Abstract
The 14-3-3 gene family members play key roles in various cellular processes. However, little is known about the numbers and roles of 14-3-3 genes in wheat. The aims of this study were to identify TaGF14 numbers in wheat by searching its whole genome through blast, to study the phylogenetic relationships with other plant species and to discuss the functions of TaGF14s. The results showed that common wheat harbored 20 TaGF14 genes, located on wheat chromosome groups 2, 3, 4, and 7. Out of them, eighteen TaGF14s are non-ε proteins, and two wheat TaGF14 genes, TaGF14i and TaGF14f, are ε proteins. Phylogenetic analysis indicated that these genes were divided into six clusters: cluster 1 (TaGF14d, TaGF14g, TaGF14j, TaGF14h, TaGF14c, and TaGF14n); cluster 2 (TaGF14k); cluster 3 (TaGF14b, TaGF14l, TaGF14m, and TaGF14s); cluster 4 (TaGF14a, TaGF14e, and TaGF14r); cluster 5 (TaGF14i and TaGF14f); and cluster 6 (TaGF14o, TaGF14p, TaGF14q, and TaGF14t). Tissue-specific gene expressions suggested that all TaGF14s were likely constitutively expressed, except two genes, i.e., TaGF14p and TaGF14f. And the highest amount of TaGF14 transcripts were observed in developing grains at 20 days post anthesis (DPA), especially for TaGF14j and TaGF14l. After drought stress, five genes, i.e., TaGF14c, TaGF14d, TaGF14g, TaGF14h, and TaGF14j, were up-regulated expression under drought stress for both 1 and 6 h, suggesting these genes played vital role in combating against drought stress. However, all the TaGF14s were down-regulated expression under heat stress for both 1 and 6 h, indicating TaGF14s may be negatively associated with heat stress by reducing the expression to combat heat stress or through other pathways. These results suggested that cluster 1, e.g., TaGF14j, may participate in the whole wheat developing stages, e.g., grain-filling (starch biosynthesis) and may also participate in combating against drought stress. Subsequently, a homolog of TaGF14j, TaGF14-JM22, were cloned by RACE and used to validate its function. Immunoblotting results showed that TaGF14-JM22 protein, closely related to TaGF14d, TaGF14g, and TaGF14j, can interact with AGP-L, SSI, SSII, SBEIIa, and SBEIIb in developing grains, suggesting that TaGF14s located on group 4 may be involved in starch biosynthesis. Therefore, it is possible to develop starch-rich wheat cultivars by modifying TaGF14s.
Keywords: Triticum aestivum, 14-3-3, phylogenetic analysis, tissue-specific expression, starch biosynthesis
Introduction
The ubiquitous 14-3-3 proteins, as one of the families of regulatory proteins, have been found in all eukaryotic organisms and tissues. The family consists of dimeric α-helical pSer/Thr binding proteins that play key roles in various cellular processes, such as signal transduction, biotic and abiotic stress responses, and carbon and nitrogen metabolism, by mediating protein–protein interactions (Aitken et al., 1992; Fulgosi et al., 2002). However, little is known about the numbers and roles of 14-3-3 genes (TaGF14s) in wheat. Therefore, it is necessary to develop elite wheat cultivars to explore the numbers and to study the functions of TaGF14s.
Different species may have different numbers of GF14s. For example, humans have seven 14-3-3 genes (Iwata et al., 2000), while Arabidopsis, rice and maize have thirteen GF14s and two pseudogenes, eight GF14s and twelve GF14s, respectively (Wu et al., 1997; Rosenquist et al., 2001; Lai et al., 2004; Sehnke et al., 2006; Yao et al., 2007; Alexandrov et al., 2009), which suggested that plants maybe have more GF14s than animal. 14-3-3 proteins, binding a range of transcription factors and signaling proteins, have roles in regulating carbon and nitrogen metabolism, plant development, and biotic and abiotic stress responses (Roberts, 2000, 2003; Fulgosi et al., 2002; Maraschin et al., 2003). For example, BdGF14f were associated Cr and cold stresses in Brachypodium distachyon (Cao et al., 2016). Different 14-3-3 protein isoforms have different roles. For example, 14-3-3A processing and 14-3-3C isoform tissue specific expression are closely related to cell fate and initiation of specific cell type differentiation (Maraschin et al., 2003). And 14-3-3 proteins were also reported to be involved in starch biosynthesis in plants. For example, Alexander and Morris (2006) identified 54 14-3-3 binding proteins by MALDI-TOF MS, and the largest category was for carbohydrate metabolism, including plastidic enzymes for starch synthesis and modification. Out of them, four enzymes, i.e., GSBSI, SSI, SSII and SBEIIa were involved in starch biosynthesis. Presently, only four GF14s have been reported in common wheat (Ikeda et al., 2000; Yao et al., 2005; Wang et al., 2008). It is unknown whether there are more GF14s in common wheat than rice and maize. Due to the roles of GF14 reported previously, it is necessary to study the 14-3-3 genes and their functions in wheat.
The allohexaploid bread wheat (Triticum aestivum, 2n = 6x = 42; genome AABBDD) is one of the largest crop worldwide. Due to two times of heterologous hybridization and two times of chromosome self-doubling, modern common wheat have a larger genome size (17 gigabase) than rice (466 megabases) and maize (2.3 gigabase) (Yu et al., 2002; Schnable et al., 2009; International Wheat Genome Sequencing Consortium [IWGSC], 2014). Because of its genome complexity and its big genome size, wheat chromosome sequencing is not possible in the last decade. However, with the advances of technologies, e.g., chromosome follow sorting and sequencing technology (next generation sequence/de novo assemble and pacbio), a reference genome of common wheat version TGACv1 is obtained by next generation sequence/de novo assembly (International Wheat Genome Sequencing Consortium [IWGSC], 2014), which is very attractive to wheat geneticists and breeders and highlights wheat genetic improvement.
