Abstract
Dystonia is characterized by involuntary muscle contractions. Its many forms are genetically, phenotypically and etiologically diverse and it is unknown whether their pathogenesis converges on shared pathways. Mutations in THAP1 [THAP (Thanatos-associated protein) domain containing, apoptosis associated protein 1], a ubiquitously expressed transcription factor with DNA binding and protein-interaction domains, cause dystonia, DYT6. There is a unique, neuronal 50-kDa Thap1-like immunoreactive species, and Thap1 levels are auto-regulated on the mRNA level. However, THAP1 downstream targets in neurons, and the mechanism via which it causes dystonia are largely unknown. We used RNA-Seq to assay the in vivo effect of a heterozygote Thap1 C54Y or ΔExon2 allele on the gene transcription signatures in neonatal mouse striatum and cerebellum. Enriched pathways and gene ontology terms include eIF2α Signaling, Mitochondrial Dysfunction, Neuron Projection Development, Axonal Guidance Signaling, and Synaptic LongTerm Depression, which are dysregulated in a genotype and tissue-dependent manner. Electrophysiological and neurite outgrowth assays were consistent with those enrichments, and the plasticity defects were partially corrected by salubrinal. Notably, several of these pathways were recently implicated in other forms of inherited dystonia, including DYT1. We conclude that dysfunction of these pathways may represent a point of convergence in the pathophysiology of several forms of inherited dystonia.
Author summary
Dystonia is a brain disorder that causes disabling involuntary muscle contractions and abnormal postures. Mutations in THAP1, a zinc-finger transcription factor, cause DYT6, but its neuronal targets and functions are unknown. In this study, we sought to determine the effects of Thap1C54Y and ΔExon2 alleles on the gene transcription signatures at postnatal day 1 (P1) in the mouse striatum and cerebellum in order to correlate function with specific genes or pathways. Our unbiased transcriptomics approach showed that Thap1 mutants revealed multiple signaling pathways involved in neuronal plasticity, axonal guidance, and oxidative stress response, which are also present in other forms of dystonia, particularly DYT1. We conclude that dysfunction of these pathways may represent a point of convergence on the pathogenesis of unrelated forms of inherited dystonia.
Introduction
Dystonia is a brain disorder that causes disabling involuntary muscle contractions and abnormal postures. When this is the only feature, it is termed isolated dystonia. The pathogenic molecular mechanisms underlying the neuronal dysfunction that leads to dystonia remain to be elucidated and current treatments are unsatisfactory. The advent of next generation sequencing is rapidly expanding the list of genes causing isolated dystonia, including dominant mutations in THAP1 (DYT6), TOR1A (DYT1), GNAL (DYT25), ANO3 (DYT24), CIZ1 (DYT23) and TUBB4A (DYT4), and recessive mutations in HPCA (DYT2), COL6A3 (DYT27) and PRKRA (DYT16) [1–6], although some of these are still pending confirmation. Apart from rare inherited defects in dopamine synthesis [7], there is no known biological pathway that causally links genetic forms of dystonia. Phenotypic similarities between some inherited dystonias, including the most common DYT1 and DYT6, may suggest a shared underlying pathogenic mechanism. Uncovering such mechanisms would be a significant milestone, and potentially widely applicable for therapeutic development.
DYT6 is caused by mutations in THAP1 [Thanatos-associated (THAP) domain-containing apoptosis-associated protein] [8,9], encoding a ubiquitously expressed transcription factor [10,11]. Similar to DYT1, caused by a mutation in TOR1A encoding TorsinA, the disease phenotype is restricted to the central nervous system despite widespread expression of the mutated protein. Disease-causing mutations in THAP1 are dispersed throughout the coding regions, but most are missense and located in the DNA-binding domain (DBD) [12]. THAP domain DNA-binding activity is zinc-dependent, and the four metal-coordinating residues of the C2CH module are crucial for functional activity [13]. Nonsense mutations, equivalent to a null allele, likely result in the generation of small mRNA species that are subject to nonsense-mediated decay [8]. Little is known about the biological function of THAP1, particularly in neurons, although there is a neuron-specific DNA-binding THAP1-like-immunoreactive species [10]. There is also an alternatively spliced form lacking Exon2 which functionally does not substitute for the full-length protein [14] and is normally present at very low levels in the brain [15].
DYT6 is inherited in an autosomal dominant manner with reduced penetrance. Few brains from DYT6 patients have been examined and, to date, they do not exhibit any characteristic neuropathologic lesions [16]. Structural and functional neuroimaging in DYT6 manifesting and non-manifesting carriers (NMCs) demonstrates abnormalities in cerebello-thalamo-cortical and cortico-striato-pallido-thalamo-cortical pathways [17]. Genetically engineered mice with heterozygote Thap1 mutations, either C54Y or ΔExon2, display structural abnormalities of the deep cerebellar nuclei and deficits on motor tasks without overt dystonia [18]. Both mutations are early embryonic lethal when homozygote [18], and in mouse embryonic stem cells, lead to decreased viability and neuroectodermal differentiation [14]. The cell cycle is the major dysregulated pathway that emerged from microarray assays of HUVECs with up- or down-regulation of THAP1 and of human lymphoblasts harboring a disease-associated intronic variant of THAP1 [19,20], but was not enriched in ES cells [14].
To study the role of Thap1 in brain, we performed unbiased transcriptomic, RNA-Seq profiling of postnatal day 1 striatal and cerebellar tissue in two genetic mouse models of THAP1/DYT6 harboring mutations that alter the DNA binding domain, either (1) Thap1C54Y, a constitutive knock in (KI) of the C54Y causative mutation in the DNA binding domain (DBD) of THAP1 and (2) Thap1-, a constitutive knockout (KO) of exon 2 (ΔExon2) [18]. This was followed by functional studies to validate dysregulated molecular pathways with a focus on those that were either “top hits” and/or overlapped with other dystonias. We, and others, [21] have found that molecular abnormalities are present in the developmental stage, but their relationship to the emergence and persistence of the phenotype remains to be determined.
Results
Differential gene expression analysis in the striatum and cerebellum of Thap1+/- and Thap1C54Y /+ vs wild-type mice
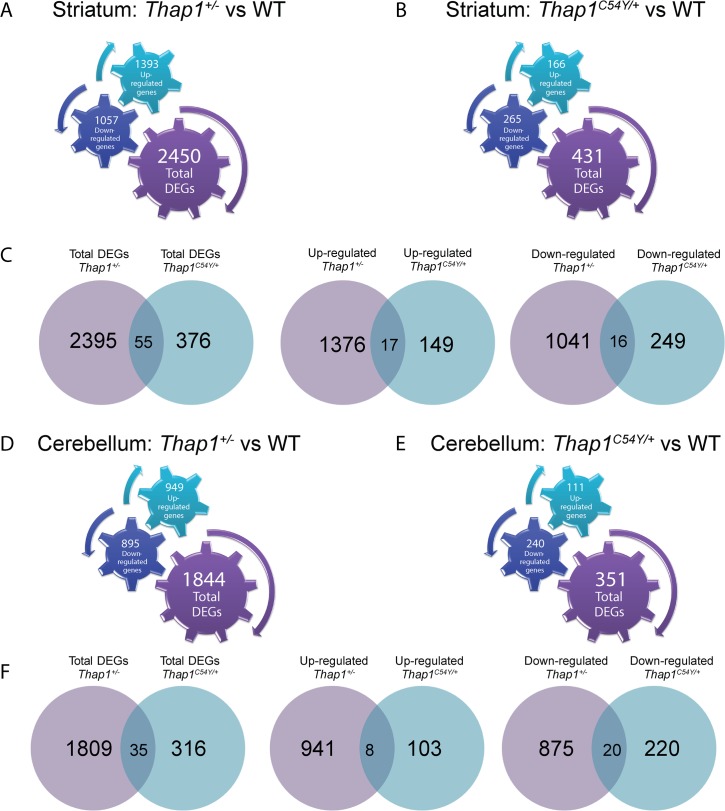
Thap1C54Y /+ KI mice carry a point mutation in one of three cysteine residues that are part of the zinc binding motif [19], altering binding of THAP1 to DNA [8,22]. Thap1 KO mice, i.e. ΔExon2, deletes part of the DBD, and is referred to herein as Thap1+/- [18]. We performed RNA-Seq analysis of cerebellar and striatal tissue dissected from postnatal day 1 (P1) Thap1+/-(ΔExon2), Thap1C54Y/+ and wild-type (WT) controls (N = 4/genotype, all males). At this age, neurons outnumber glia and the limited studies in Ruiz et al. [18] showed greater changes in mRNA levels than in the adult. There was a higher number of differentially expressed genes (DEGs) in Thap1+/- than in Thap1C54Y/+ in both structures (Fig 1 and S1–S4 Tables). 55 striatal DEGs overlapped between genotypes (Fig 1C, S5 Table) and 35 overlapped in the cerebellum (Fig 1F, S5 Table). The highest ranked DAVID Gene Ontology (GO) terms for the striatal overlapping DEGs were positive regulation of signal transduction, proteasome-mediated ubiquitin-dependent protein catabolic process and lipid storage, while those for overlapping DEGs in the cerebellum included cellular response to amino acid stimulus, and DNA-templated transcription. Among the overlapping DEGs, Cdh4 and Phf13 (up-regulated in striatum), Wibg and Rsph1 (down-regulated in striatum), Ppan (differentially regulated in cerebellum), and Nid2 (down-regulated in cerebellum) contain presumptive THABS motifs (S5 Table), supporting the notion that Thap1 may act as either an activator or repressor [13].
Fig 1. Global analysis of differential gene expression in striatum and cerebellum of Thap1+/- or Thap1C54Y/+ vs WT.
RNA-Seq was used to identify differentially expressed genes (DEGs) in the heterozygote Thap1+/- and Thap1C54Y P1 striatum and cerebellum as compared to WT. Diagrams show number of total DEGs as well as the number of up- or down-regulated genes in the (A) Thap1+/- striatum vs WT (B) Thap1C54Y striatum vs WT. (C) Venn diagrams show the number of overlapping DEGs (total, up-regulated or down-regulated) between Thap1+/- and Thap1C54Y/+ relative to WT striatum. Diagrams show number of total DEGs as well as the number of up- or down-regulated genes in the (D) Thap1+/- cerebellum vs WT (E) Thap1C54Y/+ cerebellum vs WT. (F) Venn diagrams show the number of overlapping DEGs (total, up-regulated or down-regulated) between Thap1+/- and Thap1C54Y/+ relative to WT cerebellum. Cogged gears in panels A, B, D and E represent the number and the direction of the differentially expressed genes for each genotype and brain region, as follows: up-regulated genes (turquoise color, upward right arrow); down-regulated genes (dark blue color, downward left arrow); total [number] of genes (purple color, downward right arrow).
