Wang et al. show that the Ku complex shields DNA ends from exonucleolytic digestion but facilitates endonucleolytic scission by MRX with a dependence on ATP and Sae2.
Keywords: DNA end resection, nuclease, MRX–Sae2, Ku70–Ku80, RPA, nucleosome
Abstract
The budding yeast Mre11–Rad50–Xrs2 (MRX) complex and Sae2 function together in DNA end resection during homologous recombination. Here we show that the Ku complex shields DNA ends from exonucleolytic digestion but facilitates endonucleolytic scission by MRX with a dependence on ATP and Sae2. The incision site is enlarged into a DNA gap via the exonuclease activity of MRX, which is stimulated by Sae2 without ATP being present. RPA renders a partially resected or palindromic DNA structure susceptible to MRX–Sae2, and internal protein blocks also trigger DNA cleavage. We present models for how MRX–Sae2 creates entry sites for the long-range resection machinery.
DNA double-strand breaks (DSBs) are induced by ionizing radiation (IR) and chemotherapeutic drugs and can also arise from DNA replication fork stalling or collapse (Petermann et al. 2010; Pommier et al. 2010; Cannan and Pederson 2016). If left unrepaired or repaired inappropriately, DSBs lead to loss of genetic information and chromosome rearrangements, which could result in cell transformation and tumorigenesis (Pierce et al. 2001). Interestingly, DSBs are an essential intermediate during programmed cellular processes, such as meiotic recombination, V(D)J recombination, and mating type switching in the budding yeast (Schlissel et al. 1993; Keeney et al. 1997; Haber 2012).
Nonhomologous DNA end joining (NHEJ) and homologous recombination (HR) are conserved pathways of DSB repair (Symington and Gautier 2011). In NHEJ, the Ku70–Ku80 heterodimer (referred to here as Ku) engages the DNA ends (Wu et al. 2008) and then recruits other factors to process the substrate, cumulating in end rejoining by DNA ligase IV (Palmbos et al. 2008; Weterings and Chen 2008; Chang et al. 2017). NHEJ is often associated with a gain or loss of genetic information at the break site. HR is largely error-free, as it relies on the genetically identical sister chromatid to template break repair. As such, HR becomes a major DSB repair tool only in the S and G2 phases of the cell cycle when the sister chromatid is available.
In HR, the DNA break ends undergo extensive 5′ strand resection. This yields ssDNA tails for assembling the HR machinery that harbors the recombinase Rad51 and also for activating the DNA damage checkpoint that is controlled by Mec1/ATR in conjunction with its damage sensor, Ddc2/ATRIP (Marechal and Zou 2013). Importantly, DSB end resection helps prevent NHEJ and commit cells to break repair via HR (Symington and Gautier 2011; Daley and Sung 2014). DNA end resection proceeds in two stages, with the initial step being mediated by Mre11–Rad50–Xrs2 (MRX; MRE11–RAD50–NBS1 [MRN] in mammals) and Sae2 (CtIP in mammals), which serves to create an entry site for the long-range resection machinery (Mimitou and Symington 2008; Zhu et al. 2008). In the repair of a site-specific DSB, MRX–Sae2 can incise the 5′ DNA strand ∼100–200 nucleotides (nt) away from the DNA end. During the processing of meiotic DSBs generated by the topoisomerase II-like protein Spo11, MRX–Sae2 introduces 5′ DNA strand incisions proximal to the Spo11-bound DNA ends (Garcia et al. 2011). The current model posits that MRX–Sae2 mediates a “chew-back” reaction via its 3′–5′ exonuclease activity to enlarge the initial DNA nick into a gap, which then serves as the loading pad for the Sgs1–Top3–Rmi1 (STR) complex with the nuclease/helicase Dna2 or the 5′–3′ exonuclease Exo1 for the catalysis of long-range 5′ strand resection (Garcia et al. 2011).
