ABSTRACT
Chrysogine is a yellow pigment produced by Penicillium chrysogenum and other filamentous fungi. Although the pigment was first isolated in 1973, its biosynthetic pathway has so far not been resolved. Here, we show that deletion of the highly expressed nonribosomal peptide synthetase (NRPS) gene Pc21g12630 (chyA) resulted in a decrease in the production of chrysogine and 13 related compounds in the culture broth of P. chrysogenum. Each of the genes of the chyA-containing gene cluster was individually deleted, and corresponding mutants were examined by metabolic profiling in order to elucidate their function. The data suggest that the NRPS ChyA mediates the condensation of anthranilic acid and alanine into the intermediate 2-(2-aminopropanamido)benzoic acid, which was verified by feeding experiments of a ΔchyA strain with the chemically synthesized product. The remainder of the pathway is highly branched, yielding at least 13 chrysogine-related compounds.
IMPORTANCE Penicillium chrysogenum is used in industry for the production of β-lactams, but also produces several other secondary metabolites. The yellow pigment chrysogine is one of the most abundant metabolites in the culture broth, next to β-lactams. Here, we have characterized the biosynthetic gene cluster involved in chrysogine production and elucidated a complex and highly branched biosynthetic pathway, assigning each of the chrysogine cluster genes to biosynthetic steps and metabolic intermediates. The work further unlocks the metabolic potential of filamentous fungi and the complexity of secondary metabolite pathways.
KEYWORDS: Penicillium chrysogenum, chrysogine, filamentous fungi, pigment, secondary metabolites
INTRODUCTION
Penicillium chrysogenum and several other filamentous fungi produce the yellow pigment chrysogine (1, 2). Pigments are known to protect microorganisms against adverse environmental conditions, such as ultraviolet (UV) radiation, and often these compounds also exhibit antimicrobial activity (3). The function of chrysogine has not been extensively investigated, but unlike many other pigments it lacks antimicrobial or anticancer activity (4). N-pyruvoylanthranilamide [2-(2-oxopropanamido)benzamide], a related compound produced by P. chrysogenum (5) and also identified in Colletotrichum lagenarium, is equipped with antiauxin activity (6).
Chrysogine was first isolated in 1973 by Hikino et al. (5), who observed increased production upon feeding with anthranilic acid and pyruvic acid. The putative biosynthetic gene cluster has been identified in P. chrysogenum (7, 8) and includes a nonribosomal peptide synthetase (NRPS). Recently, Wollenberg et al. showed that a dimodular NRPS is responsible for chrysogine biosynthesis in Fusarium graminearum and also suggested a putative cluster (9) homologous to the respective gene cluster of P. chrysogenum. However, the actual biosynthetic pathway has remained elusive.
NRPSs are complex multimodular enzymes that use amino acids and carboxylic acids as substrates (10). The genome of P. chrysogenum contains 10 genes that encode NRPSs (11). Nonetheless, transcriptomic analysis performed on chemostat cultures of P. chrysogenum strain Wisconsin 54-1255 and the industrially improved DS17690 strain showed that only four of these NRPS genes are expressed (11). This set includes three NRPS genes that are involved respectively in the biosynthesis of penicillins (12), roquefortines (13), and hydrophobic cyclic tetrapeptides (14). The fourth highly expressed NRPS gene (7–9) is therefore potentially involved in the biosynthesis of chrysogine, which is among the most abundant secondary metabolites produced by this fungus. Furthermore, five genes flanking the Pc21g12630 gene are also highly coexpressed, suggesting that they form a gene cluster (11).
Here, by overexpression and deletion of the core NRPS gene of the chrysogine pathway, deletion of the individual pathway gene, and by feeding experiments using chemically synthesized intermediates, we elucidate a complex and branched pathway of at least 13 compounds, assigning a function to each enzyme of the biosynthetic gene cluster.
RESULTS
Identification of chrysogine-related compounds.
In order to identify the secondary metabolites produced by the NRPS Pc21g12630, this gene was deleted from P. chrysogenum DS68530 by homologous recombination. In this strain, the penicillin cluster is removed (15, 16), facilitating further identification of other secondary metabolites, as the metabolite profile is not dominated by β-lactams. The strain with the Pc21g12630 gene deleted did not produce chrysogine and 13 other metabolites, here referred to as chrysogine-related compounds (Table 1). This identified Pc21g12630 as the NRPS responsible for chrysogine biosynthesis, and thus this gene was named chyA.
TABLE 1.