To determine if any of the DEGs are linked to dystonia or related disorders, we cross-matched them with the Emory University genetic dystonia panel (http://geneticslab.emory.edu/tests/MM550) and with the Mount Sinai Genetic Testing Laboratory Movement Disorders and Neuromuscular Disease Panel (S6 Table). We found 59 cross-matched striatal Thap1+/- DEGs and 5 Thap1C54Y/+ DEGs. Cryab and Fus appeared in both sets. In cerebellum, we identified 54 cross-matched DEGs in the Thap1+/- and 7 in Thap1C54Y/+, including 3 overlapping genes in both genotypes, Thap1, Lama1 and Sacs.
As the RNA for this study was derived from whole tissue, we utilized an RNA-sequencing transcriptome and splicing database annotating glia, neurons, and vascular cells from the mouse brain [23], in order to investigate the cell subtype origin of the DEGs in the striatum and cerebellum of the Thap1+/- or Thap1C54Y/+ vs WT mice (S7 Table). The largest cell-type fraction of both the up- and down- regulated genes in the Thap1+/- striatum were neuronal, while the greatest fraction of the up- and down- regulated genes in the cerebellum were expressed in endothelial and oligodendrocyte progenitor cells, respectively. The largest cell-type fraction of both the up- and down- regulated genes in the Thap1C54Y/+ striatum were neuronal and astrocytic in origin, while most of the up- and down- regulated genes in the cerebellum were derived from endothelial and microglial cells (S7 Table). It should be noted that P1 mice will have a greater enrichment for cell growth and gene regulatory pathways. Zhang et al. [23] showed that oligodendrocyte-lineage cell isolation does not occur until P17, the earliest time point when the full collection of oligodendrocyte-lineage cells is present. Therefore, one potential limitation of this study is age at which the transcriptomes were assayed but it is also a strength in terms of looking at altered pathways during this critical period.
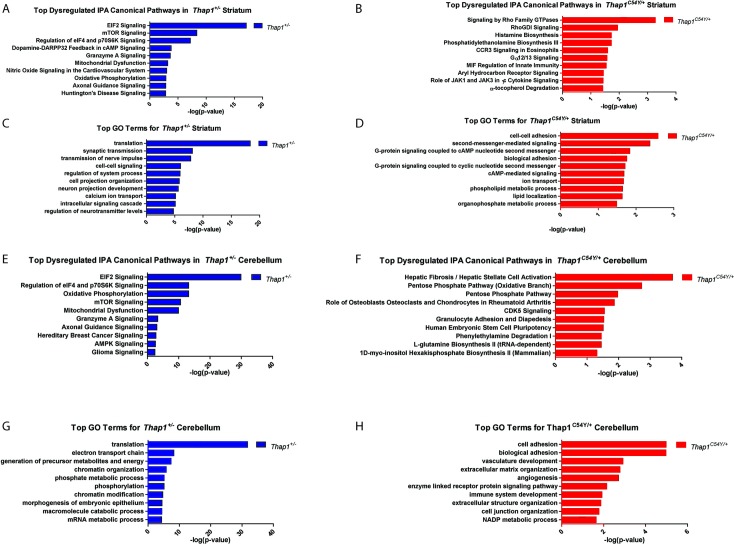
Ingenuity Pathway Analysis (IPA) and DAVID GO terms identified biological functions and pathways enriched in DEGs from each dataset. The highest ranked IPA canonical pathways and GO Terms for each set of DEGs are shown in Fig 2 and S1 and S2 Tables. There were marked genotype-dependent differences, but there were overlaps between striatum and cerebellum within each genotype. Based on what were either the most significantly enriched pathways and terms, and/or those which are related to identified abnormalities in DYT6 and DYT1 models, we chose to functionally explore the eIF2α pathway, neuron projection development, synaptic plasticity [long term depression (LTD) and potentiation (LTP)], and mitochondrial Complex I.
Fig 2. Top canonical pathways and gene ontology terms enriched in striatum and cerebellum of Thap1+/- and Thap1C54Y/+ relative to WT.
(A,B) Top canonical pathways as determined by IPA analysis, and (C,D) DAVID Gene Ontology (GO) terms show an enrichment of biological process based on the list of significant DEGs (DEseq p < 0.05) in the striatum of Thap1+/- and Thap1C54Y/+ relative to WT. (E,F) Top canonical pathways and (G,H) DAVID GO terms based on the list of significant DEGs in the cerebellum of Thap1+/- and Thap1C54Y/+ relative to WT.
To determine if any of the DEGs from the RNA-Seq datasets were bound directly by THAP1, we compared them against two publicly available THAP1 ChIP-Seq datasets, one from human ENCODE K562 cells, and the other from mouse ES cells [14]. The Thap1+/- cerebellum had the greatest number of overlapping genes when compared to both ChIP-Seq datasets, with a total of 32 overlapping genes when compared to the mouse ES cell data and 39 overlapping genes when compared to the K562 dataset (S11 Table). The highest ranking biological functions enriched in the 32-member gene set as identified by DAVID GO are: cellular macromolecules metabolic process, ribosomal small subunit assembly, and negative regulation of protein kinase activity (S11 Table). The highest ranking biological functions in the 39-member gene set as identified by DAVID GO are: cellular process, cellular metabolic process and gene expression (S11 Table).
Thap1 mutant mice show evidence of abnormal eIF2α signaling
The eIF2α signaling pathway was one of the top differentially regulated signaling pathways in the striatum and cerebellum of Thap1+/- mice. The eIF2α pathway is a key effector of the cellular response to several stressors, including the accumulation of misfolded proteins in the endoplasmic reticulum (ER), and was linked to TorsinA function soon after TOR1A was identified as the causative gene in DYT1 [24–26]. DYT16 is caused by mutations in PRKRA, a stress-activated modulator of the eIF2α kinase PKR, with evidence of abnormal phosphorylation of PKR and eIF2α in patient fibroblasts under ER stress [27,28]. In addition, a proteomics-based study identified abnormal eIF2α pathway activation in DYT1 mouse and rat brains, which correlated with assays in human brains [24]. Therefore, given the previous evidence suggesting a role of eIF2a signaling dysregulation in dystonia, and our own RNA-seq data, we investigated UPR genes and proteins to assay the baseline status of the UPR in Thap1 recombinant mice.
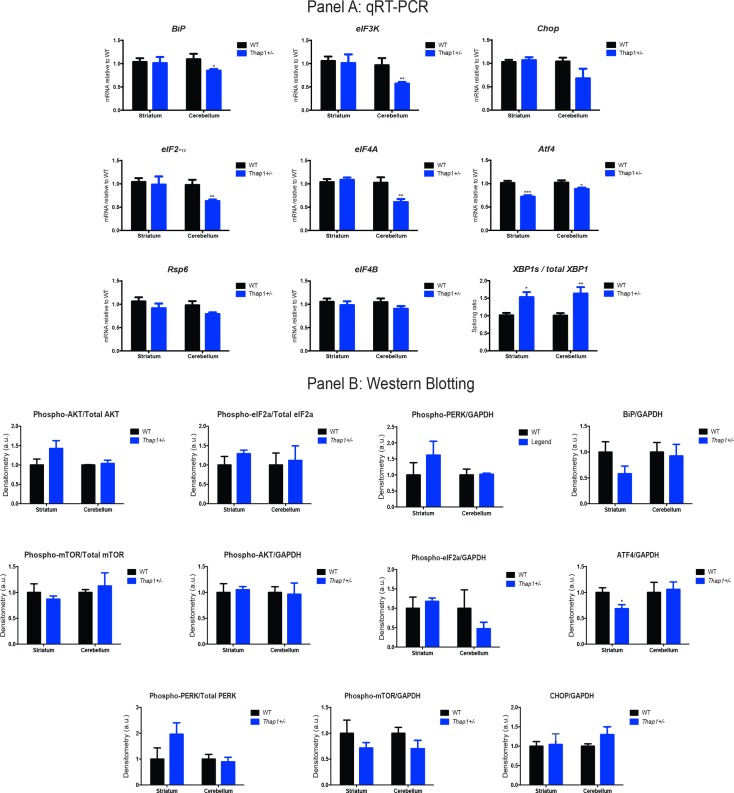
Initially, we assayed the relative mRNA levels for members of the eIF2α signaling pathway in P1 Thap1+/- cerebellum and striatum by RT-qPCR. We assessed changes in genes known to play a role in UPR or eIF2α signaling pathways using DEGs found to be dysregulated directly from the RNA-seq analysis (eIF3K, eIF2a, eIF4A, eIF4B), as well as upstream and downstream mediators of eIF2α (BiP, Chop, Rsp6, XBP1s, and total XBP1) in Fig 3A. There was dysregulated expression of most components of this signaling pathway in cerebellum (S9 Table), and in the Thap1+/- striatum significant differences were observed in Atf4 expression, and XBP1s/total XBP1 ratios (Fig 3, S9 Table). These data show that there are baseline abnormalities of the eIF2α signaling pathway; however, their contribution to the genotype-dependent phenotypes remains to be determined.
Fig 3. Key constituents of the eIF2α signaling pathway are down-regulated in the brains of Thap1+/- mice as compared to WT.
(A) mRNA expression gene profiles of key genes from the eIF2α signaling pathway were assayed with quantitative real-time PCR (RT-qPCR) using cerebellar samples. Data normalized relative to WT. Data are presented as means ± Standard Error of the Mean (SEM); n = 8 for each genotype and brain region with separate littermate WT controls, Student’s t test (*p<0.05; ** p<0.01; *** p<0.005). See also S9 Table. (B) Levels of protein expression in the striatum and cerebellum of P1 THAP1+/- mice relative to WT were assayed by western blot. Densitometry measurements (arbitrary units) are normalized to the housekeeping gene GAPDH, or for phosphoproteins, relative to GAPDH and their respective holoprotein level. Data are presented as means ± SEM; n = 4 for each genotype and region with separate, littermate WT controls, Student’s t test. *p<0.05. See also S9 Table.
