Summary
Although one‐third of the world population is infected with Mycobacterium tuberculosis, only 5–10% of the infected individuals will develop active tuberculosis (TB) disease and the rest will remain infected with no symptoms, known as latent TB infection (LTBI). Identifying biomarkers that differentiate latent and active TB disease enables effective TB control, as early detection, treatment of active TB and preventive treatment of individuals with LTBI are crucial steps involved in TB control. Here, we have evaluated the frequency of antigen‐specific memory and regulatory T (Treg) cells in 15 healthy household contacts (HHC) and 15 pulmonary TB patients (PTB) to identify biomarkers for differential diagnosis of LTBI and active TB. Among all the antigens tested in the present study, early secretory antigenic target‐6 (ESAT‐6) ‐specific CD4+ and CD8+ central memory (Tcm) cells showed 93% positivity in HHC and 20% positivity in PTB. The novel test antigens Rv0753c and Rv0009 both displayed 80% and 20% positivity in HHC and PTB, respectively. In contrast to Tcm cells, effector memory T (Tem) cells showed a higher response in PTB than HHC; both ESAT‐6 and Rv0009 showed similar positivity of 80% in PTB and 33% in HHC. PTB patients have a higher proportion of circulating antigen‐reactive Treg cells (CD4+ CD25+ FoxP3+) than LTBI. Rv2204c‐specific Treg cells showed maximum positivity of 73% in PTB and 20% in HHC. Collectively, our data conclude that ESAT‐6‐specific Tcm cells and Rv2204c‐specific Treg cells might be useful biomarkers to discriminate LTBI from active TB.
Keywords: antigen, biomarker, latent tuberculosis, memory T cells, Mycobacterium tuberculosis, regulatory T cells
Abbreviations
- CFP‐10
culture filtrate antigen
- ESAT‐6
early secretory antigenic target‐6
- HHC
healthy house‐hold contacts
- LTBI
latent tuberculosis infection
- M. tb
Mycobacterium tuberculosis
- PTB
pulmonary Tuberculosis
- QFT‐GIT
Quantiferon TB gold in tube assay
- Tcm
central memory T cells
- Tem
effector memory T cells
- Treg
regulatory T cells
Introduction
It is estimated that one‐third of the human population is infected with Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), and is therefore at risk of developing TB disease.1 In most cases, the infected individuals remain healthy, sometimes throughout their lifetime, and develop a latent TB infection (LTBI) with no sign of TB disease. A better understanding of the immunological parameters/differences between LTBI and active TB might provide a clue about the definition of correlates with protection, differential immuno‐diagnosis of LTBI and active TB, and to the development of new vaccines against the disease.
Tuberculosis is a chronic, infectious disease and prolonged exposure to mycobacterial antigens impacts the biology of antigen‐specific T cells, including the differentiation of memory and regulatory T cells. The activation and differentiation of memory cells are characterized by a number of distinct stages. During the infection, homogeneous naive T cells undergo massive proliferation and clonal expansion to form a heterogeneous population of effector and memory cells.2, 3 In this study, we used CD45RA marker for differentiating naive from memory T‐cell subsets. CD45RA is expressed on naive T cells, as well as the effector cells in both CD4 and CD8. After antigen experience, central and effector memory T cells gain expression of CD45RO and lose expression of CD45RA. During high levels of antigen burden and inflammation, generation of short‐lived effector cells occurs to control the infection and 95% of effector cells die as a result of apoptosis once the infection is cleared. The remaining pathogen‐specific effector T cells differentiate into a memory T‐cell population that can survive for a long time in contrast to effector cells. These memory cells can be divided into two functionally distinct subsets based on the expression of cell surface markers (lymph node homing receptors). They are T central memory (Tcm) and T effector memory (Tem) T cells. For differentiation of central and effector memory populations, we used chemokine receptor CCR7 as a reference standard and widely used marker. Tcm cells that express CCR7 mainly circulate in lymphoid tissues (lymph nodes, spleen and bone marrow) and blood. Tcm cells are long‐lived T cells with self‐renewal and proliferation capacity and can rapidly differentiate into effector cells upon encountering foreign antigen.4 In contrast, Tem cells lack CCR7 and mainly circulate in peripheral non‐lymphoid tissues (e.g. lung, liver, intestine), spleen and blood.5 Therefore, Tcm cells confer long‐term protection, whereas Tem cells have only limited reconstitution capacity.6
Regulatory T (Treg) cells play an important role in immune regulation to prevent autoimmune diseases and to control the immune responses by down‐regulating the effector function of CD4+ or CD8+ T cells, which lead to delayed onset of the adaptive immune response.7, 8 On the other hand, pre‐existing Treg cells have a beneficial role in the very early stage of infection that activates T cells and the subsequent secretion of pro‐inflammatory cytokines.9, 10 Several subsets of Treg cells have been identified, such as interleukin‐10 (IL‐10) ‐secreting (Tr1) or transforming growth factor‐β‐secreting (Th3) Treg cells and CD4+ CD25+ FoxP3+ Treg cells.11, 12 Among these subsets, the last one plays a major role in the regulation of immune response and the expression of CD25 is a conventional marker for CD4+ T‐cell activation.13 It has been reported that the T cells may be susceptible to the immunosuppressive effects of Treg cells.14, 15 Oldenhove et al. demonstrated that T cells isolated from CD4+ CD25+‐depleted mice produced higher levels of interferon‐γ (IFN‐γ) and low levels of IL‐4.16 To date, information about the immunoregulatory properties of antigen‐specific CD4+ CD25+ T cells in TB disease is scarce. Recent studies have revealed that a subset of CD4 T cells expressing the transcription factor called forkhead/winged helix transcription factor (FoxP3) acts as regulatory T cells. It is fundamental for the development and the function of CD4 suppressive cells and also activates anti‐inflammatory genes.17 In vitro and in vivo studies reported that, along with CD25, FoxP3 can suppress the activation, proliferation and effector functions of other T cells such as natural killer cells, natural killer T cells, B cells and antigen‐presenting cells.18 It was demonstrated earlier that these Treg cell subsets have a significant role in the differential diagnosis of latent and active TB19 and therefore further studies on these crucial regulators during M. tuberculosis infection would be informative.
In our previous report, we observed that there is an impaired T‐cell response in active TB compared with LTBI.20, 21, 22 The suppressed immune response during active TB is because Treg cells play a role in immune evasion by M. tuberculosis,23 not because of an intrinsic defect in the T‐cell response to M. tuberculosis antigens. Earlier reports suggested that evaluating memory phenotypes of responding T cells led to the identification of highly sensitive and specific biomarkers for differential diagnosis of LTBI and active TB.24 Therefore, in this study, we have evaluated the antigen‐specific frequency of memory and the regulatory response of CD4 and CD8 T cells in healthy household contacts (HHC) and patients with pulmonary TB (PTB) for the identification of biomarkers for differential TB diagnosis.
The role of a phase‐dependent antigen‐specific CD4+ and CD8+ T‐cell phenotypic profile in LTBI and active TB significantly improves the possibility of using these antigens in TB diagnostic tests. The antigens used in the present study also belong to stage‐specific antigens. The Rv2204c antigen is a hypothetical antigen, predicted to express under thiol oxidative stress conditions.25 The second antigen, Rv0753c, was identified as a probable methyl melonate semi‐aldehyde dehydrogenase (mmsA) and is an M. tuberculosis‐secreted protein that has been identified in the supernatant of culture‐filtered M. tuberculosis.26 Rv0753c is a reactivation antigen involved during the reactivation of non‐replicative dormant mycobacteria to an actively replicating form.27, 28 In addition, it also plays a critical role in the activation of dendritic cells and the initiation of the T helper type 1 (Th1) phenotype in the adaptive immune response.29 However, the immunological function of MmsA has not been elucidated, particularly with respect to the biomarker for TB diagnosis. Previous reports from our laboratory predicted that Rv0009 antigen was over‐expressed in M. tuberculosis clinical strains (S7 and S10) in an in vitro dormancy–hypoxia model.30 Hence, we are interested to analyse the potential of these phase‐specific antigens for discriminating LTBI and active TB.