In this study, we are aimed to identify TaGF14 numbers in wheat by searching the wheat whole genome through blast, to study the phylogenetic relationships with other plant species and to discuss the functions of TaGF14s.
Materials and Methods
Plant Materials
The hard white winter wheat cultivar Jimai 22, released by our lab, was used in this study and was sown in a field at the Experimental Station of Shandong Academy of Agricultural Sciences (SAAS), Jinan, Shandong Province, China. The plot size was 12 m2. Soil fertility was high. Weeds and diseases were controlled. Developing wheat ears were tagged at the onset of anthesis. Endosperm tissue was obtained from developing wheat grains (at Z71 and Z75) taken from the mid-ear region of the head (Zadoks et al., 1974).
RNA Extraction and Cloning of TaGF14-JM22
The total RNA was isolated from the developing grains or kernels at Zadok scale 71 according to the instructions of an RNeasy Plant Mini Kit (Qiagen, Germany). RNase-free DNase I (Promega, United States) was used to remove any contaminating genomic DNA. Quality and integrity of the total RNA were determined by running the appropriate amount of RNA in a formamide denaturing gel. TaGF14-JM22 was cloned from wheat according to the methods described in the Supplementary Material. The cDNA sequence of TaGF14-JM22 obtained was submitted to GenBank, and the accession number is GenBank JF957590. The 3D structure of TaGF14-JM22 was predicted using the ExPASy proteomics online server and Swiss-Model1.
Construction of the Phylogenetic Tree and Expression of the TaGF14-JM22 Genes in Developing Grains
To determine the 14-3-3 gene numbers in wheat and to construct the phylogenetic tree of 14-3-3 genes from cereal crops and Arabidopsis, the coding sequence of TaGF14-JM22, cloned from wheat in this study, was used as the query to search the NCBI database2 and the genome sequence databases of Sorghum3, wheat4, and Brachypodium5 with a cut-off parameter of E-value ≤ 1E-10 for homologous GF14s. The phylogenetic tree was constructed using the maximum likelihood method with a Poisson distribution model and 1000 bootstrap replicates by MEGA 6.0 (Tamura et al., 2013) based on the amino acid sequences of 14-3-3 proteins with a cut-off value of 50% for the condensed tree. In addition, the silicon expression profiles of TaGF14 in Root_Z13, Stem_Z30, Leaf_Z23, Spike_Z65, and the developing grains at 10, 20, and 30 days post anthesis (DPA) were obtained through WheatExp (Pearce et al., 2015) and analyzed. Data was analyzed with SAS software version 9.0. The mean expression values of every gene in different tissues were compared with each other, respectively. Duncan’s multiple range test was used to test for significant differences.
Expression and Purification
For cloning in pET29c, the TaGF14-JM22 sequence was amplified using the primers BamHI F and HindIII R. Amplicons were digested together with the pET29c vector and BamHI and HindIII enzymes at 37°C for 3 h. The digested products were purified and ligated together with T4 DNA ligase (Promega, United States) at 4°C overnight. The ligation mix was then transformed into Escherichia coli BL21 (DE3) for protein expression. The positive clones were screened for correct insertion by colony PCR and sequencing. The successful constructs were expected to express a TaGF14-JM22 fusion protein with an S-tag at the N-terminus. The recombinant proteins were purified using the S-tag rEK Purification Kit (Novagen, United States) according to the manual’s protocol. For more details, please see the Supplementary Material.
Amyloplast Isolation
The amyloplasts were isolated from the developing endosperm obtained from wheat grains (at Zadok scale 75) taken from the mid-ear region of the head as described by Tetlow et al. (2008). Starch granules were washed, and the granule-associated proteins, e.g., AGPase and GBSS, were extracted as described by Denyer et al. (1995). The protein content was measured using the Bio-Rad protein assay according to the manufacturer’s instructions and using thyroglobulin as a standard (Bio-Rad Lab., Canada).
Preparation of Peptides and Antisera
Polyclonal antibodies of starch biosynthetic enzymes were raised in rabbits against synthetic peptides, which were derived from N-terminal sequences of wheat AGP-L (CIIDMNARIGRDVVISN, Ainsworth et al., 1995), AGP-S (AIIDKNARIGENVKIIN, Rösti and Denyer, 2007), SSI (APAQSPAPTQPPLPDAG, Li et al., 1999), SSII (ARVDDDAASARQPRARRG, Li et al., 1999), GBSSI (QDLSWKGPAKNWEDV, Vrinten and Nakamura, 2000), SBEI (VSAPRDYTMATAEDGV, Rahman et al., 2001), SBEIIa (AASPGKVLVPDGESDDLAS, Rahman et al., 2001), SBEIIb (AGGPSGEVMIPDGGSG, Regina et al., 2005), DE (SVGVGEDLPEGYEQM, Bresolin et al., 2006), and SP (NYDELMGSLEGNEGYGRADYFLV, Tickle et al., 2009). The antigen was prepared by coupling the synthesized peptide to keyhole limpet haemocyanin using the heterobifunctional reagent m-maleimidobenzoyl-N-hydroxysuccinimide ester (Tetlow et al., 2008).