Production of chrysogine and related metabolites from strain DS68530a
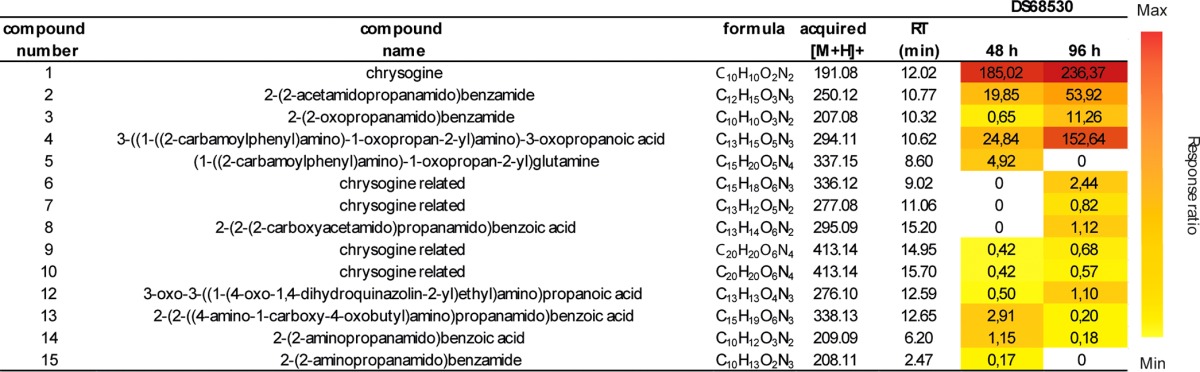
Numbers in the DS68530 columns represent the peak areas of the compounds corrected for the internal standard reserpine. The culture broth of strain DS68530 was analyzed after 48 and 96 h of growth in an SMP medium.
Compounds 1, 2, 3, 4, 8, and 13 were isolated by preparative high-performance liquid chromatography (HPLC), and their structures were determined by nuclear magnetic resonance (NMR) (see the supplemental material). Compound 1 was confirmed to be chrysogine, and compound 3 was identified as N-pyruvoylanthranilamide [2-(2-oxopropanamido)benzamide]. These compounds were first described in P. chrysogenum by Hikino et al. (5). Compound 2 was found to be N-acetylalanylanthranilamide [2-(2-acetamidopropanamido)benzamide], previously isolated from a marine Penicillium species (17). Compounds 4, 8, and 13 were identified as novel metabolites that are clearly related to chrysogine. The structures of compounds 14 [2-(2-aminopropanamido)benzoic acid] and 15 [the amidated form of compound 14, 2-(2-aminopropanamido)benzamidine] were further confirmed by the comparison of their HPLC retention times with those of the independently synthesized standards (see the supplemental material). The structures of compounds 5 and 12 were proposed based on their molecular formulas. We could not assign structures to compounds 6, 7, 9, and 10, which could not be isolated due to their low production.
Transcriptomic analysis performed on chemostat cultures of P. chrysogenum Wisconsin 54-1255 and the industrially improved DS17690 strain showed that five genes flanking chyA (Pc21g12570, Pc21g12590, Pc21g12600, Pc21g12610, and Pc21g12620) were also highly expressed, indicating that they could be part of the chrysogine gene cluster (11) (Fig. 1). Furthermore, quantitative PCR confirmed the expression of the above-listed genes in the DS68530 strain after 48 h of growth in a secondary metabolite production (SMP) medium (Fig. S2). Therefore, we tentatively assigned these as chy genes. Pc21g12640, found adjacent to the chy genes, exhibits a strong similarity to a cutinase transcription factor beta from Fusarium solani (11). Although this gene was not significantly expressed in DS68530, the possible role of Pc21g12640 as regulator of the cluster was also investigated.
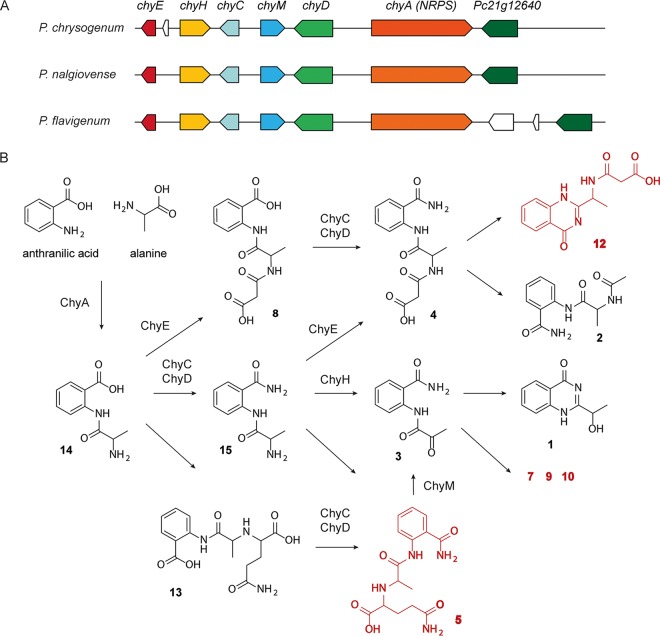
FIG 1.
Representation of the chrysogine biosynthetic gene cluster and proposed pathway. The chrysogine biosynthetic gene cluster in P. chrysogenum and two other chrysogine-producing species. Genes with the same color have >80% identity. This study identified ChyA as the NRPS, ChyE as malonyl transferase, and ChyD as amidase; ChyC participates in amidation reactions, while ChyH and ChyM are involved in oxidation reactions. The substrates of ChyA and the compounds identified in this study are depicted in black, and the putative structures and uncharacterized compounds are represented in red.
Expression of the NRPS gene chyA in a chrysogine cluster deletion strain.