Materials and methods
Study subjects
This study was approved by the institutional ethical committee of the National Institute for Research in Tuberculosis, Chennai, India. Written informed consent was obtained from all the study participants before collecting the blood. Adults (age > 18 years) with newly diagnosed active pulmonary TB and LTBI were recruited for this study. Individuals with a previous history of TB, those who underwent anti‐TB treatment or those under immunosuppressive therapy were excluded from the study. All study participants were confirmed as HIV‐negative. Peripheral blood samples (10 mL) were collected from a total of 30 participants. The demographic characteristics of the study participants are shown in Table 1.
Table 1.
Demographic characteristic of study participants with latent and active tuberculosis (TB)
| Category | Healthy household contacts (HHC) | Pulmonary tuberculosis (PTB) |
|---|---|---|
| Total number of subjects (n) | 15 | 15 |
| Median age (range) | 39 (22–52) | 43 (22–56) |
| Sex | ||
| Male, n (%) | 09 (60) | 10 (66) |
| Female, n (%) | 06 (40) | 5 (34) |
| Positivity of smear test, n (%) | 0 (0) | 15 (100) |
| Smear grade 3+, n (%) | 0 (0) | 4 (28) |
| Smear grade 2+, n (%) | 0 (0) | 3 (20) |
| Smear grade 1+, n (%) | 0 (0) | 6 (32) |
| Scanty | 0 (0) | 2 (20) |
| QFT‐GIT | ||
| Positives, n (%) | 15 (100) | 15 (100) |
| Negatives, n (%) | 0 (0) | 0 (0) |
| Indeterminate, n (%) | 0 (0) | 0 (0) |
n = number of individuals; percentage (%) are indicated in brackets; QFT‐GIT, Quantiferon TB gold in tube assay.
Healthy household contacts
Fifteen HHC were recruited from families where there was at least one newly diagnosed sputum‐positive PTB patient (index case) living in the same household for at least 3 months with the index case. Disease‐free status of HHC was confirmed by clinical examination, negative smear findings and normal chest X‐ray. All the recruited HHC were positive for the Quantiferon TB Gold‐in‐tube assay (QFT‐GIT) test (Cellestis, Qiagen, Venlo, the Netherlands) confirming their infection status with M. tuberculosis. All HHC were followed for up to 6 months and those who developed TB disease/active TB were excluded from the analysis.
Pulmonary tuberculosis patients
For this study, 15 patients with PTB were recruited from the Government Thiruvoteeswarar Hospital of Thoracic Medicine, Otteri, Chennai, India. All the recruited patients were recently diagnosed (new smear positives) as PTB patients. Two spot and one overnight sputum specimens were collected from each patient. Sputum specimens were examined for acid‐fast bacillus by fluorescence microscopy and culture that showed positivity for both smear and culture for all TB patients. Further, all the recruited PTB patients were positive for the QFT‐GIT test.
Interferon‐γ release assay
An IFN‐γ release assay (IGRA) was performed using a QFT‐GIT kit (Cellestis). Out of 10 ml, 1 ml of blood was taken from each of the three tubes pre‐coated with M. tuberculosis antigens [early secretory antigenic target‐6 (ESAT‐6), culture filtrate antigen (CFP‐10) and TB7.7] and considered as a test sample tube, phytohaemagglutinin was a positive control tube and the saline‐coated tube was a negative control. The tubes were incubated for 16–24 hr at 37° in 5% CO2 and the supernatant was collected after centrifugation. The cytokine IFN‐γ was measured in the supernatant by ELISA as per the manufacturer's instructions. The test results were interpreted as per the kit guidelines, using the software provided by the manufacturer.
In vitro stimulation of whole blood
To minimize sample consumption and also to screen larger numbers of antigens, the collected blood was diluted to the ratio of 1:1 with RPMI‐1640 medium (Sigma‐Aldrich, St Louis, MO), supplemented with glutamine (0·29 g/L), penicillin (100 U/L) and streptomycin (0·1 mg/mL). The recombinant plasmids encoding ESAT‐6 and CFP‐10 were a kind gift from Colorado State University, Fort Collins, CO, USA. The proteins Rv2204c, Rv0753c and Rv0009 were cloned, over‐expressed and purified by recombinant DNA technology as described in our earlier publications.20, 21, 22, 23 Endotoxin concentration in all recombinant protein preparations was quantified by LAL assay and ranged from 1 to 10 EU per mg of protein, which is acceptable.31 The diluted blood was stimulated with ESAT‐6, CFP‐10, Rv2204c, Rv0753c and Rv0009 at a concentration of 5 μg/mL and phytohaemagglutinin stimulation as a mitogen control at a similar concentration. Diluted blood without any stimulant served as a control. Purified co‐stimulatory antibodies, anti‐CD28 and anti‐CD49d (Becton Dickinson, San Jose, CA), were added at a final concentration of 0·5 μg/mL and the culture plate was incubated at 37°, 5% atmospheric CO2. After the incubation period of 16 hr, cells were harvested with PBS treated with BD FACS lysing solution (Becton Dickinson) to lyse the red blood cells. Finally, cells were fixed with cytofix/cytoperm buffer (BD Biosciences, San Diego, CA) and stored with 10% DMSO at −80°.
Immunostaining
The cryopreserved cells were thawed rapidly and washed with PBS. The cells were stained with fluorochrome‐conjugated antibodies for surface T‐cell markers [phycoerythrin (PE) ‐Cy7 CD3, allophycocyanin (APC) ‐Cy7 CD4, peridinin chlorophyll protein CD8, APC CD45RA, PE CCR7, APC CD25] (BD Biosciences) at a concentration of 2 μl/106 cells. Then the cells were incubated for 30 min at 4° in the dark. Following surface staining, cells were washed with PBS, fixed and permeabilized with cytofix/cytoperm buffer. After fixing, the cells were washed with perm wash solution and intracellular staining for regulatory marker FoxP3 (FITC FoxP3) was performed. Finally, stained cells were washed with PBS and acquisition was performed on a FACS Canto II flow cytometer with FACSdiva software (BD and Company, Cockeysville, MD). A total of 100 000 lymphocyte events were recorded and data were analysed with flowjo software 7·1·1 version (Tree Star Inc., Ashland, OR). Compensation was calculated from fluorochrome‐conjugated antibodies coupled with CompBeads (BD Biosciences). The boundaries were defined by setting fluorescence‐minus‐one controls for all the antibodies.
Data analysis
All analyses were performed with Graphpad Prism version 5·00 for Windows (GraphPad Software, San Diego, CA). Intergroup comparison was carried out by non‐parametric Mann–Whitney U‐test. We used a receiver operating curve (ROC) to determine the cut‐off value for each antigen. The optimal cut‐off values were chosen when the Youden's index (sensitivity + specificity − 1) was maximum. The subjects were scored as positive for the specific phenotype percentage that was greater than the cut‐off value. A p‐value <0·05 was considered statistically significant. ROC data for antigen‐specific memory and Treg cells are shown in the Supplementary material (Fig. S1). The percentage of antigen‐specific Tcm, Tem and Treg cells was obtained by subtracting the baseline values of the unstimulated control from all stimulated test samples.
Results
Demographic characteristics of the subjects enrolled
For this study, we recruited 15 HHC QFT‐GIT‐positive and 15 smear‐positive PTB cases. Their median (range) ages were: 39 (22–52) years for LTBI, 43 (22–56) years for active TB. The male/female ratio for LTBI was 9/6 and for active TB was 10/5. The demographic profiles of 15 HHC and 15 PTB are summarized in Table 1.
Frequencies of effector and memory response T cells in latent and active TB
To determine whether the memory T‐cell response to mycobacterial antigens differed between LTBI and active TB, we cultured whole blood with M. tuberculosis antigens such as ESAT‐6, CFP‐10, Rv2204c, Rv0753c and Rv0009 and examined the surface expression of CD45RA and CCR7 on CD4 and CD8 T cells, after 16 hr, by flow cytometry. We used two most commonly used surface markers (CD45RA and CCR7) for phenotypic characterization of naive cells (CD45RA+ CCR7+), central memory cells (CD45RA− CCR7+), effector memory cells (CD45RA− CCR7−) and effector cells (CD45RA+ CCR7−). The general gating strategy followed for CD4 and CD8 memory and effector responses are given in Fig. 1.
Figure 1.
