Significance
People’s capacity to generate creative ideas is central to technological and cultural progress. Despite advances in the neuroscience of creativity, the field lacks clarity on whether a specific neural architecture distinguishes the highly creative brain. Using methods in network neuroscience, we modeled individual creative thinking ability as a function of variation in whole-brain functional connectivity. We identified a brain network associated with creative ability comprised of regions within default, salience, and executive systems—neural circuits that often work in opposition. Across four independent datasets, we show that a person’s capacity to generate original ideas can be reliably predicted from the strength of functional connectivity within this network, indicating that creative thinking ability is characterized by a distinct brain connectivity profile.
Keywords: connectome, creativity, divergent thinking, fMRI
Abstract
People’s ability to think creatively is a primary means of technological and cultural progress, yet the neural architecture of the highly creative brain remains largely undefined. Here, we employed a recently developed method in functional brain imaging analysis—connectome-based predictive modeling—to identify a brain network associated with high-creative ability, using functional magnetic resonance imaging (fMRI) data acquired from 163 participants engaged in a classic divergent thinking task. At the behavioral level, we found a strong correlation between creative thinking ability and self-reported creative behavior and accomplishment in the arts and sciences (r = 0.54). At the neural level, we found a pattern of functional brain connectivity related to high-creative thinking ability consisting of frontal and parietal regions within default, salience, and executive brain systems. In a leave-one-out cross-validation analysis, we show that this neural model can reliably predict the creative quality of ideas generated by novel participants within the sample. Furthermore, in a series of external validation analyses using data from two independent task fMRI samples and a large task-free resting-state fMRI sample, we demonstrate robust prediction of individual creative thinking ability from the same pattern of brain connectivity. The findings thus reveal a whole-brain network associated with high-creative ability comprised of cortical hubs within default, salience, and executive systems—intrinsic functional networks that tend to work in opposition—suggesting that highly creative people are characterized by the ability to simultaneously engage these large-scale brain networks.
Behavioral and neuroimaging studies have begun to uncover the cognitive and neural processes that give rise to novel and useful ideas (1). Considerable research has focused on characterizing variation in creative thinking ability, often employing assessments of divergent thinking, which measure people’s ability to generate solutions to open-ended problems, such as inventing new uses for objects (2). Because divergent thinking ability moderately predicts real-world creative achievement in the arts and sciences (3), such laboratory-based assessments are thought to provide a reliable index of general creative ability. However, despite progress in the psychology and neuroscience of creativity, the field still lacks clarity on the neurocognitive characteristics that distinguish the highly creative brain (4). The present research thus aims to discover whether a specific brain connectivity profile characterizes high-creative thinking ability and to determine whether individual creativity can be reliably predicted from the strength of functional connectivity within this network.
Using functional magnetic resonance imaging (fMRI), several studies have identified discrete brain regions that support performance on verbal creativity tasks, largely localized within frontoparietal and frontotemporal cortices involved in cognitive control and semantic memory retrieval, respectively (5, 6). More recently, researchers have embraced new techniques in fMRI data analysis to examine coordinated patterns of neural activity across multiple distributed brain regions (i.e., functional connectivity) during various tasks that assess creative cognition and artistic performance, including divergent thinking, figurative language production, musical improvisation, poetry composition, and visual art production (1). This work highlights the contribution of three large-scale brain systems that dynamically interact to support creative task performance: the default mode network, comprised of cortical midline and posterior inferior parietal regions; the executive (or frontoparietal) control network, comprised of lateral prefrontal and anterior inferior parietal regions; and the salience network, comprised of bilateral insula and anterior cingulate cortex (7). Building on dual-process theories of creative cognition (8), which emphasize idea generation and evaluation processes, a recent brain network model posits that the default network supports idea generation and the executive network supports idea evaluation (1, 9), consistent with their established roles in mental simulation and executive cognition, respectively (7). Furthermore, the salience network—which contributes to the detection of behaviorally relevant stimuli and facilitates dynamic transitions between default and executive systems (10)—may identify candidate ideas stemming from generative processes within the default network and forward such information to executive systems for higher order processing (9).
A recent investigation of functional connectivity during divergent thinking (11) reported coupling between default and salience regions that preceded coupling between default and executive regions, potentially reflecting dynamic shifts in idea generation and evaluation across time. Moreover, participants that produced more original responses in a divergent thinking task showed higher global efficiency (i.e., a smaller number of paths needed to traverse between a given pair of brain regions) within a network comprised of default, salience, and executive network nodes. This pattern of functional connectivity has been observed across several task contexts requiring creative idea production, including studies with professional artists engaged in domain-specific tasks in the scanner (12). Functional coupling of these large-scale systems is particularly notable in light of past work reporting a negative or “anticorrelated” relation between the default and executive networks during task-free resting-state fMRI (13), as well as various task-based paradigms, including the well-documented finding of default network deactivation during executively demanding cognitive tasks (14).