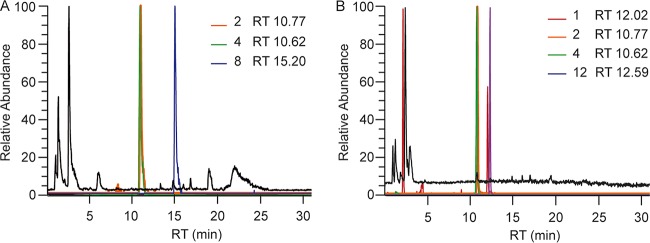
In order to identify the products of the NRPS gene chyA, a chrysogine cluster deletion strain (8) was used to overexpress the chyA gene from an episomal AMA1-based plasmid. The chyA overexpression strain produced compounds 14, 8, and 13 (Fig. 2). It is likely that compound 14 is the immediate product of the NRPS and that this compound is derived from the condensation of anthranilic acid and alanine. Compounds 8 and 13 could be derived from compound 14 by addition of a malonyl and a glutaminyl group, respectively. Our data suggest an immediate branching of the pathway, where two groups of compounds are derived from compounds 8 and 13.
FIG 2.
Chromatogram of culture broth from the chyA-expressing strain. Total ion chromatogram (TIC; black) and extracted ion chromatograms (EIC; colored) of secondary metabolites produced by the chyA-expressing strain after 48 h of growth in an SMP medium.
Metabolite profiles of chy gene deletion strains.
The expression of chyA in a chrysogine cluster deletion strain allowed identification of the product of the NRPS and metabolites produced early in the pathway. To elucidate how the initial products were further modified by the enzymes of the cluster and to resolve the complete pathway, individual chy genes knockout strains were made, and metabolite profiling was performed (Table 2).
TABLE 2.
Secondary metabolites of the chrysogine pathway in the knockout strains compared to that of the parental straina
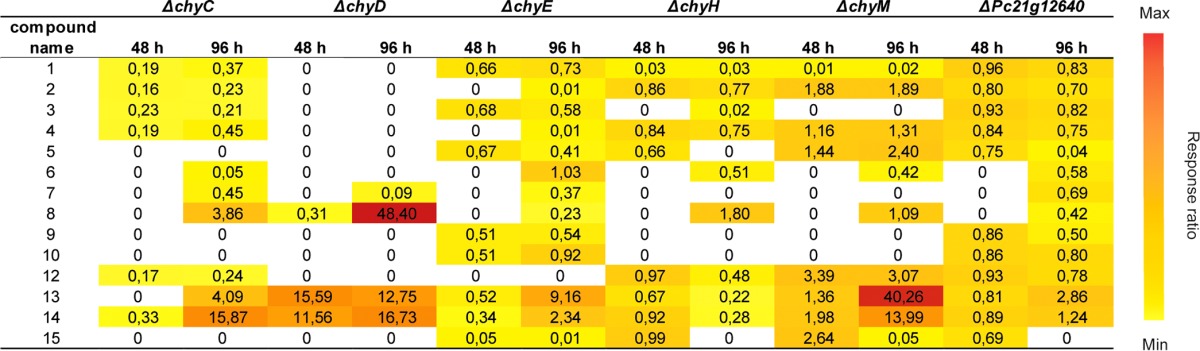
Numbers represent the peak areas of the compounds corrected for the internal standard reserpine and relative to those of the parental strain, DS68530. The culture broth of the strains was analyzed after 48 and 96 h of growth in an SMP medium.
The deletion of chyD led to a depletion of most chrysogine-related metabolites—only compounds 14, 8, and 13 were accumulated during cultivation of this mutant. This suggests that ChyD is an early enzyme in the pathway, being responsible for converting compounds 14, 8, and 13 into downstream compounds. Based on its formula, we propose that compound 14 is converted into compound 15, which is its amidated form.
The ΔchyC strain showed a metabolite profile similar to that of the ΔchyD strain, suggesting that ChyC could be also involved in the conversion of compounds 14, 8, and 13. Nonetheless, downstream compounds were still produced in small amounts in the ΔchyC strain.
In the ΔchyE strain, compounds 2, 4, 8, and 12 were not detected, or were produced in low concentrations compared to the parental strain, suggesting that these compounds belong to the same initial branch of the pathway. Based on the structures and molecular formulas available, compounds 2, 4, and 12 are derived from compound 8, with compound 4 being most likely spontaneously converted into compounds 2 and 12. Since ChyE affected the production of compound 8 and downstream compounds and accumulated compound 14 after 96 h of growth, we propose that this enzyme converts compound 14 into compound 8.
A trend opposite that of the metabolite profile of the ΔchyE strain can be observed in the ΔchyM strain. Peak areas of compounds 2, 4, 8, and 12 were comparable to those of the DS68530 strain, while compounds 1, 3, 7, 9, and 10 were absent or detected in small amounts. This indicates that these compounds are part of an independent branch of the pathway and derived from compound 13. The result is confirmed by the accumulation of compounds 14 and 13 in the ΔchyM strain. The molecular formula of compound 5 suggests that it is derived from compound 13 and that it is the precursor of compound 3, which is further converted into compounds 1, 7, 9, and 10. Because compound 3 and downstream compounds were not produced in this mutant, we propose that ChyM is responsible for the conversion of compound 5 into 3. Chrysogine (compound 1) is likely formed by a spontaneous ring closure from compound 3. Compounds 9 and 10 are isomers, having the same molecular masses but different retention times (RT) in HPLC.
