ABSTRACT
DROSHA is the catalytic subunit of the Microprocessor complex, which initiates microRNA (miRNA) maturation in the nucleus by recognizing and cleaving hairpin precursors embedded in primary transcripts. However, accumulating evidence suggests that not all hairpin substrates of DROSHA are associated with the generation of functional small RNAs. By targeting those hairpins, DROSHA regulates diverse aspects of RNA metabolism across the transcriptome, serves as a line of defense against the expression of potentially deleterious elements, and permits cell fate determination and differentiation. DROSHA is also versatile in the way that it executes these noncanonical functions, occasionally depending on its RNA-binding activity rather than its catalytic activity. Herein, we discuss the functional and mechanistic diversity of DROSHA beyond the miRNA biogenesis pathway in light of recent findings.
KEYWORDS: DROSHA, Microprocessor, MicroRNA, Noncanonical function, RNA metabolism
Characterization of DROSHA as the initiator of microRNA maturation
MicroRNAs (miRNAs) are small non-coding RNAs of ∼22 nucleotides (nt), which recognize complementary sites within target mRNAs to direct their post-transcriptional repression [1]. In mammals, each miRNA downregulates hundreds of target mRNAs [2,3], and more than 60% of mRNAs have selectively maintained at least one miRNA target site [4]. Practically, this means that virtually every biological pathway is under the control of miRNAs. In this regard, it is not surprising that dysregulation of these tiny regulators is associated with various human diseases, including cancer [5,6].
Much is now known about how metazoan miRNAs are produced. In the canonical biogenesis pathway, miRNA genes are transcribed by RNA polymerase II (Pol II) into long primary transcripts, called primary miRNAs (pri-miRNAs), containing one or more characteristic hairpin structures [7]. These miRNA hairpins are recognized and cleaved by the nuclear Microprocessor complex, a heterotrimeric complex consisting of one molecule of DROSHA, an RNase III enzyme, and two molecules of DGCR8, a double-stranded RNA (dsRNA)-binding protein, to release ∼60–80 nt precursor miRNAs (pre-miRNAs; Figs. 1, 2A & 3A) [8-13]. Pre-miRNAs are then exported to the cytoplasm with the aid of Exportin-5 (XPO5) [14-16], where they undergo further processing by a second RNase III, DICER (Fig. 3A) [17-19]. Of the resulting ∼22 base-pair (bp) miRNA duplex, one strand is preferentially loaded onto an Argonaute (AGO) family protein to form a functional miRNA-induced silencing complex (miRISC; Fig. 3A) [20-24].
Figure 1.
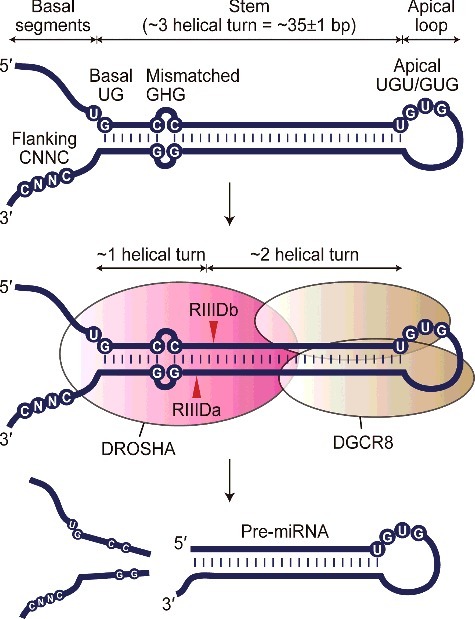
Recognition and processing of pri-miRNA by the Microprocessor complex. A representative pri-miRNA molecule is shown at the top. Pri-miRNAs are characterized by an imperfect stem of ∼3 helical turns flanked by a stretch of unstructured nucleotides at both ends, termed the basal segment and apical loop. DROSHA introduces staggered cuts ∼1 helical turn away from the basal junction and ∼2 helical turns away from the apical junction. Primary sequence motifs that are known to enhance the processing efficiency of the Microprocessor are indicated, including a basal UG motif, a flanking CNNC motif, a mismatched GHG motif, and an apical UGU/GUG motif.
Figure 2.
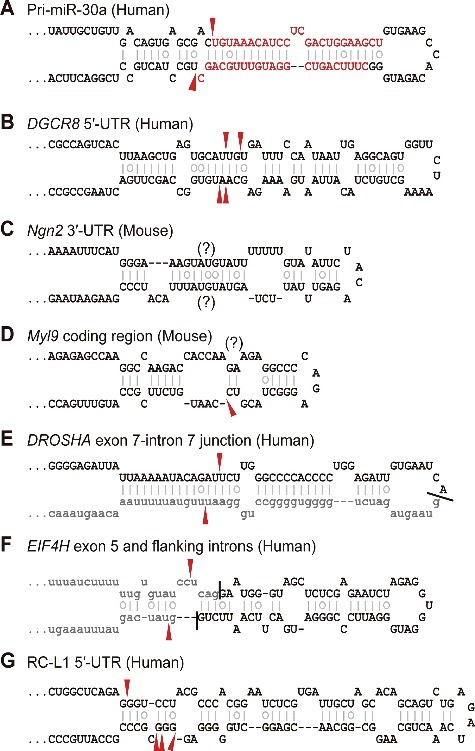
Canonical and noncanonical substrates of DROSHA. Predicted secondary structures of diverse DROSHA substrates are presented. Prediction was performed using the mfold RNA folding algorithm.[101] For pri-miR-30a, mature miRNA sequences are colored red. For the DROSHA and EIF4H hairpins, intronic sequences are shown as gray lowercase letters. The sites of DROSHA-mediated cleavage determined by characterization of in vitro or in vivo processing products are indicated by red arrowheads.
