ABSTRACT
For decades, bacterial taxonomy has been based on in vitro molecular biology techniques and comparison of molecular marker sequences to measure the degree of genetic similarity and deduce phylogenetic relatedness of novel bacterial species to reference microbial taxa. Due to the advent of the genomic era, access to complete bacterial genome contents has become easier, thereby presenting the opportunity to precisely investigate the overall genetic diversity of microorganisms. Here, we describe a high-accuracy phylogenomic approach to assess the taxonomy of members of the genus Bifidobacterium and identify apparent misclassifications in current bifidobacterial taxonomy. The developed method was validated by the classification of seven novel taxa belonging to the genus Bifidobacterium by employing their overall genetic content. The results of this study demonstrate the potential of this whole-genome approach to become the gold standard for phylogenomics-based taxonomic classification of bacteria.
IMPORTANCE Nowadays, next-generation sequencing has given access to genome sequences of the currently known bacterial taxa. The public databases constructed by means of these new technologies allowed comparison of genome sequences between microorganisms, providing information to perform genomic, phylogenomic, and evolutionary analyses. In order to avoid misclassifications in the taxonomy of novel bacterial isolates, new (bifido)bacterial taxons should be validated with a phylogenomic assessment like the approach presented here.
KEYWORDS: genomics, phylogenomics, ITS, next generation sequencing, bifidobacteria, Bifidobacterium
INTRODUCTION
Since the 1960s, bacterial taxonomy has been determined by using the DNA-DNA hybridization approach in order to measure the degree of genetic similarity between two microbial genomes (1). Another accepted method that was and is still widely used in bacterial taxonomy is comparative analysis of 16S rRNA gene-based sequences (2). Unfortunately, the DNA-DNA hybridization method suffers from reproducibility issues and does not provide an accurate measure of actual sequence identity between genomes (3). Similar limitations affect the 16S rRNA gene approach; for example, very recently diverged species that have undergone intense evolutionary pressures may possess highly similar 16S rRNA gene sequences that may nonetheless ignore a wide phylogenetic gap between such taxa (4, 5). To overcome the limitations of these techniques, a multigenic approach that relies on multiple conserved molecular markers, such as the clpC, dnaB, dnaG, dnaJ1, purF, and rpoC genes, was shown to be more reliable for species discrimination compared to single-gene phylogeny (5, 6). Having easy access to next-generation sequencers (NGS) has in recent times allowed the development of a new bioinformatics approach for phylogeny that is based on whole-genome sequencing followed by comparative genomics. Comparative genomics has proven to be accurate in strain discrimination and has been applied extensively for phylogenetic characterization of novel bacterial species, in particular those residing in complex communities, e.g., gut microbiota members (7–9).
Members of the genus Bifidobacterium are among the main representatives of the mammalian gut microbiota, particularly during the first six to 12 months following birth (10, 11). This group of microorganisms is known for a claimed ability to confer a range of health benefits to the host (12, 13, 61), although the associated genetic attributes for such beneficial or probiotic activities remain largely obscure. Bifidobacteria are widespread inhabitants of the mammalian, avian, and insect intestinal tracts (11, 13, 14, 62), yet a large part of the currently recognized bifidobacterial taxa, i.e., 49 species and 9 subspecies, has been isolated from the human gut (15, 16). Nonetheless, various ecological attempts have recently been made to survey bifidobacterial populations in other mammals (10, 17–21), which has resulted in the identification of several scientifically accepted and putative novel bifidobacterial species.
The majority of the currently proposed novel (bifido)bacterial taxa are identified through partial sequencing of several molecular marker genes and comparison to the currently recognized type strains (19–21). In the current study, we describe a methodology that is based on whole-genome comparisons and is aimed at unambiguously redefining the taxonomy of members of the genus Bifidobacterium. Notably, similar approaches based on whole-genome comparisons have been employed in the reclassification of members of the genus Bacillus (22, 23). Applying this approach, we were also able to phylogenetically characterize and position seven novel bifidobacterial strains which were isolated from animal feces, i.e., those of goose, hamster, rabbit, and monkey, and which are related to Bifidobacterium choerinum, Bifidobacterium hapali, Bifidobacterium saguini, Bifidobacterium stellenboschense, and Bifidobacterium tissieri.
RESULTS AND DISCUSSION
Pangenome reconstruction among members of the genus Bifidobacterium.
Recently, species- and genus-level comparative genomic analyses based on pangenome reconstruction have been shown to be crucial in providing information regarding the overall gene content, while also generating information on the resistome, metabolic capabilities, and mobilome of such taxonomic ranks (9, 24–28). In recent years, the number of sequenced bifidobacterial strains has increased from a few dozen to several hundred, and thus we felt it was opportune to explore the genomic biodiversity within different species of the genus Bifidobacterium (12, 15, 16, 29). Currently, the number of sequenced bifidobacterial strains for each species ranges from just one to 83 (see Table S1 in the supplemental material). In order to increase the amount of genetic data available for those bifidobacterial species for which only one or a few strains have been sequenced, we decided to decode the genomes of 13 additional bifidobacterial strains belonging to the species Bifidobacterium asteroides, Bifidobacterium pseudolongum, and Bifidobacterium thermophilum (Table 1). Furthermore, the public NCBI genomic database contains 55 type strain genomes corresponding to each known bifidobacterial (sub)species, as well as complete or draft genome sequences of 233 additional strains belonging to the species Bifidobacterium adolescentis, Bifidobacterium animalis, B. asteroides, Bifidobacterium bifidum, Bifidobacterium breve, Bifidobacterium dentium, Bifidobacterium longum, Bifidobacterium pseudocatenulatum, B. pseudolongum, and B. thermophilum (see Table S1).
TABLE 1.
