Abstract
The faster evolution of X chromosomes has been documented in several species, and results from the increased efficiency of selection on recessive alleles in hemizygous males and/or from increased drift due to the smaller effective population size of X chromosomes. Aphids are excellent models for evaluating the importance of selection in faster-X evolution because their peculiar life cycle and unusual inheritance of sex chromosomes should generally lead to equivalent effective population sizes for X and autosomes. Because we lack a high-density genetic map for the pea aphid, whose complete genome has been sequenced, we first assigned its entire genome to the X or autosomes based on ratios of sequencing depth in males (X0) to females (XX). Then, we computed nonsynonymous to synonymous substitutions ratios (dN/dS) for the pea aphid gene set and found faster evolution of X-linked genes. Our analyses of substitution rates, together with polymorphism and expression data, showed that relaxed selection is likely to be the greatest contributor to faster-X because a large fraction of X-linked genes are expressed at low rates and thus escape selection. Yet, a minor role for positive selection is also suggested by the difference between substitution rates for X and autosomes for male-biased genes (but not for asexual female-biased genes) and by lower Tajima’s D for X-linked compared with autosomal genes with highly male-biased expression patterns. This study highlights the relevance of organisms displaying alternative chromosomal inheritance to the understanding of forces shaping genome evolution.
Keywords: sex chromosome, sex-biased expression, evolutionary rates, hemizygosity, selection, drift
Introduction
Sex chromosomes are major players in evolution. Besides their role in sex determination, sex chromosomes contribute to genomic conflicts (Rice 1984; Meiklejohn and Tao 2010; Soh et al. 2014), genetic incompatibilities, and reproductive isolation (Coyne and Orr 2004; Saether et al. 2007; Kitano et al. 2009; Johnson and Lachance 2012). A pair of sex-determining chromosomes typically evolves toward reduced recombination (crossing over), which eventually causes one of the sex chromosomes to gradually lose most of the chromosomal regions (loci) present in the alternate one (Charlesworth et al. 2005). These loci will thus be found in single copy in the sex that carries the degenerate, smaller sex chromosome. When the heterogametic sex is the male, sex chromosomes are denoted X and Y (e.g., in mammals), whereas when it is the female, sex chromosomes are noted W and Z (e.g., in birds). Alleles of loci present only on the X (or Z) are more exposed to selection in individuals of the heterogametic sex, facilitating the fixation of beneficial mutations and the purging of deleterious ones (Charlesworth et al. 1987). On the other hand, because males (XY) bear and transmit a single X chromosome, the effective population size is smaller for the X compared with autosomes (Wright 1931; Caballero 1994, 1995). This can increase the rate of fixation of slightly deleterious mutations on the X by genetic drift (Kimura 1983) (the same principles apply to ZW systems, so we ignore these in the following). Consequently, X-linked genes may evolve faster than autosomes (“faster-X” evolution) due to higher levels of positive selection (rate of fixation of beneficial mutations) and/or genetic drift (rate of fixation of slightly deleterious mutations) (Vicoso and Charlesworth 2009; Mank, Vicoso et al. 2010). The faster evolution of X-linked proteins is supported by observations from a large panel of species (e.g., Drosophila, nematodes, mammals, birds, see Meisel and Connallon 2013 for a review). In some species, drift appears to play the dominant role in causing faster-X evolution (Mank, Nam et al. 2010; Avila et al. 2010), whereas positive selection appears to predominate in other species (Baines et al. 2008; Hvilsom et al. 2012; Langley et al. 2012; Mackay et al. 2012; Kousathanas et al. 2014; Sackton et al. 2014; Avila et al. 2015).
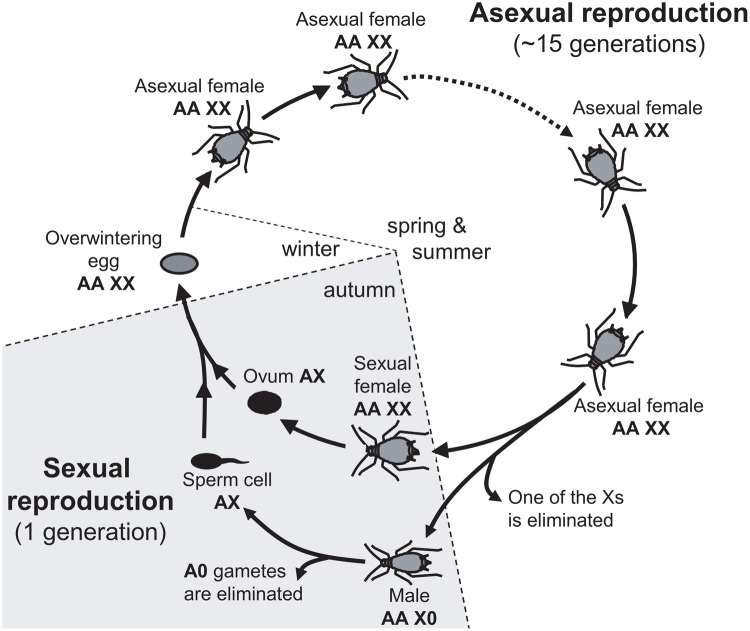
In this context, organisms with atypical sex chromosome inheritance can greatly facilitate inferences about the processes contributing to the evolution of sex chromosomes (Bachtrog et al. 2011). Aphids, which have X0 males and XX females, reproduce by cyclical parthenogenesis, such that males and sexual females constitute only a short part of their life-cycle, which is dominated by apomictic parthenogenetic (clonal) XX females (fig. 1). Males are produced asexually via the elimination of one X from the germ line (Wilson et al. 1997; Caillaud et al. 2002). As a result, X-linked recessive alleles are exposed to selection in male aphids, just like in other X0 or XY males. However, because all sexually produced aphid eggs are XX females, all progeny inherit their X from males and sexual females in equal proportions, just as with autosomes. This difference from other heterogametic systems, where progeny present even sex ratios, has deep consequences for the evolutionary trajectory of the aphid X chromosome (Jaquiéry, Stoeckel, Rispe, et al. 2012). This peculiar, autosomal-like inheritance of the X predicts similar effective population sizes for X chromosomes and autosomes. This prediction was borne out under the parameters used in the simulations performed by Jaquiéry, Stoeckel, Rispe, et al. (2012). Thus, aphids are interesting models to test causes for faster-X effects, since they are likely unaffected by confounding factors linked to the smaller effective population size of the X. Furthermore, in contrast to standard systems, variance in reproductive success between sexes, population expansion, bottlenecks, and sex-biased dispersal should not differentially affect aphid sex chromosomes and autosomes since the X is transmitted with equal probability through fathers and mothers (see Jaquiéry, Stoeckel, Rispe, et al. 2012 for further explanations). Mutation and recombination rates are also expected to be equal across chromosomes because of their similar mode of inheritance and the complete absence of crossing overs in males (Jaquiéry, Stoeckel, Rispe, et al. 2012). These similarities between X chromosomes and autosomes make aphids exceptionally useful to pinpoint the causes of faster-X evolution, since the factors mentioned above need not be accounted for. Still, a notable difference between X and autosomes in aphids is the theoretical propensity of the X to accumulate sexually antagonistic mutations beneficial for males and detrimental to asexual females, which is the consequence of cyclical parthenogenesis combined with X inheritance patterns (Jaquiéry et al. 2013). Importantly, the X should always adapt more rapidly than the autosomes, regardless of the dominance coefficient of alleles, as long as there is ongoing selection on males (Jaquiéry et al. 2013).
