Significance
In plants, DNA cytosine methylation plays a central role in diverse cellular functions, from transcriptional regulation to maintenance of genome integrity. Vast numbers of whole-genome bisulphite sequencing (WGBS) datasets have been generated to profile DNA methylation at single-nucleotide resolution, yet computational analyses vary widely among research groups, making it difficult to cross-compare findings. Here we reprocessed hundreds of publicly available Arabidopsis WGBS libraries using a uniform pipeline. We identified high-confidence differentially methylated regions and compared libraries using a hierarchical framework, allowing us to identify relationships between methylation pathways. Furthermore, by using a large number of independent wild-type controls, we effectively filtered out spontaneous methylation changes from those that are biologically meaningful.
Keywords: epigenetics, DNA methylation, Arabidopsis, computational biology
Abstract
Genome-wide characterization by next-generation sequencing has greatly improved our understanding of the landscape of epigenetic modifications. Since 2008, whole-genome bisulfite sequencing (WGBS) has become the gold standard for DNA methylation analysis, and a tremendous amount of WGBS data has been generated by the research community. However, the systematic comparison of DNA methylation profiles to identify regulatory mechanisms has yet to be fully explored. Here we reprocessed the raw data of over 500 publicly available Arabidopsis WGBS libraries from various mutant backgrounds, tissue types, and stress treatments and also filtered them based on sequencing depth and efficiency of bisulfite conversion. This enabled us to identify high-confidence differentially methylated regions (hcDMRs) by comparing each test library to over 50 high-quality wild-type controls. We developed statistical and quantitative measurements to analyze the overlapping of DMRs and to cluster libraries based on their effect on DNA methylation. In addition to confirming existing relationships, we revealed unanticipated connections between well-known genes. For instance, MET1 and CMT3 were found to be required for the maintenance of asymmetric CHH methylation at nonoverlapping regions of CMT2 targeted heterochromatin. Our comparative methylome approach has established a framework for extracting biological insights via large-scale comparison of methylomes and can also be adopted for other genomics datasets.
DNA methylation plays essential roles in regulating gene expression and maintaining genome stability. In mammals, DNA methylation is mostly restricted to CpG dinucleotides in somatic tissues, whereas non-CG methylation has been reported in pluripotent stem cells (1–3) and the mouse germ line (4, 5), as well as in the mouse cortex (6) and human brain (7, 8). Arabidopsis broadly deploys methylation in both CG and non-CG contexts (including CHG and CHH, where H can be A, T, or C) (9, 10) via the action of several DNA methyltransferases. METHYLTRANSFERASE 1 (MET1) and CHROMOMETHYLASE 3 (CMT3) maintain CG and CHG methylation, respectively; DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) targets CHH methylation via the RNA-directed DNA methylation (RdDM) machinery, whereas CHROMOMETHYLASE 2 (CMT2) carries out CHH methylation at heterochromatic regions independently of small RNA activity (11, 12). Although we have learned a great deal about the mechanisms of these methylation pathways, insights into the interactions between pathways and their biological effects are still largely unknown.
Whole-genome bisulfite sequencing (WGBS) enables the generation of global DNA methylation profiles at single-nucleotide accuracy (13, 14) and has been widely adopted for characterizing Arabidopsis methylomes (15, 16). However, experimental conditions, library preparation, and downstream bioinformatic analysis techniques can vary widely among research groups, and a means to compare and extract insight from metadata generated across these different laboratory conditions has currently been lacking. Here we collected 500 WGBS libraries and analyzed over 300 in depth from various genotypes and tissues that have been deposited in the Gene Expression Omnibus (GEO) database by the Arabidopsis community using a standardized pipeline (see Dataset S1 for the list of libraries). For each library, we defined differentially methylated regions (DMRs) with high robustness and confidence by comparison with 54 common control libraries. We clustered the libraries based on two statistical methods, named statistical measurement of overlapping of DMRs (S-MOD) and quantitative measurement of overlapping of DMRs (Q-MOD). Our analysis in different mutants revealed a previously overlooked hierarchical framework regulating non-CG methylation and established connections between different epigenetic regulators. For example, MOM1 and MORC family proteins coordinately target a small but specific subset of RNA-directed DNA methylation (RdDM) regions, whereas MET1 and CMT3 are each required for CHH methylation at unique subsets of CMT2 targeted regions. This framework could be adopted to perform large-scale methylome comparisons in other model organisms or for other types of NGS data.
Results and Discussion
Uniform Processing and Quality Check of Arabidopsis WGBS Libraries in GEO.