In the present research, we hypothesized that individual variation in the ability to simultaneously engage the default, executive, and salience brain systems may provide a neurophysiological marker of creative thinking ability. We tested this hypothesis in a sample of 163 participants engaged in a creative thinking task during fMRI, and we employed connectome-based predictive modeling (cpm)—a method recently developed to predict aspects of human behavior (e.g., cognitive abilities) from patterns of whole-brain functional connectivity (15–17)—to examine whether creative thinking ability can be reliably predicted from an individual’s unique pattern of brain connectivity. The predictive power of cpm has recently been demonstrated in studies of fluid intelligence (15) and sustained attention (18), revealing reliable prediction of these behavioral variables in participants whose data were not used in model construction (16). Here, we aimed to uncover a “creative connectome”—a whole-brain network associated with creative thinking ability—and conduct a leave-one-out cross-validation analysis (i.e., internal validation) to test whether the strength of functional connectivity within this network can reliably predict creative ability in novel participants within this dataset. To further assess the predictive power of this neural model, we conducted three external validation analyses using both task fMRI and resting-state fMRI data obtained from two different laboratories and tested whether the strength of functional connectivity within this connectome can predict creative behavior in independent samples.
Results
Network Definition and Neuroanatomy.
Functional imaging data were acquired as participants completed a classic divergent creative thinking task, where participants are presented with an object cue and are required to generate an unusual and creative use for it (Materials and Methods). The creative quality of responses produced during the fMRI task, as well as two divergent thinking tasks completed outside the scanner, was assessed by four trained raters on a five-point scale (19). Using latent variable modeling (Fig. S1), we found that creativity scores strongly correlated with self-reported creative behavior and achievement in the arts and sciences [r(161) = 0.54, P < 0.001]—consistent with past work (20)—suggesting that these scores reflected an ecologically valid marker of creative ability.
Whole-brain functional networks were constructed for each participant by extracting and correlating the task-related blood oxygen level-dependent (BOLD) signal from 268 brain regions (21). To identify network edges (i.e., functional connections) related to creative thinking ability, we correlated all edges in this network with individual creativity scores extracted via latent variable modeling and applied a statistical threshold (P < 0.01) to retain the most significant edges in the connectivity matrices (15, 16). This analysis revealed 224 edges that positively correlated with creative ability (“high-creative network”) and 603 edges that negatively correlated with creative ability (“low-creative network”; total possible edges = 35,778). Note that both low- and high-creative networks include some positive and negative connections, but the intersubject variability of these connections is either positively or negatively associated with behavior, respectively.
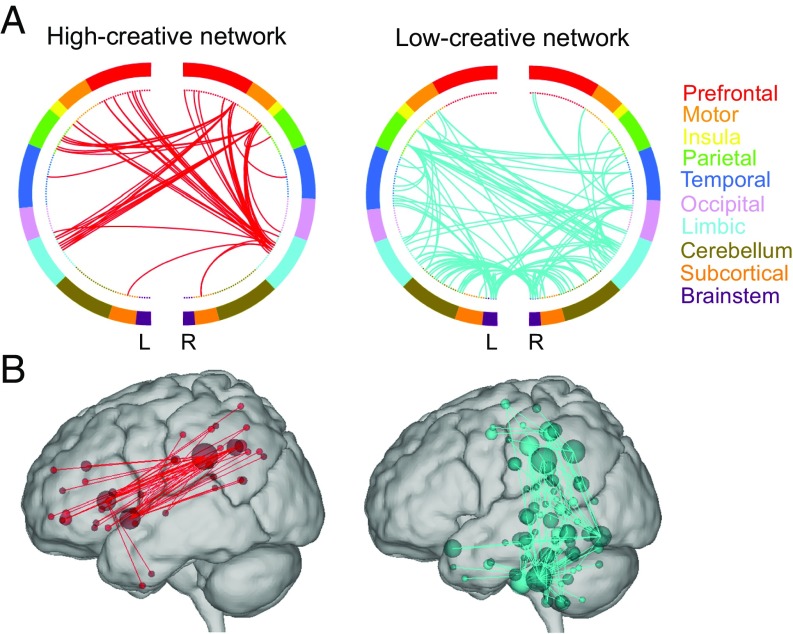
The high-creative network exhibited dense functional connections in predominantly frontal and parietal cortices (Fig. 1). Consistent with past work, the regions showing the highest degree (k ; i.e., number of functional connections) corresponded to the core hubs of the default [e.g., left posterior cingulate cortex; Brodmann area (BA) 23; k = 19], salience (e.g., left anterior insula; BA 45; k = 14), and frontoparietal/executive network (e.g., right dorsolateral prefrontal cortex; BA 9; k = 8; Table S1 and Dataset S1). Of the 25 highest degree nodes in the high-creative network, 12 were within the default network, 4 were within the salience/cingular-opercular network, and 3 were within the frontoparietal/executive network. The low-creative network showed diffuse connections across the brain, most prominently within subcortical/brainstem structures (e.g., left thalamus; k = 14), the default network (e.g., left posterior cingulate cortex; BA 23; k = 23), and cerebellum (Table S2 and Dataset S2). Of the 25 highest degree nodes in the low-creative network, 8 were within subcortical/brainstem structures, 5 were within the default network, and 4 were within the cerebellum.