SDS-PAGE and Immunoblotting
The methods of SDS-PAGE and immunoblotting were according to Tetlow et al. (2008), for more detail, please see the Supplementary Materials. Gels were stained with a colloidal Coomassie Brilliant Blue G250 kit (Neuhoff et al., 1988).
Results
Numbers of TaGF14 Genes and Phylogenetic Tree Construction
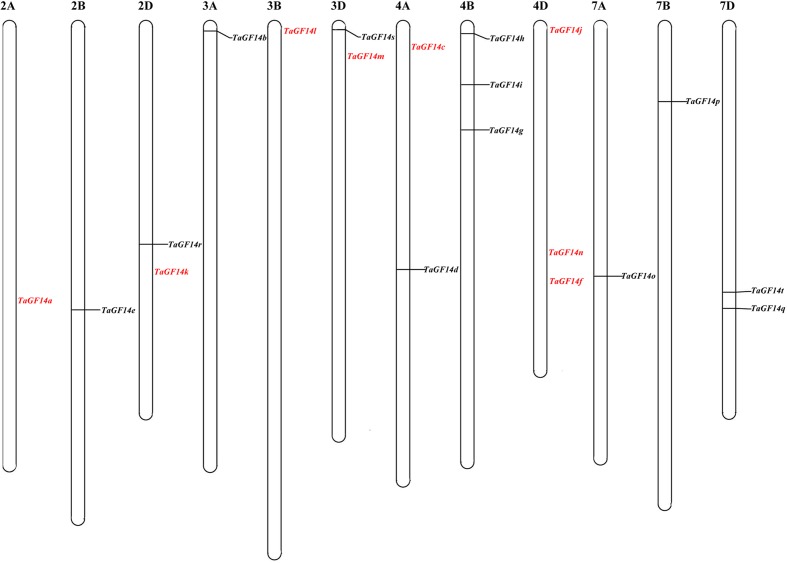
To explore the chromosomal locations and numbers of TaGF14 in wheat, the complete coding sequence of TaGF14-JM22 was used as the query to search the wheat whole-genome sequences published by IWGSC6. In total, 20 genes were obtained through Blast, and the coding sequences and chromosomal location of these genes are listed in Supplementary Table S1. In addition, these genes were located on wheat chromosome groups 2, 3, 4, and 7 (Figure 1). However, the 14-3-3 genes were not equally distributed on the wheat chromosome groups. In this study, eight genes were located on the wheat chromosome group 4 and the remaining 3 chromosome groups harbored equal numbers (four genes per group) of TaGF14 genes.
FIGURE 1.
Chromosome localization of TaGF14s based on the reference sequence (TGACv1.0) of wheat genome (International Wheat Genome Sequencing Consortium [IWGSC], 2014). The text in red color presented that the genes were not physically mapped in the reference genome.
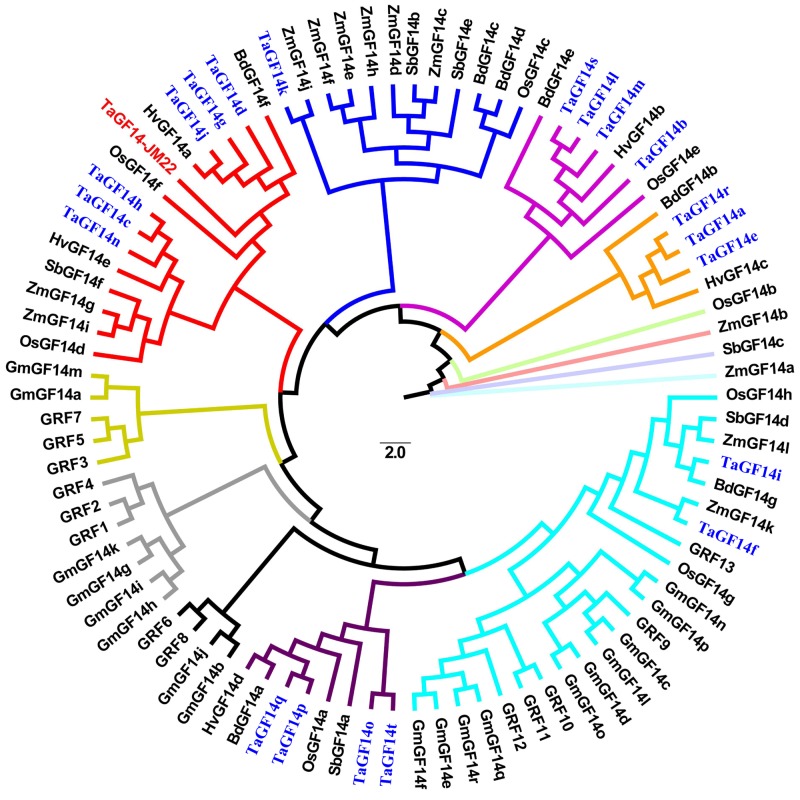
To investigate the evolutionary relationship among TaGF14-JM22 and other GF14 genes and proteins derived from Oryza sativa, B. distachyon, Zea mays, Hordeum vulgare, and Arabidopsis thaliana, phylogenetic trees were constructed using the maximum likelihood method with a Poisson model and with 1000 bootstrap replicates by MEGA 6.0 (Tamura et al., 2013) based on the amino acid sequences of 14-3-3 proteins with a cut-off value of 50% for the condensed tree (Figure 2). The results showed that the 20 wheat TaGF14s could be divided into six clusters: cluster 1, including six genes (TaGF14d, TaGF14g, TaGF14j, TaGF14h, TaGF14c, and TaGF14n); cluster 2, including one gene (TaGF14k); cluster 3, including four genes (TaGF14b, TaGF14l, TaGF14m, and TaGF14s); cluster 4, including three genes (TaGF14a, TaGF14e, and TaGF14r); cluster 5, including two genes (TaGF14i and TaGF14f); and cluster 6, including four genes (TaGF14o, TaGF14p, TaGF14q, and TaGF14t). The results also showed that eighteen TaGF14s are non-ε proteins, except two wheat GF14 genes, TaGF14i and TaGF14f, which are ε proteins (Figure 2).