Fig. 1.
—Life-cycle of the pea aphid and ploidy levels for autosomes (A) and the sex-chromosome (X) (adapted from Jaquiéry et al. 2013).
Empirical analyses on a small subset of pea aphid (Acyrthosiphon pisum) genes showed that X-linked genes evolve faster than autosomal genes (Jaquiéry, Stoeckel, Rispe, et al. 2012) and that genes expressed predominantly in males (hereafter “male-biased” genes) were predominantly locate on the X (Jaquiéry et al. 2013). Subsequent genome-wide analyses did not, however, support faster-X evolution (Purandare et al. 2014), and found a lesser degree of enrichment of male-biased genes on the X (Purandare et al. 2014; Pal and Vicoso 2015). These discrepancies likely stem from the fact that these two studies did not assign individual genes to chromosome types, but entire scaffolds, which contain assembly errors (as shown by Bickel et al. 2013 and suggested by Jaquiéry et al. 2013). Misassignment of genes to chromosomes would artificially decrease the contrast between X-linked and autosomal genes.
Here, we aimed to overcome these shortcomings in order to fully disentangle the causes for faster-X evolution in aphids. For this, we first assigned genes to the X or to autosomes at the scale of the entire genome in the pea aphid. On a large set of genes, we then combined estimates of substitution rates at the interspecific level with polymorphism data in pea aphid populations and gene expression levels in the various genders and morphs. This allowed the assessment of how relaxed selection (genetic drift) and adaptation contribute to the faster evolution of the X chromosome in this system.
Materials and Methods
Assignment of Scaffold Regions to the X and Autosomes
Full-Genome Sequencing of Females and Males
An asexual aphid mother has the same diploid autosomal genome as her sons, but has two X chromosomes instead of just one (fig. 1). We took advantage of this XX/X0 system to assign pea aphid genome sequences (Acyr 2.0, Genbank accession GCA_000142985.2, IAGC 2010) to the X or to the autosomes by comparing sequencing depth along assembled scaffolds between mapped reads from females and males of the same parthenogenetic lineage (clone). DNA from five asexual females, five winged males and five wingless males of clone P123 (Simon et al. 2011) was extracted with the Qiagen DNeasy Blood and Tissue kit, following the manufacturer’s protocol. The male wing polymorphism in this clone was used to determine the X copy that each male carried, based on the knowledge that the locus that controls this trait is X-linked (Caillaud et al. 2002) and is heterozygous in clone P123 (Frantz et al. 2010). Each individual was genotyped at seven polymorphic microsatellite markers (Peccoud et al. 2008) to confirm its clonal identity. One of those markers, which is known to be X-linked (Caillaud et al. 2002), allowed us to confirm the nature of the X copy inherited by each male. The three DNA extracts (P123 asexual females and the two types of P123 males) were sequenced on the Illumina Hiseq 2000 platform yielding 100-bp pair-end reads at ∼43× coverage for the females sample and 25–30× coverage for each male type. Reads from each sample were mapped to scaffolds of the pea aphid genome assembly (Acyr 2.0) and to genome sequences of the bacterial symbionts of this pea aphid clone using the method described in Gouin et al. (2015) using Bowtie 2 (Langmead and Salzberg 2012) with proper insert sizes and parameters set as default. Depth of coverage at each nucleotide position of the reference genome was recorded and single nucleotide polymorphisms (SNPs) were identified using GATK’s Haplotype Caller (McKenna et al. 2010; DePristo et al. 2011). The raw sequence data have been deposited in the SRA division of Genbank (project accession: ERP022905 and PRJNA385573).
Comparison of Sequencing Depth between Males and Females
The following analysis was performed in R (R Development Core Team 2015). We analyzed genome positions covered by 20–70 reads in the asexual female sample, a range chosen to eliminate regions with low-coverage and regions with suspiciously high coverage (potentially duplicated or repeat-rich regions). Since overall coverage was slightly higher for one of the male types (∼30×) than for the other (∼25×), we normalized the depth of coverage data of the second male type (multiplying coverage estimates by a 30/25 ratio). We then averaged depth of coverage at each base position over male types. The ratio of male median coverage depth to female median coverage depth was calculated on 10-kb scaffold windows sliding by 2-kb steps. A single window was used for scaffolds shorter than 10 kb. We expected the ratio of median coverage depth to be two times larger for autosomal regions than for X chromosome regions. Accordingly, this ratio had a clearly bimodal distribution (supplementary fig. S1, Supplementary Material online), with modes at 0.34 and 0.66. We assigned each 10-kb window to the X if its ratio ranged between 0.2 and 0.445, and to autosomes if it ranged between 0.53 and 1, whereas the region was tagged as “ambiguous” if it ranged between 0.445 and 0.53. Windows assigned to the same chromosome type and which were separated by less than four consecutive “ambiguous” windows were aggregated into a scaffold region we call a “block.” A whole block, including its “ambiguous” windows, was assigned to the corresponding chromosome type.
Comparison of Male and Female Genotypes
The inheritance of single nucleotide polymorphisms (SNPs) is also informative about the type of chromosome carrying a scaffold block. SNPs that are heterozygous in females but are also heterozygous in males are necessarily located on autosomes. Conversely, SNPs which are heterozygous in females but homozygous (hemizygous) in males must be on the X. This SNP-based approach is, however, expected to be less powerful than the depth of coverage-based method for genomic regions with low heterozygosity. Thus, we only used SNP data to validate X/A assignments based on depth of coverage ratio (supplementary fig. S2, Supplementary Material online). A position was determined as heterozygous if the rarest allele was represented in at least 25% of the reads, otherwise it was considered homozygous. Assignment of SNPs to chromosome types was performed according to the genotypes of males, as described earlier. SNPs showing inconsistent genotypes (e.g., females and males of one type are both heterozygous while males of the other type are homozygous) were not assigned. Assignments based on depth of coverage were then visually compared with SNP-based assignments (supplementary fig. S2, Supplementary Material online) and to a set of 305 microsatellite markers assigned to chromosomes (Jaquiéry et al. 2014; supplementary file S1, Supplementary Material online).
Assignment of Predicted Genes to the X and Autosomes
We used the 36,990 genes (v2.1) predicted from the Acyr 2.0 genome assembly available at http://bipaa.genouest.org/is/aphidbase/ (last accessed January 25, 2017). Each of these genes was determined to be X-linked or autosomal if the full length of its coding sequence (CDS) was found in a single scaffold block or was spread over several scaffold blocks assigned to the same type (either X, or A). Genes that could not be unambiguously assigned (mainly because they were located on “ambiguous” blocks) were removed from further analyses. We also excluded 589 predicted genes that corresponded to rRNA (noncoding DNA).
Sex-Biased Gene Expression
We used the eight RNAseq libraries from Jaquiéry et al. (2013) to characterize gene expression patterns between morphs. Briefly, these eight libraries correspond to whole insects, with three male libraries, three parthenogenetic female libraries and two sexual female libraries—different libraries in each morph representing biological replicates—using adults of a single clone of A. pisum (clone LSR1). Details regarding aphid rearing, library preparation, and sequencing are provided in Jaquiéry et al. (2013). Libraries were mapped to Acyr 2.0 as described previously. The number of reads covering each CDS was then counted. Read counts were normalized with the R package DESeq with default parameters (Anders and Huber 2010). For each gene, the effect of the morph (a three-level factor comprising male, sexual female, and asexual female) on expression was tested with a GLM (R package MASS, Venables and Ripley 2002) with a quasi-poisson distribution of residuals, considering the different libraries for each morph as replicates. P values were corrected for multiple testing using the Benjamini–Hochberg method implemented in R. Genes differentially expressed between morphs (P < 0.05 after adjusting for multiple testing) were then categorized according to their pattern of expression in the different morphs as described in table 1.