We devised biochemical systems to determine how MRX and Sae2 collaborate to initiate and propagate DNA end resection. Notable findings were made, specifically the following: (1) Sae2 enhances not only endonucleolytic 5′ strand scission by MRX but also the 3′–5′ chew-back reaction to generate a DNA gap. (2) The MRX–Sae2 ensemble incises the 5′ DNA strand endonucleolytically in proximity to a partially resected DNA end engaged by the ubiquitous ssDNA-binding protein RPA. (3) Sae2 licenses the scission of palindromic DNA that borders an RPA-bound ssDNA loop. (4) MRX–Sae2 acts at protein obstacles, such as a nucleosome, located at internal sites of DNA. We propose a model in which the arrest of the diffusing MRX–Sae2 ensemble at a protein obstacle leads to 5′ strand incision and 3′–5′ chew-back to enable the loading of the long-range resection machinery. Our results have implications for understanding how eukaryotic cells tolerate pathological protein-bound DNA intermediates, such as those arising from perturbations in transcription, DNA replication, and arrested topoisomerase–DNA conjugates (Hartsuiker et al. 2009; Sordet et al. 2010; Duquette et al. 2012).
Results and Discussion
Action of MRX–Sae2 at Ku-bound DNA ends
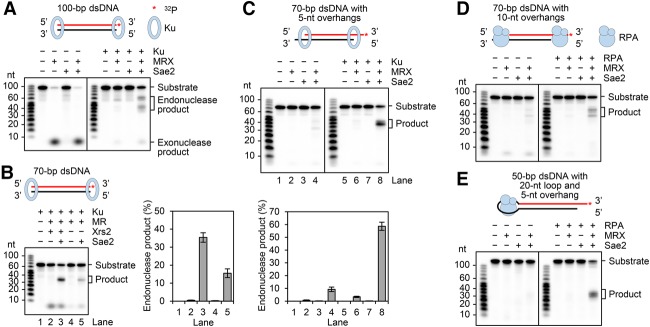
Sae2 enhances 5′ strand scission by MRX at DNA ends that harbor a biotin–streptavidin complex (Cannavo and Cejka 2014). We asked whether MRX–Sae2 also acts on Ku-occluded DNA ends. For this, we expressed MRX and Ku in yeast and Sae2 in insect cells and purified them (Supplemental Fig. S1A). We verified that our Ku preparations bind linear substrates of 70 or 100 base pairs (bp) (Supplemental Fig. S1B).
MRX released 32P exonucleolytically from the 3′-labeled end of the substrates, and Ku shielded the DNA end from exonucleolytic attack (Fig. 1A; Supplemental Fig. S1C). Importantly, the addition of Sae2 with MRX led to endonucleolytic cleavage of the Ku-occluded substrate, generating products of ∼35–40 nt from the 70-bp substrate (Supplemental Fig. S1C,D). This corresponds to cleavage ∼30–35 nt away from the 5′ terminus. In agreement with previous findings (Cannavo and Cejka 2014), pretreatment of Sae2 with λ phosphatase rendered it inactive in the DNA cleavage reaction (data not shown), indicating that Sae2 phosphorylation is indispensable for up-regulating the endonucleolytic activity of MRX.
Figure 1.
Endonuclease activity of MRX–Sae2 on Ku- and RPA-bound DNA. (A) The 100-bp 3′-labeled substrate (1 nM) was incubated with 40 nM MRX, 480 nM Sae2, and 16 nM Ku. (B) The nuclease activity of 40 nM Mre11–Rad50 (MR) on the 70-bp 3′-labeled substrate was examined with and without 40 nM Xrs2, as in A, except that 8 nM Ku was used. (C) The 3′-labeled 70-bp dsDNA containing 5-nt overhangs at the 3′ termini was tested with MRX, Sae2, and Ku as in B. In B and C, the results from three independent experiments were quantified. The error bars represent SD. (D) The 70-bp dsDNA (1 nM) that harbors 10-nt 3′ overhangs was tested with 40 nM MRX, 480 nM Sae2, and 32 nM RPA. (E) The hairpin substrate (1 nM) consisting of 50-bp dsDNA, a 20-nt loop, and a 5-nt 3′ overhang was tested with 40 nM MRX, 480 nM Sae2, and 32 nM RPA. The asterisk denotes the 32P label in the substrate in all of the figure parts. See also Supplemental Figures S1 and S2.