Figure 3.
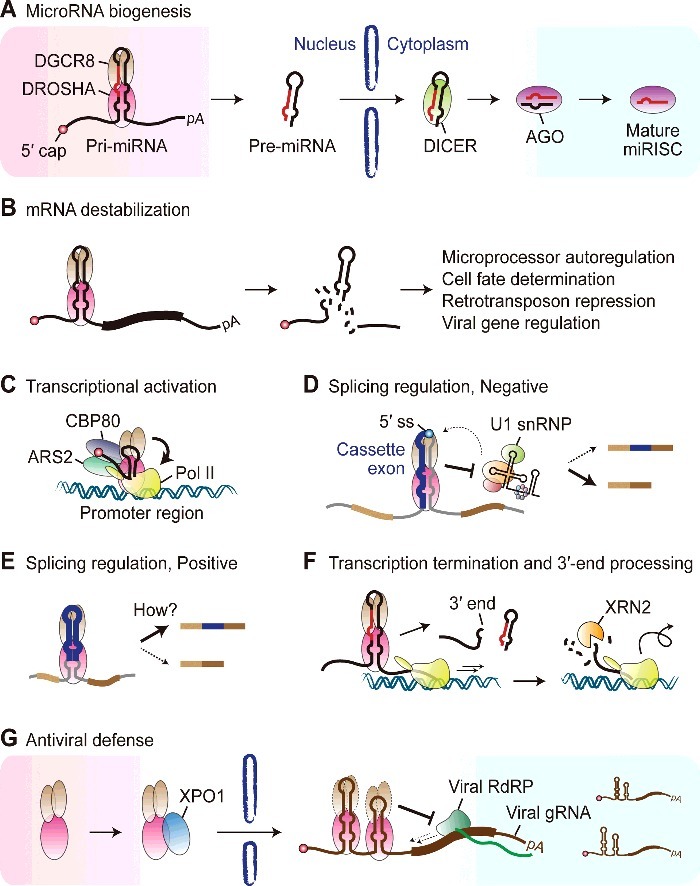
Emerging roles of DROSHA in RNA metabolism. (A) DROSHA functions as the initiator of miRNA biogenesis by cleaving pri-miRNA hairpins in the nucleus. The resulting pre-miRNAs are exported to the cytoplasm and further processed by DICER to produce mature miRNAs. (B) DROSHA destabilizes a subset of mRNAs by cleaving pri-miRNA-like hairpins, which is associated with various molecular and cellular outcomes including the homeostatic maintenance of Microprocessor activity, cell fate determination in progenitor cells, suppression of retrotransposons and viral gene regulation. (C) DROSHA is known to promote transcription of a subset of human genes, plausibly by binding to short hairpins within promoter-associated transcripts and interacting with RNA polymerase II (Pol II), CBP80 and ARS2. (D) DROSHA negatively regulates alternative splicing of its own transcript and other nascent transcripts whose splice sites overlap with pri-miRNA hairpins. For the DROSHA transcript, alternative splicing regulation by DROSHA primarily relies on its binding to a pri-miRNA-like hairpin spanning the 5′ splice site (5′ ss) and sterically blocking U1 small nuclear ribonucleoprotein (U1 snRNP). (E) DROSHA promotes splicing of a cassette exon of the EIF4H nascent transcript, which adopts a pri-miRNA-like hairpin structure, through an as-yet-uncharacterized, cleavage-independent mechanism. (F) DROSHA is involved in transcription termination and 3′-end processing of a subset of lncRNAs that serve as pri-miRNAs. DROSHA directly defines the 3′ ends of these transcripts and provides an entry site for XRN2, which is proposed to facilitate Pol II release from the template by a torpedo-like mechanism. (G) DROSHA serves as an antiviral effector against diverse positive-stranded RNA viruses by recognizing pri-miRNA-like hairpins in their genomic RNAs and conferring steric hindrance on the viral RNA-dependent RNA polymerase (RdRP). The antiviral activity of DROSHA is localized to the cytoplasm and this depends on XPO1-mediated nuclear export of DROSHA upon viral infection. It is not clear whether DGCR8 is co-exported and functions together with DROSHA in antiviral defense.
Human DROSHA contains proline-rich (P-rich) and arginine/serine-rich (RS-rich) stretches in the N-terminal region and tandem RNase III domains (RIIIDa and RIIIDb) followed by a dsRNA-binding domain (dsRBD) in the C-terminal region [25,26]. The N-terminal region of DROSHA has diverged among metazoan orthologues, and at least for human DROSHA, is dispensable for pri-miRNA processing activity in vitro [11]. However, diverse post-translational modifications within the N-terminal region of human DROSHA have been associated with subcellular localization or stability control of the protein [27-30], suggesting that this region may serve as a regulatory platform. The central and C-terminal regions of DROSHA are folded in the presence of two DGCR8 molecules to constitute the processing center of the Microprocessor [12,13], in which RIIIDa and RIIIDb dimerize intramolecularly to cleave the 3′ and 5′ strand of a pri-miRNA hairpin, respectively (Fig. 1) [11,13]. DROSHA plays a pivotal role in miRNA biogenesis, as demonstrated by the complete loss of canonical miRNA expression in DROSHA knockout cells [31].