Table 1.
Number of X-Linked and Autosomal Genes and Frequency of X-Linkage for Classes of Genes with Contrasted Patterns of Expression between Morphs
| Category of Genes | Number of X-Linked Genes | Number of Autosomal Genes | Frequency of X-Linkage | P Valuef |
|---|---|---|---|---|
| Alla | 13,726 | 19,263 | 0.42 | 10−16 |
| Low expressionb (<10 reads per kilobase) | 10,995 | 8,136 | 0.57 | 10−16 |
| Expressedc (at least 10 reads per kilobase) | 2,771 | 11,127 | 0.20 | 0.0001 |
| Unbiasedd (>10 reads per kilobase and Padj>0.1) | 697 | 3,355 | 0.17 | na |
| 2-fold male-biasede | 1,546 | 2,245 | 0.41 | 10−16 |
| 5-fold male-biasede | 962 | 948 | 0.50 | 10−16 |
| 2-fold sexual female-biasede | 448 | 1,369 | 0.25 | 10−10 |
| 5-fold sexual female-biasede | 148 | 407 | 0.27 | 10−7 |
| 2-fold asexual female-biasede | 244 | 1,023 | 0.19 | 0.10 |
| 2-fold asexual female-biasede | 93 | 423 | 0.18 | 0.68 |
All predicted genes that were assigned to the X or autosomes are included.
Genes with on an average <10 reads per kilobase of exon (average over the three morphs).
Genes with on an average ≥10 reads per kilobase of exon (average over the three morphs).
Genes with on an average ≥10 reads per kilobase of exon (average over the three morphs) and with an adjusted P value ≥ 0.1 when tested for morph-biased expression.
A gene was included in the morph-biased category (either male-, female-, or asexual-biased) if the adjusted P value for a morph effect was <0.05 and if it was at least x-fold (2 or 5) more expressed in one of the morph compared with the two other morphs.
Deviation from expectation (given by the “unbiased” category) was evaluated with a test of proportion.
Evolutionary Rates
To assess substitution rates in X-linked and autosomal genes, sequences from another aphid species were necessary. Acyrthosiphon svalbardicum was chosen to limit the risk of mutational saturation and of chromosomal rearrangements between the two species. Note that rearrangements should not increase the contrast between X and autosomes (i.e., if there is a chromosome type effect on evolutionary rates, rearrangements will only decrease the observed differences, so our tests are conservative). Asexual females of A. svalbardicum were collected in Svalbard in 2009, and were then reared in the lab under 10:14 light:dark and 15°C on Dryas octopetala. Ten females were frozen in liquid nitrogen and kept for subsequent RNA extraction using the RNeasy plant mini kit (Qiagen) according to manufacturer’s instructions. Two separate RNA extractions of five adults were performed. RNA quality was checked by Bioanalyzer (Agilent) and quantified by Nanodrop (Thermo Scientific). One sample made of a pool of 2 µg of the two independent RNA extractions was sent to GATC Company for RNA paired-end sequencing. The raw sequence data have been deposited in the SRA division of Genbank (project accession: PRJNA385897).
A de novo transcriptome assembly for A. svalbardicum was obtained following the methods of Rispe et al. (2016). Low quality parts of the reads were trimmed from the right ends with prinseq-lite (http://prinseq.sourceforge.net/; last accessed January 25, 2017) when the mean of phred score in a 20-bp window was <20. Reads longer than 20 bp after trimming were reorganized by pairs (orphans were suppressed) and assembled with Trinity (Grabherr et al. 2011) using default parameters. Coding regions were predicted using FrameDP (Gouzy et al. 2009). Reciprocal BLASTN (Altschul et al. 1990) searches between CDSs of A. svalbardicum and of A. pisum were carried out with an e-value threshold of 10−8. The following steps were performed with an R script. A reciprocal best hit criterion was used to identify putative orthologous genes between the two species. These were aligned by the pairWiseAlignment function of the Biostrings package (Pages et al. 2016). Indels were inspected to flag CDS regions where the two species did not present the same reading frame. Bases in these regions were replaced with Ns, and were trimmed by the Gblocks program (Castresana 2000; Talavera and Castresana 2007), alongside regions of unreliable alignment. We then estimated pairwise synonymous (dS) and nonsynonymous (dN) substitution rates for each gene, using the codon-based method of Li (1993), as implemented in the R package seqinR (Charif and Lobry 2007). Only the 9,696 genes (out of 9,924) with an alignment length of >90 nucleotides, dN <0.3 and dS <2 were kept. We also truncated dN/dS ratios to a maximal value of 2.5.
Estimates of Selection Intensity Based on Intraspecific Polymorphism
Polymorphism data for A. pisum were obtained from 60 genotypes originating from three alfalfa (Medicago sativa) fields located in France and Switzerland (Jaquiéry, Stoeckel, Nouhaud, et al. 2012). These fields can be considered to harbor a single large population of A. pisum (Peccoud, Ollivier, et al. 2009; Jaquiéry, Stoeckel, Nouhaud, et al. 2012). DNA was extracted from four asexual females of each clone using the method described earlier. Because the approach described below does not require reconstructing allele sequences or individual genotypes, sequencing the pooled individuals (Gautier et al. 2013) was used to save costs. After RNAse treatment on each sample and DNA dosage with Pherastar, DNA samples were pooled to attain equimolar proportions. Paired-end libraries were then sequenced on two lanes of Illumina HiSeq 2000 using the Illumina Sequencing Kit v3 (producing 100-bp reads) by Beckman Coulter Genomics (Danvers, MA). This yielded ∼85× of sequencing coverage, hence an expectation of 0.71× per individual chromosome. Reads were mapped to Acyr 2.0 and symbiont genome sequences as described previously. The two alignment (BAM) files (one per sequencing lane) were filtered from PCR duplicates using SAMtools rmdup (Li et al. 2009) and reads realigned near indels using the Genome Analysis Toolkit (McKenna et al. 2010). The raw sequence data have been deposited in the SRA division of Genbank (project accession: PRJNA385905).
The two BAM files were merged and converted as pileup format using SAMtools (options -B -Q 0 –R) (Li et al. 2009). A modified estimator of Tajima’s D (Tajima 1989) which takes into account sequencing errors (Achaz 2008) was then calculated from this mpileup with Popoolation 1.2.2 (Kofler et al. 2011), after subsampling at a uniform coverage (subsample-pileup.pl, options: –target-coverage 30 –max-coverage 120 –method withoutreplace). Computations were performed for each gene including introns (Variance-at-position.pl –pool-size 120). Tajima’s D allows evaluating the type of selection at work, since selective sweeps and/or purifying selection tend to decrease it, and balancing selection tends to increase it.