FIGURE 2.
Phylogenetic analysis of TaGF14s in common wheat with 14-3-3 proteins in other plant species. A rooted phylogenetic tree based on the sequence alignment using the MEGA 6.0 software from the CLUSTALW multiple sequence alignment. The scale represents estimated branch length. The TaGF14 genes from common wheat were marked in blue color.
Expression of TaGF14s in Wheat
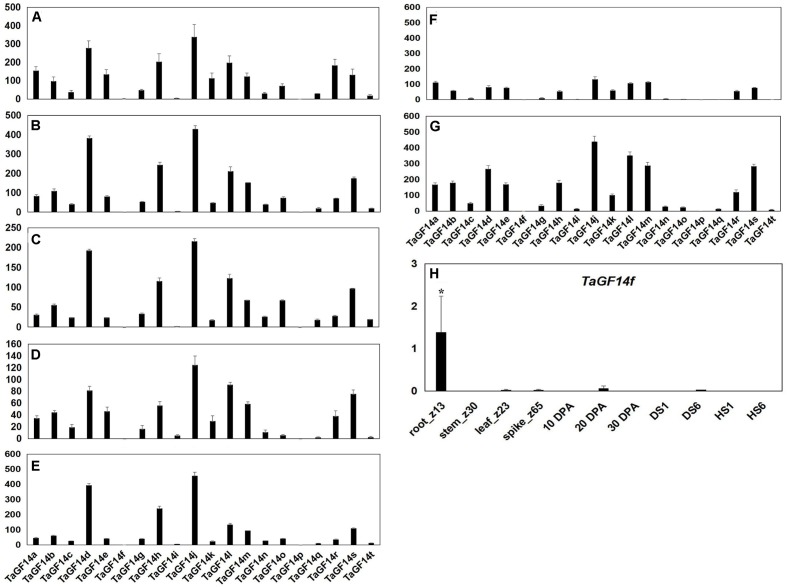
In order to investigate the gene expression levels of TaGF14 in the wheat root, stem, leaf, spike and developing grains at 10, 20, and 30 DPA, the silicon expression dataset was downloaded from WheatExp. As shown in Figure 3, it appeared that all TaGF14s were constitutively expressed, except two genes, i.e., TaGF14p, which was not observed, and TaGF14f, which showed tissue-specific expression in the root (Figure 3H and Supplementary Figure S6). In addition, TaGF14i was also likely expressed in a tissue-specific manner in stem_z30, the developing grains at 20 and 30 DPA. TaGF14o, TaGF14q, and TaGF14t, which belong to cluster 6, were expressed less in developing grains than in other investigated tissues (Supplementary Figure S6). These results indicated that TaGF14j, TaGF14l, and TaGF14i may play important role in the wheat grain-filling stage.
FIGURE 3.
Detection of TaGF14 transcripts by silicon expression profiles. (A) Root_Z13. (B) Stem_Z30. (C) Leaf_Z23. (D) Spike_Z65. (E) Developing grains_10 DPA. (F) Developing grains_20 DPA. (G) Developing grains_30 DPA. Bar represents the standard error. (H) The expression of TaGF14f was displayed in different tissues. ∗At the top of each column indicates significant difference at P = 0.05.
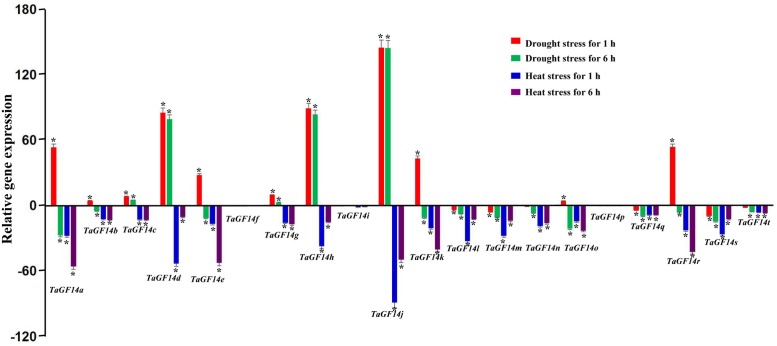
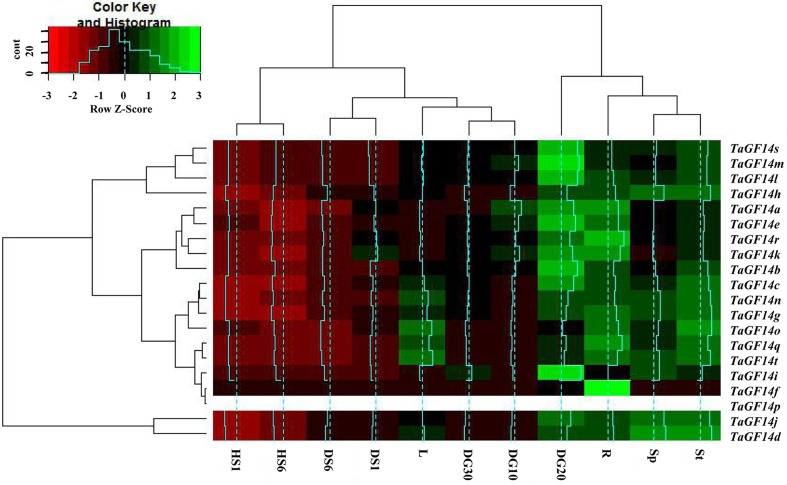
In addition, the gene expressions of TaGF14s were also determined in the wheat seedling stage treated with drought stress and heat stress. The results showed that five genes, i.e., TaGF14c, TaGF14d, TaGF14g, TaGF14h, and TaGF14j, were up-regulated expression under drought stress for both 1 and 6 h (Figure 4), suggesting these genes played vital role in combating against drought stress. However, all the TaGF14s were down-regulated expression under heat stress for both 1 and 6 h, which indicated that TaGF14s may be negatively associated with heat stress by reducing the expression to combat heat stress or through other pathways. Furthermore, the heatmap of TaGF14s were also drawn based on the gene expression data of TaGF14s. The results showed that TaGF14s in Root_Z13, Stem_Z30, Spike_Z65 and the developing grains at 20 DPA had the similar gene expression, while the rest had the similar gene expression pattern (Figure 5). And the TaGF14s clustered into three clusters based on gene expression in different samples or tissues, i.e., CL1, CL2, and CL3. And TaGF14h, TaGF14l, TaGF14m, and TaGF14s belonged to CL1. TaGF14d and TaGF14j belonged to CL3, while the rest belonged to CL2. These results suggested that TaGF14d and TaGF14j, both constitutively expressed, may participate in the whole wheat developing stages, e.g., grain-filling (starch biosynthesis) and may also participate in combating against drought stress.
FIGURE 4.
