Abstract
Proper seed development requires coordinated growth among the three genetically distinct components, the embryo, the endosperm, and the seed coat. In Arabidopsis, embryo growth rate accelerates after endosperm cellularization, which requires a chromatin-remodeling complex, the FIS2-Polycomb Repressive Complex 2 (PRC2). After cellularization, the endosperm ceases to grow and is eventually absorbed by the embryo. This sequential growth pattern displayed by the endosperm and the embryo suggests a possibility that the supply of sugar might be shifted from the endosperm to the embryo upon endosperm cellularization. Since invertases and invertase inhibitors play an important role in sugar partition, we investigated their expression pattern during early stages of seed development in Arabidopsis. Two putative invertase inhibitors (InvINH1 and InvINH2) were identified as being preferentially expressed in the micropylar endosperm that surrounds the embryo. After endosperm cellularization, InvINH1 and InvINH2 were down-regulated in a FIS2-dependent manner. We hypothesized that FIS2-PRC2 complex either directly or indirectly represses InvINH1 and InvINH2 to increase invertase activity around the embryo, making more hexose available to support the accelerated embryo growth after endosperm cellularization. In support of our hypothesis, embryo growth was delayed in transgenic lines that ectopically expressed InvINH1 in the cellularized endosperm. Our data suggested a novel mechanism for the FIS2-PRC2 complex to control embryo growth rate via the regulation of invertase activity in the endosperm.
Keywords: invertase inhibitor, sugar, embryo, endosperm, seed development, PRC2
Introduction
Angiosperm seed is the product of double fertilization (Friedman, 1998; Linkies et al., 2010). During this process, one sperm cell fuses with the egg cell to produce the embryo, while the other sperm cell fuses with the central cell to produce the endosperm (Berger et al., 2008). After fertilization, the ovule integument develops to form the seed coat (Schneitz et al., 1995; Debeaujon et al., 2007). Therefore, seed development involves coordinated growth of three distinct organs: the diploid zygotic embryo, the triploid zygotic endosperm, and the diploid maternal seed coat (Garcia et al., 2005; Yang et al., 2008; Ingram, 2010). Most angiosperms, including Arabidopsis, have nuclear endosperm (Olsen, 2004; Bhojwani, 2009). During the initial phase of nuclear endosperm development, the nuclei divide rapidly without cellularization, forming a syncytium (Boisnard-Lorig et al., 2001). The syncytial phase is followed by endosperm cellularization, which coincides with the transition in the embryo from morphogenesis phase to growth phase (Goldberg et al., 1994; Olsen, 2004; Hehenberger et al., 2012). In addition to coordinated transition in development, the embryo and the endosperm also exhibit coordinated changes in growth rate. In eudicots with transient endosperm, embryo growth accelerates after the transition, while the endosperm stops growing soon after the transition and is eventually absorbed by the expanding embryo (Goldberg et al., 1994; Olsen, 2004; Baud et al., 2008; Hehenberger et al., 2012). This sequential growth pattern suggests that nutrient supplies are shifted from the endosperm to the embryo after endosperm cellularization.
The developmental transition in the endosperm is likely responsible for the acceleration in embryo growth rate after endosperm cellularization. Works in both Arabidopsis and rice have demonstrated that a chromatin-remodeling complex produced in the endosperm, the Polycomb Repressive Complex 2 (PRC2), is required for endosperm cellularization and the acceleration in embryo growth rate (Kiyosue et al., 1999; Sørensen et al., 2001; Folsom et al., 2014). In PRC2 mutants, such as mea, fis2, fie, and msi1, the endosperm fails to cellularize and continues to proliferate, while the embryo fails to transition into the growth phase and aborts at heart stage (Ohad et al., 1996; Chaudhury et al., 1997; Kiyosue et al., 1999; Köhler et al., 2003). The PRC2 complex regulates many developmental processes by methylating histone H3 lysine 27 (H3K27me) to initiate gene silencing (Schubert et al., 2005; Schuettengruber et al., 2007; Zheng and Chen, 2011; Holec and Berger, 2012). These data indicate that PRC2-mediated gene silencing in the endosperm is involved in the regulation of nutrient allocation from the endosperm to the embryo.
Invertase plays an important role in sugar allocation during seed development. Since there is no symplastic connection between the seed coat, the endosperm, and the embryo (Stadler et al., 2005), active transport is required to move nutrients from maternal tissues to the endosperm at the chalazal interface, and from the endosperm to the embryo at the micropylar interface (Sanders et al., 2009; Pommerrenig et al., 2013; Chen et al., 2015; Sosso et al., 2015). At the chalazal interface, sucrose is unloaded from the phloem to apoplastic space, hydrolyzed by cell-wall-bound invertase to glucose and fructose, and subsequently imported into the endosperm (Cheng et al., 1996; Cheng and Chourey, 1999). Sucrose hydrolysis generates the concentration gradient that facilitates sucrose unloading from the seed coat to the endosperm at the chalazal interface (Weber et al., 1996; Sherson et al., 2003). However, it is not clear whether invertase is a part of the sugar transport mechanism at the micropylar interface.
There are two types of invertase: the acid invertase in the vacuole or the cell wall, and the neutral/alkaline invertase in the cytoplasm (Sturm, 1999). Since the acid invertases are relatively stable, their activity is mainly regulated by small proteinous invertase inhibitors (InvINHs) (Ruan et al., 2010). InvINHs and pectin methylesterase inhibitors (PMEIs) belong to the same superfamily that are characterized by four conserved cysteine residues (Camardella et al., 2000). To investigate whether invertases and InvINHs are involved in sugar transport across the micropylar interface, we analyzed the spatial and temporal expression pattern of invertase and the members of InvINH/PMEI superfamily during seed development. We identified two putative InvINHs (InvINH1 and InvINH2) that were specifically expressed in the micropylar endosperm. Moreover, both genes were silenced by FIS2-PRC2 complex after endosperm cellularization. Finally, ectopic expression data suggested that InvINH1 inhibited embryo growth.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 and fis2-8 mutant (Wang et al., 2006) plants were housed in a walk-in Environmental Room (Norlake Scientific, Hudson, WI, United States) at 22°C under 16-h light/8-h dark long-day condition. Seeds were stratified for 4 days at 4°C before germination. Plants were grown in Pro-Mix BX soil (Premier Horticulture, Quakertown, PA, United States) and fertilized with Peters 20-20-20 (Scotts-Sierra Horticultural Products Company, Marysville, OH, United States).