Interestingly, with the 3′ end-labeled 100-bp substrate, two clusters of endonuclease products (∼55–65 nt and ∼35–45 nt, marked as cluster 1 and cluster 2, respectively, in Supplemental Fig. S1D) were generated by MRX, corresponding to cleavage sites ∼35–45 and ∼55–65 nt away from the 5′ terminus, respectively. The addition of Sae2 led to (1) an enhancement of product formation within both clusters and (2) products more heterogeneous in size. The effect of Sae2 is specific, as its human ortholog CtIP is devoid of such an attribute (Supplemental Fig. S1E). Given that MRX–Sae2 can also efficiently process protein obstacles at internal sites of the DNA substrate (see below), we surmise that the cluster 2 products from the 100-bp substrate (Supplemental Fig. S1D) stem from MRX acting next to a second molecule of Ku or an MRX complex that abuts the Ku-occluded end (see Fig. 4A, below, for model). By testing Ku-bound plasmid-length linear dsDNA, we verified that MRX–Sae2 incises only the 5′-terminated DNA strand (Supplemental Fig. S1F).
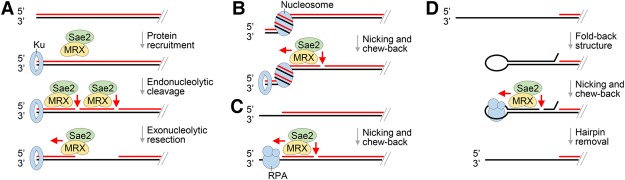
Figure 4.
Models for MRX action in DNA end resection and processing of protein obstacles. (A) In DSB end resection, Ku, while protecting the DNA end from exonucleolytic digestion, promotes 5′ strand incision by MRX–Sae2. The resulting DNA nick is enlarged exonucleolytically in the 3′–5′ direction by MRX–Sae2. (B) Within the context of chromatin, MRX–Sae2 endonucleolytically cleaves the 5′ strand of DNA that borders a nucleosome. The resulting DNA nick is expected to be subject to 3′–5′ chew-back to generate a DNA gap. (C) Binding of RPA to a partially resected DNA end activates 5′ strand scission by MRX–Sae2, which would lead to a gap upon 3′–5′ chew-back. (D) The fold-back structure, which arises from annealing between inverted repeats in ssDNA, is removed by MRX–Sae2 in collaboration with RPA via nicking and chew-back action.
Importantly, we found that even though the Mre11–Rad50 (MR) complex can carry out endonucleolytic cleavage, the addition of Xrs2 leads to a marked stimulation of reaction efficiency (Fig. 1B). As expected, MRX–Sae2 that harbors the Mre11 (H125L/D126V) nuclease-dead mutant (Bressan et al. 1998) lacks DNA cleavage activity (Supplemental Fig. S1G). Neither Sae2 nor CtIP showed any nuclease activity alone (Fig. 1A; Supplemental Fig. S1E). Endonucleolytic scission by MRX–Sae2 is strictly dependent on ATP hydrolysis, as revealed by testing MRX variants that harbor a mutation (K40A, K40E, or K40R) in the conserved Rad50 Walker A motif (Supplemental Fig. S1H; Chen et al. 2005) and by substitution of ATP with the nonhydrolyzable analog ATPγS or AMP-PNP (Supplemental Fig. S1I).
The introduction of a 5-nt 3′ overhang renders the DNA ends refractory to the exonuclease activity of MRX (Fig. 1C; Paull and Gellert 1998), but we consistently observed endonucleolytic cleavage by MRX–Sae2 in the absence of Ku (Fig. 1C). Stimulation of nucleolytic scission still occurred upon the inclusion of Ku (Fig. 1C; Supplemental Fig. S2A). Altogether, these results indicate that while a protein-free DNA end is subject to 5′ strand cleavage by MRX–Sae2, engagement by Ku enhances its susceptibility to endonucleolytic action.
Endonucleolytic scission activated by RPA association with ssDNA
The 3′ ssDNA tail stemming from end resection is expected to be bound by the abundant ssDNA-binding protein RPA (Lisby et al. 2004; Wang and Haber 2004). We asked whether such an RPA-bound structure is subject to endonucleolytic processing by MRX–Sae2. For this, we verified that whereas RPA does not bind the 70-bp DNA substrate with 5-nt 3′ overhangs, it stably engages a related substrate with 3′ overhangs of 10 nt (Supplemental Fig. S2B). Importantly, while preincubation of the former substrate with RPA had no stimulatory effect on 5′ strand scission (data not shown), such a treatment led to a strong enhancement of MRX–Sae2 activity on the latter substrate (Fig. 1D). Similar observations were made with a substrate harboring 3′ overhangs of 30 nt (Supplemental Fig. S2C,D).
