Abstract
Understanding the genetic basis of the switch from asexual to sexual lifestyles in response to sometimes rapid environmental changes is one of the major challenges in fungal ecology. Light appears to play a critical role in the switch—but fungal genomes harbor diverse light sensors. Fungal opsins are homologous to bacterial green-light sensory rhodopsins, and their organismal functions in fungi have not been well understood. Three of these opsin-like proteins were widely distributed across fungal genomes, but homologs of the Fusarium opsin-like protein CarO were present only in plant-associated fungi. Key amino acids, including potential retinal binding sites, functionally diverged on the phylogeny of opsins. This diversification of opsin-like proteins could be correlated with life-history associated differences among fungi in their expression and function during morphological development. In N. crassa and related species, knockout of the opsin NOP-1 led to a phenotype in the regulation of the asexual-sexual switch, modulating response to both light and oxygen conditions. Sexual development commenced early in Δnop-1 strains cultured in unsealed plates under constant blue and white light. Furthermore, comparative transcriptomics showed that expression of nop-1 is light-dependent and that the Δnop-1 strain abundantly expresses genes involved in oxidative stress response, genes enriched in NAD/NADP binding sites, genes with functions in proton trans-membrane movement and catalase activity, and genes involved in the homeostasis of protons. Based on these observations, we contend that light and oxidative stress regulate the asexual-sexual switch via light-responsive and ROS pathways in model fungus N. crassa and other fungi.
Keywords: fungi, asxual-sexual switch, light response, oxidative stress, ecology
Introduction
Like plants, most terrestrial fungi cannot move freely to choose their surroundings. Instead, they rapidly modify their growth and development to respond to environmental changes. The genetics underlying fungal responses to environmental factors, including fungal-plant associations, cellular oscillations, and the global carbon cycle, have become classic cases in molecular ecology (Kendrick 2011). One long lasting challenge in molecular ecology, however, has been to understand the mechanisms behind the fungal asexual-sexual switch, which apparently regulates two morphologically distinct phases in fungal life history (Stajich et al. 2009). Typically, the asexual phase is a fast-growing stage that produces a large number of mitotic spores composed from relatively fragile structures. The asexual-sexual switch is a one-way developmental shift to sexual reproduction from asexual growth—a developmental process which otherwise would lead to production of asexual spores called conidia. Reproduction of ascomycetes in the sexual phase usually produces smaller numbers of meiotic spores via a longer process within well-developed and protective fruiting bodies, often in response to relatively harsh conditions, associated with low levels of light, temperature, humidity, and other stressors. Accordingly, the one-way asexual-sexual switch may be quantitatively and qualitatively induced by diverse interacting environmental stimuli (Rodriguez-Romero et al. 2010).
Among all physical elements in a natural environment, ambient light can serve as a direct indicator and predictor for daily and seasonal changes of temperature, humidity, and voltage, and other environmental fluctuations that affect the growth, development, and status of the asexual-sexual switch in many fungi (Rodriguez-Romero et al. 2010; Schumacher 2017). Three molecules that putatively bind chromophores that absorb blue, red, and green light, respectively, have been identified in fungal genomes, and the blue light sensors have been well studied in different fungal models (Ahrendt et al. 2017; Avalos & Estrada 2010; Chen & Loros 2009; Rodriguez-Romero et al. 2010). Blue light sensory genes wc-1 (white collar-1) and wc-2 (not a chromophore binding protein)—in addition to playing key roles in the regulation of the internal circadian oscillation (Dunlap & Loros 2004)—have been demonstrated to play a central role in light response and asexual reproduction in N. crassa (Chen et al. 2009). Nitrogen starvation and blue light irradiation inhibit conidiation and significantly increase the numbers of sexual reproductive structures (protoperithecia before crossing and perithecia after crossing) produced in N. crassa (Innocenti et al. 1983; Kritsky et al. 2002). Furthermore, protoperithecium production is increased under a very low light intensity, compared to exposure to constant light or to culture in complete darkness (Tan et al. 2004). This increase is consistent with the observation that perithecia (the sexual structures that develop from protoperithecia after crossing) of Neurospora species form in locations that are not completely blocked from light (Pandit & Maheshwari 1996). Indeed, blue light has also been identified as important in late sexual development in N. crassa (Harding & Melles 1983). Moreover, photoreceptors (including WCC, CRY-1, NOP-1, and PHY-2) were suggested to play a complex role in the regulation of conidiation genes in Neurospora crassa (Olmedo et al. 2010a; Olmedo et al. 2010b). Research on several other models has contributed to our understanding of asexual and sexual development in fungi—especially research on the long-standing genetic model species Aspergillus nidulans. Light responses mediated by phytochromes were first characterized using A. nidulans (Bayram et al. 2010; Blumenstein et al. 2005; Brandt et al. 2008; Corrochano 2007; Purschwitz et al. 2009; Purschwitz et al. 2008; Rohrig et al. 2013). All these findings support the contention that light regulates fungal ecology via the balance between asexual and sexual components of fungal life history. Potential sensors for all three primary colors have been reported in major ascomyceteous lineages, including many plant-associated fungi (Wang et al. 2016). Plant-associated fungi have evolved developmental programs and life cycles that are adapted to host growth and development, and host plants influence the light environment for these fungi (Wang et al. 2009). The intensity and spectrum of sunlight shifts as it passes through barriers, providing a means to use light spectra to sense the biotic and abiotic environment. Plants differentially filter light conditions in their environment: overhanging foliage strips incident sunlight of red and blue wavelengths, yet permits far-red, and to a lesser extent green light, to pass through to the area beneath, providing a cue as to the nearby natural environment to organisms able to discern relative light intensity across the spectrum (Schmitt & Wulff 1993; Wang & Folta 2013).
Distinct from the blue- and red-light sensing proteins (the phytochromes; Rockwell & Lagarias 2010), rhodopsins constitute a third kind of putative light-sensory molecule in fungi (Fischer et al. 2016). In prokaryotes, these seven-transmembrane-helix photoreceptors are green-light absorbing pigments composed of retinal chromophore and associated integral membrane opsin protein (Brown & Jung 2006). One type of rhodopsin that was discovered first in halophile archaea is often called “archeal opsins” (Ruiz-Gonzalez & Marin 2004). There are two classes of archeal opsins: one class is a component of sensory rhodopsin (sR), and functions as sensor for phototaxis. The other class is a component of transport rhodopsin (tR) or bacteriorhodopsin (bR), functioning as a proton-extruding pump and a chloride uptake pump, respectively (Ihara et al. 1999). Molecular functions of rhodopsins are strikingly well maintained when microbial opsins are expressed in eukaryotic cells (Avelar et al. 2014). Indeed, their ability to convert light into ion flux has made them promising tools for visualizing neural activity (Looger 2012).
The gene nop-1 (new eukaryotic opsin-1) was first identified as a putative green-light photoreceptor in the eukaryote N. crassa (Bieszke et al. 1999a; Bieszke et al. 1999b). Actually, nop-1 is so similar to sR and bR archeal opsins that its predicted function as a sensory or as a bacteriorhodopsin is ambiguous (Bieszke et al. 1999a; Chen et al. 2010). However, no clear knockout phenotype for nop-1 has been observed in N. crassa, despite numerous efforts (Bluhm et al. 2008; Rodriguez-Romero et al. 2010). Accumulating evidence suggests that light sensing by nop-1 in N. crassa is likely associated with responses to oxidative stress, which affects conidiation in many fungal species (Cabrera et al. 2015). A N. crassa nop-1 mutant has exhibited elevated resistance to oxidative reagents peroxide and menadione and slightly altered colony morphology, and knockout of the nop-1 gene has affected several conidiation genes (Aguirre et al. 2005; Bieszke et al. 2007; Bieszke et al. 1999a; Bieszke et al. 1999b). Expression of Sop1, a Nop-1 homolog in Sclerotinia sclerotiorum, can be induced with H2O2 (Heller et al. 2012; Temme et al. 2012; Temme & Tudzynski 2009), and recently it was found to be involved in oxidative stress responses and to be essential for growth and for sclerotial development (Lyu et al. 2015; Schumacher 2017).
A second opsin gene was identified in the ascomycete Leptosphaeria maculans (Idnurm & Howlett 2001) as a proton pump (Waschuk et al. 2005). In plants and fungi, proton pumps in the plasma membrane are involved in creating proton gradients that drive secondary transport processes and initiate cellular responses to the environment (Palmgren 2001; Pedersen et al. 2007; Serrano et al. 1986). Recently opsins in Fusarium fujikuroi and Phaeosphaeria nodorum were also reported to function as a green light-driven proton pump. In F. fujikuroi, the proton pump CarO is highly expressed in conidia, and the introduction of a highly active expression construct for CarO represses germination of conidia (Fan et al. 2011; Garcia-Martinez et al. 2015).
To provide insights into the genes and gene regulatory reactions to environmental signals that regulate the key biological and developmental processes operating in the ecology of fungal light reception and life cycle regulation, we performed analyses of genome content and transcriptomic expression. We examined the expression of genes for opsin-like proteins within saprobic Neurospora species and pathogenic Fusarium species during their asexual and sexual development, and profiled genome-wide expression patterns for wild type and nop-1 knockout strains subsequent to a range of light exposures, specifically comparing influential metabolic and signaling pathways. Lastly, we cultured Δnop-1 under diverse conditions guided by our findings to identify a phenotype and a putative organismal function of nop-1 in N. crassa.
Materials and Methods
Strains and growth condition
N. crassa strains FGSC4200 (mat a) and FGSC2489 (mat A) were purchased the Fungal Genetic Stock Center (FGSC) for genome-wide transcriptome profiling during asexual and sexual development (McCluskey et al. 2010). Knockout strains FGSC15897 (mat A) for nop-1, and FGSC11552 (mat a) and FGSC11553 (mat A) for orp-1 were acquired from FGSC as part of the N. crassa knockout collection. For assays of asexual development, strains were grown on Bird Medium (Metzenberg 2004) covered by a cellophane membrane (Fisher Scientific) at 26°C under constant light. For assays of sexual development, the strains were grown on Carrot Agar (CA) (Klittich & Leslie 1988) at 26 C under a constant light condition. Crosses were performed by applying 2 ml mat a conidia suspension in 1.5% Tween 60 to the surface of the mat A protoperithecia plates. Additional details of the conditions for growth and sampling for asexual and perithecial development experiments were as previously described (Wang et al. 2012a; Wang et al. 2012b).