Finally, the ΔchyH strain showed a metabolite profile similar to that of ΔchyM, suggesting that both enzymes are needed for the formation of the same compounds. Nonetheless, the ΔchyH strain did not accumulate compounds 13 and 5, suggesting that ChyH forms compounds 1, 3, 7, 9, and 10 through an independent path. In the analysis of the mutant strains, we could not assign the position of compound 6 in the pathway. Based on the molecular formula, compound 6 could be an unstable precursor of compound 13.
Metabolite profile and gene expression in a strain with a deletion of a putative transcription factor.
Pc21g12640 encodes a putative transcription factor and, because of its chromosomal location in the vicinity of the chrysogine biosynthetic gene cluster, it would be plausible that it acts as a local regulator of this pathway. Although Pc21g12640 is not significantly expressed in the DS68530 strain, transcription factors can regulate transcription even when present at very low levels. Therefore, to investigate its possible role as a regulator of the chrysogine cluster, Pc21g12640 was deleted from strain DS68530. Nonetheless, the ΔPc21g12640 strain did not show any significant changes in the chrysogine-related metabolite profiles compared to those of the parental strain (Table 2). Similarly, quantitative PCR (qPCR) indicated that the deletion of Pc21g12640 did not significantly affect the expression of the genes of the chrysogine cluster (Fig. S2). Thus, Pc21g12640 is not part of the chrysogine biosynthetic gene cluster.
Feeding of the ΔchyA strain with compounds 14 and 15.
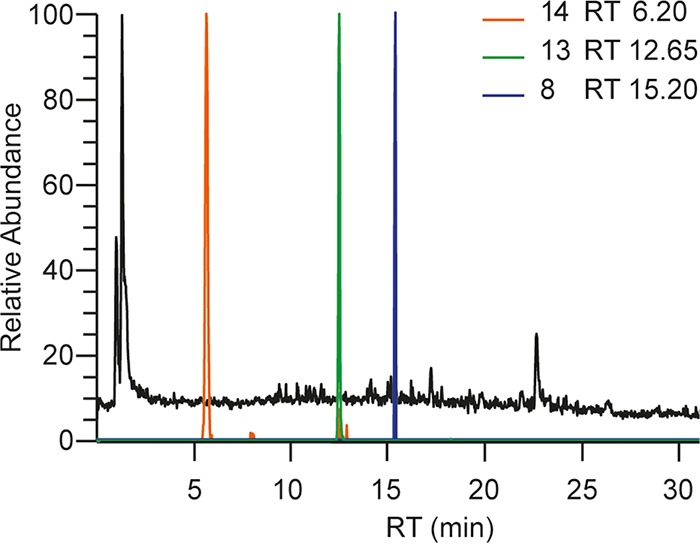
In order to further investigate the role of compounds 14 and 15 as potential NRPS products, the ΔchyA strain was fed with chemically synthesized variants of these. Based on the formula, compound 15 is the amidated form of 14.
We showed above that the expression of chyA in the chrysogine cluster deletion strain resulted in the production of compounds 14, 8, and 13. The ΔchyA strain fed with compound 14 produced compounds 2, 4, and 8, while 13 and downstream compounds were not detected (Fig. 3A). This result suggests that the conversion of compound 14 into 8 is faster than its conversion into 13. The feeding with compound 15 resulted in the production of metabolites that are derived from compounds 8 (compounds 2, 4, 12) and 13 (compound 1; Fig. 3B). As compound 15 is very similar to compound 14, we suggest that compound 15 undergoes the same reactions, being converted into compound 4 by ChyE and into compound 5 by a transaminase. Since ΔchyH affected the production of compound 3 and downstream metabolites without any accumulation of compound 5, we propose that ChyH is involved in the biosynthesis of compound 3 from 15. Therefore, the late metabolites can be formed from two different paths.
FIG 3.
Chromatogram of culture broth from the ΔchyA strain fed with compound 14 or 15. TIC (black) and EIC (colored) of secondary metabolites produced by the ΔchyA strain fed with compound 14 (A) or 15 (B) after 48 h from the feeding.
Distribution and diversity of chrysogine gene clusters in Penicillium species.
Since the above studies characterized the chrysogine biosynthetic gene cluster, the distribution of this gene cluster in other Penicillium species was investigated (Fig. 1). The chy genes and Pc21g12640 from P. chrysogenum were subjected to a BLAST analysis against the genomes of two known chrysogine producers (2), P. nalgiovense and P. flavigenum, recently sequenced by Nielsen et al. (18). These genomes contain a chrysogine gene cluster with a similar gene organization, while a Pc21g12580 homolog is missing, supporting the notion that this gene is not essential for chrysogine biosynthesis. Interestingly, P. flavigenum has two extra genes located near the NRPS gene, suggesting that it may produce additional chrysogine-related metabolites.