Fig. 1.
Depictions of the high- and low-creative networks. Circle plots (A) and glass brains (B) were thresholded to show the highest degree (k) nodes in the networks (high-creative k = 10, low-creative k = 18). Colors within the circle plots correspond to lobes of the brain. L, left hemisphere; R, right hemisphere.
Internal Validation: Prediction of Creative Ability Using Task fMRI Data.
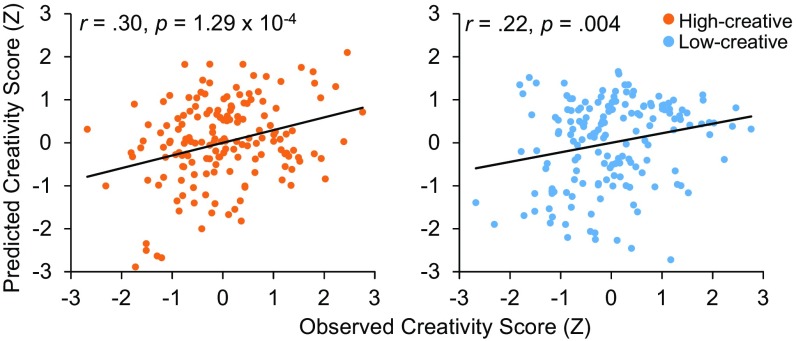
We employed a leave-one-out cross-validation analysis (i.e., internal validation) to test whether the brain connectivity model (i.e., strength of functional connectivity within the high- and low-creative networks, respectively) (15 and 18) could reliably predict creative thinking ability in novel participants (17). (Note that high- and low-creative networks can differ in each round of cross-validation since they are defined on a different set of 162 individuals and tested on the left-out 163rd participant.) Significant correlations emerged between the model-predicted and observed creativity scores in both the high-creative network [r(161) = 0.30, P = 1.29 × 10−4] and low-creative network [r(161) = 0.22, P = 0.004] (Fig. 2); a Steiger’s Z test for differences between two dependent correlations revealed that the correlations were not significantly different from one another (z = 0.83, P = 0.40) (22).
Fig. 2.
Correlation between predicted and observed creativity scores, standardized for visualization.
To account for the nonindependence of the leave-one-out folds, we conducted a permutation test by randomly shuffling the creativity scores 5,000 times and rerunning the prediction pipeline, to create a null distribution of r values for both high- and low-creative networks (18). The P values of the empirical correlation values, based on their corresponding null distribution, were computed by the following formula: (1 + the number of permutated r values greater than or equal to the empirical r)/1,001. The permutation test results revealed significant P values for the empirical r values for both high-creative (P = 0.001) and low-creative (P = 0.019) networks.
As a test of model specificity, we reran the prediction model using functional connectivity data from the noncreative control task [i.e., object characteristics task (OCT)] and the same creative ability scores. Here, the model construction procedure was the same as above (i.e., trained on n − 1 participants’ creativity data), but the model was tested on the left-out participant’s OCT connectivity data instead of their alternate uses task (AUT) connectivity data. The results revealed a weak, nonsignificant correlation between the observed and predicted creative-ability score for the high-creative network [r(161) = 0.05, P = 0.47], but a moderate and significant correlation between the observed and predicted creative-ability score for the low-creative network [r(161) = 0.19, P = 0.01]. Permutation tests using 1,000 iterations yielded a nonsignificant effect for the high-creative network (P = 0.25) and a significant effect for the low-creative network (P = 0.01), indicating that a model trained on the creative task data and tested on the control task data is moderately predictive of low-creative ability.
In sum, the internal validation confirmed that individual differences in creative thinking ability can be reliably predicted in novel participants based on the strength of functional connectivity within task-related brain networks. Next, we further tested the predictive potential of the high- and low-creative network models by conducting three external validation analyses using fMRI data obtained from three independent samples of participants. Note that all three analyses testing associations with creative behavior use the same high- and low-creative network models derived above.
External Validation 1: Prediction of Creative Ability Using Novel Task fMRI Data.
We began by testing whether strength of connectivity within the high- and low-creative networks defined in the full set of 163 participants from the internal validation sample could predict creative thinking ability in a sample of Austrian participants (n = 39) who performed a different divergent thinking task during fMRI (SI Materials and Methods). The high- and low-creative networks derived from the model training dataset above were applied to fMRI data acquired during an experiment that required participants to continuously generate creative object uses over an extended period (60 s). High- and low-creative network strength values were thus computed for each participant by summing correlation coefficients within the high- and low-creative networks. Results revealed a significant prediction of creativity scores (i.e., composite ideational fluency and originality) for the high-creative network [r (37) = 0.35, P = 0.03], but not the low-creative network [r (37) = −0.04, P = 0.78] (Fig. S2), indicating that participants with stronger functional connectivity within the high-creative network tended to generate more original ideas; a Steiger’s Z test revealed that the correlations were not significantly different from one another (z = 1.64, P = 0.10). This external validation extends the internal validation by demonstrating the generalizability of the high-creative model to a novel and somewhat culturally distinct sample.