Results from genetic studies suggest that MRX–Sae2 functions with RPA to eliminate hairpin structures that arise from fold-back hybridization of repeated sequences in ssDNA, an activity that prevents the formation of palindromic duplications and dicentric chromosomes (Deng et al. 2015). To determine whether MRX can resolve these potentially pathological structures, we constructed a hairpin substrate that harbors a 20-nt loop region capable of binding RPA (Supplemental Fig. S2B) and asked whether MRX–Sae2 is able to incise the DNA with and without RPA being present. The results revealed that while MRX–Sae2 has little reactivity toward the substrate, the addition of RPA leads to endonucleolytic scissions on the duplex DNA region that abuts the RPA-bound DNA loop (Fig. 1E; Supplemental Fig. S2E). We note that MRX–Sae2 also incises a hairpin harboring a 10-nt ssDNA loop when RPA is present (data not shown). Thus, RPA binding to a partially resected end or a DNA loop marks the neighboring duplex DNA region for endonucleolytic scission by MRX–Sae2.
Licensing of strand scission at a nucleosome and other internal obstacles by Sae2
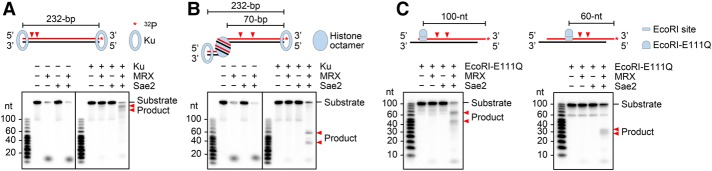
In sgs1Δ exo1Δ cells that are impaired for long-range resection, MRX–Sae2 incises the 5′ DNA strand in ∼100-nt increments from the DSB end (Mimitou and Symington 2008; Zhu et al. 2008). Moreover, studies in meiotic cells have suggested that MRX–Sae2 acts on an internucleosomal DNA region at +1 and +2 nucleosomes proximal to the DSB end (Mimitou et al. 2017). In view of these findings, it was of considerable interest to determine whether MRX–Sae2 acts on the 5′ DNA strand next to a nucleosome. For this purpose, we used purified histone octamer (Supplemental Fig. S3A) and step salt dialysis to assemble a nucleosomal template using 3′ end-labeled 232-bp dsDNA that harbors the 601-nucleosome positioning sequence (Supplemental Fig. S3B). Next, the nucleosomal substrate was incubated with Ku to prevent the release of the 3′ end label by MRX. As expected, in the absence of nucleosome, MRX–Sae2 endonucleolytically cleaved the 5′ strand within the proximity of the Ku-occluded end (Fig. 2A). Importantly, the nucleosome induced novel scission sites with a strict dependence on Sae2 (Fig. 2B). Two clusters of endonuclease products (∼55–60 and ∼35–40 nt) were generated, corresponding to cleavage sites ∼10–15 and ∼30–35 nt away from the nucleosome boundary (Fig. 2B). These results indicate that the placement of a nucleosome activates 5′ strand cleavage by MRX in a Sae2-dependent fashion. The distance between the two scission clusters was ∼20 nt, which is in congruence with results obtained using the 100-bp dsDNA substrate with Ku-occluded ends (Fig. 1A). As expected, MRX variants that harbor either the Mre11 (H125L/D126V) nuclease-dead mutant or a mutation (K40A, K40E, or K40R) in the Rad50 Walker A motif lack the ability to endonucleolytically cleave the nucleosomal substrate (Supplemental Fig. S3C).
Figure 2.
Licensing of 5′ strand cleavage at a nucleosome and other internally bound obstacles by Sae2. (A) The 232-bp dsDNA (1 nM) was incubated with 40 nM MRX, 480 nM Sae2, and 64 nM Ku. (B) Reactions were carried out as in A except with the 232-bp dsDNA containing a site-specific nucleosome. The nucleosomal substrate was generated using a 2:1 molar ratio of histone octamer to DNA. (C) The two 95-bp substrates with 5-nt 3′ overhangs (2 nM each) containing an EcoRI site at either the DNA end or an internal site were tested with 24 nM EcoRI-E111Q, 40 nM MRX, and 480 nM Sae2. The asterisk denotes the 32P label in the substrate in all of the figure parts. See also Supplemental Figure S3.
