ABSTRACT
Interspecific hybridization is a valuable tool for developing and improving brewing yeast in a number of industry-relevant aspects. However, the genomes of newly formed hybrids can be unstable. Here, we exploited this trait by adapting four brewing yeast strains, three of which were de novo interspecific lager hybrids with different ploidy levels, to high ethanol concentrations in an attempt to generate variant strains with improved fermentation performance in high-gravity wort. Through a batch fermentation-based adaptation process and selection based on a two-step screening process, we obtained eight variant strains which we compared to the wild-type strains in 2-liter-scale wort fermentations replicating industrial conditions. The results revealed that the adapted variants outperformed the strains from which they were derived, and the majority also possessed several desirable brewing-relevant traits, such as increased ester formation and ethanol tolerance, as well as decreased diacetyl formation. The variants obtained from the polyploid hybrids appeared to show greater improvements in fermentation performance than those derived from diploid strains. Interestingly, it was not only the hybrid strains, but also the Saccharomyces cerevisiae parent strain, that appeared to adapt and showed considerable changes in genome size. Genome sequencing and ploidy analysis revealed that changes had occurred at both the chromosome and single nucleotide levels in all variants. Our study demonstrates the possibility of improving de novo lager yeast hybrids through adaptive evolution by generating stable and superior variants that possess traits relevant to industrial lager beer fermentation.
IMPORTANCE Recent studies have shown that hybridization is a valuable tool for creating new and diverse strains of lager yeast. Adaptive evolution is another strain development tool that can be applied in order to improve upon desirable traits. Here, we apply adaptive evolution to newly created lager yeast hybrids by subjecting them to environments containing high ethanol levels. We isolated and characterized a number of adapted variants which possess improved fermentation properties and ethanol tolerance. Genome analysis revealed substantial changes in the variants compared to the original strains. These improved variant strains were produced without any genetic modification and are suitable for industrial lager beer fermentations.
KEYWORDS: adaptive evolution, beer, ethanol, flavor, hybridization, yeast
INTRODUCTION
Yeast breeding and hybridization have in recent years been shown to be promising tools for developing and improving brewing yeast in a number of industry-relevant aspects (1–4). These include improving fermentation rates, sugar use, and aroma compound production. However, the genomes of newly formed hybrids tend to be unstable, and these may undergo substantial structural changes after a hybridization event (5–9). As yeast is commonly reused for multiple consecutive fermentations in industrial breweries (even over 10 times depending on brewery), it is vital that the genomes of any newly developed yeast strains remain stable to ensure product quality during industrial use. Yeast encounter a range of challenges during brewery fermentations, such as low oxygen availability, osmotic stress, CO2 accumulation, nutrient limitation, and ethanol toxicity (10), which may contribute to faster changes in the genome size (11, 12). Yeast strains with larger genomes (i.e., with a higher ploidy level) in particular have been shown to show greater changes in genome size under such conditions (9, 11–15). This may especially cause concerns when a rare mating approach is used for hybridization (3, 16). However, genome stabilization can be achieved, for example, by growing newly formed hybrids for 30 to 70 generations under fermentative conditions (2, 9). Phenotypic changes may occur, however, during the genome stabilization process, altering the properties of the original hybrid (6).
To influence the changes occurring during the stabilization process, an adaptive or experimental evolution approach could be applied. Studies have demonstrated that adaptive evolution can be used, for example, to obtain strains with increased tolerance to ethanol (14, 17–19), high-gravity wort (20, 21), lignocellulose hydrolysates (22), and extreme temperatures (23, 24), as well as improved consumption of various sugars (25–27). Numerous recent studies utilizing experimental evolution have also provided valuable information on what genetic changes take place in the yeast strains during adaptation to various stresses (5, 8, 14, 15, 22–24, 28, 29). Evolution experiments with yeast hybrids have shown that various changes may occur during adaptation, including a partial loss of one of the parental subgenomes, loss of heterozygosity and selection of superior alleles, and the formation of fusion genes following translocations (5, 8, 22, 28). Studies have also revealed that the ploidy of the yeast has an effect on adaptability, with tetraploid strains appearing to adapt more rapidly than diploid strains (12, 15, 30). Taking this into consideration, we sought to not only stabilize but simultaneously adapt a range of our newly created lager yeast hybrids to conditions normally encountered during brewery fermentations. One of the main stresses brewing yeast encounter during the fermentation process is that of increasing ethanol concentrations (10), particularly as interest from the industry toward very high-gravity fermentations (i.e., those with wort containing over 250 g of extract/liter, resulting in beer with alcohol contents above 10% [vol/vol]) has increased in recent years (31).
In this study, we therefore exposed 3 de novo lager yeast hybrids of different ploidy and a common S. cerevisiae ale parent strain to 30 consecutive batch fermentations in media containing 10% ethanol in an attempt to retrieve variant strains with increased tolerance to ethanol. Following the adaptation stage, isolates were screened and selected based on their ability to ferment both wort sugars efficiently in the presence of ethanol and high-gravity wort. Eight variant strains, two from each of the original strains, were ultimately selected and compared in 2-liter-scale wort fermentations. Analysis of the fermentations and resulting beers revealed that the majority of the variants appeared to outperform the original strains in regard to fermentation rate. Furthermore, the majority of the variants produced beers with higher concentrations of desirable aroma-active esters and lower concentrations of many undesirable aroma compounds, such as higher alcohols and diacetyl. The genomes of the variant strains were also sequenced, and genome analysis revealed that changes had occurred at both the chromosome and single nucleotide levels.
RESULTS
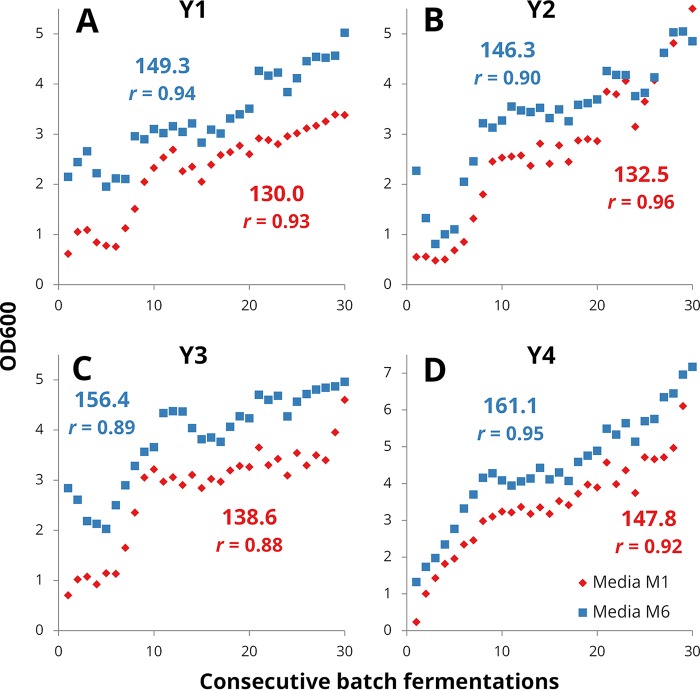
Three different de novo lager yeast hybrids, generated in previous studies by our lab (3, 32), along with an S. cerevisiae ale parent strain (common to all three hybrids), were subjected to the adaptation process (Table 1). The ploidy of the interspecific hybrids varied from around 2.4N to 4N. These four unevolved yeast strains (wild type), referred to as Y1 to Y4, as shown in Table 1, were grown for 30 consecutive batch fermentations in two different media containing 10% ethanol in an attempt to generate ethanol-tolerant variants with improved fermentation properties (Fig. 1A). The first medium, M1, contained 2% maltose as a fermentable carbon source, while the second, M6, contained 1% maltose and 1% maltotriose. These sugars were chosen because they are the main sugars in all-malt wort. Over the 30 fermentations, approximately 130 to 161 yeast generations were achieved, depending on the yeast strain and growth medium (Fig. 2). The optical density at the end of each batch fermentation increased from around 2.5- to over 10-fold depending on the yeast strain, suggesting adaptation to the high ethanol concentration (Fig. 2). Isolates from each adaptation line were obtained after 10, 20, and 30 fermentations (these evolved isolates are referred to as variants) by randomly selecting from among the fastest growing colonies on agar plates containing solidified versions of the same adaptation medium (Fig. 1B).
TABLE 1.
Yeast strains used in the study
| Strain working code | Species | Information | Measured ploidy (SD) | Source or reference |
|---|---|---|---|---|
| Unevolved wild-type strains | ||||
| Y1 | S. cerevisiae | S. cerevisiae ale strain (VTT-A81062) | 1.95 (0.15) | VTT Culture Collection |
| Y2 | S. cerevisiae × S. eubayanus | Tetraploid interspecific hybrid between strain Y1 and S. eubayanus type strain VTT-C12902; known as “hybrid H1” in the source study | 4.03 (0.25) | 32 |
| Y3 | S. cerevisiae × S. eubayanus | Triploid interspecific hybrid between strain Y1 and S. eubayanus type strain VTT-C12902; known as “hybrid B3” in the source study | 2.98 (0.22) | 3 |
| Y4 | S. cerevisiae × S. eubayanus | Interspecific hybrid containing DNA from strain Y1, S. cerevisiae WLP099 (White Labs, Inc.), and the S. eubayanus type strain VTT-C12902; known as “hybrid T2” in the source study | 2.38 (0.24) | 32 |
| Evolved variant strains | ||||
| Y1_M1 | S. cerevisiae | Variant obtained from Y1; isolated after 30 fermentations from medium M1, replicate A | 2.02 (0.21) | Isolated in this study |
| Y1_M6 | S. cerevisiae | Variant obtained from Y1; isolated after 30 fermentations from medium M6, replicate A | 3.64 (0.17) | Isolated in this study |
| Y2_M1 | S. cerevisiae × S. eubayanus | Variant obtained from Y2; isolated after 30 fermentations from medium M1, replicate B | 3.47 (0.26) | Isolated in this study |
| Y2_M6 | S. cerevisiae × S. eubayanus | Variant obtained from Y2; isolated after 30 fermentations from medium M6, replicate B | 3.57 (0.31) | Isolated in this study |
| Y3_M1 | S. cerevisiae × S. eubayanus | Variant obtained from Y3; isolated after 30 fermentations from medium M1, replicate B | 3.03 (0.27) | Isolated in this study |
| Y3_M6 | S. cerevisiae × S. eubayanus | Variant obtained from Y3; isolated after 30 fermentations from medium M6, replicate B | 3.19 (0.23) | Isolated in this study |
| Y4_M1 | S. cerevisiae × S. eubayanus | Variant obtained from Y4; isolated after 30 fermentations from medium M1, replicate A | 2.27 (0.25) | Isolated in this study |
| Y4_M6 | S. cerevisiae × S. eubayanus | Variant obtained from Y4; isolated after 20 fermentations from medium M6, replicate B | 2.27 (0.24) | Isolated in this study |
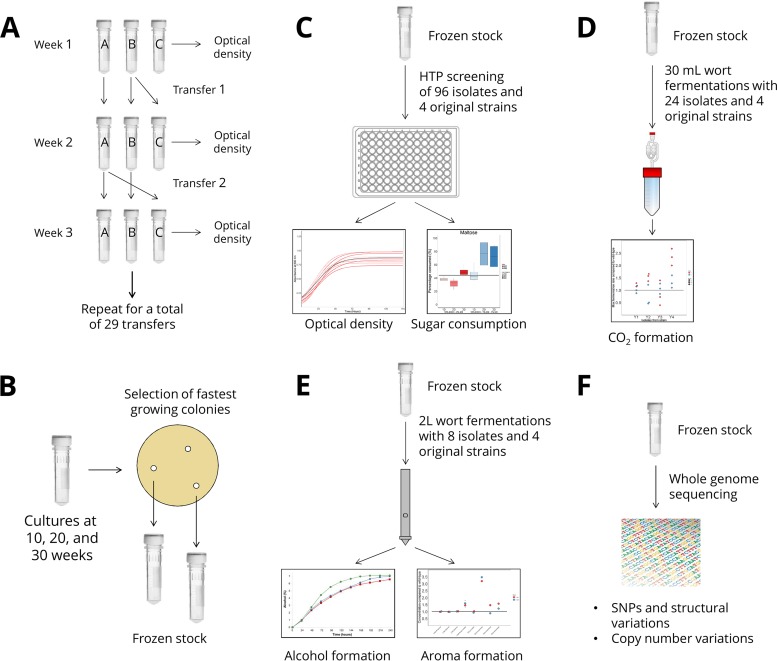
FIG 1.