External Validation 2: Prediction of Creative Ability Using Novel Task fMRI Data.
A second external validation analysis was conducted with a new sample of Austrian participants who completed a divergent thinking task during fMRI (n = 54) (SI Materials and Methods). The high-creative and low-creative networks were again applied to each participant’s fMRI data to test whether creative thinking ability could be predicted from strength of connectivity within these networks. The strength of connectivity within the high-creative network was significantly correlated with behavioral performance (i.e., composite ideational fluency and originality) [r(52) = 0.28, P = 0.03] (Fig. S2). Performance was not significantly correlated with strength within the low-creative network [r(52) = 0.02, P = 0.91]. A Steiger’s Z test revealed that the correlations were not significantly different from one another (z = 1.57, P = 0.11), consistent with the previous external validation analysis.
To test whether the model selectively predicts performance on the cognitive task of interest (i.e., divergent thinking) and not tasks that do not require divergent thinking, we ran a second analysis with functional connectivity matrices extracted from fMRI data during the control task in this experiment (i.e., generating adjectives in response to word cues). The model was sensitive to the cognitive process engaged: Neither high-creative network strength [r(52) = 0.12, P = 0.38] nor low-creative network strength [r(52) = 0.03, P = 0.84] was significantly related to divergent thinking performance when based on functional connectivity data from the (noncreative) control task. Stated differently, the high- and low-creative networks applied to fMRI data during the control task were not significantly related to creative thinking ability. Likewise, when performance data from the noncreative control task was considered (i.e., mean number of adjectives generated), neither high-creative network strength [r(52) = 0.11, P = 0.41] nor low-creative network strength [r(52) = 0.13, P = 0.33] was related to performance: The high- and low-creative networks applied to fMRI data during the noncreative control task were not significantly related to performance on the noncreative control task. Together, these findings provide evidence for the specificity of the high-creative network in predicting creative thinking ability.
External Validation 3: Prediction of Creative Ability Using Resting-State fMRI Data.
As an even more powerful test of external validity, we examined whether strength of connectivity within the high- and low-creative networks could predict creative thinking ability using task-free resting-state fMRI data. Given past work showing a correspondence between resting-state and task-induced network architecture (23), we hypothesized that strength of functional connectivity within the high- and low-creative networks at rest would predict creative ability, albeit to a lesser extent than with task-state connectivity data given the model’s apparent sensitivity to task-congruent neural activity.
We obtained resting-state fMRI data from a large sample of Chinese participants (n = 405) who completed a battery of divergent thinking tasks outside of the scanner (SI Materials and Methods). The high-creative and low-creative networks were again applied to the resting-state fMRI data of the participants. Creativity scores correlated significantly with high-creative network strength [r(403) = 0.13, P = 0.008], as well as low-creative network strength [r(403) = 0.11, P = 0.03] (Fig. S2), indicating that the model can reliably predict creative performance from intrinsic functional brain connectivity. (Note that the positive relationship between creative performance and low-creativity network strength is unexpected. Given the failure of the low-creativity network to generalize to external validation sets 1 and 2, this network may be a less reliable predictor of creativity than the high-creativity network.) A Steiger’s Z test revealed that the correlations were not significantly different from one another (z = 0.79, P = 0.42). As a test of model sensitivity, we assessed whether high- and low-creative network strength could predict performance on a test of fluid intelligence. Neither high-creative [r(403) = 0.04, P = 0.39] nor low-creative [r(403) = 0.04, P = 0.40)] network strength correlated with intelligence scores, thus demonstrating that the neural models selectively predict creative thinking ability and not simply higher cognitive ability.
Discussion
Using a recently developed technique in fMRI data analysis—connectome-based predictive modeling (15–17)—we uncovered whole-brain networks associated with performance on a classic assessment of general creative thinking ability. This high-creative network exhibits dense functional connections between core nodes of the default, executive, and salience systems—networks that typically work in opposition—suggesting that the creative brain is marked by a tendency to simultaneously engage these large-scale circuits to a greater degree than the less creative brain. Critically, we demonstrate the robustness of the model in predicting creative ability across three independent datasets obtained from two different laboratories, including two task-based fMRI samples and one task-free resting-state sample. Taken together, the findings provide evidence that creative thinking ability can be reliably predicted from an individual’s unique brain connectivity profile.
The neuroanatomy of the high-creative network is remarkably similar to patterns of functional connectivity reported in recent studies of creative cognition (1). Specifically, we found that the regions showing the greatest number of significantly correlated functional connections corresponded to the hubs of three large-scale brain networks: default (posterior cingulate cortex), executive (right dorsolateral prefrontal cortex), and salience (left anterior insula). These regions showed increased functional connectivity in a recent fMRI study of divergent thinking (11), which also reported a positive correlation between individual creativity scores and global efficiency within a set of regions consisting of the core nodes of these three networks, consistent with the connectivity profile associated with high-creative ability in the present study. Notably, neuroimaging studies of artistic performance have also reported functional coupling among regions within these networks (12, 24, 25), suggesting that the ability to coactivate brain systems that tend to work in opposition at rest and during demanding cognitive tasks (14) may reflect a domain-general mechanism of creative information processing.