Phylogenetic analyses
Multiple BLAST searches were conducted in the NCBI database for homologs of N. crassa nop-1 (NCU10055) and orp-1 (NCU01735) in other fungal genomes, with particular focus on representative genomes in major ascomycetous groups. Two opsins were preliminarily identified in the lichenized fungi Cladonia grayi by Drs. Muller and Armaleo at Duke University, and based on conservation of sequence similarity, opsin-like genes were predicted in GenBank accessions for two powdery mildew genomes (Spanu et al. 2010). We also included additional rhodopsin sequences from bacteria, green algae, and plants. Sequence of a predicted fungal opsin-like protein in a rice species was acquired from NCBI; this sequence is distantly related to ascomyceteous NOP-1 homologs, and its presence or absence had no impact on the phylogeny of the rest of the fungal opsin-like proteins. Amino acid sequences were aligned using SATé-II (Liu et al. 2012) specifying MAFFT as the aligner, MUSCLE as the merger, and RaxML as the tree estimator under WAG model. Alignment scores were estimated using 10 iterations of SATé-II-ML for three independent runs, and the alignment with the best score was retained for further analysis. The robustness of branching topologies was estimated with 1000 bootstrap maximum likelihood searches using PhyML (Guindon et al. 2010) under a WAG model. The phylogenetic informativeness of the opsin genes among fungal lineages was quantified using PhyDesign (Lopez-Giraldez & Townsend 2011). Rates for each site of the best nucleotide alignment resulting from SATé-II were estimated by Hyphy (Pond et al. 2005) with the general time-reversible model, using the maximum likelihood phylogeny scaled in molecular evolutionary units.
Analyses of molecular evolutionary conservation
Evolutionary conservation of the sequence and structure of three fungal opsins (homologs of NOP-1 and ORP-1 in N. crassa, and of CarO in F. graminearum) was calculated for each subfamily individually with ConSurf (Ashkenazy et al. 2010) using the best SATé alignment and associated RaxML tree. Estimation of the evolutionary conservation was performed without use of a reference PDB model using a Bayesian calculation specifying the WAG substitution model provided by ConSeq in the ConSurf online server (Ashkenazy et al. 2010).
Functional divergence analysis
To assess changes in the selection pressure occurring during the divergence of these genes, we specified the random site models (M0, M1a, M2a, M3, M7, M8a and M8), and clade model C in the PAML4 software package (Yang 2007; Yang & Nielsen 2000) to estimate the pairwise ratio of nonsynonymous and synonymous divergence (ω = dN/dS). Clade model C was used to test for differences in selection among clades, and to determine what division of clades best fit the data, applying the new M2a_rel model (Weadick & Chang 2011). Varied starting values of κ and υ were tested to ensure robust convergence of the maximum likelihood estimator. To identify the best fitting model of selection, a Likelihood Ratio Test (LRT) was applied to all pair-wise nested model comparisons. Additionally, the Akaike Information Criterion (AIC) was applied to compare all selective models (Schott et al. 2014).
To predict amino acid residues important to functional differences that have arisen, we evaluated site-specific rate changes between two clades on our inferred phylogenetic tree (e.g. Gu 1999). This analysis of functional divergence the proteins between paralog clades was performed using DIVERGE v2.0 (Gu & Vander Velden 2002; Gu et al. 2013).
Characterizing expression and regulation network of opsins during asexual and sexual development
Genome-wide expression of Neurospora crassa and related species has been profiled recently (Lehr et al. 2014; Sikhakolli et al. 2012; Trail et al. 2017; Wang et al. 2012a; Wang et al. 2012b; Wang et al. 2014). For microarray gene expression profiling, RNA preparation, microarray preparation, hybridization, and scanning were performed as previously described (Clark et al. 2008). Normalized data were analyzed using Bayesian Analysis of Gene Expression Levels (BAGEL, Townsend 2004; Townsend & Hartl 2002) and reported as supplemental data. For transcriptomic sequencing of sexual development, the tally of cDNA reads that aligned with each gene for each sample was processed with LOX v1.6 (Zhang et al. 2010). Genome-wide gene expression data deposited in Gene Expression Omnibus (GEO accession GSE101412) was analyzed to assess associations among nop-1, oxidative stress pathways, conidiation, and sexual reproduction. The RNAseq data were sampled at four conidia germination and hyphal growth stages during N. crassa growth on Bird medium (permissive for asexual development only) and the same four developmental stages for maple sap medium (supporting both asexual and sexual development). The maple sap medium was prepared using only maple sap water (VerticalTM, Feronia Forests) and 2% agar. From this GEO data, expression of nop-1, selected genes involved in oxidative pathways, two conidiation genes, as well as three genes involved in early perithecial development was used to assess their interactions using Bayesian network inference (Wang et al. 2014; Ziebarth et al. 2013).
Phenotyping opsins in N. crassa
Opposite mating types of knockout strains for each opsin were crossed, as were opposite mating types of knockout and wild type strains (FGSC4200, FGSC2489) on synthetic complete medium (SCM; Clark et al. 2008). All crosses were performed as three replicates in each growth condition. All strains were consistently cultured at room temperature (26 C), but were exposed to six light conditions including complete dark, constant white light, constant red light, far-red light, and blue light, each colored light provided as the sole light source. Light conditions were provided by an E-30LED incubator (Percival Scientific), except the green light condition, which was assayed by filtering light from a green-LED light bulb using a plastic green light filter (Edmund Optics). Two airflow conditions were tested, using unsealed or tightly double-layered parafilm-sealed plates (90 mm in diameter). Abundance of protoperithecia was assayed using a 100 × 100 × 15 mm gridded petri dish (Fisher Scientific). Plates with 40 ml SCM medium seeded with 3000 conidia (validated by Hemacytometer counts) were incubated for five days. Numbers of protoperithecia were manually counted for four 13 mm × 13 mm blocks in the center of each plate, with two biological replicates. Sensitivity of strains to the H+-ATPase inhibitor during sexual development was tested with SCM plates containing 100 ng/ml oligomycin under constant light condition. Phenotypes for both wild type and Δnop-1 knockout strains during sexual development were also investigated under light/dark and sealed/unsealed conditions with treatment of menadione (100μM), NAC (10mM) as well as H2O2 (10mM). These chemicals were added to SCM medium before the inoculation and to the crossing-cultures at 48, 72, and 96 hours after inoculation.
Data from 10 microarrays—initially performed to reveal temporal patterns in light response (Chen et al. 2009)—including measurements for wild type and Δnop-1 knockouts at five sample points, were acquired from the Gene Expression Omnibus database (GEO accession GSE8932), restructured, and reanalyzed to focus on the impact of removing nop-1 on genome-wide expression. Signal intensity of MRAT (the median of the set of background corrected single pixel ratios for all pixels with the spot) was compared between reference and samples, and these ratios were supplied as input for BAGEL. For the purpose of analysis, data from proximate light conditions were conservatively treated as replicates, including constant dark (DD) on its own, short light exposure for 15 and 30 minutes (LL15, 30) as replicates, and long light exposure for 60 and 120 minutes (LL60, 120) as replicates. Similar analyses were conducted on a different subset of the Chen et al. 2009 data (on knockouts of phytochromes, WC-1, and WC-2) (Wang et al. 2016).
Complementation of the NOP-1 knockout mutant
Co-segregation experiments were performed to ensure that the intended deletion is responsible for the mutant phenotype (Chinnici et al. 2014; Fu et al. 2011). Twenty-four individual ascospore progenies were isolated with hygromycin resistance from crosses between Δnop-1 and wild type stains. Fully co-segregated deletion of nop-1 and earlier protoperithecia production under light condition was observed. A plasmid pCB1532-nop-1 was constructed, including the nop-1 gene (1048 bp), and its upstream (1566 bp) and downstream (559 bp) regions. This incorporated DNA fragment including the nop-1 coding sequence and putative regulatory regions was amplified from the wild-type strain (FGSC2489, matA) using primer pair Nop-1F-BamHI: CGCGGATCCATATCGATTTAGGGTATTTAGG and Nop-1R-EcoRI: CCGGAATTCGTCCACATGCCTTTGATCGTC (BamHI and EcoRI sites are underlined). The amplified DNA was digested by BamHI and EcoRI, then ligated into the plasmid pCB1532, purchased from FGSC, which features a sulfonylurea resistant allele of the Magnaporthe grisea as a selective marker (Sweigard et al. 1997). The resulting construct, pCB1532-nop-1, was transformed into the NOP-1 knockout strain using protoplast transformation (Royer & Yamashiro 1992). Transformed protoplasts were selected for by culturing on potato dextrose agar (PDA), followed by serial transfers onto Bird-medium to induce formation of homokaryons. To select for sulfonylurea resistance 15 μg/ml Chlorimuron ethyl (Roche) was added to culture media.
Genotyping the knockout strains
All KO strains used in this study were produced and verified by Southern blotting within the NIH Neurospora Genome knock-out project (Dunlap et al. 2007). We independently verified nop-1 and orp-1 gene knockouts using a highly effective PCR genotyping protocol (Lichius et al. 2012). For genotyping, genomic DNA was extracted from mutants and wild types (as controls) and analyzed by multiplex PCR (Henegariu et al. 1997).
Results
Four clades of opsin-like proteins exist in the Ascomycota
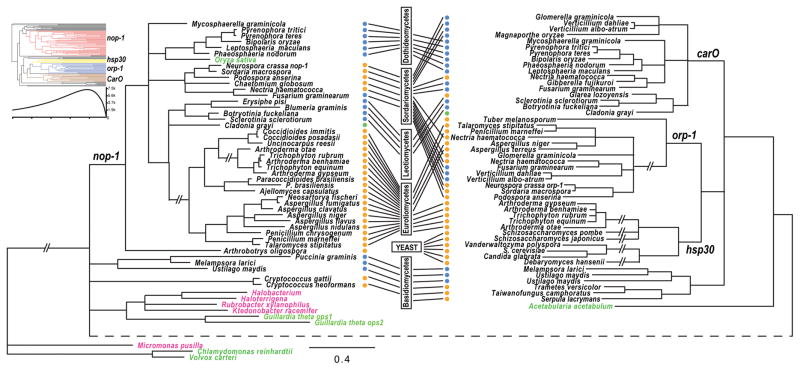
BLAST searches of sequenced fungal genomes yielded 81 opsin-like proteins (Table S1), identified from major ascomyceteous groups. Four clades of opsin-like proteins were identified in ascomycetes that presumably identify paralogous gene pairs between clades. Corresponding homologous gene sets within clades are named herein as NOP-1, HSP30, ORP-1, and CarO (Fig. 1). No NOP-1 homolog was found in unicellular fungi, including sampled yeast genomes in both Saccharomycetes and Schizosaccharomycetes, or in the underground Tuber melanosporum. The phylogeny inferred from homologs of NOP-1 was generally consistent with the systematics inferred from multilocus phylogenies for the sampled ascomycetes (Schoch et al. 2009).
Figure 1.
Phylogeny of fungal opsins and opsin-like proteins inferred from comparing amino acid sequences by maximum likelihood with PhyML under a WAG model. Branches without strong support (bootstrap ≤ 50%) were collapsed. Taxa for opsins from prokaryotic organisms (pink). Taxa for opsins from green algae and other plants (green). Taxon ecology indicated by dots: blue (plant associated), green (lichens) and orange (saprotrophic/animal pathogens). Boxes and lines indicate current fungal classification of these taxa into six major fungal groups. The double slash symbols (—//—) indicate extremely long internodes shortened to efficiently display the tree. The dashed internode across the bottom of the figure was stretched to facilitate side-by-side comparison of the nop-1 clade with the other three clades. Top-left: phyloinformativeness of fungal opsin-like protein was measured and presented with color-coded four clades, nop-1 (red), hsp30 (yellow), orp-1 (blue) and CarO (brown), in an ultrametric phylogeny. High phylogenetic informativeness during recent evolution is consistent with the poor resolution of the phylogeny among early lineages of fungal opsins.