Experimental overview. (A) Thirty consecutive batch fermentations were carried out with four different yeast strains and two different ethanol-containing media in duplicate adaptation lines. (B) An initial set of isolates were obtained by selecting fast-growing colonies on solidified versions of the adaptation media. (C) High-throughput (HTP) screening of all the isolates was performed in a malt extract-based screening medium containing ethanol. The best-performing isolates were chosen for further screening based on maltose and maltotriose consumption. (D) Small-scale wort fermentations were performed with selected isolates to ensure they were able to ferment wort efficiently and perform in media without exogenous ethanol. (E) Two-liter-scale wort fermentations replicating industrial conditions were performed with 8 variant strains (Table 1), and vital aroma compounds of the resulting beers were analyzed. (F) The genomes of the 8 variant strains were sequenced and compared to those of the wild-type strains. For more information, see Materials and Methods.
FIG 2.
The optical densities at the end of each consecutive batch fermentation with yeast strains Y1 (A), Y2 (B), Y3 (C), and Y4 (D) in the two different ethanol-containing media (red diamonds, medium M1 [10% ethanol, 2% maltose]; blue squares, medium M6 [10% ethanol, 1% maltose, 1% maltotriose]). The cumulative number of yeast generations after the 30th batch fermentation and Pearson's correlation coefficient (r) between the optical densities and number of consecutive fermentations are presented above in blue and below in red for the yeast grown in media M6 and M1, respectively.
Screening of isolates reveals improved sugar consumption and fermentation rates.
The 96 isolates that were obtained from the adaptation fermentations were then subjected to high-throughput screening in a malt-based medium containing ethanol and sorbitol (Fig. 1C). The ethanol and sorbitol were added to replicate the stresses to which the yeast is exposed during brewery fermentations. The majority of the evolved variant strains grew similarly to the wild-type strains, and all strains were able to reach stationary-growth phase during the 144-h cultivation period (see Fig. S1 in the supplemental material). As the objective was to select variant strains with an enhanced fermentation rate, rather than enhanced growth, we also monitored the sugar concentrations in the media at three time points.
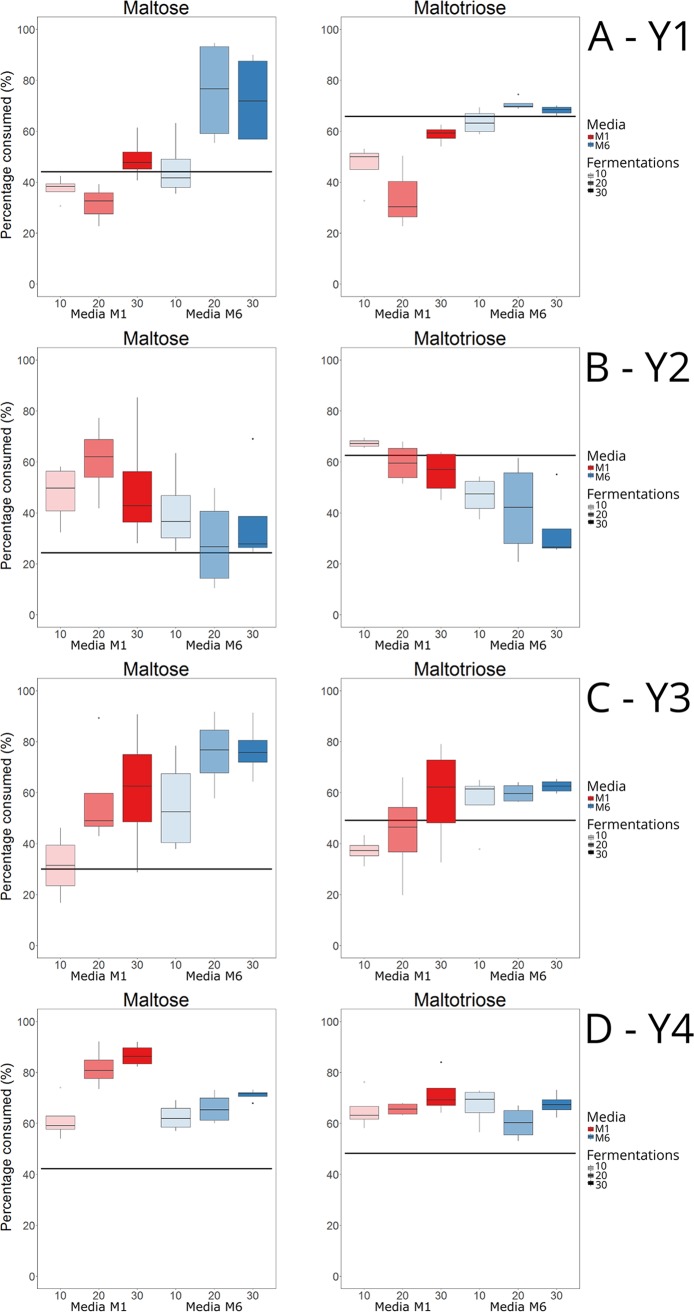
There were considerable differences in the amounts of maltose and maltotriose consumed between the wild-type and variant strains after 144 h of fermentation (Fig. 3). There was no obvious pattern between the consumption of the different sugars, the isolation time points (i.e., the amount of consecutive batch fermentations), and the two different adaptation media among the variant strains. In many cases, the largest consumption of both maltose and maltotriose was observed with variants that had been isolated after 30 batch fermentations. However, with variants obtained from yeast strain Y2, the average maltose and maltotriose consumption of variants obtained after 30 batch fermentations was lower than those isolated at earlier stages (Fig. 3B). Nevertheless, the variant strain derived from Y2 with the highest maltose consumption was obtained after 30 batch fermentations. Several variant strains from all four wild-type strains (Y1 to Y4) showed higher sugar consumption than the wild-type strains. In total, 83% of the variants consumed more maltose, and 60% consumed more maltotriose than the wild-type strains during the screening fermentations (consumptions of up to 94% maltose and 84% maltotriose were observed in the variants). Interestingly, all variants that consumed more maltotriose than the wild-type strains also consumed more maltose. Excluding maltotriose from the adaptation media did not appear to have any negative effect on maltotriose consumption in the variants, as the variant strains derived from Y2, Y3, and Y4 with the highest maltotriose consumption were obtained from the adaptation media lacking maltotriose. Six variants per wild-type strain were selected for further screening in small-scale wort fermentations, based on the highest sugar consumptions and the requirement that they were derived from separate adaptation lines and isolation time points.
FIG 3.
The percentages of maltose and maltotriose consumed by isolates (selected after 10, 20, and 30 consecutive batch fermentations) of the yeast strains Y1 (A), Y2 (B), Y3 (C), and Y4 (D) after 144 h of fermentation in the screening media (6.2% malt extract, 10% sorbitol, 5% ethanol) during high-throughput screening in 96-well plates. The black line depicts the amount of sugar consumed by the wild-type strain (average calculated from 12 to 16 replicate fermentations). For each of the three isolation points (10, 20, and 30 consecutive batch fermentations), four isolates were selected per yeast strain per medium (a total of 24 isolates per parent strain). Three replicate fermentations were carried out for each isolate.
Small-scale wort fermentations were used as a final screening step to ensure that the selected variants were also able to ferment wort efficiently and perform in media without exogenous ethanol (Fig. 1D). A 15 °Plato (°P) high-gravity wort, i.e., a wort similar to what is used in the brewing industry, was used for the fermentations. They revealed that 17 out of the 24 tested variants outperformed the wild-type strains from which they were derived (Fig. 4) in regard to the maximum fermentation rate that was observed. Of these 17 variants, 13 also reached a significantly higher (P < 0.05 as determined by two-tailed Student's t test) final alcohol level after the 9 days of fermentation (data not shown). Variants which had been adapted in the medium containing 2% maltose as the sugar source (medium M1) appeared to outperform those obtained from the medium containing both 1% maltose and maltotriose (medium M6). One isolate per yeast strain and medium (for a total of 8 isolates) were selected for a more thorough characterization in 2-liter-scale wort fermentations. These isolates (listed in Table 1) were selected based on the highest fermentation rates, and those that had undergone 30 batch fermentations were also preferentially selected.
FIG 4.