The low-creative network was largely comprised of subcortical, cerebellar, and sensorimotor brain regions. Although we did not have a priori hypotheses about the low-creative network, a few points are worth noting. Regarding neuroanatomy, the regions showing the most connections within the network are less commonly reported in neuroimaging studies of creative cognition (5, 6), suggesting that they are less relevant to successful performance on creative thinking tasks. One possibility, then, is that low-creative ability is characterized by increased interactions among brain regions that do not reliably support creative cognition. Although the network included some regions of the default and salience systems, a closer examination of the connectivity pattern showed that these systems were not functionally linked—as in the high-creative network—but rather showed greater coupling with subcortical, sensorimotor, and cerebellar regions. Previous evidence has found that these regions support procedural (habitual) responses, which suggests that low-creative people might be retrieving cached or previously learned responses (26). In light of past work linking default activity to strong semantic associations (27) and automatic response tendencies (28), low-creative ability may be characterized by an inability to transcend salient conceptual knowledge when drawing on memory representations within the default network. Thus, when attempting to generate original ideas, low-creative individuals may activate common, cached semantic associations that are, in turn, not effectively regulated by the executive network, which may function to inhibit prepotent response tendencies and redirect search processes in the highly creative brain (29).
Moreover, although the low-creative network showed significant performance prediction in the original dataset, the external validation analyses only revealed significant correlations with creative thinking ability in one of the three samples (i.e., external validation 3; resting-state fMRI)—and this relationship was in the unexpected direction—suggesting that the network may be less reliable in its prediction of low-creative ability. It is important to note, however, that, across all datasets, the numerically different correlations between creativity scores and high- versus low-creative network strength were not significantly different from one another (and our samples were underpowered to detect such differences). Nevertheless, the high-creative network showed stable prediction of creative ability across datasets, and the neuroanatomy of this network mirrors previous fMRI studies of creative cognition reporting functional interactions among default, executive, and salience networks (1), suggesting that it may be a more reliable neural marker of creative ability in future work. It is also worth noting that, although we found stronger prediction of creative thinking ability using task-based fMRI data, this pattern also emerged using resting-state data, indicating that the brain-behavior correlations are driven in part by stable trait-level variation in functional connectivity. Thus, both intrinsic network structure as well as task-induced network reorganization likely capture important individual variation (30).
Coordination of the default, executive, and salience networks is consistent with a recent model of brain dynamics supporting creative cognition (1). According to this framework, the default network contributes to the generation of ideas via flexible and spontaneous combinatory mechanisms involved in memory retrieval and mental simulation. The salience network, in turn, functions to identify candidate ideas—potentially useful information generated via the default network—and forward such information to frontoparietal executive systems for high-order processing (e.g., idea evaluation, elaboration, or revision). Although the model implies a serial progression of idea generation and evaluation, recent evidence suggests that executive systems may interact with ongoing generative processes within the default network by imposing constraints on performance and maintaining higher order goals (1). Goal maintenance may thus benefit creative performance by guiding and constraining spontaneous cognition to meet specific creative goals (31).
Creativity remains a complex construct that will require considerable further research to uncover its many manifestations in the brain. Unlike some aspects of cognition that have been reliably localized to specific brain regions, complex constructs such as creativity are likely a product of similarly complex neural mechanisms that engage the whole brain. Thus, in addition to pursuing classic questions related to where creativity occurs in the brain, a promising direction for future research will be to focus on how the brain thinks creatively. Network-based approaches are particularly well-suited to address such questions because they can accommodate the complex interplay of multiple neurocognitive processes (e.g., memory retrieval, mental simulation, and cognitive control). We encourage future research to further explore how the brain thinks creatively across different task contexts and to develop behavioral and neural interventions (e.g., using brain stimulation) to enhance creative performance by targeting large-scale networks and their dynamic interactions.
Materials and Methods
Participants.
The total sample consisted of 163 participants recruited from the University of North Carolina at Greensboro (UNCG) and the surrounding community (113 women, mean age = 22.50 y, SD = 5.79) and specifically over-sampled art, music, and science majors to increase the sample’s population of creative domains. Participants were recruited as part of a larger study on individual differences in creativity (which involved numerous laboratory and ecological measures and procedures not discussed here) and were paid up to $100 for their time. All participants were right-handed with normal or corrected-to-normal vision and reported no history of neurological disorder, cognitive disability, or medication that affects the central nervous system. Participants provided written informed consent. The study was approved by the UNCG Institutional Review Board.
fMRI Task Paradigm.
Participants completed a creativity task and a control task in an event-related design during functional imaging: an alternate uses task (AUT) of divergent thinking and an object characteristics task (OCT). The task paradigm was similar to protocols used in past research (11, 32). In the AUT, participants were presented with a common object cue and asked to imagine a new and unusual use for it; in the OCT, participants were presented an object cue and asked to think of typical object characteristics. Participants received thorough training on both tasks and completed several practice trials before scanning.
