Abstract
Metabolic changes are linked to epigenetic reprogramming and play important roles in several tumor types. PGC-1α is a transcriptional coactivator controlling mitochondrial biogenesis and is linked to oxidative phosphorylation. We provide evidence that melanoma models with elevated PGC-1α levels are characteristic of the proliferative phenotype and are sensitive to bromodomain and extra-terminal domain (BET) inhibitor treatment. A super-enhancer region highly occupied by the BET family member BRD4 was identified for the PGC-1α gene. BET inhibitor treatment prevented this interaction, leading to a dramatic reduction of PGC-1α expression. Accordingly, BET inhibition diminished respiration and mitochondrial function in cells. In vivo, melanoma models with high PGC-1α expression strongly responded to BET inhibition by reduction of PGC-1α and impaired tumor growth. Altogether, our findings identify epigenetic regulatory elements that define a subset of melanomas with high sensitivity to BET inhibition, which opens up the opportunity to define melanoma patients most likely to respond to this treatment, depending on their tumor characteristics.
Introduction
About 75 000 new cases of melanoma were diagnosed in the United States in 2014 and the incidence rate is expected to rise in the next years.1, 2 Early stages can be treated with surgery but advanced tumors are usually highly metastatic and difficult to address, due to the poor response to radio- and chemotherapy. Few treatment options were available until recent years, but this has dramatically changed with the discovery that about 50% of melanomas harbor mutations in the BRAF kinase, which activate the MAPK pathway.3 Inhibitors blocking mutated BRAF and MEK are now approved for melanoma treatment,4 but despite high initial response, most patients will develop resistance within 6–8 months.5 Another major breakthrough was the finding that checkpoint regulators such as CTLA4 and PD-1 are essential for the immune response against melanoma, which is a highly mutagenic and antigenic tumor,4 and different monoclonal antibodies addressing these targets have been approved.4 However, cases of intrinsic or acquired resistance are frequently observed and there is still a great need for more effective therapies.5
At the genetic level, melanoma is extremely complex with thousands of documented alterations, including mutations of BRAF or NRAS3, 6 and aberrant expression of SOX family members,7 but only a few of these represent driver events. These alterations often affect cellular metabolism, and melanomas can shift from oxidative phosphorylation (OXPHOS) to aerobic glycolysis to meet increased energetic and anabolic needs.6 This is exacerbated by BRAF mutations as documented by the ensuing reduced number and function of mitochondria, increased glucose uptake and elevated serum lactate levels measured in patients.8 Enzymes associated with both the glycolytic and the OXPHOS pathways are elevated in early- and late-stage melanomas, suggesting that tumors show high plasticity when it comes to their energy and building block demands.6, 8 Indeed, a number of studies show that non-glycolytic pathways and increased mitochondrial activity are essential for melanoma and that targeting mitochondrial respiration can dramatically inhibit melanoma growth.9 Energy production by mitochondria is partially controlled by the melanocyte-specific microphthalmia-associated transcription factor (MITF) and the transcriptional cofactor peroxisome proliferator-activated receptor-gamma coactivator-1α (PGC-1α, encoded by the PPARGC1A gene), which drive metabolic reprogramming in melanocytes.10, 11 Importantly, this pathway is suppressed by mutated BRAF but can be re-activated by its inhibitor, so that an OXPHOS program can ultimately lead to therapy resistance.10
Expression regulation of oncogenic players and tumor suppressors by epigenetic mechanisms has been evidenced.12, 13 It is controlled largely by the local chromatin conformation and the interaction between histones and gene regulatory regions. Histone acetylation, which loosens the contact of histones with DNA and is read by bromodomain proteins, is an essential aspect of this regulation.14 The bromodomain and extra-terminal domain (BET) family protein BRD4 promotes transcription elongation of a number of oncogenes and is essential in numerous tumor types.14, 15 However, BET bromodomain inhibitors are only effective in certain subgroups of solid tumors so that predictive response biomarkers are urgently needed. In melanoma, different levels of anti-proliferative activity are observed for the BET inhibitor I-BET151 in vitro,16, 17 but, importantly, the BRAF or NRAS mutational status is not predictive of response.18, 19
In this study, we investigated the impact of BET bromodomain inhibition on epigenetic regulatory elements that define the phenotype and metabolic subtypes of melanoma. We provide evidence for a direct role of BRD4 binding at super-enhancers that drive the expression of PGC-1α and SOX10, a transcription factor involved in melanocyte development. We furthermore found that expression of PGC-1α was associated with the response to BET inhibitors, most likely due to the fact that melanoma cell proliferation relied on elevated levels of PGC-1α to regulate bioenergetic pathways.