Wheat TaGF14s expression induced by drought and heat. Red, green, blue, and purple column represent drought stress for 1 h, drought stress for 6 h, heat stress for 1 h, and heat stress for 6 h. Bar represents the standard error. ∗At the top of each column indicates significant difference at P = 0.05.
FIGURE 5.
Heatmap of TaGF14s drawn with software R program (gplot) based on its expression data in different tissues or samples treated with drought and heat stresses. St, Stem_Z30. Sp, Spike_Z65. R, Root_Z13. L, Leaf_Z23. DG10, Developing grains_10 DPA. DG20, Developing grains_20 DPA. DG30, Developing grains_30 DPA.
Cloning and Sequence Analysis of TaGF14-JM22
To validate the gene function of TaGF14j, a homologous gene, TaGF14-JM22 was cloned and used for further analysis. The full-length cDNA of TaGF14-JM22, containing 786 nucleotides, was obtained using the RACE-PCR technique (Supplementary Table S3 and Supplementary Figures S1, S2) and submitted to GenBank (Accession number: JF957590). Multiple alignments showed that this sequence shared high identity with 14-3-3 proteins from other species ranging from 31 to 98% (Supplementary Figure S3), e.g., 98% identity with Brachypodium (BdGF14f) and Oryza (OsGF14f) and 31% identity with Oryza (OsGF14h). TaGF14-JM22 was predicted to encode 261 amino acids (AA), with a predicted molecular mass of 29.27 kDa and an isoelectric point (pI) of 4.82. Structure analysis revealed that the predicted TaGF14-JM22 protein contained two 14-3-3 protein signatures and six functional motifs (Supplementary Table S2), such as a cAMP- (or cGMP-) dependent protein kinase phosphorylation site and a tyrosine kinase phosphorylation site, which were highly conserved in 14-3-3 homologs. Based on a WoLF PSORT analysis7, TaGF14-JM22 was located in the plasma membrane or nuclear plasma membrane. In addition, the three-dimensional (3D) structure prediction was analyzed by comparative protein modeling. The coding sequence of TaGF14-JM22 was submitted to the Swiss-Model online server8, and six 14-3-3-like proteins with sequence similarities of 90.60, 90.34, 90.17, 85.83, 84.86, and 84.52% were selected as templates to build models. Subsequently, nine models were generated using the abovementioned 14-3-3 proteins as models for the Swiss-Model homology modeling (Supplementary Figure S5). In addition, the QMEAN Z-score evaluations for the models were -1.12, -0.95, -1.50, -1.03, -0.50, -0.97, -1.33, -1.39, and -1.75, respectively, showing that the predicted models were of good quality. Furthermore, the phylogenetic results indicated that TaGF14-JM22, cloned in this study and belonging to non-ε protein, was closely related to three wheat genes (TaGF14d, TaGF14g, and TaGF14j) as well as OsGF14f and HvGF14f (Figure 2 and Supplementary Figure S3).
Validation the Function of TaGF14-JM22 in Developing Grains
To validate TaGF14-JM22 similar to TaGF14j participating in starch biosynthesis in developing grains, the coding sequence of TaGF14-JM22 was sub-cloned into pET29c. After induction by 1 mM IPTG at 37°C for 1, 3, 5, and 7 h, the highest expression occurred with 1 mM IPTG in both 5 and 7 h inductions at 37°C. SDS-PAGE was used for induction and purification of the TaGF14-JM22 protein. The protein with the highest abundance was found in the E. coli extracts. The molecular mass of the induced protein was about 29 kDa, which was in accordance with the predicted amino acid sequence (Supplementary Figure S4).