The task paradigm consisted of a jittered fixation cross (4–6 s), a cue indicating the upcoming condition (“create” or “object”; 3 s), a thinking period presenting an object cue in text (12 s), and a verbal response period requiring participants to verbalize their response into an MRI-compatible microphone (5 s). In the AUT, participants were asked to use the thinking period to imagine creative uses for the object and then speak their most creative response into the microphone during the response period (29, 33); in the OCT, participants were asked to think of the most common physical characteristic of the object. Consistent with past work, an experimenter recorded verbal responses in real time for subsequent analysis of idea quality.
Behavioral Assessment.
In-scanner performance on the AUT was assessed by recording verbal responses via MRI-compatible microphone, which were subsequently coded for creative quality. Participants also completed two AUTs as part of the postscan behavioral assessment to obtain data on their performance in a standard testing environment. Consistent with conventional procedures, participants were given 2 min to continuously generate alternate uses for each of two objects (i.e., box and rope), which were subsequently coded for creative quality by four trained raters along with the in-scanner responses, using a 1 (not at all creative) to 5 (very creative) scale; raters were instructed to consider uncommonness, remoteness, and cleverness when coding responses, but to provide a single holistic score for each (11, 19, 34, 35). The raters scored all in-scanner responses (23 trials) and laboratory-based responses (two tasks). For in-scanner responses, the average of the scores for each rater was computed, resulting in good interrater reliability [intraclass correlation (ICC) = 0.78]; for laboratory-based responses, the ratings for each task were similarly averaged, also resulting in good rater agreement in the box (ICC = 0.69) and rope tasks (ICC = 0.75). Finally, a battery of questionnaires was administered to assess creative achievement and everyday creative behavior: the Creative Achievement Questionnaire (CAQ) (36); the Biographical Inventory of Creative Behavior (BICB) (37); and the Inventory of Creative Activities and Achievements (ICAA) (38).
Latent variable modeling was used to model the creativity ratings of the in-scanner and laboratory-based AUT responses. We specified a higher order latent variable model using Mplus 7.2. The higher order creativity factor was indicated by two lower order latent factors: in-scanner ratings and laboratory-based ratings. The in-scanner factor was indicated by four observed variables (i.e., the average creativity ratings of the four raters). The laboratory-based factor was indicated by two lower-order factors representing the two laboratory-based tasks, which were both in turn indicated by four observed variables (i.e., the average creativity ratings of the four raters; see Fig. S1). A “creative behavior and achievement” variable was also modeled, indicated by four observed variables, to assess the latent correlation between creative thinking ability and real-world creative behavior and accomplishment. For model identification, the paths of all indicators were constrained to equality, and the variances of the latent variables were fixed to 1.
MRI Data Acquisition and Preprocessing.
Participants completed the tasks in a single fMRI run. Whole-brain imaging was performed on a 3T Siemens Magnetom MRI system (Siemens Medical Systems) using a 16-channel head coil. BOLD-sensitive T2*-weighted functional images were acquired using a single shot gradient-echo echo-planar imaging (EPI) pulse sequence [repetition time (TR) = 2,000 ms, echo time (TE) = 30 ms, flip angle = 78°, 32 axial slices, 3.5 × 3.5 × 4.0 mm, distance factor 0%, field of view (FoV) = 192 × 192 mm, interleaved slice ordering] and corrected online for head motion. The first two volumes were discarded to allow for T1 equilibration effects. Visual stimuli were presented using E-Prime and viewed through a mirror attached to the head coil. In addition to functional imaging, a high resolution T1 scan was acquired for anatomic normalization. Functional volumes were slice time-corrected and realigned using the Statistical Parametric Mapping (SPM) 12 package (six motion parameters), coregistered and normalized to the Montreal Neurological Institute (MNI) template brain, and smoothed with an 8-mm3 isotropic Gaussian kernel. To determine whether creativity scores (i.e., latent variable factor scores) were associated with participant head motion during functional imaging, we correlated these scores with mean frame-to-frame displacement. Results showed that creativity scores were marginally but not significantly related to head motion (r = −0.14, P = 0.07).
Task-related functional connectivity was assessed using the CONN toolbox (https://www.nitrc.org/projects/conn) (39) in MATLAB. For each participant, CONN implemented CompCor, a method for identifying principal components associated with segmented white matter (WM) and cerebrospinal fluid (CSF). In a first-level analysis, CompCor components and first-order derivatives of motion were entered as confounds and regressed from the BOLD signal. Additional preprocessing steps included high-pass filtering, linear detrending, and regression of outlying functional volumes (>97th percentile in normative sample; global-signal z-value threshold = 5, subject-motion mm threshold = 0.09) identified using the artifact removal toolbox (ART) (https://www.nitrc.org/projects/artifact_detect/). We also regressed the task structure corresponding to the onsets and durations of the verbal response periods to account for expected artifacts related to participant vocalization. Because CompCor can account for subject movement effects and other sources of noise in the BOLD signal, the global signal was not regressed.
Functional Network Construction.
