Abstract
Antibiotic resistance is one of the greatest current threats to human health, and without significant action we face the chilling prospect of a world without effective antibiotics. Although continued effort toward the development of new antibiotics, particularly those with novel mechanisms of action, remains crucial, this alone probably will not be enough to prevail, and it is imperative that additional approaches are also explored. One such approach is the identification of adjuvants that augment the activity of current antibiotics. This approach has the potential to render an antibiotic against which bacteria have developed resistance once again effective, to broaden the spectrum of an antibiotic, and to lower the required dose of an antibiotic. In this viewpoint we discuss some of the advantages and disadvantages of the use of adjuvants, and describe various approaches to their identification.
Antibiotic resistance in pathogenic bacteria, particularly the six bacterial species termed the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), is one of the greatest threats to human health facing the world today.1 In its 2014 report “Antimicrobial Resistance: Global Report on Surveillance” the World Health Organization (WHO) states that the problem is “so serious that it threatens the achievements of modern medicine. A post-antibiotic era—in which common infections and minor injuries can kill—is a very real possibility for the 21st century”.2 Indeed, in August 2016 the first U.S. case of a patient with an infection that was resistant to every available antimicrobial drug was reported. The K. pneumoniae isolate responsible for the infection, which ultimately resulted in the death of the patient, harbored the New Delhi metallo-β-lactamase (NDM-1) gene and was additionally resistant to all aminoglycosides and polymyxins tested.3 Many of the lifesaving medical practices we take for granted today, such as surgery, premature infant care, cancer chemotherapy, and transplantation medicine, are feasible only with the existence of effective antibiotic therapy.4 Projections indicate that if antibiotic resistance continues to rise at the current rate, drug-resistant infections will account for over 10 million deaths annually by 2050, with an estimated global economic cost reaching U.S. $100 trillion.5
Without any doubt, something must be done. The continuance of effective antibiotic therapy in the wake of the diminishing efficacy of our current armamentarium of antibacterial drugs will require, in addition to significant improvements in antibiotic stewardship,5b either (1) a constant supply of new drugs to replace those being rendered obsolete or (2) a means of prolonging the lifespan of current antibiotics.
The former is not likely to transpire without significant government investment and subsidies as a result of the divestment of pharmaceutical companies from new antibiotic drug discovery.6 The reasons for this divestment stem from both economic and scientific hurdles. From an economic standpoint, new antibiotics have a poor projected return on investment compared to drugs targeted at chronic diseases such as heart disease and high blood pressure, chiefly because antibiotic regimens are typically much shorter in duration and are often curative. Additionally, in efforts to limit resistance acquisition, new antibiotics are increasingly being held in reserve for cases in which no older antibiotics are effective, which further reduces profitability.7 There are also considerable scientific challenges facing the discovery of new antibiotics, exemplified by the fact that only two novel antibiotic classes have been introduced to the clinic since the “golden age” of antibiotic discovery ended in the 1970s. The vast majority of antibiotics that have been approved since then, or are currently in development, are derivatives of previously approved scaffolds, resistance mechanisms against which many bacteria already possess. Furthermore, even the development of such derivatives is slowing as a result of the low-hanging fruit being taken and discouragement of their development by regulators.4
In vitro high-throughput screening of synthetic chemical libraries for the identification of novel antibiotics has been profoundly unsuccessful, particularly for the discovery of antibacterial agents that are active against Gram-negative bacteria.6a,8 The reasons for this lack of success are multifactorial and are described in detail by Tommasi et al.6a Contributory factors include the metrics defined by pharmaceutical companies as defining a hit or lead compound; for example, until recently, many campaigns sought only to identify broad-spectrum agents that exhibit in vitro activity against several isozymes of the target enzyme or protein from both Gram-positive and Gram-negative bacteria. Another factor is the lack of correlation between activity in a biochemical assay and whole-cell activity, and perhaps the most significant factor, particularly in the case of Gram-negative active compounds, is the nature of the compounds that make up most small-molecule synthetic libraries. These libraries typically consist predominantly of compounds that were designed and developed in the combinatorial chemistry era for screens against mammalian targets. These libraries typically occupy a narrow chemical space, and most known antibacterial agents fall outside this space.8,9
Several of these issues have been recognized and are being addressed. The development of more rapid and sensitive diagnostic tests means the use of narrow-spectrum antibiotics is now more feasible, and the EMA and the FDA have recently included narrow-spectrum antibacterial compounds in a list of treatments that qualify for a shorter route to registration.10 The use of whole cell phenotypic screening overcomes the limitation of a lack of translation of activity from biochemical assays, although this type of screening does necessitate follow-up mechanism of action studies. Finally, screening natural products gives access to much needed increased chemical diversity, although unfortunately can often lead to the identification of known scaffolds.11 The search for new standalone antibacterial agents, particularly those with novel mechanisms of action, continues to be hugely important in the fight against drug-resistant bacterial infections. However, the challenges described above, in addition to the almost certainty that resistance to such agents will emerge, means that it is imperative that additional approaches are also explored.