Microarray Data Analysis
Microarray data was downloaded from Gene Expression Omnibus database (accession no. GSE12404). The expression values were normalized with the GeneChip Robust Multiarray Averaging method (GC-RMA) implemented as a Bioconductor package under the R platform1 (Wu et al., 2004). Hierarchical cluster analysis (average linkage and Euclidean distance as similarity measure) was performed using Cluster 3.0 on log2-transformed expression values (de Hoon et al., 2004). The result of cluster analysis was visualized using Java TreeView (Saldanha, 2004).
The gene IDs for eight acid invertases (Glycoside Hydrolase Family 32) and nine neutral/alkaline invertases (Glycoside Hydrolase Family 100) were obtained from Carbohydrate-Active enZYmes Database2 (Sturm et al., 1999; Lammens et al., 2009). The gene IDs for 125 members of the plant InvINH/PMEI superfamily were obtained from SUPERFAMILY database3 (Wilson et al., 2009). Cluster analysis was conducted on 107 genes that are present on Affymetrix Arabidopsis ATH1 Genome Array, including 17 invertase genes and 90 InvINH/PMEI genes.
Bioinformatics Analyses
The following online programs were used to predict the subcellular localizations of InvINH1 and InvINH2: PSORT4 (Nakai and Kanehisa, 1991), MultiLoc25 (Blum et al., 2009), and YLoc6 (Briesemeister et al., 2010). All three programs were run with the default setting for plant proteins.
Genevestigator7 (Hruz et al., 2008) was used to analyze the expression pattern of InvINH1 and InvINH2. The data selection includes a compendium of 5,825 wild-type A. thaliana samples profiled on the Affymetrix Arabidopsis ATH1 Genome Array platform. The Anatomy tool and Perturbations tool from the CONDITION SEARCH toolset were used to analyze the expression level of InvINH1 and InvINH2, which are represented by the same Affymetrix probe (248823_s_at).
RNA Isolation and Quantitative RT-PCR
Total RNAs were extracted from roots, stems, rosette leaves, closed floral buds from stage 0 to 12 (Smyth et al., 1990), and young siliques at 3 and 5 days after pollination (dap) following a modified hot borate extraction method (Wan and Wilkins, 1994). The first-strand cDNAs were synthesized with the RETROscript kit (Ambion, Inc., Austin, TX, United States). Quantitative PCR (qPCR) was performed with the SYBR Select Master Mix and the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, United States). The primers used for qPCR are as follows: InvINH1, forward 5′-ctgagtgctgctttggatgta-3′, reverse 5′-gttctcgttggtaatcggagac-3′; InvINH2 forward 5′-aagacccgcaatcgtcatac-3′, reverse 5′-gtcgatgctagggccaaatc-3′; and Actin2 forward 5′-tccctcagcacattccagcagat-3′, reverse 5′-aacgattcctggacctgcctcatc-3′. CT values were normalized against Actin2. The use of Actin2 as the reference gene for qPCR analysis has been previously described (Wang et al., 2010). Most tissues were analyzed with three biological replicates except for silique tissues, which were analyzed with two biological replicates and two technical replicates. The mRNA level of InvINH1 or InvINH2 in 3-dap wild-type siliques was set as the reference point (100%) to calculate the relative mRNA levels in other tissues following the -ΔΔCT method (Livak and Schmittgen, 2001).
Constructs and Plant Transformation
The 5′ flanking region of InvINH1 or InvINH2 was cloned into the binary vector pBN-GFP (Wang et al., 2006) to create promoter fusion constructs. In brief, the 5′ flanking regions of InvINH1 (1172 bp) and InvINH2 (2238 bp), including the entire 5′ intergenic region and the coding region encoding the first seven amino acids, were amplified from Col-0 genomic DNA using Phusion Polymerase (Thermo Fisher, Waltham, MA, United States). The 5′ UTR region and the first seven amino acids were included in the promoter fusion to ensure proper translation of the GFP gene. Since the N-terminal signal peptide for InvINH1 and InvINH2 was predicated to be 20-amino-acid long, the inclusion of the first seven amino acids is unlikely to change the subcellular localization of the GFP protein. The primers used for amplification are as follows: InvINH1 forward primer (5′-aatgtctagagctgaaatgaaactacatgtgc-3′), InvINH2 forward primer (5′-cgtttctagacgtctccgattaccaacga-3′), and a common reverse primer (5′-gagaaggatcccaatgaaaccaagaacttcat-3′). The amplified fragments were cloned in frame into the pBN-GFP vector between the XbaI and BamHI sites.
The ectopic expression construct for InvINH1 was generated by cloning the ZOU promoter and InvINH1 coding region into the binary vector pBN (Wang et al., 2006). Since InvINH1 has no intron, both InvINH1 coding region and Zou promoter were amplified from Col-0 genomic DNA using Phusion Polymerase (Thermo Fisher). The Zou promoter region (-2009 bp to +18 bp) was amplified with forward primer (5′-tgattacgccaagcttgtgttacgttgtaacgaattt-3′) and reverse primer (5′-tgctcaccatggatccctcttgagcattagtcatattg-3′), then cloned into pBN vector between the HindIII and BamHI site, resulting in construct pBN-pZOU. Next, the InvINH1 coding region (525 bp) was amplified with forward primer (5′-attaggatccatgaagttcttggtttcattggt-3′) and reverse primer (5′-gataggtaccttacaacatattagtaaaagccaaagga-3′), then cloned into pBN-pZOU in between the BamHI and KpnI sites, resulting in construct pBN-pZou-InvINH1. All constructs were verified by sequencing.
Arabidopsis plant transformation was carried out as described previously (Wang et al., 2006). In brief, Agrobacterium tumefaciens strain GV3101 pMP90 (Koncz and Schell, 1986) carrying the appropriate binary vector was used to perform the standard floral dip method (Clough and Bent, 1998). Transgenic seedlings were selected on 0.5x Murashige and Skoog (MS) media containing 35 μg/ml Kanamycin. The presence of the transgene in T1 plants was confirmed using PCR.
Image Collection and Processing
Seeds were dissected out of the silique as described previously (Wang et al., 2010). To isolate the embryos, seeds were punctured with a dissecting needle and then gently pressed to release the embryos. GFP expression pattern in whole-mount seeds were obtained with a Zeiss LSM 700 inverted confocal microscope (Carl Zeiss, Oberkochen, Germany). Dissected embryos were imaged with Nikon C-DS stereoscopic microscope (Nikon, Tokyo, Japan) equipped with an AxioCam Icc1 digital camera imaging kit (Carl Zeiss). Image processing was performed with Adobe Photoshop CS (Adobe Systems Inc., San Jose, United States).