We retrieved 503 Arabidopsis whole-genome bisulfite sequencing libraries from the National Center for Biotechnology Information GEO database (17), which included a wide spectrum of genotypes, tissues, and treatments (Dataset S1). Considering the variation in sequencing depth and quality among these libraries, we developed a uniform procedure to process all libraries and assess their quality (Fig. 1A; see Materials and Methods for more details). We excluded libraries with low efficiency of bisulfite conversion, libraries with low coverage, libraries that were not in the reference Col-0 background, and libraries that represent duplicated GEO entries (Fig. 1 B and C and Dataset S2). In total, these quality control steps filtered out 189 libraries. The remaining 314 high-quality WGBS libraries, including 54 designated as “control” libraries (this set includes all libraries of wild-type leaf or seedling tissue—these are the most common tissue types submitted for WGBS analysis) and 260 “test” libraries (this set includes all non-WT genotypes, treatments, or nonleaf/seedling tissue types), were selected for further analysis.
Fig. 1.
Quality check and data processing of Arabidopsis WGBS libraries in GEO. (A) Summary of the pipeline for processing Arabidopsis WGBS libraries. (B) Percentage of unconverted-C from reads that mapped to nucleic and chloroplastic genome of each library. Libraries in the shaded area were discarded from further analysis due to the low bisulfite conversion rate. (C) Distribution of total data size and average genome coverage of sequencing reads of each library. Libraries in the shaded area were discarded from further analysis due to the low genome coverage.
Identification of High-Confidence Differentially Methylated Regions.
To establish connections among the different WGBS libraries, we first evaluated the changes in methylation [differentially methylated regions (DMRs)] for each genotype/tissue/treatment by comparing each of the 260 test libraries to each of the 54 control libraries (see Materials and Methods for more details and Datasets S3 and S4). In brief, we defined six types of DMR (hyper- or hypo-CG/CHG/CHH) and performed DMR calling with 100-bp bin resolution for each test library. A DMR is only valid when the test library differs from at least 33 out of the 54 control libraries (see Materials and Methods for more details; SI Appendix, Figs. S1 and S2; and Dataset S3). These DMRs were designated as “high-confidence” DMRs (hcDMRs) and are listed in Datasets S5–S10, and the hcDMR calling pipeline is available for download at https://github.com/yu-z/hcDMR_caller. By design, hcDMRs should filter out spontaneous DMRs that occur in wild-type plants (18, 19) through comparison with a large number of control libraries. To validate the hcDMRs, we sought to complement a mutant because true DMRs arising as a consequence of the genetic knockout should be restored after the reintroduction of a functional copy of the gene, whereas DMRs that arise spontaneously should not. We chose to complement morc6 because this mutant is known to cause very modest changes in methylation (20, 21), allowing us to assess both the accuracy and sensitivity of the hcDMR calling method. We reintroduced a FLAG tagged version of MORC6 into the morc6 mutant background and performed RNAseq in the first (T1) generation alongside wild-type and morc6 mutant controls. This confirmed the functional activity of the MORC6 transgene because nearly all genes derepressed in the morc6 mutant were restored to wild-type levels (SI Appendix, Fig. S3). Next, we performed WGBS on the complementation line and found that whereas only 3% of hcDMRs were not complemented (9/311), ∼26% of DMRs derived by comparing morc6 to a matched wild-type control (22) failed to regain methylation by 10% or more (355/1,391) (Fig. 2 A and B). The increased rate of false positives using the matched control method could be the result of the different life histories of the laboratory strain wild-type and morc6 T-DNA plant. Each acquires independent spontaneous methylation changes through generations, which are filtered out using the hcDMR method, but would be identified using a matched control method because the methylation differences are real, although unconnected to the underlying genetic mutation. These results indicate that our pipeline is both sensitive and robust, identifying true DMRs even for a mutant with relatively weak methylation defects.
Fig. 2.
Validation of hcDMRs and comparison with other DMR calling strategy. (A) Boxplot for methylation levels at morc6 (lib_GSM1375965) defined hypo-CHH hcDMRs (n = 311). Lines connect levels of methylation at individual hcDMRs in the genotypes indicated (WT = lib_GSM1375966). (B) Boxplot for methylation levels at morc6 (lib_GSM1375965) matched wild-type (WT = lib_GSM1375966) defined hypo-CHH DMRs (n = 1,391). Note that lines at a near-horizontal or descending incline from “morc6” to “MORC6 in morc6” indicate a lack of complementation in the MORC6-FLAG T1 line. (C) Venn diagrams for comparison of number of DMRs identified in three independent nrpe1 samples, generated by using one WT from the same experiment (Left) and a group of at least 33 WTs (hcDMRs; Right). The numbers outside the Venn diagrams show the percentage of overlapped DMRs (intersect/total number of DMRs) in each sample. Middle illustrates the overlap of intersect set of DMRs identified by these two methods.
Validation and Comparison of hcDMRs to Existing DMR Calling Methods.