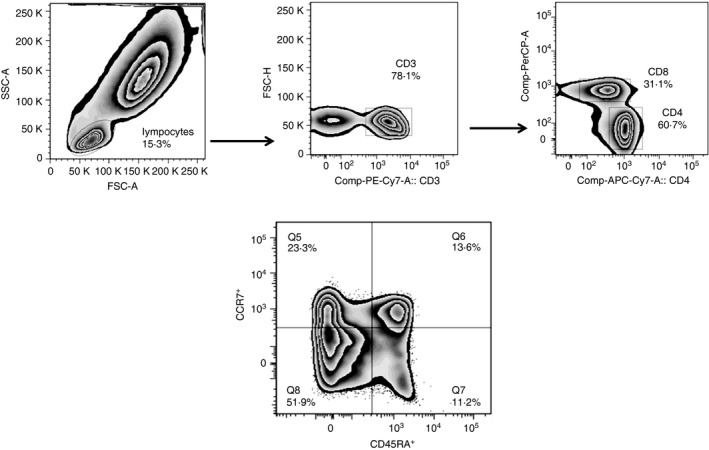
Gating strategy of effector and memory T cells. Gating strategy followed to evaluate the phenotypes of effector and memory CD4+ and CD8+ T cells in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB). Lymphocyte populations were first selected based on size and granularity to analyse the expression of CD3+, CD4+ and CD8+. Then, the expression of CD45RA and CCR7 markers was analysed on the selected population. The corresponding percentages of central memory (Tcm) (CCR7+ CD45RA −), effector memory (Tem) (CCR7− CD45RA −), effector (CCR7− CD45RA +) and naive (CCR7+ CD45RA +) T cells among the CD4 and CD8+ T cells were determined and plotted. The numbers indicate the percentages of the different cell subsets among the CD4+ and CD8+ populations. The same gating strategy was followed for all antigens in this study.
Increased expression of central memory T cells in latent TB
The predominant cell phenotype responding to antigenic stimulation was CD4+ Tcm (CD3+ CD4+ CD45RA − CCR7+) cells in LTBI compared with the PTB (Fig. 2a). Although we detected a higher frequency of CD4+ T cells that express CCR7 alone in response to M. tuberculosis antigens in HHC than in PTB, a significant difference was observed only for ESAT‐6, Rv0753c and Rv0009. We analysed whether these antigen‐specific memory markers can be used for differential diagnosis of LTBI and active TB by performing ROC analysis and determined the cut‐off value for each antigen‐specific cytokine based on Youden's index. The p‐values, area under the curve (AUC) values and positivity in HHC and PTB for each of these antigens are shown in Table 2. The values above the cut‐off were considered as positive in both groups. The discriminative capacity of antigen‐specific CD4+ Tcm cells was shown by AUC values (Table 2). Among the antigens tested here, ESAT‐6‐specific CD4+ Tcm cells showed a 0·84 AUC value with 95% CI. It showed 93% (14/15) positivity in HHC and only 20% (3/15) positivity in the PTB. The test antigen Rv0753c exhibited positivity of 73% (11/15) in HHC and 33% (5/15) in the PTB. Hence, compared with test antigens, standard antigen, ESAT‐6 showed better discrimination between HHC and PTB.
Figure 2.
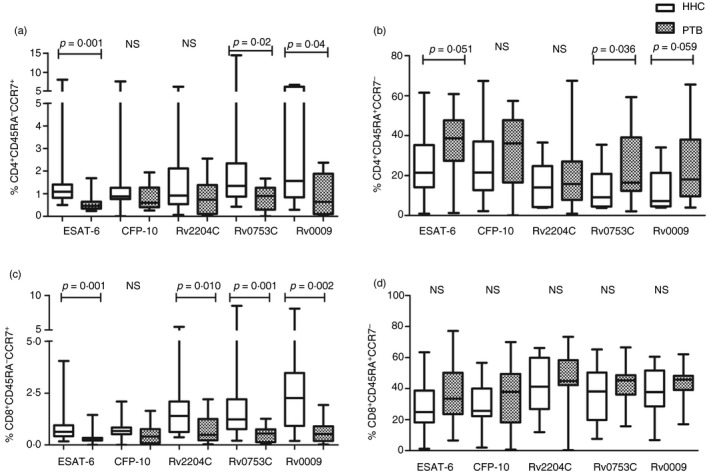
Percentage of effector and memory CD4+ and CD8+ T cells in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB). The percentages of effector and memory T cells among the CD4+ and CD8+ T cells of 15 individuals with latent tuberculosis infection (LTBI) and 15 PTB participants are represented as box plots with whiskers. The bottom and top lines of the boxes are the first and third centiles, the middle line inside the box is the median, and the whiskers represent the range. (a) CD4+ CCR7+ CD45RA − central memory T (Tcm) cells, (b) CD4+ CCR7− CD45RA + effector T cells, (c) CD8+ CCR7+ CD45RA − Tcm cells, (d) CD8+ CCR7− CD45RA + effector T cells. Differences between the groups were compared using a Mann–Whitney U‐test. Statistical differences with p < 0·05 are considered significant and shown in the plot. NS, non‐significant.
Table 2.
Positivity for CD4+ and CD8+ effector and memory phenotypes in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB)
| Antigen | Phenotype | p‐value | AUC | CI 95% | Positivity in HHC (%) | Positivity in PTB (%) |
|---|---|---|---|---|---|---|
| ESAT‐6 | CD4+ CD45RA− CCR7+ | 0·0015 | 0·84 | 0·666–1·013 | 93·3 (14/15) | 20 (3/15) |
| Rv0753c | CD4+ CD45RA− CCR7+ | 0·0225 | 0·7444 | 0·568–0·920 | 73·3 (11/15) | 33·3 (5/15) |
| Rv0009 | CD4+ CD45RA− CCR7+ | 0·0443 | 0·7156 | 0·530–0·900 | 86·6 (13/15) | 46·6 (7/15) |
| ESAT‐6 | CD8+ CD45RA− CCR7+ | 0·0013 | 0·8444 | 0·689–0·999 | 93·3 (14/15) | 20 (3/15) |
| Rv2204c | CD8+ CD45RA− CCR7+ | 0·0107 | 0·7733 | 0·605–0·940 | 86·6 (13/15) | 40 (6/15) |
| Rv0753c | CD8+ CD45RA− CCR7+ | 0·0012 | 0·8467 | 0·707–0·986 | 80 (12/15) | 20 (3/15) |
| Rv0009 | CD8+ CD45RA− CCR7+ | 0·0024 | 0·8244 | 0·673–0·975 | 80 (12/15) | 20 (3/15) |
| Rv0753c | CD4+ CD45RA+ CCR7+ | 0·0294 | 0·7333 | 0·547–0·918 | 40 (6/15) | 80 (12/15) |
| ESAT‐6 | CD4+ CD45RA+ CCR7− | 0·050 | 0·7089 | 0·513–0·904 | 33·3 (5/15) | 80 (12/15) |
| Rv0753c | CD4+ CD45RA+ CCR7− | 0·0362 | 0·7244 | 0·535–0·913 | 33·3 (5/15) | 80 (12/15) |
| Rv0009 | CD4+ CD45RA+ CCR7− | 0·0591 | 0·7022 | 0·513–0·891 | 40 (6/15) | 80 (12/15) |
Central memory (Tcm) cell, CCR7+ CD45RA−; effector memory (Tem) cell,CCR7− CD45RA−; effector cell (CCR7− CD45RA+); and naive T cells, CCR7+ CD45RA+.
Interestingly, compared with CD4 memory cells, the percentage of CD8 Tcm cells (CD3+ CD8+ CD45RA− CCR7+) was significantly high for all the antigens used except for CFP‐10 (Fig. 2c). Among all the antigens, the highest levels of significance were obtained for Rv0753c and ESAT‐6 antigen stimulations in HHC compared with the PTB (p < 0·001). The other test antigen, Rv2204c, also showed significance levels of p < 0·01 with high CD8+ Tcm subtype in HHC. We analysed whether the differences in the percentage of this phenotype could be used to distinguish individuals with active TB from those with LTBI (Table 2). Similar to CD4 Tcm cells, CD8 Tcm cells also discriminated between HHC and PTB with the AUC > 0·75. Among the antigens used here, the AUC was maximal for Rv0753c (0·846) followed by ESAT‐6 (0·844) and Rv0009 (0·82) with 95% CI. Similar to CD4 Tcm cells, ESAT‐6‐specific CD8 Tcm cells exhibited maximum positivity of 93% (14/15) in HHC and only 20% (3/15) in the PTB. The test antigens Rv0753c and Rv0009 both showed 80% (12/15) positivity in HHC and 20% (3/20) positivity in the PTB.