Results
Expression Profiling of Invertases and InvINHs/PMEIs during Seed Development
In Arabidopsis, embryo growth rate accelerates after endosperm cellularization (Goldberg et al., 1994; Baud et al., 2008; Hehenberger et al., 2012). To investigate whether the change in embryo growth rate is correlated with any change in invertase activity, we analyzed the temporal and spatial expression pattern of invertase and InvINHs by performing hierarchical cluster analysis on a published seed microarray dataset (Belmonte et al., 2013). The dataset includes six developmental time points from pre-globular stage to mature green embryo stage (Figure 1). Each stage contains five to six distinct seed compartments captured with laser capture microdissection (Figure 1). The list of genes included in our analysis was described in Section “Materials and Methods.” Since it is difficult to distinguish InvINHs from PMEIs based on sequence conservation alone (Hothorn et al., 2004), we included all members of the InvINH/PMEI superfamily in our analysis.
FIGURE 1.
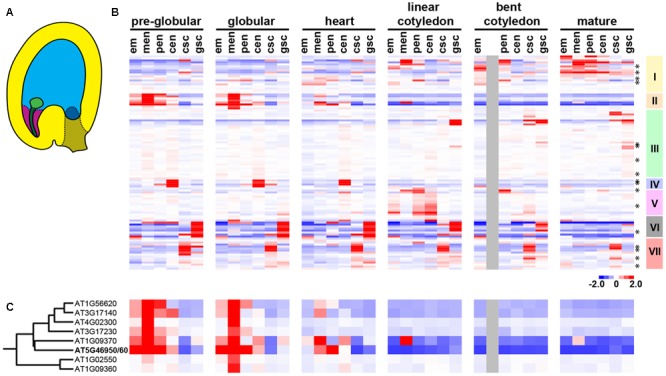
The expression pattern of invertase and members of the InvINH/PMEI superfamily during seed development. (A) Illustration depicting a globular stage seed containing embryo proper (light green), suspensor (dark green), chalazal seed coat (brown), general seed coat (yellow), as well as micropylar (purple), peripheral (light blue), and chalazal (dark blue) endosperm. (B) Hierarchical clustering of 35 seed samples (in columns) and 107 genes (in rows), including 17 invertases (indicated by asterisks) and 90 InvINH/PMEIs. The early developmental transition in the embryo and the endosperm occurs at the heart stage. Developmental stages are indicated by the stage of embryo development. Each stage includes six seed compartments arranged from left to right as embryo proper (em), micropylar endosperm (men), peripheral endosperm (pen), chalazal endosperm (cen), chalazal seed coat (csc), and general seed coat (gsc). The color represents gene expression level relative to the median level of expression across all samples. Red, high expression; blue, low expression; gray, data not available. Seven clusters of co-regulated genes (I–VII) were identified based on the gene dendrogram. (C) A zoomed-in heatmap for Cluster II genes with the data points arranged in the same sequence as (B).
Hierarchical cluster analysis revealed seven co-regulated gene clusters that share distinct spatial and temporal expression patterns during seed developments (Figure 1). Some of the co-regulated clusters were specific to seed compartments located at the interface of nutrient transfer, such as chalazal endosperm (Figure 1B, Cluster IV), chalazal seed coat (Figure 1B, Cluster VII), general seed coat (Figure 1B, Cluster VI), and micropylar endosperm (Figure 1B, Cluster II). Among these four seed compartments, chalazal endosperm, chalazal seed coat, and general seed coat are located at the seed coat/endosperm interface, suggesting that clusters IV, VI, and VII genes might be involved in nutrient transfer from the maternal tissues to the endosperm during seed development (Li and Berger, 2012). Since we were interested in the nutrient transfer mechanism that regulates embryo growth rate, we focused on Cluster II genes that were specifically expressed in the micropylar endosperm surrounding the embryo (Figure 1B). Cluster II contains nine members of the plant InvINH/PMEI superfamily represented by eight Affymetrix probes (Figure 1C). After the transition at heart-stage (Figure 1), cluster II genes were down-regulated in the endosperm (Figure 1B), which is expected to cause an increase in invertase or pectin methylesterase activity after endosperm cellularization.
InvINH1 and InvINH2 Were Specifically Expressed in Reproductive Tissues
To validate the microarray data, two genes from Cluster II were selected for additional expression analysis. These two genes were tentatively named as InvINH1 (At5g46960) and InvINH2 (At5g46950). InvINH1 and InvINH2 are represented by the same Affymetrix probe due to 93.5% DNA sequence identity between the two genes. Both InvINH1 and InvINH2 proteins are 174 amino acids long and are 88.5% identical to each other. We used three different methods to predict the subcellular location of InvINH1 and InvINH2. The probability of InvINH1 and InvINH2 to be localized to the extracellular space is 75.2, 99.9, and 68% according to PSORT (Nakai and Kanehisa, 1991), Yloc (Briesemeister et al., 2010), and MultiLoc2 (Blum et al., 2009), respectively. The PSORT program predicted that both InvINH1 and InvINH2 have a 20-amino-acid-long N-terminal signal peptide. In addition, InvINH1 has been experimentally confirmed to be a secreted protein associated with the plant cell wall (Irshad et al., 2008), which supports the possibility of InvINH1 to act as an inhibitor of cell-wall-associated enzymes.
To determine whether InvINH1 and InvINH2 are specifically expressed in the seed, we used qRT-PCR to analyze the mRNA level of InvINH1 and InvINH2 in both vegetative tissues (roots, stems, and rosette leaves) and reproductive tissues (closed floral buds, 3-dap siliques, and 5-dap siliques). Among the reproductive tissues, the 3-dap siliques contained globular-stage embryo and syncytial endosperm, while the 5-dap siliques contained early-torpedo stage embryo and cellularized endosperm. Since InvINH1 and InvINH2 are 93.5% identical in DNA sequence, gene-specific qRT-PCR primers were first designed to distinguish InvINH1 from InvINH2. In wild-type plants, the expression level of InvINH1 and InvINH2 were dramatically higher in reproductive tissues than in vegetative tissues (Figure 2). InvINH1 was primarily expressed in closed floral buds and 3-dap siliques, while InvINH2 was primarily expressed in 3-dap siliques (Figure 2). Both InvINH1 and InvINH2 were strongly down-regulated in 5-dap siliques compared to 3-dap siliques (Figure 2). Therefore, both the qRT-PCR and the microarray data suggested that InvINH1 and InvINH2 were expressed in the seeds during the syncytial phase and are down-regulated after endosperm cellularization.
FIGURE 2.