The monophyletic group including CarO, ORP-1, and HSP30 in ascomycetes and some opsin-like proteins shared within basidiomycetes (Fig. 1) implies a single origin for these proteins. Furthermore, the homologs of CarO and ORP-1 and homologs of ORP-1 and HSP30 were found co-existing in genomes of some fungal lineages but were solitary in others. This pattern of coexistence demonstrates that the diversification of ORP-1 and HSP30 followed the emergence of CarO in ascomycetes via multiple duplications and losses (Fig. 1). Homologs of CarO were found exclusively in plant pathogens, except two species: the lichen Cladonia grayi, and a leotiomycetes Glarea lozoyensis that is likely related to plant-associated fungi or endophytic Hymenoscyphus species (Bills et al. 1999; Wang et al. 2006). Profiles of phylogenetic informativeness for these sequences demonstrated a low power to resolve phylogeny previous to the divergence of the four clades of fungal opsins and opsin-like proteins. Consistent with that predicted power, the relationships among opsins in basidiomycetes and that from prokaryotes and green algae were not well resolved (Fig. 1).
Structure and functional sites are generally conserved in opsin-like proteins
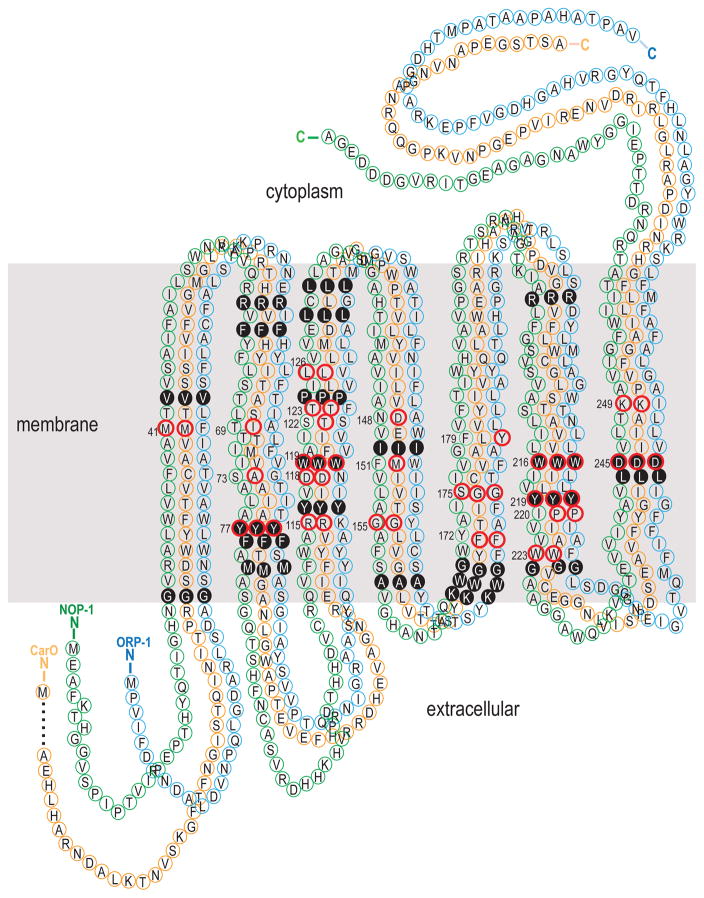
Amino acid sites buried within the membrane were generally found to be conserved for all homologs of NOP-1, ORP-1, and CarO. All twenty-two buried sites, including five putative retinal binding pocket residues, are composed of the same amino acids present in these three genes in Fusarium graminearum (Fig. 2, S1). Among the 17 retinal binding pocket sites that are conserved among the archaeal transport and sensory rhodopsins, eight were conserved between NOP-1 and CarO in both position and amino acid state, with five sites only conserved for CarO (Fig. 2). Likelihood ratio test of positive selection (sites models in PAML) suggested divergent non-synonymous/synonymous rate ratios (ω = dN/dS) in each opsin-like protein group, but statistically significant positive selection was detected only for orthologs of hsp-30 (M8 vs. M8a, P < 0.05, Table S2). The best-fitting partition was the clade-specific one (Hsp-30/CarO/Orp-1/Nop-1) suggested by the Clade model C test. Clades of hsp-30 exhibited higher non-synonymous/synonymous rate ratios, implying less functional constraint on this opsin-like protein (Table S3). The Clade model D (CmD) (Bielawski & Yang 2004; Weadick & Chang 2012a, b) assumes no constraints on the ω estimates for any site classes, and was used to clarify possible bias caused by sequence convergence No differences in results were obtained comparing CmC and CmD analyses. Analysis of functional divergence of amino acids identified critical sites that are conserved in each of the four opsin-like proteins but divergent between clades (Table S4). Two aspartic acid residues within the retinal binding pocket sites (118 and 148, Fig. 2) and other transmembrane buried sites (Fig. 2), could be responsible for divergence of function between ORP-1 and HSP-30 compared to NOP-1 and CarO. Fewer sites exhibited site-specific rate shifts between the generally fast evolving HSP-30 and the generally conserved NOP-1 and CarO.
Figure 2.
Homology-based predicted secondary topology of the NOP-1 (green), ORP-1 (blue) and CarO (orange) protein in Fusarium graminearum. The seven transmembrane α-helices and helix boundaries are inferred by comparison to bacteriorhodopsin (Bieszke et al. 1999b; Grigorieff et al. 1996). Numbers indicate retinal-binding pocket residues that are conserved among the archaeal transport and sensory rhodopsins (Henderson et al. 1990; Hoff et al. 1997). A red circle around a numbered amino acid site indicates that the amino acid is shared with bacteriorhodopsin (i.e. the site is conserved). A solid black circle around a numbered amino acid site indicates that the amino acid is identical among the three fungal opsins.
Knockout of nop-1 affects expression of genes involved in oxidative stress
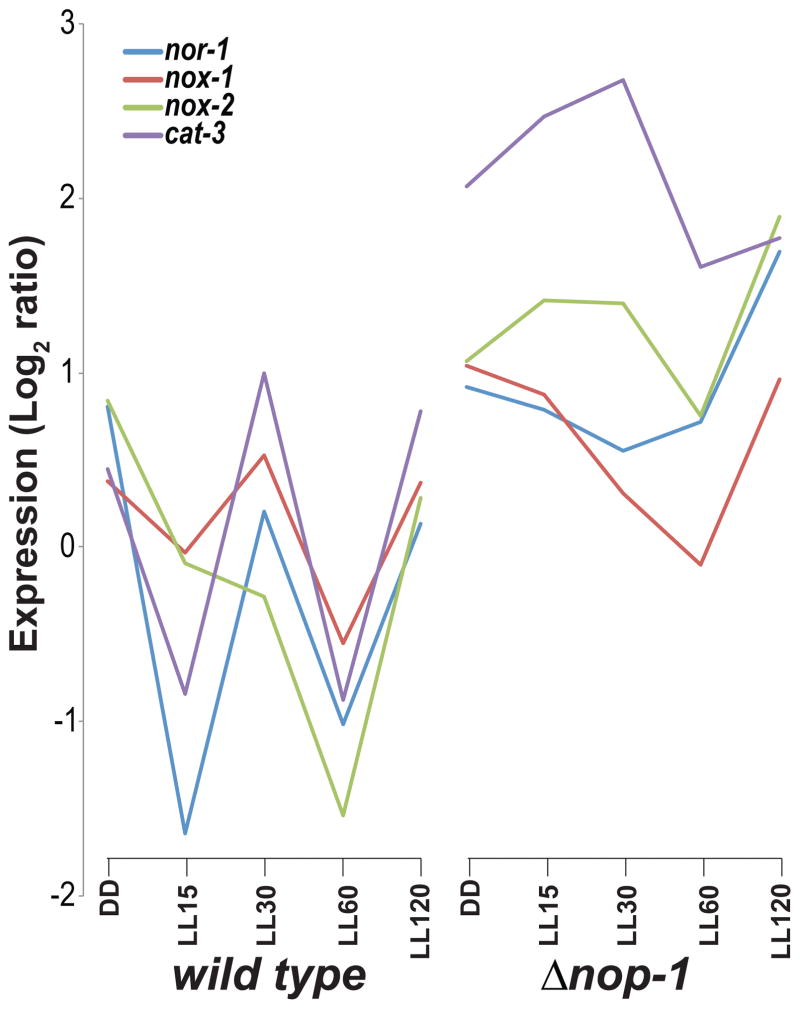
The genome-wide light-inducible response was characterized for a knockout of the predicted light sensor nop-1. Gene expression in wild type and Δnop-1 strains cultured in the dark, exposed to 15, 30, 60 or 120 minutes of light (Chen et al. 2009) was previously measured via a spotted oligomer microarray targeting known ORFs in N. crassa. Reanalysis of these data identified significantly higher expression (P < 0.05 and higher than 3-fold change) of 736 genes in Δnop-1 than in wild type after 2 h of exposure to light. After light stimulation, light-responsive genes including blue-light responsive crp and vvd exhibited much higher up-regulation in Δnop-1 compared to wild type. Genome-wide, one hundred sixty-one genes exhibited statistically significant 5-fold or higher expression in Δnop-1 than in wild type. Of these highly upregulated genes, 143 genes were functionally annotated at FunCat (MIPS), and these genes were significantly enriched for metabolism, secondary metabolism, FAD/FMN binding, NAD/NADP binding, transport facilities, electrochemical potential-driven transport, proton driven symporter, catalase reaction, homeostasis of protons, and oxidative stress response (P < 0.01). These pathways were also enriched (P < 0.05) for the 778 genes identified as exhibiting significantly higher (P < 0.01) expression in Δnop-1 than wild type after a short exposure to white light. In response to a long exposure to light, many genes responding to oxidative stress continued to be significantly (P < 0.05) higher-expressed in the nop-1 knockout strain compared to wild type. These genes include nox-2 (encoding NADH oxidase-2), nor-1 (encoding NADH oxidase regulator-1), and cat-3 (encoding ROS-detoxifying enzyme catalase-3). The expression pattern of these three genes was highly similar in wild type but dissimilar among genes in the nop-1 knockout (Fig. 3). Of 15 genes reported to be responsive to the oxidative stressor menadione (Sasano et al. 2010), three gst (glutathione S-transferase) and 11 mig (menadione-induced gene) genes were highly expressed (10 significantly; P < 0.05) in Δnop-1 compared to the wild type strain, when exposed to light—the exception being mig-4 (Table S5). Expression of mating type locus mat A, matA-specific pheromone precursor ccg-4, and two pheromone receptors pre-1 and pre-2 were also significantly higher (P < 0.05) in Δnop-1 (Table S5).