Pri-miRNAs appear to be the canonical substrates of DROSHA. Inspection of Caenorhabditis elegans, Drosophila, and human pri-miRNAs revealed the typical structure of these molecules, in which an imperfect stem of ∼3 helical turns is flanked by unpaired basal segments at the base and an apical loop at the top (Figs. 1 & 2A) [32,33]. DROSHA introduces staggered cuts ∼1 helical turn away from the basal junction and ∼2 helical turns away from the apical junction (Figs. 1 & 2A) [8,33-36]. In addition to these structural determinants, several primary sequence motifs in pri-miRNAs were found to promote DROSHA-mediated processing, including a UG motif at the basal junction, a CNNC motif ∼17–18 nt downstream of the 3′ cleavage site, a mismatched GHG motif in the middle of the basal stem (where H is any nucleotide except G), and a UGU/GUG motif in the 5′ end of the apical loop (Fig. 1) [12,37-40]. More recently, N6-methyladenosine (m6A) RNA modifications emerged as an epigenetic determinant of pri-miRNA processing [41-43].
Noncanonical functions of DROSHA beyond nuclear pri-miRNA processing
In addition to understanding pri-miRNAs and their recognition and cleavage by DROSHA, several genome-wide approaches aimed at uncovering the cellular functions of DROSHA have revealed the existence of its non-miRNA substrates [44-46]. Given that DROSHA-mediated cleavage is accompanied by substantial destabilization of pri-miRNAs [8,10,11,47,48], transcriptomic changes upon DROSHA depletion were measured to identify any additional substrates [49-52]. mRNAs that are derepressed by DROSHA deficiency, but not DICER deficiency, tend to possess hairpin structures that closely resemble those of pri-miRNAs [49,51], suggesting that DROSHA-mediated destabilization of mRNAs involves their direct cleavage. Instead of measuring steady-state transcript abundance, cleavage products can be directly quantified using parallel analysis of RNA ends (PARE) or degradome sequencing, which aims to sequence the 5′ ends of RNA fragments bearing 5′ monophosphate termini [53,54]. Degradome sequencing in mammalian cells identified a number of DROSHA-dependent cleavage sites within mRNAs [55,56]. In addition, application of a crosslinking, immunoprecipitation, and sequencing (CLIP-seq) technique to the Microprocessor components demonstrated the presence of substrates that give rise to very little or no detectable small RNAs [57-59]. These findings raise the question of whether noncanonical targeting by DROSHA is widespread, and if so, what are its functions? Below, we describe the reported roles of DROSHA beyond nuclear pri-miRNA processing.
Ribosomal RNA biogenesis
The earliest characterization of DROSHA indicated a putative role in ribosomal RNA biogenesis [26], consistent with the previously reported functions of bacterial and yeast RNase III enzymes [60,61]. A significant fraction of nuclear DROSHA protein is translocated to the nucleolus during the S phase [26], and antisense inhibition of DROSHA leads to the accumulation of precursors of 5.8 S rRNA [26,62]. However, levels of mature rRNAs are not evidently changed following DROSHA depletion [26,62,63], suggesting that DROSHA may play a minor role, if any, in the maturation of rRNAs.
mRNA destabilization
The best-characterized function of DROSHA outside of the miRNA pathway is post-transcriptional destabilization of mRNAs through cleaving pri-miRNA-like hairpins embedded within them (Fig. 3B). The herald of this paradigm was the DGCR8 mRNA, which contains two evolutionarily conserved hairpin structures in its 5′-untranslated region (5′-UTR) and coding region, respectively (Fig. 2B) [64]. These hairpins are cleaved by DROSHA in vitro and in cells, which imparts instability to the reporter construct in a DROSHA-dependent manner [50,65]. DROSHA-mediated destabilization of the DGCR8 mRNA is proposed to serve as a means of maintaining constant Microprocessor activity, and its deep conservation in mammals and Drosophila may reflect its importance [49-51,56,65]. Notably, the DGCR8 mRNA hairpins seem unlikely to serve as pri-miRNAs, because the corresponding small RNAs could only be detected, if at all, by deep sequencing [50,51]. For example, the ∼60 nt pre-miRNA-like fragment generated from the 5′-UTR hairpin is not processed down to small RNAs in cells[50,65] despite its full potential to produce small RNAs in vitro [65], and this processing failure is attributed in part to its strict nuclear retention [50].