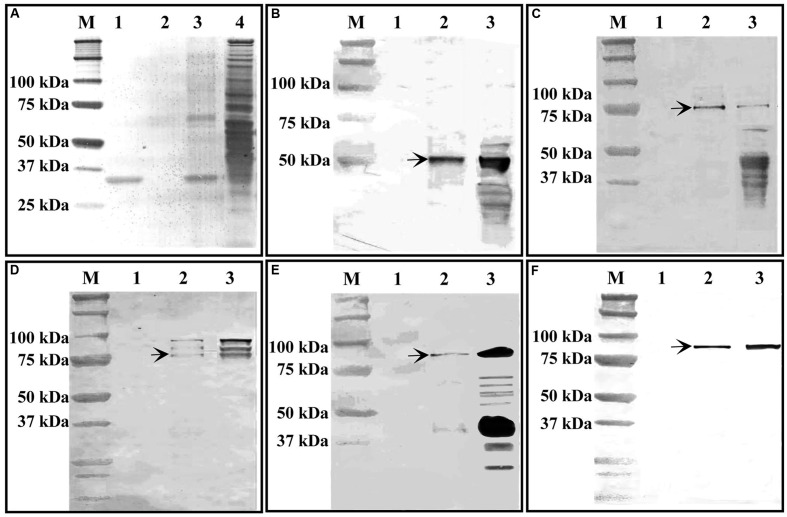
The purified recombinant TaGF14-JM22 protein was bound to S-protein agarose as a biochemical bait and then incubated with wheat amyloplast extract. Protein–protein interactions between the TaGF14-JM22 protein and ten key starch biosynthetic enzymes from amyloplasts, i.e., AGP-L, AGP-S, SSI, SSII, GBSSI, SBEI, SBEIIa, DE, SBEIIb, and SP, were investigated and analyzed by SDS-PAGE and western blotting. As shown in Figure 6, BSA, as a control, could not bind any starch biosynthetic enzymes, but protein–protein interactions between the TaGF14-JM22 protein and starch biosynthetic enzymes were observed. AGP-L, SSI, SSII, SBEIIa, and SBEIIb interacted with the TaGF14-JM22 protein (Figure 6). However, AGP-S, GBSSI, SBEI, DE, and SP could not interact with the TaGF14-JM22 protein. These results suggested that TaGF14-JM22 indeed participated in starch biosynthesis by binding to biosynthetic enzymes.
FIGURE 6.
Interaction between GF14-JM22 protein and starch biosynthetic enzymes revealed by immunoblotting assays. M, Protein markers. (A) Gel image of SDS-PAGE. 1, Recombinant GF14-JM22 protein binding to S-protein agarose resin. 2, Blank control. 3, Recombinant GF14-JM22 protein through affinity chromatography. 4, Wheat amyloplast extracts. (B–F), Gel images of SH2-antibody, SSI-antibody, SSII-antibody, SBEIIa-antibody and SBEIIb-antibody, respectively. 1, Blank control. 2, Recombinant GF14-JM22 protein immunoblotting through affinity chromatography. 3, Wheat amyloplast extracts immunoblotting.
Discussion
The family of 14-3-3 proteins is one of the families of regulatory proteins in plants (Aitken et al., 1992). Previous studies showed that plants have more 14-3-3 genes than animals. For example, human has seven 14-3-3 genes, while Arabidopsis and maize have thirteen 14-3-3 genes and twelve 14-3-3 genes, respectively (Wu et al., 1997; Iwata et al., 2000; Rosenquist et al., 2001). However, the number of 14-3-3 genes in common wheat and their relationships with other species are still unknown. In the present study, it was determined by Blastn against the whole genome sequences of Chinese Spring wheat released by International Wheat Genome Sequencing Consortium [IWGSC], 2014. The results indicated that common wheat harbored 20 GF14s (Supplementary Table S1 and Figure 2), which was much more than rice and Brachypodium (Wu et al., 1997; Iwata et al., 2000; Rosenquist et al., 2001). Of all the genes, eight (40%) were located on wheat chromosome group 4 (Supplementary Table S1). In addition, the phylogenetic tree was constructed based on 14-3-3 protein sequences, which revealed that most of the TaGF14s, including five clusters (clusters 1–5), are non-ε proteins, except cluster 6 (TaGF14f and TaGF14i) which are ε proteins (Figure 2 and Supplementary Table S1).