Importantly, a streptavidin–biotin complex located at an internal site also triggers endonucleolytic scission of the neighboring DNA strand by MRX–Sae2 (Supplemental Fig. S3D). Similarly, the placement of EcoRI-E111Q (King et al. 1989; Wright et al. 1989), a catalytically inactive EcoRI variant capable of target site recognition (Supplemental Fig. S3E), also engendered sensitivity of the 5′ DNA strand to endonucleolytic cleavage by MRX–Sae2 (Fig. 2C). Collectively, the results above suggest that any stably bound protein obstacle can activate 5′ DNA strand cleavage by MRX–Sae2. We envision that DNA damage signaling would help recruit the MRX–Sae2 ensemble to DNA lesions within the cellular setting, where it would incise the 5′ strand adjoining a stably bound protein molecule, such as Ku at a DNA end.
DNA nick processing activity of Mre11 and MRX complexes
The current model posits that the DNA nick introduced by MRX–Sae2 is enlarged into a gap via the 3′–5′ exonuclease activity of Mre11 (Garcia et al. 2011; Daley and Sung 2014). That 3′–5′ exonucleolytic chew-back occurs in our reconstituted system was revealed using a Ku-bound 100-bp dsDNA that was 32P-labeled on the 5′ terminus of one of the strands. As shown in Supplemental Figure S4A, incubation of the substrate with MRX–Sae2 generated two clusters of nuclease products ∼10–15 and ∼20–25 nt in length. Since the initial endonucleolytic scission on the same substrate occurs ∼35–45 and ∼55–65 nt away from the 5′ terminus, as determined using 3′ end-labeled DNA (Fig. 1A), the result (Supplemental Fig. S4A) provides evidence for an ability of MRX–Sae2 to mediate significant 3′–5′ chew-back from the DNA nick that it generates.
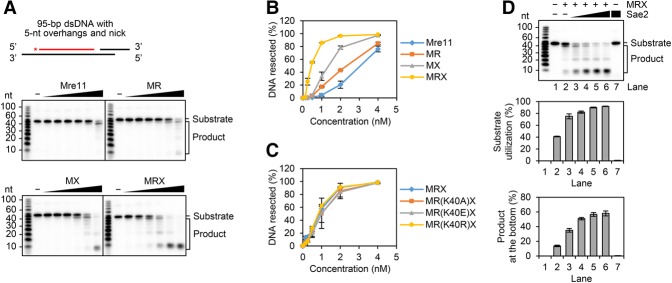
To gain mechanistic insights into the exonucleolytic chew-back reaction, we employed a 95-bp dsDNA substrate that harbors a 5′ 32P label in one of the strands, a defined nick 55 nt away from the label, and also 5-nt overhangs at both 3′ termini to render the DNA ends refractory to exonuclease attack. We tested Mre11 alone or in combination with Rad50, Xrs2, and/or Sae2. The results revealed that while Mre11 alone is able to digest the DNA nick exonucleolytically, its chew-back activity is enhanced by Rad50 or Xrs2, with the fully assembled MRX complex being significantly more adept than either of the MR and Mre11–Xrs2 (MX) subcomplexes in this regard (Fig. 3A,B). Interestingly, the exonucleolytic chew-back reaction is not affected by mutations within the Walker A motif of the Rad50 subunit (Fig. 3C). Taken together, these results provide evidence that Mre11 in complex with Rad50 and Xrs2 can efficiently process a DNA nick in the 3′-to-5′ direction and that, unlike the endonucleolytic scission of DNA (Supplemental Fig. S1H), this process does not require the ATP binding or hydrolytic function of Rad50 (Fig. 3C).
Figure 3.