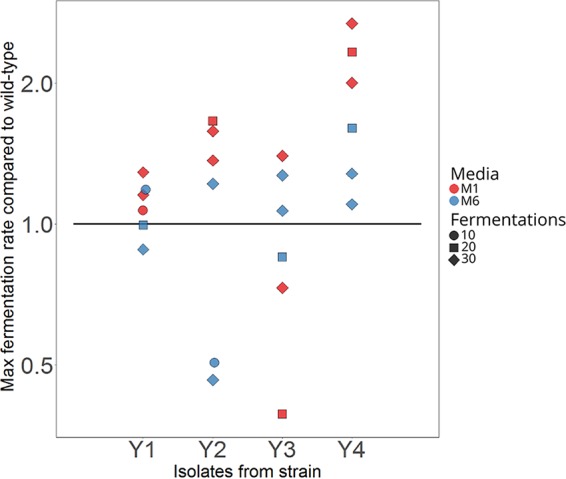
The maximum (Max) fermentation rates of 24 isolates compared to those of their wild-type strains during small-scale fermentations in 15 °P all-malt wort at 15°C. Isolates were selected based on sugar consumption during high-throughput screening and were from the two different growth media (M1 and M6 in red and blue, respectively) and three different isolation points (10, 20, and 30 consecutive batch fermentations, shown by circles, squares, and diamonds, respectively). The black line depicts the maximum fermentation rate of the wild-type strains. Duplicate fermentations were carried out for each isolate.
Enhanced performance confirmed in 2-liter-scale wort fermentations.
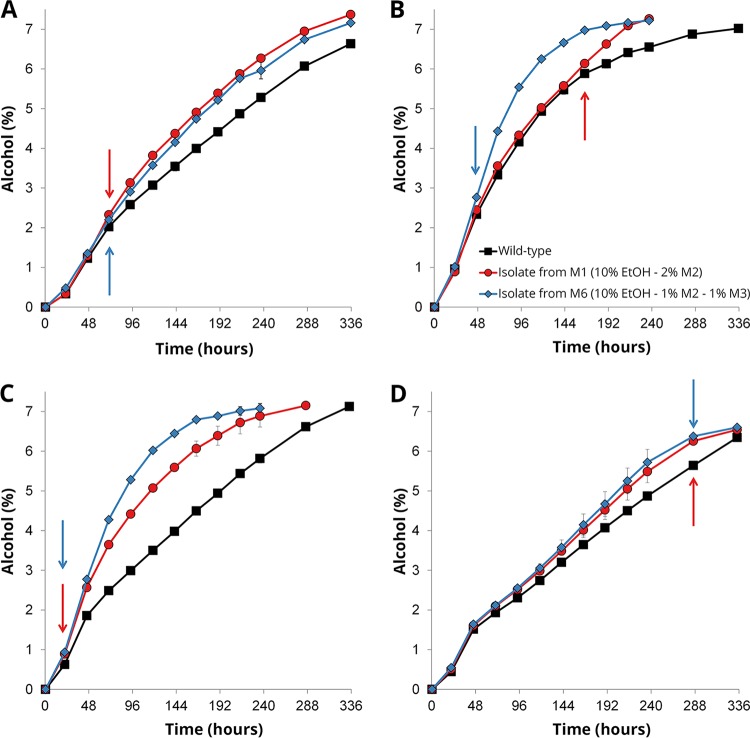
In order to examine how the variant strains (Table 1) perform in a brewery environment, 2-liter-scale tall-tube fermentations were carried out in high-gravity 15 °P all-malt wort at 15°C (Fig. 1E). These conditions were chosen to replicate those of industrial lager fermentations. All eight variant strains appeared to outperform their respective wild-type strains during these fermentations in regard to fermentation rate (Fig. 5 and Table S2). Time points after which a significant difference (P < 0.05 as determined by Student's t test) was observed between the variant and the wild-type strain are marked with arrows in the plot in Fig. 5. The largest differences in fermentation compared to the wild-type strains were observed with the variants of Y2, i.e., the tetraploid interspecific S. cerevisiae × Saccharomyces eubayanus hybrid, and those of Y3, i.e., the triploid interspecific S. cerevisiae × S. eubayanus hybrid. For most strains, the differences between variant and wild-type strains seemed to appear after approximately 48 h of fermentation. Before this time point, it is mainly the monosaccharides that are consumed from the wort and the alcohol level is still below 2% (vol/vol) (Fig. S2). The sugar profiles during fermentation also revealed that improved maltose consumption appears to be one of the main causes for the increased fermentation rate of the variant strains. Three of the variants, derived from strains Y2 and Y4, only showed a difference compared to the wild-type strain late in fermentation. These observations suggest that the observed differences may be due to the variant strains possessing an enhanced ability to ferment maltose and maltotriose or to tolerate increasing ethanol concentrations in the wort.
FIG 5.
The alcohol content (% alcohol by volume [ABV]) in the beers fermented on a 2-liter-scale from 15 °P wort at 15°C with wild-type (black squares) and variant (red circles and blue diamonds) strains derived from yeast strains Y1 (A), Y2 (B), Y3 (C), and Y4 (D). Values are means from two independent fermentations, and error bars, where visible, represent the standard deviation. Arrows indicate the time point after which a significant difference was observed between the variant and wild-type strains, as determined by two-tailed Student's t test (P < 0.05).
We also wanted to compare the aroma profiles of the beers produced with the variant strains with those produced with the wild-type strains to ensure that the adaptation process had not introduced any negative side effects to the resulting beer. Genetic hitchhiking is common during adaptive evolution (33), and here, we only screened and selected for an increased fermentation rate. Analysis of the aroma-active higher alcohols and esters in the beers revealed that the variant strains, in general, produced equal or smaller amounts of unwanted higher alcohols and equal or larger amounts of desirable esters compared to the wild-type strains (Fig. 6). The concentrations of 3-methylbutyl acetate, possessing a banana-like flavor (34), and ethyl esters, possessing fruity and apple-like flavors (34), in particular appeared to increase in the variant strains. We also monitored the concentrations of diacetyl, an important unwanted off-flavor in lager beer fermentations (35), and the results revealed that five out of eight variant strains had produced significantly lower concentrations of diacetyl than the wild-type strains, while the other three produced concentrations that were equal to the wild-type strains. Hence, the results revealed that the adaptation process had yielded not only variant strains with improved fermentation performance in wort, but also, inadvertently, strains that produced more desirable aroma profiles. In addition, no further genome loss or major rearrangements were detected in the eight variant strains over 80 generations of growth (Fig. S3).
FIG 6.
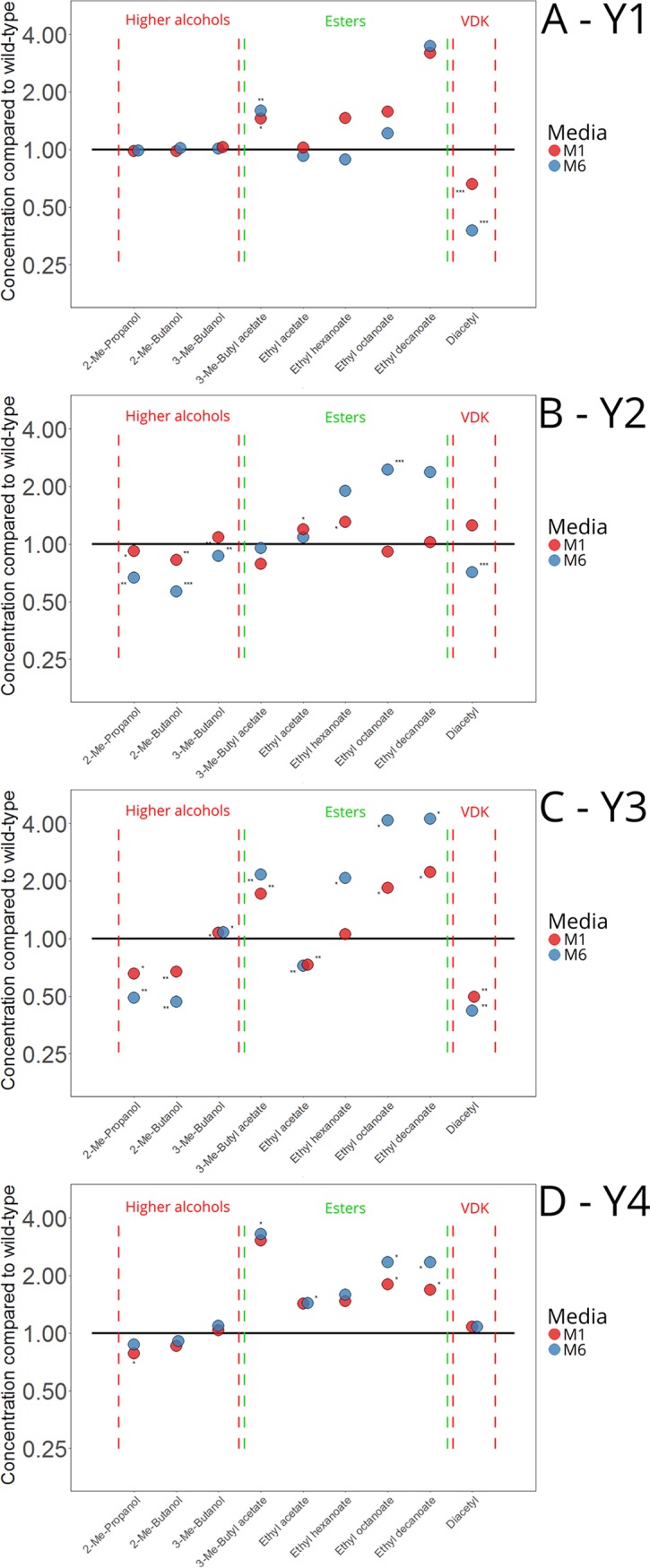
The concentrations of nine yeast-derived aroma compounds in the beers fermented with the variant strains relative to those fermented with the wild-type strains Y1 (A), Y2 (B), Y3 (C), and Y4 (D). Values are means from two independent fermentations, and asterisks depict a significant difference in the variant compared to the wild-type, as determined by two-tailed Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The black lines depict the concentrations produced by the wild-type strains. The compounds are displayed by type: higher alcohols, esters, and vicinal diketones (VDK). The types are colored by whether the compounds are generally considered undesirable (red) or desirable (green) in lager beer (34). Me, methyl.
Ethanol tolerance and accumulation capacity of variant strains.