General features of sequenced bifidobacterial strains
| Species | Strain | Average coverage (fold) | No. of contigs | Genome length (bp) | Average GC content (%) | No. of predicted ORFs | No. of tRNAs | No of rRNAsa | Biological origin | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| B. asteroides | 1460B | 104.83 | 38 | 2,121,817 | 60.46 | 1,653 | 45 | 2 | Honeybee hindgut | PCHJ00000000 |
| B. pseudolongum | 1370B | 89.46 | 17 | 1,902,036 | 62.97 | 1,571 | 52 | 4 | Feces of pig | PCHI00000000 |
| B. pseudolongum | 1520B | 115.03 | 17 | 2,008,481 | 63.12 | 1,632 | 52 | 4 | Feces of hamster | PCHH00000000 |
| B. pseudolongum | 1549B | 118.25 | 48 | 1,990,203 | 63.19 | 1,686 | 55 | 4 | Feces of Brahma chicken | PCHG00000000 |
| B. pseudolongum | 1595B | 147.96 | 16 | 1,936,418 | 63.02 | 1,593 | 53 | 4 | Feces of pig | PCHF00000000 |
| B. pseudolongum | 1691B | 65.49 | 59 | 2,148,724 | 63.06 | 1,810 | 52 | 4 | Feces of hippopotamus | PCHE00000000 |
| B. pseudolongum | 1734B | 84.15 | 25 | 2,111,856 | 63.27 | 1,778 | 62 | 4 | Feces of wallaby | PCHD00000000 |
| B. pseudolongum | 1619B | 235.22 | 31 | 2,050,408 | 63.27 | 1,722 | 53 | 3 | Feces of llama | PCHC00000000 |
| B. pseudolongum | 1744B | 101.65 | 51 | 2,143,581 | 63.05 | 1,801 | 55 | 4 | Feces of bear | PCHB00000000 |
| B. pseudolongum | 1747B | 56.17 | 52 | 2,143,079 | 63.18 | 1,830 | 60 | 4 | Feces of giraffe | PCHA00000000 |
| B. pseudolongum | 1524B | 99.95 | 23 | 2,062,414 | 63.15 | 1,700 | 52 | 4 | Feces of hamster | PCGZ00000000 |
| B. thermophilum | 1542B | 89.45 | 36 | 2,359,132 | 60.28 | 1,820 | 47 | 3 | Feces of pig | PCGY00000000 |
| B. thermophilum | 1543B | 115.32 | 16 | 2,316,133 | 60.41 | 1,751 | 50 | 3 | Feces of pig | PCGX00000000 |
Predicted number of rRNA loci.
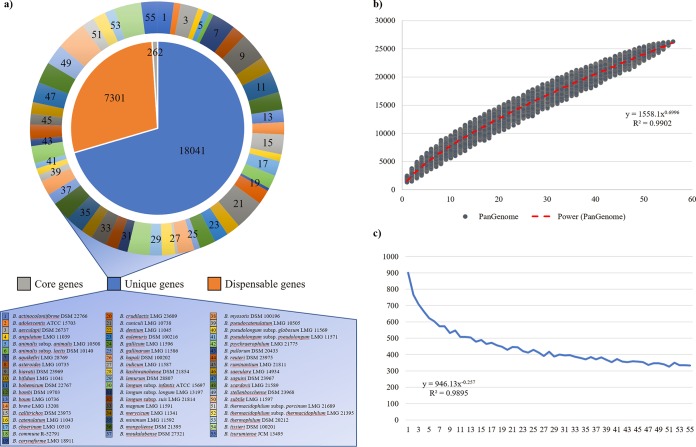
These gathered data sets were then employed for the genomic approach to study of the phylogeny of the genus Bifidobacterium. Thus, a pangenome analysis of available type strains was undertaken to determine putative orthologous genes between the 55 (sub)species of the genus Bifidobacterium sequenced to date. The analysis resulted in the identification of 26,201 clusters of orthologous genes (COGs), representing the pangenome of the Bifidobacterium genus. The collected COGs allowed the identification of genes that are shared between the genomes of the 55 bifidobacterial type strains, i.e., the core genome of the Bifidobacterium genus (Fig. 1). Furthermore, dispensable genes present in two or more strains and unique genes retrieved in a single type strain were unveiled (Fig. 1). The pangenome size, when plotted versus the number of included bifidobacterial genomes, shows that the power trendline has yet to reach a plateau and data sets from the last strains added to the analysis still substantially expand the total gene pool by circa 330 genes per added genome (Fig. 1). Therefore, according to these data, the pangenome of the genus Bifidobacterium can be considered open (25) (Fig. 1). This open pangenome profile is typically associated with genera in which the constituent species occupy multiple environments with mixed microbial communities and have extended their total set of genes through horizontal gene transfer events, e.g., Escherichia (9, 30).
FIG 1.
Pangenome of the genus Bifidobacterium. (a) Number of core genes (gray), unique genes (blue), and dispensable genes (orange) identified in the pangenome analysis (internal pie chart). The number of unique genes of each type strain of the genus sequenced to date is highlighted in different colors (external pie chart). (b) Representation of the pangenome size based on sequential addition of the 55 type strains genomes. (c) Average of new genes upon sequential addition of the bifidobacterial type strain genomes.
In order to assess intraspecies variability, we selected bifidobacterial species for which at least five sequenced genomes are available in the NCBI databases, or which were decoded as part of the current study. Specifically, a total of 256 different strains were employed for the reconstruction of 10 species-specific bifidobacterial pangenomes (see Fig. S1 and S2 in the supplemental material). Notably, and similar to those of the pangenome of the overall Bifidobacterium genus (Fig. 1), the power trendlines do not reach a plateau when species-specific pangenome sizes are plotted versus the number of included genomes (see Fig. S1 and S2). Furthermore, the species-specific pangenome analyses also revealed that the average number of new genes added by inclusion of additional genomes tend to decrease for 7 out of 10 analyzed species. In contrast, the comparative genomics analysis of B. animalis, B. breve, and B. longum revealed that the pangenome expansion seems to remain stable at a minimum of circa 43, 75, and 101 new genes added for each further iteration, respectively (see Fig. S1), suggesting that the biodiversity of these species has been extensively explored. In this regard, it is worth mentioning that a portion of these “new” genes corresponds to truncated genes predicted at the edges of contigs within partially sequenced genomes. In this context, the pangenome's expansion trend in B. breve and B. longum reflects the high number of strains sequenced for both these two species (12, 16), whereas the limited pangenome expansion of B. animalis is probably due to the monophyletic origin of the subspecies B. animalis subsp. lactis, as previously suggested (29).