Figure 3.
Disruption of nop-1 affected expression of NADH oxidases that control cell differentiation and growth in N. crassa. Expression of NADH oxidases genes showed highly correlated regulation in a wild type strain, but expression of nox-2 and cat-3 was up-regulated immediately following light stimulation in the knockout strain Δnop-1. Log2 ratio of expression level was estimated from microarray results (Chen et al. 2009). Expression was measured under constant darkness (DD) and in response to light exposure (5–120 minutes, LL5 to LL120).
Expression of genes coding for opsin-like proteins is up-regulated during sexual development
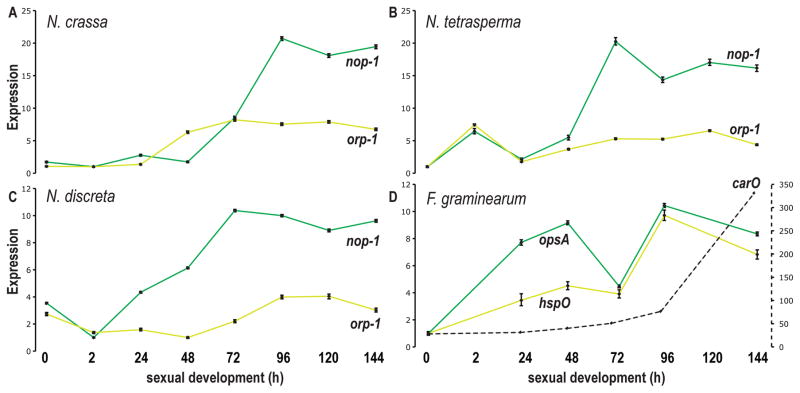
Expression of genes coding for NOP-1, ORP-1, and CarO was assayed during sexual development in three Neurospora species and two Fusarium species (Lehr et al. 2014; Sikhakolli et al. 2012, Fig. 4). In Neurospora, both nop-1 and orp-1 were generally up-regulated with maturation of perithecia. While nop-1 expression steadily increased to more than 10-fold its initial level, orp-1 increased to nearly 5-fold of its initial level at 48 h after crossing, then was approximately stably expressed. Orthologs of nop-1 and orp-1 in F. graminearum (annotated as opsA and hspO respectively), exhibited up-regulation of expression across perithecial development, with a drop at 72 h in F. graminearum. Expression of carO, which is an opsin-like protein with orthologs only found in genomes of plant pathogens and plant associated fungi, exhibited a dramatic 340-fold up-regulation in samples after 96 h from induction of the sexual cycle. Expression of nop-1 exhibited no significant change during asexual development of N. crassa under a constant white light condition (Wang et al. 2012a).
Figure 4.
Expression of fungal opsins across perithecial development from 0 h (protoperithecia) to 144 h (mature perithecia). Expression was measured for A) N. crassa nop-1 (green) and orp-1 (yellow), B) their orthologs in Neurospora tetrasperma, C) their orthologs in Neurospora discreta, and D) their orthologs opsA and hspO and paralog carO (black, dashed) in F. graminearum. The right-hand Y-axis indicates gene expression levels for carO, whose expression changes across sexual development much more than does opsA and hspO.
A nop-1 knockout in N. crassa exhibited a phenotype in production of perithecia when exposed to white and blue light
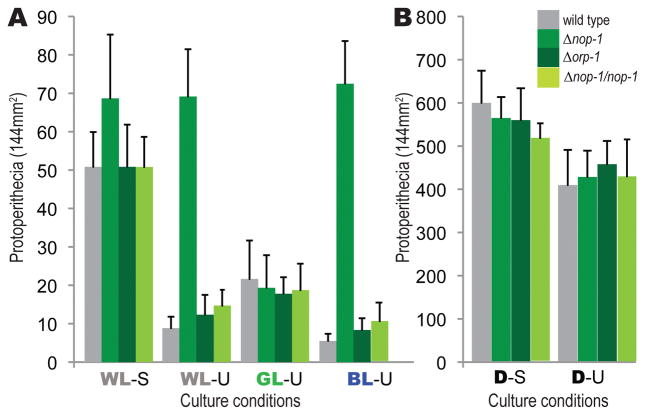
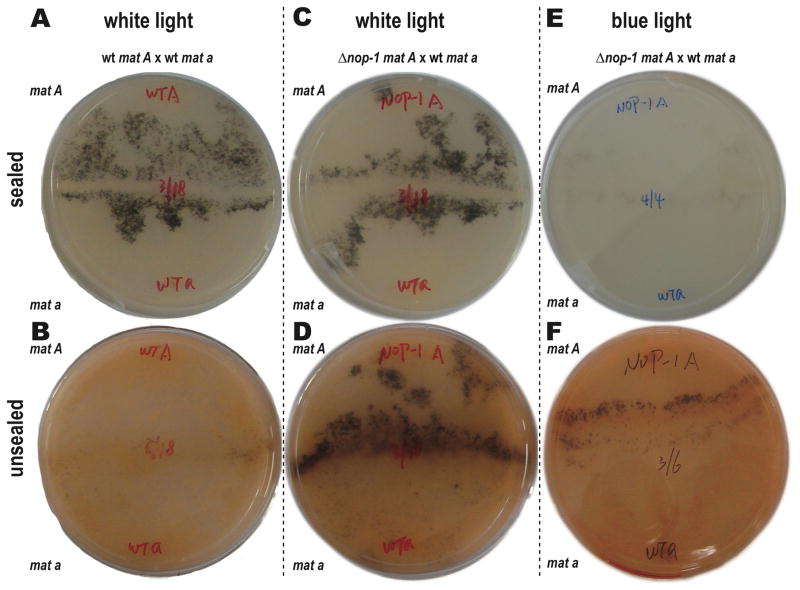
For Δnop-1, an abundance of protoperithecia was produced when exposed to white and blue light regardless of the airflow (Fig. 5). Unlike the wild type strain (Fig. 6A–B), Δnop-1 exhibited the same quantity of perithecia between sealed and unsealed plates when exposed to low white light. Moreover, when Δnop-1 was crossed with wild type while exposed to low-intensity white light, hyphae on the Δnop-1 side of the crossing plate exhibited earlier perithecial development (Fig. 6C–D; Fig. S2). When exposed to intense blue light, initiation of sexual development in protoperithecial production occurred at equal velocity between Δnop-1 and wild type in tightly sealed plates, but occurred earlier for Δnop-1 in unsealed plates (Fig. 6E–F). We observed higher protoperithecia production similarly for all strains under green light than white or blue light. Additional H+-ATPase inhibitor oligomycin in the medium showed no impacts to the observed phenotypes of both wild type and Δnop-1 strains under different airflow conditions.
Figure 5.
Abundance of protoperithecia for Δnop-1, Δorp-1, wild type strains (mat A) and complemented strains of Δnop-1 after six days of incubation on SCM medium under different light and air flow conditions. Numbers of protoperithecia were recorded as the detectable protoperithecia per 13 mm × 13 mm square, with four replicate squares in three replicate plates. Error bars represent one standard deviation. A. Abundance of protoperithecia under constant white light (WL), green light (GL), or blue light (BL) with parafilm-sealed (S) or unsealed (U) plates. B. Abundance of protoperithecia under constant darkness (D) with plates parafilm-sealed (S) or unsealed (U).
Figure 6.
Phenotype of Δnop-1 after six days of incubation on SCM medium. A wild type cross produced perithecia earlier when cultured under constant white light on a sealed plate (A) than when cultured under constant white light on an unsealed plate (B). A mat A Δnop-1 cross with mat a wild type exhibited wild type perithecial development when cultured under constant white light on a sealed plate (C), but exhibited earlier and more abundant production of perithecia for Δnop-1 when cultured under constant white light on an unsealed plate (D). A mat A Δnop-1 cross with mat a wild type exhibited wild type perithecial development when cultured under constant blue light on a sealed plate (E), but exhibited earlier and more abundant production of perithecia when cultured under constant blue light on an unsealed plate (F). Conidiation (visible as orange pigment) is much more abundant on unsealed plates in all comparisons.
Crosses of the wild type strains, Δnop-1, and Δorp-1 knockout strains cultured in the dark or exposed to constant red, far red, blue, and low and high white light conditions yielded robust and replicable phenotypic results, all demonstrating that low light intensity disposes N. crassa toward initiation of sexual reproduction (Fig. 5). Low airflow, likely associated with low oxygen level, was also found to induce early production of protoperithecia, examining all strains under two air flow conditions achieved by either tightly sealing plates with parafilm or by leaving plates covered but unsealed. For the wild type, a sealed plate and darkness promoted protoperithecial production and early perithecial development, but white and blue light promoted asexual development and repressed sexual reproduction. We observed no distinct phenotype for Δnop-1 knockout during sexual development with treatments of menadione (a ROS-generating quinone), hydrogen peroxide (H2O2), and N-acetyl-cysteine (NAC, an antioxidant) under dark or light, sealed or unsealed conditions, and adding these reagents with short-term effectiveness caused uncontrollable physical disturbance to the five-day protoperithecia development. No phenotypes in the Δnop-1 or Δorp-1 knockout strains were observed when cultured in complete darkness, nor were any observed during asexual development of cultures grown on conidiation medium when exposed to any light conditions, nor were any observed in late perithecial development. No phenotypes differing from wild type were observed for orp-1 knockout strains.
The plasmid pCB1532-nop-1 complemented the nop-1 knockout mutant. Integration of the complementary DNA fragment into the transformant genomes was verified by PCR. Of 14 PCR-verified transformants obtained, eight displayed wild-type protoperithecial production when cultured under white light exposed to normal or low airflow.
Reconstructed gene networks positioned nop-1 as tightly associated with the oxidative pathway in regulating the asexual-sexual switch in Neurospora crassa
N. crassa produces only asexual conidia on Bird medium, but produced both asexual conidia and sexual perithecia on our maple sap medium. The asexual-sexual switch is regulated by various environmental factors via a complex network, suggested by comparison of gene expression from a medium permissive for asexual development only and a medium that supports the asexual-sexual switch. Inferred networks using expression data (Table S6) from asexual growth exhibited different topologies when expression data was derived from culturing on Bird media and on maple sap media, including associations between nop-1 and genes involved in asexual development, sexual development, and oxidative stress pathways (Fig. S3). For cultures on artificial Bird medium, the inferred network suggested coordinated expression regulation between oxidative pathways and conidiation (asexual development). Genes annotated for perithecial production (sexual development) were sparsely connected within the network, implying less robustly coordinated regulation of the process. For cultures on natural maple sap medium, the inferred network showed dense associations among well-defined subnetworks for oxidative pathways, conidiation and initiation of perithecial development.