As the variant strains (Table 1) were derived from repeated exposure to high ethanol concentrations and they performed better particularly toward the end of high-gravity wort fermentations, we wanted to test and compare their ethanol tolerance and accumulation capacities to those of the wild-type strains. All wild-type and variant strains were able to grow on yeast extract-peptone-maltose (YPM) agar supplemented with 9% (vol/vol) ethanol, but differences in growth were revealed for certain strains on YPM agar supplemented with 11% (vol/vol) ethanol (Fig. 7). The variant strains derived from Y4 in particular showed better growth at 11% ethanol than the wild-type strain (Fig. 7D). For the variants derived from the other strains (Y1 to Y3), there were no or less-obvious differences in their abilities to grow in the presence of 11% ethanol. For strain Y2, the variant derived from adaptation medium M1 (10% ethanol and 2% maltose), Y2_M1, also appeared to grow better than the wild-type strain at 11% (vol/vol) ethanol (Fig. 7B). The ethanol accumulation capacity, which measures both the osmotolerance and ethanol tolerance of a strain, was significantly higher for both variant strains of Y2 and Y4 than for their wild-type strains, while no significant differences were observed for strain Y3. Surprisingly, the ethanol accumulation capacities of the variants of Y1 were significantly lower than that of the wild-type strain, despite both variant strains appearing to grow slightly better on YPM agar supplemented with 11% ethanol (Fig. 7A).
FIG 7.
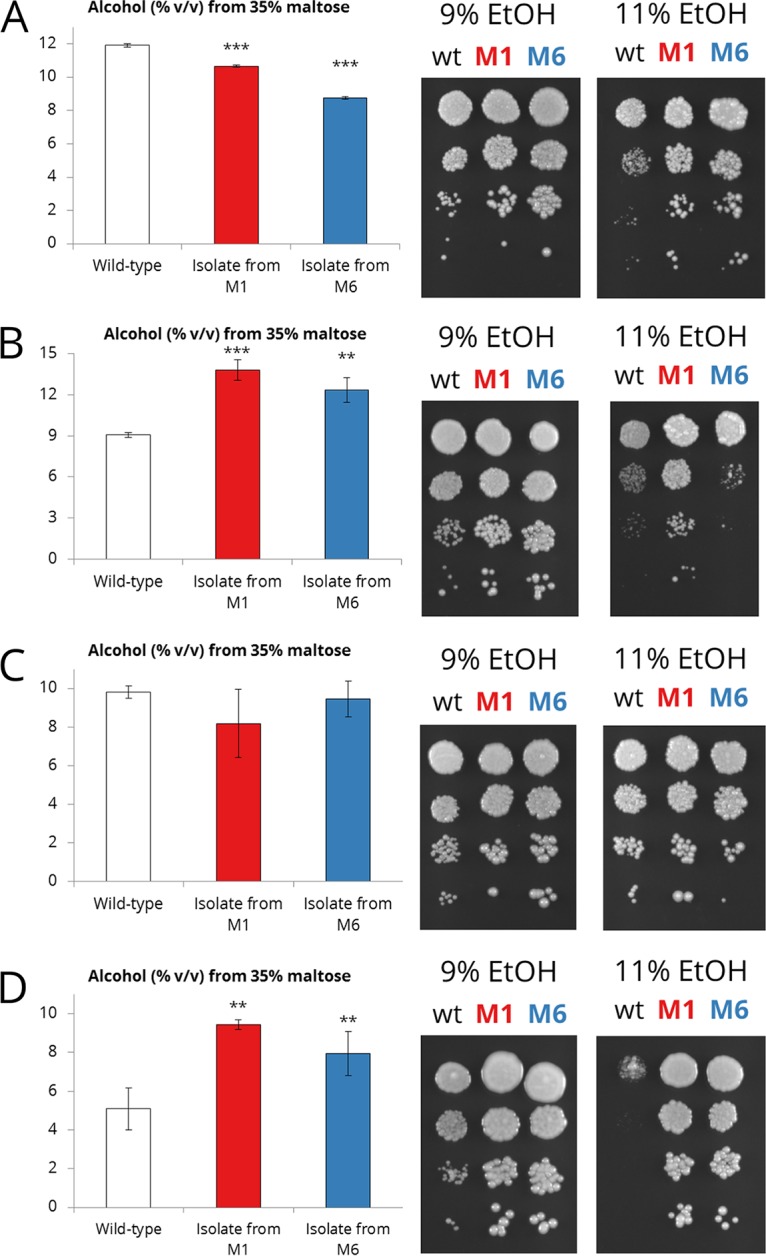
The ethanol accumulation capacity from 35% maltose and the ability to grow on media containing 9% and 11% ethanol of the wild-type (wt) and variant strains derived from yeast strains Y1 (A), Y2 (B), Y3 (C), and Y4 (D). Values are the means from two independent cultures, error bars, where visible, represent the standard deviation, and asterisks depict a significant difference in the variant compared to the wild-type, as determined by two-tailed Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). v/v, vol/vol.
Sequencing reveals large-scale changes in genomes.
In order to investigate what genetic changes had occurred in the variant strains during the adaptation process, whole-genome sequencing and estimation of ploidy by flow cytometry were performed (Fig. 1F). Ploidy analysis revealed that relatively large changes in genome size had occurred for many of the variant strains (Table 1). The genome of the variant derived from the diploid S. cerevisiae strain Y1 and adaptation medium M6, Y1_M6, had almost doubled in size, while the genomes of both variants derived from the tetraploid interspecies hybrid Y2 (Y2_M1 and Y2_M6) had decreased by approximately 0.5N. Smaller changes were observed in the genome sizes of the variants derived from the triploid and diploid interspecies hybrids Y3 and Y4.
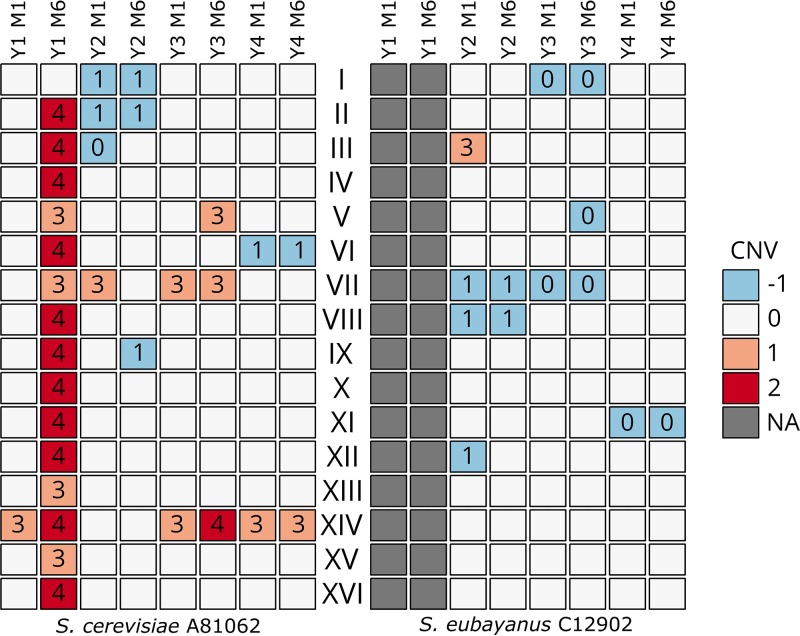
Whole-genome sequencing of the wild-type and variant strains (average coverage ranged from 152× to 1,212×) revealed both chromosome duplications and losses across all variant strains (Fig. 8 and Table S4). As indicated by the ploidy analysis, the largest changes in chromosome copy numbers were observed in the variant derived from strain Y1 and adaptation medium M6 (Y1_M6), where the majority of the chromosomes were now present in two extra copies. The variants derived from interspecies hybrids (Y2 to Y4) had, on average, gained 1.7 and lost 3.3 chromosomes. A greater amount of chromosome copy number changes was also observed in the variants derived from the polyploid hybrids (6.5, 5.5, and 3 for Y2, Y3, and Y4, respectively). In regard to the two subgenomes of the hybrid variants, there were significantly more (P < 0.05) chromosome duplications in the S. cerevisiae subgenome (average, 1.5 per variant) than in the S. eubayanus subgenome (average, 0.17 per variant). In the S. cerevisiae subgenome, there was no significant difference between the amounts of chromosome duplications and losses (average, 1.33 per variant). In contrast, the S. eubayanus subgenome had experienced significantly more (P < 0.003) chromosome losses (average, 2 per variant) than duplications.
FIG 8.
Chromosome copy number variations (CNV) in the S. cerevisiae A81062 (left) and S. eubayanus C12902 (right) subgenomes of the variant strains compared to the wild-type strains (the numbers inside the cells indicate the estimated absolute chromosome copy number). A blue color indicates a chromosome loss, while a red color indicates a chromosome duplication compared to the wild-type strain (e.g., −1 corresponds to one less chromosome in the variant compared to the wild-type strain). NA, not available. The chromosome copy numbers of the wild-type strains are listed in Table S3 in the supplemental material.
Common chromosome copy number changes were seen in several variants, as the S. cerevisiae-derived chromosomes VII and XIV were amplified in four and six variants, respectively, while the S. eubayanus-derived chromosome VII had been lost in four variants (Fig. 8). Interestingly, the S. cerevisiae-derived chromosome VII carries several copies of α-glucoside transporters, a single copy of MAL31, and two copies of MAL11/AGT1, and a moderate positive correlation (Pearson's correlation coefficient [r] = 0.68) was observed between the maximum maltose consumption rates during the 2-liter-scale wort fermentations and the combined copy numbers of known genes related to maltose transport (Fig. S4 and Table S5).
The genomes of the variant strains varied not only at the chromosomal level, as several unique single nucleotide polymorphisms (SNPs) and insertions and deletions (indels) were also observed. A total of 109 unique mutations were identified in the eight variant strains (Table S1). Of these, 64.2% mutations were intergenic, 8.3% were synonymous, and 27.5% were nonsynonymous. In addition, 21% of the mutations were hemi- or homozygous. The nonsynonymous mutations caused both missense mutations and frameshift mutations (Table 2 and S1), and at least one was present in all variant strains. Interestingly, nonsynonymous mutations in three genes (IRA2, HSP150, and MNN4) were found in multiple variants. In the case of IRA2, an inhibitory regulator of the RAS-cAMP pathway (36) which contained nonsynonymous mutations in three of the variants, both the S. cerevisiae and S. eubayanus orthologues were affected. In addition to the unique mutations that were observed in the variant strains, several of the variant strains had undergone a loss of heterozygosity in large regions of several S. cerevisiae-derived chromosomes (Fig. S5 to S16). The left arms of chromosomes X and XII, as well as the right arm of chromosome XV, for example, were affected in multiple variants. No unique translocations or complex structural variations were identified in the variant strains. The lack of chromosome chimerization and presence of mainly heterozygous SNPs in the variant strains suggest that meiosis has not occurred during the adaptation process, and instead, genome stabilization occurred during vegetative growth through mitotic recombination and chromosome losses (37).