The eIF2α signaling pathway is involved in translational regulation, and notably, the DEGs were enriched for a second translational control pathway mediated by mTOR. These kinase cascades regulate protein function via phosphorylation and protein levels. We assayed components of both pathways at basal level by western blotting of protein lysates derived from P1 Thap1+/- striatum and cerebellum. Consistent with the RNA-Seq and RT-qPCR data, the key effector of the eIF2α pathway, ATF4, was reduced by 25% in the striatum (Fig 3, right panel). Overall, the data lend further support to the presence of dysfunction of the eIF2α pathway in DYT6 brain. Notably, screening designed to identify genes implicated in the response to ER stress in human B cells via genetic interactions identified THAP1 as the top hit and all changes were at the level of protein interactions [29].
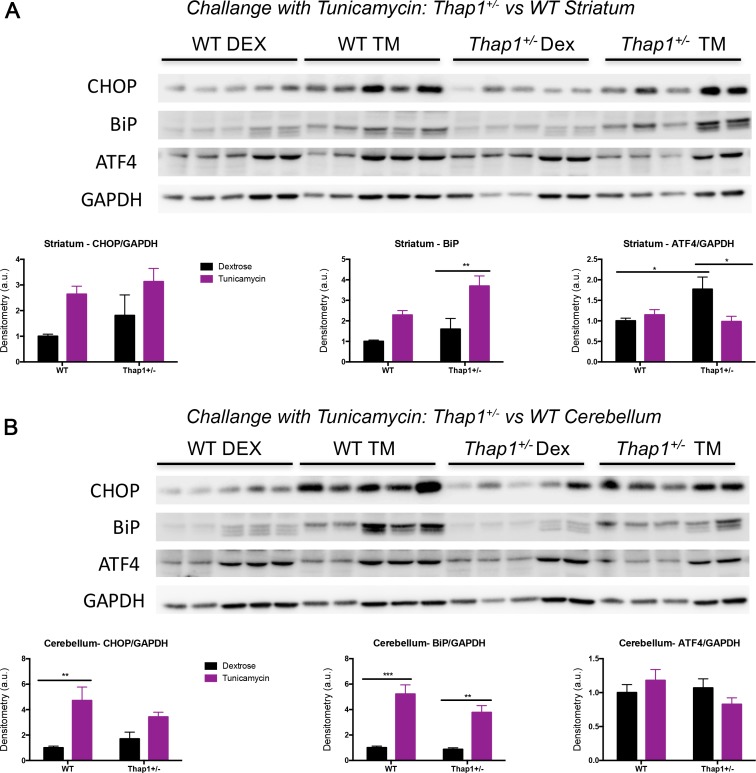
To assay the function of the eIF2α pathway in the presence of a Thap1 mutation, we challenged P4 Thap1+/- and WT pups with tunicamycin, which induces ER stress and the unfolded protein response (UPR) in liver, cerebral cortex, and cerebellum at this age [30]. There was a clear engagement of the main ER chaperone, BiP, in striatum of Thap1+/- mice, and cerebellum of both genotypes (Fig 4). We detected genotype-dependent differences in the expression levels of ATF4 at basal level (dextrose-only controls) in the striatum of Thap1+/- mice as compared WT mice, and the most notable genotype-dependent difference following challenge with tunicamycin was a decrease in ATF4 striatal expression in Thap1+/- but not in WT mice (Fig 4). No differences were observed amongst the different groups when we assessed p-eIF2α/eIF2α and p-eIF2α/GAPDH expression levels (S3 Fig). Therefore, we could not find consistent up or down changes in the UPR with Thap1 baseline status and tunicamycin treatments. Nonetheless, our data suggest a dysregulation of the eIF2α signaling pathway. Therefore, we went on to perform physiologic/functional experiments in Figs 5–7.
Fig 4. Tunicamycin challenge in P4 Thap1+/- and WT pups increases eiF2α signaling pathway proteins and reveals genotype-dependent differences in striatum and cerebellum.
Western blot analysis of (A) striatal and (B) cerebellar lysates from Thap1+/- and WT littermates for BiP, ATF4 and CHOP were performed 24 hrs after subcutaneous tunicamycin (TM) diluted in 150mM dextrose (or dextrose-only control; DEX). Protein expression levels represent normalization to the housekeeping gene GAPDH. Data are presented as means ± SEM; n = 5 for each genotype and region, data normalized to WT (dextrose-only) controls. Statistical differences were assessed by two-way ANOVAs with Tukey’s post hoc tests. *p < 0.05; **p < 0.01; ***p < 0.001. See also S9 Table.
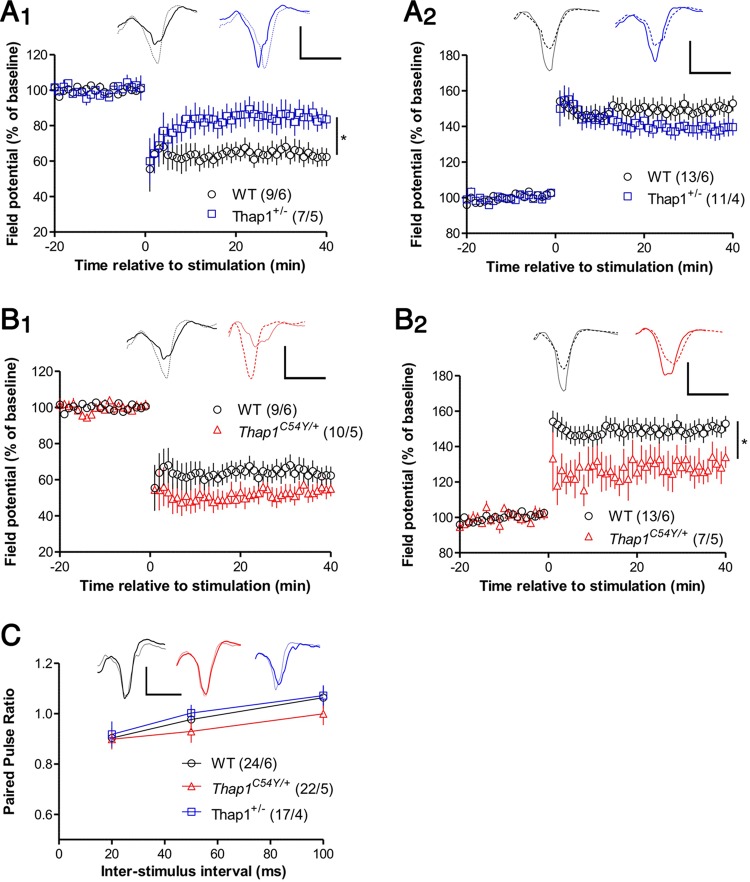
Fig 5. Cortico-striatal synaptic plasticity is altered in Thap1+/- and Thap1C54Y/+ derived slices.
(A) Thap1+/- mice are deficient in synaptically-induced LTD in dorsolateral striatum compared to wildtype controls (A1; p < .05), while LTP in the dorsomedial region is intact (A2). Representative excitatory postsynaptic potential (EPSP) traces in this and panel B were averaged over the baseline period (thin line) and over the final 5 min of recording (thick line), color coded to the graph. Calibration for these and all other traces: 1 mV / 5 ms. (B) In Thap1C54Y/+ mice, LTD was not significantly reduced (B1), but LTP was deficient (B2; p < .05). Note that wildtype data are the same as for panel A, and that all genotypes were analyzed together. (C) Paired-pulse ratio was not altered in Thap1+/- and Thap1C54Y/+ mice. The traces show averaged EPSPs recorded at inter-stimulus interval = 50 ms (thin and thick lines show responses to first and second stimuli, respectively). All graphs show group means ± SEM, and the number of slices/mice for each group are shown in parentheses. Data were analyzed by ANOVAs performed over the final 5 minutes of recording (panels A and B) or on averaged paired-pulse data for each interval (panel C), followed where appropriate by Newman-Keuls post-hoc tests. *p<0.05 See also S9 Table.
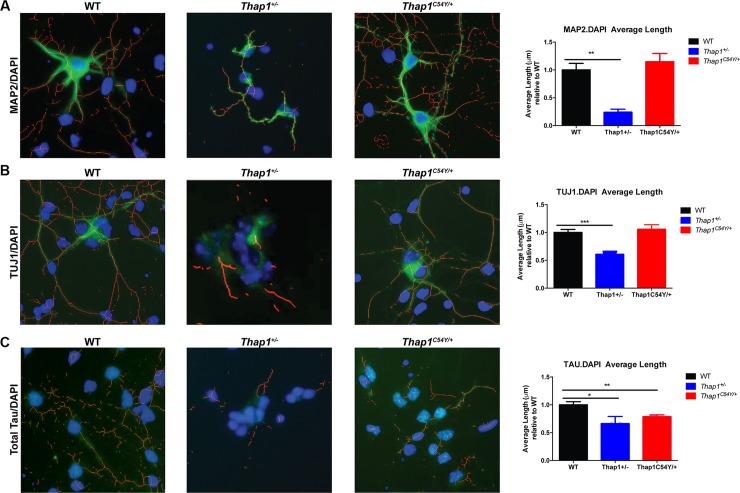
Fig 7. Neurite length in Thap1+/- and Thap1C54Y/+ E16 primary striatal neurons in vitro is decreased relative to neurons from WT mice.
Traces and measures of neurite length using NeuriteTracer show that Thap1+/- striatal neurons have shorter processes as compared to WT as calculated after immunostaining with (A) MAP2, (B) TUJ1 or (C) Total Tau. Thap1C54Y/+ striatal neurons exhibit a milder phenotype and only have shorter axonal processes (Tau immunostaining) as compared to WT. The total number of nuclei (DAPI stain) was independent of genotype after plating equal numbers of neurons in all wells. Neurons were derived from 3 independent platings. Data are presented as means ± SEM, minimum of 25 neurons per well. Statistical differences were assessed by ANOVA with Student’s post hoc t-test. *p < 0.05; **p < 0.01; ***p < 0.001. See also S9 Table.
IPA pathways related to oxidative stress (i.e. Mitochondrial Dysfunction and Oxidative Phosphorylation) were significantly dysregulated in Thap1+/- striatum and cerebellum, and abnormalities in these pathways can contribute to the UPR and to synaptic plasticity [31,32]. Specifically, mitochondrial complex I deficiency (OMIM:252010_3) was enriched (FDR < 0.05) in Thap1+/- cerebellum using Harmonizome [33]. However, we found no genotype-dependent differences in complex I activity in either the striatum (t = 0.89, df = 5, p = 0.42) or cerebellum (t = 0.36, df = 5, p = 0.74).