Discussion
We have demonstrated a history of diversification of putative green light sensor opsins in higher fungi. Four families of opsin-like proteins were identified within ascomyceteous genomes. They were the HSP-30 gene family, which is found mainly in single cellular yeast species; the CarO gene family, which is shared among only plant pathogens and plant-associated fungi, and the orthologs of two Neurospora opsins—NOP-1 and ORP-1—which are shared among exclusively multicellular ascomycetes;. We have further demonstrated the active regulation of opsin expression during early sexual development in Neurospora and closely related fungi. Guided by this regulation of expression, we identified the knockout phenotype for the putative green-light sensor nop-1 in N. crassa. In wild-type N. crassa under constant light, low oxygen levels induced by sealing the culture plate promotes sexual development. In contrast, in a Δnop-1 strain, there is no difference in sexual development times between cultures in sealed as compared to unsealed plates. Thus, NOP-1 could play a role in regulating asexual-sexual development in response to different environmental signals including light and ROS level in N. crassa.
Opsin-like proteins in higher fungi have been suggested to originate from a lateral gene transfer from haloarchaea into the ancestor of ascomycetes and basidiomycetes (Ruiz-Gonzalez & Marin 2004). Compared to the transport and sensory rhodopsins of the haloarcheal ancestor, nearly all of the 22 retinal binding pocket residues are conserved in the CarO and NOP-1 gene families. A few binding pockets are lost throughout ORP-1 lineages. No opsins similar to nop-1, orp-1 or carO were identified in the genomes of deeply divergent fungal lineages (some of these fungi maintain another opsin-like protein homologous with animal opsins (Fuller et al. 2016; Idnurm et al. 2010). The relationships among opsins in early basidiomycetes and that from prokaryotes and green algae were not resolved. This pattern of well-resolved nodes for recent divergences and lack of resolution for deep divergences is consistent with phylogenetic informativeness profiles (Lopez-Giraldez & Townsend 2011; Townsend 2007), which indicated high utility for recent evolutionary events in fungal opsin divergence, but dropped markedly near the time of the point of diversification of the four families of fungal opsins.
Within the genomes of all major ascomyceteous groups, orthologs of NOP-1 were present, and the evolution of NOP-1 generally was consistent with the species phylogeny. As suggested by the phylogeny presented here, the orthologous gene sets CarO, ORP-1, and HSP30 all derived from a single origin following the divergence of NOP-1 in higher fungi Dikarya. Their subsequent diversification appears to have been driven in part by the ecology of their lineage. Orthologs of CarO were found only in plant-associated fungi, as well as in the lichen Cladonia grayi. In contrast, orthologs of NOP-1 and ORP-1 were found in saprobic fungi, as well as several Sordariomycetes plant pathogens that also maintain a copy of a CarO ortholog. The fast-evolving HSP-30 gene family is not conserved at retinal binding sites; indeed, secondary structures we predicted based on S. cerevisiae HSP-30 amino acid sequence suggested that the typical seven-transmembrane helices of the opsins are not conserved within the HSP-30 family. Orthologs of HSP-30 have only been identified in yeasts and Arthroderma, a group of fungi often isolated from skin of human and other animals. Conceivably, sensing of temperature, rather than light, could be the critical function of HSP-30 genes for these fungi.
Environmental factors have not been distinctly connected to the phenotypic functions of opsin-like proteins. Consistent with previous observations under constant light in N. crassa, our results included up-regulated expression of nop-1 during asexual development, and no observable phenotype for the nop-1 knockout strain in asexual reproduction. However, except for hsp-30, expression of opsin-like proteins was all up-regulated during sexual development in Neurospora and closely related Fusarium species under constant white light, implying that expression of these genes is related to sexual development. The gene nop-1 is not expressed during submerged growth of N. crassa in liquid Vogel’s minimal medium—a low air-exchange condition that suppresses conidiation and sexual differentiation (Bieszke et al. 2007; Bieszke et al. 1999a). Limiting air exchange with a parafilm seal and exposing the culture to constant blue or white light, we observed precocious sexual development in Δnop-1 compared to wild type. Light and ROS or oxygen conditions are critical for fungal development, and in many fungal species darkness and low air exchange are physical conditions that induce sexual reproduction (Bohm et al. 2012; O’Gorman et al. 2009). In N. crassa, oxygen, not carbon dioxide, was found to be able to affect asexual development (Iigusa et al. 2005). Initiation of sexual (protoperithecial) development occurs rapidly and abundantly for N. crassa on sealed SCM plates under a regime of constant darkness. In contrast, we have demonstrated that constant white light or blue light promote conidiation and suppress sexual reproduction in N. crassa, especially when cultured on unsealed plates. So far, fungal light-responsive genes have been observed to have functions and phenotypes responding to blue and red light—the two strongest lights in natural environment. Research on several fungal models suggests that blue-light responses, mediated by WCC orthologs, are a core (or hub) regulatory factor in fungal development. Light responsive pathways acted on by phytochromes or opsins may play secondary roles in fungal development. Observation of NOP-1 phenotypes occurring in response to light outside of its peak absorption implies non-peak absorption of blue light by NOP-1 is playing a coordinated developmental regulatory role mediated by blue-light responsive pathways.
Genome-wide expression analysis has provided precise estimates of the effect of light on the N. crassa transcriptome, including a demonstration that extended exposure to light led to significant oxidative stress to the organism (Wu et al. 2014). The knockout of nop-1 affects expression for many genes involved in sexual development, including pheromone precursors and receptors, genes in oxidative stress responses, and NADPH oxidases that control cell differentiation and sexual growth in N. crassa. The latter two stress pathways signals are mediated through MAPK signaling to affect transcription factors, which further awaken cellular responses (Bahn et al. 2007). Up-regulation of expression was observed for oxidative stress related genes in wild type and knockouts of phytochromes, white collar genes and NOP-1, but only in the NOP-1 knockout was a significantly higher expression than wild type observed for oxidative stress related genes. Among those cellular responses are the increased expression of genes with function a hypotonic shock, high osmolarity, and starvation stress—all expressed higher in Δnop-1 than in wild type, especially under light exposure. Six genes with annotated function in MAPK signaling pathways, including two pheromone receptors, a SSU81 related protein, a putative MAPKK kinase, a protein kinase C-like protein, and a hypothetical protein (NCU01489) were significantly abundant compared to wild type (P < 0.05). Pathway analysis and individual gene analysis are consistent with the involvement of N. crassa NOP-1 protein in sexual development being governed by conditions of light and oxidative stress. The combination of low expression of nop-1 in early sexual development in wild type, with earlier protoperithecial production under light in Δnop-1, suggests a negative regulatory role for NOP-1.
How light could mechanistically regulates asexual development in N. crassa via reactive oxygen species (ROS) remains unknown (Peraza & Hansberg 2002). Production of perithecia is stimulated by brief exposure to blue light, which in N. crassa is accompanied by formation of ROS during sexual development (Belozerskaya et al. 2012). The light-responsive transcription factor BcLTF1 was found to be required to cope with oxidative stress in plant pathogen Botrytis cinerea (Schumacher et al. 2014). Indeed, functions of homologs of BcLTF1 (SUB-1, submerged protoperithecia-1 in Neurospora) are required for normal sexual reproduction in Aspergillus and Neurospora (Colot et al. 2006; Han et al. 2001; Nowrousian et al. 2012; Smith et al. 2010; Szewczyk & Krappmann 2010). Expression of sub-1 was significantly up-regulated in the Δnop-1 strain when cultured under constant light. Across the life cycle of wild-type N. crassa, sub-1 is most highly expressed in protoperithecia during sexual development (Wang et al. 2014). Our results imply that light sensing and ROS response might be coupled with NOP-1 to regulate the initiation of sexual development in N. crassa. This coupling is suggested by significantly up-regulated expression of oxidative stress responsive genes in the Δnop-1 strain after an extended exposure to light. ROS are formed in fungi with metabolic activity such as the respiratory chain in mitochondria and activity of intracellular enzymes such as NADPH-oxidases (NOX). NOX produces ROS, which in turn affect diverse cellular functions including cell differentiation and redox-dependent signaling (Aguirre et al. 2005; Bedard et al. 2007; Cano-Dominguez et al. 2008; Idnurm & Heitman 2005; Jiang et al. 2011; Sasano et al. 2010; Takemoto et al. 2007). Removing NOX-1, NOX-2, or their regulatory subunit NOR-1 caused abnormal sexual development, and elimination of NOX-1 or NOR-1 resulted female sterility in N. crassa (Cano-Dominguez et al. 2008). In response to stimulation by white light, genes nor-1, nox-1, nox-2, and cat-3 exhibited highly similar patterns of expression in wild type strains, but this cohesion of expression was sundered in the Δnop-1, especially during the first hour of exposure to light. In addition, four genes, including sre, cat-3, ccp and ccs, recently were identified with knockout phenotypes of hypersensitive to menadione (Zhu et al. 2013), and all these genes were expressed at a much higher level in nop-1 mutant than in wild type strain. Phenotypes of the nop-1 knockout strain in protoperithecia production and expression of oxidative stress genes imply that oxidative stress might couple the light response and ROS response and negatively regulate production of protoperithecia in N. crassa sexual development. Similarly, SOP1, NOP-1 homolog in Sclerotinia sclerotiorum, was reported to play many roles in responses to oxidative and osmotic stresses and in fungicide resistance and sclerotial development (Lyu et al. 2015). NOP-1 probably is of pleiotropic function during the asexual-sexual switch in N. crassa.
NOP-1 has been identified as binding a green light-absorbing pigment (λmax = 534 nm) with a spectral shape and bandwidth typical for archaeal rhodopsins, and a photochemical reaction cycle similar to that of the phototaxis receptor sensory rhodopsin (Bieszke et al. 1999a; Bieszke et al. 1999b; Furutani et al. 2006). Flash photolysis of NOP-1 yielded results consistent with a slow-cycling pigment that is typical for sensory rhodopsins, but strong conservation of homologous amino acids that are involved in the proton-conducting pathway in halobacterial rhodopsins suggests that NOP-1 should possess some proton-pumping activity (Furutani et al. 2006). Accordingly, the retinal binding sites have not evolved over a deep time scale spanning NOP-1 diversification and going back to the Leptosphaeria opsin that has been demonstrated to exhibit proton pump ability (Fan et al. 2007). A comparatively slowly cycling proton pump without accompanying transducer proteins might be the product of evolution to regulate the comparatively slow process of sexual development in Neurospora and related fungi. A nop-1 knockout strain exhibited a slight difference in conidiation in the presence of the mitochondrial ATPase inhibitor oligomycin under constant light (Bieszke et al. 1999a). We observed no impact on sexual development with the addition of oligomycin during sexual development in wild type and the Δnop-1 mutant; however, this negative result does not discount a hypothetical proton pump function for NOP-1, considering the negative regulation of NOP-1 during sexual initiation and the alternative H+ pumps that might exist in N. crassa. We observed no sexual-developmental phenotypes in wild type or Δnop-1 knockout strains with treatments of menadione, NAC, or H2O2 under light/dark and sealed/unsealed conditions. However, these chemicals are of short effectiveness, and are sensitive to light, humidity, and heat, and have been validated for use in studies of rapid asexual development in Neurospora crassa (Cabrera et al. 2015; Videira et al. 2009). NOP-1 roles in response to ROS in the asexual-sexual switch in N. crassa should be investigated by manipulation of ROS levels through genetic means, such as in knockouts of sod-1, cat-1, cat-3 or ras-1 (Belden et al. 2007).