TABLE 2.
Nonsynonymous mutations discovered in the variant strainsa
| Variant strain | Gene(s) with: |
|
|---|---|---|
| Missense mutation | Frameshift mutation | |
| Y1_M1 | MNN4 (Scer) | |
| Y1_M6 | PEX11 (Scer), TPO1 (Scer) | |
| Y2_M1 | TOD6 (Seub) | BSC1 (Scer)*, COS9 (Scer)*, DAL81 (Scer), HSP150 (Scer)*, RKM3 (Scer)*, UTH1 (Scer) |
| Y2_M6 | YIM1 (Scer), YMC1 (Scer) | MHP1 (Scer)* |
| Y3_M1 | CBP1 (Scer), HAP4 (Scer), IRA2 (Scer), LPL1 (Scer) | MNN4 (Scer), RAT1 (Seub)* |
| Y3_M6 | GET2 (Scer), PRP40 (Scer), EAP1 (Seub)* | FIT3 (Scer), JHD2 (Seub)* |
| Y4_M1 | IRA2 (Scer) | |
| Y4_M6 | BST1 (Seub), SFL1 (Seub), YOR292C (Seub) | HSP150 (Scer), IRA2 (Seub) |
Genes that were affected by mutations in several different variants are in bold. Whether the S. cerevisiae (Scer) or S. eubayanus (Seub) orthologue was affected is indicated in parentheses after the gene name. An asterisk (*) denotes that the mutation was either homo- or hemizygous rather than heterozygous. The positions and nucleotide changes of mutations are listed in Table S1 in the supplemental material.
DISCUSSION
Here, we demonstrate how adaptive evolution can be applied to newly created interspecific lager yeast hybrids, in order to further improve their fermentation traits, and reveal the genetic changes that have occurred in the variant strains during adaptation. By performing 30 consecutive batch fermentations in media supplemented with 10% ethanol, we aimed to generate and select ethanol-tolerant variants of four different brewing yeast strains, 3 of which were interspecific lager hybrids between S. cerevisiae and S. eubayanus. While experimental evolution is typically carried out in chemo- or turbidostats to allow for constant growth in defined nutrient availability (38), we chose here to use serial batch cultures for simplicity and to mimic the growth cycle the yeast encounters in repeated use in brewery fermentations (10). Our results show that the amount of yeast produced during each 1-week fermentation cycle is positively correlated with the number of consecutive batch fermentations, indicating that the strains adapted to the high ethanol concentration in the growth media. Here, approximately 130 to 160 yeast generations were achieved with 30 batch fermentations. Previous studies on adaptive evolution for ethanol tolerance have shown that an increase can be achieved after 140 to 480 generations (14, 17–19), with evidence of increased fitness already after 40 generations in media containing ethanol (14). As the yeast is not constantly in exponential growth, it is expected that the batch fermentation process used here is more time-consuming than a continuous setup, where similar results have been achieved in less time (19).
A two-step screening process was used to ensure that variants exhibiting improved fermentation both in wort and in the presence of ethanol were selected from the adapted population. While growth in the presence of ethanol has been shown to be weakly positively correlated with ethanol production (39), we chose to monitor and select based on sugar consumption instead of growth, since we were interested in improving fermentation. As was revealed from the initial high-throughput screening, the majority of the strains that were isolated throughout the adaptation process outperformed the wild-type strains in regard to the consumption of maltose and maltotriose in the presence of ethanol. However, variants arising from experimental evolution may in some cases exhibit antagonistic pleiotropy, where evolved variants show better fitness only in the environment in which they were selected (14). To prevent this, we performed a final screening step in small-scale wort fermentations. As was revealed during these small-scale fermentations, several isolates did in fact perform worse than the wild-type strains in wort, despite outperforming the wild-type strains in the ethanol-containing media used during high-throughput screenings.
The 2-liter-scale fermentations revealed that all eight of the tested variant strains outperformed the wild-type strains from which they were derived in regard to the fermentation rate. The exact mechanisms for this improvement were not elucidated, but the results seem to suggest that both improved ethanol tolerance and maltose use could have contributed, particularly as differences between variant and wild-type strains seemed to appear as fermentation progressed. The results revealed that many of the variants exhibited improved ethanol tolerance and accumulation capacity, while isolates showed considerable improvements in maltose consumption during high-throughput screening and wort fermentation. In previous studies, where brewing strains have been adapted to very high-gravity wort conditions, variant strains have exhibited increased expression of α-glucoside transporters and genes involved in amino acid synthesis (20, 21). Here, several changes in the genomes of the variant strains were observed that could potentially contribute to the improved fermentation performance and ethanol tolerance. No SNPs, structural variations, or gene-level copy number changes were observed for the genes encoding α-glucoside transporters in the variant strains. However, whole-chromosome copy number duplications of the S. cerevisiae-derived chromosome VII, containing MAL31 and MAL11/AGT1, were observed in several variants, and a moderate positive correlation was observed between the maximum maltose consumption rates and the combined copy numbers of genes related to maltose transport (MALx1, MTT1, and MPH2). The simultaneous copy number decrease of the S. eubayanus-derived chromosome VII in most of the variant strains suggests that the adaptation environment selected preferentially for the S. cerevisiae chromosome. Interestingly, nonsynonymous mutations in IRA2 were observed in three of the variant strains. This gene negatively regulates the RAS-cAMP pathway (36), which in turn is involved in regulating metabolism, cell cycle, and stress resistance (40, 41). Adaptive mutations in this gene have been reported previously (42) after experimental evolution in glucose-limited media, with ira2 deletion strains exhibiting increased fitness. Mutations in IRA2 have also been reported for strains evolved for increased xylose fermentation (43).
Genome analysis of ethanol-tolerant variants has revealed that ethanol tolerance is a complex process affected by several different mechanisms, including general stress response, intracellular signaling, and cell wall and membrane composition and organization (14, 19, 44). Here, we only identified nonsynonymous mutations in one gene, UTH1, which has previously been reported to enhance ethanol tolerance. In turbidostat evolution experiments in high-ethanol media, Avrahami-Moyal et al. (19) found mutations in UTH1 in a fraction of the evolved clones and showed that the deletion of this gene enhanced ethanol tolerance. While testing the direct effect of the nonsynonymous mutations listed in Table 2 on ethanol tolerance by reverse engineering was outside the scope of this particular study, we feel it would be valuable to confirm their role in response to ethanol stress. In fact, several of the genes that were affected here (BST1, CBP1, DAL81, EAP1, HAP4, HSP150, IRA2, MHP1, RAT1, RKM3, SFL1, TOD6, and YIM1) were also found to contain mutations in the evolved clones that were isolated by Voordeckers et al. (14) following exposure to increasing ethanol concentrations. In addition to SNPs and indels, copy number variations are commonly reported in adapted strains (14, 15, 45). While it is thought that chromosome copy number changes allow for a rapid route of adaptation, they have a nonspecific effect on the phenotype (14). Here, we observed several common chromosome losses and duplications. The S. cerevisiae-derived chromosome XIV was amplified in several of the variant strains, and interestingly, this chromosome has been reported through quantitative trait locus (QTL) mapping to carry genes (MKT1, SWS2, and APJ1) associated with increased ethanol tolerance (44).
The variants not only fermented faster, but in most cases also produced greater amounts of desirable esters and smaller amounts of unwanted off-flavors than the wild-type strains. This was unexpected, as we only selected for fermentation, and genetic hitchhiking is common during adaptive evolution (33). In previous studies on brewing yeasts adapted to high-gravity conditions, Ekberg et al. (21) reported increased concentrations of unwanted diacetyl, while Blieck et al. (20) observed slight increases in higher-alcohol and diacetyl concentrations. As the aroma profile was not monitored during the screening process, it is vital to ensure that it is satisfactory for any selected variants. Genome analysis did not reveal any obvious causes for the increase in ester formation and decrease in diacetyl formation, as no SNPs, indels, or gene-level copy number changes affected genes that have previously been reported to be linked with the formation of these compounds. Some genes, such as ATF2 on chromosome VII and ILV6 on chromosome III, were affected by chromosome-level copy number changes and could therefore have altered expression levels. Furthermore, most of the observed mutations were intergenic, and they could therefore have an indirect effect on these phenotypes by affecting gene regulation. Transcript analyses of adapted yeast strains have revealed altered gene expression, e.g., in sugar transport or carbon metabolism, as a central adaptive response (21, 30, 46–48). Analysis of gene expression, along with its effects on intracellular metabolic fluxes, could be explored in future studies to further clarify the phenotypic changes observed here in the adapted variant strains. The loss of heterozygosity has also been reported to be a method of adaptation in hybrid strains (28), and here, for example, we observed a loss of heterozygosity on the right arm of the S. cerevisiae-derived chromosome XV in multiple variant strains. This particular region contains ATF1, the gene encoding the main alcohol acetyltransferase responsible for acetate ester synthesis (49, 50), which in the S. cerevisiae A81062 genome contains four heterozygous SNPs, one of which is nonsynonymous. The two alleles of ATF1 may therefore have slightly different functionality.
Interestingly, the greatest improvements in fermentation compared to the wild-type strains were observed with the polyploid interspecific hybrids. An increased ploidy level may allow for more rapid adaptation (12, 15, 30), presumably from gaining beneficial mutations at higher rates, along with chromosome losses and aneuploidy. The genome sizes of both of the variants derived from the tetraploid hybrid Y2 had decreased, while they had increased slightly for those derived from the triploid hybrid Y3. Aneuploidy and convergence toward a diploid state have commonly been reported during evolutionary engineering (11, 14, 15). Surprisingly, the largest change in genome size was observed for one of the variants derived from the diploid S. cerevisiae parent. The results indicate that under these adaptation conditions, it was not only hybrid strains that possessed an unstable genome, and adapted variants from an industrial ale strain could be obtained without any prior mutagenesis. Evolutionary engineering studies involving interspecific hybrids have indicated that either of the parental subgenomes may be preferentially retained, while the other is lost (5, 8, 28, 51). Although this occurs even under nonstressful conditions, the application of selective pressure can cause more drastic changes (51). Piotrowski et al. (5), for example, showed that growing S. cerevisiae × Saccharomyces uvarum hybrids at high temperatures resulted in the loss of the heat-sensitive S. uvarum subgenome. Here, we saw a greater loss of the S. eubayanus subgenome in the variants derived from interspecific hybrids. It is therefore tempting to speculate that repeating the adaptation process at a lower temperature would have retained more of the S. eubayanus subgenome in the variants, and this could be a target for future studies. In fact, the natural lager yeast hybrids of Saaz-type have retained a larger fraction of the S. eubayanus subgenome than the S. cerevisiae subgenome (52, 53), and it is still unclear whether exposure to cold temperatures has had any effect on its evolution.