Overall, these data indicate that availability of a very large number, perhaps as many as thousands, of strains representing populations residing in a wide range of different hosts will be pivotal to obtain a comprehensive overview of the biodiversity that characterizes these common gut commensals.
Phylogenetic and phylogenomic analyses of bifidobacterial type strains.
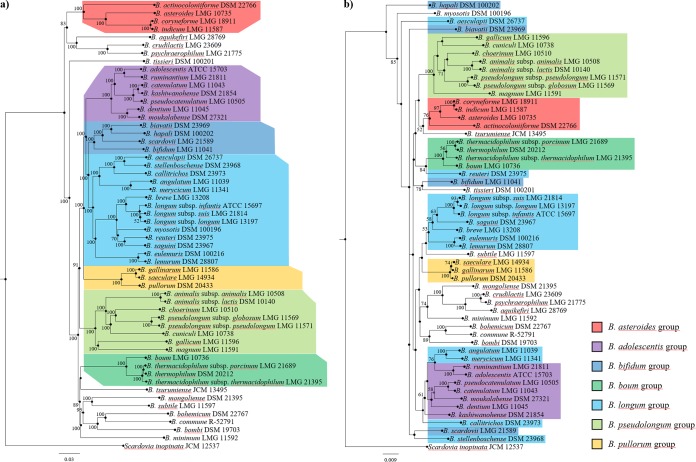
While 16S rRNA gene-based comparative analysis has for many years been considered to represent the gold standard for phylogenetic investigations, the recent advent of NGS and associated bioinformatic tools has led to the introduction of novel genome-wide approaches, such as the core-genome-based supertree (31, 32). We therefore decided to compare the phylogeny of the type strains of the genus Bifidobacterium reconstructed through alignment of the 16S rRNA gene and concatenated alignment of the bifidobacterial core genome. The 16S rRNA genes were retrieved from the 55 type strains of each taxon belonging to the genus Bifidobacterium and were used to construct a phylogenetic tree (Fig. 2). Moreover, the predicted 26,201 clusters of orthologous genes identified by comparative genomics analysis of the bifidobacterial type strains allowed the identification of 262 shared COGs, representing the core bifidobacterial genome coding sequences (Fig. 2). After exclusion of paralogs, concatenation of the remaining 236 core protein sequences was used to build a Bifidobacterium phylogenomic tree (Fig. 2). These analyses showed that 34 and 52 nodes were supported by bootstrap values greater than 50 for the 16S rRNA gene- and core gene-based trees, respectively (Fig. 2). These data clearly support the notion that increasing sequence lengths used in the phylogenomic tree leads to improved robustness of the results. Furthermore, the phylogenomic analyses are based on a core genome, which is represented by amino acid sequences. Remarkably, these protein sequences involve a variation of 20 amino acids, while the sequence of the 16S rRNA gene is based on only four bases. The use of amino acid sequences clearly enhances the robustness of the resulting phylogenomic tree.
FIG 2.
Phylogenetic and phylogenomic trees of the genus Bifidobacterium. (a) Proteomic tree based on the concatenation of 262 bifidobacterial core genes identified in the pangenome analysis of the 55 type strains. Phylogenetic groups are highlighted in different colors. (b) 16S rRNA gene-based tree of the 55 type strains of the genus. Bifidobacterial species are colored according to their phylogenetic membership group. Each tree was constructed by the neighbor-joining method, and the genome sequence of Scardovia inopinata JCM 12537 was used as outgroup. Bootstrap percentages above 50 are shown at node points, based on 1,000 replicates of each phylogenetic tree.
The bifidobacterial phylogenomic tree clearly delineates the presence of seven previously described bifidobacterial phylogenetic groups, i.e., the B. adolescentis, B. asteroides, B. bifidum, Bifidobacterium boum, B. longum, B. pseudolongum, and Bifidobacterium pullorum groups (33, 34). Notably, the number of bifidobacterial taxa included does not affect the consistency of the core gene-based tree (Fig. 2) (9, 33). Comparison of the phylogenomic and 16S rRNA gene-based trees revealed discrepancies for the B. bifidum and B. longum groups (Fig. 2), while the phylogeny of the B. adolescentis, B. asteroides, B. boum, B. pseudolongum, and B. pullorum groups is conserved in both trees. In detail, the four members of the B. bifidum group cluster together only in the core gene-based tree, while they are scattered across the 16S rRNA gene-based tree. In contrast, the core of the B. longum group is consistent in both trees, represented by B. longum subsp. suis LMG 21814, B. longum subsp. longum LMG 13197, B. longum subsp. infantis ATCC 15697, Bifidobacterium saguini DSM 23967, B. breve LMG 13208, Bifidobacterium eulemuris DSM 100216, and Bifidobacterium lemurum DSM 28807 (Fig. 2). Nevertheless, the B. longum group identified in the core gene-based tree includes six additional taxa compared to that in the 16S rRNA gene-based tree.
The presented comparison between the phylogenetic reconstruction based on alignment of the 16S rRNA gene and the concatenated alignment of the bifidobacterial core genome suggests that the latter permits the reconstruction of a more robust and consistent overview of bifidobacterial evolution and may for this reason be considered the preferential approach for genus- and species-wide phylogenetic investigations.
Evaluation of intraspecies variability between bifidobacterial genomes.
The 256 bifidobacterial genomes collected above were also used to perform whole-genome comparisons, focusing specifically on the species B. adolescentis, B. animalis, B. asteroides, B. bifidum, B. breve, B. dentium, B. longum, B. pseudocatenulatum, B. pseudolongum, and B. thermophilum. The genomes of bifidobacterial taxa belonging to the same species were subjected to average nucleotide identity (ANI) evaluation. These analyses revealed consistent species classification in seven out of 10 phylogenetic groups, highlighting ANI values ranging from 95.31% to 99.98% (see Table S2 and Fig. S3–S6 in the supplemental material). In this regard, it should be noted that two strains displaying an ANI value of <95% are considered to belong to two distinct species (35, 36). Thus, all the strain pairs that belong to the species B. adolescentis, B. animalis, B. bifidum, B. breve, B. dentium, B. longum, and B. pseudocatenulatum possess ANI values higher than 95%, confirming taxonomic assignment to the same species (35, 36). Conversely, the genomes belonging to the species B. asteroides, B. pseudolongum, and B. thermophilum exhibit cases of ANI values lower than 93.3%, which would cast doubt on the correct taxonomic classification of several strains previously assigned to these species (see Table S2).