Increased expression of effector and naive T cells in active TB
Another phenotype, effector (CD45RA+ CCR7−) T cells, showed a higher response in PTB than HHC (Fig. 2b). Interestingly, only CD4+ effector T cells (CD3+ CD4+ CD45RA+ CCR7−) showed a significant difference for ESAT‐6 and Rv0753c stimulation. Both the antigens showed similar positivity of 80% (12/15) in PTB and 33% (5/15) in HHC (Table 2). On the other hand, none of the antigens showed a significant difference between HHC and PTB for CD8 Tem cells (Fig. 2d).
However, the median levels of CD4+ naive T cells (CD3+ CD4+ CD45RA+ CCR7+) were high in PTB compared with HHC; the difference was not significant except for Rv0753c antigen‐specific CD4+ naive T cells. The AUC of Rv0753c‐specific CD4 naive T cells was 0·733 with a positivity of 80% (12/15) in PTB and 40% (6/15) in HHC (Table. 2). Though all the antigens showed a higher frequency of CD4+/CD8+ Tem cells (CD3+ CD4+/CD8+ CD45RA− CCR7−) in PTB than HHC, the difference was not statistically significant (p > 0·05).
Frequency of Treg cells in subjects with latent and active TB
The frequencies of different Treg cell marker combinations (CD25 and FoxP3) were analysed in the blood of HHC and PTB cases. The gating strategy followed for CD4 and CD8 regulatory responses are shown in Fig. 3.
Figure 3.
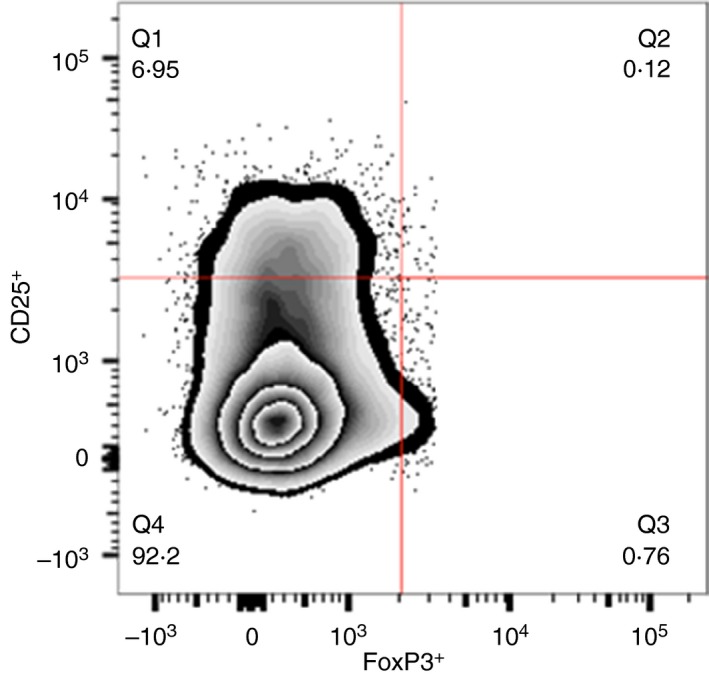
Representative flow cytometry plot for CD25 and FoxP3 expression by regulatory T (Treg) cells. This figure shows the representative flow cytometry plot of CD4+ Treg cells expressing CD25 and FoxP3 from a patient with active tuberculosis (TB). The numbers in the quadrants indicate the percentages of cells expressing CD25 and/or FoxP3 among the CD4+ population. Q1, CD3+ CD4+ CD25+ FoxP3− (CD4+ FoxP3− Treg cells); Q2, CD3+ CD4+ CD25+ FoxP3+ (CD4+ Treg cells); Q3, CD3+ CD4+ CD25− FoxP3+ (CD4+ FoxP3+ Treg cells); Q4, CD3+ CD4+ CD25− FoxP3− T‐cell subset. The same gating strategy was followed for all antigens. [Colour figure can be viewed at wileyonlinelibrary.com]
Increased percentage of CD4+ or CD8+ CD25+ and CD4+ CD25+ FoxP3+ T cells in active TB
In the current study, we considered CD3+ CD4+/CD8+ CD25+ FoxP3− as FoxP3− Treg cells. The median levels and the range of FoxP3− Treg cells were higher in PTB than HHC against all antigens. Results depicted in Fig. 4(a) indicate that the frequency of FoxP3− Treg cells within the CD4+ population was significantly increased in patients with PTB compared with HHC (P < 0·05). A statistically significant difference was observed for ESAT‐6‐, Rv2204c‐ and Rv0009‐specific CD4+ FoxP3− Treg cells. Subsequently, we analysed whether the differences in the percentage of CD4+ FoxP3− Treg cells could be used to distinguish LTBI from patients with active TB. The AUC of various antigen‐specific phenotypes were above 0·6, suggesting that these antigens may have discriminative capacity between HHC and PTB. The p‐value, AUC with 95% CI values and positivity in HHC and PTB of ESAT‐6, Rv2204c and Rv0009 are shown in Table 3. Among these antigens, maximum positivity of 80% (12/15) in PTB was observed for Rv0009, this antigen also showed moderate positivity of 33% (5/15) in HHC (Table 3). Regarding CD8+ FoxP3− Treg cells, ESAT‐6 exhibited a significant difference between HHC and PTB (Fig. 4c). This antigen identified nine PTB cases out of 15 (60% positivity) whereas in HHC, none of them was positive (0% positivity).
Figure 4.
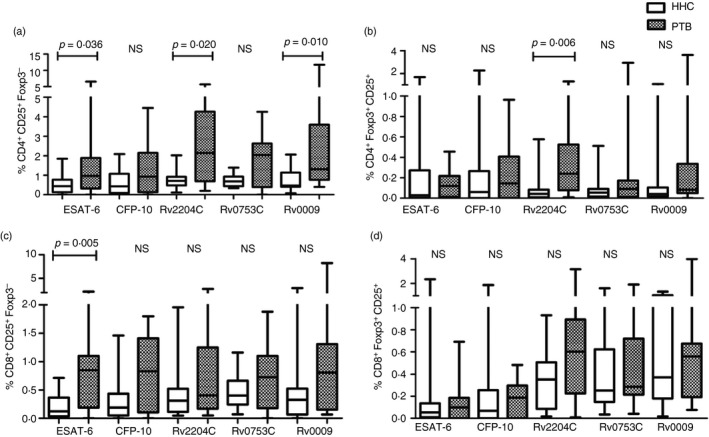
Frequency of CD4+ and CD8+ regulatory T (Treg) cells in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB). The percentages of regulatory T cells (CD25 and FoxP3) among the CD4+ and CD8+ T cells of 15 individuals with latent tuberculosis infection (LTBI) and 15 patients with PTB are represented as box plots with whiskers: the bottom and top lines of the boxes are the first and third centiles, middle line indicates median value, and the whiskers represent the range. (a) CD4+ CD25+ FoxP3− (CD4+ FoxP3− Treg cells), (b) CD4+ CD25+ FoxP3+ (CD4+ Treg cells), (c) CD8+ CD25+ FoxP3− (CD8+ FoxP3− Treg cells), and (d) CD8+ CD25+ FoxP3+ (CD8+ Treg cells). Differences between the groups were compared by non‐parametric Mann–Whitney U‐test. Statistical differences with p‐values are shown in the graph. NS, non‐significant.
Table 3.