In conclusion, adaptive evolution in high-ethanol media was successfully used to generate stable and superior variant strains from 4 different brewing strains, 3 of which were de novo interspecific lager yeast hybrids. These adapted variants outperformed the strains from which they were derived from during wort fermentation, and the majority also possessed several desirable brewing-relevant traits, such as increased ester formation and ethanol tolerance, and decreased diacetyl formation. While not tested here, it is likely that many of the adapted variant strains would also outperform the wild-type strains in very high-gravity wort, i.e., wort containing over 250 g of extract · liter−1, as these fermentations require good tolerance toward both high osmotic pressure and ethanol concentrations (10), which several of the variant strains demonstrated by their ethanol accumulation capacity. Our study demonstrates the possibility of improving de novo lager yeast hybrids through adaptive evolution, and these superior and stable variants are promising candidates for industrial lager beer fermentation. However, to ensure their industrial suitability, the variant strains should still be tested in multiple consecutive industrial-scale fermentations, and finished beers should be tasted by a trained sensory panel (21, 25).
MATERIALS AND METHODS
Yeast strains.
A list of strains used in this study can be found in Table 1. Three different de novo lager yeast hybrids, generated in previous studies by our lab (3, 32), along with an S. cerevisiae ale parent strain (common to all three hybrids) were subjected to the adaptation process. These four unevolved strains (Y1 to Y4) are referred to as wild types. Eight evolved variant strains (two from each of the four wild-type strains) were isolated and subjected to phenotypic and genetic analysis. The ploidy of all the strains was determined by flow cytometry, as described previously (32).
Adaptation in a high-ethanol environment.
The adaptation process was carried out in batch fermentations to mimic consecutive industrial brewery fermentations. Yeasts were grown in sterile 2-ml screw-cap microcentrifuge tubes (catalog no. 10025-754; VWR) containing 1 ml of growth medium. Four different yeast strains (Y1, Y2, Y3, and Y4) were used for the adaptation experiment (see Table 1 for more information). These were grown in two different adaptation media: M1 (1% yeast extract, 2% peptone, 2% maltose, 10% ethanol) and M6 (1% yeast extract, 2% peptone, 1% maltose, 1% maltotriose, 10% ethanol). Each batch fermentation was inoculated to a starting optical density at 600 nm (OD600) of 0.1 with yeast from the previous batch fermentation. The first batch fermentations were inoculated from precultures that were grown overnight in YPM medium (1% yeast extract, 2% peptone, 2% maltose). Tubes were incubated statically for 7 days at 18°C. Three replicate tubes or adaptation lines (A, B, and C) were used for each yeast strain and medium (A and B were never mixed). In order to avoid contamination, the optical density at the end of each batch fermentation was measured only from the third replicate (C), which was subsequently discarded following the OD600 measurement. Prior to OD600 measurement, the tubes were centrifuged, and the supernatant was replaced by an equal volume of 10 mM EDTA to disperse flocculated cells. The number of cell generations in each batch fermentation was approximated from the initial (0.1) and final OD600 as log2(OD600 final/0.1). After 10, 20, and 30 consecutive batch fermentations, 10-μl aliquots of the cell populations were spread onto agar plates containing solidified versions of the growth media (2% agar added) for isolation of variants showing rapid growth. The agar plates were incubated at 18°C until colonies started emerging, and the two largest colonies from each plate were selected for further screening (for a total of four isolates per yeast strain, per medium, per isolation time point). An overview of the adaptation process and initial isolation step is depicted in Fig. 1A and B, respectively.
Screening.
The isolates were initially screened on 96-well plates using a Beckman Coulter liquid-handling robot to select for fast-fermenting variants. Strains were grown in Nunc 96-well polystyrene round-bottom microwell plates (catalog no. 268200; Thermo Scientific), in a 150-μl volume at 14°C, with 1,200 rpm agitation in a Thermo Scientific Cytomat plate hotel (1-mm throw). Precultures were prepared by inoculating 10-μl aliquots of cell suspension from frozen stocks into 140 μl of medium consisting of 6.2% malt extract (Senson Oy, Finland) in the plates. The precultures were incubated for 4 days until all strains had reached stationary phase. The preculture plates were centrifuged, and pellets were resuspended in 50 mM citrate buffer (pH 7.2) to deflocculate the yeast. Ten-microliter aliquots of these suspensions were used to inoculate 140 μl of screening medium for the experimental cultures. The isolates were grown in a screening medium consisting of 6.2% malt extract (Senson Oy), 5% ethanol, and 10% sorbitol. The extract content of this medium was approximately 5 °P (50 g/liter). The ethanol was added to the screening medium to replicate the conditions under which the yeast is exposed toward the end of brewery fermentations, while the sorbitol was added to replicate the increased osmotic pressure to which the yeast is exposed in the beginning of brewery fermentations when sugar-rich wort is used. Each isolate was grown in triplicate, while wild-type strains were grown in at least 12 replicates. The strains and replicates were distributed randomly on the 96-well plates. The fermentations were monitored by measuring the optical density at 595 nm every 3 h using the DTX 880 multimode detector (Beckman Coulter) associated with the robot, and by drawing samples for high-performance liquid chromatography (HPLC) analysis after 48, 96, and 144 h. This screening step is depicted in Fig. 1C. Three isolates per yeast strain and medium (for a total of 24 isolates) were selected for further screening in small-scale wort fermentations based on (i) the highest sugar consumption after 144 h and (ii) the requirement of isolates being from separate adaptation lines and isolation time points.
To ensure that the isolates were also able to ferment actual wort efficiently, a final screening step was conducted by carrying out a set of small-scale wort fermentations. The small-scale fermentations were carried out in plastic 50-ml centrifuge tubes capped with a glycerol-filled airlock. The 24 isolates selected from the previous screening step and the 4 wild-type strains were grown overnight in 50 ml of YPM at 18°C. The precultured yeast was then inoculated into 30 ml of 15 °P all-malt wort at a rate of 15 × 106 viable cells · ml−1. Fermentations were carried out in duplicate at 15°C for 9 days, and these were monitored daily by measuring the mass lost as CO2. This screening step is depicted in Fig. 1D. The maximum fermentation rate of each strain was determined, and one isolate per yeast strain and medium (for a total of 8 isolates) was selected based on (i) the highest fermentation rate and (ii) its being obtained after a larger number of batch fermentations. These eight isolates are listed in Table 1 and were further characterized in 2-liter-scale wort fermentations.
Two-liter-scale wort fermentations.
The eight variant strains were characterized in fermentations performed in a 15 °P high-gravity wort at 15°C. Yeast was propagated essentially as described previously (3), with the use of a “generation 0” fermentation prior to the actual experimental fermentations. The experimental fermentations were carried out in duplicate, in 2-liter cylindroconical stainless steel fermenting vessels, containing 1.5 liter of wort medium. The 15 °P wort (69 g of maltose, 17.4 g of maltotriose, 15.1 g of glucose, and 5.0 g of fructose per liter) was produced at the VTT Pilot Brewery from barley malt. Yeast was inoculated at a rate of 15 × 106 viable cells · ml−1. The wort was oxygenated to 15 mg · liter−1 prior to pitching (oxygen indicator model 26073 and sensor 21158; Orbisphere Laboratories, Switzerland). The fermentations were carried out at 15°C until an apparent attenuation of 80% (corresponding to approximately 7% [vol/vol] alcohol) was reached, or for a maximum of 14 days. Wort samples were drawn regularly from the fermentation vessels aseptically and placed directly on ice, after which the yeast was separated from the fermenting wort by centrifugation (9,000 × g, 10 min, 1°C). Samples for yeast-derived flavor compound analysis were drawn from the beer when fermentations were ended.
Chemical analysis.
The concentrations of fermentable sugars (maltose and maltotriose) were measured by HPLC using a Waters 2695 separation module and Waters system interphase module liquid chromatograph coupled with a Waters 2414 differential refractometer (Waters Co., Milford, MA, USA). A Rezex RFQ-Fast Acid H+ (8%) LC column (100 × 7.8 mm; Phenomenex, USA) was equilibrated with 5 mM H2SO4 (Titrisol, Merck, Germany) in water at 80°C, and samples were eluted with 5 mM H2SO4 in water at a 0.8 ml · min−1 flow rate.
The alcohol level (% vol/vol) of samples was determined from the centrifuged and degassed fermentation samples using an Anton Paar density meter DMA 5000 M with Alcolyzer beer ME and pH ME modules (Anton Paar GmbH, Austria).
Yeast-derived higher alcohols and esters were determined by headspace gas chromatography with flame ionization detector (HS-GC-FID) analysis. Four-milliliter samples were filtered (0.45 μm pore size) and incubated at 60°C for 30 min, and then 1 ml of gas phase was injected (split mode, 225°C, split flow of 30 ml · min−1) into a gas chromatograph equipped with an FID detector and headspace autosampler (Agilent 7890 series; Palo Alto, CA, USA). Analytes were separated on a HP-5 capillary column (50 m by 320 μm by 1.05 μm column; Agilent, USA). The carrier gas was helium (constant flow of 1.4 ml · min−1). The temperature program was 50°C for 3 min, 10°C · min−1 to 100°C, 5°C · min−1 to 140°C, 15°C · min−1 to 260°C, and then isothermal for 1 min. Compounds were identified by comparison with authentic standards and were quantified using standard curves. 1-Butanol was used as an internal standard.
Total diacetyl (free and acetohydroxy acid form) was measured according to Analytica-EBC method 9.24.2 (54). Samples were heated to 60°C and kept at this temperature for 90 min. Heating to 60°C results in the conversion of α-acetolactate to diacetyl. The samples were then analyzed by headspace gas chromatography using a gas chromatograph equipped with a micro-electron capture detector (μECD) detector and headspace autosampler (Agilent 7890 series; Palo Alto, CA, USA). Analytes were separated on a HP-5 capillary column (50 m by 320 μm by 1.05 μm column; Agilent, USA). 2,3-Hexanedione was used as an internal standard.