The McDonald and Kreitman (1991) approach, which compares fixed mutations to polymorphic mutations in CDS, was adopted to further evaluate selection pressures on these different categories of genes, using the DoS estimate (Direction of Selection, Stoletzki and Eyre-Walker 2011). Positive, null, and negative values of DoS, respectively, suggest adaptive evolution, neutral evolution, and purifying selection. Fixed mutations between species were counted from alignments we previously generated for A. svalbardicum and A. pisum CDSs. We restricted the analysis to regions of reliable alignments, as given by the Gblocks txts outputs. In these regions, we called SNPs on the BAM files with LoFreq (Wilm et al. 2012), which offers a good compromise between speed, sensitivity, and accuracy in pools of multiple individuals (Huang et al. 2015). We used SAMtools mpileup (Li et al. 2009) to assess depth of coverage at all positions in these regions, polymorphic or not. We instructed mpileup to discard reads with mapping quality 0. The following was done in R. We discarded all positions covered by less than three reads (both BAM files combined). At each SNP, the number of polymorphic mutations was the number of different bases (alleles) found in the pea aphid population minus one. A fixed difference was counted if no base was shared at a position between the pea aphid population and A. svalbardicum. The number of polymorphic nonsynonymous mutations per codon was taken as the number of amino acids found in the pea aphid populations for that codon minus one. To count the number of fixed nonsynonymous differences per codon, we considered that a codon might differ between the two species by up to three mutations. Any of these may involve a change in protein sequence that we cannot ascertain without knowledge on the order of appearance of the mutations. We adopted parsimony and considered the minimum number of mutations required between the two codons. If several codons were present in the pea aphid population (due to a SNP), we considered the minimum number of coding changes that any pair of codons between the pea aphid and A. svalbardicum involves. For all these counts, we discarded rare codons showing more than one SNP, because the actual codons (and amino acids) present in the pea aphid population cannot be determined without phasing. We counted the following for each gene: the number of polymorphic nonsynonymous changes (Pn), the number of all polymorphic changes minus Pn (which is the number of polymorphic synonymous mutations, noted Ps), the number of fixed nonsynonymous differences (Dn), the number of all fixed differences minus Dn (which is the number of fixed synonymous changes, noted Ds). DoS was then calculated as Dn/(Dn + Ds)−Pn/(Pn + Ps) for each gene. Similarly, we measured α (the proportion of amino acid substitution driven by positive selection) as (Stoletzki and Eyre-Walker 2011). For both indices, we considered only genes whose average depth of coverage, as given by mpileup, was between 20 and 150 (expected coverage was ∼85) to avoid including genes presenting multiple collapsed copies that could artificially inflate polymorphism.
Statistical Analyses
Differences in expression levels between X-linked and autosomal genes in the different morphs, as well as differences in dN/dS, dN, and dS between X-linked and autosomal genes were tested with Mann–Whitney U tests. The latter analysis was done on all genes, and on genes grouped based on an average expression over the three morphs. To evaluate evolutionary forces responsible for faster-X evolution, we then compared dN/dS, Tajima’s D, DoS, and α between X-linked and autosomal genes for classes of genes with different expression patterns (unbiased, male-biased, sexual female-biased, and asexual female-biased genes). For biased genes, we considered different fold changes in expression (2- to 5-fold, and > 5-fold). Statistical significance was evaluated with Mann–Whitney U tests. Finally, we tested the factors affecting log-transformed dN/dS using a complete linear model. Included variables in model 1 were CDS size, CAI (the codon adaptation index, calculated with CAIcal, Puigbò et al. 2008), τ (a measure of morph specificity in expression, Yanai et al. 2005), mean expression level (averaged over the three morphs), and chromosome. To test whether dN/dS measures were significantly higher for X-linked male-expressed genes than for autosomal genes (expected if selection is more efficient on X due to the hemizygosity of this chromosome in males), we constructed a second model (model 2) including the following variables: CDS size, CAI, τ, expression level in asexual female, expression level in sexual females, expression level in males, chromosome, and the interaction between the last two terms. Significance was tested with permutations in the R package lmPerm (Wheeler and Torchiano 2016).
Results
Gene Assignment to the X and Autosomes
Based on the depth of coverage ratio based on reads from males versus females, 64% of the nucleotides assembled in the pea aphid reference genome (Acyr 2.0) were assigned to autosomes and 31% to the X chromosome, whereas only 5% could not be assigned (supplementary fig. S1, Supplementary Material online). Genotypes of males at SNPs that were heterozygous in the female generally confirmed the assignment from coverage depth data (supplementary fig. S2, Supplementary Material online), though confirmation was not possible in regions lacking such SNPs. These estimates roughly correspond to the expected size of the X chromosome in the pea aphid, which represents ∼30% of the chromosome content based on karyotypes (Mandrioli and Borsatti 2007). This assignment revealed a high rate of misassembly in Acyr 2.0: 56% of scaffolds ≥150 kb (which represent 80% of the assembly length) contained blocks assigned to both chromosome types (supplementary fig. S3 and table S1, Supplementary Material online). Based on assigned scaffold blocks, 19,263 predicted genes were located on autosomes and 13,726 on the X chromosome, whereas 4,001 genes could not be unambiguously assigned. The X chromosome contained a higher fraction of predicted genes than expected from its relative size (42%, test of proportion, P < 10−15).
Gene Evolutionary Rates
We assessed substitution rates by comparing pea aphid gene sequences to transcripts sequenced from a related species (A. svalbardicum). We found that X-linked genes had almost a twice higher nonsynonymous substitution rate, dN (mean dNX= 0.034; dNA= 0.019, Mann–Whitney U = 4,839,860, P <10−15, n = 9,096) and only slightly higher synonymous substitution rate, dS (mean dSX=0.101; dSA= 0.085, Mann–Whitney U = 6,190,137, P <10−6), compared with autosomal genes. As a result, the evolution of X-linked genes involves more protein-sequence changes (in proportion) than the evolution of autosomal genes (mean dNX/dSX = 0.390; dNA/dSA = 0.237; Mann–Whitney U = 5,026,170, P <10−15, fig. 2A and supplementary table S2, Supplementary Material online).
Fig. 2.
—Evolutionary rates for autosomal and X-linked genes and gene expression in males, sexual and asexual females. (A) Evolutionary rates (dN/dS) are shown for all genes (expressed or not in Acyrthosiphon pisum) and for genes expressed at different levels (when averaged over male, sexual and asexual females): lowly expressed genes (i.e., covered by < 100 reads per kilobase of exon model); moderately expressed (from 100 to 1,000 reads per kilobase), highly expressed genes (>1000 reads per kilobase). The number of genes per category is shown above each boxplot. (B) Expression level for X-linked (n = 13,613) and autosomal genes (n = 18,812) in males, sexual females and asexual females. It should be noted that males carry only one X chromosome per cell and females carry two. Significance was tested with Mann–Whitney U tests.