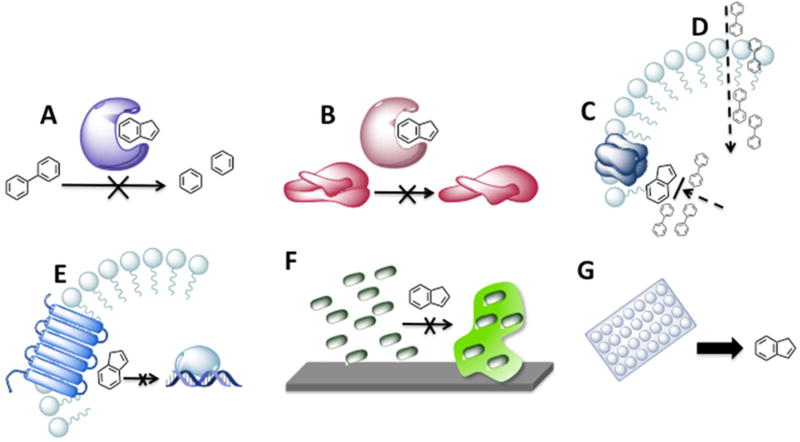
One such approach, which forms the basis of point 2 earlier, is a means of prolonging the lifespan of current antibiotics, namely, the development of antibiotic adjuvants.12 Antibiotic adjuvants are compounds that do not themselves kill bacteria but instead enhance the effect of an antibiotic, for example, by inhibiting a mechanism of resistance. Examples of adjuvant targets are depicted in Figure 1. Adjuvants have potential application in a number of situations, with the most obvious being the case of formerly susceptible bacteria that have acquired resistance. Additionally, adjuvants that suppress intrinsic resistance could expand the activity spectrum of antibiotics, for example, enabling the use of current Gram-positive selective antibiotics for the treatment of Gram-negative infections. Finally, adjuvants that increase bacterial susceptibility to antibiotics for which toxicity is an issue, such as colistin,13 would allow these antibiotics to be efficacious at lower doses, thereby mitigating potential side effects. The identification of nonbactericidal adjuvant compounds holds several advantages over the development of new antibiotics, perhaps the most significant being that evolutionary pressure on bacteria to evolve resistance to a compound that does not exert bactericidal or growth inhibitory effects appears to be abated,14 whereas it is known that optimally designed combination antibiotic therapies have the potential to slow resistance evolution.15 Another advantage is a result of the fact that truly novel antibiotic targets are rare given both the finite number of essential genes and the extensive exploration that this approach has already received. In contrast, the relative infancy of the adjuvant approach means that there are likely a greater number of both undiscovered targets and previously unidentified chemical scaffolds.
Figure 1.
Examples of adjuvant mechanisms of action and discovery: (A) inhibition of antibiotic modification; (B) inhibition of target modification; (C) inhibition of efflux; (D) enhancement of antibiotic uptake; (E) inhibition of signaling pathways that mediate antibiotic resistance; (F) inhibition of biofilm formation, which leads to increased antibiotic tolerance; (G) target-blind whole cell screening of previously approved drugs for adjuvant activity.
The adjuvant approach is not without challenges. Some of the challenges discussed above that plague the development of new antibiotics, such as identifying compounds with the necessary physicochemical properties to access bacterial targets, also apply to the identification of adjuvants. In addition, as with any combination therapy, there is the potential for deleterious drug–drug interactions, and compatible pharmacokinetic and pharmacodynamics properties between the antibiotic and adjuvant are necessary to enable an effective codosing regimen. Although combination therapy for the treatment of bacterial infections is well-precedented, for example, in the treatment of tuberculosis16 and some Gram-negative infections,17 developing a regimen with optimal temporal and spatial delivery of the antibiotic–adjuvant combination may prove more complicated than for combination therapies in which the targets are independent of one another.