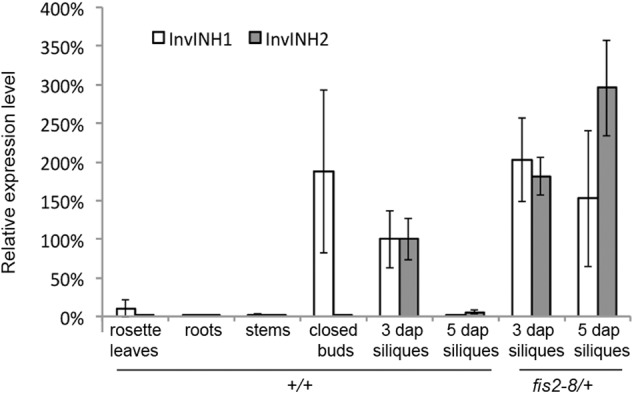
The expression pattern of InvINH1 and InvINH2 in wild type (+/+) and fis2-8/+ mutant plants. The mRNA level of InvINH1 and InvINH2 was determined by qRT-PCR in rosette leaves, roots, stems, closed floral buds, and whole siliques at 3 and 5 dap. The expression level in wild-type 3-dap siliques was set as 100%, which was used as a reference point to calculate the relative expression level in other tissues.
In support of our qRT-PCR data, the silique was identified as the structure with the highest expression level for InvINH1 and InvINH2 among the 87 anatomical structures annotated by Genevestigator (Supplementary Figure S1A). Genevestigator analysis also revealed that the expression of InvINH1 and InvINH2 were regulated by additional developmental cues and environmental stimuli (Supplementary Figure S1B). For example, InvINH1 and InvINH2 were up-regulated during germination (Narsai et al., 2011) and upon Pseudomonas syringae inoculation (GEO accession GSE5520, GSE18978). The down-regulation of InvINH1 and InvINH2 was observed in the endosperm/seed coat fraction from germinating seeds upon ABA treatment (GEO accession GSE5751), and in seedlings upon exposure to sucrose (Stokes et al., 2013). Moderate expression was also detected in senescent leaves (Supplementary Figure S1A). In general, InvINH1 and InvINH2 are expressed in tissues that undergo active sugar reallocation, such as the endosperm, germinating seeds, and senescent leaves. Collectively, theses data suggested that InvINH1 and InvINH2 are potentially involved in other biological processes in addition to their function during early endosperm development.
InvINH1 and InvINH2 Were Specifically Expressed in Syncytial Micropylar Endosperm
To investigate the spatial and temporal expression pattern of InvINH1 and InvINH2 during seed development, we generated promoter-GFP fusions for these two genes and analyzed GFP expression pattern in stable transgenic plants. The InvINH1 promoter-GFP signal was detected in both the female gametophyte and the syncytial endosperm (Figures 3A–K). Briefly, the InvINH1 promoter-GFP activity was detectable in stage FG4 female gametophyte (4-nucleate, Christensen et al., 1997, Figure 3A). In the mature female gametophyte at 1 day after emasculation, the promoter activity was only present in the central cell (Figure 3B). After fertilization, the promoter-GFP signal was more prevalent in the syncytial endosperm from endosperm stage I (one nucleus) to stage VIII (∼100 nuclei, Boisnard-Lorig et al., 2001, Figures 3C–J). After endosperm cellularization at stage IX (Figure 3K), the GFP signal decreased dramatically and was no longer detectable in seeds containing heart stage embryos (Figure 3L). In seeds that contained globular-stage embryo and syncytial endosperm, the GFP signal was primarily detected in the micropylar endosperm, with weak expression in the periphery endosperm and no expression in the chalazal endosperm (Figure 3J). Promoter-GFP expression pattern similar to that of InvINH1 was also observed for InvINH2 (Figures 3M–X), except that InvINH2 promoter-GFP signal was not detectable in the female gametophyte at 1 day after emasculation (Figure 3N). The observed GFP expression pattern was consistent among the 13 analyzed T1 lines for pInvINH1-GFP, as well as among the 11 analyzed T1 lines for pInvINH2-GFP. Therefore, both InvINH1 and InvINH2 were preferentially expressed in the micropylar endosperm prior to endosperm cellularization, which was in good agreement with the microarray and qRT-PCR data.
FIGURE 3.
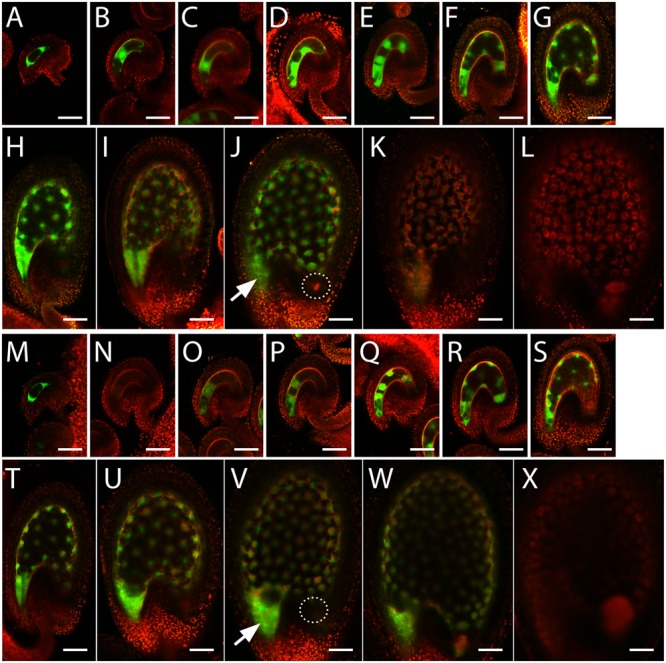
InvINH1 and InvINH2 promoter activity in the female gametophyte and the endosperm. Promoter-GFP fusions for InvINH1 (A–L) and InvINH2 (M–X) were analyzed by confocal microscopy in the ovules isolated from stage 12 flowers (Smyth et al., 1990) containing stage FG4 (Christensen et al., 1997) female gametophyte (A,M) and from flowers at 1 day after emasculation (B,N). The GFP signals in the fertilized seeds were analyzed at endosperm stage I (C,O), II (D,P), III (E,Q), IV (F,R), V (G,S), VI (H,T), VII (I,U), VIII (J,V), and IX (K,W) (Boisnard-Lorig et al., 2001), as well as in seeds containing heart stage embryos (L,X). The embryo stage in (L,X) was verified by dissection. GFP (green) and auto-fluorescent signals (red) were imaged simultaneously and merged. All the images were oriented with the micropylar ends on the left and the chalazal ends on the right. Arrow indicates micropylar endosperm. Dashed circle indicates the location of chalazal endosperm. Bar = 50 μm.