DROSHA-mediated cleavage also contributes to the clearance of a subset of mRNAs in progenitor cells, which appears to be essential for cell fate determination and differentiation [52,66-68]. In embryonic neural stem cells, DROSHA destabilizes the mRNA of the proneural factor Neurogenin2 (Ngn2) by cleaving pri-miRNA-like hairpins within its 3′-UTR (Fig. 2C) [66,69]. The loss of this repression largely explains the precocious differentiation of progenitors caused by DROSHA deficiency, because forced expression of Ngn2 leads to the same phenotype, and double depletion of DROSHA and Ngn2 restores normal progenitor status [66]. The Tbr1 mRNA is another substrate for DROSHA in neuronal progenitors and its downregulation is critical for corticogenesis [68]. In adult hippocampal stem cells, DROSHA targets the NF1B mRNA to prevent oligodendrogenesis and to promote neurogenesis [67]. A similar mechanism operates in hematopoietic stem cells, where DROSHA-mediated destabilization of the Myl9 and Todr1 mRNAs is necessary for dendritic cell development and myelopoiesis (Fig. 2D) [52]. It will be interesting to investigate whether this type of regulation is at play in other progenitor cells during their differentiation.
In addition to these biologically relevant examples, many other mRNAs are subject to DROSHA-mediated cleavage [49,51,56-59]. However, with the exception of the DGCR8 mRNA, candidate mRNAs identified from one study tend to overlap poorly with those from another, probably due to differences in subjects and the experimental approaches employed. DROSHA-mediated cleavage of these mRNAs leads to their destabilization, but the extent to which they are derepressed upon DROSHA depletion is modest compared to pri-miRNAs [49,51,56,57,59]. Indeed, mRNA hairpins are cleaved less efficiently than pri-miRNAs in vitro [50-52,57,59,65], and are proposed to have suboptimal structures and sequences [59]. This may be because these hairpins have evolved to modulate the expression of their host transcripts, rather than to serve as a source for downstream effectors. Alternatively, but not mutually exclusively, this may represent a strategy that allows robust cleavage to occur only under certain conditions. For example, the Ngn2 hairpin deviates significantly from optimal pri-miRNA hairpins in terms of its structure and sequence (Fig. 2C), but it is readily cleaved by DROSHA in neural stem cells, leading to degradation of more than 80% of the Ngn2 mRNA [66]. TDP-43, a neural activity-responsive factor, was reported to associate with DROSHA[10,70] and to be required for DROSHA-mediated destabilization of the Ngn2 mRNA [69]. This suggests that trans-acting factors may modulate DROSHA-mediated cleavage of mRNA hairpins in order to achieve spatiotemporally regulated destabilization of specific mRNAs.
Regulation of transcription and RNA processing
Microprocessor-mediated processing of pri-miRNAs occurs co-transcriptionally, as evidenced by the chromatin association of DROSHA [71,72]. Interestingly, genome-wide analysis of its chromatin footprints found that DROSHA binds not only miRNA genes but also the 5′ ends of many other genes [73]. This binding is reportedly mediated by short hairpins within promoter-associated transcripts that are structurally distinct from pri-miRNAs or pri-miRNA-like hairpins found in mRNAs. Surprisingly, the consequence of this binding is neither cleavage nor destabilization of the corresponding transcripts, but rather transcriptional activation. Furthermore, the function of DROSHA as a transcriptional activator is independent of its catalytic activity and is mediated through its N-terminal region, which appears to interact with Pol II and CBP80 (Fig. 3C).
Consistent with the notion that nuclear RNA processing events are closely coupled to one another [74], it has been demonstrated that DROSHA can play dual roles in both pri-miRNA (or pri-miRNA-like hairpin) cleavage and other RNA processing. DGCR8 CLIP-seq revealed hundreds of cassette exons bound by DGCR8, and depletion of DROSHA or DGCR8 causes the accumulation of splice isoforms containing these exons [57]. Bioinformatics analysis identified dozens of loci in which pri-miRNA hairpins are juxtaposed with exon-intron junctions such that the Microprocessor and splicing machinery functionally antagonize each other [75,76]. Splicing regulation by DROSHA was recently corroborated by the discovery of a pri-miRNA-like hairpin located across a specific exon-intron junction of the human DROSHA gene itself (Fig. 2E) [59,77,78]. The DROSHA hairpin, likely derived from transposable elements, is structurally conserved among placental mammals but seems to be functionally divergent. For example, the human DROSHA hairpin is cleaved by DROSHA and tilts splicing of the adjacent exon toward skipping in a DROSHA-dependent manner, while the mouse DROSHA hairpin lacks these activities [77]. Interestingly, the suppressive effect of DROSHA on splicing of its own transcript does not require the catalytic activity of DROSHA, but it involves sterically hindering the splicing machinery from recognizing its cognate splice site, thereby expanding the repertoire of its mode of action (Fig. 3D) [77]. DROSHA can also act as a positive regulator of alternative splicing independently of its cleavage function (Figs. 2F & 3E) [79]. Although the underlying mechanism is poorly understood, documented interactions between the Microprocessor and spliceosome components may provide a clue [10,80,81].
Transcription termination and 3′-end processing is another process subject to DROSHA-mediated regulation (Fig. 3F). In the canonical pathway of Pol II transcriptional termination, endonucleolytic cleavage near a polyadenylation signal is followed by degradation of downstream cleavage products by the 5′-3′ exonuclease XRN2, and Pol II is released from the template when XRN2 catches up to it [82]. The very same exonuclease is recruited to the sites of DROSHA-mediated cleavage [71,83,84], suggesting that a similar ‘torpedo-like’ mechanism may be operational downstream of pri-miRNA processing. Indeed, abrogating pri-miRNA processing results in increased Pol II density at the downstream region and extensive transcriptional readthrough [83,85]. In this alternative pathway, DROSHA appears not only to provide an entry site for XRN2, but also to directly define the 3′ end of the transcript [85]. Interestingly, genome-wide analysis suggested that transcriptional termination by DROSHA is restricted to long non-coding RNAs (lncRNAs) that serve as pri-miRNAs [85]. It remains to be elucidated how miRNA hairpins within lncRNAs are distinguished from those embedded in protein-coding transcripts and are specifically destined for the transcription termination signals.