DISCUSSION
Chrysogine was isolated from the culture broth of P. chrysogenum in 1973 (5) and was found to be produced also by other filamentous fungi (1, 2). Chrysogine biosynthesis is mediated by a dimodular NRPS that we recently identified in P. chrysogenum (7, 8) and that was also shown to be responsible for chrysogine biosynthesis in Fusarium graminearum (9). Although the biosynthetic gene cluster was suggested, the role of the enzymes in the pathway has thus far not been characterized. In this work, we assigned a function to each enzyme of the cluster and elucidated a complex pathway, validating the compound structures by NMR. The pathway is highly branched, with some enzymes involved in multiple steps of the biosynthesis (Fig. 1).
The NRPS ChyA is a 260-kDa dimodular enzyme which is predicted to contain two adenylation domains. The increased production of chrysogine upon feeding with anthranilic acid and pyruvic acid (5) suggests that these molecules are possible substrates of the NRPS. However, here we identify compound 14 as the direct product of ChyA, showing that the NRPS, in addition to anthranilic acid, utilizes alanine instead of pyruvic acid. However, alanine is readily derived from pyruvic acid by transamination which explains why pyruvic acid stimulates chrysogine production. NRPSsp, NRPSpredictor2, and SEQL-NRPS (19–21) were used for predicting the substrates of ChyA, but results were inconclusive. For the first adenylation domain, phenylalanine, a hydrophobic aliphatic amino acid, 2,3-dihydroxy-benzoic acid, or salicylic acid was predicted, while for the second adenylation domain, proline or a hydrophobic aliphatic amino acid was suggested. This shows that with fungal NRPS genes, predictions can be unreliable, necessitating experimental validation. Compound 14 acts as a substrate for several enzymes, which immediately results in a split in the pathway by forming compounds 8, 13, and 15, the latter being the amidated form of compound 14. Two independent groups of compounds are derived from compounds 8 and 13. Since compound 15 undergoes the same reactions as 14, the more distal metabolites in the pathway can be formed via either of the converging branches.
Transcriptomic data (11) suggested that chyA and five flanking genes could form a cluster. These genes are coexpressed under a set of conditions, whereas expression profiles in the flanking regions of the putative gene cluster vary. Metabolic profiling of the mutant strain indicated that ChyE is a malonyl transferase that can convert compounds 14 and 15 into compounds 8 and 4, respectively. Interestingly, the expression of chyA in a chrysogine cluster deletion strain showed that compound 14 can be converted into compound 8 without involvement of any of the enzymes of the cluster; this conversion likely involves a transferase. In line with this observation, the deletion of chyE did not lead to a complete depletion of compound 8 and downstream metabolites, although it significantly decreased the amounts produced. These data suggest that chyE is part of the biosynthetic cluster, as it is coexpressed together with the other genes (11) and its deletion affects chrysogine metabolite production, but one or more other transferases can catalyze the same reactions. The orthologous gene in Fusarium species is not involved in chrysogine biosynthesis, showing a different expression pattern compared to the genes of the cluster (9).
In addition, compound 13 was formed by the strain that solely expresses chyA, likely through the involvement of a transaminase, whose encoding gene is not part of the gene cluster. Based on sequence alignment, no genes encoding a transaminase have been identified in the immediate vicinity of the chrysogine genes, but the genome contains many transaminases.
Our data indicate that ChyD is an amidase, being responsible for the amidation of the carboxylic acid moiety of compounds 14, 8, and 13, in line with the bioinformatics prediction of ChyD as an asparagine synthetase that amidates aspartate to form asparagine. The ΔchyC strain showed a metabolite profile similar to that of the ΔchyD strain, suggesting that ChyC is involved in the same reactions as ChyD. Indeed, downstream compounds were still produced in small amounts in the ΔchyC strain. For this reason, we speculate that ChyC plays a more minor role in the amidation reactions compared to ChyD, whose deletion completely abolished the production of the late metabolites. Protein alignment does not provide sufficient information to assign a specific function to ChyC. ChyH and ChyM are predicted to be involved in oxidation reactions and to form compound 3 from compounds 15 and 5, respectively. Compound 3 originates from two further branches in the pathways, yielding chrysogine and compounds 7, 9, and 10.
Regulatory genes are usually clustered with secondary metabolite biosynthetic genes (22). Therefore, we hypothesized that the putative transcription factor Pc21g12640 can regulate the expression of the chrysogine genes, since Pc21g12640 is located downstream of chyA. Nonetheless, metabolite profiling and qPCR of the deletion strain gave no indications that Pc21g12640 is involved in the regulation of the chy genes. This conclusion is supported by the absence of the transcription factor in Fusarium and the other filamentous fungi investigated by Wollenberg et al. (9), although the orthologous gene is present in the genome of other Penicillium species (Fig. 1).
As already shown for some other fungal secondary metabolite clusters (22, 23), it is possible that the chrysogine biosynthetic genes are regulated by other transcription factors. Moreover, epigenetic regulation has been suggested for the chrysogine cluster. Shwab et al. (24) first demonstrated that secondary metabolite genes can be regulated by chromatin remodeling, for example, by histone acetylation. In P. chrysogenum DS68530, the deletion of the histone deacetylase gene hdaA resulted in a significant downregulation of expression of the chy genes and subsequent reduction of chrysogine biosynthesis (F. Guzmán-Chávez, O. Salo, and M. Samol, unpublished data).