Ethanol tolerance and accumulation capacity.
As several of the wild-type and variant strains flocculated strongly, we were unable to reliably determine ethanol tolerance in liquid cultures based on optical density measurements. Therefore, we assessed ethanol tolerance based on the ability to grow on yeast extract-peptone-dextrose (YPD) agar plates supplemented with various levels of ethanol. Overnight precultures of all the strains were grown in YPM at 25°C. The yeast was then pelleted and resuspended in 50 mM citrate buffer (pH 7.2) to deflocculate the yeast. The cell concentration was measured with a NucleoCounter YC-100 (ChemoMetec, Denmark), after which suspensions were diluted to contain approximately 105, 104, and 103 cells · ml−1. Five-microliter aliquots of the suspensions of each strain were spotted onto agar plates containing YPD supplemented with 9% and 11% ethanol. Plates were sealed with Parafilm, placed in ziplock bags, and incubated at 25°C for up to 21 days.
The ethanol accumulation capacity of the strains was also assessed as described by Gallone et al. (55), with modifications. Overnight precultures of all the strains were grown in YP-4% maltose at 25°C. The yeast was then pelleted and resuspended to an OD600 of 20 in 50 mM citrate buffer (pH 7.2) to deflocculate the yeast. Thirty-five milliliters of YP-35% maltose was then inoculated with the yeast strains to an initial OD600 of 0.5. Fermentations took place in 100-ml Erlenmeyer flasks capped with glycerol-filled airlocks. Flasks were incubated at 18°C with gentle shaking (100 rpm) for 28 days. The mass loss was monitored to estimate when fermentation finished. After the fermentations had finished, the cultures were centrifuged, after which the alcohol content of the supernatants was measured with an Anton Paar density meter DMA 5000 M with Alcolyzer beer ME and pH ME modules (Anton Paar GmbH, Austria).
Genetic stability of variant strains.
The genetic stability of the eight variant strains (Table 1) was assessed by culturing them repeatedly in YP-4% maltose at 18°C for over 80 generations (2, 9). After this, DNA was extracted from two randomly chosen isolates from each variant strain. DNA fingerprints were produced for each isolate and the eight variant strains with PCR, using delta12 (5′-TCAACAATGGAATCCCAAC-3′) and delta21 (5′-CATCTTAACACCGTATATGA-3′) primers for interdelta DNA analysis (56). The DNA fingerprints of the isolates obtained after 80 generations were compared with those of the variant strains, and the variants were deemed genetically stable if the fingerprints were identical.
Genome sequencing and analysis.
Wild-type strains Y1 and Y2 have been sequenced in previous studies (3, 32), and reads for these strains were obtained from NCBI-SRA (accession numbers SRX1423875 and SRX2459842, respectively). For this study, wild-type strains Y2 to Y4 and the eight variant strains were sequenced by Biomedicum Genomics (Helsinki, Finland). In brief, DNA was initially isolated using Qiagen 100/G Genomic-tips (Qiagen, The Netherlands), after which an Illumina TruSeq LT paired-end 150-bp library was prepared for each strain, and sequencing was carried out with a NextSeq 500 instrument. Paired-end reads from the NextSeq 500 sequencing were analyzed for quality with FastQC (57) and trimmed and filtered with Cutadapt (58). Alignment of the reads was carried out using SpeedSeq (59). Reads of S. cerevisiae Y1 (VTT-A81062) and its variants were aligned to a previously assembled reference genome (available under BioProject no. PRJNA301545) of the strain (3), while reads of hybrid strains Y2 to Y4 and their variants were aligned to concatenated reference sequences of S. cerevisiae VTT-A81062 and S. eubayanus FM1318 (60), as described previously (3). The quality of the alignments was assessed with QualiMap (61). Variant analysis was performed on aligned reads using FreeBayes (62). Variants in wild-type and variant strains were called simultaneously (multisample). Prior to variant analysis, alignments were filtered to a minimum MAPQ of 50 with SAMtools (63). Structural variation analysis was performed with LUMPY (64), Manta (65), and Scalpel (66). Variants that were unique to the variant strains (i.e., not present in the wild-type strain) were obtained with SnpSift (67). Annotation and effect prediction of the variants were performed with SnpEff (68). The filtered and annotated variants were finally manually inspected in IGV (69). Copy number variations of chromosomes, as well as genes related to maltose transport (MALx1, MTT1, and MPH2), were estimated based on coverage with CNVkit (70). The median coverage over 10,000-bp windows was calculated with BEDTools (71).
Data visualization and analysis.
Data and statistical analyses were performed with R (http://www.r-project.org/). Flow cytometry data were analyzed with the ‘flowCore' (72) and ‘mixtools' (73) packages. Growth curves from the high-throughput screening cultivations were produced based on optical density measurements using the logistic model in the ‘grofit' package (74). Scatter, box, and heatmap plots were produced in R. The ‘Circos-like' plots shown in Fig. S5 to S16 were produced with the ‘circlize' package (75). The significance between variant wild-type strains was tested by Student's t test (two-tailed, unpaired, and unequal variances).
Accession number(s).
The Illumina reads generated in this study have been submitted to the NCBI SRA under BioProject number PRJNA408119 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
Supplementary Material
ACKNOWLEDGMENTS
We thank Sue James for valuable comments during the study, Virve Vidgren for performing DNA extractions, Eero Mattila for wort preparation and other assistance in the VTT Pilot Brewery, and Aila Siltala for skilled technical assistance.
This work was supported by the Alfred Kordelin Foundation, Svenska Kulturfonden-The Swedish Cultural Foundation in Finland, Suomen Kulttuurirahasto, SABMiller (ABInBev), and the Academy of Finland (academy project 276480).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02302-17.
REFERENCES
- 1.Krogerus K, Magalhães F, Vidgren V, Gibson B. 2015. New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol 42:769–778. doi: 10.1007/s10295-015-1597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertens S, Steensels J, Saels V, De Rouck G, Aerts G, Verstrepen KJ. 2015. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl Environ Microbiol 81:8202–8214. doi: 10.1128/AEM.02464-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krogerus K, Arvas M, De Chiara M, Magalhães F, Mattinen L, Oja M, Vidgren V, Yue JX, Liti G, Gibson B. 2016. Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl Microbiol Biotechnol 100:7203–7222. doi: 10.1007/s00253-016-7588-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steensels J, Meersman E, Snoek T, Saels V, Verstrepen KJ. 2014. Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl Environ Microbiol 80:6965–6975. doi: 10.1128/AEM.02235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piotrowski JS, Nagarajan S, Kroll E, Stanbery A, Chiotti KE, Kruckeberg AL, Dunn B, Sherlock G, Rosenzweig F. 2012. Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol Biol 12:46. doi: 10.1186/1471-2148-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunicka-Styczyńska A, Rajkowska K. 2011. Physiological and genetic stability of hybrids of industrial wine yeasts Saccharomyces sensu stricto complex. J Appl Microbiol 110:1538–1549. doi: 10.1111/j.1365-2672.2011.05009.x. [DOI] [PubMed] [Google Scholar]
- 7.Louis VL, Despons L, Friedrich A, Martin T, Durrens P, Casarégola S, Neuvéglise C, Fairhead C, Marck C, Cruz JA, Straub M-L, Kugler V, Sacerdot C, Uzunov Z, Thierry A, Weiss S, Bleykasten C, De Montigny J, Jacques N, Jung P, Lemaire M, Mallet S, Morel G, Richard G-F, Sarkar A, Savel G, Schacherer J, Seret M-L, Talla E, Samson G, Jubin C, Poulain J, Vacherie B, Barbe V, Pelletier E, Sherman DJ, Westhof E, Weissenbach J, Baret PV, Wincker P, Gaillardin C, Dujon B, Souciet J-L. 2012. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3 (Bethesda) 2:299–311. doi: 10.1534/g3.111.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn B, Paulish T, Stanbery A, Piotrowski J, Koniges G, Kroll E, Louis EJ, Liti G, Sherlock G, Rosenzweig F. 2013. Recurrent rearrangement during adaptive evolution in an interspecific yeast hybrid suggests a model for rapid introgression. PLoS Genet 9:e1003366. doi: 10.1371/journal.pgen.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez-Través L, Lopes CA, Barrio E, Querol A. 2014. Stabilization process in Saccharomyces intra and interspecific hybrids in fermentative conditions. Int Microbiol 17:213–224. doi: 10.2436/20.1501.01.224. [DOI] [PubMed] [Google Scholar]
- 10.Gibson BR, Lawrence SJ, Leclaire JPR, Powell CD, Smart KA. 2007. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev 31:535–569. doi: 10.1111/j.1574-6976.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- 11.Gerstein AC, Chun H-JE, Grant A, Otto SP. 2006. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet 2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y-J, Swamy KBS, Leu J-Y. 2016. Experimental evolution reveals interplay between Sch9 and polyploid stability in yeast. PLoS Genet 12:e1006409. doi: 10.1371/journal.pgen.1006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez-Través L, Lopes CA, Barrio E, Querol A. 2012. Evaluation of different genetic procedures for the generation of artificial hybrids in Saccharomyces genus for winemaking. Int J Food Microbiol 156:102–111. doi: 10.1016/j.ijfoodmicro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Voordeckers K, Kominek J, Das A, Espinosa-Cantú A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y, Zhu B, Marchal K, DeLuna A, Van Noort V, Jelier R, Verstrepen KJ. 2015. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet 11:e1005635. doi: 10.1371/journal.pgen.1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selmecki AM, Maruvka YE, Richmond P a, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, Pellman D. 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunge N, Nakatomi Y. 1972. Genetic mechanisms of rare matings of the yeast Saccharomyces cerevisiae heterozygous for mating type. Genetics 70:41–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Xu Y. 2014. Adaptive evolution of Saccharomyces cerevisiae with enhanced ethanol tolerance for Chinese rice wine fermentation. Appl Biochem Biotechnol 173:1940–1954. doi: 10.1007/s12010-014-0978-z. [DOI] [PubMed] [Google Scholar]
- 18.Stanley D, Fraser S, Chambers PJ, Rogers P, Stanley GA. 2010. Generation and characterisation of stable ethanol-tolerant mutants of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 37:139–149. doi: 10.1007/s10295-009-0655-3. [DOI] [PubMed] [Google Scholar]