Positivity for various CD4+ and CD8+ T regulatory subtypes in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB)
| Antigen | Phenotype | p‐value | AUC | CI 95% | Positivity in HHC (%) | Positivity in PTB (%) |
|---|---|---|---|---|---|---|
| ESAT‐6 | CD4+ FoxP3− CD25+ | 0·0362 | 0·7244 | 0·539–0·909 | 20 (3/15) | 66·6 (10/15) |
| Rv2204c | CD4+ FoxP3− CD25+ | 0·0202 | 0·7489 | 0·556–0·941 | 13·3 2/15) | 66·6 (10/15) |
| Rv0009 | CD4+ FoxP3− CD25+ | 0·0107 | 0·7733 | 0·604–0·942 | 33·3 5/15) | 80 (12/15) |
| ESAT‐6 | CD8+ FoxP3− CD25+ | 0·0051 | 0·8 | 0·628–0·971 | 0 (0/15) | 60 (9/15) |
| Rv2204c | CD4+ FoxP3+ CD25+ | 0·0066 | 0·7911 | 0·623–0·958 | 20 (3/15) | 73·3 (11/15) |
| Rv0009 | CD8+ FoxP3+ CD25− | 0·031 | 0·7089 | 0·512–0·904 | 26·6 4/15) | 73·3 (11/15) |
| Rv2204c | CD4+ FoxP3− CD25− | 0·0034 | 0·8133 | 0·659–0·967 | 73·3 11/15) | 20 (3/15) |
| Rv0753c | CD4+ FoxP3− CD25− | 0·0084 | 0·7822 | 0·613–0·951 | 60 (9/15) | 6·6 (1/15) |
| Rv0009 | CD4+ FoxP3− CD25− | 0·0114 | 0·7711 | 0·598–0·943 | 93·3 (14/15) | 40 (6/15) |
We considered CD3+ CD4+/CD8+ CD25+ FoxP3− as FoxP3− regulatory T (Treg) cells, CD3+ CD4+/CD8+ CD25+ FoxP3+ subset as Treg cells, CD3+ CD4+/CD8+ CD25− FoxP3+ subset as FoxP3+ Treg cells.
In this study, we considered the CD3+ CD4+/CD8+ CD25+ FoxP3+ subset as Treg cells. A substantial fraction of the CD4+ T cells from patients with active TB expressed both CD25+ and FoxP3+ compared with LTBI (Fig. 4b). The proportion of antigen‐specific CD4+ Treg cells expanded by ESAT‐6, CFP‐10, Rv0753c and Rv0009 but did not significantly differ between PTB and LTBI. However, there was a significant increase in the proportion of Rv2204c‐specific CD4+ Treg cells in active TB compared with LTBI. This antigen showed maximum positivity of 73% (11/15) in PTB and 20% (3/15) in HHC (Table 3). None of the antigens tested here showed a significant difference between HHC and PTB in the percentage of CD8+ Treg cells (Fig. 4d).
Percentage of CD25− FoxP3+ expressing CD8 T cells is increased in active TB
We defined the CD3+ CD4+/CD8+ CD25− FoxP3+ phenotype as FoxP3+ Treg cells. The proportion of antigen‐specific FoxP3 marker was similar in the two groups except for Rv0009 (Fig. 5a). Statistically, significant differences in percentages of Rv0009‐specific CD8+ FoxP3+ Treg cells were observed between active TB and LTBI (P < 0·05). The AUC value of CD8+ FoxP3+ is 0·708 with 95% CI. This antigen identified 11 PTB cases with 73·3% positivity and four HHC with 26·6% positivity (Table 3).
Figure 5.
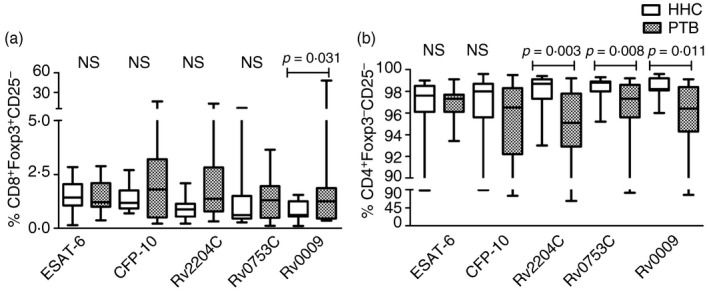
Frequency of CD25− FoxP3+ and CD25− FoxP3− T‐cell subsets in healthy household contacts (HHC) and patients with pulmonary tuberculosis (PTB). The percentages of regulatory T‐cell subsets (CD25 and FoxP3) of 15 individuals with latent tuberculosis infection (LTBI) and 15 patients with PTB are given as whisker box plots. The bottom and top lines indicate first and third centiles, the middle line represents median value, and the whiskers represent the range. (a) CD8+ FoxP3+ CD25− (CD8+ FoxP3+ Treg cells), (b) CD4+ CD25− FoxP3− T cells. Statistical differences were calculated by non‐parametric Mann–Whitney U‐test and the p‐values are given in the graph. NS, non‐significant.
Interestingly, the cells negative for both markers CD4+ CD25− and FoxP3− (CD3+ CD4+ CD25− FoxP3−) had shown a significant difference between LTBI and active TB against Rv2204c, Rv0753c and Rv0009 stimulations (Fig. 5b). The AUC values ranged from 0·77 to 0·813 for three antigens (Table 3). We observed the ROC curve with an AUC of 0·813 with a p‐value of 0·003, for the Rv2204c‐specific CD4+ CD25− FoxP3− phenotype. This antigen displayed 73·3% (11/15) positivity in HHC and 20% (3/15) positivity in the PTB (Table 3).
Discussion
It is estimated that 2 billion people have been infected with M. tuberculosis, which represents a potential source of future active TB. Identifying potential biomarkers that differentiate latent and active TB are urgently needed for TB control. Recently, IGRA has been widely used in TB diagnosis because it shows higher sensitivity and specificity compared with the tuberculin skin test (TST). However, like TST, IGRA cannot distinguish between LTBI and active TB, which limits its usage in countries with high levels of endemicity.32 Hence, identification of potential biomarkers that can differentiate LTBI and active TB would be beneficial.
Many studies have focused on the response to different M. tuberculosis antigens expressed during the different phases of infection.26, 33, 34, 35, 36 Therefore, identifying new antigens involved at different phases of TB infection and analysing antigen‐specific T‐cell responses in LTBI and active TB significantly improves antigen‐based TB diagnostic tests. Further, exploring immune response to new antigens might enlarge our knowledge in TB diagnostics. The main limitation of using new antigens for immunodiagnosis is the lack of specificity for M. tuberculosis. Although, the selected antigens are also not M. tuberculosis specific, the cytokine response to these antigens was not influenced by bacillus Clamette–Guérin (BCG) vaccination as observed in our previous studies.20, 21, 22 This might be due to the fact that BCG fails to establish a long‐lived memory response against these antigens.
Many studies focus on exploring functional CD4 and CD8 responses because these subsets are impacted by disease stage and bacterial burden. However, limited data are available on the surface markers of these subsets that are crucial for T‐cell characterization (memory and regulatory). In the current study, we analysed antigen‐specific memory and regulatory phenotypic profiles in latent and active TB for differential diagnosis of latent and active TB.
It has been shown that the functional and phenotypic properties of Tcm and Tem cells of CD4 and CD8 are deferentially expressed in LTBI and active TB.37 So, the subsets of memory cells express different cell‐surface markers based on the phase of the disease. In this study, we also evaluated the phenotypic properties of Tcm and Tem cells in LTBI and active TB on overnight antigenic stimulation.