Secondary metabolite pathways can provide a wide range of compounds from the initial scaffold molecule. Moreover, the same compounds can be produced through different paths. Branched secondary metabolite pathways have been described before in P. chrysogenum (13). The chrysogine pathway is even more branched than the previously described roquefortine pathway, and in this case, chrysogine is the final product of one ramification. As a pigment, chrysogine could contribute to protecting the cell from UV light. No antimicrobial activity has been found for this metabolite (4) nor for N-acetylalanylanthranilamide (compound 2), which was also identified in a marine fungus (17). The functions of the other metabolites in the cell remain unknown. Nonetheless, the approaches used in this work and the established methods can provide a blueprint for the elucidation of novel secondary metabolite pathways that potentially specify unknown bioactive compounds. Moreover, the understanding of the biosynthetic mechanisms can help to develop new molecules by feeding with chemically modified intermediates.
MATERIALS AND METHODS
Fungal strains, media and culture conditions.
P. chrysogenum DS68530 was kindly provided by DSM Sinochem Pharmaceuticals. Strain DS68530 lacks the penicillin gene cluster and the hdfA gene (15, 16). For RNA extraction and metabolite analysis, strains were pregrown in YGG medium (25) for 24 h. Next, 3 ml of culture inoculum was transferred into 22 ml of secondary metabolite production (SMP) medium (13), and growth was continued for the time indicated. The Pc21g12630 (chyA) overexpression strain was grown in SMP medium lacking urea and CH3COONH4 and supplemented with 2 g/liter acetamide for plasmid maintenance. The ΔchyA strain was fed with 300 μM compound 14 or 15 after 48 h of growth. All cultivations were performed as 25-ml cultures in 100-ml Erlenmeyer flasks shaken at 200 rpm and 25°C.
Construction of deletion and overexpression plasmids.
Plasmids for the deletion of the chrysogine genes were built by PCR amplification of 1 to 2 kbp of the 5′ and 3′ flanking regions of each gene, using genomic DNA (gDNA) from the DS68530 strain as the template. All primers used in this study are listed in Tables 3 and 4, and the constructed plasmids are shown in the supplemental material.
TABLE 3.
Oligonucleotide primers used for amplifying the 5′ and 3′ flanking regions of the targeted genes and for qPCR
TABLE 4.
Oligonucleotide primers for amplification and checking of integration, gene absence, and deletion cassette amplificationa

Primers used for amplification of PpcbC, chyA, and TpenDE for cloning into pDSM-JAK108; amplification of the amdS cassette for in vivo homologous recombination into pDSM108_AV1; checking the correct integration of the amdS cassette into pDSM108_AV1; and checking the absence of the genes in the knockout strains and amplification of the deletion cassettes into the genome. PCR products were sent for sequencing by using primers phleo_seq and amdS_seq in order to check the purity of the strains.
For the deletion of Pc21g12630 (chyA), Pc21g12570 (chyE), Pc21g12590 (chyH), Pc21g12610 (chyM), and Pc21g12640 genes, the Multisite Gateway Three-Fragment Vector Construction kit (Invitrogen) was used. PCR products were inserted into the donor vectors pDONR4-R1 and pDONR2-R3 by the BP Clonase II reaction. The resulting plasmids were mixed with the vector carrying the selection marker (pDONR-amdS or pDONR-phleo), the destination vector pDESTR4-R3, and the LB Clonase II mixture to form the final constructs. The acetamidase gene amdS (25, 26) was employed as a marker for the deletion of chyH, chyM, and Pc21g12640 genes, while the phleomycin resistance gene was used for selecting chyA and chyE deletion strains. The modular cloning (MoClo) system (27) was used for building Pc21g12600 (chyC) and Pc21g12620 (chyD) deletion vectors containing an amdS marker cassette.
Due to its strength, the pcbC promoter was chosen for overexpression of chyA, followed by the penDE terminator. All genetic elements were amplified from P. chrysogenum DS68530 gDNA, and the chyA expression cassette was built in subsequent steps of digestions and ligation, using pCM251 (Euroscarf) as the backbone vector. The promoter and terminator were digested with BamHI, PmeI, and NotI enzymes for cloning into pCM251. chyA was inserted into the resulting pCM251 plasmid after digestion with AscI and PmeI. The expression cassette was digested with NotI for insertion into pDSM-JAK108 (28) to form pDSM108_AV1. pDSM-JAK108 contains the AMA1 (autonomous maintenance in Aspergillus) (29) sequence, the DsRed gene for visualization of the cells, and the essential gene tif35. In this study, the tif35 gene on the plasmid was replaced with an amdS cassette by in vivo homologous recombination in P. chrysogenum. The amdS cassette containing 100-bp flanks homologous to pDSM108_AV1 was obtained by oligonucleotide extension-PCR, using pDONR-amdS as the template.
Transformation and purification procedures.