Causes of Faster-X Evolution
High ratios of nonsynonymous to synonymous rates, dN/dS, can result from a decreased influence of selection—and thus an increased influence of drift—on amino acid substitutions (i.e., relaxed negative selection) and/or from more efficient selection of adaptive changes in the protein sequence (i.e., increased positive selection). To distinguish between these two hypotheses, we used gene expression levels as proxies for possible impacts on fitness, although we recognize that this proxy suffers limitations (Wall et al. 2005; Zhang and He 2005; Hart et al. 2014). The positive correlation between α (the proportion of amino-acid substitutions driven by positive selection) and expression demonstrates the relevance of this proxy in our data set (supplementary fig. S4, Supplementary Material online). Expression levels of X-linked genes were significantly lower than those of autosomal genes in all three aphid morphs: males, sexual females, and parthenogenetic females (fig. 2B). Expression level averaged over the 13,726 X-linked genes ranged from ∼52 reads per kilobase in sexual and asexual females to 155 reads per kilobase in males, whereas expression level averaged over the 19,263 autosomal genes varied from 575 (in asexual females) to 648 reads per kilobase in males. Genes that are not expressed or expressed at a low level may have a reduced effect on the phenotype and may therefore accumulate nonsynonymous substitutions faster (reduced purifying selection), an hypothesis supported by the increase of α with expression (supplementary fig. S4, Supplementary Material online). We indeed observed that, for both X and autosomes, dN/dS ratios decrease with increasing expression levels (averaged over the three morphs, P < 10−15 in table 2, model 1, supplementary fig. S4, Supplementary Material online), and that the contrast between X and autosomes tends to decline for highly expressed genes (fig. 2A and supplementary table S2, Supplementary Material online). The regression model also revealed that dN/dS decreases with gene length (P < 10−11), increases with τ (a measure of morph specificity in expression, P < 10−15) and CAI (P < 10−15), and highlighted a significant chromosome effect (P = 0.00015), dN/dS being higher for X-linked genes (model 1, table 2). A slight contrast in dN ratios is still maintained for highly expressed genes, though (supplementary table S2, Supplementary Material online). Therefore, low expression levels of X-linked genes may not entirely account for faster-X evolution in aphids.
Table 2.
Results from the Linear Models Examining the Below Variables on Log-Transformed dN/dS
| Variables | Estimate | P Value |
|---|---|---|
| Model 1 | ||
| CDS size | −1.0×10−5 | 10−11 |
| CAI | 0.33 | <10−15 |
| τ | 0.21 | <10−15 |
| Log(Mean expression+1) | −0.019 | <10−15 |
| Chromosome | −0.012 | 0.00015 |
| Model 2 | ||
| CDS size | −9.4×10−6 | 10−9 |
| CAI | 0.19 | 10−11 |
| τ | 0.14 | <10−15 |
| Log(Asexual female expression+1) | −0.047 | <10−15 |
| Log(Sexual female expression+1) | 0.011 | 10−6 |
| Log(Male expression+1) | 0.013 | 10−8 |
| Chromosome | −0.011 | 0.0007 |
| Log(Male expression+1): Chromosome | −0.0012 | 0.37 |
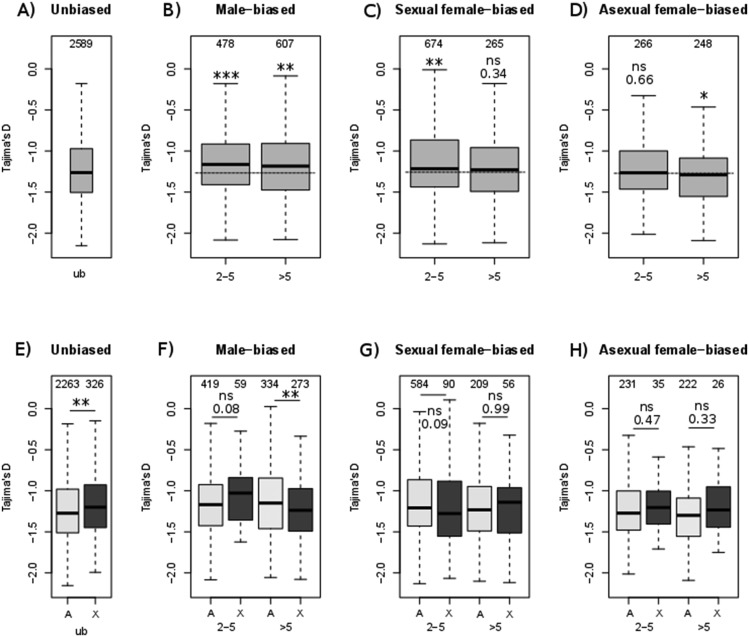
Selection may also be relaxed in genes that are predominantly expressed in rare morphs (males and sexual females), which constitute a minor fraction of the annual life cycle of aphids, which is dominated by asexual females. Relaxed selection on mutations affecting male-biased genes (Brisson and Nuzhdin 2008; Purandare et al. 2014), combined with the tendency of such genes to locate on the X (Jaquiéry et al. 2013; Pal and Vicoso 2015) could contribute to faster-X evolution. However, the influence of X-linkage could not properly be evaluated in Purandare et al. (2014) because misassembled scaffolds, rather than individual genes, were assigned to chromosomes. Our new data set of X-linked and autosomal genes unambiguously confirmed that the X is largely enriched for genes overexpressed in males, and to a smaller extent for those overexpressed in sexual females (table 1). Like Purandare et al. (2014), we observed higher dN/dS ratios in genes overexpressed in the rarer morphs (i.e., males and sexual females, fig. 3B and C) than in genes overexpressed in the common morph (parthenogenetic females, fig. 3D) when considering X-linked and autosomal genes together. The global analysis (model 2, table 2) further demonstrated that dN/dS decreases with expression in asexual females (P < 10−15), but increases with expression in males and sexual females (P < 10−8 and 10−6, respectively) as well as with morph specificity in expression τ (P < 10−15). Tajima’s D also tends to increase in genes overexpressed in the sexual morphs compared with unbiased genes (significantly so for all male-biased and for 2- to 5-fold female-biased genes, fig. 4B), but not in genes overexpressed in the common morph (where D is significantly lower compared with unbiased genes, fig. 4D), a pattern compatible with relaxed selection on genes expressed mainly in the rare morphs. Supporting this hypothesis, α (the proportion of amino acid substitutions driven by positive selection) is significantly lower for male- and sexual female-biased genes compared with unbiased genes, but not in genes overexpressed in the common morph (supplementary fig. S6, Supplementary Material online). However, the DoS did not differ significantly between these categories of genes, except for strongly female-biased genes (supplementary fig. S5, Supplementary Material online).
Fig. 3.
—Substitution rates of genes (dN/dS, measured between Acyrthosiphon pisum and A. svalbardicum) according to the ratios of expression levels between morphs. Panels (A–D) consider all genes together (X-linked and autosomal), and panels (E–H) consider X-linked (dark gray) and autosomal (light gray) genes separately. The number of genes in each class is shown above each boxplot. Only genes supported by at least 100 reads per kilobase of exon model were retained. ub: unbiased genes (Padj > 0.1 for morph effect on expression), 2–5: levels of gene expression are two to five times higher in the specified morph (Padj <0.05 for morph effect), >5: levels of gene expression are at least five times higher in the specified morph (Padj < 0.05). Significance of differences: ns: P > 0.05; *P < 0.05, **P < 0.01, ***P < 0.001 (Mann–Whitney U tests). For panels (B–D), differences correspond to comparisons with genes of the “unbiased” category, whereas X and autosomes were compared in panels (F–H).
Fig. 4.
—Tajima’s D according to the ratios of gene expression levels between morphs. Panels (A–D) consider all genes together (X-linked and autosomal) and panels (E–H) consider X-linked (dark gray) and autosomal (light gray) genes separately. Terms are defined as in figure 3. Dashed lines show median values for unbiased genes. Significance of differences was tested with Mann–Whitney U tests.
When analyses were done by chromosome type, dN/dS ratios of X-linked genes were significantly higher than those of autosomal genes for both sexual female- and male-biased genes (fig. 3E–H), but not for asexual females. Contrastingly, Tajima’s D differed between chromosome types only for strongly male-biased genes (being lower for X-linked genes, suggesting more positive selection, fig. 4F) and for unbiased genes (being larger for X-linked genes, possibly revealing more balanced selection, fig. 4E). No signal was detected between chromosome types based on the DoS index or α (supplementary figs. S5 and S6, Supplementary Material online).