We observed significantly elevated levels in the frequencies of CD4+ and CD8+ Tcm cells in HHC relative to PTB with all antigenic stimulations. Among these stimulations, the highest levels of significance were obtained for CD8+ Tcm cells when compared with the CD4 Tcm phenotype. The percentage of positivity shown by ESAT‐6‐, Rv0753c‐ and Rv0009‐specific Tcm CD8 T cells seems to be promising in discriminating LTBI and PTB due to their higher positivity (80%) in HHC. Hence, including these antigens in the TB diagnostic test might improve the test performances, if similar results are obtained on the larger study population. On the other hand, effector memory T cells were higher in PTB than HHC. It has been previously reported that TST‐positive individuals induce CD4 Tcm cells in response to ESAT‐6 and CFP‐10.38, 39 Moreover, prolonged re‐stimulation of peripheral blood mononuclear cells isolated from individuals with LTBI preferentially induces a central memory phenotype. The discriminative expansion of Tcm cells in response to RD1 antigens in TST‐positive individuals with cured TB and in healthy individuals in response to BCG has also been reported.39, 40 In agreement with previous studies, our report also showed that CD4+ Tcm cells were associated with latent infection, whereas Tem cells were associated with active TB.39, 40, 41, 42 Overall, the association of Tem cells and active TB may reflect the active M. tuberculosis replication, whereas Tcm cells in LTBI indicate successful M. tuberculosis replication control. Earlier, we observed that significant levels of CD4 and CD8 secreting IFN‐γ in LTBI might be associated with a central memory phenotype that leads to maintenance of latency and protection against reactivation (unpublished data). On the other hand, Tem cells show immediate effector functions, maintaining pre‐formed cytotoxic granules for rapid cytolysis of infected host cells.43 The loss of CCR7 in effector T cells allows T‐cell accumulation at the site of infection and subsequent onset of the adaptive immune response to replicating bacteria occurs.44
The differentiation of T cells into a distinct functional population determines the quality of the immune response.45 It has been reported that the relative proportions and frequencies of these distinct T‐cell subsets (effector and memory) are correlated with pathogen burden and antigen load in viral infections46 and TB infection.47 In the present study, effector and effector memory T cells are predominant in PTB, which could be due to the higher antigenic load in active TB, unlike LTBI where the antigenic load is often presumed to be low and Tcm cells were predicted to be predominant in our results. The relative proportion of antigen‐specific Tcm and Tem cell subsets might correlate with protection by their distinct cytokine secretion profiles. Effector T cells secrete only IFN‐γ, Tem cells secrete both IFN‐γ and IL‐2, Tcm cells secrete only IL‐2 cytokine and these cytokines have immunomodulatory functions during M. tuberculosis infection.
Tuberculosis is a chronic disease during which bacilli evade the immune system to persist within the host and there is a need to find and understand the mechanism of immunopathology of M. tuberculosis. The immune system possesses a regulatory mechanism in which Treg cells play essential roles in establishing and sustaining self‐tolerance and immune homeostasis as well as regulating the host immune response to infection. Though extensive research has been performed on the role of Treg cells in recent years, it is not completely understood whether their differential expression can be used as a biomarker for differential diagnosis of LTBI and active TB. In this study, we have analysed the most frequently used classical regulatory markers like surface marker CD25 and intracellular transcription factor FoxP3. The FoxP3 is considered as a fundamental factor for the development and function of Treg cells and also acts as a most reliable molecular marker for Treg cells.17 CD25 is an activation marker for T cells but, it can also be used as a marker for Treg cells. It is well known that Treg cells suppress the proliferation of Th cells by increasing expression of the IL‐2 co‐receptor CD25, which allows Treg cells to act as a sink for IL‐2. This leads to IL‐2 deprivation and the inhibition of T‐cell proliferation.48 The CD4+ CD25+ Treg cells have the specific role of limiting strong Th1 responses induced by microbial antigens during infections to prevent excessive inflammation and tissue damage in the host.11 Hence, in this study, along with FoxP3, a key regulator of Treg cell development and function, CD25 can be used as a phenotype marker for Treg cells. Therefore, we included these two markers for analysing antigen‐specific Treg cells and evaluated their possible role in discriminating LTBI and active TB.
We observed an increased frequency of Rv2204c antigen‐specific CD4+ Treg cells in the peripheral blood of patients with active TB compared with HHC. Consistent with our observation, several studies also reported that CD4+ Treg cells were significantly higher in peripheral blood mononuclear cells of patients with active TB compared with individuals with LTBI23, 49 despite few reports stating no significant differences in the percentage of CD4+/CD8+ Treg cells between active TB and LTBI.50 This varying observation could be due to the difference in the source and nature of the antigens used, incubation period, and ethnicity of the recruited population.
The immune status of PTB reflects the pathogenesis of the disease and phenotypes like Treg are often found to be higher in PTB for most of the predicted antigens. In contrast to LTBI, the increase in Treg cells in PTB is often correlated with dampening of the Th1 response, which is still considered to be important for disease control.51, 52 Due to the immunological imbalance in active TB, increased inflammation causes the higher frequency of circulating Treg cells in peripheral blood.53, 54 It is unclear whether differentiation and expansion of Treg cells during TB infection lead to disease progression. However, Treg cells play a role in the immunosuppression of TB51, 55 and other chronic infectious diseases,11, 56 and subsequently lead to disease progression. A possible mechanism of immune suppression by Treg cells is producing transforming growth factor‐β and/or IL‐10 cytokines, which depresses IFN‐γ production, an important mediator required for anti‐TB immunity.49, 53 Our earlier observation predicted21, 22 an increased expression of IL‐10 in active TB that may be associated with the suppression of Th1 and Th17 immune responses,53, 57 inhibition of antigen processing and presentation58 and inhibition of granuloma formation.59 It has been also demonstrated that there is an increased IFN‐γ response after depletion of Treg cells in active TB.60 Similar to this, our previous study21, 22, 23 reported a decreased IFN‐γ response in patients with active TB who had increased levels of Treg cells in their circulation. Further, it is evidenced that Treg cells delay the activation of CD4+ and CD8+ cells in lymph nodes, which in turn causes delayed migration of these cells to the site of infection (lungs) during TB.8 Also, it was reported that Treg cells allow the establishment of M. tuberculosis infection by delaying the onset of an adaptive immune response.61, 62 On the other hand, the lower levels of Treg cells in LTBI could be due to the migration of Treg cells to the lungs during early infection.63 Due to these distinct features in active TB and LTBI, Treg cells could be considered as a potential biomarker to differentiate latent infection and TB disease. It has already been reported that latency antigen heparin‐binding haemagglutinin‐specific Treg cells showed significant differences between latent and active TB.64 Similarly, our selected antigens also showed maximum discrimination between LTBI and active TB.
The main objective of our study was to differentiate latent infection and active TB disease, which is a prerequisite to control the TB burden in endemic settings like India. Exposure to environmental mycobacteria is considered to be widespread (0·7–34%) in India65, 66 and pre‐existing immune responses might cross‐react/interfere with the assessment of antigen‐specific immune responses in healthy controls. Hence, for the present preliminary analysis, healthy community controls were not included. Also, lack of a reference standard test to differentiate healthy controls and healthy LTBI contacts further impedes the recruitment of healthy controls. In addition, BCG vaccination coverage in India is nearly 100%, hence it is highly challenging to recruit a sufficient number of uninfected (healthy) or non‐BCG‐vaccinated controls in an endemic setting like India. However, to enrich our understanding of the spectrum of immune responses during TB infection and to assess antigen‐specific immune responses, discrimination between controls and infected patients, including healthy controls, for in vitro analysis would be beneficial.
Conclusion
Taken collectively, our results demonstrate the association of central memory T cells with LTBI and effector T‐cell association with active TB. ESAT‐6‐, Rv0753c‐ and Rv0009‐specific central memory CD8 T cells showed > 80% positivity in HHC and 20% positivity in PTB. Hence, this phenotype might be useful to discriminate LTBI from active TB. Further, our data indicate that patients with active TB have a higher proportion of circulating Rv2204c antigen‐reactive CD4+ Treg cells than individuals who are LTBI. However, additional studies are required to validate our observation in the larger study populations using these phenotypic markers for the differential diagnosis of latent and active TB. Further, prospective studies to follow up individuals who are LTBI to assess their risk of TB progression by measuring the specific memory and regulatory phenotypes of these antigens.
Disclosures
The authors declare that they have no competing interests.
Supporting information
Figure S1. Receiver operating characteristics curve analysis of memory and regulatory T cells.
Acknowledgements
The authors wish to thank all the study participants. Dr Balaji Pathakumari and Dr Santhi Devasundaram express their gratitude to the Indian Council of Medical Research (ICMR), New Delhi, India, for providing the Senior Research Fellowship. We thank Dr Subash Babu, NIH‐NIRT for providing flow cytometry facilities.