Maintenance of genome integrity and antiviral defense
The DNA-damage response (DDR) is a signaling pathway that has evolved to cope with various genotoxic stresses confronted by cells. In addition to a multitude of protein components comprising the cascade, a class of small RNAs distinct from miRNAs have been implicated in the DDR pathway in various organisms [86-89]. In mammals, proper formation of DDR foci following exogenous or oncogene-induced genotoxic insults requires small RNAs generated at sites of DNA damage in a DROSHA- and DICER-dependent manner, termed DNA-damage RNAs (DDRNAs) [87]. Similar to miRNAs, DDRNAs depend on AGO proteins to elicit their functions in DNA repair [88]. However, DDRNAs are distinguished from miRNAs by being independent of GW182-like proteins, the downstream effectors of miRNA-mediated post-transcriptional repression, and in their probable nuclear localization [87]. The mechanistic details of their biogenesis and functions await further investigation, and in particular, how DNA lesions occurring randomly throughout the genome are capable of providing adequate substrates for DROSHA remains to be established.
Another threat to the maintenance of genome stability is transposable elements. Although most transposable elements found in the human genome are currently inactive, a small fraction of retrotransposons still retain their activity and contribute to evolution and human diseases [90]. To prevent uncontrolled transposition of these elements, which would likely be detrimental to cells, several restriction mechanisms have evolved at multiple levels of gene expression. For example, a class of small RNAs, called PIWI-interacting RNAs (piRNAs), mediate transcriptional silencing of transposons in the germlines of mammals and insects [91]. The role for the Microprocessor as a post-transcriptional repressor of retrotransposons was hinted at by DGCR8 CLIP-seq, which revealed that around one third of identified binding sites correspond to repetitive elements, mainly LINE-1 and SINE (including Alu) retrotransposons [57]. Indeed, DROSHA negatively regulates LINE-1 and Alu retrotransposition by cleaving pri-miRNA-like hairpin structures embedded in their cognate transcripts (Figs. 2G & 3B) [92].
Accumulating evidence suggests that the protective functions of DROSHA extend to antiviral defense. DROSHA targets two pri-miRNA hairpins within the 3′-UTR of the mRNA for Kaposin B (KapB), a protein encoded by Kaposi's sarcoma-associated herpesvirus (KSHV), and decreased KapB expression mediated by DROSHA is associated with latent replication of the virus (Fig. 3B) [93]. Interestingly, DROSHA expression is reduced during lytic infection with concomitant derepression of KapB, suggesting that the virus may have evolved to utilize Microprocessor activity as a means of regulating its own life cycle. On the other hand, diverse RNA viruses including Sindbis virus (SINV) trigger translocation of DROSHA into the cytoplasm, where it cleaves viral genomic RNA [94,95]. Surprisingly, a recent study demonstrated that cytoplasmic DROSHA exerts its antiviral activity primarily through clamping viral RNA hairpins and conferring steric hindrance on the viral RNA-dependent RNA polymerases (RdRPs), rather than cleaving them (Fig. 3G) [96].
Concluding remarks
Extensive efforts over the past decade have greatly expanded the functional repertoire of DROSHA beyond nuclear pri-miRNA processing (Fig. 3A). From a molecular standpoint, DROSHA regulates diverse aspects of RNA metabolism across the transcriptome, ranging from post-transcriptional control of RNA stability (Fig. 3B) [49-52,57,59,65-68,92] to transcriptional activation (Fig. 3C) [73], alternative splicing (Fig. 3D & E) [57,75-77,79], and 3′-end processing and transcriptional termination (Fig. 3F) [83-85]. Meanwhile, at the cellular level, DROSHA defends against genotoxic stresses[87] and the expression of potentially deleterious elements such as retrotransposons (Fig. 3B) [92] and viruses (Fig. 3G) [93-96]. From a physiological stance, DROSHA ensures appropriate cell fate decisions in progenitor cells and allows their timely differentiation (Fig. 3B) [52,66-68]. Notably, multiple types of cancer exhibit aberrant expression of the Microprocessor components, and the resulting perturbation of the miRNA pool has been thought to contribute to tumorigenesis [6]. Further studies are needed to reveal whether dysregulation of other biological processes controlled by DROSHA plays a role in human diseases, and if so, to what extent.
The mechanistic repertoire of DROSHA has also been expanded, as illustrated by its recently-discovered cleavage-independent functions (Figs. 3C-E, G) [73,77,79,96]. Interestingly, cleavage-independent functions are not the exclusive property of DROSHA, and may represent a common regulatory paradigm for RNase III enzymes. The Escherichia coli RNase III has been reported to promote the translation of phage genes independently of its cleavage function [97]. Transcriptome-wide mapping of DICER-binding sites in human cells and C. elegans found that more than three quarters of identified loci are not associated with apparent small RNA production, and these ‘passive sites’ instead confer stability and granular localization on target RNAs [98]. Although the mechanistic details underlying cleavage-independent functions remain to be elucidated, DROSHA appears to employ at least two strategies: first, DROSHA may recruit positive regulators, possibly through its N-terminal region, to stimulate biological processes of interest such as transcription (Fig. 3C) [73] and splicing (Fig. 3E) [79] and second, DROSHA may bind to its substrates and catalyze little or no cleavage activity, but rather impede the access of other proteins to exert its inhibitory roles (Fig. 3D, G) [77,96].