Synaptic plasticity is impaired in Thap1+/- and Thap1C54Y/+ mouse striatum
Synaptic plasticity is a specific neuronal function predicted by IPA and GO to be disrupted in the striatum of Thap1+/- mice, particularly long-term depression (LTD) and the related phenomenon of synaptic depotentiation (S1 Table). This association is again reminiscent of the deficit in striatal LTD in mouse models of DYT1 [34,35], suggesting that this might be a convergent feature among different types of dystonia and might even be related to dysfunction of the eIF2α pathway [26]. Although differential gene expression for Thap1C54Y/+ striatum was less predictive of a synaptic plasticity phenotype (S2 Table), we tested whether persistent synaptically-induced forms of plasticity at glutamatergic synapses–both LTD and long-term potentiation (LTP)–were altered in the striatum of both mouse lines. Extracellular recordings in acute slices from Thap1+/- mice stimulated with high-frequency synaptic stimulation (HFS) revealed greatly reduced LTD, similar to that reported for DYT1 mouse models (Fig 5A1, S9 Table). In contrast, LTP was normal in Thap1+/- striatum (Fig 5A2). The plasticity phenotype was different for the Thap1C54Y/+ mice: LTD showed a non-significant trend towards enhancement (Fig 5B1, S9 Table), but LTP was reduced (Fig 5B2, S9 Table).
The potential for plasticity at inhibitory synapses could distort the interpretation of our striatal field recordings. For example, reduced LTD at excitatory synapses is also consistent with increased GABA-A mediated currents. To address this possibility, we repeated the LTD experiment in the presence of the GABA-A antagonist gabazine (S2 Fig), and confirmed that LTD was reduced in slices from Thap1+/- mice. Moreover, the Thap1C54Y/+ mutation led to significantly enhanced LTD, in agreement with the trend observed in the absence of gabazine (Fig 5B1, S9 Table).
These results indicate that synaptic plasticity is susceptible to the deletion of Thap1 Exon2 or expression of a DYT6-related mutation of Thap1, while the specific nature of the disruption differs between these two manipulations. Note that paired-pulse ratio was not affected in either the Thap1+/- or Thap1C54Y/+ striatum (Fig 5C, S9 Table), indicating that presynaptic function was normal. However, basal synaptic efficiency was enhanced in Thap1C54Y/+ mice [stimulus strength to evoke 1 mV field excitatory postsynaptic potential (EPSP): 0.88 ± 0.11 μA (n = 24) for wildtype, 0.87 ± 0.10 μA (n = 21) for Thap1+/-, and 0.49 ± 0.05 μA (n = 23) for Thap1C54Y/+; S9 Table]. The increased efficiency in Thap1C54Y/+ striatum would be consistent with elevated synapse number. Alternatively, postsynaptic function could be up-regulated in Thap1C54Y/+ mice. Since striatal LTP at excitatory synapses is expressed postsynaptically [36], basal upregulation of postsynaptic function might limit the extent to which these synapses can be further potentiated, consistent with the reduced LTP seen in these mice.
These extracellular recordings reveal plasticity phenotypes that must be expressed by a substantial fraction of MSNs. Future patch-clamp experiments, performed on MSNs expressing either the D1 or D2 receptor, will be useful to determine if the Thap1+/- and Thap1C54Y/+ mutations differentially affect LTD or LTP in MSN subtypes.
Thap1+/- deficiency in mGluR-LTD is normalized by inhibiting the eIF2α phosphatase
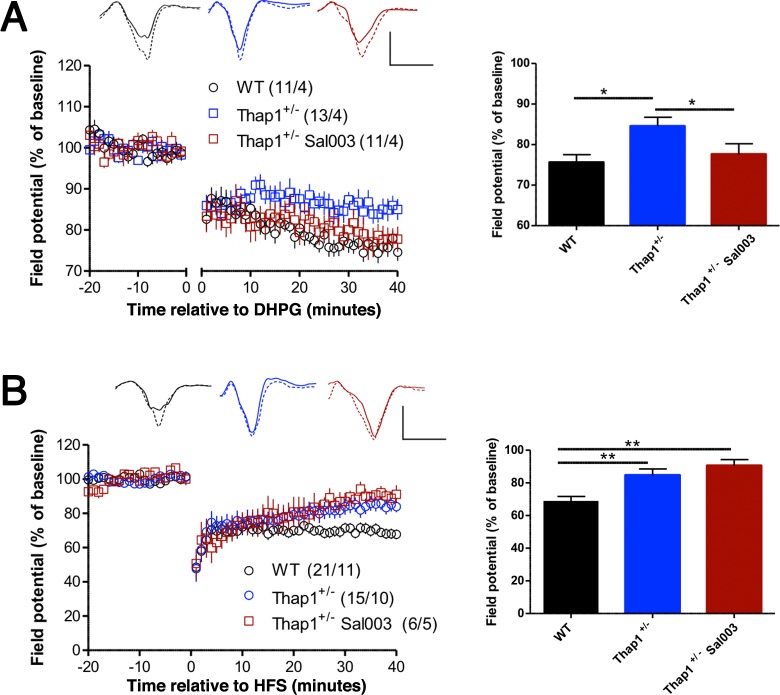
Normal regulation of eIF2α by phosphorylation is required for multiple forms of LTD. In the striatum, inhibition of the eIF2α kinase PERK prevents synaptically-induced LTD [26], and eIF2α has been implicated in hippocampal LTD induced by pharmacological stimulation of metabotropic glutamate receptors (mGluR-LTD) [37,38]. To test whether the LTD deficits in Thap1+/- striatum could be due to dysregulation of eIF2α, we evaluated if abnormal LTD were rescued by Sal003, a selective inhibitor of the eIF2α phosphatase (Fig 6). We found differential effects of Sal003 on mGluR-LTD (induced by the group 1 agonist DHPG) and synaptically-induced LTD (induced by high-frequency stimulation; HFS). Sal003 restored mGluR-LTD to wildtype levels (Fig 6A, S9 Table), in agreement with the finding that eIF2α phosphorylation is required for this form of plasticity in the hippocampus [37]. However, Sal003 had no effect on HFS-induced LTD (Fig 6B, S9 Table). This finding contrasts with the ability of Sal003 to restore LTD in DYT1 mutant mice [26], indicating that Thap1 deletion can interfere with LTD by affecting signaling mechanisms in addition to eIF2α dysregulation. Since HFS-induced LTD in striatum depends on the synaptic activation of mGluRs [39,40], it is possible that, independent of its effect on eIF2α signaling,Thap1 deletion interferes with the ability of HFS to activate postsynaptic mGluRs. In fact, we identified numerous dysregulated genes that participate in synaptic transmission and conduction of nerve impulses in Thap1+/- striatum (S1 Table).
Fig 6. Inhibition of eIF2α phosphatase rescues mGluR-LTD, but not synaptically-induced LTD.
The summary graphs in the right panels show mean ± SEM for the final 5 min of recording. (A) LTD after treatment with group 1 agonist DHPG (100 μM, applied during the gap in the graph) was reduced in Thap1+/- slices, and pretreatment with Sal003 (20 μM) (eIF2α phosphatase inhibitor) rescued LTD in Thap1+/- slices. (B) In high frequency stimulation (HFS)-induced LTD, Sal003 (10 μM) failed to reverse the deficit observed in Thap1+/- slices. Numbers in parentheses indicate number of slices/number of mice. Representative traces are shown during baseline period (solid lines) and at the end of the recording period (dashed lines). Calibrations: 1 mV / 5 ms. Asterisks indicate p < .05 (*) or p < .01 (**) by ANOVAs followed by Newman-Keuls post-hoc tests. See also S9 Table.
Neurite outgrowth is decreased in Thap1+/- and Thap1C54Y/+ striatal neurons in vitro
Neurite development was another enriched pathway identified across regions in Thap1+/- mice. TorsinA has a proposed role in neuritogenesis, and diffusion tensor imaging abnormalities detected by MRI are seen in different brain regions of patients with DYT6, DYT1 and other dystonias [41–43]. Signaling by the Rho family of GTPases is the top pathway in the Thap1C54Y/+ striatum, and this pathway plays a critical role in neuritogenesis and axonal pathfinding [44]. To investigate the influence of Thap1 on this process, we assayed neurite development in striatal MSNs in vitro. We cultured striatal E16 (embryonic day 16) neurons from individual Thap1+/- and WT embryos and quantified neurite length after 24 hours. Thap1+/- striatal neurons exhibited shorter processes as compared to WT controls. The phenotype was present but less severe in Thap1C54Y/+ striatal neurons (Fig 7, S9 Table). The total number of cells was equal between the genotypes, suggesting normal survival of the heterozygote mutant neurons following plating at equal densities.
Discussion
We used an unbiased RNA-Seq approach to identify dysregulated genes and pathways in mice harboring either a Thap1 C54Y (disease causing) or a ΔExon2, i.e. null allele. A major goal was to determine if these pathways and resultant phenotypes overlap with abnormalities observed in other dystonias, particularly DYT1 [45], due to their clinical, electrophysiological, structural and functional neuroimaging similarities. RNA-Seq was performed in two brain regions, striatum and cerebellum, consistently implicated in dystonia pathogenesis [46–50], and at a developmental time point when Thap1 levels and transcriptional abnormalities peak [10,15,18], supporting the notion that dystonia is a developmental disorder [51,52]. These initial experiments yielded data consistent with the proposed hypothesis, further supported by ensuing functional validation.
Thap1+/- and Thap1C54Y/+ mice are not equivalent
The data herein support our previous conclusion that the C54Y mutation does not equal a DNA-binding loss-of-function mutation. First, there were a far greater number of DEGs in Thap1+/- than Thap1C54Y/+ mice (Fig 1). Second, fold changes were overall lower in Thap1C54Y/+ in comparison to Thap1+/- (S5 Table). Third, only a small number of DEGs were altered in both genotypes (S5 Table). For both mutations, the log2 values are low for what would be predicted for a transcription factor [53], perhaps in the presence of the C54Y mutation due to compensatory auto-up-regulation of Thap1 [18,54]. Reports of dystonia patients with homozygous THAP1 mutations with non-manifesting heterozygous parents [55,56] highlight dosage dependent effects of THAP1 mutations.