More specifically, within the species B. thermophilum, the chromosomal sequence of strain JCM 1207 generates ANI values of 90.16% and 90.28% compared to the genome sequences of B. thermophilum DSM 20210 and B. thermophilum DSM 20212, respectively (see Table S2). These values are the lowest retrieved between strains of the same species in this intraspecies analysis, highlighting that the JCM 1207 strain should not be classified as a B. thermophilum species (see Fig. S6 in the supplemental material). Further ANI analyses encompassing all bifidobacterial type strains showed that B. thermophilum JCM 1207 possesses a higher genome sequence identity with B. boum LMG 10736, sharing an ANI value of 94.9% (33), which is very close to the threshold for species discrimination (35, 36).
Besides, different clusters were detected within the species B. pseudolongum (see Fig. S6). In particular, the genome sequence of B. pseudolongum subsp. pseudolongum LMG 11571 displays high ANI values compared to strains 1370B (99.21%) and 1595B (99.21%), both sequenced in the current study. Nonetheless, the chromosomal sequences of these three strains generate corresponding ANI values lower than 93.99% compared to 11 other B. pseudolongum strains, which among each other display ANI values above 95.81% (see Table S2). These data suggest that B. pseudolongum subsp. pseudolongum LMG 11571, along with strains 1370B and 1595B, may represent a distinct bifidobacterial species.
The most intriguing data, due to the high genomic diversity observed for the six analyzed strains, were retrieved for members of the species B. asteroides. In fact, the genomes belonging to 1460B, Bin2, Hma3, and Bin7 strains produced ANI values that are below the species threshold level compared to the genome of the type strain B. asteroides DSM 20089 (see Table S2) and resulted in four clusters, i.e., four putative distinct bifidobacterial species (as reported in Fig. S3).
Further inspection of genomic sequence identity at subspecies level revealed that the chromosomes of the 30 B. animalis strains analyzed constitute two clear clusters, encompassing seven B. animalis subsp. animalis and 23 B. animalis subsp. lactis, with strains A6, 08, 11, and RH without a (stated) subspecies classification falling in the latter group (see Fig. S3). Moreover, the low genome variability observed in the B. animalis subsp. lactis strains confirms the previously reported observation that this subspecies is a strict monophyletic taxon (29, 37, 38).
Interestingly, the intraspecies analysis performed on B. longum highlights several inconsistencies in the subspecies classifications (see Fig. S5). In this context, a clear cluster composed of 19 B. longum subsp. infantis strains was identified, as well as a smaller cluster representative of B. longum subsp. suis. Notably, the latter cluster includes the genomes of AGR2137, Su859, CMCC P0001, BXY01, and JDM301 strains previously classified as B. longum subsp. longum. Remarkably, strains named CMCC P0001, BXY01, and JDM301 displayed ANI values above 99.89% compared to each other, confirming their previously described association (12). Nonetheless, genome comparisons between the seven identified B. longum subsp. suis generated ANI values above 98%, and for this reason no distinct phylogenetic groups could be identified based on genomic identity approaches (see Table S2). The largest B. longum cluster contains 57 B. longum subsp. longum strains, of which three strains, named 157F, CCUG 52486, and CECT 7210, had erroneously been classified as B. longum subsp. infantis strains.
Overall, ANI evaluation of all 258 available bifidobacterial genomes indicated that 16 strains should be taxonomically reclassified, while it also revealed the putative existence of six novel bifidobacterial species.
Genome sequencing as the most current standard for taxonomic classification of bifidobacteria.
The pipeline for accurate bifidobacterial taxonomic classification described here was also exploited to precisely taxonomically classify putative novel bifidobacterial taxa isolated as part of a recently published study aimed at the exploration of the biodiversity of bifidobacterial communities of 291 animals, including goose, hamster, rabbit, and monkeys (10) (Table 2). Subsequent selection on mupirocin medium (39) allowed the isolation of strains belonging to members of the genus Bifidobacterium, which were further characterized by amplification and sequencing of the 16S rRNA gene and internal transcribed spacer (ITS). While 16S rRNA gene sequence comparisons have been used in (bifido)bacterial taxonomy for decades, the ability to distinguish closely related bifidobacterial taxa using ITS sequences has recently been described (40). 16S rRNA gene sequence analysis revealed that seven isolated strains, named Rab10A, Ham19E, Goo31D, Tam1G, Tam10B, Uis4E, and Uis1B, showed 16S rRNA sequence-based identity values that ranged from 96.2% to 98.6% with respect to known bifidobacterial type strains listed in Table 2, while the hypervariable ITS sequence displayed values ranging from 65.7% to 89% (Table 2). Notably, these data highlight a high degree of sequence diversity between these seven isolates and the known bifidobacterial species, especially for the hypervariable ITS sequence, thus suggesting that they present novel bifidobacterial species.
TABLE 2.