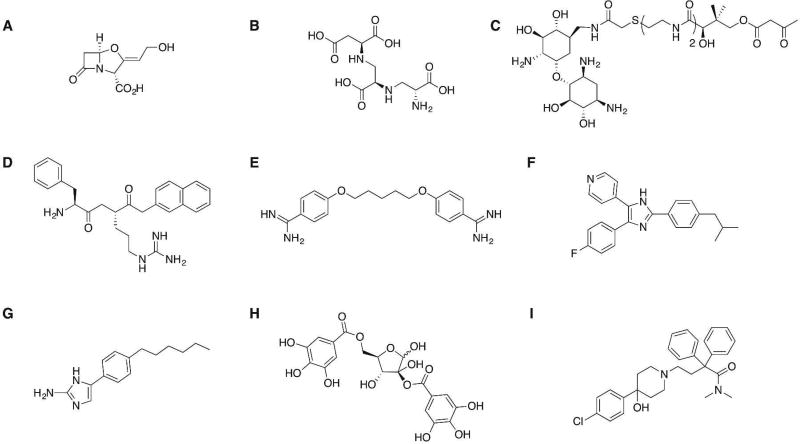
Several compounds that enhance the effects of antibiotics via a number of different mechanisms have been reported, and examples are shown in Figure 2. The classical, and only clinically approved, example of small-molecule antibiotic adjuvants are the β-lactamase inhibitors. β-Lactamase inhibitors, in combination with β-lactam antibiotics, have been successfully employed for over 30 years for the treatment of both Gram-positive and Gram-negative infections and have been extensively reviewed.18 The continued development of new β-lactamase inhibitors to extend the spectrum of activity, for example, to include metallo-β-lactamase (MBL) enzymes19 for which there are no current clinically approved inhibitors, remains an important area of development; however, there are numerous other resistance mechanisms that represent potential adjuvant targets that have been much less well explored. Conceivably any product, pathway, or phenotype that contributes to decreased antibiotic susceptibility represents a potential adjuvant target, many of which have yet to be fully probed.
Figure 2.
Examples of compounds that enhance antibiotic activity: (A) FDA-approved serine β-lactamase inhibitor clavulanic acid;18c (B) MBL inhibitor aspergillomarasmine A (AMA) restores meropenem activity against several Gram-negative species;19 (C) AME inhibitor exhibits synergism with kanamycin A against E. faecium;38 (D) efflux pump inhibitor Phe-Arg-β-naphthylamide (PAβN) increases the susceptibility of P. aeruginosa to levofloxacin;39 (E) pentamidine perturbs the Gram-negative membrane and sensitizes the bacteria to antibiotics whose activity is ordinarily limited to Gram-positive bacteria;26a (F) inhibitor of BlaR1 phosphorylation in MRSA increases susceptibility to β-lactam antibiotics;27 (G) 2-aminoimidazole derivative interferes with TCS signaling in A. baumannii to suppress colistin resistance;14b (H) quorum sensing inhibitor Hamamelitannin increases the sensitivity of S. aureus biofilms to several classes of antibiotics;30b (I) previously approved opioid-receptor agonist loperamide potentiates the effects of tetracycline antibiotics against several Gram-negative bacteria.34b
Along the same lines as β-lactamase enzymes, other antibiotic-modifying enzymes are also potential targets. One prominent class is the aminoglycoside-modifying enzyme (AME) class, which comprises the major mechanism of resistance to aminoglycoside antibiotics. Despite the identification of several classes of AME inhibitors,20 none have yet been clinically approved. Other antibiotic-modifying enzymes that are potential adjuvant targets include chloramphenicol acetyltransferases, macrolide kinases, and macrolide glycosyltransferases.21 Similarly, enzymes that confer resistance by modifying the antibiotic target, such as ErmC methyltransferases that impart resistance to the macrolide–lincosamide–streptoin-B class of antibiotics,12b,22 represent another class of potential adjuvant targets.