To extend our comparison of hcDMRs to the standard method for DMR detection of comparing matched wild-type controls, we utilized three nrpe1 mutant libraries which have been sequenced by independent laboratories alongside wild-type controls from the same studies (23–25). Although the total number of DMRs identified using the laboratory matched control method was higher compared with hcDMRs, the intersect of DMRs among these three nrpe1 libraries was much lower than that of the hcDMR method (∼30% in laboratory matched compared with ∼70% in hcDMRs) (Fig. 2C). This indicates that a high proportion of laboratory matched control identified DMRs may represent false positives. Additionally, the hcDMR method identified 5,296 DMRs that are commonly shared among all three nrpe1 samples, whereas the matched control method identified only 2,901 common DMRs (Fig. 2C), suggesting that hcDMRs more accurately reflect DMRs that result from the mutation. In conclusion, the hcDMR method allows us to extract and identify a robust set of DMRs from a mutant genotype that is broadly independent of the laboratory of origin.
Statistical and Quantitative Measurement of Overlapping of DMRs and Library Clustering.
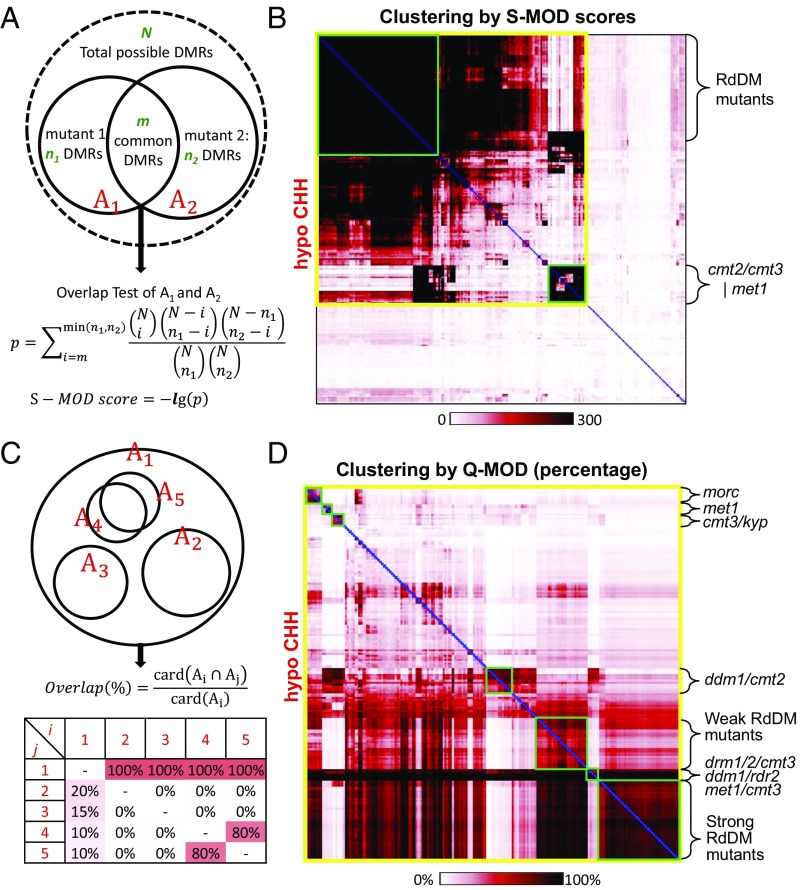
Having established hcDMRs for each library, we sought to use this information to gain insight into the relatedness and relationships between the libraries. To evaluate the degree of overlap between libraries, we first calculated the probability of obtaining the observed number of shared DMRs once the total number of potential DMRs and DMRs identified in each library has been taken into account (see Materials and Methods for more details). This probability was calculated in a pairwise manner for each test library against every other test library, and we refer to this method as S-MOD (Materials and Methods and Fig. 3A). Thus, for the 260 mutant libraries, we obtained a 260 × 260 symmetric matrix containing pairwise S-MOD scores (Fig. 3 A and B; SI Appendix, Figs. S4–S9; and Dataset S11), with higher scores indicating stronger statistical significance. The matrix was clustered using hierarchical clustering, which groups samples with high correlation. The S-MOD approach was sufficient to cluster libraries into major groups representing the key methylation pathways, such as the CMT2 (CHH methylation at heterochromatic regions) and RdDM (DRM2-mediated CHH methylation) pathways (11) (Fig. 3B and SI Appendix, Figs. S4–S9). Next, to quantitatively dissect the relationships between different mutants, we used the percentage of overlapped hcDMRs between test libraries to assist in finer clustering of the matrix (Fig. 3 C and D; SI Appendix, Figs. S10–S15; and Dataset S12). We refer to this method as Q-MOD. Using Q-MOD, we were able obtain matrix clustering that revealed a clear separation between weak and strong RdDM mutants (Fig. 3D and SI Appendix, Fig. S15).
Fig. 3.