The cytoplasmic functions of DROSHA are another important subject for future studies. Antiviral activity of DROSHA toward RNA viruses is localized to the cytoplasm [94-96], and this relies on Exportin-1 (XPO1)-mediated nuclear export of DROSHA upon viral infection (Fig. 3G) [95] Interestingly, a mutant version of DROSHA that fails to associate with DGCR8 suppresses viral replication as efficiently as wild-type DROSHA, suggesting the possibility of an alternative DROSHA complex in the cytoplasm [96]. In addition to viral entry, heat shock and oxidative stress are also known to cause cytoplasmic re-localization of DROSHA [30]. In fact, a small fraction of DROSHA protein is present in the cytoplasm even under normal conditions, due to alternative splicing [77,99,100], although its biological role is largely unknown. It will be interesting to investigate whether cytoplasmic DROSHA has any distinct substrates, interacting partners, and functions.
Acknowledgment
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST; No. 2016R1A2B4010472), Republic of Korea.
References
- [1].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lim LP, Lau NC, Garrett-Engele P, et al.. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–73. doi: 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- [3].Baek D, Villen J, Shin C, et al.. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Friedman RC, Farh KK, Burge CB, et al.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482(7385):347–55. doi: 10.1038/nature10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–33. doi: 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee Y, Kim M, Han J, et al.. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60. doi: 10.1038/sj.emboj.7600385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee Y, Ahn C, Han J, et al.. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. [DOI] [PubMed] [Google Scholar]
- [9].Denli AM, Tops BB, Plasterk RH, et al.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- [10].Gregory RI, Yan KP, Amuthan G, et al.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120 [DOI] [PubMed] [Google Scholar]
- [11].Han J, Lee Y, Yeom KH, et al.. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–27. doi: 10.1101/gad.1262504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nguyen TA, Jo MH, Choi YG, et al.. Functional Anatomy of the Human Microprocessor. Cell. 2015;161(6):1374–87. doi: 10.1016/j.cell.2015.05.010 [DOI] [PubMed] [Google Scholar]
- [13].Kwon SC, Nguyen TA, Choi YG, et al.. Structure of Human DROSHA. Cell. 2016;164(1–2):81–90. doi: 10.1016/j.cell.2015.12.019 [DOI] [PubMed] [Google Scholar]
- [14].Yi R, Qin Y, Macara IG, et al.. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–91. doi: 10.1261/rna.5167604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lund E, Guttinger S, Calado A, et al.. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599 [DOI] [PubMed] [Google Scholar]
- [17].Grishok A, Pasquinelli AE, Conte D, et al.. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- [18].Hutvagner G, McLachlan J, Pasquinelli AE, et al.. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- [19].Ketting RF, Fischer SE, Bernstein E, et al.. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–9. doi: 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mourelatos Z, Dostie J, Paushkin S, et al.. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–8. doi: 10.1101/gad.974702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–16. doi: 10.1016/S0092-8674(03)00801-8 [DOI] [PubMed] [Google Scholar]
- [22].Schwarz DS, Hutvagner G, Du T, et al.. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/S0092-8674(03)00759-1 [DOI] [PubMed] [Google Scholar]
- [23].Liu J, Carmell MA, Rivas FV, et al.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–41. doi: 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- [24].Meister G, Landthaler M, Patkaniowska A, et al.. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–97. doi: 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- [25].Filippov V, Solovyev V, Filippova M, et al.. A novel type of RNase III family proteins in eukaryotes. Gene. 2000;245(1):213–21. doi: 10.1016/S0378-1119(99)00571-5 [DOI] [PubMed] [Google Scholar]
- [26].Wu H, Xu H, Miraglia LJ, et al.. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275(47):36957–65. doi: 10.1074/jbc.M005494200 [DOI] [PubMed] [Google Scholar]
- [27].Tang X, Zhang Y, Tucker L, et al.. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38(19):6610–9. doi: 10.1093/nar/gkq547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tang X, Li M, Tucker L, et al.. Glycogen synthase kinase 3 beta (GSK3beta) phosphorylates the RNAase III enzyme Drosha at S300 and S302. PLoS One. 2011;6(6):e20391. doi: 10.1371/journal.pone.0020391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tang X, Wen S, Zheng D, et al.. Acetylation of drosha on the N-terminus inhibits its degradation by ubiquitination. PLoS One. 2013;8(8):e72503. doi: 10.1371/journal.pone.0072503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang Q, Li W, She H, et al.. Stress induces p38 MAPK-mediated phosphorylation and inhibition of Drosha-dependent cell survival. Mol Cell. 2015;57(4):721–34. doi: 10.1016/j.molcel.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim YK, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A. 2016;113(13):E1881–9. doi: 10.1073/pnas.1602532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lim LP, Lau NC, Weinstein EG, et al.. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008. doi: 10.1101/gad.1074403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Han J, Lee Y, Yeom KH, et al.. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043 [DOI] [PubMed] [Google Scholar]
- [34].Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9(1):112–23. doi: 10.1261/rna.2780503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24(1):138–48. doi: 10.1038/sj.emboj.7600491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280(30):27595–603. doi: 10.1074/jbc.M504714200 [DOI] [PubMed] [Google Scholar]
