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Zhou X, Xu Y, Yao M, Xie F, Gai J, Li Y, Yang S. 2016. Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 248, 82–91. (doi:10.1016/j.plantsci.2016.04.010) [DOI] [PubMed] [Google Scholar]
- 44.Yao ZF, Liang CY, Zhang Q, Chen ZJ, Xiao BX, Tian J, Liao H. 2014. SPX1 is an important component in the phosphorus signalling network of common bean regulating root growth and phosphorus homeostasis. J. Exp. Bot. 65, 3299–3310. (doi:10.1093/jxb/eru183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, et al. 2014. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl Acad. Sci. USA 111, 14 953–14 958. (doi:10.1073/pnas.1404680111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi J, Hu H, Zhang K, Zhang W, Yu Y, Wu Z, Wu P. 2014. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 65, 859–870. (doi:10.1093/jxb/ert424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lv Q, et al. 2014. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. (doi:10.1105/tpc.114.123208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi WJ, Manfield IW, Muench SP, Baker A. 2017. AtSPX1 affects the AtPHR1–DNA-binding equilibrium by binding monomeric AtPHR1 in solution. Biochem. J. 474, 3675–3687. (doi: 10.1042/BCJ20170522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puga MI, et al. 2014. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl Acad. Sci. USA 111, 14 947–14 952. (doi:10.1073/pnas.1404654111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao Z, Tian J, Liao H. 2014. Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann. Bot. 114, 477–488. (doi:10.1093/aob/mcu147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang WB, Ding WW, Xiao K, Li XJ. 2016. Molecular characterization and expression profile of TaSPX129 in wheat under abiotic stress. J. Agric. Univ. Hebei. 39, 41–45 (In Chinese). (doi:10.13320/j.cnki.jauh.2016.0031) [Google Scholar]
- 52.Wei XN, Fan RC, Xu SC, Kang ZS, Zhang XQ. 2011. Function analysis of Taspx gene for high temperature resistance to strip rust in wheat cultivar Xiaoyan54. In Proc. Natl Conf. Plant Biol, Nanning, Guangxi, China, p. 131 (in Chinese). [Google Scholar]
- 53.Zhou Z, Wang Z, Lv Q, Jing S, Zhong Y, Ping W, Mao C. 2015. SPX proteins regulate Pi homeostasis and signaling in different subcellular level. Plant Signal Behav. 10, e1061163 (doi:10.1080/15592324.2015.1061163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nilsson L, Müller R, Nielsen TH. 2007. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 30, 1499–1512. (doi:10.1111/j.1365-3040.2007.01734.x) [DOI] [PubMed] [Google Scholar]
- 55.Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. 2010. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. (doi:10.1111/j.1365-313X.2010.04170.x) [DOI] [PubMed] [Google Scholar]
- 56.Shang WJ, Jia LH, Shi L, Lin DL, Liu N, Zheng WM. 2016. Screening and expression analysis of genes responded to low phosphate in wheat root. J. Nanjing Agric. Univ. 21, 1–10 (In Chinese). [Google Scholar]
- 57.Shukla V, Kaur M, Aggarwal S, Bhati KK, Kaur J, Mantri S, Pandey AK. 2016. Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci. Rep. 6, 39293 (doi:10.1038/srep39293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang G, Ding G, Shi L, Cai H, Xu F. 2012. Characterization of phosphorus starvation-induced gene BnSPX3 in Brassica napus. Plant Soil 350, 339–351. (doi:10.1007/s11104-011-0913-9) [Google Scholar]
- 59.Yang GZ. 2011. Isolation and identification of phosphorus starvation-induced gene BnSPX3, BnIPS1 and their promoters in Brassica napus. Doctoral dissertation, Huazhong Agric University, Wuhan, People's Republic of China: (In Chinese). [Google Scholar]
- 60.Möller K. 2013. Improving the phosphorus efficiency of organic farming systems. Core Organic Newsletter.
- 61.Varshney RK, Glaszmann JC, Leung H, Ribaut JM. 2010. More genomic resources for less-studied crops. Trends Biotechnol. 28, 452–460. (doi:10.1016/j.tibtech.2010.06.007) [DOI] [PubMed] [Google Scholar]
- 62.Gupta RM, Musunuru K. 2014. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Invest. 124, 4154–4161. (doi:10.1172/JCI72992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. 2002. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14, 889–902. (doi:10.1105/tpc.000745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poirier Y, Thoma S, Somerville C, Schiefelbein J. 1991. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97, 1087–1093. (doi:10.1104/pp.97.3.1087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ribot C, Wang Y, Poirier Y. 2008. Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227, 1025–1036. (doi:10.1007/s00425-007-0677-x) [DOI] [PubMed] [Google Scholar]
- 66.Nakanishi H, Okumura N, Umehara Y, Nishizawa NK, Chino M, Mori S. 1993. Expression of a gene specific for iron deficiency (Ids3) in the roots of Hordeum vulgare. Plant Cell Physiol. 34, 401–410. (doi:org/10.1093/oxfordjournals.pcp.a078434) [PubMed] [Google Scholar]
- 67.Sell S, Hehl R. 2005. A fifth member of the tomato 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase gene family harbours a leucine zipper and is anaerobically induced. DNA Seq. 16, 80–82. (doi:10.1080/10425170500050817) [DOI] [PubMed] [Google Scholar]
- 68.Kang X, Ni M. 2006. Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18, 921–934. (doi:10.1105/tpc.105.037879) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.