General features of novel bifidobacterial strains
| Strain | Average coverage (fold) | No. of contigs | Genome length (bp) | Average GC content (%) | No. of predicted ORFs | tRNA | rRNAa | 16S identity (%, species) | ITS identity (%, species) | ANI value (%, species) | GGDCb value (%, species) | Biological origin |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rab10A | 104.3 | 72 | 2,276,351 | 65.45 | 1,825 | 52 | 4 | 98.36, B. choerinum | 87.2, B. choerinum | 91.8, B. choerinum | 41.30, B. choerinum | European rabbit |
| Ham19E | 108.6 | 44 | 2,155,882 | 62.53 | 1,733 | 53 | 4 | 97.32, B. choerinum | 68.6, B. animalis | 87.81, B. choerinum | 28.50, B. choerinum | European hamster |
| Goo31D | 115.4 | 9 | 2,166,761 | 64.3 | 1,681 | 52 | 4 | 97.65, B. choerinum | 84.7, B. choerinum | 91.47, B. choerinum | 39.30, B. choerinum | Domestic goose |
| Tam1G | 55.52 | 62 | 2,639,899 | 56.13 | 2,215 | 59 | 4 | 99, B. saguini | 89, B. saguini | 94.55, B. saguini | 55.20, B. saguini | Emperor tamarin |
| Tam10B | 138 | 68 | 3,111,005 | 62.46 | 2,522 | 60 | 4 | 96.4, B. tissieri | 72, B. hapali | 89, B. tissieri | 31.60, B. tissieri | Emperor tamarin |
| Uis1B | 66.93 | 80 | 2,789,387 | 61.91 | 2,281 | 60 | 3 | 96.2, B. tissieri | 65.7, B. biavatii | 88.04, B. hapali | 28.50, B. hapali | Pygmy marmoset |
| Uis4E | 139.27 | 44 | 2,820,211 | 65.81 | 2,247 | 62 | 6 | 96.8, B. stellenboschense | 81.3, B. stellenboschense | 93.45, B. stellenboschense | 50.20, B. stellenboschense | Pygmy marmoset |
Predicted number of rRNA loci.
GGDC, Genome-to-Genome Distance Calculator.
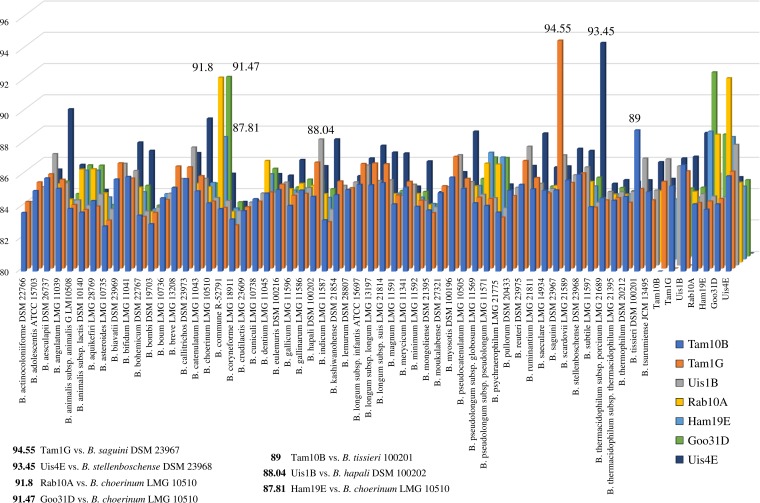
In order to get insights into genome-wide genetic relatedness between the putative novel species and currently known taxa of the genus Bifidobacterium, the genomes of these seven isolates were sequenced. The reconstructed genome length ranged from 2,155,882 to 3,111,005 bp, with an average fold coverage ranging from 55.52 to 139.27 (Table 2). Using the ANI system based on whole genome sequence comparisons, the seven sequenced strains were compared with the currently recognized bifidobacterial type strains (9, 41). Interestingly, the seven isolates exhibited ANI values below the threshold for species recognition compared to all 55 available type strains, with the highest ANI value obtained against B. saguini DSM 23967 (94.55%) (Fig. 3) (36, 42). Furthermore, an ANI analysis involving only these seven strains revealed that the highest ANI value (91.76%) was obtained between the genomes of Rab10A and Ham19E (Fig. 3). Genome-to-Genome Distance Calculator (GGDC) analysis, which is based on in silico DNA-DNA hybridization (DDH) of genome-to-genome comparison (43), was employed to validate the ANI results based on the genomic relatedness between bifidobacterial taxa. The seven sequenced strains' genomes compared with the closest related type strains' genomes exhibited estimated DDH values below 70%, ranging from 28.5% to 55.2% between pairs Uis1B/B. hapali DSM 100202 and Tam1G/B. saguini DSM 23967, respectively (Table 2).
FIG 3.
Graphical representation in three-dimensional columns of the average nucleotide identity (ANI) between novel bifidobacterial taxa. The x axis displays the 62 bifidobacterial taxa used in the comparison (55 type strains and seven novel taxa), the y axis reflects the ANI percentages between strains, and the z axis displays the seven novel taxa. Thus, each column represents the ANI value observed between a given bifidobacterial species pair. The higher ANI values, involving Rab10A, Ham19E, Goo31D, Tam1G, Tam10B, Uis4E, and Uis1B, are highlighted with different colors, showing ANI values below 95%.
Taken together, ANI and GGDC analyses clearly indicate that Rab10A, Ham19E, Goo31D, Tam1G, Tam10B, Uis4E, and Uis1B belong to novel bifidobacterial species (Fig. 3). Recently, based on this genomic approach, Tam10B was formally accepted by the International Committee on Systematic Bacteriology (ICSB) as a novel species of the genus Bifidobacterium, and was accordingly named Bifidobacterium vansinderenii (44).
Phylogenomic approach for the evaluation of novel bifidobacterial taxa.