The deletion plasmids (1.5 μg) were linearized and transformed into P. chrysogenum DS68530 protoplasts using a standard protocol (30). pDSM108_AV1 (1 μg) was linearized by digestion with the MluI enzyme and cotransformed with the amdS cassette (1 μg). The transformants were plated on respective selective media transformation agar [T agar] (25) and grown at 25°C for 5 days. For strain purification, the colonies were transferred to minimal selective solid media (selective agar [S agar]) and sporulation media (25). Rice batches were prepared for inoculation of conidia and long-term storage.
Analysis of the gene deletion strains.
The absence of the deleted genes was verified by PCR with gDNA isolated from the knockout strains after 48 h of growth, using an adapted yeast gDNA extraction protocol (31). Primers binding outside the homologous flanking regions were used for amplification of the targeted fragment, after which the PCR products were further verified by sequencing (Macrogen, UK). To verify the correct integration of the amdS cassette into pDSM108-AV1, colony PCR was performed on red colonies (bearing the AMA1 plasmid, as seen by the DsRed marker on the plasmid).
RNA extraction, cDNA, amplification and qPCR analysis.
Total RNA was isolated from the DS68530 and ΔPc21g12640 strains after 48 h of growth in SMP medium, by using the TRIzol (Invitrogen) extraction method with additional DNase treatment (Turbo DNA-free kit, Ambion). For the cDNA synthesis, 500 ng of RNA was used (iScript cDNA synthesis kit, Bio-Rad). The γ-actin gene was used for normalization. The expression levels were measured in technical duplicates with a MiniOpticon system (Bio-Rad) using the Bio-Rad CFX manager software, which automatically determines the threshold cycle (Ct) values by regression. The SensiMix SYBR Hi-ROX kit (Bioline) was used as the master mix for qPCR. The reactions were run as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s.
Metabolite profiling.
All the strains used were grown in triplicates for metabolite analysis. Samples were collected after 48 h from the chyA overexpression strain and after 48 and 96 h from the deletion mutants and the parental strain. Samples were taken before the feeding of ΔchyA, immediately after the feeding, and then after 48 h. All the samples from the different experiments were centrifuged for 10 min, after which the supernatant was filtered with 0.2 μm polytetrafluoroethylene (PTFE) syringe filters and stored at −80°C. The analysis of secondary metabolites was performed with an Accella1250 HPLC system coupled with the ES-MS Orbitrap Exactive (Thermo Fisher Scientific, CA), following the method described by Salo et al. (32).
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by a grant from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7/2007-2013, under grant 607332. The financial support from the Dutch Organization for Scientific Research (NWO VIDI grant 723.014.001 for W.S.) is gratefully acknowledged.
We thank DSM Sinochem Pharmaceuticals (Delft, the Netherlands) for kindly providing the DS68530 strain.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02246-17.
REFERENCES
- 1.Frisvad JC, Smedsgaard J, Larsen TO, Samson RA. 2004. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol 49:201–241. [Google Scholar]
- 2.Frisvad JC, Samson RA. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium: a guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol 49:1–174. [Google Scholar]
- 3.Pagano MC, Dhar PP. 2015. Fungal pigments: an overview, p 173–189. In Gupta VK, Mach RL, Sreenivasaprasad S (ed), Fungal Biomolecules: Sources, Applications and Recent Developments. John Wiley & Sons, Chichester, West Sussex, UK. [Google Scholar]
- 4.Yunianto P, Rusman Y, Saepudin E, Priyono W, Sumaryono W. 2014. Alkaloid (meleagrine and chrysogine) from endophytic fungi (Penicillium sp.) of Annona squamosa L. Pak J Biol Sci 17:667–674. doi: 10.3923/pjbs.2014.667.674. [DOI] [PubMed] [Google Scholar]
- 5.Hikino H, Nabetani S, Takemoto T. 1973. Structure and biosynthesis of chrysogine, a metabolite of Penicillium chrysogenum. Yakugaku Zasshi 93:619–623. doi: 10.1248/yakushi1947.93.5_619. [DOI] [PubMed] [Google Scholar]
- 6.Kimura Y, Inoue T, Tamura S. 1973. Isolation of 2-pyruvoylaminobenzamide as an antiauxin from Colletotrichum lagenarium. Agric Biol Chem 37:2213–2214. [Google Scholar]
- 7.Salo O, Ries M, Medema MH, Lankhorst PP, Vreeken RJ, Bovenberg RAL, Driessen AJM. 2015. Genomic mutational analysis of the impact of the classical strain improvement program on β-lactam producing Penicillium chrysogenum. BMC Genomics 16:937. doi: 10.1186/s12864-015-2154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pohl C, Kiel JAKW, Driessen AJM, Bovenberg RAL, Nygård Y. 2016. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol 5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 9.Wollenberg RD, Saei W, Westphal KR, Klitgaard CS, Nielsen LK, Lysøe E, Gardiner DM, Wimmer R, Sondergaard TE, Sørensen JL. 2017. Chrysogine biosynthesis is mediated by a two-module nonribosomal peptide synthetase. J Nat Prod 80:2131–2135. doi: 10.1021/acs.jnatprod.6b00822. [DOI] [PubMed] [Google Scholar]