Since visible light is a signal for stress, and oxidative stress is known to induce asexual development (Fuller et al. 2015), oxidative stress could be the link between NOP-1 associated proton pumping activity and modulation of asexual-sexual switch by light. Genes highly up-regulated in Δnop-1 after light exposure were significantly enriched for FAD/FMN binding, NAD/NADP binding, transport facilities, electrochemical potential-driven transport, proton driven symporter, catalase reaction, homeostasis of protons, and oxidative stress response (P < 0.01). Phenotypes observed for Δnop-1 integrating aspects of both light and airflow imply that initiation of sexual development in N. crassa is regulated by a complex network of interactions, with potentially multiple sensory modules responding to environmental signals. N. crassa perithecia have been discovered under the surface of plant epidermal tissues, with well-developed beaks protruding through the cracked tissue (Pandit & Maheshwari 1996); correspondingly, proper perithecia beak development in N. crassa requires light stimulation. Coupling ROS and light responses could ensure rapid production of perithecia in a timely manner in response to challenging conditions.
A dramatic up-regulation (>300×) of carO was observed in late sexual development in F. graminearum, when ascospores are fully mature and forcibly released (Sikhakolli et al. 2012). Unlike N. crassa, parafilming plates inhibits perithecium formation and encourages vegetative growth in F. graminearum. Recently CarO is characterized as a light-driven proton pump and highly expressed in light-formed conidia in F. fujikuroi, and knockout strain of carO showed a faster development in conidia germlings (Garcia-Martinez et al. 2015). Given the high expression level of carO observed in F. graminearum during the later sexual reproduction, expression level of CarO in ascospores and how it would affect germination of ascospores under different environmental conditions require further investigation. Expression of carO ortholog ops1 is also regulated by light in the pathogen Bipolaris oryzae; in B. oryzae, ops-1 is weakly expressed in mycelia when cultured in the dark but exhibited increased expression after near-UV irradiation (Kihara et al. 2009).
ROS is known to mediate communication between fungal endophytes and plant pathogens (Scott et al. 2007; Tanaka et al. 2006). Recent data suggests that the superoxide-generating NADPH oxidases in fungi could be indirectly involved in pathogen-host communication via fungal differentiation processes that are necessary for virulence (Heller & Tudzynski 2011; Tudzynski et al. 2012). Unlike in N. crassa, expression of the NOP-1 ortholog in some of these pathogens, including Leptosphaeria, Fusarium, and Bipolaris species, was high in mycelia cultured in the dark, and exhibited no response to light exposure (Estrada & Avalos 2009; Idnurm & Howlett 2001; Kihara et al. 2009), implying operation in a function independent of light exposure.
In conclusion, light sensor proteins play important roles in perception of environmental signals and in guiding fungal development. In particular, four opsins or opsin-like proteins have evolved in phylogeny of filamentous fungi that are homologous to the green-light sensors of bacterial rhodopsins. Evolution of these proteins is correlated to unicellularity and multicellularity and to ecology: members of the HSP30 orthologous gene set are generally found in unicellular yeasts but not multicellular hyphomycetes, and members of the CarO orthologous gene set are generally found in plant-associated species such as plant pathogens and endophytes. Based on transcriptomic profiles of both wild type and knockout strains, we propose a model in which NOP-1 couples response to light with oxidative stress response to ROS, negatively regulating initiation of sexual development in the fungal model N. crassa (Fig. 7). This model is supported by inferred network showing tight association among subnetworks of nop-1, oxidative stress responses, asexual development, and sexual development based on N. crassa growth on natural maple sap medium. This model can be further validated with systems biology approaches combing wild type transcriptomics and genetics data and knockout phenotypes under controlled environmental conditions for lights and oxidative stress during the asexual-sexual switch in Neurospora and related fungi.
Figure 7.
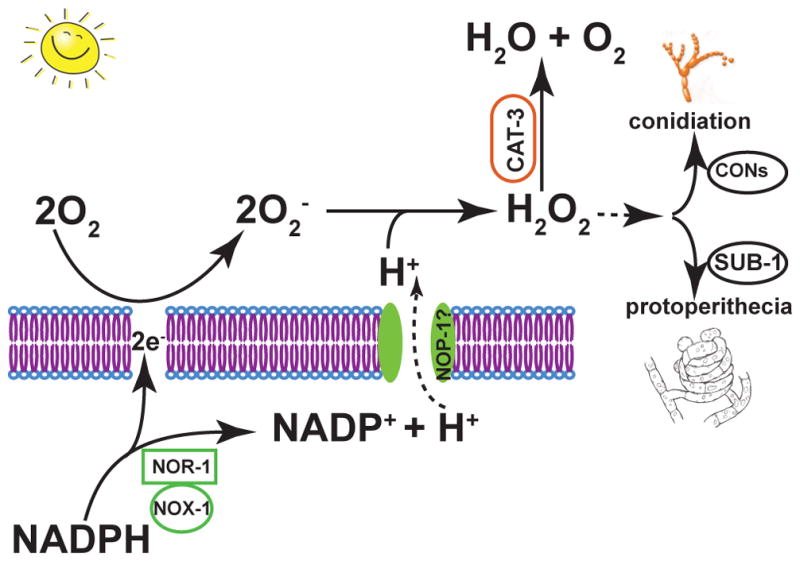
A model in which superoxide and hydrogen peroxide mediate the asexual-sexual balance via diverse transcription factors in N. crassa, and levels of these ROS elements are regulated by coupled proton pump function of NOP-1 and NADPH oxidation by NOX and NOR-1
Supplementary Material
Figure S1: Amino acid sequences conservation and secondary structure predicted for ascomycetous orthologs of nop-1, orp-1, and carO using Consurf (Ashkenazy et al. 2010). Page 1: Results for nop-1 orthologs, mapped to the N. crassa nop-1 protein sequence. Page 2: Results for nop-1, mapped to the F. graminearum nop-1 protein sequence. Page 3: Results for orp-1, mapped to the F. graminearum orp-1 protein sequence. Page 4: Results for carO, mapped to the F. graminearum carO protein sequence.
Figure S2: Triplet crossing plates between the wild type strains (A and B) and between the nop-1 knockout and the wild type strains (C and D) when cultured under constant light with sealed (A and C) and unsealed plates (B and D). Wild type crosses produced perithecia earlier when cultured under constant white light on sealed plates (A) than when cultured under constant white light on unsealed plates (B). matA Δnop-1 cross with mata wild type exhibited wild type perithecial development when cultured under constant white light on sealed plates (C), but exhibited earlier and more abundant production of perithecia for Δnop-1 when cultured under constant white light on unsealed plates (D). Conidiation (visible as orange pigment) is much more abundant on unsealed plates in all comparisons
Figure S3: Bayesian networks inferred from expression of nop-1 and selected genes involved in oxidative stress pathways, asexual development, and initiation of perithecia. Edges with posterior probability higher than 50% (presence in greater than 50% of 100 models produced) are depicted. (A) For cultures on Bird medium, the inferred network coordinated expression regulation between oxidative pathways and conidiation (asexual development). Genes annotated for perithecial production (sexual development) were sparsely connected within the network, implying less robustly coordinated regulation of the process. (B) For cultures on natural maple sap medium, the inferred network showed dense associations among well-defined subnetworks for oxidative pathways, conidiation and initiation of perithecial development.
Table S1. Genbank accession and annotated opsin-like proteins analyzed in this study.
Table S2. Likelihood ratio test of positive selection (sites model in PAML) analyses on subsets of the fungal opsins gene tree in figure 1.
Table S3. Results of Clade Model C analyses on fungal opsins Bayesian tree.
Table S4. Results of DIVERGE 2.0 analysis on fungal opsin-like proteins.
Table S5: Relative gene expression analysis of 10 microarrays from Chen et al. (2009). Abbreviations: WT: wild type, nop: nop-1 knockout, Ref: pooled RNA reference, D: dark (DD), S: short light exposure (15–30 minutes, LL15, LL30), and L: long light exposure (60–120 minutes, LL60 and LL120).
Table S6: Expression of nop-1 and selected genes involved in oxidative stress pathways, conidiation (asexual development), and initiation of perithecia (sexual development).
Acknowledgments
We are grateful to the Neurospora community for their courtesy in making strains available for this study. We thank the Broad Institute and MIPS for making N. crassa gene and genomic data available for oligonucleotide prediction. Drs. Muller and Armaleo at Duke University annotated opsins in the lichenized fungi Cladonia grayi. All authors have declared that no competing interests exist. This study was supported by funding to JCD and JPT from The National Institutes of Health P01 grant GM068067, to JCD from GM118021, by funding from the National Science Foundation (Grant numbers MCB 0923794 and IOS 1456482), by funding from National Institute of Food and Agriculture (2015-67013-22932) to JPT and FT, and Michigan AgBioResearch to FT and by funding from the National Science Foundation (Grant numbers MCB 0923797 and IOS 1457044) to JPT. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Data accessibility
Whole-genome RNA sequencing data for Neurospora and Fusarium species across sexual development and whole-genome microarray data for Neurospora crassa vegetative growth and asexual development are available at Gene Expression Omnibus database (GEO accession GSE8932, GSE61865, GSE60257, GSE26209, GSE22658 and GSE101412). Results of BAGEL analysis with GSE8932 were provided as supplemental data and are available for publics at the Filamentous Fungal Gene Expression Database (http://bioinfo.townsend.yale.edu). Sequences used for phylogenetics analyses were downloaded from NCBI database, and their accession numbers were provided in supplemental files. Data were also available at Dryad (doi:10.5061/drayad.58gn0).