Another adjuvant target that has received a significant amount of attention due to its ubiquity and involvement in resistance to many different antibiotic classes in both Gram-positive and Gram-negative bacteria is the efflux pump family. Several inhibitors of various efflux pumps have been identified, and although this class of adjuvants appears to hold true promise for clinical development, additional work needs to be done to improve toxicity profiles and determine the exact mechanism of inhibition.23 Efflux pumps confer resistance by maintaining the intracellular concentration of the antibiotic below levels at which it is efficacious, and this can also be effected by restricting the uptake of antibiotics into the cell. In Gram-negative bacteria, the outer membrane limits antibiotic uptake by imposing a significant permeation barrier. Many Gram-negative bacteria are intrinsically resistant to several classes of antibiotics that are active against Gram-positive bacteria, such as macrolides and glycopeptides, despite the presence of the target, simply because the antibiotic cannot access said target.24 Facilitating access to the antibiotic target therefore represents a potential adjuvant pathway.25 An adjuvant that enables entry of Gram-positive selective antibiotics into the Gram-negative bacterial cell has the potential to render these antibiotics clinically useful across a much broader spectrum of bacteria.26
Signaling and regulatory pathways used by bacteria to detect antibiotics and activate resistance mechanisms also represent potential adjuvant targets. Examples include inhibition of BlaR1 and MecR1 mediated detection of β-lactam antibiotics in S. aureus,27 interference with the SOS DNA repair and mutagenesis pathway,28 and interference with one of the many two-component signaling system (TCS) pathways that play a role in antibiotic resistance.29
In addition to the resistance mechanisms described above, drug tolerance mechanisms are also potential adjuvant targets. Bacterial drug tolerance can be conferred by the adoption of a biofilm phenotype, and there are many examples of antibiofilm agents that potentiate the effects of antibiotics,30 including the recombinant human DNase I,31 marketed as Pulmozyme, which is approved for the treatment of pulmonary disease in cystic fibrosis patients.32 Drug tolerance can also be a result of the formation of persister cells,33 and this phenotype therefore represents another potential adjuvant target.
Finally, combination screening of antibiotics with synthetic or natural product libraries or with libraries of drugs that are approved for alternative indications, can also be used to identify adjuvants.19,34 Follow-up studies are required to determine the mechanism of action of compounds identified from target-blind screens such as this, but the existence of well-characterized toxicology and pharmacology profiles for previously approved drugs has been estimated to reduce the cost of bringing the drug to market by up to 40%.35
It is important to note that the list of adjuvant approaches described above is not complete, and there are likely many as yet undiscovered pathways that could be targeted. The development of methods to identify such targets will play an extremely important role in realizing the full potential of this field. Methods that have been used to identify new adjuvant targets include both the use of Tn-seq to identify resistance genes36 and chemical genetic strategies using antisense-based genetic knockdown to identify factors that potentiate the effects of antibiotics.37
In conclusion, there is no doubt that the antibiotic resistance situation is dire and every potential avenue must be utilized if we are to overcome this crisis. Although the development of new antibiotics remains crucial, and improvements in antibiotic stewardship are essential, the adoption of complementary approaches such as the development of adjuvants that combat antibiotic resistance represents a powerful and underexploited weapon.
Acknowledgments
The authors would like to thank the National Institutes of Health for financial support (GM055769 and AI106733).
Footnotes
The authors declare the following competing financial interest(s): C.M. is co-founder of Agile Sciences, a biotechnology company developing 2-aminoimidazole-based antibiotic adjuvants.