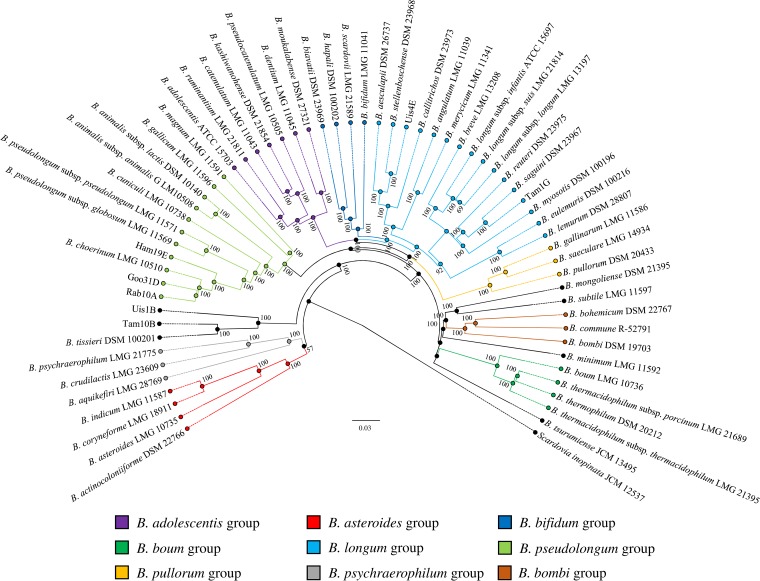
The availability of genome sequences of the seven putative novel bifidobacterial species also allowed updating of the phylogeny of the genus Bifidobacterium. A comparative genomics analysis was undertaken to determine putative orthologous genes between the 55 sequenced type strains of the genus Bifidobacterium and the seven putative novel taxa, resulting in the identification of 27,868 BifCOGs (Bifidobacterium-specific clusters of orthologous genes). Analysis of the predicted BifCOGs identified 259 COGs that were shared among all these genomes, representing the core bifidobacterial genome coding sequences (core BifCOGs). This core BifCOG collection represents an updated version of a previously published core genome of the genus Bifidobacterium (9, 33). The concatenation of 232 core BifCOG protein sequences (note that 27 core BifCOGs were excluded, as they constitute paralogs within the bifidobacterial pangenome) was used to build a Bifidobacterium phylogenomic tree (Fig. 4).
FIG 4.
Phylogenomic tree of the genus Bifidobacterium based on the concatenation of the deduced amino acid sequences of 232 core genes of Goo31D, Ham19E, Rab10A, Tam1G, Uis4E, and Uis1B and members of the genus Bifidobacterium. The amino-acid-deduced core gene-based tree shows the division intoseven phylogenetic groups of the Bifidobacterium genus as represented by different colors. The phylogenetic tree was constructed by the neighbor-joining method, with genome sequence of Scardovia inopinata JCM 12537 as outgroup. Bootstrap percentages above 50 are shown at node points, based on 1,000 replicates of the phylogenetic tree.
The updated bifidobacterial phylogenomic tree confirmed the seven bifidobacterial phylogenetic groups previously described (33, 34). Moreover, the seven putative novel species appear to be distributed across the whole tree. As expected through the ANI analysis, Rab10A, Ham19E, and Goo31D cluster in the proximity of B. choerinum LMG 10510 and fall within the B. pseudolongum group. Interestingly, Rab10A, Ham19E, and Goo31D were isolated from three different animals, i.e., rabbit, hamster, and goose, respectively, while B. choerinum LMG 10510 is an isolate from piglet feces. Thus, these data indicate that the B. pseudolongum group is currently the most variable phylogenetic bifidobacterial group in terms of ecological niches represented by animal species, in which group strains were isolated from chickens, geese, hamsters, oxen, pigs, rabbits and rats. Nonetheless, one has to keep in mind that related bifidobacterial species appear to be widespread among mammals (10). Moreover, strains Tam1G and Uis4E are included in the B. longum group and show high phylogenetic relatedness with B. saguini DSM 23967 and B. stellenboschense DSM 23968, respectively (Fig. 4). Furthermore, Tam10B and Uis1B cluster together with B. tissieri DSM 100201.
Phylogenomic analysis indicates that 13 bifidobacterial taxa do not belong to any previously described phylogenetic group (Fig. 4). Nevertheless, based on the relatedness among these bifidobacterial type strains, two new phylogenetic clusters are proposed, namely, the Bifidobacterium psychraerophilum group and the Bifidobacterium bombi group (Fig. 4). The results presented highlight once again the potential of the phylogenomic approach to establish detailed phylogenetic reconstruction of the entire Bifidobacterium genus and detailed taxonomic characterization of novel bacterial species.
Conclusions.
Next-generation sequencing has significantly influenced microbial taxonomy by giving access to genome sequences of essentially all known bacterial taxa. In fact, the deciphered genome sequences of bifidobacterial type strains now provide genetic information for genomic, phylogenomic, and evolutionary analyses, and facilitate the determination of the gene contribution of each isolated microorganism. Here, 301 sequenced strains belonging to the genus Bifidobacterium were compared to perform inter- and intraspecies analyses aimed at redefining bifidobacterial taxonomy and unveiling discrepancies in species assignment. Overall, ANI analyses identified inconsistencies in classification of a total of 16 strains within the B. asteroides, B. pseudolongum, and B. thermophilum species, unveiling the existence of six additional clusters of strains that may represent novel putative bifidobacterial species. Furthermore, we validated the potential of the phylogenomics approach in the identification of novel species through the sequencing of new bifidobacterial isolates. Seven of these strains may be characterized as novel bifidobacterial species, showing genomic compositions related to those of B. choerinum LMG 10510, B. hapali DSM 100202, B. saguini DSM 23967, B. stellenboschense DSM 23968, and B. tissieri DSM 100201, while maintaining ANI values below the threshold for species assignment. The phylogenomic analysis of these seven putative novel species also revealed their localization into the updated phylogeny of the genus Bifidobacterium, with three strains belonging to the B. pseudolongum group and two novel taxa falling into the B. longum group. In light of these results, we propose to implement the current taxonomic scheme for the classification of novel bifidobacterial taxa through a (phylo)genomic assessment of a proposed new taxon.
MATERIALS AND METHODS
Bifidobacterial genome sequences.
We retrieved complete and partial genome sequences of 55 Bifidobacterium type strains from the National Center for Biotechnology Information (NCBI) public database, in additional to genomes of a further 233 taxa that belong to this genus (see Table S1). Furthermore, 13 bifidobacterial genomes were sequenced to perform intraspecies analyses, creating a pool of 301 bifidobacterial genomes. We also analyzed the genome sequences of seven novel bifidobacterial taxa deposited at DDBJ/ENA/GenBank, available under accession numbers MVOG00000000, MVOH00000000, NEWD00000000, NMWT00000000, NMWU00000000, NMWV00000000, and NMYC00000000 (Table 2).
Isolation of bifidobacterial species.