Clustering of test libraries with overlapping DMRs by S-MOD method and Q-MOD method. (A) Summary of S-MOD calculation for pairwise relatedness between libraries based on overlapping hcDMRs. (B) Clustering of S-MOD scores between pairs of 260 test libraries at hypo-CHH hcDMRs. Submatrices with yellow borderline indicate libraries that have significant overlap of hcDMRs between any other libraries, whereas green borderlines indicate subgroups of high relatedness. (C) Summary of Q-MOD calculation for pairwise relatedness between libraries based on overlapping hcDMRs. (D) Q-MOD clustering at hypo-CHH hcDMRs after filtering out libraries by S-MOD score >100 maximum cutoff. With Q-MOD, finer-scale groupings, such as between weak vs. strong RdDM mutants, can be observed.
Nonredundancy Within CHH Methylation Pathways and Within CHG Demethylation Pathways and a Connection Between MOM1 and MORC.
Previous studies have shown that the CMT2 and RdDM pathways act nonredundantly to control genome-wide CHH methylation in Arabidopsis (11, 12). This is consistent with the results from both our S-MOD and Q-MOD analyses, which revealed that the overlap between hypo-CHH DMRs observed in cmt2 and RdDM mutants was minimal (SI Appendix, Fig. S16A). We also noticed interesting patterns of nonoverlap looking at hyper-CHG DMR clustering. Ectopic gains of CHG methylation over gene bodies versus transposable elements (TEs) are prevented by distinct pathways, and our analysis confirmed this nonredundancy (SI Appendix, Fig. S16B). The Arabidopsis H3K9 demethylase, IBM1, removes H3K9me2 in gene bodies to prevent the establishment of CHG methylation (26–29), and our analysis showed that loss-of-function ibm1 mutants contain over 109,000 hyper-CHG hcDMRs (SI Appendix, Fig. S16B). In contrast to gene bodies, heterochromatic regions in Arabidopsis are marked by the H2A variant H2A.W, which is encoded by the gene HTA6. hta6 loss-of-function mutants result in TE derepression and elevated levels of CHG methylation (30). Consistent with the clear separation between these two distinct pathways, Q-MOD analysis showed that the overlap of hyper-CHG DMRs between ibm1 and hta6 was minimal (SI Appendix, Fig. S16B). Pollen samples, which include microspore, sperm cell, and vegetative nucleus, ranked very highly based on the total number of hyper-CHG DMRs (SI Appendix, Fig. S16B). These pollen hyper-CHG DMRs almost exclusively overlapped with the hta6 hyper-CHG DMRs and not with the ibm1. In line with our observation, the expression level of HTA6 in pollen is among the lowest compared with that from other types of Arabidopsis tissue (31) (SI Appendix, Fig. S17). This suggests that during pollen development, TEs rather than protein-coding genes tend to be hypermethylated in the CHG context, which is partially due to the down-regulation of HTA6. This is also consistent with the observation that pollen cells undergo epigenetic reprogramming to reinforce TE silencing via small RNA reactivation (32).
In addition, we observed extensive clustering of root tissue samples, indicating a distinct methylation profile for this tissue type, and confirmed extensive hypermethylation in the CHG and CHH contexts in columella cells (Dataset S12) as described (33). On the other hand, we did not detect any distinct clustering patterns for stress treated samples, perhaps suggesting that any methylation changes observed are not consistent between treatments.
We also observed a connection between the methylation profile of mom1 and morc mutants. MOM1 is a plant-specific transcriptional silencer (34, 35). It acts synergistically with MORC6 to silence the transcription of a group of TEs (22). Genome-wide bisulfite sequencing suggested that MOM1 has minimum effects on DNA methylation: TEs that are suppressed by MOM1 display no changes in DNA methylation in mom1 mutants (15, 34, 36, 37). However, our large-scale BS-seq analysis revealed that mom1 indeed affects DNA methylation at a small number of loci (53 hypo-CHH, 271 hypo-CHG, and 721 hypo-CG). These loci show significant overlap with those of morc1/2/4/5/6/7 (20) at a subset of RdDM loci (SI Appendix, Fig. S16 C and D), which is especially evident for non-CG methylation (SI Appendix, Fig. S16 C and D). For example, the strong RdDM mutants, nrpe1 and rdr2, represent over 80% of the hypo-CHH DMR found in mom1 (SI Appendix, Fig. S16C). Similarly, although morc1/2/4/5/6/7 only results in 1% of the genome-wide hypo-CHH DMRs, they account for 75% of the hypo-CHH DMRs found also in mom1 (SI Appendix, Fig. S16 C and D).
MET1 and CMT3 Are Independently Required for the Maintenance of Asymmetric CHH Methylation at CMT2 Target Sites.