The 14-3-3 proteins play important roles in diverse cellular processes by mediating protein-protein interactions in plants (Fulgosi et al., 2002). Previous studies indicated that HvGF14a was a protein induced by powdery mildew fungus, suggesting that it was involved in plant resistance to fungus infection in H. vulgare (Brandt et al., 1992). In Brachypodium, BdGF14f was significantly induced by Cr and cold stress (Cao et al., 2016). In addition, OsGF14f was constitutively expressed in rice (Yao et al., 2007). Previous studies indicated that starch was synthesized through the coordinated interactions of a suite of biosynthetic enzymes in plants (Zeeman et al., 2010). However, whether 14-3-3 as a regulatory protein involved in starch biosynthesis was known in wheat. And very little was also known about the functions of 14-3-3 proteins in wheat. In this study, TaGF14-JM22, which is similar to TaGF14d, TaGF14g and TaGF14j, the most highly expressed genes among the twenty TaGF14s in developing wheat grains, was used to investigate the protein-protein interactions between 14-3-3s and ten key starch biosynthetic enzymes, i.e., AGP-L, AGP-S, SSI, SSII, GBSSI, SBEI, SBEIIa, DE, SBEIIb, and SP, during grain filling by a immunoblotting assay. The results showed that five enzymes, i.e., AGP-L, SSI, SSII, SBEIIa, and SBEIIb, interacted with the TaGF14-JM22 protein, while the rest of the enzymes did not (Figure 6), suggesting that TaGF14d, TaGF14g, and TaGF14j may be involved in starch biosynthesis through protein–protein interactions. In barley, 14-3-3 proteins were reported to interact with four starch biosynthetic enzymes, i.e., GSBSI, SSI, SSII and SBEIIa in developing grains (Alexander and Morris, 2006), which were also clearly detected in our study. In addition, two starch biosynthetic enzymes, i.e., DE and SBEIIb, also interacted with 14-3-3 proteins in developing wheat grains, which was firstly reported in this study and may be unique to wheat, considering the fact that wheat harbors more 14-3-3s than other species (Figures 2, 6). In addition, the results showed that TaGF14s in Leaf_Z23, the developing grains at 10 and 30 DPA, drought stress and heat stress had the similar gene expressions, which can be explained by the fact that wheat production was usually affected by heat and drought stress, especially in the grain-filling stages (Skylas et al., 2002; Farooq et al., 2011). And in Figures 2, 3, the results showed that TaGF14-JM22, TaGF14d, TaGF14g, and TaGF14j were closely related to HvGF14a (a pathogen-related protein), BdGF14f, induced by Cr and cold stress, and OsGF14f, and were constitutively expressed in wheat (Yao et al., 2007; Cao et al., 2016). Therefore, we speculated that TaGF14d, TaGF14g, and TaGF14j may also have the similar functions, e.g., resistance to pathogen and Cr stress, with other plant species. Furthermore, Arabidopsis GRF6 was linked to the “stay green” phenotype and drought tolerance by cotton transformation experiments (Yan et al., 2004). Our results indicated that the TaGF14s, closely related to GRF6, on group 7 that belong to cluster 6 may also be linked to the “stay green” phenotype (Figure 2). So, in our next project, the functions of TaGF14s, especially TaGF14s located on group 4 and group 7 will be further analyzed by gene overexpressing or gene silencing in wheat.
Author Contributions
JS and JG conceived and designed the experiments. JS, JG, SD, and HL performed the experiments. JG, CL, XsC, and JS analyzed the data. DC, AL, XyC, SZ, ZZ, and JL contributed reagents/materials/analysis tools. JG and JS wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the financial support of the NSF of China (31471487), the Transgenic Special Item (2016ZX08002003), the National Key Research and Development Program of China (2017YFD0100600) and State Key of Crop Biology (2017KF03).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00012/full#supplementary-material
References
- Ainsworth C., Hosein F., Tarvis M., Weir F., Burrell M., Devos K. M., et al. (1995). Adenosine diphosphate glucose pyrophosphorylase genes in wheat: differential expression and gene mapping. Planta 197 1–10. 10.1007/BF00239933 [DOI] [PubMed] [Google Scholar]
- Aitken A., Collinge D. B., Heusden B. P. H. V., Isobe T., Roseboom P. H., Rosenfeld G., et al. (1992). 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem. Sci. 17 498–501. 10.1016/0968-0004(92)90339-B [DOI] [PubMed] [Google Scholar]
- Alexander R. D., Morris P. C. (2006). A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics 6 1886–1896. 10.1002/pmic.200500548 [DOI] [PubMed] [Google Scholar]
- Alexandrov N. N., Brover V. V., Freidin S., Troukhan M. E., Tatarinova T. V., Zhang H., et al. (2009). Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 69 179–194. 10.1007/s11103-008-9415-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt J., Thordal-Christensen H., Vad K., Gregersen P. L., Collinge D. B. (1992). A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J. 2 815–820. [PubMed] [Google Scholar]
- Bresolin N. S., Li Z., Kosar-Hashemi B., Tetlow I. J., Chatterjee M., Rahman S., et al. (2006). Characterisation of disproportionating enzyme from wheat endosperm. Planta 224 20–31. 10.1007/s00425-005-0187-7 [DOI] [PubMed] [Google Scholar]
- Cao H., Xu Y., Yuan L., Bian Y., Wang L., Zhen S., et al. (2016). Molecular characterization of the 14-3-3 gene family in Brachypodium distachyon L. reveals high evolutionary conservation and diverse responses to abiotic stresses. Front. Plant Sci. 7:1099. 10.3389/fpls.2016.01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer K., Hylton C. M., Jenner C. F., Smith A. M. (1995). Identification of multiple isoforms of soluble and granule-bound starch synthase in developing wheat endosperm. Planta 196 256–265. 10.1007/BF00201382 [DOI] [Google Scholar]
- Farooq M., Bramley H., Palta J. A., Siddique K. H. M. (2011). Heat stress in wheat during reproductive and grain-filling phases. Crit. Rev. Plant Sci. 30 491–507. 10.1080/07352689.2011.615687 [DOI] [Google Scholar]