Effect of Rad50, Xrs2, and Sae2 on the nick chew-back activity of Mre11. (A) The 5′-labeled 95-bp dsDNA (4 nM) with 5-nt 3′ overhangs and a nick 55 nt away from the labeled end was tested with Mre11, MR, MX, and MRX (0.125, 0.25, 0.5, 1, 2, and 4 nM). The asterisk denotes the 32P label in the substrate. (B) The results from three independent experiments shown in A were quantified. The error bars represent SD. (C) The substrate used in A was tested with MRX and MRX variants (0.125, 0.25, 0.5, 1, 2, and 4 nM) that harbor the Rad50 K40A, K40E, or K40R mutant protein. The results were quantified and graphed as in B. (D) The substrate in A was tested with 0.8 nM MRX and 30, 60, 120, and 240 nM Sae2. The results from three independent experiments were quantified in two different ways (either as percentage of substrate that had been used or focusing on the product at the gel bottom). The error bars indicate SD. See also Supplemental Figure S4.
Enhancement of DNA nick processing activity of Mre11 and MRX by Sae2
Whether Sae2 affects the exonucleolytic action of MRX at a DNA nick is not known. We therefore investigated the effect of Sae2 on the 3′–5′ nick chew-back activity of MRX. As shown in Figure 3D, a robust stimulatory effect of the chew-back reaction was seen upon the addition of Sae2. Moreover, we verified that human CtIP does not affect MRX chew-back activity, indicating that Sae2 functions with MRX in a species-specific manner (Supplemental Fig. S4B). Interestingly, we found that Sae2 also enhances chew-back by Mre11 without Rad50 and Xrs2 being present (Supplemental Fig. S4C). Collectively, the results provide evidence for an important role of Sae2 in DNA gap creation from the DNA nick generated via the endonucleolytic action of MRX.
DSB end resection serves several distinct purposes; namely, to activate the Mec1/ATR-dependent checkpoint via RPA recruitment, prevent repair by NHEJ, and generate a ssDNA template for assembling the HR repair machinery consisting of the recombinase Rad51 (Zou and Elledge 2003; Sung and Klein 2006; Symington and Gautier 2011). Here, we show that Ku shields DSB ends from exonucleolytic attack but promotes the endonucleolytic scission of the 5′ DNA strand (Fig. 1A; Supplemental Fig. S1C). Consistent with published work that employed DNA substrates with their ends blocked by streptavidin (Cannavo and Cejka 2014), the 5′ strand endonucleolytic scission of DNA with Ku-occluded ends is strongly enhanced by Sae2 and requires ATP binding and hydrolysis by Rad50 (Supplemental Fig. S1H). It remains to be determined how Rad50-mediated ATP hydrolysis and Sae2 together induce the requisite changes in DNA and MRX conformation conducive for 5′ strand scission. In addition, we presented evidence that, with the inclusion of 100 mM KCl in the reaction, optimal strand scission activity requires Xrs2 (Fig. 1B).
It has been suggested that MRX–Sae2 incises DNA next to the first or second nucleosome away from meiotic DSBs introduced by the Spo11 protein complex (Mimitou et al. 2017). We now show that MRX–Sae2 endonucleolytically cleaves the 5′ DNA strand bordering a nucleosome (Fig. 4B), thus providing direct biochemical results to support the above model. Importantly, we found that endonucleolytic DNA scission by MRX–Sae2 can also be triggered by a streptavidin–biotin linkage (Supplemental Fig. S3D) or an enzymatically inactive form of the EcoRI restriction enzyme (Fig. 2C) placed internally. Overall, our study highlights Sae2 as a general licensing factor for 5′ DNA strand cleavage by MRX at either terminal or internal protein obstacles. We note that these results have implications as to how the MRX–Sae2 ensemble and its equivalent in other eukaryotes process covalent DNA adducts of topoisomerases (Hartsuiker et al. 2009; Pommier et al. 2010; Aparicio et al. 2016; Hoa et al. 2016). Using its endonuclease activity, MRX–Sae2 could remove trapped topoisomerase–DNA adducts at either DSBs or single-strand breaks, which represent terminal and internal protein obstacles, respectively.