Whole-brain networks were computed for each participant using CONN. Consistent with past work employing cpm (15, 18), we used the Shen brain atlas, which consists of 268 regions of interest (ROIs) of 2-mm dimensions and provides whole-brain coverage of the cerebral cortex, cerebellum, and brainstem (21). BOLD signal was extracted from each ROI during the thinking period of the AUT (23 trials, 12 s; collapsing across trials), and bivariate correlations were computed between each pair of ROIs, resulting in a 268 × 268 correlation matrix for each participant. Note that the same anatomical parcellation and procedure were used to construct functional networks for all external validation analyses described here.
Connectome-Based Predictive Modeling.
The main analysis employed cpm to estimate participants’ creative thinking ability from whole-brain, task-related functional connectivity. Cpm is a recently developed method for identifying functional brain connections related to a behavior variable of interest, which are then used to predict behavior in novel participants (i.e., participants whose data were not used in model creation) (17). The cpm procedure was recently described in a series of studies reporting its application to cognitive variables such as fluid intelligence and attention control (18), but we briefly summarize the cpm processing pipeline here. The MATLAB syntax used for cpm is freely available online (https://www.nitrc.org/projects/bioimagesuite/), as is the connectome visualization software (bisweb.yale.edu/build/connviewer.html).
In a first step, a vector of behavioral values (i.e., a single creativity factor score for each participant) was correlated with each edge (i.e., correlation of mean BOLD signals between a given pair of brain regions) in the functional connectivity matrix of each participant. Next, a threshold was applied to the matrix to retain only edges that were significantly positively and negatively correlated with behavior (P < 0.01). Cpm was then applied to a participant’s data by summing the edge strength (i.e., correlation coefficients) in the positive and negative tails of correlation distribution; the frequency distributions of behavioral and connectivity values were checked for normality to meet assumptions for Pearson correlations. Next, a linear regression model was specified to estimate the relationship between the model predicted behavior score and the observed behavior score. Finally, the model was applied to new participants in a level-one-out cross-validation, such that the model was trained on n − 1 participants’ connectivity matrices and behavior scores, and tested on the left-out participant. Note that feature selection (i.e., network edges retained) also occurs within the leave-one-out loop, resulting in slightly different networks and predictive models for each iteration. The predictive power of the resulting model is reflected in the magnitude and statistical significance of the Pearson correlation between the model predicted and observed behavior scores. For a thorough description of cpm, see Shen et al. (17).
External Validation.
One hundred sixty-three different positive and negative networks and linear models—one for each round of leave-one-out cross-validation—were defined during internal validation. To generate a final model to apply to completely independent samples, we defined final high- and low- creativity networks using data from all 163 training subjects. The “final” high creativity network included 224 edges, and the final low creativity network included 603 edges. These networks are visualized in Fig. 1. The networks were highly similar to networks identified in leave-one-out folds during internal validation: On average, edges in the final high-creative network appeared in 151 of the 163 positive masks generated during leave-one-out cross-validation, and edges in the final low-creative network appeared in 154 of the 163 negative masks. Next, we calculated strength in these networks for each subject in three completely independent datasets. We correlated network strength and behavior in these novel samples, revealing that, in each case, individuals with higher high-creative networks were more creative.
Supplementary Material
Acknowledgments
This research was supported by Grant RFP-15-12 from the Imagination Institute (www.imagination-institute.org), funded by the John Templeton Foundation. Q.C. and J.Q. were supported by National Science Foundation of China Grants 31571137 and 31470981. The opinions expressed in this publication are those of the authors and do not necessarily reflect the view of the Imagination Institute or the John Templeton Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. O.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713532115/-/DCSupplemental.