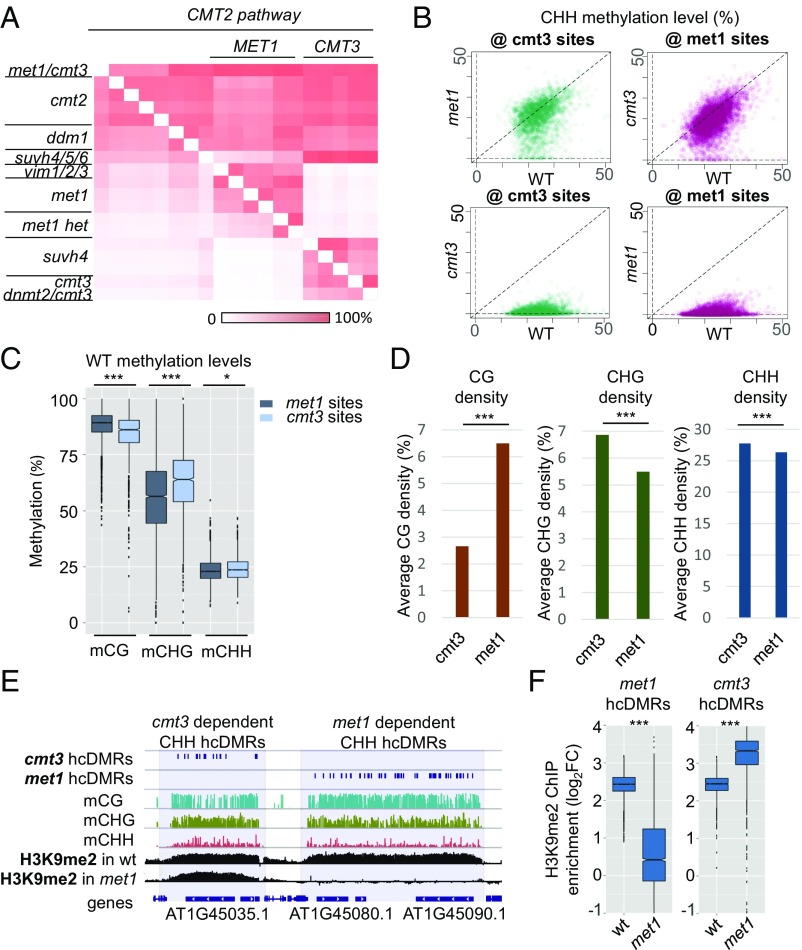
We noticed that the mutants of the primary CG and CHG methyltransferases, MET1 and CMT3, share significant and largely nonoverlapping hypo-CHH DMRs with cmt2 (Fig. 4A). Although we previously noted nonoverlapping hypo-CHH DMRs in cmt3 and met1 (15), the extent of interdependence and the relationship with CMT2 remain to be investigated. Using the libraries generated previously (15), we found that of the 21,782 hypo-CHH DMRs in cmt2, 4,867 are shared by met1 (∼22%, hereafter referred to as “met1 subset”), and 2,290 are shared by cmt3 (∼11%, hereafter referred to as “cmt3 subset”), whereas only 119 are shared by all three (∼0.5%). Because this lack of overlap between the met1 and cmt3 subsets may result from the artificial selection of DMR cutoffs, we directly compared CHH methylation levels in cmt3 and met1. The vast majority of cmt3 sites were unaltered in met1, and vice versa, indicating that many of these sites are truly independent, requiring either MET1 or CMT3 for CHH methylation maintenance (Fig. 4B). Comparison of WT methylation levels at these sites revealed that met1 subset loci had higher levels of mCG and that cmt3 subset loci had higher levels of mCHG (Fig. 4C). Furthermore, the CG density at met1 sites is more than twice of that at cmt3 sites (6.5% vs. 2.7%), whereas conversely, the CHG density is greater at cmt3 sites than met1 sites (6.9% vs. 5.5% at met1 sites) (Fig. 4D). These higher densities are consistent with the well-established in vivo function of these enzymes because MET1 and CMT3 are primarily associated with maintenance of CG and CHG levels, respectively, throughout the genome.
Fig. 4.
MET1 and CMT3 are independently required for the maintenance of asymmetric CHH methylation at CMT2 target sites. (A) Q-MOD clustering of cmt2 hypo-CHH block. met1 and cmt3 related mutants show independent overlaps with cmt2 hypo-CHH hcDMRs. (B) Scatterplots for methylation levels in WT (lib_GSM980986) vs. met1 (lib_GSM981031) or cmt3 (lib_GSM981003) at the met1 or cmt3 defined hcDMR subset of cmt2 (lib_GSM981002) sites. Methylation levels at cmt3 subset sites (@ cmt3 sites, n = 2,290) are largely unaffected in met1, and methylation at met1 subset sites (@ met1 sites, n= 4,867) are largely unaffected in cmt3 (Upper). Lower is control, showing CHH methylation level loss at the met1 and cmt3 sites in their respective mutant backgrounds. (C) Levels of WT methylation in CG, CHG, and CHH at cmt3 and met1 subset sites. Significant differences indicated by P value (Wilcoxon rank sum test); ***P < 2.2e−16 and *P = 0.000196. (D) Average cytosine context density at cmt3 and met1 subset sites. Density is given in percentage (5% indicates five sites per 100 bp of sequence). ***P < 2.2e−16 (Wilcoxon rank sum test). (E) Browser shot showing cmt3 and met1 (cmt2 subset) hypo-CHH hcDMRs at distinct adjacent TEs. The TE requiring met1 for CHH methylation maintenance also shows loss of H3K9me2 in the met1 background. (F) H3K9me2 levels in WT vs. met1 over met1 subset hypo-CHH hcDMRs (Left) and over cmt3 subset hypo-CHH hcDMRs (Right). ***P < 2.2e−16 (Wilcoxon rank sum test).