Fecal samples were collected from several zoological parks as described previously (10). In order to acquire the fecal material as fresh as possible and to be sure of its origin, it was collected immediately following defecation. Fecal samples consisted of 6 to 10 g of fresh material, which was cooled to 4°C immediately after collection and transferred to the Laboratory of Probiogenomics of Parma University (Parma, Italy). Bifidobacterial isolation from stool samples was performed starting from 1 g of fecal sample mixed with 9 ml of phosphate-buffered saline (PBS) solution. Serial dilutions and subsequent plating were performed using the de Man-Rogosa-Sharpe (MRS) agar, supplemented with 50 μg/ml mupirocin (Delchimica, Italy) and 0.05% (wt/vol) l-cysteine hydrochloride. Bifidobacterial cultures were incubated for 48 h at 37°C in a chamber (Concept 400; Ruskin) with anaerobic atmosphere (composed of 2.99% H2, 17.01% CO2, and 80% N2). Colonies were randomly picked and restreaked to isolate purified bacterial strains. All colonies were subjected to DNA isolation and characterized as previously described by Turroni et al. (45) and Ventura et al. (46).
DNA extraction and amplification of 16S rRNA and ITS sequences.
Fecal samples maintained at −80°C were subjected to DNA extraction using the QIAamp DNA Stool minikit following the manufacturer's instructions (Qiagen). Partial 16S rRNA gene sequences were amplified from extracted DNA using the primer pair Probio_Uni/Probio_Rev, which targets the V3 region of the 16S rRNA gene sequence (47), while partial ITS sequences were amplified using the Probio-bif_Uni/Probiobif_Rev primer pair, which targets the hypervariable region of the bifidobacterial ITS sequences (40). Results were then subjected to a BLAST search against the GenBank database.
Genome sequencing and assemblies.
DNA extracted from the bifidobacterial isolates was subjected to whole-genome sequencing using a MiSeq system (Illumina, UK) at GenProbio srl (Parma, Italy) following the supplier's protocol (Illumina, UK). Fastq files of the paired-end reads obtained from targeted genome sequencing of the isolated strains were used as input for the genome assemblies through the MEGAnnotator pipeline (48). The MIRA program (version 4.0.2) was used for de novo assembly of each bifidobacterial genome sequence (49).
Genome annotation.
Protein-encoding open reading frames (ORFs) were predicted using Prodigal (50). tRNA genes were identified using tRNAscan-SE v1.4 (51), while rRNA genes were detected using RNAmmer v1.2 (52). Gene annotation was defined by means of RAPSearch2 (Reduced Alphabet based Protein similarity Search 2) (53) in a nonredundant protein database provided by the National Center for Biotechnology Information (NCBI) and a hidden Markov model (HMM) search (http://hmmer.org/) of the manually curated Pfam-A protein family database (54). Results were inspected by Artemis (55), which was used for genome analyses of predicted genes and for manual editing where necessary.
Pangenome and identification of shared and unique genes.
Genomes of bifidobacterial type strains (see Table S1), together with bifidobacterial taxa that belong to the same species (Table 1; see also S1), as well as the seven sequenced genomes of novel strains (Table 2) were subjected to a pangenome calculation using PGAP (Pan-Genomes Analysis Pipeline) (56). In order to reduce genome content redundancy, all analyses included a single type strain of each (sub)species. The ORF content of all assessed genomes was organized into functional gene clusters using the GF (gene family) method, which involves comparison of each protein to all other proteins using BLAST analysis, followed by clustering into protein families named as Bifidobacterium-specific clusters of orthologous genes (BifCOGs; cutoff E value of 1 × 10−5 and 50% identity across at least 80% of both protein sequences), using MCL (a graph-theory-based Markov cluster algorithm) (57). Pangenome profiles were built using an optimized algorithm incorporated into PGAP software, based on a presence/absence matrix that included all identified BifCOGs in the analyzed genomes. Consequently, the unique protein families for each bifidobacterial genome were classified. Protein families shared between all genomes, named core BifCOGs, were defined by selecting the families that contained at least one single protein member for each genome.
Phylogenomic comparison between strains.
The concatenated core genome sequences of the genus Bifidobacterium were aligned using MAFFT (Multiple Alignment using Fast Fourier Transform) (58), and the corresponding phylogenomic tree was constructed using the neighbor-joining method in Clustal W version 2.1 (59). The core genome tree was built using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). For each genome pair, a value for the average nucleotide identity (ANI) was calculated using the program JSpecies version 1.2.1 (35). Clusters based on ANI values between bacterial strains were constructed using Multiple Experiment Viewer (MeV) software (60). The Genome-to-Genome Distance Calculator (GGDC) version 2.1 was employed to estimate the DNA-DNA hybridization (DDH) between bifidobacterial taxa using the recommended “Formula 2” (identities/high-scoring segment pairs length) (43).
Accession number(s).
The 13 bifidobacterial genome sequences have been deposited at DDBJ/ENA/GenBank under the accession numbers reported in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) to D.V.S. (in conjunction with Science Foundation Ireland [SFI], grant 15/JP-HDHL/3280) and to M.V. (in conjunction with MIUR, Italy). We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. L.M. is supported by Fondazione Cariparma, Parma, Italy. D.V.S. is a member of the APC Microbiome Institute, funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan (grant SFI/12/RC/2273).
Part of this research was conducted using the high performance computing (HPC) facility of the University of Parma.
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02249-17.