References
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Antimicrobial Resistance – Global Report on Surveillance, 2014
- 3.Chen L, Todd R, Kiehlbauch J, Walters M, Kallen A. Notes from the Field: Pan-Resistant New Delhi Metallo-Beta-Lactamase-Producing Klebsiella pneumoniae – Washoe County, Nevada, 2016. MMWR Morb. Mortal. Wkly. Rep. 2017;66(1):33. doi: 10.15585/mmwr.mm6601a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 5.(a) O’Neill J. Review on AMR. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Wellcome Trust; 2014. [Google Scholar]; (b) Goff DA, Kullar R, Goldstein EJ, Gilchrist M, Nathwani D, Cheng AC, Cairns KA, Escandon-Vargas K, Villegas MV, Brink A, van den Bergh D, Mendelson M. A global call from five countries to collaborate in antibiotic stewardship: united we succeed, divided we might fail. Lancet Infect. Dis. 2017;17(2):e56–e63. doi: 10.1016/S1473-3099(16)30386-3. [DOI] [PubMed] [Google Scholar]
- 6.(a) Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discovery. 2015;14(8):529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]; (b) Martens E, Demain AL. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017;70:520. doi: 10.1038/ja.2017.30. [DOI] [PubMed] [Google Scholar]
- 7.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P&T Community. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 8.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discovery. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 9.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 10.Bax R, Green S. Antibiotics: the changing regulatory and pharmaceutical industry paradigm. J. Antimicrob. Chemother. 2015;70(5):1281–1284. doi: 10.1093/jac/dku572. [DOI] [PubMed] [Google Scholar]
- 11.Cox G, Sieron A, King AM, De Pascale G, Pawlowski AC, Koteva K, Wright GD. A Common Platform for Antibiotic Dereplication and Adjuvant Discovery. Cell Chem. Biol. 2017;24(1):98–109. doi: 10.1016/j.chembiol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 12.(a) Wright GD. Antibiotic Adjuvants: Rescuing Antibiotics from Resistance. Trends Microbiol. 2016;24(11):862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]; (b) Pieren M, Tigges M. Adjuvant strategies for potentiation of antibiotics to overcome antimicrobial resistance. Curr. Opin. Pharmacol. 2012;12(5):551–5555. doi: 10.1016/j.coph.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Hartzell JD, Neff R, Ake J, Howard R, Olson S, Paolino K, Vishnepolsky M, Weintrob A, Wortmann G. Nephrotoxicity associated with intravenous colistin (colistimethate sodium) treatment at a tertiary care medical center. Clin. Infect. Dis. 2009;48(12):1724–1728. doi: 10.1086/599225. [DOI] [PubMed] [Google Scholar]
- 14.(a) Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discovery. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]; (b) Harris TL, Worthington RJ, Hittle LE, Zurawski DV, Ernst RK, Melander C. Small Molecule Downregulation of PmrAB Reverses Lipid A Modification and Breaks Colistin Resistance. ACS Chem. Biol. 2014;9(1):122–127. doi: 10.1021/cb400490k. [DOI] [PubMed] [Google Scholar]
- 15.Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 2013;14(4):243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Mitchison D, Davies G. The chemotherapy of tuberculosis: past, present and future. Int. J. Tuberc Lung Dis. 2012;16(6):724–732. doi: 10.5588/ijtld.12.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fischbach MA. Combination therapies for combating antimicrobial resistance. Curr. Opin. Microbiol. 2011;14(5):519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Tamma PD, Cosgrove SE, Maragakis LL. Combination therapy for treatment of infections with gram-negative bacteria. Clin Microbiol Rev. 2012;25(3):450–70. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Petrosillo N, Ioannidou E, Falagas ME. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin. Microbiol. Infect. 2008;14(9):816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 18.(a) Drawz SM, Papp-Wallace KM, Bonomo RA. New beta-lactamase inhibitors: a therapeutic renaissance in an MDR world. Antimicrob. Agents Chemother. 2014;58(4):1835–1846. doi: 10.1128/AAC.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Papp-Wallace KM, Bonomo RA. New beta- Lactamase Inhibitors in the Clinic. Infect Dis Clin North Am. 2016;30(2):441–464. doi: 10.1016/j.idc.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Drawz SM, Bonomo RA. Three decades of beta-lactamase inhibitors. Clin Microbiol Rev. 2010;23(1):160–201. doi: 10.1128/CMR.00037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bush K. A resurgence of beta-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents. 2015;46(5):483–493. doi: 10.1016/j.ijantimicag.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 19.King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510(7506):503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist. Updates. 2010;13(6):151–71. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Labby KJ, Garneau-Tsodikova S. Strategies to overcome the action of aminoglycoside-modifying enzymes for treating resistant bacterial infections. Future Med. Chem. 2013;5(11):1285–1309. doi: 10.4155/fmc.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wright GD. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Delivery Rev. 2005;57(10):1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets: Infect. Disord. 2004;4(3):193–202. doi: 10.2174/1568005043340777. [DOI] [PubMed] [Google Scholar]