However, these features do not explain why CHH methylation levels are reduced in met1 or cmt3 because CMT2 itself remains the most likely candidate for deposition of CHH at these loci (11, 12). We hypothesized that the loss of CHH methylation may be occurring indirectly, through loss of H3K9me2, which is needed for CMT2 function (11). Using a met1 H3K9me2 ChIP–Chip data set (38), we found that TEs with met1-dependent CHH methylation indeed experienced a striking coincident reduction in H3K9me2 (Fig. 4 E and F). At cmt3 sites, H3K9me2 levels are somewhat increased in the met1 background. This is consistent with the ectopic gains of H3K9me2 previously observed in the met1 background (38) and perhaps suggests that although global levels of H3K9me2 are maintained, there may be an antagonistic relationship between H3K9me2 levels at cmt3 vs. met1 subset sites. The coincident loss of CHH and H3K9me2 is consistent with a model whereby symmetric cytosine methylation at a subset of CMT2 target sites is required to maintain sufficient levels of H3K9me2 for CMT2 function. In support of this hypothesis, the met1/cmt3 double mutant shows a near-complete loss of CHH methylation (see ref. 15 and Fig. 4A). Although the interdependence of CHG methylation and H3K9me2 can be explained through the well-established feedback loop between CMT3 and KRYPTONITE/SUHV4 (39, 40), the connection between CG methylation and H3K9me2 at these sites is less obvious. Although the hypo-CHH DMRs in the suvh4 mutant reside clearly within the cmt3 subset block, the suvh4/5/6 mutant resides in the cmt2 block along with the met1/cmt3 double mutant (Fig. 4A). This perhaps suggests that SUVH5 and/or SUVH6 may be responsible for linking CG methylation to H3K9me2 at the met1 subset loci. A recent informatics study of DNA methylation patterns predicted that the different SUVHs might show distinct trinucleotide context preferences for binding to methylated DNA (41). Ultimately, future work will be required to elucidate the functional connection between symmetric methylation and H2K9me2 at CMT2 targeted heterochromatin.
Conclusion
Spontaneous changes in DNA methylation are known to occur at many sites throughout the Arabidopsis genome (18, 19). Here we effectively filter out such unstable regions through comparison of each library to a large number of published wild-type controls. These hcDMRs therefore represent an ideal starting point for researchers attempting to interpret methylation changes in their experimental Arabidopsis thaliana line of interest. Using a two-step statistical framework for clustering hcDMRs, we have constructed a hierarchical network for genes controlling DNA methylation in Arabidopsis (SI Appendix, Fig. S20). We also observed detailed relationships between different DNA methylation mutants, including a nonoverlapping requirement for MET1 and CMT3 for CHH methylation at a subset of CMT2 sites, suggesting a potential link between symmetric methylation and H3K9me2 at heterochromatic loci. Hierarchical clustering therefore provides predictive power, and our work demonstrates that large-scale mining of genomics data can uncover biologically meaningful connections in this big-data era.
Materials and Methods
Bioinformatic Analysis.
BS-seq analysis and DMR calling.
Raw sequencing data (SRA files) of 503 Arabidopsis WGBS libraries were downloaded from GEO (Dataset S1). RNA-seq and BS-seq data from MORC6 transgenic materials are deposited to GEO under accession GSE98872 (reviewer access token: odwdowmwtvitpwv).
BS-seq reads from each library were mapped to Arabidopsis TAIR10 genome using BSMAP (42), allowing only uniquely mapped reads, with up to 4% mismatches. We also applied a CHH filter, which discarded reads with three or more consecutive CHH sites to remove reads with low conversion efficiency (13, 15). Fractional DNA methylation levels were computed by #C/(#C + #T) using 100-bp bin. Libraries with a high proportion of unconverted-C in chloroplast (>1%) and low genome coverage (<5) were discarded. Libraries that are not in Col-0 background or have duplicated GEO entries were also filtered out from further analysis.