References
- 1.Beaty RE, Benedek M, Silvia PJ, Schacter DL. Creative cognition and brain network dynamics. Trends Cogn Sci. 2016;20:87–95. doi: 10.1016/j.tics.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Runco MA, Acar S. Divergent thinking as an indicator of creative potential. Creat Res J. 2012;24:66–75. [Google Scholar]
- 3.Plucker JA. Is the proof in the pudding? Reanalyses of Torrance’s (1958 to present) longitudinal data. Creat Res J. 1999;12:103–114. [Google Scholar]
- 4.Abraham A. The promises and perils of the neuroscience of creativity. Front Hum Neurosci. 2013;7:246. doi: 10.3389/fnhum.2013.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonen-Yaacovi G, et al. Rostral and caudal prefrontal contribution to creativity: A meta-analysis of functional imaging data. Front Hum Neurosci. 2013;7:465. doi: 10.3389/fnhum.2013.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu X, et al. A meta-analysis of neuroimaging studies on divergent thinking using activation likelihood estimation. Hum Brain Mapp. 2015;36:2703–2718. doi: 10.1002/hbm.22801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zabelina DL, Andrews-Hanna JR. Dynamic network interactions supporting internally-oriented cognition. Curr Opin Neurobiol. 2016;40:86–93. doi: 10.1016/j.conb.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Sowden PT, Pringle A, Gabora L. The shifting sands of creative thinking: Connections to dual-process theory. Think Reason. 2014;21:40–60. [Google Scholar]
- 9.Jung RE, Mead BS, Carrasco J, Flores RA. The structure of creative cognition in the human brain. Front Hum Neurosci. 2013;7:330. doi: 10.3389/fnhum.2013.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uddin LQ. Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci. 2015;16:55–61. doi: 10.1038/nrn3857. [DOI] [PubMed] [Google Scholar]
- 11.Beaty RE, Benedek M, Kaufman SB, Silvia PJ. Default and executive network coupling supports creative idea production. Sci Rep. 2015;5:10964. doi: 10.1038/srep10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellamil M, Dobson C, Beeman M, Christoff K. Evaluative and generative modes of thought during the creative process. Neuroimage. 2012;59:1783–1794. doi: 10.1016/j.neuroimage.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Fox MD, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anticevic A, et al. The role of default network deactivation in cognition and disease. Trends Cogn Sci. 2012;16:584–592. doi: 10.1016/j.tics.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn ES, et al. Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg MD, Finn ES, Scheinost D, Constable RT, Chun MM. Characterizing attention with predictive network models. Trends Cogn Sci. 2017;21:290–302. doi: 10.1016/j.tics.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X, et al. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc. 2017;12:506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenberg MD, et al. A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci. 2016;19:165–171. doi: 10.1038/nn.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvia PJ, et al. Assessing creativity with divergent thinking tasks: Exploring the reliability and validity of new subjective scoring methods. Psychol Aesthetics Creativity Arts. 2008;2:68–85. [Google Scholar]
- 20.Jauk E, Benedek M, Neubauer AC. The road to creative achievement: A latent variable model of ability and personality predictors. Eur J Pers. 2014;28:95–105. doi: 10.1002/per.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X, Tokoglu F, Papademetris X, Constable RT. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage. 2013;82:403–415. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull. 1980;87:245–251. [Google Scholar]
- 23.Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S, et al. Brain activity and connectivity during poetry composition: Toward a multidimensional model of the creative process. Hum Brain Mapp. 2015;36:3351–3372. doi: 10.1002/hbm.22849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinho AL, de Manzano Ö, Fransson P, Eriksson H, Ullén F. Connecting to create: Expertise in musical improvisation is associated with increased functional connectivity between premotor and prefrontal areas. J Neurosci. 2014;34:6156–6163. doi: 10.1523/JNEUROSCI.4769-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dezfouli A, Balleine BW. Habits, action sequences and reinforcement learning. Eur J Neurosci. 2012;35:1036–1051. doi: 10.1111/j.1460-9568.2012.08050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatansever D, et al. Varieties of semantic cognition revealed through simultaneous decomposition of intrinsic brain connectivity and behaviour. Neuroimage. 2017;158:1–11. doi: 10.1016/j.neuroimage.2017.06.067. [DOI] [PubMed] [Google Scholar]
- 28.Vatansever D, Menon DK, Stamatakis EA. Default mode contributions to automated information processing. Proc Natl Acad Sci USA. 2017;114:12821–12826. doi: 10.1073/pnas.1710521114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaty RE, Christensen AP, Benedek M, Silvia PJ, Schacter DL. Creative constraints: Brain activity and network dynamics underlying semantic interference during idea production. Neuroimage. 2017;148:189–196. doi: 10.1016/j.neuroimage.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hearne LJ, Mattingley JB, Cocchi L. Functional brain networks related to individual differences in human intelligence at rest. Sci Rep. 2016;6:32328. doi: 10.1038/srep32328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chrysikou EG, Weber MJ, Thompson-Schill SL. A matched filter hypothesis for cognitive control. Neuropsychologia. 2014;62:341–355. doi: 10.1016/j.neuropsychologia.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fink A, et al. The creative brain: Investigation of brain activity during creative problem solving by means of EEG and FMRI. Hum Brain Mapp. 2009;30:734–748. doi: 10.1002/hbm.20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benedek M, et al. To create or to recall? Neural mechanisms underlying the generation of creative new ideas. Neuroimage. 2014;88:125–133. doi: 10.1016/j.neuroimage.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fink A, et al. Training of verbal creativity modulates brain activity in regions associated with language- and memory-related demands. Hum Brain Mapp. 2015;36:4104–4115. doi: 10.1002/hbm.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benedek M, et al. Creating metaphors: The neural basis of figurative language production. Neuroimage. 2014;90:99–106. doi: 10.1016/j.neuroimage.2013.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carson SH, Peterson JB, Higgins DM. Reliability, validity, and factor structure of the creative achievement questionnaire. Creat Res J. 2005;17:37–50. [Google Scholar]
- 37.Batey M, Furnham A, Safiullina X. Intelligence, general knowledge and personality as predictors of creativity. Learn Individ Differ. 2010;20:532–535. [Google Scholar]
- 38.Diedrich J, et al. Assessment of real-life creativity: The inventory of creative activities and achievements (ICAA) Psychol Aesthetics Creativity Arts. doi: 10.1037/aca0000137. [DOI] [Google Scholar]
- 39.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: A functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.