References cited
- Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Ahrendt SR, Medina EM, Chang CA, Stajich JE. Exploring the binding properties and structural stability of an opsin in the chytrid Spizellomyces punctatus using comparative and molecular modeling. PeerJ. 2017;5:e3206. doi: 10.7717/peerj.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J, Estrada AF. Regulation by light in Fusarium. Fungal Genet Biol. 2010;47:930–938. doi: 10.1016/j.fgb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Avelar GM, Schumacher RI, Zaini PA, et al. A rhodopsin-guanylyl cyclase gene fusion functions in visual perception in a fungus. Curr Biol. 2014;24:1234–1240. doi: 10.1016/j.cub.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Xue C, Idnurm A, et al. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- Bayram O, Braus GH, Fischer R, Rodriguez-Romero J. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol. 2010;47:900–908. doi: 10.1016/j.fgb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Bedard K, Lardy B, Krause KH. NOX family NADPH oxidases: not just in mammals. Biochimie. 2007;89:1107–1112. doi: 10.1016/j.biochi.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Larrondo LF, Froehlich AC, et al. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007;21:1494–1505. doi: 10.1101/gad.1551707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 2004;59:121–132. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- Bieszke J, Li L, Borkovich K. The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Current Genetics. 2007;52:149–157. doi: 10.1007/s00294-007-0148-8. [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Braun EL, Bean LE, et al. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci U S A. 1999a;96:8034–8039. doi: 10.1073/pnas.96.14.8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry. 1999b;38:14138–14145. doi: 10.1021/bi9916170. [DOI] [PubMed] [Google Scholar]
- Bills GF, Platas G, Pelaez F, Masurekar P. Reclassification of a pneumocandin-producing anamorph, Glarea lozoyensis gen. et sp. nov., previously identified as Zalerion arboricola. Mycol Res. 1999;103:179–192. [Google Scholar]
- Bluhm BH, Dhillon B, Lindquist EA, et al. Analyses of expressed sequence tags from the maize foliar pathogen Cercospora zeae-maydis identify novel genes expressed during vegetative, infectious, and reproductive growth. BMC Genomics. 2008;9:523. doi: 10.1186/1471-2164-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstein A, Vienken K, Tasler R, et al. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Bohm J, Hoff B, O’Gorman CM, et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci U S A. 2012;110:1476–1481. doi: 10.1073/pnas.1217943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt S, von Stetten D, Gunther M, Hildebrandt P, Frankenberg-Dinkel N. The fungal phytochrome FphA from Aspergillus nidulans. J Biol Chem. 2008;283:34605–34614. doi: 10.1074/jbc.M805506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown LS, Jung KH. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem Photobiol Sci. 2006;5:538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- Cabrera IE, Pacentine IV, Lim A, et al. Global analysis of predicted G protein-coupled receptor genes in the filamentous fungus, Neurospora crassa. G3 (Bethesda) 2015;5:2729–2743. doi: 10.1534/g3.115.020974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Dominguez N, Alvarez-Delfin K, Hansberg W, Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 2008;7:1352–1361. doi: 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Dunlap JC, Loros JJ. Neurospora illuminates fungal photoreception. Fungal Genet Biol. 2010;47:922–929. doi: 10.1016/j.fgb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Loros JJ. Neurospora sees the light: light signaling components in a model system. Commun Integr Biol. 2009;2:448–451. doi: 10.4161/cib.2.5.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 2009;28:1029–1042. doi: 10.1038/emboj.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnici JL, Fu C, Caccamise LM, Arnold JW, Free SJ. Neurospora crassa female development requires the PACC and other signal transduction pathways, transcription factors, chromatin remodeling, cell-to-cell fusion, and autophagy. PLoS One. 2014;9:e110603. doi: 10.1371/journal.pone.0110603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TA, Guilmette JM, Renstrom D, Townsend JP. RNA extractioin, probe preparation, and competitve hybridization for transcriptional profiling using Neurospora crassa long-oligomer DNA microarrays. Fungal Genetics Reports. 2008;55:18–28. [Google Scholar]
- Colot H, Park G, Turner G, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano LM. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Borkovich KA, Henn MR, et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ. The Neurospora circadian system. J Biol Rhythms. 2004;19:414–424. doi: 10.1177/0748730404269116. [DOI] [PubMed] [Google Scholar]
- Estrada A, Avalos J. Regulation and targeted mutation of opsA, coding for the NOP-1 opsin orthologue in Fusarium fujikuroi. J Mol Biol. 2009;387:59–73. doi: 10.1016/j.jmb.2009.01.057. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shi L, Brown LS. Structural basis of diversification of fungal retinal proteins probed by site-directed mutagenesis of Leptosphaeria rhodopsin. FEBS Lett. 2007;581:2557–2561. doi: 10.1016/j.febslet.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Fan Y, Solomon P, Oliver RP, Brown LS. Photochemical characterization of a novel fungal rhodopsin from Phaeosphaeria nodorum. Biochim Biophys Acta. 2011;1807:1457–1466. doi: 10.1016/j.bbabio.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Fischer R, Aguirre J, Herrera-Estrella A, Corpochano LM. The complexity of fungal vision. Microbiol Spectrum. 2016;4 doi: 10.1128/microbiolspec.FUNK-0020-2016. FUNK-0020–2016. [DOI] [PubMed] [Google Scholar]
- Fu C, Iyer P, Herkal A, et al. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot Cell. 2011;10:1100–1109. doi: 10.1128/EC.05003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller KK, Dunlap JC, Loros JJ. Fungal light sensing at the bench and beyond. Adv Genet. 2016;96:1–51. doi: 10.1016/bs.adgen.2016.08.002. [DOI] [PubMed] [Google Scholar]
- Fuller KK, Loros JJ, Dunlap JC. Fungal photobiology: visible light as a signal for stress, space and time. Curr Genet. 2015;61:275–288. doi: 10.1007/s00294-014-0451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani Y, Sumii M, Fan Y, et al. Conformational coupling between the cytoplasmic carboxylic acid and the retinal in a fungal light-driven proton pump. Biochemistry. 2006;45:15349–15358. doi: 10.1021/bi061864l. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J, Brunk M, Avalos J, Terpitz U. The CarO rhodopsin of the fungus Fusarium fujikuroi is a light-driven proton pump that retards spore germination. Sci Rep. 2015;5:7798. doi: 10.1038/srep07798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18:500–501. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- Gu X, Zou Y, Su Z, et al. An update of DIVERGE software for functional divergence analysis of protein family. Mol Biol Evol. 2013;30:1713–1719. doi: 10.1093/molbev/mst069. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Han KH, Han KY, Yu JH, et al. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol. 2001;41:299–309. doi: 10.1046/j.1365-2958.2001.02472.x. [DOI] [PubMed] [Google Scholar]
- Harding RW, Melles S. Genetic analysis of phototropism of Neurospora crassa perithecial beaks using white collar and albino mutants. Plant Physiol. 1983;72:996–1000. doi: 10.1104/pp.72.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J, Ruhnke N, Espino JJ, et al. The mitogen-activated protein kinase BcSak1 of Botrytis cinerea is required for pathogenic development and has broad regulatory functions beyond stress response. Mol Plant Microbe Interact. 2012;25:802–816. doi: 10.1094/MPMI-11-11-0299. [DOI] [PubMed] [Google Scholar]
- Heller J, Tudzynski P. Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu Rev Phytopathol. 2011;49:369–390. doi: 10.1146/annurev-phyto-072910-095355. [DOI] [PubMed] [Google Scholar]
- Henderson R, Baldwin JM, Ceska TA, et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Henegariu O, Heerema NA, Dlouhy SR, Vance GH, Vogt PH. Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques. 1997;23:504–511. doi: 10.2144/97233rr01. [DOI] [PubMed] [Google Scholar]
- Hoff WD, Jung KH, Spudich JL. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophysics Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Howlett BJ. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome. 2001;44:167–171. doi: 10.1139/g00-113. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Verma S, Corrochano LM. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet Biol. 2010;47:881–892. doi: 10.1016/j.fgb.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara K, Umemura T, Katagiri I, et al. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol. 1999;285:163–174. doi: 10.1006/jmbi.1998.2286. [DOI] [PubMed] [Google Scholar]
- Iigusa H, Yoshida Y, Hasunuma K. Oxygen and hydrogen peroxide enhance light-induced carotenoid synthesis in Neurospora crassa. Febs Letters. 2005;579:4012–4016. doi: 10.1016/j.febslet.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Innocenti FD, Pohl U, Russo VE. Photoinduction of protoperithecia in Neurospora crassa by blue light. Photochem Photobiol. 1983;37:49–51. doi: 10.1111/j.1751-1097.1983.tb04432.x. [DOI] [PubMed] [Google Scholar]
- Jiang T, Zhang XF, Wang XF, Zhang DP. Arabidopsis 3-ketoacyl-CoA thiolase-2 (KAT2), an enzyme of fatty acid beta-oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 2011;52:528–538. doi: 10.1093/pcp/pcr008. [DOI] [PubMed] [Google Scholar]
- Kendrick B. Fungi: ecological importance and impact on humans. eLS 2011 [Google Scholar]
- Kihara J, Tanaka N, Ueno M, Arase S. Cloning and expression analysis of two opsin-like genes in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol Lett. 2009;295:289–294. doi: 10.1111/j.1574-6968.2009.01609.x. [DOI] [PubMed] [Google Scholar]
- Klittich C, Leslie JF. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi) Genetics. 1988;118:417–423. doi: 10.1093/genetics/118.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritsky MS, Russo VE, Filippovich SY, Afanasieva TP, Bachurina GP. The opposed effect of 5-azacytidine and light on the development of reproductive structures in Neurospora crassa. Photochem Photobiol. 2002;75:79–83. doi: 10.1562/0031-8655(2002)075<0079:toeoaa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Lehr NA, Wang Z, Li N, et al. Gene expression differences among three Neurospora species reveal genes required for sexual reproduction in Neurospora crassa. PLoS One. 2014;9:e110398. doi: 10.1371/journal.pone.0110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichius A, Lord KM, Jeffree CE, et al. Importance of MAP kinases during protoperithecial morphogenesis in Neurospora crassa. PLoS One. 2012;7:e42565. doi: 10.1371/journal.pone.0042565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Warnow TJ, Holder MT, et al. SATe-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol. 2012;61:90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- Looger LL. Running in reverse: rhodopsins sense voltage. Nat Methods. 2012;9:43–44. doi: 10.1038/nmeth.1817. [DOI] [PubMed] [Google Scholar]
- Lopez-Giraldez F, Townsend JP. PhyDesign: an online application for profiling phylogenetic informativeness. BMC Evol Biol. 2011;11:152. doi: 10.1186/1471-2148-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X, Shen C, Fu Y, et al. The microbial opsin homolog Sop1 is involved in Sclerotinia sclerotiorum development and environmental stress response. Front Microbiol. 2015;6:1504. doi: 10.3389/fmicb.2015.01504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- Metzenberg RL. Bird medium: an alternative to Vogel medium. Fungal Genet Newslet. 2004;51:19–20. [Google Scholar]