Alternatively, the faster evolution of X-linked male-biased genes compared with autosomal male-biased genes could result from the fact that the former are present in a hemizygous state in males. Nonsynonymous mutations on the X are thus more exposed to selection in males, since they are not masked by potentially dominant alleles, such that adaptive mutations on the X should more rapidly and more likely reach fixation than adaptive mutations on autosomes. This hypothesis predicts that the contrast between substitution rates of X-linked genes and autosomal genes will be highest for male-biased genes, and lowest for sexual- and asexual female-biased genes, because in these morphs, the X is always diploid and adaptive mutations can be recessive. We indeed observed these patterns (fig. 3F), although the interaction between male expression level and chromosome in model 2 (table 2) was not significant. Nevertheless, the significantly lower Tajima’s D for X-linked male-biased genes compared with autosomal genes provides some support to this hypothesis (fig. 4F, P < 0.01) as do DoS and α (though not significantly, supplementary figs. S5F and S6F, Supplementary Material online).
Discussion
Here, we performed a genome-wide identification of X-linked genes, enabling us to locate a large number (13,726) and proportion (42%) of predicted genes on the X chromosome. We demonstrated that these genes tend to evolve faster than autosomal genes, on an average, confirming earlier results based on a much smaller set of genes (Jaquiéry, Stoeckel, Rispe, et al. 2012). We found that faster-X evolution mainly results from the low expression of a large fraction of X-linked genes, which likely have a lesser effect on phenotypes and may accumulate nonsynonymous mutations at a higher rate. The enrichment of the X chromosome with genes expressed in the rare male and sexual female morphs (which show signs of relaxed selection) might also contribute to faster-X evolution. Lastly, some of our analyses suggested that higher exposure of recessive X-linked alleles to selection in hemizygous males might also contribute to faster-X evolution via more efficient positive selection, although this hypothesis requires further testing.
We demonstrated clear faster-X evolution in the pea aphid based on a large set of X-linked and autosomal genes. The nonsynonymous to synonymous substitution ratio (dN/dS) for X-linked genes is 1.69 times greater than for autosomal genes. This clearly places aphids among species showing strong contrast between the evolution of X-linked and autosomal genes, as the dN/dS for X-linked genes is between ∼0.9 and ∼1.8 times that of autosomes in most species studied (i.e., Drosophila, mammals, birds and moths, review in Meisel and Connallon 2013; see also Sackton et al. 2014). In addition, both X and autosomes exhibit higher dN/dS for sex-biased genes, as previously shown in aphids (Purandare et al. 2014) and in several other organisms (Torgerson and Singh 2003; Ellegren and Parsch 2007; Parsch and Ellegren 2013; Kousathanas et al. 2014).
Remarkably, the pea aphid has the same effective population size for the X and autosomes under the demographic scenario investigated in Jaquiéry, Stoeckel, Rispe, et al. (2012), such that hemizygosity in males should be the only differentiating factor affecting the evolution of genes located on different chromosome types. However, our analyses revealed another key difference between X-linked and autosomal genes, in that the former are, on an average, four to ten times less expressed than the latter (fig. 2B), and expectedly show higher rates of substitution (supplementary fig. S4, Supplementary Material online). Such negative correlations between substitution rates and expression levels have already been observed in several species (Drummond et al. 2005; Nguyen et al. 2015; Zhang and Yang 2015). Therefore, enrichment of the X with lowly expressed genes is likely to explain, to a large extent, the faster-X evolution in aphids.
Gene expression differs between chromosome types in another dimension, as the X is enriched in genes that are mostly expressed in the rare morphs (i.e., males and sexual females) (Jaquiéry et al. 2013; Pal and Vicoso 2015). Such genes should evolve under more relaxed constraints as they are exposed to the selective environment only during a short period of the aphid life cycle (Brisson and Nuzhdin 2008; Purandare et al. 2014). Enrichment of the X chromosome with genes expressed in the rare male morph (which show signs of relaxed selection based on α) might also contribute to faster-X evolution in the pea aphid.
This leaves hemizygosity of the X chromosome in males, which exposes all X-linked alleles expressed in males to the selective environment, as another contributing cause. This hypothesis finds some support in the contrast in dN/dS between X-linked genes and autosomal genes, which is larger for male-biased genes than for sexual and asexual females-biased genes (fig. 3F–H), and in the lower Tajima’s D for male-biased X-linked genes than for genes on autosomes (fig. 4F and G). The DoS or α also tend to support the hypothesis of positive selection on the X, but the effect of chromosome type proved not significant.
The mean expression levels of X-linked genes measured from the whole bodies were strikingly lower than those of autosomal genes, in all morphs studied. The difference we found is more pronounced than in previous observations (Jaquiéry et al. 2013; Pal and Vicoso 2015) probably due to more reliable gene assignments to chromosomes (we found that 10% of the 3,712 genes used in Jaquiéry et al. 2013 had been misassigned because of scaffold misassembly). Lower expression of X-linked genes compared with autosomal genes is observed in mammals (Nguyen et al. 2015), but not in Drosophila (Zhang and Presgraves 2016). To our knowledge, no other taxon displays such a strong contrast between the X and autosomal gene expression levels. This raises the question of why genes on the X are so lowly expressed in this species. We cannot rule out that the lower average expression of X-linked genes measured on whole bodies reflects expression patterns that are more tissue/organ-specific than those of autosomal genes. Another hypothesis from a theoretical model (Jaquiéry et al. 2013) predicts that the X chromosome is more easily invaded than autosomes by sexually antagonistic alleles beneficial to males and deleterious to females. Such evolution may have favored a global decrease in gene expression of this chromosome in the common morph (the asexual females) for which it could be harmful. Indeed, an analysis of the structure of the chromatin has revealed that the chromatin of the X is less accessible in females than in males, suggesting the existence of a global mechanism of regulation (Richard et al. 2017). Pseudogenization on the X chromosome would have ensued if genetic variation in lowly expressed genes has little effect on fitness. Yet, this chromosome carries one third of the genome and contains a higher fraction of genes than predicted by its relative size. Insights into the respective role of sexual antagonism or the breadth of gene expression on the peculiar expression patterns observed here could be gained by studying expression of X and autosomal genes in different male and female tissues. Particularly, transcriptomes of tissues subject to different sex-specific selection pressures (Parisi 2003; Khil et al. 2004; Yang et al. 2006; Huylmans and Parsch 2015) could help examine this hypothesis.
Assignments of scaffold blocks to chromosomes revealed widespread errors in the pea aphid genome assembly (Acyr 2.0). More than half of scaffolds >150 kb are clear chimeras of X and autosomes. This is a minimal estimate for the rate of misassembly, since our method only detects breakpoints between X and autosomes. Consequently, we confidently conclude that the genome of the pea aphid presents considerable assembly problems, to a degree that goes far beyond what current assembly pipelines typically yield (Salzberg et al. 2004; Muggli et al. 2015). Although the cause of these errors remains undetermined, they have important drawbacks for genomic studies on a species that is currently considered the model aphid, in particular those relying on the physical organization of the genome, ranging from high-resolution genome scans to studies of chromatin conformation, and genomic rearrangements. The results presented here should not be affected by misassembly because we were able to unambiguously assign almost 90% of the 36,990 predicted genes. Such a high number of genes compared with other arthropod genomes (Adams 2000; Colbourne et al. 2011; Mathers et al. 2017) could indicate errors in gene prediction in the official gene consensus set. There is, however, no doubt that many functional groups show an unusual high level of gene duplication in the pea aphid (IAGC 2010). Most importantly, we see no reason why prediction errors would be more common on the X than on autosomes, and so this should not affect our conclusions.