The current model for the transition between short-range and long-range resection posits that MRX–Sae2 acts exonucleolytically at the DNA scission site to generate a gap for the loading of either Exo1 or the STR–Dna2 ensemble (Garcia et al. 2011). In support of this model, we found that MRX–Sae2 can efficiently convert a DNA nick into a gap via its 3′–5′ exonuclease activity. In this regard, our work furnishes valuable mechanistic details regarding this 3′–5′ chew-back reaction. Specifically, we show that while Mre11 alone is able to process the DNA nick, the efficiency of the reaction is enhanced by Rad50, Xrs2, or Sae2, and optimal activity is realized only when all of the latter components are present (Fig. 3A,D). Even though we reproducibly observed that Sae2 enhances the exonucleolytic activity of Mre11 and MX in the presence of 100 mM KCl, it remains possible that under other reaction conditions, Sae2 would synergize only with the full MRX complex. Interestingly, even though the endonucleolytic scission of 5′ DNA strand is strictly coupled to ATP hydrolysis by Rad50, the chew-back reaction has no such requirement. Moreover, unlike the endonucleolytic action, the exonucleolytic chew-back reaction occurs normally with the dephosphorylated form of Sae2 (data not shown). We note that the combined endonucleolytic and exonucleolytic action of MRX–Sae2 may contribute to the removal of Ku from DNA ends.
Unexpectedly, we found that the engagement of a partially resected structure by the ubiquitous ssDNA-binding protein RPA renders the neighboring 5′ DNA strand susceptible to endonucleolytic scission by MRX–Sae2 (Fig. 1D). Based on this finding, we propose that in cases where the long-range resection machinery has become disengaged from a partially resected DNA end or DNA gap and renewed access may be restricted by RPA or another DNA-binding protein, MRX–Sae2 would readily create a new entry point for the long-range resection machinery through its combined DNA-nicking and chew-back action (Fig. 4C). Also important is our finding that the engagement of a ssDNA loop by RPA renders the neighboring duplex DNA region susceptible to the endonucleolytic scission by MRX–Sae2 (Fig. 1E). This attribute of MRX–Sae2 is likely germane for preventing the formation of palindromic duplications and dicentric chromosomes stemming from intramolecular hybridization of short inverted repeats present in ssDNA (Fig. 4D; Deng et al. 2015).
Collectively, the specific roles of the MRX–Sae2 nuclease ensemble in processing DSB ends engaged by physiological protein obstacles, such as Ku, RPA, or a nucleosome, are recapitulated in our reconstituted systems. Our findings provide mechanistic details for understanding how MRX–Sae2 orchestrates its DNA-nicking and chew-back action in the initiation of DNA end processing to create entry sites for the long-range resection machinery at DNA-bound protein obstacles.
Materials and methods
DNA substrates
Oligonucleotide-based DNA substrates, plasmid-length DNA substrates, and the nucleosomal substrate were prepared as described in the Supplemental Material.
Protein expression and purification
The procedures devised for the expression and purification of MRX, Sae2, and EcoRI-E111Q are detailed in the Supplemental Material. Ku was expressed and purified as reported before (Krasner et al. 2015). RPA was expressed and purified as described previously (Van Komen et al. 2006). The histone octamer was purified to near homogeneity according to our published procedure (Kwon et al. 2008). Streptavidin was purchased from Invitrogen.
Nuclease reactions
The standard nuclease reaction was performed at 30°C in 12 µL of reaction buffer (25 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 100 µg/mL BSA, 2 mM ATP, 20 mM creatine phosphate, 30 µg/mL creatine kinase) containing 100 mM KCl (final concentration). The DNA substrate was preincubated with the indicated concentrations of Ku, RPA, streptavidin, or EcoRI-E111Q for 15 min followed by the addition of MRX and Sae2. After a 30-min incubation, the reaction was terminated by treatment with 0.5% SDS and 1 mg/mL proteinase K for 10 min at 37°C. After adding an equal volume of 2× loading buffer (95% formamide, 20 mM EDTA, 0.01% bromophenol blue), the reaction mixtures were held for 7 min at 95°C before being analyzed in a denaturing polyacrylamide gel containing 7 M urea in TAE buffer. Gels were fixed in 50% methanol and 20% PEG 400 for 20 min, dried onto positively charged nylon membrane (GE Healthcare), and subjected to phosphorimaging analysis.