References
- 1. World Health Organization (2015) Global tuberculosis report. WHO/HTM/TB/2015.22. Geneva, Switzerland: World Health Organization. [WWW document] URL http://www.who.int/tb/publications/global_report/en/ [accessed on 2016]
- 2. Stemberger C, Neuenhahn M, Buchholz VR, Busch DH. Origin of CD8+ effector and memory T cell subsets. Cell Mol Immunol 2007; 4:399–405. [PubMed] [Google Scholar]
- 3. MacLeod MK, Kappler JW, Marrack P. Memory CD4 T cells: generation, reactivation and re‐assignment. Immunology 2010; 130:10–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lanzavecchia A, Sallusto F. Understanding the generation and function of memory T cell subsets. Curr Opin Immunol 2005; 17:326–32. [DOI] [PubMed] [Google Scholar]
- 5. Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science 2001; 291:2413–7. [DOI] [PubMed] [Google Scholar]
- 6. Klebanoff CA, Gattinoni L, Torabi‐Parizi P, Kerstann K, Cardones AR, Finkelstein SE et al Central memory self/tumor‐reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci USA 2005; 102:9571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakaguchi S, Sakaguchi N. Regulatory T cells in immunologic self‐tolerance and autoimmune disease. Int Rev Immunol 2005; 24:211–26. [DOI] [PubMed] [Google Scholar]
- 8. Shafiani S, Tucker‐Heard G, Kariyone A, Takatsu K, Urdahl KB. Pathogen‐specific regulatory T cells delay the arrival of effector T cells in the lung during early tuberculosis. J Exp Med 2010; 207:1409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ozeki Y, Sugawara I, Udagawa T, Aoki T, Osada‐Oka M, Tateishi Y et al Transient role of CD4+CD25+ regulatory T cells in mycobacterial infection in mice. Int Immunol 2010; 22:179–89. [DOI] [PubMed] [Google Scholar]
- 10. Gratz IK, Rosenblum MD, Maurano MM, Paw JS, Truong HA, Marshak‐Rothstein A et al Cutting edge: self‐antigen controls the balance between effector and regulatory T cells in peripheral tissues. J Immunol 2014; 192:1351–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol 2005; 6:353–60. [DOI] [PubMed] [Google Scholar]
- 12. Coenen JJ, Koenen HJ, Emmer PM, van Rijssen E, Hilbrands LB, Joosten I. Allogeneic stimulation of naturally occurring CD4+CD25+ T cells induces strong regulatory capacity with increased donor‐reactivity. Transpl Immunol 2007; 17:237–42. [DOI] [PubMed] [Google Scholar]
- 13. O'Garra A, Vieira P. Regulatory T cells and mechanisms of immune system control. Nat Med 2004; 10:801–5. [DOI] [PubMed] [Google Scholar]
- 14. Levings MK, Sangregorio R, Roncarolo MG. Human CD25+ CD4+ T regulatory cells suppress naive and memory T cell proliferation and can be expanded in vitro without loss of function. J Exp Med 2001; 193:1295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakaguchi S, Hori S, Fukui Y, Sasazuki T, Sakaguchi N, Takahashi T. Thymic generation and selection of CD25+CD4+ regulatory T cells: implications of their broad repertoire and high self‐reactivity for the maintenance of immunological self‐tolerance. Novartis Found Symp 2003; 252:6–16; discussion 16‐23, 106‐114. [PubMed] [Google Scholar]
- 16. Oldenhove G, de Heusch M, Urbain‐Vansanten G, Urbain J, Maliszewski C, Leo O et al CD4+ CD25+ regulatory T cells control T helper cell type 1 responses to foreign antigens induced by mature dendritic cells in vivo . J Exp Med 2003; 198:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nat Immun 2003; 4:330–6. [DOI] [PubMed] [Google Scholar]
- 18. Sakaguchi S. Regulatory T cells in the past and for the future. Eur J Immunol 2008; 38:901–37. [DOI] [PubMed] [Google Scholar]
- 19. Rozot V, Vigano S, Mazza‐Stalder J, Idrizi E, Day CL, Perreau M et al Mycobacterium tuberculosis‐specific CD8+ T cells are functionally and phenotypically different between latent infection and active disease. Eur J Immunol 2013; 43:1568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prabhavathi M, Pathakumari B, Raja A. IFN‐γ/TNF‐α ratio in response to immuno proteomically identified human T‐cell antigens of Mycobacterium tuberculosis – The most suitable surrogate biomarker for latent TB infection. J Infect 2015; 71:238–49. [DOI] [PubMed] [Google Scholar]
- 21. Pathakumari B, Anbarasu D, Parthasarathy RT, Raja A. PpiA antigen specific immune response is a potential biomarker for latent tuberculosis infection. Tuberculosis 2015; 95:736–43. [DOI] [PubMed] [Google Scholar]
- 22. Pathakumari B, Prabhavathi M, Raja A. Evaluation of cytokine and chemokine response elicited by Rv2204c and Rv0753c to detect latent tuberculosis infection. Cytokine 2015; 76:496–504. [DOI] [PubMed] [Google Scholar]
- 23. Hougardy JM, Place S, Hildebrand M, Drowart A, Debrie AS, Locht C et al Regulatory T cells depress immune responses to protective antigens in active tuberculosis. Am J Respir Crit Care Med 2007; 176:409–16. [DOI] [PubMed] [Google Scholar]
- 24. Pollock KM, Whitworth HS, Montamat‐Sicotte DJ, Grass L, Cooke GS, Kapembwa MS et al T‐cell immunophenotyping distinguishes active from latent tuberculosis. J Infect Dis 2013; 208:952–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dosanjh NS, Rawat M, Chung JH, Av‐Gay Y. Thiol specific oxidative stress response in Mycobacteria. FEMS Microbiol Lett 2005; 249:87–94. [DOI] [PubMed] [Google Scholar]
- 26. Deenadayalan A, Heaslip D, Rajendiran AA, Velayudham BV, Frederick S, Yang HL et al Immunoproteomic identification of human T cell antigens of Mycobacterium tuberculosis that differentiate healthy contacts from tuberculosis patients. Mol Cell Proteomics 2010; 9:538e49 [PMID: 20031926]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kassa D, Ran L, Geberemeskel W, Tebeje M, Alemu A, Selase A et al Analysis of immune responses against a wide range of Mycobacterium tuberculosis antigens in patients with active pulmonary tuberculosis. Clin Vaccine Immunol 2012; 19:1907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Serra‐Vidal MM, Latorre I, Franken KL, Diaz J, de Souza‐Galvao ML, Casas I et al Immunogenicity of 60 novel latency‐related antigens of Mycobacterium tuberculosis . Front Microbiol 2014; 5:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim JS, Kim WS, Choi HH, Kim HM, Kwon KW, Han SJ et al Mycobacterium tuberculosis MmsA, a novel immunostimulatory antigen, induces dendritic cell activation and promotes Th1 cell‐type immune responses. Cell Immunol 2015; 298:115–25. [DOI] [PubMed] [Google Scholar]
- 30. Devasundaram S, Khan I, Kumar N, Das S, Raja A. The influence of reduced oxygen availability on gene expression in laboratory (H37Rv) and clinical strains (S7 and S10) of Mycobacterium tuberculosis . J Biotechnol 2015; 210:70–80. [DOI] [PubMed] [Google Scholar]
- 31. Casey R, Blumenkrantz D, Millington K, Montamat‐Sicotte D, Kon OM, Wickremasinghe M et al Enumeration of functional T‐cell subsets by fluorescence‐immunospot defines signatures of pathogen burden in tuberculosis. PLoS One 2010; 5:e15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Metcalfe JZ, Everett CK, Steingart KR, Cattamanchi A, Huang L, Hopewell PC et al Interferon‐γ release assays for active pulmonary tuberculosis diagnosis in adults in low‐ and middle‐income countries: systematic review and meta‐analysis. J Infect Dis 2011; 204(Suppl. 4):S1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leyten EM, Lin MY, Franken KL, Friggen AH, Prins C, van Meijgaarden KE et al Human T‐cell responses to 25 novel antigens encoded by genes of the dormancy regulon of Mycobacterium tuberculosis . Microbes Infect 2006; 8:2052–60. https://doi.org/10.1016/j.micinf.2006.03.018 [DOI] [PubMed] [Google Scholar]