- [37].Auyeung VC, Ulitsky I, McGeary SE, et al.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152(4):844–58. doi: 10.1016/j.cell.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mori M, Triboulet R, Mohseni M, et al.. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156(5):893–906. doi: 10.1016/j.cell.2013.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fang W, Bartel DP. The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol Cell. 2015;60(1):131–45. doi: 10.1016/j.molcel.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Roden C, Gaillard J, Kanoria S, et al.. Novel determinants of mammalian primary microRNA processing revealed by systematic evaluation of hairpin-containing transcripts and human genetic variation. Genome Res. 2017;27(3):374–384. doi: 10.1101/gr.208900.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Alarcon CR, Lee H, Goodarzi H, et al.. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–5. doi: 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Alarcon CR, Goodarzi H, Lee H, et al.. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162(6):1299–308. doi: 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Knuckles P, Carl SH, Musheev M, et al.. RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat Struct Mol Biol. 2017;24(7):561–569. doi: 10.1038/nsmb.3419 [DOI] [PubMed] [Google Scholar]
- [44].Macias S, Cordiner RA, Caceres JF. Cellular functions of the microprocessor. Biochem Soc Trans. 2013;41(4):838–43. doi: 10.1042/BST20130011 [DOI] [PubMed] [Google Scholar]
- [45].Johanson TM, Lew AM, Chong MM. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013;3(10):130144. doi: 10.1098/rsob.130144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Burger K, Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat Rev Mol Cell Biol. 2015;16(7):417–30. doi: 10.1038/nrm3994 [DOI] [PubMed] [Google Scholar]
- [47].Chang TC, Pertea M, Lee S, et al.. Genome-wide annotation of microRNA primary transcript structures reveals novel regulatory mechanisms. Genome Res. 2015;25(9):1401–9. doi: 10.1101/gr.193607.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jeong G, Lim YH, Kim YK. Precise mapping of the transcription start sites of human microRNAs using DROSHA knockout cells. BMC Genomics. 2016;17(1):908. doi: 10.1186/s12864-016-3252-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kadener S, Rodriguez J, Abruzzi KC, et al.. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15(4):537–45. doi: 10.1261/rna.1319309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Han J, Pedersen JS, Kwon SC, et al.. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chong MM, Zhang G, Cheloufi S, et al.. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24(17):1951–60. doi: 10.1101/gad.1953310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Johanson TM, Keown AA, Cmero M, et al.. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat Immunol. 2015;16(11):1134–41. doi: 10.1038/ni.3293 [DOI] [PubMed] [Google Scholar]
- [53].German MA, Pillay M, Jeong DH, et al.. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26(8):941–6. doi: 10.1038/nbt1417 [DOI] [PubMed] [Google Scholar]
- [54].Addo-Quaye C, Eshoo TW, Bartel DP, et al.. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18(10):758–62. doi: 10.1016/j.cub.2008.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shin C, Nam JW, Farh KK, et al.. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38(6):789–802. doi: 10.1016/j.molcel.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Karginov FV, Cheloufi S, Chong MM, et al.. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38(6):781–8. doi: 10.1016/j.molcel.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macias S, Plass M, Stajuda A, et al.. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19(8):760–6. doi: 10.1038/nsmb.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Seong Y, Lim DH, Kim A, et al.. Global identification of target recognition and cleavage by the Microprocessor in human ES cells. Nucleic Acids Res. 2014;42(20):12806–21. doi: 10.1093/nar/gku957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim B, Jeong K, Kim VN. Genome-wide Mapping of DROSHA Cleavage Sites on Primary MicroRNAs and Noncanonical Substrates. Mol Cell. 2017;66(2):258–269 e5. doi: 10.1016/j.molcel.2017.03.013 [DOI] [PubMed] [Google Scholar]
- [60].Dunn JJ, Studier FW. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Elela SA, Igel H, Ares M Jr.. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85(1):115–24. doi: 10.1016/S0092-8674(00)81087-9 [DOI] [PubMed] [Google Scholar]
- [62].Liang XH, Crooke ST. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39(11):4875–89. doi: 10.1093/nar/gkr076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chong MM, Rasmussen JP, Rudensky AY, et al.. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205(9):2005–17. doi: 10.1084/jem.20081219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pedersen JS, Bejerano G, Siepel A, et al.. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2(4):e33. doi: 10.1371/journal.pcbi.0020033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Triboulet R, Chang HM, Lapierre RJ, et al.. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15(6):1005–11. doi: 10.1261/rna.1591709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Knuckles P, Vogt MA, Lugert S, et al.. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat Neurosci. 2012;15(7):962–9. doi: 10.1038/nn.3139 [DOI] [PubMed] [Google Scholar]
- [67].Rolando C, Erni A, Grison A, et al.. Multipotency of Adult Hippocampal NSCs In Vivo Is Restricted by Drosha/NFIB. Cell Stem Cell. 2016;19(5):653–662. doi: 10.1016/j.stem.2016.07.003 [DOI] [PubMed] [Google Scholar]