Embryo Growth Was Inhibited by Ectopically Expressed InvINH1
Based on the specific expression pattern of InvINH1 and InvINH2, we hypothesized that their function is to inhibit cell-wall-bound enzymes, such as invertase or pectin methylesterase, that are located at the embryo-endosperm interphase prior to endosperm cellularization. To investigate whether the down-regulation of InvINH1 after endosperm cellularization is connected to the acceleration in embryo growth rate, we ectopically expressed InvINH1 after endosperm cellularization using the ZOU promoter, which is preferentially active in the micropylar endosperm at both the syncytial stage and the cellularized stage (Yang et al., 2008). Since InvINH1 and InvINH2 share similar expression pattern (Figure 3) and 93.5% similarity in DNA sequence, there is a possibility that these two genes are functionally redundant. Moreover, it is difficult to generate a double mutant to address the redundancy issue, because these two genes are only 2 kb apart. Therefore, we decided to use the ectopic expression approach to investigate the function of InvINH1 during seed development. We generated transgenic plants carrying pZOU-InvINH1 and analyzed seed morphology in 21 independent T1 lines. Embryo morphology was analyzed at the late-bent-cotyledon stage. Six out of the 21 T1 lines had more than 25% delayed embryos within a given silique (Figure 4A). Within the same silique from a T1 hemizygous transgenic plant, the normal embryos were at the late-bent-cotyledon stage, while the delayed embryos ranged from early torpedo to mid-bent-cotyledon stage (Figure 4C). The delay in embryo growth was transient. At maturation stage, most of the seeds contained normal size embryos. Since the delay in embryo growth was not observed in all the lines, we next investigated if the expression level of the transgene is variable among the T1 lines. We analyzed the mRNA level of InvINH1 at 5dap in six selected T1 lines that represented different degrees of delayed embryo phenotype (Figure 4D). In general, there was a good correlation between the expression level of InvINH1 and the severity of the phenotype. As the control, we generated and analyzed 20 T1 lines carrying the promoter-only pZOU transgene (Figure 4B). Seeds containing delayed embryos were observed occasionally. However, none of the pZOU lines contained more than 25% delayed seeds (Figure 4B). We also observed semi-sterility lines containing close to 50% undeveloped ovules for both pZou and pZou-InvINH1 transgene (Figures 4A,B). Semi-sterility is caused by chromosomal rearrangement during T-DNA mediated transformation (Nacry et al., 1998). Since semi-sterility was observed for both pZou and pZou-InvINH1, this phenotype was not associated with the ectopic expression of InvINH1. Collectively, our data indicated that prolonged InvINH1 expression is sufficient to delay embryo growth.
FIGURE 4.
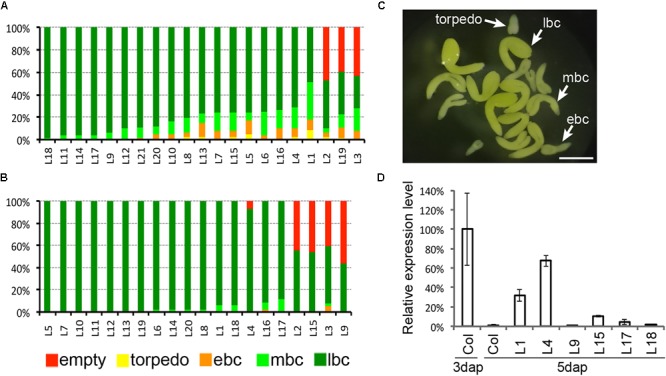
Prolonged expression of InvINH1 delayed embryo growth. (A) The embryo phenotype of 21 individual T1 plants carrying pZOU-InvINH1 transgene. (B) The embryo phenotype of 20 individual T1 plants carrying pZou transgene. (C) Dissected embryos from the same silique of a T1 plant carrying pZOU-InvINH1. (D) Ectopic expression of InvINH1 in pZOU-InvINH1 transgenic plants. The InvINH1 mRNA level in whole siliques was determined by qRT-PCR at 5 dap. The expression level in wild-type (Col-0) siliques at 3 dap was used as a reference point (100%) to calculate the relative expression level in other samples. Around 50–60 seeds were dissected per line to determine the embryo phenotype. empty, undeveloped ovules; ebc, early-bent-cotyledon stage; mbc, mid-bent-cotyledon stage; lbc, late-bent-cotyledon stage (lbc). Bar = 500 μm.
InvINH1 and InvINH2 Were Up-Regulated in fis2 Mutant
Since embryo growth was severely suppressed in mea, fis2, fie, and msi1 mutant (Ohad et al., 1996; Chaudhury et al., 1997; Kiyosue et al., 1999; Köhler et al., 2003), we next investigated whether the embryo abortion phenotype in these mutants could be attributed to any changes in InvINH1 and InvINH2 expression level. We used qRT-PCR to compare the mRNA level of InvINH1 and InvINH2 between the wild-type and fis2-8 mutant siliques (Figure 2). In wild-type plants, InvINH1 and InvINH2 were expressed in 3-dap siliques containing syncytial endosperm, and down-regulated in 5-dap siliques containing cellularized endosperm (Figure 2). In fis2 mutant plants, the expression level of InvINHs1 and InvINH2 were up-regulated at both 3 and 5 dap (Figure 2). More specifically, roughly 2-fold up-regulation was detected at 3 dap, while at least 80-fold up-regulation was detected at 5 dap. The elevation in InvINH1 and InvINH2 expression level at 5 dap was not simply the consequence of extended syncytial stage in fis2 mutant, because elevated expression was detected as early as 3 dap when there was no morphological difference between the wild-type and the mutant seeds. Our data indicated that FIS2-PRC2 complex was required to directly or indirectly silence the expression of InvINH1 and InvINH2, which is expected to increase the activity of cell-wall-bound enzymes, such as invertase or pectin methylesterase after endosperm cellularization.
Discussion
Proper seed development requires coordinated growth among the embryo, the endosperm, and the seed coat (Garcia et al., 2005; Yang et al., 2008; Ingram, 2010). To determine if embryo growth rate is correlated with invertase activity, we analyzed the spatial and temporal expression pattern of invertases and InvINH/PMEI-related genes during Arabidopsis seed development. Our analysis revealed distinct gene clusters that were specifically expressed at the interface between the seed coat and the endosperm, as well as between the endosperm and the embryo. Among these genes, two putative InvINHs (InvINH1 and InvINH2) were specifically expressed in the syncytial endosperm surrounding the embryo. After endosperm cellularization, the down-regulation of InvINH1 and InvINH2 was dependent on FIS2 function. Moreover, embryo growth was suppressed by the ectopic expression of InvINH1 in the cellularized micropylar endosperm. Collectively, our data suggested a novel mechanism for the FIS2-PRC2 complex to control embryo growth rate through the repression of InvINH1 in the micropylar endosperm.