To test for 3′–5′ exonucleolytic chew-back activity, the indicated concentration of Mre11, MR, MX, or MRX was incubated with the DNA substrate with an internal nick in 12 µL of reaction buffer (25 mM Tris-HCl at pH 7.5, 1 mM MnCl2, 1 mM DTT, 100 µg/mL BSA) containing 100 mM KCl (final concentration). After a 30-min incubation at 30°C, the reaction was terminated, and the digested DNA species were analyzed by denaturing polyacrylamide gel electrophoresis as described above.
Electrophoretic mobility shift assays
DNA substrates were incubated with the indicated concentration of Ku, RPA, or EcoRI-E111Q for 15 min at 30°C in 12 µL of the buffer used for the nuclease reactions (see above). The reactions were mixed with 4 µL of loading buffer (20 mM Tris-HCl at pH 7.5, 40% glycerol, 2 mM EDTA, 0.2% orange G) before electrophoresis in a 4% native polyacrylamide gel in TAE buffer on ice. Gels were dried onto filter paper and subjected to phosphorimaging analysis.
Supplementary Material
Acknowledgments
We are grateful to members of the Sung laboratory for helpful discussions, and Petr Cejka for the Sae2 expression vector. This work was supported by National Institutes of Health grants ES007061 and ES015632.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.307900.117.
References
- Aparicio T, Baer R, Gottesman M, Gautier J. 2016. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II–DNA adducts. J Cell Biol 212: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannan WJ, Pederson DS. 2016. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol 231: 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514: 122–125. [DOI] [PubMed] [Google Scholar]
- Chang HHY, Pannunzio NR, Adachi N, Lieber MR. 2017. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 18: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo KM, Van Komen S, Roh DH, Krejci L, Lewis LK, Resnick MA, Sung P, Tomkinson AE. 2005. Effect of amino acid substitutions in the rad50 ATP binding domain on DNA double strand break repair in yeast. J Biol Chem 280: 2620–2627. [DOI] [PubMed] [Google Scholar]
- Daley JM, Sung P. 2014. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol 34: 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng SK, Yin Y, Petes TD, Symington LS. 2015. Mre11–Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol Cell 60: 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Zhu Q, Taylor ER, Tsay AJ, Shi LZ, Berns MW, McGowan CH. 2012. CtIP is required to initiate replication-dependent interstrand crosslink repair. PLoS Genet 8: e1003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. 2011. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM. 2009. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell 33: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Shimizu T, Zhou ZW, Wang ZQ, Deshpande RA, Paull TT, Akter S, Tsuda M, Furuta R, Tsusui K, et al. 2016. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol Cell 64: 580–592. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- King K, Benkovic SJ, Modrich P. 1989. Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease. J Biol Chem 264: 11807–11815. [PubMed] [Google Scholar]
- Krasner DS, Daley JM, Sung P, Niu H. 2015. Interplay between Ku and replication protein A in the restriction of Exo1-mediated DNA break end resection. J Biol Chem 290: 18806–18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y, Seong C, Chi P, Greene EC, Klein H, Sung P. 2008. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J Biol Chem 283: 10445–10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Marechal A, Zou L. 2013. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5: a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Yamada S, Keeney S. 2017. A global view of meiotic double-strand break end resection. Science 355: 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE. 2008. Recruitment of Saccharomyces cerevisiae Dnl4–Lif1 complex to a double-strand break requires interactions with Yku80 and the Xrs2 FHA domain. Genetics 180: 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M. 2001. Double-strand breaks and tumorigenesis. Trends Cell Biol 11: S52–S59. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Leo E, Zhang H, Marchand C. 2010. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 17: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev 7: 2520–2532. [DOI] [PubMed] [Google Scholar]
- Sordet O, Nakamura AJ, Redon CE, Pommier Y. 2010. DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle 9: 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Klein H. 2006. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol 7: 739–750. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Macris M, Sehorn MG, Sung P. 2006. Purification and assays of Saccharomyces cerevisiae homologous recombination proteins. Methods Enzymol 408: 445–463. [DOI] [PubMed] [Google Scholar]
- Wang X, Haber JE. 2004. Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol 2: E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. 2008. The endless tale of non-homologous end-joining. Cell Res 18: 114–124. [DOI] [PubMed] [Google Scholar]
- Wright DJ, King K, Modrich P. 1989. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J Biol Chem 264: 11816–11821. [PubMed] [Google Scholar]
- Wu D, Topper LM, Wilson TE. 2008. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.