- Nowrousian M, Teichert I, Masloff S, Kuck U. Whole-genome sequencing of Sordaria macrospora mutants identifies developmental genes. G3 (Bethesda) 2012;2:261–270. doi: 10.1534/g3.111.001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gorman CM, Fuller H, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Olmedo M, Ruger-Herreros C, Corrochano LM. Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa. Genetics. 2010a;184:651–658. doi: 10.1534/genetics.109.109975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M, Ruger-Herreros C, Luque EM, Corrochano LM. A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet Biol. 2010b;47:352–363. doi: 10.1016/j.fgb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. PLANT PLASMA MEMBRANE H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- Pandit A, Maheshwari R. Life-history of Neurospora intermedia in a sugar cane field. J Biosci. 1996;21:57–79. [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- Peraza L, Hansberg W. Neurospora crassa catalases, singlet oxygen and cell differentiation. Biol Chem. 2002;383:569–575. doi: 10.1515/BC.2002.058. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Muller S, Fischer R. Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the White Collar protein LreB. Mol Genet Genomics. 2009;281:35–42. doi: 10.1007/s00438-008-0390-x. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Muller S, Kastner C, et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Lagarias JC. A brief history of phytochromes. Chemphyschem. 2010;11:1172–1180. doi: 10.1002/cphc.200900894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Romero J, Hedtke M, Kastner C, et al. Fungi, Hidden in Soil or Up in the Air: Light Makes a Difference. Annu Rev Microbiol. 2010;64:585–610. doi: 10.1146/annurev.micro.112408.134000. [DOI] [PubMed] [Google Scholar]
- Rohrig J, Kastner C, Fischer R. Light inhibits spore germination through phytochrome in Aspergillus nidulans. Curr Genet. 2013;59:55–62. doi: 10.1007/s00294-013-0387-9. [DOI] [PubMed] [Google Scholar]
- Royer JC, Yamashiro CT. Generation of transformable spheroplasts from mycelia, macroconidia, microconidia and germinating ascospores of Neurospora crassa. Fungal Genet Newslett. 1992;39:76–79. [Google Scholar]
- Ruiz-Gonzalez MX, Marin I. New insights into the evolutionary history of type 1 rhodopsins. J Mol Evol. 2004;58:348–358. doi: 10.1007/s00239-003-2557-8. [DOI] [PubMed] [Google Scholar]
- Sasano Y, Takahashi S, Shima J, Takagi H. Antioxidant N-acetyltransferase Mpr1/2 of industrial baker’s yeast enhances fermentation ability after air-drying stress in bread dough. Int J Food Microbiol. 2010;138:181–185. doi: 10.1016/j.ijfoodmicro.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Schmitt J, Wulff RD. Light spectral quality, phytochrome and plant competition. Trends Ecol Evol. 1993;8:47–51. doi: 10.1016/0169-5347(93)90157-K. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Sung GH, Lopez-Giraldez F, et al. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, Lopez-Fernandez H, Chang BS. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol Biol Evol. 2014;31:1149–1165. doi: 10.1093/molbev/msu064. [DOI] [PubMed] [Google Scholar]
- Schumacher J. How light affects the life of Botrytis. Fungal Genet Biol. 2017;106:26–41. doi: 10.1016/j.fgb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Simon A, Cohrs KC, Viaud M, Tudzynski P. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genet. 2014;10:e1004040. doi: 10.1371/journal.pgen.1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott B, Takemoto D, Tanaka A. Fungal endophyte production of reactive oxygen species is critical for maintaining the mutualistic symbiotic interaction between Epichloe festucae and perennial ryegrass. Plant Signal Behav. 2007;2:171–173. doi: 10.4161/psb.2.3.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R, Kielland-Brandt MC, Fink GR. Yeast plasma membrane ATPase is essential for growth and has homology with (Na++ K+), K+- and Ca2+-ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- Sikhakolli UR, Lopez-Giraldez F, Li N, et al. Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genet Biol. 2012;49:663–673. doi: 10.1016/j.fgb.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Smith KM, Sancar G, Dekhang R, et al. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora white collar complex. Eukaryot Cell. 2010;9:1549–1556. doi: 10.1128/EC.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330:1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- Stajich J, Berbee M, Blackwell M, et al. The Fungi. Current Biology. 2009;19:R840–R845. doi: 10.1016/j.cub.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard J, Chumley FG, Carroll A, Farrall L, Valent B. A series of vectors for fungal transformation. Fungal Genet Newslett. 1997;44:52–53. [Google Scholar]
- Szewczyk E, Krappmann S. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot Cell. 2010;9:774–783. doi: 10.1128/EC.00375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–1076. doi: 10.1016/j.fgb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Tan Y, Merrow M, Roenneberg T. Photoperiodism in Neurospora crassa. J Biol Rhythms. 2004;19:135–143. doi: 10.1177/0748730404263015. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18:1052–1066. doi: 10.1105/tpc.105.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme N, Oeser B, Massaroli M, et al. BcAtf1, a global regulator, controls various differentiation processes and phytotoxin production in Botrytis cinerea. Mol Plant Pathol. 2012;13:704–718. doi: 10.1111/j.1364-3703.2011.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme N, Tudzynski P. Does botrytis cinerea Ignore H(2)O(2)-induced oxidative stress during infection? Characterization of Botrytis activator protein 1. Mol Plant Microbe Interact. 2009;22:987–998. doi: 10.1094/MPMI-22-8-0987. [DOI] [PubMed] [Google Scholar]
- Townsend JP. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics. 2004;5:54. doi: 10.1186/1471-2105-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP. Profiling phylogenetic informativeness. Syst Biol. 2007;56:222–231. doi: 10.1080/10635150701311362. [DOI] [PubMed] [Google Scholar]
- Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:RESEARCH0071. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trail F, Wang Z, Stefanko K, Cubba C, Townsend JP. The ancestral levels of transcription and the evolution of sexual phenotypes in filamentous fungi. PLoS Genet. 2017;13:e1006867. doi: 10.1371/journal.pgen.1006867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudzynski P, Heller J, Siegmund U. Reactive oxygen species generation in fungal development and pathogenesis. Curr Opin Microbiol. 2012;15:653–659. doi: 10.1016/j.mib.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Videira A, Kasuga T, Tian C, et al. Transcriptional analysis of the response of Neurospora crassa to phytosphingosine reveals links to mitochondrial function. Microbiology. 2009;155:3134–3141. doi: 10.1099/mic.0.029710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Folta KM. Contributions of green light to plant growth and development. Am J Bot. 2013;100:70–78. doi: 10.3732/ajb.1200354. [DOI] [PubMed] [Google Scholar]
- Wang Z, Binder M, Schoch CL, et al. Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol Phylogenet Evol. 2006;41:295–312. doi: 10.1016/j.ympev.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Wang Z, Johnston PR, Yang ZL, Townsend JP. Evolution of reproductive morphology in leaf endophytes. PLoS One. 2009;4:e4246. doi: 10.1371/journal.pone.0004246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kin K, Lopez-Giraldez F, Johannesson H, Townsend J. Sex-specific gene expression during asexual development of Neurospora crassa. Fungal Genet Biol. 2012a;49:533–543. doi: 10.1016/j.fgb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lehr N, Trail F, Townsend J. Differential impact of nutrition on developmental and metabolic gene expression during fruiting body development in Neurospora crassa. Fungal Genet Biol. 2012b;49:405–413. doi: 10.1016/j.fgb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li N, Li J, et al. The fast-evolving phy-2 gene modulates sexual development in response to light in the model fungus Neurospora crassa. MBio. 2016;7:e02148. doi: 10.1128/mBio.02148-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Lopez-Giraldez F, Lehr N, et al. Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryot Cell. 2014;13:154–169. doi: 10.1128/EC.00248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschuk S, Bezerra A, Shi L, Brown L. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci U S A. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 2011;29:1297–1300. doi: 10.1093/molbev/msr311. [DOI] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. Complex patterns of divergence among green-sensitive (RH2a) African cichlid opsins revealed by Clade model analyses. BMC Evol Biol. 2012a;12:206. doi: 10.1186/1471-2148-12-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. An improved likelihood ratio test for detecting site-specific functional divergence among clades of protein-coding genes. Mol Biol Evol. 2012b;29:1297–1300. doi: 10.1093/molbev/msr311. [DOI] [PubMed] [Google Scholar]
- Wu C, Yang F, Smith KM, et al. Genome-wide characterization of light-regulated genes in Neurosproa crassa. G3. 2014 doi: 10.1534/g3.114.012617. G3.114.012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lopez-Giraldez F, Townsend JP. LOX: inferring Level Of eXpression from diverse methods of census sequencing. Bioinformatics. 2010;26:1918–1919. doi: 10.1093/bioinformatics/btq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yu X, Xie B, et al. Transcriptomic profiling-based mutant screen reveals three new transcription factors mediating menadione resistance in Neurospora crassa. Fungal Biol. 2013;117:422–430. doi: 10.1016/j.funbio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Ziebarth JD, Bhattacharya A, Cui Y. Bayesian Network Webserver: a comprehensive tool for biological network modeling. Bioinformatics. 2013;29:2801–2803. doi: 10.1093/bioinformatics/btt472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Amino acid sequences conservation and secondary structure predicted for ascomycetous orthologs of nop-1, orp-1, and carO using Consurf (Ashkenazy et al. 2010). Page 1: Results for nop-1 orthologs, mapped to the N. crassa nop-1 protein sequence. Page 2: Results for nop-1, mapped to the F. graminearum nop-1 protein sequence. Page 3: Results for orp-1, mapped to the F. graminearum orp-1 protein sequence. Page 4: Results for carO, mapped to the F. graminearum carO protein sequence.
Figure S2: Triplet crossing plates between the wild type strains (A and B) and between the nop-1 knockout and the wild type strains (C and D) when cultured under constant light with sealed (A and C) and unsealed plates (B and D). Wild type crosses produced perithecia earlier when cultured under constant white light on sealed plates (A) than when cultured under constant white light on unsealed plates (B). matA Δnop-1 cross with mata wild type exhibited wild type perithecial development when cultured under constant white light on sealed plates (C), but exhibited earlier and more abundant production of perithecia for Δnop-1 when cultured under constant white light on unsealed plates (D). Conidiation (visible as orange pigment) is much more abundant on unsealed plates in all comparisons
Figure S3: Bayesian networks inferred from expression of nop-1 and selected genes involved in oxidative stress pathways, asexual development, and initiation of perithecia. Edges with posterior probability higher than 50% (presence in greater than 50% of 100 models produced) are depicted. (A) For cultures on Bird medium, the inferred network coordinated expression regulation between oxidative pathways and conidiation (asexual development). Genes annotated for perithecial production (sexual development) were sparsely connected within the network, implying less robustly coordinated regulation of the process. (B) For cultures on natural maple sap medium, the inferred network showed dense associations among well-defined subnetworks for oxidative pathways, conidiation and initiation of perithecial development.
Table S1. Genbank accession and annotated opsin-like proteins analyzed in this study.
Table S2. Likelihood ratio test of positive selection (sites model in PAML) analyses on subsets of the fungal opsins gene tree in figure 1.
Table S3. Results of Clade Model C analyses on fungal opsins Bayesian tree.
Table S4. Results of DIVERGE 2.0 analysis on fungal opsin-like proteins.
Table S5: Relative gene expression analysis of 10 microarrays from Chen et al. (2009). Abbreviations: WT: wild type, nop: nop-1 knockout, Ref: pooled RNA reference, D: dark (DD), S: short light exposure (15–30 minutes, LL15, LL30), and L: long light exposure (60–120 minutes, LL60 and LL120).
Table S6: Expression of nop-1 and selected genes involved in oxidative stress pathways, conidiation (asexual development), and initiation of perithecia (sexual development).