InvINH1 was recently reported as a PMEI (PMEI12) that conferred pathogen resistance by inhibiting PME and strengthening the cell wall during infection (Lionetti et al., 2017). However, this report didn’t directly demonstrate that InvINH1/PMEI12 inhibited pectin methylesterase. InvINHs and PMEIs employ similar scaffold to inhibit two very different enzymes, invertase and pectin methylesterase (Hothorn et al., 2010). However, the conservation in tertiary structure between InvINHs and PMEIs is not reflected in any significant conservation in the primary sequence (Hothorn et al., 2010). Even though several studies have attempted to identify conserved sequence motifs that distinguish InvINHs from PMEIs (Hothorn et al., 2004, 2010; Di Matteo et al., 2005), there are still exemptions to the rule. For example, the PKF motif was suggested as a distinguishing feature for InvINHs (Hothorn et al., 2010). However, this motif is not present in AtC/VIF2, which has been shown to inhibit cell-wall bound invertase in vitro (Link et al., 2004). With only a handful of functionally characterized InvINHs and PMEIs, it is still unreliable to distinguish InvINHs from PMEIs based on their primary sequence. Therefore, whether InvINH1 is InvINH or PMEI still remains to be determined by direct enzymatic assay.
InvINH1-mediated suppression of embryo growth may occur via two different mechanisms depending on whether InvINH1 targets invertase or pectin methylesterase. As a pectin methylesterase inhibitor, InvINH1 likely restricts embryo growth via the modification of cell wall composition, since pectin methylesterase is a cell-wall modification enzyme that dimethylesterify cell wall polygalacturonans (Micheli, 2001). As an invertase inhibitor, InvINH1 likely restricts embryo growth by slowing down the flow of sucrose from the endosperm to the embryo before endosperm cellularization. Several lines of evidence suggested that the transport of sucrose from the endosperm to the embryo is important for embryo growth. Based on [14C]sucrose tracing experiment, sugar is likely exported as sucrose from the endosperm into the apoplastic space that surrounds the embryo (Morley-Smith et al., 2008). In addition, embryo growth rate was suppressed in mutants that lack functional sucrose transporters in the endosperm and the seed coat (Baud et al., 2005; Chen et al., 2015). Since invertase hydrolyzes sucrose and facilitates sugar transport (Ruan et al., 2010), InvINH1-mediated inhibition of invertase activity could explain the slow embryo growth rate before endosperm cellularization.
The discovery of InvINH1 and InvINH2 provided a missing link between FIS2-PRC2-mediated developmental transition in the endosperm and the accelerated embryo growth that follows. Several attempts have been made to identify the genes targeted by FIS2-PRC2 complex during endosperm cellularization (Tiwari et al., 2010; Weinhofer et al., 2010). However, it has been difficult to tease out the syncytial program from other developmental programs that are suppressed by the FIS2-PRC2 complex, such as the flowering and embryonic programs (Makarevich et al., 2006; Weinhofer et al., 2010). InvINH1 is the first structural gene from the collection of known FIS2-PRC2 targets that might offer an explanation why embryo growth during syncytial endosperm phase is limited. In addition to being up-regulated in fis mutants such as mea and fis2, InvINH1 and InvINH2 were also up-regulated in interploidy crosses with excess paternal genome (Erilova et al., 2009; Tiwari et al., 2010). Since paternal-excess cross leads to prolonged syncytial stage (Scott et al., 1998), both these studies and our data suggested that InvINH1 and InvINH2 are specifically associated with the syncytial endosperm program. Furthermore, our data suggested that the expression of InvINH1 during the syncytial stage may be connected to the slow embryo growth rate observed before endosperm cellularization (Baud et al., 2008).
It remains to be determined whether InvINH1 and InvINH2 are direct or indirect targets of the FIS2-PRC2 complex. The FIS2-PRC2 complex maintains gene silencing and genomic imprinting of several endosperm-expressed genes through the methylation of H3K27 (Köhler et al., 2005; Baroux et al., 2006; Fitz Gerald et al., 2009). However, InvINH1 and InvINH2 have not been identified as imprinted genes (Gehring et al., 2011; Hsieh et al., 2011; McKeown et al., 2011; Wolff et al., 2011), nor have they displayed significant enrichment of methylated H3K27 (Weinhofer et al., 2010). Therefore, InvINH1 and InvINH2 may not be directly targeted by FIS2-PRC2 complex. Instead, additional regulators may exist to connect InvINH1 and InvINH2 to the FIS2-PRC2 regulatory network. Future studies aimed at identifying the upstream regulators of InvINH1 and InvINH2 will provide a more definitive answer to this question.
Author Contributions
DW designed research and wrote the paper. DW, BZ, and MD performed research and analyzed data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer T-FH and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported by grant to DW (#1656556) from the National Science Foundation (NSF). BZ and MD were supported by the ASPIRE program (NSF, #0714553) and the CURE program (NSF, #1436759), respectively.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00061/full#supplementary-material
In silico expression analysis of InvINH1 and InvINH2. (A) The relative expression level was illustrated as a heatmap containing 87 anatomical features that were hierarchically organized by the Anatomy tool from Genevestigator. (B) The Perturbations tool from Genevestigator was used to generate the heatmap displaying the experimental stimuli that induced greater than 1.5-fold changes in the expression level of InvINH1 and InvINH2. InvINH1 and InvINH2 are represented by the same Affymetrix probe, 248823_s_at.