In conclusion, faster-X evolution of proteins in the pea aphid seems to be primarily explained by relaxed selection on lowly expressed genes, a class of genes more frequent on the X chromosome than on autosomes. We found little evidence that the exposure of X-linked recessive alleles to selection in hemizygous males plays an additional role in faster-X evolution, but this hypothesis deserves further investigation on more specific data sets. Importantly, the pea aphid forms a species complex, including races and cryptic species at different stages of divergence (Peccoud, Ollivier, et al. 2009; Peccoud, Simon, et al. 2009; Peccoud et al. 2015). This complex offers an excellent opportunity to investigate the tempo and mechanisms of chromosome evolution through comparative genomics. In particular, characterizing gene expression in morphs of closely and distantly related lineages could help disentangling the role of drift and selection in the low expression of the X and its masculinization.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Agence Nationale de la Recherche (grants ANR-09-GENM-017-001 to Denis Tagu, ANR-11-BSV7-005-01 to Denis Tagu and ANR-11-BSV7-007 to J.C.S.); INRA-AIP BioRessources (project Poly-Express to J.C.S. and Denis Tagu); the Fondation pour la Recherche sur la Biodiversité (grant AAP-IN-2009-020 to J.C.S.); the Institut Polaire Francais Paul-Emile Victor (IPEV project 426 Arctaphid to Maurice Hullé and J.C.S.); the Genoscope (project 62 AAP 2009/2010 to J.C.S.) and the Swiss National Science Foundation (grants PBLAA-122658 and PA00P3-139720 to J.J.). We would like to thank the GenOuest platform for the use of its cluster to perform the computational analyses, and two anonymous referees for their constructive comments on a previous draft of this manuscript.
Literature Cited
- Achaz G. 2008. Testing for neutrality in samples with sequencing errors. Genetics 179(3):1409–1424.http://dx.doi.org/10.1534/genetics.107.082198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195.http://dx.doi.org/10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11(10):R106.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila V, Campos JL, Charlesworth B.. 2015. The effects of sex-biased gene expression and X-linkage on rates of adaptive protein sequence evolution in Drosophila. Biol Lett. 11(4):20150117..http://dx.doi.org/10.1098/rsbl.2015.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila V, et al. 2014. Faster-X effects in two Drosophila lineages. Genome Biol Evol. 6(10):2968–2982.http://dx.doi.org/10.1093/gbe/evu229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, et al. 2011. Are all sex chromosomes created equal? Trends Genet. 27(9):350–357. [DOI] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J.. 2008. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 25(8):1639–1650.http://dx.doi.org/10.1093/molbev/msn111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel RD, Dunham JP, Brisson JA.. 2013. Widespread selection across coding and noncoding DNA in the pea aphid genome. G3 (Bethesda) 3(6):993–1001.http://dx.doi.org/10.1534/g3.113.005793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Nuzhdin SV.. 2008. Rarity of males in pea aphids results in mutational decay. Science 319(5859):58..http://dx.doi.org/10.1126/science.1147919 [DOI] [PubMed] [Google Scholar]
- Caballero A. 1994. Developments in the prediction of effective population size. Heredity 73(6):657–679.http://dx.doi.org/10.1038/hdy.1994.174 [DOI] [PubMed] [Google Scholar]
- Caballero A. 1995. On the effective size of populations with separate sexes, with particular reference to sex-linked genes. Genetics 139(2):1007–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Boutin M, Braendle C, Simon J-C.. 2002. A sex-linked locus controls wing polymorphism in males of the pea aphid, Acyrthosiphon pisum (Harris). Heredity 89(5):346–352.http://dx.doi.org/10.1038/sj.hdy.6800146 [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.http://dx.doi.org/10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR.. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. Biological and Medical Physics, Biomedical Engineering. New York: Springer Verlag; p. 207–232. [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH.. 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 130(1):113–146. [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G.. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128.http://dx.doi.org/10.1038/sj.hdy.6800697 [DOI] [PubMed] [Google Scholar]
- Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331(6017):555–561.http://dx.doi.org/10.1126/science.1197761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA.. 2004. Speciation. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43(5):491–498.http://dx.doi.org/10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J.. 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 8(9):689–698.http://dx.doi.org/10.1038/nrg2167 [DOI] [PubMed] [Google Scholar]
- Frantz A, Plantegenest M, Simon JC.. 2010. Host races of the pea aphid Acyrthosiphon pisum differ in male wing phenotypes. Bull Entomol Res. 100(1):59–66. [DOI] [PubMed] [Google Scholar]
- Gautier M, et al. 2013. Estimation of population allele frequencies from next-generation sequencing data: pool-versus individual-based genotyping. Mol Ecol. 22(14):3766–3779.http://dx.doi.org/10.1111/mec.12360 [DOI] [PubMed] [Google Scholar]
- Gouin A, et al. 2015. Whole-genome re-sequencing of non-model organisms: lessons from unmapped reads. Heredity 114(5):494–501.http://dx.doi.org/10.1038/hdy.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzy J, Carrere S, Schiex T.. 2009. FrameDP: sensitive peptide detection on noisy matured sequences. Bioinformatics 25(5):670–671.http://dx.doi.org/10.1093/bioinformatics/btp024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652.http://dx.doi.org/10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J.. 2014. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 10:733.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HW, Mullikin JC, Hansen NF.. 2015. Evaluation of variant detection software for pooled next-generation sequence data. BMC Bioinformatics 16(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huylmans AK, Parsch J.. 2015. Variation in the X: autosome distribution of male-biased genes among Drosophila melanogaster tissues and its relationship with dosage compensation. Genome Biol Evol. 7(7):1960–1971.http://dx.doi.org/10.1093/gbe/evv117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvilsom C, et al. 2012. Extensive X-linked adaptive evolution in central chimpanzees. Proc Natl Acad Sci U S A. 109(6):2054–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genome Consortium 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, et al. 2013. Masculinization of the X chromosome in the pea aphid. PLoS Genet. 9(8):e1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, et al. 2014. Genetic control of contagious asexuality in the pea aphid. PLoS Genet. 10(12):e1004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquiéry J, Stoeckel S, Nouhaud P, et al. 2012. Genome scans reveal candidate regions involved in the adaptation to host plant in the pea aphid complex. Mol Ecol. 21:5251–5264. [DOI] [PubMed] [Google Scholar]
- Jaquiéry J, Stoeckel S, Rispe C. et al. 2012. Accelerated evolution of sex chromosomes in aphids, an X0 system. Mol Biol Evol. 29(2):837–847. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Lachance J.. 2012. The genetics of sex chromosomes: evolution and implications for hybrid incompatibility. Ann N Y Acad Sci. 1256:E1–E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD.. 2004. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 36(6):642–646. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge: University Press. [Google Scholar]
- Kitano J, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461(7267):1079–1083.http://dx.doi.org/10.1038/nature08441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, et al. 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One 6(1):e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousathanas A, Halligan DL, Keightley PD.. 2014. Faster-X adaptive protein evolution in house mice. Genetics 196(4):1131–1143.http://dx.doi.org/10.1534/genetics.113.158246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192(2):533–598.http://dx.doi.org/10.1534/genetics.112.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359.http://dx.doi.org/10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 36(1):96–99.http://dx.doi.org/10.1007/BF02407308 [DOI] [PubMed] [Google Scholar]