The remaining 314 high-quality WGBS libraries, including 54 designated wild-type control (Col-0 leaf or seedling) and 260 test libraries, were selected for DMR analysis. Standard DMR calling between test and control libraries was performed as previously described (15), where the difference in CG, CHG, and CHH methylation in each bin needed to be at least 0.4, 0.2, and 0.1, respectively (15). DMRs were defined as 100-bp different methylated genomic fragments.
To identify the number of shared control sets required for robust DMR designation, we assessed DMR calling in each control library (in addition to test type libraries) against the other 53 control libraries (Dataset S3). For each 100-bp bin, setting the cutoff for DMR designation to one control library results in a large number of DMRs being called from both the test and the control libraries (SI Appendix, Fig. S1), many of which are spontaneous or false positive DMRs. As the cutoff increases, the number of DMRs in each of the control libraries drops significantly, whereas the number of DMRs from the test libraries remains substantial. However, requiring a cutoff of 54—meaning that the locus in the test library must be different from every other control library—appeared too stringent because most libraries only retained a small number of DMRs using this criterion (Dataset S3). To find the balance between stringency and false positive DMR designation, we calculated the deceleration of number of DMRs identified in control libraries as the increase of the number of supporting control libraries using the equation , where nc is the average of number of DMRs identified from 54 control libraries, which are supported by at least c control libraries (c ≥ 3). In all six contexts (hypo-/hyper-CG/CHG/CHH), the deceleration rate of the amount of DMRs being called becomes steady at around 33 control sets (SI Appendix, Fig. S2). Thus, a hcDMR is only defined when the test library bin differs from at least 33 out of the 54 control libraries. Note that it is not necessarily the same 33 libraries (of 54) that are taken as supported controls for each bin. Although these stringent criteria afford a reduced the rate of false positives, this comes at a trade-off with false negatives (Fig. 2 A and B). We recorded both the number of “Goodbin” (i.e., 100-bp bin with sufficient coverage, ≥5) and the number of DMRs for all libraries in CG, CHG, and CHH contexts. For the matched WT hypo-CHH DMRs (Fig. 2), we used the same 100-bp bins and 0.1 methyl ratio change cutoff, then applied Fisher’s exact test requiring an adjusted P value of <0.01 for DMR designation. Our pipeline for calling hcDMRs can be downloaded from https://github.com/yu-z/hcDMR_caller.
Comparative analysis of DMRs across libraries.
Given two test libraries, we compare them by testing the dependence of DMR sets A1 and A2 from the two libraries. The null hypothesis to be tested against is that A1 and A2 are independent samples from the population of all of the libraries; the alternative hypothesis is that A1 and A2 are dependent samples. We use the number of overlapping DMRs n between A1 and A2 as test statistic with performance of hypergeometic test. The null hypothesis will be rejected when the test statistics are high. The statistical significance for the overlap of DMRs between two test libraries were calculated using the equation , in which n1 and n2 are the number of DMRs in the two libraries being compared, m is the number of common DMRs shared by the two libraries, and N is defined as the size of the union of all possible DMRs identified in the whole dataset of test libraries. Hence, the p_value indicates the level of their dependence between two libraries.
The p_value between libraries was then transformed into an S-MOD score using the equation S-MOD score = −log(p_value). Pairwise S-MOD scores between all test libraries were stored in a symmetric matrix, of which the horizontal and vertical coordinate are library IDs. Before Q-MOD clustering, we filtered out libraries in which the S-MOD score was below 100 in all pairwise comparisons, indicating that the library has low relatedness to all other libraries (this was performed separately for hyper-/hypo-CG/CHG/CHH). We chose 100 as the S-MOD cutoff score because it can filter out most libraries with no/weak relatedness and libraries with low number (<40) of DMRs (SI Appendix, Fig. S18). Also, to offset the effect of library quality on number of hcDMRs (libraries with lower sequencing depth typically yield fewer hcDMRs), the total number of hcDMRs in the denominator was adjusted to remove hcDMRs that do not have sufficient coverage in both mutant libraries (SI Appendix, Fig. S19). Q-MOD overlap percentages were calculated using the equation , where Ai and Aj represent the set of DMRs of the two test libraries being compared. The total number of hcDMRs in the denominator was adjusted to remove hcDMRs that do not have sufficient coverage in both test libraries (Goodbin filter). The matrix of overlap percentage was clustered using the Ward method in R.
Experimental Procedures.
Transgenic plant material and constructs.
Wild-type and morc6-3 mutant lines (gene AT1G19100, GABI_599B06, aka crh6-5) are from the ecotype Columbia (Col-0) and were grown under 16-h light, 8-h dark cycles. The pAtMORC6::AtMORC6-FLAG construct was previously described (43). Agrobacterium tumfaciens AGLO strain carrying this construct were used to transform morc6-3 using the floral dip method. For Western blot, HRP-coupled FLAG-specific antibody (A8592; Sigma) was used.
RNA-seq.