- [68].Marinaro F, Marzi MJ, Hoffmann N, et al.. MicroRNA-independent functions of DGCR8 are essential for neocortical development and TBR1 expression. EMBO Rep. 2017;18(4):603–618. doi: 10.15252/embr.201642800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Di Carlo V, Grossi E, Laneve P, et al.. TDP-43 regulates the microprocessor complex activity during in vitro neuronal differentiation. Mol Neurobiol. 2013;48(3):952–63. doi: 10.1007/s12035-013-8564-x [DOI] [PubMed] [Google Scholar]
- [70].Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109(9):3347–52. doi: 10.1073/pnas.1112427109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Morlando M, Ballarino M, Gromak N, et al.. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15(9):902–9. doi: 10.1038/nsmb.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Morlando M, Dini Modigliani S, Torrelli G, et al.. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31(24):4502–10. doi: 10.1038/emboj.2012.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gromak N, Dienstbier M, Macias S, et al.. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013;5(6):1499–510. doi: 10.1016/j.celrep.2013.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Pawlicki JM, Steitz JA. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol. 2010;20(1):52–61. doi: 10.1016/j.tcb.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mattioli C, Pianigiani G, Pagani F. A competitive regulatory mechanism discriminates between juxtaposed splice sites and pri-miRNA structures. Nucleic Acids Res. 2013;41(18):8680–91. doi: 10.1093/nar/gkt614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Melamed Z, Levy A, Ashwal-Fluss R, et al.. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol Cell. 2013;50(6):869–81. doi: 10.1016/j.molcel.2013.05.007 [DOI] [PubMed] [Google Scholar]
- [77].Lee D, Nam JW, Shin C. DROSHA targets its own transcript to modulate alternative splicing. RNA. 2017;23(7):1035–1047. doi: 10.1261/rna.059808.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mechtler P, Johnson S, Slabodkin H, et al.. The evidence for a microRNA product of human DROSHA gene. RNA Biol. 2017:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Havens MA, Reich AA, Hastings ML. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014;10(5):e1004312. doi: 10.1371/journal.pgen.1004312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kataoka N, Fujita M, Ohno M. Functional association of the Microprocessor complex with the spliceosome. Mol Cell Biol. 2009;29(12):3243–54. doi: 10.1128/MCB.00360-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Agranat-Tamir L, Shomron N, Sperling J, et al.. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014;42(7):4640–51. doi: 10.1093/nar/gkt1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol. 2015;16(3):190–202. doi: 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- [83].Ballarino M, Pagano F, Girardi E, et al.. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29(20):5632–8. doi: 10.1128/MCB.00664-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wagschal A, Rousset E, Basavarajaiah P, et al.. Meziane O and others. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150(6):1147–57. doi: 10.1016/j.cell.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Dhir A, Dhir S, Proudfoot NJ, et al.. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol. 2015;22(4):319–27. doi: 10.1038/nsmb.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lee HC, Chang SS, Choudhary S, et al.. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459(7244):274–7. doi: 10.1038/nature08041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Francia S, Michelini F, Saxena A, et al.. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488(7410):231–5. doi: 10.1038/nature11179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wei W, Ba Z, Gao M, et al.. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149(1):101–12. doi: 10.1016/j.cell.2012.03.002 [DOI] [PubMed] [Google Scholar]
- [89].Michalik KM, Bottcher R, Forstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40(19):9596–603. doi: 10.1093/nar/gks711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Beck CR, Garcia-Perez JL, Badge RM, et al.. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Czech B, Hannon GJ. One Loop to Rule Them All: The Ping-Pong Cycle and piRNA-Guided Silencing. Trends Biochem Sci. 2016;41(4):324–37. doi: 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Heras SR, Macias S, Plass M, et al.. The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol. 2013;20(10):1173–81. doi: 10.1038/nsmb.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci U S A. 2011;108(27):11229–34. doi: 10.1073/pnas.1105799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shapiro JS, Langlois RA, Pham AM, et al.. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18(7):1338–46. doi: 10.1261/rna.032268.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Shapiro JS, Schmid S, Aguado LC, et al.. Drosha as an interferon-independent antiviral factor. Proc Natl Acad Sci U S A. 2014;111(19):7108–13. doi: 10.1073/pnas.1319635111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Aguado LC, Schmid S, May J, et al.. RNase III nucleases from diverse kingdoms serve as antiviral effectors. Nature. 2017;547(7661):114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Altuvia S, Locker-Giladi H, Koby S, et al.. RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1987;84(18):6511–5. doi: 10.1073/pnas.84.18.6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rybak-Wolf A, Jens M, Murakawa Y, et al.. A variety of dicer substrates in human and C. elegans. Cell. 2014;159(5):1153–67. doi: 10.1016/j.cell.2014.10.040 [DOI] [PubMed] [Google Scholar]
- [99].Dai L, Chen K, Youngren B, et al.. Cytoplasmic Drosha activity generated by alternative splicing. Nucleic Acids Res. 2016;44(21):10454–10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Link S, Grund SE, Diederichs S. Alternative splicing affects the subcellular localization of Drosha. Nucleic Acids Res. 2016;44(11):5330–43. doi: 10.1093/nar/gkw400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]