References
- Baroux C., Gagliardini V., Page D. R., Grossniklaus U. (2006). Dynamic regulatory interactions of polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev. 20 1081–1086. 10.1101/gad.378106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Dubreucq B., Miquel M., Rochat C., Lepiniec L. (2008). Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. Arab. Book Am. Soc. Plant Biol. 6:e0113. 10.1199/tab.0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Wuillème S., Lemoine R., Kronenberger J., Caboche M., Lepiniec L., et al. (2005). The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant J. Cell Mol. Biol. 43 824–836. 10.1111/j.1365-313X.2005.02496.x [DOI] [PubMed] [Google Scholar]
- Belmonte M. F., Kirkbride R. C., Stone S. L., Pelletier J. M., Bui A. Q., Yeung E. C., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. U.S.A. 110 E435–E444. 10.1073/pnas.1222061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Hamamura Y., Ingouff M., Higashiyama T. (2008). Double fertilization - caught in the act. Trends Plant Sci. 13 437–443. 10.1016/j.tplants.2008.05.011 [DOI] [PubMed] [Google Scholar]
- Bhojwani S. P. B. S. S. (2009). The Embryology Of Angiosperms, 5E. Available at: https://books.google.com.np/books?id=dD5NvUGbnaYC&hl=en [Google Scholar]
- Blum T., Briesemeister S., Kohlbacher O. (2009). MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformat. 10:274. 10.1186/1471-2105-10-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisnard-Lorig C., Colon-Carmona A., Bauch M., Hodge S., Doerner P., Bancharel E., et al. (2001). Dynamic analyses of the expression of the HISTONE::YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13 495–509. 10.1105/tpc.13.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briesemeister S., Rahnenführer J., Kohlbacher O. (2010). Going from where to why–interpretable prediction of protein subcellular localization. Bioinforma. Oxf. Engl. 26 1232–1238. 10.1093/bioinformatics/btq115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camardella L., Carratore V., Ciardiello M. A., Servillo L., Balestrieri C., Giovane A. (2000). Kiwi protein inhibitor of pectin methylesterase amino-acid sequence and structural importance of two disulfide bridges. Eur. J. Biochem. 267 4561–4565. 10.1046/j.1432-1327.2000.01510.x [DOI] [PubMed] [Google Scholar]
- Chaudhury A. M., Ming L., Miller C., Craig S., Dennis E. S., Peacock W. J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94 4223–4228. 10.1073/pnas.94.8.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Q., Lin I. W., Qu X.-Q., Sosso D., McFarlane H. E., Londoñ;o A., et al. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27 607–619. 10.1105/tpc.114.134585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.-H., Chourey P. S. (1999). Genetic evidence that invertase-mediated release of hexoses is critical for appropriate carbon partitioning and normal seed development in maize. Theor. Appl. Genet. 98 485–495. 10.1007/s001220051096 [DOI] [Google Scholar]
- Cheng W. H., Taliercio E. W., Chourey P. S. (1996). The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8 971–983. 10.1105/tpc.8.6.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C. A., King E. J., Jordan J. R., Drews G. N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10 49–64. 10.1007/s004970050067 [DOI] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- de Hoon M. J. L., Imoto S., Nolan J., Miyano S. (2004). Open source clustering software. Bioinforma. Oxf. Engl. 20 1453–1454. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- Debeaujon I., Lepiniec L., Pourcel L., Routaboul J.-M. (2007). “Seed coat development and dormancy,” in Annual Plant Reviews: Seed Development, Dormancy and Germination Vol. 27 eds Bradford K. J., Nonogaki H. (Hoboken, NJ: Blackwell Publishing Ltd; ) 25–49. 10.1002/9780470988848.ch2 [DOI] [Google Scholar]
- Di Matteo A., Giovane A., Raiola A., Camardella L., Bonivento D., Lorenzo G. D., et al. (2005). Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell Online 17 849–858. 10.1105/tpc.104.028886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erilova A., Brownfield L., Exner V., Rosa M., Twell D., Mittelsten Scheid O., et al. (2009). Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLOS Genet. 5:e1000663. 10.1371/journal.pgen.1000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz Gerald J. N., Hui P. S., Berger F. (2009). Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Dev. Camb. Engl. 136 3399–3404. 10.1242/dev.036921 [DOI] [PubMed] [Google Scholar]
- Folsom J. J., Begcy K., Hao X., Wang D., Walia H. (2014). Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165 238–248. 10.1104/pp.113.232413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman W. E. (1998). The evolution of double fertilization and endosperm: an “historical” perspective. Sex. Plant Reprod. 11 6–16. 10.1007/s004970050114 [DOI] [Google Scholar]
- Garcia D., Gerald J. N. F., Berger F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17 52–60. 10.1105/tpc.104.027136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Missirian V., Henikoff S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLOS ONE 6:e23687. 10.1371/journal.pone.0023687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., de Paiva G., Yadegari R. (1994). Plant embryogenesis: zygote to seed. Science 266 605–614. 10.1126/science.266.5185.605 [DOI] [PubMed] [Google Scholar]
- Hehenberger E., Kradolfer D., Kohler C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139 2031–2039. 10.1242/dev.077057 [DOI] [PubMed] [Google Scholar]
- Holec S., Berger F. (2012). Polycomb group complexes mediate developmental transitions in plants. Plant Physiol. 158 35–43. 10.1104/pp.111.186445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Van den Ende W., Lammens W., Rybin V., Scheffzek K. (2010). Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc. Natl. Acad. Sci. U.S.A. 107 17427–17432. 10.1073/pnas.1004481107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn M., Wolf S., Aloy P., Greiner S., Scheffzek K. (2004). Structural insights into the target specificity of plant invertase and pectin methylesterase inhibitory proteins. Plant Cell 16 3437–3447. 10.1105/tpc.104.025684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinforma. 2008:420747. 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.-F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M. J., et al. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108 1755–1762. 10.1073/pnas.1019273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G. C. (2010). Family life at close quarters: communication and constraint in angiosperm seed development. Protoplasma 247 195–214. 10.1007/s00709-010-0184-y [DOI] [PubMed] [Google Scholar]
- Irshad M., Canut H., Borderies G., Pont-Lezica R., Jamet E. (2008). A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol. 8:94. 10.1186/1471-2229-8-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Ohad N., Yadegari R., Hannon M., Dinneny J., Wells D., et al. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96 4186–4191. 10.1073/pnas.96.7.4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Hennig L., Bouveret R., Gheyselinck J., Grossniklaus U., Gruissem W. (2003). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22 4804–4814. 10.1093/emboj/cdg444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C., Page D. R., Gagliardini V., Grossniklaus U. (2005). The Arabidopsis thaliana MEDEA polycomb group protein controls expression of PHERES1 by parental imprinting. Nat. Genet. 37 28–30. 10.1038/ng1495 [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. 10.1007/BF00331014 [DOI] [Google Scholar]
- Lammens W., Le Roy K., Schroeven L., Van Laere A., Rabijns A., Van den Ende W. (2009). Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J. Exp. Bot. 60 727–740. 10.1093/jxb/ern333 [DOI] [PubMed] [Google Scholar]
- Li J., Berger F. (2012). Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 195 290–305. 10.1111/j.1469-8137.2012.04182.x [DOI] [PubMed] [Google Scholar]