REFERENCES
- 1.Wayne LG. 1988. International Committee on Systematic Bacteriology: announcement of the report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralbl Bakteriol Mikrobiol Hyg A 268:433–434. [DOI] [PubMed] [Google Scholar]
- 2.Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- 3.Rossello-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 4.Fox GE, Wisotzkey JD, Jurtshuk P Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 5.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 6.Cooper JE, Feil EJ. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol 12:373–377. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi M, Sentausa E, Raoult D, Fournier PE. 2014. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol 64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 9.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M. 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J 11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Callaghan A, Bottacini F, O'Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ventura M, Turroni F, Lugli GA, van Sinderen D. 2014. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric 94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 14.Whitman W, Goodfellow M, Kampfer P, Busse H-J, Trujillo M, Ludwig W, Suzuki K-I, Parte A (ed). 2012. The actinobacteria. Bergey's manual of systematic bacteriology, vol. 5 Springer-Verlag, New York, NY. [Google Scholar]
- 15.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 16.Bottacini F, O'Connell Motherway M, Kuczynski J, O'Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Endo A, Futagawa-Endo Y, Schumann P, Pukall R, Dicks LM. 2012. Bifidobacterium reuteri sp. nov., Bifidobacterium callitrichos sp. nov., Bifidobacterium saguini sp. nov., Bifidobacterium stellenboschense sp. nov. and Bifidobacterium biavatii sp. nov. isolated from faeces of common marmoset (Callithrix jacchus) and red-handed tamarin (Saguinus midas). Syst Appl Microbiol 35:92–97. doi: 10.1016/j.syapm.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Tsuchida S, Takahashi S, Nguema PP, Fujita S, Kitahara M, Yamagiwa J, Ngomanda A, Ohkuma M, Ushida K. 2014. Bifidobacterium moukalabense sp. nov., isolated from the faeces of wild west lowland gorilla (Gorilla gorilla gorilla). Int J Syst Evol Microbiol 64:449–455. doi: 10.1099/ijs.0.055186-0. [DOI] [PubMed] [Google Scholar]
- 19.Modesto M, Michelini S, Stefanini I, Sandri C, Spiezio C, Pisi A, Filippini G, Biavati B, Mattarelli P. 2015. Bifidobacterium lemurum sp. nov., from faeces of the ring-tailed lemur (Lemur catta). Int J Syst Evol Microbiol 65:1726–1734. doi: 10.1099/ijs.0.000162. [DOI] [PubMed] [Google Scholar]
- 20.Michelini S, Oki K, Yanokura E, Shimakawa Y, Modesto M, Mattarelli P, Biavati B, Watanabe K. 2016. Bifidobacterium myosotis sp. nov., Bifidobacterium tissieri sp. nov. and Bifidobacterium hapali sp. nov., isolated from faeces of baby common marmosets (Callithrix jacchus L.). Int J Syst Evol Microbiol 66:255–265. doi: 10.1099/ijsem.0.000708. [DOI] [PubMed] [Google Scholar]
- 21.Michelini S, Modesto M, Pisi AM, Filippini G, Sandri C, Spiezio C, Biavati B, Sgorbati B, Mattarelli P. 2016. Bifidobacterium eulemuris sp. nov. isolated from the faeces of the black lemur (Eulemur macaco). Int J Syst Evol Microbiol 66:1567–1576. doi: 10.1099/ijsem.0.000924. [DOI] [PubMed] [Google Scholar]
- 22.Stropko SJ, Pipes SE, Newman JD. 2014. Genome-based reclassification of Bacillus cibi as a later heterotypic synonym of Bacillus indicus and emended description of Bacillus indicus. Int J Syst Evol Microbiol 64:3804–3809. doi: 10.1099/ijs.0.068205-0. [DOI] [PubMed] [Google Scholar]
- 23.Dunlap CA. 2015. Phylogenomic analysis shows that ‘Bacillus vanillea’ is a later heterotypic synonym of Bacillus siamensis. Int J Syst Evol Microbiol 65:3507–3510. doi: 10.1099/ijsem.0.000444. [DOI] [PubMed] [Google Scholar]
- 24.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. 2011. The Salmonella enterica pan-genome. Microb Ecol 62:487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouli L, Merhej V, Fournier PE, Raoult D. 2015. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect 7:72–85. doi: 10.1016/j.nmni.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kweon O, Kim SJ, Blom J, Kim SK, Kim BS, Baek DH, Park SI, Sutherland JB, Cerniglia CE. 2015. Comparative functional pan-genome analyses to build connections between genomic dynamics and phenotypic evolution in polycyclic aromatic hydrocarbon metabolism in the genus Mycobacterium. BMC Evol Biol 15:21. doi: 10.1186/s12862-015-0302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milani C, Duranti S, Lugli GA, Bottacini F, Strati F, Arioli S, Foroni E, Turroni F, van Sinderen D, Ventura M. 2013. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl Environ Microbiol 79:4304–4315. doi: 10.1128/AEM.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daubin V, Gouy M, Perriere G. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res 12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daubin V, Gouy M, Perriere G. 2001. Bacterial molecular phylogeny using supertree approach. Genome Inform 12:155–164. [PubMed] [Google Scholar]
- 33.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ventura M, Canchaya C, Del Casale A, Dellaglio F, Neviani E, Fitzgerald GF, van Sinderen D. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int J Syst Evol Microbiol 56:2783–2792. doi: 10.1099/ijs.0.64233-0. [DOI] [PubMed] [Google Scholar]
- 35.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc Lond B Biol Sci 361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Briczinski EP, Loquasto JR, Barrangou R, Dudley EG, Roberts AM, Roberts RF. 2009. Strain-specific genotyping of Bifidobacterium animalis subsp. lactis by using single-nucleotide polymorphisms, insertions, and deletions. Appl Environ Microbiol 75:7501–7508. doi: 10.1128/AEM.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ventura M, Reniero R, Zink R. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl Environ Microbiol 67:2760–2765. doi: 10.1128/AEM.67.6.2760-2765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson PJ, Fitzgerald GF, Stanton C, Ross RP. 2004. The evaluation of a mupirocin-based selective medium for the enumeration of bifidobacteria from probiotic animal feed. J Microbiol Methods 57:9–16. doi: 10.1016/j.mimet.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 41.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M. 2017. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 43.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duranti S, Mangifesta M, Lugli GA, Turroni F, Anzalone R, Milani C, Mancabelli L, Ossiprandi MC, Ventura M. 2017. Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). Int J Syst Evol Microbiol 67:3987–3995. doi: 10.1099/ijsem.0.002243. [DOI] [PubMed] [Google Scholar]
- 45.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. 2009. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 46.Ventura M, Zink R. 2002. Rapid identification, differentiation, and proposed new taxonomic classification of Bifidobacterium lactis. Appl Environ Microbiol 68:6429–6434. doi: 10.1128/AEM.68.12.6429-6434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 49.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WE, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res 14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Tang H, Ye Y. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 60.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378. [DOI] [PubMed] [Google Scholar]
- 61.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2016. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.