- 23.Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014;453(2):254–267. doi: 10.1016/j.bbrc.2014.05.090. [DOI] [PubMed] [Google Scholar]
- 24.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 25.(a) Livermore DM. Antibiotic uptake and transport by bacteria. Scand. J. Infect. Dis. Suppl. 1990;74:15–22. [PubMed] [Google Scholar]; (b) Lambert PA. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 2002;92(Suppl):46S–54S. [PubMed] [Google Scholar]
- 26.(a) Stokes JM, MacNair CR, Ilyas B, French S, Cote JP, Bouwman C, Farha MA, Sieron AO, Whitfield C, Coombes BK, Brown ED. Pentamidine sensitizes Gram-negative pathogens to antibiotics and overcomes acquired colistin resistance. Nat. Microbiol. 2017;2:17028. doi: 10.1038/nmicrobiol.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zabawa TP, Pucci MJ, Parr TR, Jr, Lister T. Treatment of Gram-negative bacterial infections by potentiation of antibiotics. Curr. Opin. Microbiol. 2016;33:7–12. doi: 10.1016/j.mib.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Boudreau MA, Fishovitz J, Llarrull LI, Xiao QB, Mobashery S. Phosphorylation of BlaR1 in Manifestation of Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus and Its Abrogation by Small Molecules. ACS Infect. Dis. 2015;1(10):454–459. doi: 10.1021/acsinfecdis.5b00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam MK, Alhhazmi A, DeCoteau JF, Luo Y, Geyer CR. RecA Inhibitors Potentiate Antibiotic Activity and Block Evolution of Antibiotic Resistance. Cell Chem. Biol. 2016;23(3):381–391. doi: 10.1016/j.chembiol.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Worthington RJ, Blackledge MS, Melander C. Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med. Chem. 2013;5(11):1265–1284. doi: 10.4155/fmc.13.58. [DOI] [PubMed] [Google Scholar]
- 30.(a) Worthington RJ, Richards JJ, Melander C. Small molecule control of bacterial biofilms. Org. Biomol. Chem. 2012;10(37):7457–74. doi: 10.1039/c2ob25835h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brackman G, Breyne K, De Rycke R, Vermote A, Van Nieuwerburgh F, Meyer E, Van Calenbergh S, Coenye T. The Quorum Sensing Inhibitor Hamamelitannin Increases Antibiotic Susceptibility of Staphylococcus aureus Biofilms by Affecting Peptidoglycan Biosynthesis and eDNA Release. Sci. Rep. 2016;6:20321. doi: 10.1038/srep20321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2009;64(2):317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- 32.Cramer GW, Bosso JA. The role of dornase alfa in the treatment of cystic fibrosis. Ann. Pharmacother. 1996;30(6):656–661. doi: 10.1177/106002809603000614. [DOI] [PubMed] [Google Scholar]
- 33.Lewis K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 34.(a) Ejim L, Farha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright GD. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 2011;7(6):348–350. doi: 10.1038/nchembio.559. [DOI] [PubMed] [Google Scholar]; (b) Taylor PL, Rossi L, De Pascale G, Wright GD. A Forward Chemical Screen Identifies Antibiotic Adjuvants in Escherichia coli. ACS Chem. Biol. 2012;7(9):1547–1555. doi: 10.1021/cb300269g. [DOI] [PubMed] [Google Scholar]
- 35.Chong CR, Sullivan DJ., Jr New uses for old drugs. Nature. 2007;448(7154):645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- 36.Gallagher LA, Shendure J, Manoil C. Genomescale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2(1):e00315–10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SH, Jarantow LW, Wang H, Sillaots S, Cheng H, Meredith TC, Thompson J, Roemer T. Antagonism of chemical genetic interaction networks resensitize MRSA to beta-lactam antibiotics. Chem. Biol. 2011;18(11):1379–1389. doi: 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 38.Gao F, Yan X, Shakya T, Baettig OM, Ait-Mohand-Brunet S, Berghuis AM, Wright GD, Auclair K. Synthesis and structure-activity relationships of truncated bisubstrate inhibitors of aminoglycoside 6′-N-acetyltransferases. J. Med. Chem. 2006;49(17):5273–5281. doi: 10.1021/jm060732n. [DOI] [PubMed] [Google Scholar]
- 39.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, Blais J, Cho D, Chamberland S, Renau T, Leger R, Hecker S, Watkins W, Hoshino K, Ishida H, Lee VJ. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]