THAP1 mutations occur in all domains of the protein, but genotype-phenotype correlations have proven difficult to establish due to the small number of patients with each mutation. The C54Y protein, unlike the WT protein, does not bind to the Tor1a promoter [22], but it may bind DNA at other sites and perhaps aberrantly expands DNA binding beyond THAB motifs [57] and/or alter cofactor binding [58]. Thus, some DYT6 mutations represent a partial or total loss of function, whereas others could lead to a combination of haploinsufficiency and gain of function, accounting for the genotype-dependent DEG disparity. Shared transcriptomic and phenotypic features, despite the many differences between mice carrying the C54Y and null alleles, also support this possibility. Thus, there was overlap of key disordered biological processes and biofunctions in the striatum and cerebellum between genotypes. These results indicate that the choice of animal model must be carefully considered in DYT6 research, as different mutations can yield divergent results, although sometimes leading to dysregulation of the same pathways and processes.
EIF2α signaling dysfunction: A link between DYT6 and other dystonias?
These new data together with published DYT1 studies [24,26], suggest a point of convergence of neuronal dysfunction on the eIF2α pathway in DYT6 and DYT1. To a lesser extent, other translational control pathways (mTOR and eIF4/p70S6K) are also implicated in the dysregulation produced by mutations in Thap1. Notably, as a transcription factor, THAP1 regulates TOR1A transcription in artificial systems, but this has not been verified in vivo [18,22,59].
A proteomics-based study identified abnormal eIF2α pathway activation in DYT1 mouse and rat brain, which correlated with assays in human brain [24]. A second group designed an RNAi-based functional genomic screening in HEK293T cells that also implicated the eIF2α pathway in DYT1 biology. Moreover, pharmacological manipulation of eIF2α signaling restored absent cortico-striatal LTD in DYT1 knock-in mice [26]. Together, these reports support the presence of abnormal eIF2α signaling in DYT1 brain and its possible causal link to DYT1 synaptic defects. EIF2α signaling provides a potential point of merger with another, rarer form of primary dystonia. DYT16 is caused by mutations in PRKRA, an activator of the eIF2α kinase PKR, with evidence of abnormal eIF2α phosphorylation in patient fibroblasts [27,28]. Coding variants in ATF4, a direct target of eIF2α, were found in patients with focal dystonia [26] and lastly, a recent gene-expression analysis in adult cerebellar tissue from a mouse model of DYT11 dystonia also identified genes associated with protein translation among the top down-regulated mRNAs [60].
We report eIF2α-pathway-related molecular and electrophysiological findings in DYT6 mice that have some similarities with those in DYT1, including abnormalities in baseline ATF4 and in LTD. Taken together, these reports suggest that efforts in dystonia research should include the unravelling of the mechanisms underlying these observations, addressing causality and reversibility.
Thap1 mutations alter synaptic plasticity and neuritogenesis
Multiple dysregulated pathways highlighted in the GO and IPA analyses of the RNA-Seq data may contribute to the deficits in synaptic plasticity and neuritogenesis described herein. These include eIF2α signaling, which in addition to being a key component of ER stress responses, regulates important physiological events under homeostatic conditions, including synaptic plasticity [61–65]. ATF4 plays a specific role in neuronal plasticity, postsynaptic development, mushroom spine density, memory, neuronal survival, caspase activation and dopaminergic neuron degeneration [66–69]. Selective knockdown of ATF4 impairs hippocampal LTP in vitro and in vivo [68] and as noted above, pharmacological inhibition of eIF2α dephosphorylation rescues cortico-striatal LTD defects in DYT1 KI mice [26]. Notably, eIF2α signaling was not among the top functional pathways in C54Y mice, and these mice exhibited reduced LTP but normal LTD. This highlights the presence of a synaptic plasticity defect in both dystonia models, but with genotype-driven differences as discussed earlier. Furthermore, these data suggest intriguing similarities between the different dystonia models that deserve further study, as there are other IPA and GO-enriched pathways in both genotypes that impinge on synaptic plasticity. These include LTD and LTP themselves in the ΔExon2 mice, the mTOR translation control pathway [70,71], Dopamine-DARPP-32 feedback in cAMP signaling [72], and G-protein/second messenger/cAMP gene groups.
Finally, we identify deficits in neuritogenesis in vitro as a possible convergence point between Tor1a and Thap1 mutations. Mutant TorsinA inhibits neurite extension in cultured cells [73] and DYT1 mice have thinner and less complex dendrites in Purkinje cells and striatal medium spiny neurons [42,43]. The deficit in neuritogenesis is present in both ΔExon2 and C54Y striatal neurons. Many of the same highlighted GO and IPA pathways that could alter synaptic plasticity may also contribute to defective neuritogenesis, including the overlapping GO neurite projection terms and IPA Axonal Guidance Signaling, eIF2α/ATF4 pathway [74], Rho GTPase signaling, and G-protein and cAMP signaling pathways [75]. The fact that in 9-week-old mice, basal synaptic efficiency was enhanced in corticostriatal inputs of Thap1C54Y mice yet normal in Thap1+/- mice, suggests that any effects on neuritogenesis that persist into adulthood might be offset by additional synaptic changes. Future studies will need to address the temporal and biological relationship between the neurodevelopmental and plasticity phenotypes uncovered in this study.
Additional insights into THAP1/DYT6 neurobiology
Other disordered pathways previously implicated in DYT1 and observed in the study reported herein are related to mitochondrial dysfunction [76,77] and lipid metabolism [78,79]. Interestingly, many of the Thap1 DEGs which are included in the Emory University and Mount Sinai Genetic Testing Movement Disorders and Neuromuscular Disease Panels are also linked to those biological processes, suggesting that lipid metabolism and mitochondrial function may deserve further investigation in different forms of dystonia. Furthermore, torsins have recently been implicated as essential regulators of cellular lipid metabolism [78].
In conclusion, using an unbiased transcriptomic analysis in two brain regions from two mouse models of DYT6, we identified eIF2α dysregulation as a potential point of convergence between different forms of dystonia, possibly through its influence on key homeostatic neurodevelopmental events. The identification of similar eIF2α dysregulation and synaptic plasticity defects as previously described in DYT1 mice and rats in the DYT6 animals is a key convergence of biological mechanisms among inherited dystonias, perhaps adding the group of translational dysregulation-associated dystonias (DYT1, DYT3, DYT6, and perhaps DYT16 [24,26,80] to those linked to dopamine dysfunction (DYT5, DYT11, DYT25) [7]. Moreover, the disordered post-synaptic DARPP-32/G-protein/cAMP signaling system(s) potentially overlaps with other dystonias, particularly DYT25 caused by mutations in GNAL [8], suggesting that there are multiple pathways which may contribute to this phenotype. The consolidation of multiple types of dystonia into specific pathogenic mechanisms could facilitate focused research into the etiology of dystonia and the rational design of targeted therapies applicable to this group of movement disorders.
Materials and methods
Ethics statement
Experimental procedures were carried out in compliance with the United States Public Health Service's Policy on Humane Care and Use of Experimental Animals and were approved by the Institutional Animal Care and Use Committee (IACUC) at Icahn School of Medicine at Mount Sinai (Protocol #07–0483).
Animals
The Thap1C54Y/+ and Thap1+/- mice used in this study were congenic C57Bl. The generation of the original mice has been previously provided in detail in Ruiz et al. [18] and as the mutations are early embryonic lethal, breeding strategy was always heterozygote X WT. Animals were maintained on a 12:12 light: dark cycle with ad libitum access to food and water throughout the course of the entire experiment.
Real-time qPCR, library preparation, and sequencing
Postnatal day 1 (P1) pups were euthanized by decapitation, the brain was dissected, snap-frozen striatal and cerebellar tissues were homogenized in QIAzol Lysis Reagent (Qiagen). Total RNA purification was performed with the miRNeasy mini kit (Qiagen), and was carried out according to the manufacturer’s instructions. Five hundred nanograms of RNA were reversed-transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA, USA). The cDNA solution was subjected to real-time qPCR in a Step-One Plus system (Applied Biosystems) using the PerfeCTa SYBR Green FastMix ROX (Quanta BioSciences). Quantitative PCR consisted of 40 cycles, 15 s at 95°C and 30 s at 60°C each, followed by dissociation curve analysis. Primer sequences can be found in S8 Table.
Total RNA from P1 striatal and cerebellar tissues (2–3 μg/ sample) were submitted for further processing to the Genomics Core Facility at the Icahn School of Medicine at Mount Sinai. Sequencing libraries were prepared with the TruSeq RNA Sample Prep Kit v2 protocol (Illumina, San Diego, CA, USA). Briefly, ribosomal RNA was removed using the Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Illumina), the remaining RNA fragmented and cDNA synthesized with random hexamers, end-repaired and ligated with appropriate adaptors for sequencing. After size selection and purification using AMPure XP beads (Beckman Coulter, CA, USA), 6 bp barcode bases were introduced at one end of the adaptors. The size and concentration of the RNA-Seq libraries was measured by Bioanalyzer and Qubit fluorometry (Life Technologies, Grand Island, NY, USA), and the rRNA-depleted libraries sequenced on the Illumina HiSeq 2500 System with 100 nucleotide paired-end reads.
RNA-Seq data analysis
Fastq files were aligned to the Ensembl release 88 (GRCm38.75) version of Human Reference genome (mm10) [81], using STAR read aligner [82]. Accepted mapped reads were summarized separately to gene and exon levels using the featureCounts function of subread [83,84], and used to generate gene and exon count matrices. We examined gene expression data for each sample, and found that the total number of mapped reads [Total Reads (Mean): 39,708,979.5; Uniquely Mapped Reads (Mean): 36,244,152.41] were similar across all samples (S10 Table). There were no obvious outlier samples on visual inspection of principal component analysis or hierarchically clustering of gene expression, and all samples were retained for downstream analyses.
Differential gene expression analysis
For each of the primary comparisons within the study, the assembled count matrix was filtered to remove transcripts without any counts in any sample. Counts were adjusted by total library size and normalized using DESeq2 [85]. P-values were adjusted using the Benjamini-Hochberg method [86]. Due to the small number of DEGs present in striatum and cerebellum of Thap1C54Y/+ mice, p-value of 0.05 was used as cut off.
Mitochondrial respiratory complex I activity
Mitochondrial respiratory complex I activity assay (ab109721, Abcam) was performed, utilizing striatal and cerebellar protein lysates from P1 Thap1+/-(ΔExon2) and WT mice (n = 4 per genotype), according to manufacturer's instructions.
Treatment with tunicamycin
Two subcutaneous injections of 3 μg/g tunicamycin (T7765, Sigma-Aldrich) diluted in 150mM dextrose (or dextrose-only control) were applied two hours apart on postnatal day 4 as described [30]. Mice were euthanized 24 hours later, the striatum and cerebellum dissected and snap frozen.