RNA was extracted from unopened floral bud tissue using TRIzol (Thermo Fisher) and the Direct-Zol RNA MiniPrep kit (R2050; Zymo Research) including in-column DNase treatment. Seventy-five nanograms total RNA was used as input for the TruSeq Stranded mRNA Library Prep Kit for Neoprep (NP-202-1001; Illumina). Libraries were sequenced on a HiSeq 2000 (Illumina). Reads were aligned with TopHat, including the fr-firststrand parameter. Cufflinks was used to generate count data using annotation from TAIR10 that was fed into the DEseq2 package in R for differential expression analysis.
BS-seq.
Genomic DNA from leaf tissue from a T1 (transgenic generation one) pAtMORC6:AtMORC6-FLAG in morc6-3 plant was isolated using DNeasy Plant Mini kit (69106; Qiagen). Five hundred nanograms genomic DNA starting material was sheared using the Covaris instrument. Libraries were generated using the KAPA hyper prep kit (KK8502) with EZ DNA methylation lightning kit for bisulfite conversion (D5030; Zymo Research) and MyTaq HS mix (BioLine BIO-25045) for amplification. Libraries were sequenced on a HiSeq 2000 (Illumina), and reads were aligned to the TAIR10 genome using BSMAP.
Supplementary Material
Acknowledgments
The authors thank the UCLA-FAFU Joint Research Center on Plant Proteomics for institutional support and Life Sciences Editors for assistance with manuscript editing. Y.Z. was supported by National Natural Science Foundation of China Grant 31700192 and Southern University of Science and Technology Presidential Postdoctoral Fellowship. C.J.H. was supported by an European Molecular Biology Organization Long-Term Fellowship (ALTF 1138-2014). H.W. was supported by National Natural Science Foundation of China Grant 31501031. S.E.J. is an investigator of the Howard Hughes Medical Institute. The group of J.Z. is supported by the Thousand Talents Program for Young Scholars and by the Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2016ZT06S172). Work in the S.E.J. laboratory was supported by NIH Grant GM60398.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98872).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716300115/-/DCSupplemental.
References
- 1.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomizawa S, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie W, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lister R, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varley KE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 11.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153:193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakatsu T, et al. 1001 Genomes Consortium Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, et al. NCBI GEO: Archive for functional genomics data sets–Update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 19.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris CJ, et al. Arabidopsis AtMORC4 and AtMORC7 form nuclear bodies and repress a large number of protein-coding genes. PLoS Genet. 2016;12:e1005998. doi: 10.1371/journal.pgen.1005998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu ZW, et al. Two components of the RNA-directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in Arabidopsis. PLoS Genet. 2016;12:e1006026. doi: 10.1371/journal.pgen.1006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moissiard G, et al. Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc Natl Acad Sci USA. 2014;111:7474–7479. doi: 10.1073/pnas.1406611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinh TT, et al. DNA topoisomerase 1α promotes transcriptional silencing of transposable elements through DNA methylation and histone lysine 9 dimethylation in Arabidopsis. PLoS Genet. 2014;10:e1004446, and erratum (2015) 11:e1005452. doi: 10.1371/journal.pgen.1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, et al. DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc Natl Acad Sci USA. 2013;110:8290–8295. doi: 10.1073/pnas.1300585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei M, et al. Regulatory link between DNA methylation and active demethylation in Arabidopsis. Proc Natl Acad Sci USA. 2015;112:3553–3557. doi: 10.1073/pnas.1502279112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, et al. RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc Natl Acad Sci USA. 2013;110:15467–15472. doi: 10.1073/pnas.1315399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei M, et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc Natl Acad Sci USA. 2014;111:527–532. doi: 10.1073/pnas.1320106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miura A, et al. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 30.Yelagandula R, et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell. 2014;158:98–109. doi: 10.1016/j.cell.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 32.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants. 2016;2:16058. doi: 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amedeo P, Habu Y, Afsar K, Mittelsten Scheid O, Paszkowski J. Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature. 2000;405:203–206. doi: 10.1038/35012108. [DOI] [PubMed] [Google Scholar]
- 35.Mlotshwa S, et al. Transcriptional silencing induced by Arabidopsis T-DNA mutants is associated with 35S promoter siRNAs and requires genes involved in siRNA-mediated chromatin silencing. Plant J. 2010;64:699–704. doi: 10.1111/j.1365-313X.2010.04358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokthongwattana C, et al. MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 2010;29:340–351. doi: 10.1038/emboj.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habu Y, et al. Epigenetic regulation of transcription in intermediate heterochromatin. EMBO Rep. 2006;7:1279–1284. doi: 10.1038/sj.embor.7400835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deleris A, et al. Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151:167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du J, et al. Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol Cell. 2014;55:495–504. doi: 10.1016/j.molcel.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouil Q, Baulcombe DC. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 2016;12:e1006526. doi: 10.1371/journal.pgen.1006526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xi Y, Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moissiard G, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.