- Mackay TFC, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178.http://dx.doi.org/10.1038/nature10811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrioli M, Borsatti F.. 2007. Analysis of heterochromatic epigenetic markers in the holocentric chromosomes of the aphid Acyrthosiphon pisum. Chromosome Res. 15(8):1015–1022.http://dx.doi.org/10.1007/s10577-007-1176-4 [DOI] [PubMed] [Google Scholar]
- Mank JE, Nam K, Ellegren H.. 2010. Faster-Z evolution is predominantly due to genetic drift. Mol Biol Evol. 27(3):661–670.http://dx.doi.org/10.1093/molbev/msp282 [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B.. 2010. Effective population size and the faster-X effet: empirical results and their interpretation. Evolution 64(3):663–674.http://dx.doi.org/10.1111/j.1558-5646.2009.00853.x [DOI] [PubMed] [Google Scholar]
- Mathers TC, et al. 2017. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 18(1):27.http://dx.doi.org/10.1186/s13059-016-1145-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M.. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351(6328):652–654.http://dx.doi.org/10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McKenna A, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–1303.http://dx.doi.org/10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y.. 2010. Genetic conflict and sex chromosome evolution. Trends Ecol Evol. 25(4):215–223.http://dx.doi.org/10.1016/j.tree.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T.. 2013. The faster-X effect: integrating theory and data. Trends Genet. 29(9):537–544.http://dx.doi.org/10.1016/j.tig.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggli MD, Puglisi SJ, Ronen R, Boucher C.. 2015. Misassembly detection using paired-end sequence reads and optical mapping data. Bioinformatics 31(12):i80–i88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-P, Galtier N, Nabholz B.. 2015. Gene expression, chromosome heterogeneity and the fast-X effect in mammals. Biol Lett. 11(2):20150010..http://dx.doi.org/10.1098/rsbl.2015.0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages H, Aboyoun P, Gentleman R, DebRoy S.. 2016. Biostrings: string objects representing biological sequences, and matching algorithms. R package version 2.40.2. Available at: https://www.rdocumentation.org/packages/Biostrings/versions/2.40.2, last accessed January 25, 2017.
- Pal A, Vicoso B.. 2015. The X chromosome of hemipteran insects: conservation, dosage compensation and sex-biased expression. Genome Biol Evol. 7(12):3259–3268.http://dx.doi.org/10.1093/gbe/evv215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299(5607):697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Ellegren H.. 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet. 14(2):83–87.http://dx.doi.org/10.1038/nrg3376 [DOI] [PubMed] [Google Scholar]
- Peccoud J, et al. 2008. Host range expansion of an introduced insect pest through multiple colonizations of specialized clones. Mol Ecol. 17(21):4608–4618.http://dx.doi.org/10.1111/j.1365-294X.2008.03949.x [DOI] [PubMed] [Google Scholar]
- Peccoud J, et al. 2015. Genetic characterisation of new host-specialised biotypes and novel associations with bacterial symbionts in the pea aphid complex. Insect Conserv. Divers. 8(5):484–492.http://dx.doi.org/10.1111/evo.12478 [Google Scholar]
- Peccoud J, Ollivier A, Plantegenest M, Simon J-C.. 2009. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci U S A. 106(18):7495–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccoud J, Simon J-C, McLaughlin HJ, Moran NA.. 2009. Post-Pleistocene radiation of the pea aphid complex revealed by rapidly evolving endosymbionts. Proc Natl Acad Sci U S A. 106:16315–16320.http://dx.doi.org/10.1073/pnas.0905129106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P, Bravo IG, Garcia-Vallve S.. 2008. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Dir. 3:38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purandare SR, Bickel RD, Jaquiery J, Rispe C, Brisson JA.. 2014. Accelerated evolution of morph-biased genes in pea aphids. Mol Biol Evol. 31(8):2073–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2015. A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.R-project.org, last accessed January 25, 2017. [Google Scholar]
- Rice WR. 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38(4):735–742.http://dx.doi.org/10.1111/j.1558-5646.1984.tb00346.x [DOI] [PubMed] [Google Scholar]
- Richard G, et al. 2017. Dosage compensation and sex-specific epigenetic landscape of the X chromosome in the pea aphid. Epigenet Chromatin 10:30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispe C, et al. 2016. De novo transcriptome assembly of the grapevine phylloxera allows identification of genes differentially expressed between leaf- and root-feeding forms. BMC Genomics 17(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, et al. 2014. Positive selection drives faster-Z evolution in silkmoths. Evolution 68(8):2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether SA, et al. 2007. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science 318(5847):95–97.http://dx.doi.org/10.1126/science.1141506 [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Church D, DiCuccio M, Yaschenko E, Ostell J.. 2004. The genome assembly archive: a new public resource. PLoS Biol. 2(9):e285.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J-C, et al. 2011. Facultative symbiont infections affect aphid reproduction. PLoS One 6(7):e21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQS, et al. 2014. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159(4):800–813.http://dx.doi.org/10.1016/j.cell.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A.. 2011. Estimation of the neutrality index. Mol Biol Evol. 28(1):63–70.http://dx.doi.org/10.1093/molbev/msq249 [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:485–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.http://dx.doi.org/10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Singh RS.. 2003. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol Biol Evol. 20(10):1705–1709.http://dx.doi.org/10.1093/molbev/msg193 [DOI] [PubMed] [Google Scholar]
- Venables WN, Ripley BD.. 2002. Modern applied statistics with S. 4th ed.New York: Springer. [Google Scholar]
- Vicoso B, Charlesworth B.. 2009. Effective population size and the faster-X effect: an extended model. Evolution 63(9):2413–2426.http://dx.doi.org/10.1111/j.1558-5646.2009.00719.x [DOI] [PubMed] [Google Scholar]
- Wall DP, et al. 2005. Functional genomic analysis of the rates of protein evolution. Proc Natl Acad Sci U S A. 102(15):5483–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler B, Torchiano M.. 2016. lmPerm: permutation tests for linear models. R Package version 2.1.0. Available at: https://cran.r-project.org/web/packages/lmPerm/index.html, last accessed January 25, 2017.
- Wilm A, et al. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40(22):11189–11201.http://dx.doi.org/10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ACC, Sunnucks P, Hales DF.. 1997. Random loss of X chromosome at male determination in an aphid, Sitobion near fragariae, detected using an X-linked polymorphic microsatellite marker. Genet Res. 69:233–236. [Google Scholar]
- Wright S. 1931. Evolution in mendelian populations. Genetics 16(2):97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659.http://dx.doi.org/10.1093/bioinformatics/bti042 [DOI] [PubMed] [Google Scholar]
- Yang X, et al. 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16(8):995–1004.http://dx.doi.org/10.1101/gr.5217506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, He X.. 2005. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 22(4):1147–1155.http://dx.doi.org/10.1093/molbev/msi101 [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang J-R.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420.http://dx.doi.org/10.1038/nrg3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Presgraves DC.. 2016. Drosophila X-linked genes have lower translation rates than autosomal genes. Mol Biol Evol. 33(2):413–428.http://dx.doi.org/10.1093/molbev/msv227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.