- Link M., Rausch T., Greiner S. (2004). In Arabidopsis thaliana, the invertase inhibitors AtC/VIF1 and 2 exhibit distinct target enzyme specificities and expression profiles. FEBS Lett. 573 105–109. 10.1016/j.febslet.2004.07.062 [DOI] [PubMed] [Google Scholar]
- Linkies A., Graeber K., Knight C., Leubner-Metzger G. (2010). The evolution of seeds. New Phytol. 186 817–831. 10.1111/j.1469-8137.2010.03249.x [DOI] [PubMed] [Google Scholar]
- Lionetti V., Fabri E., De Caroli M., Hansen A. R., Willats W. G. T., Piro G., et al. (2017). Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 173 1844–1863. 10.1104/pp.16.01185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods San Diego Calif. 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., et al. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7 947–952. 10.1038/sj.embor.7400760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown P. C., Laouielle-Duprat S., Prins P., Wolff P., Schmid M. W., Donoghue M. T., et al. (2011). Identification of imprinted genes subject to parent-of-origin specific expression in Arabidopsis thaliana seeds. BMC Plant Biol. 11:113. 10.1186/1471-2229-11-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6 414–419. 10.1016/S1360-1385(01)02045-3 [DOI] [PubMed] [Google Scholar]
- Morley-Smith E. R., Pike M. J., Findlay K., Köckenberger W., Hill L. M., Smith A. M., et al. (2008). The transport of sugars to developing embryos is not via the bulk endosperm in oilseed rape seeds. Plant Physiol. 147 2121–2130. 10.1104/pp.108.124644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P., Camilleri C., Courtial B., Caboche M., Bouchez D. (1998). Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 149 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K., Kanehisa M. (1991). Expert system for predicting protein localization sites in gram-negative bacteria. Proteins 11 95–110. 10.1002/prot.340110203 [DOI] [PubMed] [Google Scholar]
- Narsai R., Law S. R., Carrie C., Xu L., Whelan J. (2011). In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol. 157 1342–1362. 10.1104/pp.111.183129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Margossian L., Hsu Y. C., Williams C., Repetti P., Fischer R. L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. U.S.A. 93 5319–5324. 10.1073/pnas.93.11.5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O.-A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell Online 16 S214–S227. 10.1105/tpc.017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerrenig B., Popko J., Heilmann M., Schulmeister S., Dietel K., Schmitt B., et al. (2013). SUCROSE TRANSPORTER 5 supplies Arabidopsis embryos with biotin and affects triacylglycerol accumulation. Plant J. 73 392–404. 10.1111/tpj.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y.-L., Jin Y., Yang Y.-J., Li G.-J., Boyer J. S. (2010). Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol. Plant 3 942–955. 10.1093/mp/ssq044 [DOI] [PubMed] [Google Scholar]
- Saldanha A. J. (2004). Java Treeview—extensible visualization of microarray data. Bioinformatics 20 3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Sanders A., Collier R., Trethewy A., Gould G., Sieker R., Tegeder M. (2009). AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. Cell Mol. Biol. 59 540–552. 10.1111/j.1365-313X.2009.03890.x [DOI] [PubMed] [Google Scholar]
- Schneitz K., Hülskamp M., Pruitt R. E. (1995). Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7 731–749. 10.1046/j.1365-313X.1995.07050731.x [DOI] [Google Scholar]
- Schubert D., Clarenz O., Goodrich J. (2005). Epigenetic control of plant development by Polycomb-group proteins. Curr. Opin. Plant Biol. 8 553–561. 10.1016/j.pbi.2005.07.005 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128 735–745. 10.1016/j.cell.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Scott R. J., Spielman M., Bailey J., Dickinson H. G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Dev. Camb. Engl. 125 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sherson S. M., Alford H. L., Forbes S. M., Wallace G., Smith S. M. (2003). Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J. Exp. Bot. 54 525–531. 10.1093/jxb/erg055 [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen M. B., Chaudhury A. M., Robert H., Bancharel E., Berger F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11 277–281. 10.1016/S0960-9822(01)00072-0 [DOI] [PubMed] [Google Scholar]
- Sosso D., Luo D., Li Q.-B., Sasse J., Yang J., Gendrot G., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47 1489–1493. 10.1038/ng.3422 [DOI] [PubMed] [Google Scholar]
- Stadler R., Lauterbach C., Sauer N. (2005). Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 139 701–712. 10.1104/pp.105.065607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes M. E., Chattopadhyay A., Wilkins O., Nambara E., Campbell M. M. (2013). Interplay between sucrose and folate modulates auxin signaling in Arabidopsis. Plant Physiol. 162 1552–1565. 10.1104/pp.113.215095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A. (1999). Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 121 1–8. 10.1104/pp.121.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A., Hess D., Lee H.-S., Lienhard S. (1999). Neutral invertase is a novel type of sucrose-cleaving enzyme. Physiol. Plant. Den. 107 159–165. 10.1034/j.1399-3054.1999.100202.x [DOI] [Google Scholar]
- Tiwari S., Spielman M., Schulz R., Oakey R. J., Kelsey G., Salazar A., et al. (2010). Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 10:72. 10.1186/1471-2229-10-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C. Y., Wilkins T. A. (1994). A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal. Biochem. 223 7–12. 10.1006/abio.1994.1538 [DOI] [PubMed] [Google Scholar]
- Wang D., Tyson M. D., Jackson S. S., Yadegari R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 13244–13249. 10.1073/pnas.0605551103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang C., Hearn D. J., Kang I.-H., Punwani J. A., Skaggs M. I., et al. (2010). Identification of transcription-factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biol. 10:110. 10.1186/1471-2229-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Borisjuk L., Wobus U. (1996). Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J. 10 823–834. 10.1046/j.1365-313X.1996.10050823.x [DOI] [Google Scholar]
- Weinhofer I., Hehenberger E., Roszak P., Hennig L., Köhler C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLOS Genet. 6:e1001152. 10.1371/journal.pgen.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D., Pethica R., Zhou Y., Talbot C., Vogel C., Madera M., et al. (2009). SUPERFAMILY–sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37 D380–D386. 10.1093/nar/gkn762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P., Weinhofer I., Seguin J., Roszak P., Beisel C., Donoghue M. T. A., et al. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLOS Genet. 7:e1002126. 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R. A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99 909–917. 10.1198/016214504000000683 [DOI] [Google Scholar]
- Yang S., Johnston N., Talideh E., Mitchell S., Jeffree C., Goodrich J., et al. (2008). The endosperm-specific ZHOUPI gene of Arabidopsis thaliana regulates endosperm breakdown and embryonic epidermal. Development 135 3501–3509. 10.1242/dev.026708 [DOI] [PubMed] [Google Scholar]
- Zheng B., Chen X. (2011). Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 14 123–129. 10.1016/j.pbi.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In silico expression analysis of InvINH1 and InvINH2. (A) The relative expression level was illustrated as a heatmap containing 87 anatomical features that were hierarchically organized by the Anatomy tool from Genevestigator. (B) The Perturbations tool from Genevestigator was used to generate the heatmap displaying the experimental stimuli that induced greater than 1.5-fold changes in the expression level of InvINH1 and InvINH2. InvINH1 and InvINH2 are represented by the same Affymetrix probe, 248823_s_at.