- Fulgosi H., Soll J., de Faria Maraschin S., Korthout H. A., Wang M., Testerink C. (2002). 14-3-3 proteins and plant development. Plant Mol. Biol. 50 1019–1029. 10.1023/A:1021295604109 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Koizumi N., Kusano T., Sano H. (2000). Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase, and to WPK4-phosphorylated nitrate reductase. J. Biol. Chem. 275 31695–31700. 10.1074/jbc.M004892200 [DOI] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium [IWGSC] (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092. 10.1126/science.1250092 [DOI] [PubMed] [Google Scholar]
- Iwata N., Yamamoto H., Sasaki S., Itoh F., Suzuki H., Kikuchi T., et al. (2000). Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 gene in human hepatocellular carcinoma. Oncogene 19 5298–5302. 10.1038/sj.onc.1203898 [DOI] [PubMed] [Google Scholar]
- Lai J., Dey N., Kim C. S., Bharti A. K., Rudd S., Mayer K. F., et al. (2004). Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res. 14 1932–1937. 10.1101/gr.2780504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Rahman S., Kosar-Hashemi B., Mouille G., Appels R., Morell M. K. (1999). Cloning and characterization of a gene encoding wheat starch synthase I. Theor. Appl. Genet. 98 1208–1216. 10.1007/s001220051186 [DOI] [Google Scholar]
- Maraschin S. D. F., Lamers G. E., de Pater B. S., Spaink H. P., Wang M. (2003). 14-3-3 isoforms and pattern formation during barley microspore embryogenesis. J. Exp. Bot. 54 1033–1043. 10.1093/jxb/erg098 [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988). Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9 255–262. 10.1002/elps.1150090603 [DOI] [PubMed] [Google Scholar]
- Pearce S., Vazquezgross H., Herin S. Y., Hane D., Wang Y., Gu Y. Q., et al. (2015). WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol. 15:299. 10.1186/s12870-015-0692-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S., Regina A., Li Z., Mukai Y., Yamamoto M., Kosar-Hashemi B., et al. (2001). Comparison of starch-branching enzyme genes reveals evolutionary relationships among isoforms. Characterization of a gene for starch-branching enzyme IIa from the wheat genome donor Aegilops tauschii. Plant Physiol. 125 1314–1324. 10.1104/pp.125.3.1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A., Kosar-Hashemi B., Li Z., Pedler A., Mukai Y., Yamamoto M., et al. (2005). Starch branching enzyme IIb in wheat is expressed at low levels in the endosperm compared to other cereals and encoded at a non-syntenic locus. Planta 222 899–909. 10.1007/s00425-005-0032-z [DOI] [PubMed] [Google Scholar]
- Roberts M. R. (2000). Regulatory 14-3-3 protein–protein interactions in plant cells. Curr. Opin. Plant Biol. 3 400–405. 10.1016/S1369-5266(00)00103-5 [DOI] [PubMed] [Google Scholar]
- Roberts M. R. (2003). 14-3-3 proteins find new partners in plant cell signalling. Trend. Plant Sci. 8 218–223. 10.1016/S1360-1385(03)00056-6 [DOI] [PubMed] [Google Scholar]
- Rosenquist M., Alsterfjord M., Larsson C., Sommarin M. (2001). Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 127 142–149. 10.1104/pp.127.1.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösti S., Denyer K. (2007). Two paralogous genes encoding small subunits of ADP-glucose pyrophosphorylase in maize, Bt2 and L2, replace the single alternatively spliced gene found in other cereal species. J. Mol. Evol. 65 316–327. 10.1007/s00239-007-9013-0 [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Sehnke P. C., Laughne B., Cardasis H., Powell D., Ferl R. J. (2006). Exposed loop domains of complexed 14-3-3 proteins contribute to structural diversity and functional specificity. Plant Physiol. 140 647–660. 10.1104/pp.105.073916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylas D. J., Cordwell S. J., Hains P. G., Larsen M. R., Basseal D. J., Walsh B. J., et al. (2002). Heat shock of wheat during grain filling: proteins associated with heat-tolerance. J. Cereal Sci. 35 175–188. 10.1006/jcrs.2001.0410 [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I. J., Beisel K. G., Cameron S., Makhmoudova A., Liu F., Bresolin N. S., et al. (2008). Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 146 1878–1891. 10.1104/pp.108.116244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickle P., Burrell M. M., Coates S. A., Emes M. J., Tetlow I. J. (2009). Characterization of plastidial starch phosphorylase in Triticum aestivum L. endosperm. J. Plant Physiol. 166 1465–1478. 10.1016/j.jplph.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Vrinten P. L., Nakamura T. (2000). Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol. 122 255–264. 10.1104/pp.122.1.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Ma Q. H., Lin Z. B., He P., Liu J. Y. (2008). Cloning and characterization of a cDNA encoding 14-3-3 protein with leaf and stem-specific expression from wheat. DNA Seq. 19 130–136. 10.1080/10425170701447515 [DOI] [PubMed] [Google Scholar]
- Wu K., Rooney M. F., Ferl R. J. (1997). The Arabidopsis 14-3-3 multigene family. Plant Physiol. 114 1421–1431. 10.1104/pp.114.4.1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., He C., Wang J., Mao Z., Holaday S. A., Allen R. D., et al. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a “stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45 1007–1014. 10.1093/pcp/pch115 [DOI] [PubMed] [Google Scholar]
- Yao Y., Du Y., Jiang L., Liu J. Y. (2007). Interaction between ACC synthase 1 and 14-3-3 proteins in rice: a new insight. Biochem. 72 1003–1007. 10.1134/S000629790709012X [DOI] [PubMed] [Google Scholar]
- Yao Y., Ni Z., Zhang Y., Chen Y., Ding Y., Han Z., et al. (2005). Identification of differentially expressed genes in leaf and root between wheat hybrid and its parental inbreds using PCR-based cDNA subtraction. Plant Mol. Biol. 58 367–384. 10.1007/s11103-005-5102-x [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G. K. S., Li S., Liu B., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Zadoks J. C., Chang T. T., Konzak C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14 415–421. 10.1111/j.1365-3180.1974.tb01084.x [DOI] [Google Scholar]
- Zeeman S. C., Kossmann J., Smith A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61 209–234. 10.1146/annurev-arplant-042809-112301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.