Western blotting
Snap-frozen striatal and cerebellar tissues were homogenized in RIPA buffer with N-ethylmaleimide and protease/phosphatase inhibitors as described previously [24,87]. Protein concentrations were determined using the BCA assay (23225, ThermoFisher Scientific), 30 μg protein lysates were resolved in 10% or 12% Bis/Trisacrylamide gels (BioRad), transferred to nitrocellulose membranes and western blot completed and quantified using the following antibodies: ATF4 / CREB-2 (1:200; sc-200, Santa Cruz), BiP (1:1000; 3177, Cell Signaling), PERK (1:1000; 5683, Cell Signaling), p-PERK (1:1000; 3179, Cell Signaling), eIF2α (1:200; sc-11386, Santa Cruz), p-eIF2α (1:1000; 9721, Cell Signaling), CHOP (1:500; sc-7351, Santa Cruz), mTOR (1:1000; 9862, Cell Signaling), p-eIF4B (Ser406) (1:1000; 5399, Cell Signaling) and GAPDH (1:1000; sc-32233, Santa Cruz). All primary antibodies were incubated in TBS-Tween 5% milk for overnight at 4°C. Membranes were then washed in TBS-Tween. Secondary antibodies anti-rabbit IgG–HRP (PI-1000, Vector Laboratories) or anti-mouse IgG-HRP (PI-2000, Vector Laboratories) were used (1: 5,000) in TBS-Tween 5% milk for 1 hour at room temperature. Immunoreactive proteins were visualized using either Pierce ECL Western Blotting Substrate (32106, ThermoScientific) or Amersham ECL Prime Western Blotting Detection Reagent (RPN2232, GE Health) on a Fujifilm LAS4000 imaging device.
Electrophysiology
Nine-week-old mice were anesthetized with isoflurane, their brains rapidly removed from the skull and placed in ice-cold modified solution (aCSF) containing (in mM): 215 sucrose, 2.5 KCl, 1.6 NaH2PO4, 4 MgSO4, 1 CaCl2, 4 MgCl2, 20 glucose, 26 NaHCO3 (pH = 7.4, equilibrated with 95% O2 and 5% CO2) and 250 μm thick coronal slices containing the striatum prepared with a Vibratome VT1000S (Leica Microsystems), incubated at 31°C for 30 min and then at room temperature for ≥ 1h in normal aCSF containing (in mM): 120 NaCl, 3.3 KCl, 1.2 Na2HPO4, 26 NaHCO3, 1.3 MgSO4, 1.8 CaCl2, 11 glucose (pH = 7.4 equilibrated with 95% O2 and 5% CO2). Hemi-slices were transferred to a recording chamber constantly oxygenated and perfused with aCSF at ~4mL/min using a peristaltic pump (Masterflex C/L); experiments were performed at 28.0 ± 0.1°C. Recordings were acquired with a GeneClamp 500B amplifier (Axon Instruments) and Digidata 1440A (Molecular Devices). All signals were low-pass filtered at 2 kHz and digitized at 10 kHz. For field EPSP recordings, a patch pipette was fabricated on a micropipette puller (Sutter Instruments), filled with normal aCSF, and placed in the dorsomedial striatum for LTP or dorsolateral striatum for LTD. A concentric bipolar electrode (FHC) was positioned immediately above the corpus callosum. Before and after HFS, the stimulus intensity was set evoke an EPSP that was 50% of the maximal obtainable response. During HFS, stimulus intensity was increased to evoke a maximal response. LTD or LTP was induced by the following high-frequency stimulation (HFS) protocol: four 1-s duration, 100 Hz trains, separated by 10 s. mGluR LTD was induced by a 5-min bath application of 100 μM DHPG [(S)-3,5-Dihydroxyphenylglycine]. Sal003, when present, was applied for 5–10 min before DHPG, and washed out with DHPG. Gabazine, when present, was applied at 10 μM for at least 20 min before the delivery of HFS, and remained in the superfusate for the remainder of the recording. Square-wave current pulses (100 μs pulse width) were delivered through a stimulus isolator (Isoflex, AMPI). Paired-pulse ratio was measured by delivering two stimuli at 20, 50, 100 ms inter-stimulus intervals. Each inter-stimulus interval was repeated three times and the resulting EPSPs were averaged.
Quantification of neurite outgrowth
Primary striatal cultures were prepared as described [10], fixed with 4% paraformaldehyde 48 hours after plating, and processed for immunostaining following our published protocol [14] using the following primary antibodies: Tau (1:500, MN1000, ThermoFisher Scientific), TUJ1 (1:250, sc-51670, Santa Cruz), and MAP2 (1:1,000; AB5622, Millipore). Cells were visualized under an Olympus IX51 inverted fluorescent microscope.
NeuriteTracer, a neurite tracing plugin for the freely available image-processing program ImageJ was used to analyze fluorescence microscopy images of neurites and nuclei of cultured primary neurons. The plugin was used to count neuronal nuclei, and traces and measure neurite length as described [88]. Ten randomly selected images of each neuronal culture type were processed. DAPI staining was employed as a nuclear stain. The average length was obtained by dividing the total length of the traces by the number of nuclear counts.
Statistical analyses
GraphPad software (GraphPad Prism 5) was used to perform Student's t-tests for the qPCR and western blot densitometry, two-way ANOVAs followed by Tukey’s post hoc tests were used to analyze the tunicamycin western blot densitometry. Statistical differences in neurite length / outgrowth were assessed by ANOVA with Student’s post hoc t-test. Statistical significance was deemed to be achieved if P < 0.05. Values are presented as mean ± SEM. Electrophysiological data were analyzed by one-way ANOVAs followed, where appropriate, by Newman-Keuls post hoc tests.
Accession numbers
All next generation sequencing data are deposited in NCBI-Gene Expression Omnibus database and are accessible through GEO Series accession number GSE98839. The accession number for human ENCODE ChIP-Seq data from K562 cells used in the manuscript for THAP1 is GEO: GSM803408. The accession number for the mouse embryonic stem (ES) cells ChIP-Seq data for Thap1 used in the manuscript is GEO: GSE86911.
Supporting information
Full western blots of striatal and cerebellar lysates obtained from Thap1+/- and WT littermates for expression of the indicated proteins. * denotes nonspecific band. CHOP bands are compared to the lower GAPDH panel whereas all others are compared to the highest GAPDH panel.
(TIF)
The GABA-A antagonist gabazine (10μM) was present throughout the recordings. Left: Time-course; representative traces show baseline (dashed lines) and 40 min post HFS (solid lines), with colors corresponding to the time-course graph. Calibration: 1 mV / 5 ms. Right: Summary data over final 5 min of recording (mean ± sem). LTD, measured over the final 5 min, was reduced in slices from Thap+/- mice, and enhanced in slices from Thap1C54Y/+ mice (ANOVA followed by Neman-Keuls post hoc tests). See also S9 Table.
(TIF)
Western blot analysis of (A) striatal and (B) cerebellar lysates from Thap1+/- and WT littermates for p-eIF2α/eIF2α, and p-eIF2α/GAPDH were performed 24 hrs after subcutaneous tunicamycin (TM) diluted in 150mM dextrose (or dextrose-only control; DEX). Data are presented as means ± SEM; n = 3 for each genotype and region; data normalized to WT (dextrose-only) controls. Statistical differences were assessed by two-way ANOVAs with Tukey’s post hoc tests. See also S9 Table.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
In silico analysis was used to identification putative THABS motifs in 1KB of sequence upstream of the translation start codon. Putative THABs motif indicated by “+” symbol.
(XLSX)
(XLSX)
(XLSX)
Primer sequences for the constituents of eIF2α signaling pathway, and the top 5 DEGs from the striatum and cerebellum of Thap1+/- or Thap1C54Y/+ relative to WT.
(XLSX)
(XLSX)
(XLSX)
Cross-match lists of DEGs found to be dysregulated in either of the Thap1 mutant striatum or cerebellum (Thap1C54Y/+ or Thap1+/- vs WT) with THAP1 publicly available ChIP-Seq datasets from (A) Thap1 mouse embryonic stem cells, and (B) human ENCODE K562 cells. The highest ranking biological functions as identified by DAVID GO for shared genes between the Thap1+/- cerebellum and the (C) Thap1 mouse embryonic stem cells, and (D) human ENCODE K562 cells are shown.
(XLSX)
Data Availability
All next generation sequencing data are deposited in NCBI-Gene Expression Omnibus database and are accessible through GEO Series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) accession number GSE98839. The accession number for human ENCODE ChIP-Seq data from K562 cells used in the manuscript for THAP1 is GEO: GSM803408. The accession number for the mouse embryonic stem (ES) cells ChIP-Seq data for Thap1 used in the manuscript is GEO: GSE86911.