- 19.Avrahami-Moyal L, Engelberg D, Wenger JW, Sherlock G, Braun S. 2012. Turbidostat culture of Saccharomyces cerevisiae W303-1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1. FEMS Yeast Res 12:521–533. doi: 10.1111/j.1567-1364.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blieck L, Toye G, Dumortier F, Verstrepen KJ, Delvaux FR, Thevelein JM, Van Dijck P. 2007. Isolation and characterization of brewer's yeast variants with improved fermentation performance under high-gravity conditions. Appl Environ Microbiol 73:815–824. doi: 10.1128/AEM.02109-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekberg J, Rautio J, Mattinen L, Vidgren V, Londesborough J, Gibson BR. 2013. Adaptive evolution of the lager brewing yeast Saccharomyces pastorianus for improved growth under hyperosmotic conditions and its influence on fermentation performance. FEMS Yeast Res 13:335–349. doi: 10.1111/1567-1364.12038. [DOI] [PubMed] [Google Scholar]
- 22.Peris D, Moriarty RV, Alexander WG, Baker E, Sylvester K, Sardi M, Langdon QK, Libkind D, Wang Q-M, Bai F-Y, Leducq J-B, Charron G, Landry CR, Sampaio JP, Gonçalves P, Hyma KE, Fay JC, Sato TK, Hittinger CT. 2017. Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol Biofuels 10:78. doi: 10.1186/s13068-017-0763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Malo M, García-Rios E, Melgar B, Sanchez MR, Dunham MJ, Guillamón JM. 2015. Evolutionary engineering of a wine yeast strain revealed a key role of inositol and mannoprotein metabolism during low-temperature fermentation. BMC Genomics 16:537. doi: 10.1186/s12864-015-1755-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caspeta L, Chen Y, Nielsen J. 2016. Thermotolerant yeasts selected by adaptive evolution express heat stress response at 30°C. Sci Rep 6:27003. doi: 10.1038/srep27003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brickwedde A, van den Broek M, Geertman J-MA, Magalhães F, Kuijpers NGA, Gibson B, Pronk JT, Daran J-MG. 2017. Evolutionary engineering in chemostat cultures for improved maltotriose fermentation kinetics in Saccharomyces pastorianus lager brewing yeast. Front Microbiol 8:1690. doi: 10.3389/fmicb.2017.01690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wouter Wisselink H, Toirkens MJ, Wu Q, Pronk JT, Van Maris AJA. 2009. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 75:907–914. doi: 10.1128/AEM.02268-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scalcinati G, Otero JM, Van Vleet JRH, Jeffries TW, Olsson L, Nielsen J. 2012. Evolutionary engineering of Saccharomyces cerevisiae for efficient aerobic xylose consumption. FEMS Yeast Res 12:582–597. doi: 10.1111/j.1567-1364.2012.00808.x. [DOI] [PubMed] [Google Scholar]
- 28.Smukowski Heil CS, DeSevo CG, Pai DA, Tucker CM, Hoang ML, Dunham MJ. 2017. Loss of heterozygosity drives adaptation in hybrid yeast. Mol Biol Evol 34:1596–1612. doi: 10.1093/molbev/msx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hope EA, Amorosi CJ, Miller AW, Dang K, Heil CS, Dunham MJ. 2017. Experimental evolution reveals favored adaptive routes to cell aggregation in yeast. Genetics 206:1153–1167. doi: 10.1534/genetics.116.198895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott AL, Richmond PA, Dowell RD, Selmecki AM. 2017. The influence of polyploidy on the evolution of yeast grown in a sub-optimal carbon source. Mol Biol Evol 34:2690–2703. doi: 10.1093/molbev/msx205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puligundla P, Smogrovicova D, Obulam VSR, Ko S. 2011. Very high gravity (VHG) ethanolic brewing and fermentation: a research update. J Ind Microbiol Biotechnol 38:1133–1144. doi: 10.1007/s10295-011-0999-3. [DOI] [PubMed] [Google Scholar]
- 32.Krogerus K, Seppänen-Laakso T, Castillo S, Gibson B. 2017. Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb Cell Fact 16:66. doi: 10.1186/s12934-017-0679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buskirk SW, Peace RE, Lang GI. 2017. Hitchhiking and epistasis give rise to cohort dynamics in adapting populations. Proc Natl Acad Sci U S A 114:8330–8335. doi: 10.1073/pnas.1702314114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pires EJ, Teixeira JA, Brányik T, Vicente AA. 2014. Yeast: the soul of beer's aroma–a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl Microbiol Biotechnol 98:1937–1949. doi: 10.1007/s00253-013-5470-0. [DOI] [PubMed] [Google Scholar]
- 35.Krogerus K, Gibson BR. 2013. 125th anniversary review: diacetyl and its control during brewery fermentation. J Inst Brew 119:86–97. [Google Scholar]
- 36.Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, Toh-e A. 1990. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol 10:4303–4313. doi: 10.1128/MCB.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sipiczki M. 2008. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res 8:996–1007. doi: 10.1111/j.1567-1364.2008.00369.x. [DOI] [PubMed] [Google Scholar]
- 38.Gresham D, Dunham MJ. 2014. The enduring utility of continuous culturing in experimental evolution. Genomics 104:399–405. doi: 10.1016/j.ygeno.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snoek T, Picca Nicolino M, Van den Bremt S, Mertens S, Saels V, Verplaetse A, Steensels J, Verstrepen KJ. 2015. Large-scale robot-assisted genome shuffling yields industrial Saccharomyces cerevisiae yeasts with increased ethanol tolerance. Biotechnol Biofuels 8:32. doi: 10.1186/s13068-015-0216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thevelein JM, de Winde JH. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 42.Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Hérissant L, Blundell JR, Levy SF, Fisher DS, Sherlock G, Petrov DA. 2016. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell 166:1585–1596.e22. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato TK, Tremaine M, Parreiras LS, Hebert AS, Myers KS, Higbee AJ, Sardi M, McIlwain SJ, Ong IM, Breuer RJ, Avanasi Narasimhan R, McGee MA, Dickinson Q, La Reau A, Xie D, Tian M, Reed JL, Zhang Y, Coon JJ, Hittinger CT, Gasch AP, Landick R. 2016. Directed evolution reveals unexpected epistatic interactions that alter metabolic regulation and enable anaerobic xylose use by Saccharomyces cerevisiae. PLoS Genet 12:e1006372. doi: 10.1371/journal.pgen.1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swinnen S, Schaerlaekens K, Pais T, Claesen J, Hubmann G, Yang Y, Demeke M, Foulquíe-Moreno MR, Goovaerts A, Souvereyns K, Clement L, Dumortier F, Thevelein JM. 2012. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res 22:975–984. doi: 10.1101/gr.131698.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, Raghuraman MK, Brewer BJ, Dunham MJ. 2014. The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. G3 (Bethesda) 4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao KC, Sherlock G. 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet 40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rollero S, Mouret J-R, Sanchez I, Camarasa C, Ortiz-Julien A, Sablayrolles J-M, Dequin S. 2016. Key role of lipid management in nitrogen and aroma metabolism in an evolved wine yeast strain. Microb Cell Fact 15:32. doi: 10.1186/s12934-016-0434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong K-K, Vongsangnak W, Vemuri GN, Nielsen J. 2011. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc Natl Acad Sci U S A 108:12179–12184. doi: 10.1073/pnas.1103219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mason AB, Dufour JP. 2000. Alcohol acetyltransferases and the significance of ester synthesis in yeast. Yeast 16:1287–1298. doi:. [DOI] [PubMed] [Google Scholar]
- 50.Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. 2003. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol 69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopandic K, Pfliegler WP, Tiefenbrunner W, Gangl H, Sipiczki M, Sterflinger K. 2016. Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation. Appl Microbiol Biotechnol 100:6331–6343. doi: 10.1007/s00253-016-7481-0. [DOI] [PubMed] [Google Scholar]
- 52.Dunn B, Sherlock G. 2008. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res 18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuno M, Kajitani R, Ryusui R, Morimoto H, Kodama Y, Itoh T. 2015. Next-generation sequencing analysis of lager brewing yeast strains reveals the evolutionary history of interspecies hybridization. DNA Res 23:67–80. doi: 10.1093/dnares/dsv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.European Brewery Convention. 2004. 9.24.2. Vicinal biketones in beer: gas chromatographic method. In Analytica EBC. Verlag Hans Carl Getränke-Fachverlag, Nürnberg, Germany. [Google Scholar]
- 55.Gallone B, Steensels J, Baele G, Maere S, Verstrepen KJ, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, Merlevede A, Roncoroni M, Voordeckers K, Miraglia L, Teiling C, Steffy B, Taylor M, Schwartz A, Richardson T, White C. 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166:1397.e16–1410.e16. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Legras JL, Karst F. 2003. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol Lett 221:249–255. doi: 10.1016/S0378-1097(03)00205-2. [DOI] [PubMed] [Google Scholar]
- 57.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 58.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10. [Google Scholar]
- 59.Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM. 2015. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods 12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baker E, Wang B, Bellora N, Peris D, Hulfachor AB, Koshalek JA, Adams M, Libkind D, Hittinger CT. 2015. The genome sequence of Saccharomyces eubayanus and the domestication of lager-brewing yeasts. Mol Biol Evol 32:2818–2831. doi: 10.1093/molbev/msv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 62.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907.
- 63.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Layer RM, Chiang C, Quinlan AR, Hall IM. 2014. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. 2016. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 66.Fang H, Bergmann EA, Arora K, Vacic V, Zody MC, Iossifov I, O'Rawe JA, Wu Y, Jimenez Barron LT, Rosenbaum J, Ronemus M, Lee Y, Wang Z, Dikoglu E, Jobanputra V, Lyon GJ, Wigler M, Schatz MC, Narzisi G. 2016. Indel variant analysis of short-read sequencing data with Scalpel. Nat Protoc 11:2529–2548. doi: 10.1038/nprot.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. 2012. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet 3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talevich E, Shain AH, Botton T, Bastian BC. 2016. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. 2009. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benaglia T, Chauveau D, Hunter DR, Young D. 2009. mixtools: an R package for analyzing finite mixture models. J Stat Softw 32:1–29. doi: 10.18637/jss.v032.i06. [DOI] [Google Scholar]
- 74.Kahm M, Hasenbrink G, Lichtenberg-Frate H, Ludwig J, Kschischo M. 2010. grofit: fitting biological growth curves. J Stat Softw 33:1–21. doi: 10.18637/jss.v033.i07.20808728 [DOI] [Google Scholar]
- 75.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.