- 10.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 11.van den Berg M, Albang R, Albermann K, Badger J, Daran J, Driessen A, Garcia-Estrada C, Fedorova N, Harris D, Heijne W, Joardar V, Kiel J, Kovalchuk A, Martín J, Nierman W, Nijland J, Pronk J, Roubos J, van der Klei I, van Peij N, Veenhuis M, von Döhren H, Wagner C, Wortman J, Bovenberg R. 2008. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol 26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez S, Fierro F, Casqueiro J, Mart JF. 1999. Gene organization and plasticity of the β-lactam genes in different filamentous fungi. Antonie Van Leeuwenhoek 75:81–94. doi: 10.1023/A:1001861025070. [DOI] [PubMed] [Google Scholar]
- 13.Ali H, Ries MI, Nijland JG, Lankhorst PP, Hankemeier T, Bovenberg RAL, Vreeken RJ, Driessen AJM. 2013. A branched biosynthetic pathway is involved in production of roquefortine and related compounds in Penicillium chrysogenum. PLoS One 8:e65328. doi: 10.1371/journal.pone.0065328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali H, Ries MI, Lankhorst PP, van der Hoeven RAM, Schouten OL, van Peij NME, Bovenberg RAL, Vreeken RJ, Driessen AJM. 2014. A non-canonical NRPS is involved in the synthesis of fungisporin and related hydrophobic cyclic tetrapeptides in Penicillium chrysogenum. PLoS One 9:e98212. doi: 10.1371/journal.pone.0098212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoff B, Kamerewerd J, Sigl C, Zadra I, Kück U. 2010. Homologous recombination in the antibiotic producer Penicillium chrysogenum: strain ΔPcku70 shows up-regulation of genes from the HOG pathway. Appl Microbiol Biotechnol 85:1081–1094. doi: 10.1007/s00253-009-2168-4. [DOI] [PubMed] [Google Scholar]
- 16.Snoek ISI, van der Krogt ZA, Touw H, Kerkman R, Pronk JT, Bovenberg RAL, van den Berg MA, Daran JM. 2009. Construction of an hdfA Penicillium chrysogenum strain impaired in non-homologous end-joining and analysis of its potential for functional analysis studies. Fungal Genet Biol 46:418–426. doi: 10.1016/j.fgb.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Onuki H, Miyashige H, Hasegawa H, Yamashita S. 1998. NI15501A, a novel anthranilamide derivative from a marine fungus Penicillium sp. J Antibiot (Tokyo) 51: 442–444. doi: 10.7164/antibiotics.51.442. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen JC, Grijseels S, Prigent S, Ji B, Dainat J, Nielsen KF, Frisvad JC, Workman M, Nielsen J. 2017. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat Microbiol 2:17044. doi: 10.1038/nmicrobiol.2017.44. [DOI] [PubMed] [Google Scholar]
- 19.Prieto C, García-estrada C, Lorenzana D, Martín JF. 2012. NRPSSP: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28:426–427. doi: 10.1093/bioinformatics/btr659. [DOI] [PubMed] [Google Scholar]
- 20.Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. 2011. NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knudsen M, Søndergaard D, Tofting-Olesen C, Hansen FT, Brodersen DE, Pedersen CNS. 2016. Computational discovery of specificity-conferring sites in non-ribosomal peptide synthetases. Bioinformatics 32:325–329. doi: 10.1093/bioinformatics/btv600. [DOI] [PubMed] [Google Scholar]
- 22.Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol 11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 23.Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, Hertweck C, Brakhage AA, Biology I, Kno H. 2010. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide. Appl Environ Microbiol 76:8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle SKN. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell 6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovalchuk A, Weber SS, Nijland JG, Bovenberg RAL, Driessen AJM. 2012. Fungal ABC transporter deletion and localization analysis. Methods Mol Biol 835:1–16. doi: 10.1007/978-1-61779-501-5_1. [DOI] [PubMed] [Google Scholar]
- 26.Kolar M, Punt PJ, van den Hondel CAMJJ, Schwab H. 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62:127–134. doi: 10.1016/0378-1119(88)90586-0. [DOI] [PubMed] [Google Scholar]
- 27.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. 2011. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bovenberg RAL, Kiel JAKW, Wenzel TJ, Los AP. April 2014. Vector-host system. US patent 2014/0106398 A1.
- 29.Gems D, Johnstone IL, Clutterhuck AJ. 1991. An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene 98:61–67. doi: 10.1016/0378-1119(91)90104-J. [DOI] [PubMed] [Google Scholar]
- 30.Weber SS, Kovalchuk A, Bovenberg RAL, Driessen AJM. 2012. The ABC transporter ABC40 encodes a phenylacetic acid export system in Penicillium chrysogenum. Fungal Genet Biol 49:915–921. doi: 10.1016/j.fgb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Harju S, Fedosyuk H, Peterson KR. 2004. Rapid isolation of yeast genomic DNA: Bust n' Grab. BMC Biotechnol 4:8. doi: 10.1186/1472-6750-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salo O, Guzmán-Chávez F, Ries MI, Lankhorst PP, Bovenberg RAL, Vreeken RJ, Driessen AJM. 2016. Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl Environ Microbiol 82:3971–3978. doi: 10.1128/AEM.00350-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.