Funding Statement
This work was supported by the National Institutes of Health grant R01NS081282-04 to MEE and LJO, the Stanley Fahn Award of the Dystonia Medical Research Foundation to PGA and the Huffington Foundation to TPZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Charlesworth G, Bhatia KP, Wood NW. The genetics of dystonia: new twists in an old tale. Brain. 2013;136: 2017–2037. doi: 10.1093/brain/awt138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo A, Erro R, Lohmann K. Novel Dystonia Genes: Clues on Disease Mechanisms and the Complexities of High-Throughput Sequencing. Mov Disord Off J Mov Disord Soc. 2016;31: 471–477. doi: 10.1002/mds.26600 [DOI] [PubMed] [Google Scholar]
- 3.Xiao J, Vemula SR, LeDoux MS. Recent advances in the genetics of dystonia. Curr Neurol Neurosci Rep. 2014;14: 462 doi: 10.1007/s11910-014-0462-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrucci S, Valente EM. Genetic Issues in the Diagnosis of Dystonias. Front Neurol. 2013;4 doi: 10.3389/fneur.2013.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jochim A, Zech M, Gora-Stahlberg G, Winkelmann J, Haslinger B. The clinical phenotype of early-onset isolated dystonia caused by recessive COL6A3 mutations (DYT27). Mov Disord Off J Mov Disord Soc. 2016;31: 747–750. doi: 10.1002/mds.26501 [DOI] [PubMed] [Google Scholar]
- 6.Lohmann K, Klein C. Update on the Genetics of Dystonia. Curr Neurol Neurosci Rep. 2017;17: 26 doi: 10.1007/s11910-017-0735-0 [DOI] [PubMed] [Google Scholar]
- 7.Wijemanne S, Jankovic J. Dopa-responsive dystonia—clinical and genetic heterogeneity. Nat Rev Neurol. 2015;11: 414–424. doi: 10.1038/nrneurol.2015.86 [DOI] [PubMed] [Google Scholar]
- 8.Fuchs T, Gavarini S, Saunders-Pullman R, Raymond D, Ehrlich ME, Bressman SB, et al. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat Genet. 2009;41: 286–288. doi: 10.1038/ng.304 [DOI] [PubMed] [Google Scholar]
- 9.Xiao J, Zhao Y, Bastian RW, Perlmutter JS, Racette BA, Tabbal SD, et al. Novel THAP1 sequence variants in primary dystonia. Neurology. 2010;74: 229–238. doi: 10.1212/WNL.0b013e3181ca00ca [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ortiz-Virumbrales M, Ruiz M, Hone E, Dolios G, Wang R, Morant A, et al. Dystonia type 6 gene product Thap1: identification of a 50 kDa DNA-binding species in neuronal nuclear fractions. Acta Neuropathol Commun. 2014;2: 139 doi: 10.1186/s40478-014-0139-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard J-P. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003;22: 2432–2442. doi: 10.1038/sj.onc.1206271 [DOI] [PubMed] [Google Scholar]
- 12.Xiromerisiou G, Houlden H, Scarmeas N, Stamelou M, Kara E, Hardy J, et al. THAP1 mutations and dystonia phenotypes: genotype phenotype correlations. Mov Disord Off J Mov Disord Soc. 2012;27: 1290–1294. doi: 10.1002/mds.25146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard J-P. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102: 6907–6912. doi: 10.1073/pnas.0406882102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilo F, Zakirova Z, Nolan K, Wagner R, Sharma R, Hogan M, et al. THAP1: Role in Mouse Embryonic Stem Cell Survival and Differentiation. Stem Cell Rep. 2017; doi: 10.1016/j.stemcr.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Xiao J, Gong S, Clara JA, LeDoux MS. Neural expression of the transcription factor THAP1 during development in rat. Neuroscience. 2013;231: 282–295. doi: 10.1016/j.neuroscience.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paudel R, Li A, Hardy J, Bhatia KP, Houlden H, Holton J. DYT6 Dystonia: A Neuropathological Study. Neurodegener Dis. 2016;16: 273–278. doi: 10.1159/000440863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niethammer M, Carbon M, Argyelan M, Eidelberg D. Hereditary dystonia as a neurodevelopmental circuit disorder: Evidence from neuroimaging. Neurobiol Dis. 2011;42: 202–209. doi: 10.1016/j.nbd.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz M, Perez-Garcia G, Ortiz-Virumbrales M, Méneret A, Morant A, Kottwitz J, et al. Abnormalities of motor function, transcription and cerebellar structure in mouse models of THAP1 dystonia. Hum Mol Genet. 2015;24: 7159–7170. doi: 10.1093/hmg/ddv384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, Loreau E, et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109: 584–594. doi: 10.1182/blood-2006-03-012013 [DOI] [PubMed] [Google Scholar]
- 20.Vemula SR, Xiao J, Zhao Y, Bastian RW, Perlmutter JS, Racette BA, et al. A rare sequence variant in intron 1 of THAP1 is associated with primary dystonia. Mol Genet Genomic Med. 2014;2: 261–272. doi: 10.1002/mgg3.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yellajoshyula D, Liang C-C, Pappas SS, Penati S, Yang A, Mecano R, et al. The DYT6 Dystonia Protein THAP1 Regulates Myelination within the Oligodendrocyte Lineage. Dev Cell. 2017;42: 52–67.e4. doi: 10.1016/j.devcel.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavarini S, Cayrol C, Fuchs T, Lyons N, Ehrlich ME, Girard J-P, et al. Direct interaction between causative genes of DYT1 and DYT6 primary dystonia. Ann Neurol. 2010;68: 549–553. doi: 10.1002/ana.22138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci Off J Soc Neurosci. 2014;34: 11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beauvais G, Bode N, Watson J, Wen HM, Kawano H, Glenn KA, et al. Disruption of protein processing in the endoplasmic reticulum of DYT1 knock in mice implicates novel pathways in dystonia pathogenesis. J Neurosci. 2016;40: 10245–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hewett JW, Tannous B, Niland BP, Nery FC, Zeng J, Li Y, et al. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci U S A. 2007;104: 7271–7276. doi: 10.1073/pnas.0701185104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rittiner JE, Caffall ZF, Hernández-Martinez R, Sanderson SM, Pearson JL, Tsukayama KK, et al. Functional Genomic Analyses of Mendelian and Sporadic Disease Identify Impaired eIF2α Signaling as a Generalizable Mechanism for Dystonia. Neuron. 2016;92: 1238–1251. doi: 10.1016/j.neuron.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bragg DC, Armata IA, Nery FC, Breakefield XO, Sharma N. Molecular pathways in dystonia. Neurobiol Dis. 2011;42: 136–147. doi: 10.1016/j.nbd.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaughn LS, Bragg DC, Sharma N, Camargos S, Cardoso F, Patel RC. Altered activation of protein kinase PKR and enhanced apoptosis in dystonia cells carrying a mutation in PKR activator protein PACT. J Biol Chem. 2015;290: 22543–22557. doi: 10.1074/jbc.M115.669408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nayak RR, Bernal WE, Lee JW, Kearns MJ, Cheung VG. Stress-induced changes in gene interactions in human cells. Nucleic Acids Res. 2014;42: 1757–1771. doi: 10.1093/nar/gkt999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Wang X, Ke Z-J, Comer AL, Xu M, Frank JA, et al. Tunicamycin-induced Unfolded Protein Response in the Developing Mouse Brain. Toxicol Appl Pharmacol. 2015;283: 157–167. doi: 10.1016/j.taap.2014.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485: 507–511. doi: 10.1038/nature11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattson MP, Gleichmann M, Cheng A. Mitochondria in Neuroplasticity and Neurological Disorders. Neuron. 2008;60: 748–766. doi: 10.1016/j.neuron.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016 doi: 10.1093/database/baw100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundmann K, Glöckle N, Martella G, Sciamanna G, Hauser T-K, Yu L, et al. Generation of a novel rodent model for DYT1 dystonia. Neurobiol Dis. 2012;47: 61–74. doi: 10.1016/j.nbd.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 35.Martella G, Tassone A, Sciamanna G, Platania P, Cuomo D, Viscomi MT, et al. Impairment of bidirectional synaptic plasticity in the striatum of a mouse model of DYT1 dystonia: role of endogenous acetylcholine. Brain. 2009;132: 2336–2349. doi: 10.1093/brain/awp194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia Y, Gall CM, Lynch G. Presynaptic BDNF promotes postsynaptic long-term potentiation in the dorsal striatum. J Neurosci Off J Soc Neurosci. 2010;30: 14440–14445. doi: 10.1523/JNEUROSCI.3310-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Prisco GV, Huang W, Buffington SA, Hsu C-C, Bonnen PE, Placzek AN, et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat Neurosci. 2014;17: 1073–1082. doi: 10.1038/nn.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinh MA, Ma T, Kaphzan H, Bhattacharya A, Antion MD, Cavener DR, et al. The eIF2α kinase PERK limits the expression of hippocampal metabotropic glutamate receptor-dependent long-term depression. Learn Mem. 2014;21: 298–304. doi: 10.1101/lm.032219.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calabresi P, Maj R, Pisani A, Mercuri NB, Bernardi G. Long-term synaptic depression in the striatum: physiological and pharmacological characterization. J Neurosci Off J Soc Neurosci. 1992;12: 4224–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung KW, Choi S, Lovinger DM. Activation of group I mGluRs is necessary for induction of long-term depression at striatal synapses. J Neurophysiol. 2001;86: 2405–2412. doi: 10.1152/jn.2001.86.5.2405 [DOI] [PubMed] [Google Scholar]
- 41.Carbon M, Eidelberg D. Abnormal structure-function relationships in hereditary dystonia. Neuroscience. 2009;164: 220–229. doi: 10.1016/j.neuroscience.2008.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song C-H, Bernhard D, Bolarinwa C, Hess EJ, Smith Y, Jinnah HA. Subtle microstructural changes of the striatum in a DYT1 knock-in mouse model of dystonia. Neurobiol Dis. 2013;54: 362–371. doi: 10.1016/j.nbd.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song C-H, Bernhard D, Hess EJ, Jinnah HA. Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol Dis. 2014;62: 372–380. doi: 10.1016/j.nbd.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall A, Lalli G. Rho and Ras GTPases in Axon Growth, Guidance, and Branching. Cold Spring Harb Perspect Biol. 2010;2: a001818 doi: 10.1101/cshperspect.a001818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17: 40–48. doi: 10.1038/ng0997-40 [DOI] [PubMed] [Google Scholar]
- 46.Fremont R, Tewari A, Angueyra C, Khodakhah K. A role for cerebellum in the hereditary dystonia DYT1. eLife. 2017;6: e22775 doi: 10.7554/eLife.22775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.LeDoux MS. Movement Disorders: Genetics and Models. 2nd ed. Boston: Academic Press; 2014. [Google Scholar]