- 34. Delogu G, Chiacchio T, Vanini V, Butera O, Cuzzi G, Bua A et al Methylated HBHA produced in M. smegmatis discriminates between active and non‐active tuberculosis disease among RD1‐responders. PLoS One 2011; 6:e18315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Black GF, Thiel BA, Ota MO, Parida SK, Adegbola R, Boom WH et al Immunogenicity of novel DosR regulon‐encoded candidate antigens of Mycobacterium tuberculosis in three high‐burden populations in Africa. Clin Vaccine Immunol 2009; 16:1203–12. https://doi.org/10.1128/CVI.00111-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Commandeur S, van Meijgaarden KE, Prins C, Pichugin AV, Dijkman K, van den Eeden SJ et al An unbiased genome‐wide Mycobacterium tuberculosis gene expression approach to discover antigens targeted by human T cells expressed during pulmonary infection. J Immunol 2013; 190:1659–71. https://doi.org/10.4049/jimmunol.1201593 [DOI] [PubMed] [Google Scholar]
- 37. Petruccioli E, Petrone L, Vanini V, Sampaolesi A, Gualano G, Girardi E et al IFNγ/TNFα specific‐cells and effector memory phenotype associate with active tuberculosis. J Infect 2013; 66:475–86. [DOI] [PubMed] [Google Scholar]
- 38. Rueda CM, Marin ND, Garcia LF, Rojas M. Characterization of CD4 and CD8 T cells producing IFN‐γ in human latent and active tuberculosis. Tuberculosis 2010; 90:346–53. [DOI] [PubMed] [Google Scholar]
- 39. Goletti D, Butera O, Bizzoni F, Casetti R, Girardi E, Poccia F. Region of difference 1 antigen‐specific CD4+ memory T cells correlate with a favorable outcome of tuberculosis. J Infect Dis 2006; 194:984–92. [DOI] [PubMed] [Google Scholar]
- 40. Mueller H, Detjen AK, Schuck SD, Gutschmidt A, Wahn U, Magdorf K et al Mycobacterium tuberculosis‐specific CD4+, IFNγ +, and TNFα + multifunctional memory T cells coexpress GM‐CSF. Cytokine 2008; 43:143–8. [DOI] [PubMed] [Google Scholar]
- 41. Wang X, Cao Z, Jiang J, Niu H, Dong M, Tong A et al Association of mycobacterial antigen‐specific CD4+ memory T cell subsets with outcome of pulmonary tuberculosis. J Infect 2010; 60:133–9. https://doi.org/10.1016/j.jinf.2009.10.048 [DOI] [PubMed] [Google Scholar]
- 42. Riano F, Arroyo L, París S, Rojas M, Friggen AH et al T cell responses to DosR and Rpf proteins in actively and latently infected individuals from Colombia. Tuberculosis 2011; 92:148–59. [DOI] [PubMed] [Google Scholar]
- 43. Woodland David L, Kohlmeier Jacob E. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol 2009; 9:153–61. [DOI] [PubMed] [Google Scholar]
- 44. Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol 2005; 6:895–901. [DOI] [PubMed] [Google Scholar]
- 45. Seder RA, Darrah PA, Roederer M. T‐cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 2008; 8:247–58. [DOI] [PubMed] [Google Scholar]
- 46. Sester U et al PD‐1 expression and IL‐2 loss of cytomegalovirus‐specific T cells correlates with viremia and reversible functional anergy. Am J Transplant 2008; 8:1486–97. [DOI] [PubMed] [Google Scholar]
- 47. Millington KA, Innes JA, Hackforth S, Hinks TS, Deeks JJ, Dosanjh DP et al Dynamic relationship between IFN‐γ and IL‐2 profile of Mycobacterium tuberculosis‐specific T cells and antigen load. J Immunol 2007; 178:5217–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmidt A, Nina O, Krammer PH. Molecular mechanisms of Treg‐mediated T cell suppression. Front Immunol 2012; 3:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ribeiro‐Rodrigues R, Resende Co T, Rojas R, Toossi Z, Dietze R, Boom WH et al A role for CD4+CD25+ T cells in regulation of the immune response during human tuberculosis. Clin Exp Immunol 2006; 144:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Semple PL, Binder AB, Davids M, Maredza A, van Zyl‐Smit RN, Dheda K. Regulatory T cells attenuate mycobacterial stasis in alveolar and blood‐derived macrophages from patients with tuberculosis. Am J Respir Crit Care Med 2013; 187:1249–58. [DOI] [PubMed] [Google Scholar]
- 51. Li Li, Lao Sui‐hua, Chang‐you Wu. Increased frequency of CD4+ CD25high Treg cells inhibit BCG‐specific induction of IFN‐γ by CD4+ T cells from TB patients. Tuberculosis 2007; 87:526–34. [DOI] [PubMed] [Google Scholar]
- 52. Chen X, Zhou B, Li M, Deng Q, Wu X, Le X et al CD4+ CD25+ FoxP3+ regulatory T cells suppress Mycobacterium tuberculosis immunity in patients with active disease. Clin Immunol 2007; 123:50–9. [DOI] [PubMed] [Google Scholar]
- 53. Marin ND, París SC, Vélez VM, Rojas CA, Rojas M, García LF. Regulatory T cell frequency and modulation of IFN‐γ and IL‐17 in active and latent tuberculosis. Tuberculosis 2010; 90:252–61. https://doi.org/10.1016/j.tube.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 54. Guyot‐Revol V, Innes JA, Hackforth S, Hinks T, Lalvani A. Regulatory T cells are expanded in blood and disease sites in patients with tuberculosis. Am J Respir Crit Care Med 2006; 173:803–10. [DOI] [PubMed] [Google Scholar]
- 55. Kursar M, Koch M, Mittrucker HW, Nouailles G, Bonhagen K, Kamradt T et al Cutting edge: regulatory T cells prevent efficient clearance of Mycobacterium tuberculosis . J Immunol 2007; 178:2661–5. [DOI] [PubMed] [Google Scholar]
- 56. Mills KH. Regulatory T cells: friend or foe in immunity to infection? Nat Rev Immunol 2004; 4:841–55. [DOI] [PubMed] [Google Scholar]
- 57. Pitt JM, Stavropoulos E, Redford PS, Beebe AM, Bancroft GJ, Young DB et al Blockade of IL‐10 signaling during bacillus Calmette–Guérin vaccination enhances and sustains Th1, Th17, and innate lymphoid IFN‐γ and IL‐17 responses and increases protection to Mycobacterium tuberculosis infection. J Immunol 2012; 189:4079e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bobadilla K, Sada E, Jaime ME, Gonzalez Y, Ramachandra L, Rojas RE et al Human phagosome processing of Mycobacterium tuberculosis antigens is modulated by interferon‐γ and interleukin‐10. Immunology 2013; 138:34e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cyktor JC, Carruthers B, Kominsky RA, Beamer GL, Stromberg P, Turner J. IL‐10 inhibits mature fibrotic granuloma formation during Mycobacterium tuberculosis infection. J Immunol 2013; 190:2778e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Place S, Verscheure V, de San N, Hougardy JM, Schepers K, Dirix V et al Heparin‐binding, hemagglutinin‐specific IFN‐γ synthesis at the site of infection during active tuberculosis in humans. Am J Respir Crit Care Med 2010; 182:848–54. [DOI] [PubMed] [Google Scholar]
- 61. Cooper AM. Cell‐mediated immune responses in tuberculosis. Annu Rev Immunol 2009; 27:393–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ottenhoff TH. The knowns and unknowns of the immunopathogenesis of tuberculosis. Int J Tuberc Lung Dis 2012; 16:1424–32. [DOI] [PubMed] [Google Scholar]
- 63. Burl S, Hill PC, Jeffries DJ, Holland MJ, Fox A, Lugos MD et al FOXP3 gene expression in a tuberculosis case contact study. Clin Exp Immunol 2007; 149:117–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hougardy JM, Schepers K, Place S, Drowart A, Lechevin V, Verscheure V et al Heparin‐binding‐hemagglutinin‐induced IFN‐γ release as a diagnostic tool for latent tuberculosis. PLoS One 2007; 2:e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paramasivan CN, Govindan D, Prabhakar R, Somasundaram PR, Subbammal S, Tripathy SP. Species level identification of non‐tuberculous mycobacteria from South Indian BCG trial area during 1981. Tubercle 1985; 66:9–15. [DOI] [PubMed] [Google Scholar]
- 66. Myneedu VP, Verma AK, Bhalla M, Arora V, Reza S, Sah GC, Behera D. Occurrence of non‐tuberculous mycobacterium in clinical samples—a potential pathogen. Ind J Tub 2013; 60:71–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Receiver operating characteristics curve analysis of memory and regulatory T cells.