- 48.Neychev VK, Fan X, Mitev VI, Hess EJ, Jinnah HA. The basal ganglia and cerebellum interact in the expression of dystonic movement. Brain J Neurol. 2008;131: 2499–2509. doi: 10.1093/brain/awn168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prudente CN, Hess EJ, Jinnah HA. Dystonia as a Network Disorder: What is the Role of the Cerebellum? Neuroscience. 2014;260: 23–35. doi: 10.1016/j.neuroscience.2013.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White JJ, Sillitoe RV. Genetic silencing of olivocerebellar synapses causes dystonia-like behaviour in mice. Nat Commun. 2017;8: 14912 doi: 10.1038/ncomms14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang C-C, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124: 3080–3092. doi: 10.1172/JCI72830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sciamanna G, Tassone A, Martella G, Mandolesi G, Puglisi F, Cuomo D, et al. Developmental Profile of the Aberrant Dopamine D2 Receptor Response in Striatal Cholinergic Interneurons in DYT1 Dystonia. PLOS ONE. 2011;6: e24261 doi: 10.1371/journal.pone.0024261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao F, Foat BC, Bussemaker HJ. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. BMC Bioinformatics. 2004;5: 31 doi: 10.1186/1471-2105-5-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erogullari A, Hollstein R, Seibler P, Braunholz D, Koschmidder E, Depping R, et al. THAP1, the gene mutated in DYT6 dystonia, autoregulates its own expression. Biochim Biophys Acta. 2014;1839: 1196–1204. doi: 10.1016/j.bbagrm.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 55.Houlden H, Schneider SA, Paudel R, Melchers A, Schwingenschuh P, Edwards M, et al. THAP1 mutations (DYT6) are an additional cause of early-onset dystonia. Neurology. 2010;74: 846–850. doi: 10.1212/WNL.0b013e3181d5276d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider SA, Ramirez A, Shafiee K, Kaiser FJ, Erogullari A, Brüggemann N, et al. Homozygous THAP1 mutations as cause of early-onset generalized dystonia. Mov Disord Off J Mov Disord Soc. 2011;26: 858–861. doi: 10.1002/mds.23561 [DOI] [PubMed] [Google Scholar]
- 57.Campagne S, Muller I, Milon A, Gervais V. Towards the classification of DYT6 dystonia mutants in the DNA-binding domain of THAP1. Nucleic Acids Res. 2012;40: 9927–9940. doi: 10.1093/nar/gks703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hollstein R, Reiz B, Kötter L, Richter A, Schaake S, Lohmann K, et al. Dystonia-causing mutations in the transcription factor THAP1 disrupt HCFC1 cofactor recruitment and alter gene expression. Hum Mol Genet. 2017; doi: 10.1093/hmg/ddx187 [DOI] [PubMed] [Google Scholar]
- 59.Kaiser FJ, Osmanoric A, Rakovic A, Erogullari A, Uflacker N, Braunholz D, et al. The dystonia gene DYT1 is repressed by the transcription factor THAP1 (DYT6). Ann Neurol. 2010;68: 554–559. doi: 10.1002/ana.22157 [DOI] [PubMed] [Google Scholar]
- 60.Xiao J, Vemula SR, Xue Y, Khan MM, Carlisle FA, Waite AJ, et al. Role of major and brain-specific Sgce isoforms in the pathogenesis of myoclonus-dystonia syndrome. Neurobiol Dis. 2017;98: 52–65. doi: 10.1016/j.nbd.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational Control of Long-Lasting Synaptic Plasticity and Memory. Neuron. 2009;61: 10–26. doi: 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Prisco GV, Huang W, Buffington SA, Hsu C-C, Bonnen PE, Placzek AN, et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat Neurosci. 2014;17: 1073–1082. doi: 10.1038/nn.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W, Placzek AN, Prisco GVD, Khatiwada S, Sidrauski C, Krnjević K, et al. Translational control by eIF2α phosphorylation regulates vulnerability to the synaptic and behavioral effects of cocaine. eLife. 2016;5: e12052 doi: 10.7554/eLife.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kazemi S, Mounir Z, Baltzis D, Raven JF, Wang S, Krishnamoorthy J-L, et al. A Novel Function of eIF2α Kinases as Inducers of the Phosphoinositide-3 Kinase Signaling Pathway. Mol Biol Cell. 2007;18: 3635–3644. doi: 10.1091/mbc.E07-01-0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trinh MA, Ma T, Kaphzan H, Bhattacharya A, Antion MD, Cavener DR, et al. The eIF2α kinase PERK limits the expression of hippocampal metabotropic glutamate receptor-dependent long-term depression. Learn Mem. 2014;21: 298–304. doi: 10.1101/lm.032219.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hetz C, Mollereau B. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15: 233–249. doi: 10.1038/nrn3689 [DOI] [PubMed] [Google Scholar]
- 67.Liu J, Pasini S, Shelanski ML, Greene LA. Activating transcription factor 4 (ATF4) modulates post-synaptic development and dendritic spine morphology. Front Cell Neurosci. 2014;8 doi: 10.3389/fncel.2014.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasini S, Corona C, Liu J, Greene LA, Shelanski ML. Specific downregulation of hippocampal ATF4 reveals a necessary role in synaptic plasticity and memory. Cell Rep. 2015;11: 183–191. doi: 10.1016/j.celrep.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun X, Liu J, Crary JF, Malagelada C, Sulzer D, Greene LA, et al. ATF4 protects against neuronal death in cellular PD models by maintaining levels of parkin. J Neurosci Off J Soc Neurosci. 2013;33: 2398–2407. doi: 10.1523/JNEUROSCI.2292-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buffington SA, Huang W, Costa-Mattioli M. Translational Control in Synaptic Plasticity and Cognitive Dysfunction. Annu Rev Neurosci. 2014;37: 17–38. doi: 10.1146/annurev-neuro-071013-014100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hoeffer CA, Klann E. mTOR Signaling: At the Crossroads of Plasticity, Memory, and Disease. Trends Neurosci. 2010;33: 67 doi: 10.1016/j.tins.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calabresi P, Gubellini P, Centonze D, Picconi B, Bernardi G, Chergui K, et al. Dopamine and cAMP-regulated phosphoprotein 32 kDa controls both striatal long-term depression and long-term potentiation, opposing forms of synaptic plasticity. J Neurosci Off J Soc Neurosci. 2000;20: 8443–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hewett JW, Zeng J, Niland BP, Bragg DC, Breakefield XO. Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol Dis. 2006;22: 98–111. doi: 10.1016/j.nbd.2005.10.012 [DOI] [PubMed] [Google Scholar]
- 74.Roffé M, Hajj GNM, Azevedo HF, Alves VS, Castilho BA. IMPACT Is a Developmentally Regulated Protein in Neurons That Opposes the Eukaryotic Initiation Factor 2α Kinase GCN2 in the modulation of Neurite Outgrowth. J Biol Chem. 2013;288: 10860–10869. doi: 10.1074/jbc.M113.461970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emery AC, Eiden MV, Eiden LE. Separate cyclic AMP sensors for neuritogenesis, growth arrest, and survival of neuroendocrine cells. J Biol Chem. 2014;289: 10126–10139. doi: 10.1074/jbc.M113.529321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bode N, Massey C, Gonzalez-Alegre P. DYT1 Knock-In Mice Are Not Sensitized against Mitochondrial Complex-II Inhibition. PLoS ONE. 2012;7: e42644 doi: 10.1371/journal.pone.0042644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin JN, Bair TB, Bode N, Dauer WT, Gonzalez-Alegre P. Transcriptional and proteomic profiling in a cellular model of DYT1 dystonia. Neuroscience. 2009;164: 563–572. doi: 10.1016/j.neuroscience.2009.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grillet M, Dominguez Gonzalez B, Sicart A, Pöttler M, Cascalho A, Billion K, et al. Torsins Are Essential Regulators of Cellular Lipid Metabolism. Dev Cell. 2016;38: 235–247. doi: 10.1016/j.devcel.2016.06.017 [DOI] [PubMed] [Google Scholar]
- 79.Teleman AA. Role for Torsin in Lipid Metabolism. Dev Cell. 2016;38: 223–224. doi: 10.1016/j.devcel.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 80.Shi-Chen Ou D, Lee S-B, Chu C-S, Chang L-H, Chung B, Juan L-J. Transcriptional activation of endoplasmic reticulum chaperone GRP78 by HCMV IE1-72 protein. Cell Res. 2011;21: 642–653. doi: 10.1038/cr.2011.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44: D574–580. doi: 10.1093/nar/gkv1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29: 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41: e108 doi: 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30: 923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15: 550 doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57: 289–300. [Google Scholar]
- 87.Gordon KL, Glenn KA, Bode N, Wen HM, Paulson HL, Gonzalez-Alegre P. The ubiquitin ligase F-box/G-domain protein 1 promotes the degradation of the disease-linked protein torsinA through the ubiquitin-proteasome pathway and macroautophagy. Neuroscience. 2012;224: 160–171. doi: 10.1016/j.neuroscience.2012.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pool M, Thiemann J, Bar-Or A, Fournier AE. NeuriteTracer: a novel ImageJ plugin for automated quantification of neurite outgrowth. J Neurosci Methods. 2008;168: 134–139. doi: 10.1016/j.jneumeth.2007.08.029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full western blots of striatal and cerebellar lysates obtained from Thap1+/- and WT littermates for expression of the indicated proteins. * denotes nonspecific band. CHOP bands are compared to the lower GAPDH panel whereas all others are compared to the highest GAPDH panel.
(TIF)
The GABA-A antagonist gabazine (10μM) was present throughout the recordings. Left: Time-course; representative traces show baseline (dashed lines) and 40 min post HFS (solid lines), with colors corresponding to the time-course graph. Calibration: 1 mV / 5 ms. Right: Summary data over final 5 min of recording (mean ± sem). LTD, measured over the final 5 min, was reduced in slices from Thap+/- mice, and enhanced in slices from Thap1C54Y/+ mice (ANOVA followed by Neman-Keuls post hoc tests). See also S9 Table.
(TIF)
Western blot analysis of (A) striatal and (B) cerebellar lysates from Thap1+/- and WT littermates for p-eIF2α/eIF2α, and p-eIF2α/GAPDH were performed 24 hrs after subcutaneous tunicamycin (TM) diluted in 150mM dextrose (or dextrose-only control; DEX). Data are presented as means ± SEM; n = 3 for each genotype and region; data normalized to WT (dextrose-only) controls. Statistical differences were assessed by two-way ANOVAs with Tukey’s post hoc tests. See also S9 Table.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
In silico analysis was used to identification putative THABS motifs in 1KB of sequence upstream of the translation start codon. Putative THABs motif indicated by “+” symbol.
(XLSX)
(XLSX)
(XLSX)
Primer sequences for the constituents of eIF2α signaling pathway, and the top 5 DEGs from the striatum and cerebellum of Thap1+/- or Thap1C54Y/+ relative to WT.
(XLSX)
(XLSX)
(XLSX)
Cross-match lists of DEGs found to be dysregulated in either of the Thap1 mutant striatum or cerebellum (Thap1C54Y/+ or Thap1+/- vs WT) with THAP1 publicly available ChIP-Seq datasets from (A) Thap1 mouse embryonic stem cells, and (B) human ENCODE K562 cells. The highest ranking biological functions as identified by DAVID GO for shared genes between the Thap1+/- cerebellum and the (C) Thap1 mouse embryonic stem cells, and (D) human ENCODE K562 cells are shown.
(XLSX)
Data Availability Statement
All next generation sequencing data are deposited in NCBI-Gene Expression Omnibus database and are accessible through GEO Series (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) accession number GSE98839. The accession number for human ENCODE ChIP-Seq data from K562 cells used in the manuscript for THAP1 is GEO: GSM803408. The accession number for the mouse embryonic stem (ES) cells ChIP-Seq data for Thap1 used in the manuscript is GEO: GSE86911.