Results
BET inhibition has a differential impact on melanoma growth
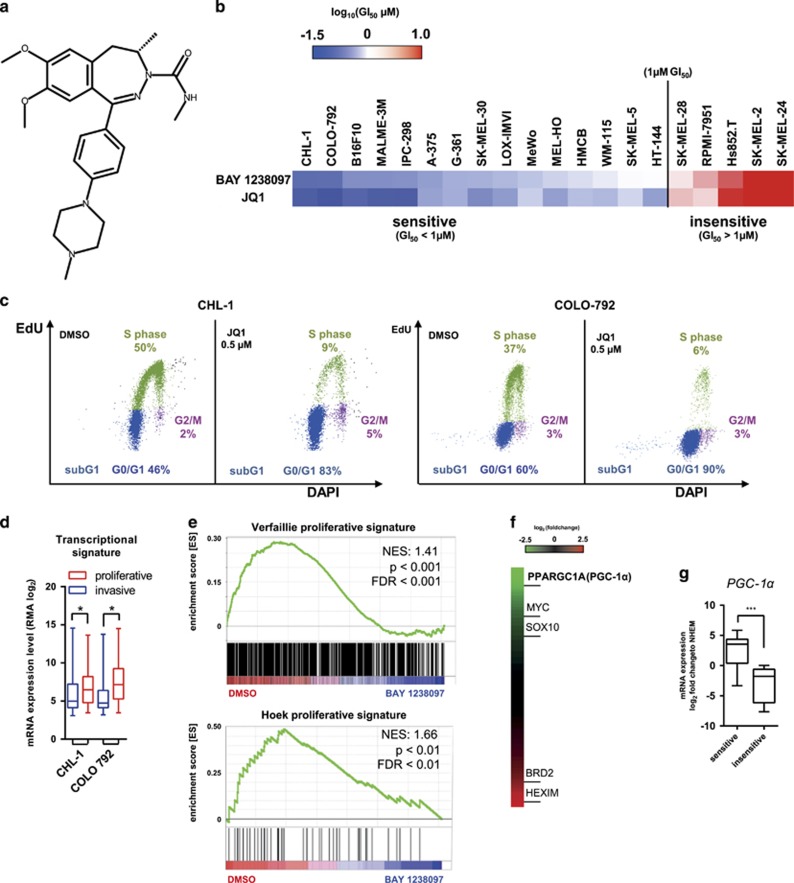
The newly identified BAY 1238097 (Figure 1a)20 has a similar BET bromodomain 1 (BD1) binding profile as the established tool compound JQ121 and a 3- to 5-fold weaker activity at BET bromodomain 2 (BD2; Supplementary Table 1a). The anti-proliferative activity of BAY 1238097 and JQ1 was determined in a panel of 20 melanoma cell lines. Treatment with either compound resulted in similar anti-proliferative effects, giving a broad range of GI50 values from below 100 nM to >10 μM (Figure 1b and Supplementary Table 1b). Representative growth inhibition curves are shown in Supplementary Figure 1a. Similarly, a colony formation assay performed with BAY 1238097 showed a strong inhibitory activity in CHL-1 cells, but far less so in SK-MEL-28 cells (Supplementary Figure 1b). No correlation was observed between the response to BET inhibitors and the mutation status of BRAF or NRAS, two oncogenes frequently altered in melanoma (Supplementary Table 1b). The BET family member BRD4 was shown to have key roles in maintaining the proliferative state of tumor cells,22 however its basal expression in melanoma cell lines varied only slightly (Supplementary Figure 1c), indicating that this was not determining sensitivity to BET inhibitors either. The impact of BET inhibition on the cell cycle was determined in the sensitive CHL-1 and COLO-792 models following 24 h treatment with 500 nM JQ1. For both cell lines a decrease of the S phase and an increase of the G0/G1 phase population was observed (Figure 1c), in accordance with previous findings in other solid tumors.21, 23
Figure 1.
Differential in vitro impact of BET inhibitors on melanoma models. (a) Chemical structure of BAY 1238097. (b) Color-coded ranking of GI50 values of BAY 1238097 and JQ1. (c) Cell cycle distribution following JQ1 treatment for 24 h. (d) Comparison of proliferative and invasive transcriptional signatures25 of CHL-1 and COLO-792 cells. Data are from GSE36133.26 Whiskers denote min to max. (e) Enrichment plot of proliferative signatures comparing the phenotypes of DMSO-treated control (n=4) and BAY 1238097-treated (n=4) CHL-1 cells. (f) Heatmap of differential gene expression following BAY 1238097 GI90 treatment of CHL-1 cells for 4 h. (g) Comparison of PGC-1α gene expression between sensitive (n=15) and insensitive cells (n=5) classified according to the data shown in b. Levels were normalized to expression in normal human epidermal melanocytes. Whiskers denote min to max. FDR, false discovery rate; NES, normalized enrichment score.
PGC-1α-expressing models are enriched in the BET inhibitor-sensitive melanoma subgroup
Gene expression profiles characteristic of metastatic melanomas have been identified.24 Also, two melanoma subgroups have recently been defined, based on proliferative vs invasive gene signatures.25 A comparison of basal gene expression levels (GSE36133)26 revealed that the vast majority of cell lines we tested, including the CHL-1 and COLO-792 models, had transcriptional signatures characteristic of the melanoma proliferative phenotype (Figure 1d and Supplementary Figure 1d). To quantify the effects of BET inhibitors on gene response, we performed global gene expression analysis of the CHL-1 cell line, which showed the highest sensitivity to BET inhibitors. Cells were analyzed after 4 h treatment with the GI90 concentration of BAY 1238097, using Affymetrix microarrays. Gene Set Enrichment Analysis (GSEA) revealed that among the genes downregulated after BAY 1238097 treatment (Supplementary Table 2), there was an overrepresentation of proliferative signature genes (Figure 1e). Genes with significantly reduced expression after BET inhibition included the transcription factors MYC (as previously reported)27 and SOX10, a recently identified master regulator of the proliferation phenotype25 (Figure 1f). Expression of HEXIM1 and BRD2 was markedly upregulated, confirming reports in previous studies (Figure 1f and Supplementary Table 2). Most surprisingly, expression of the metabolic regulator PGC-1α was strikingly repressed after 4 h of BET inhibitor treatment (Figure 1f). Interestingly, elevated expression of PGC-1α, which defines a subset of melanomas with a distinct metabolic profile, was associated with BET inhibitor sensitivity (Figure 1g). This was not the case for MYC, which has been linked to BET inhibitor response in other tumor models,23 nor for MITF, a melanoma oncogene, or for SOX10 expression (Supplementary Figure 1e).
PGC-1α expression is driven by a BRD4-bound super-enhancer
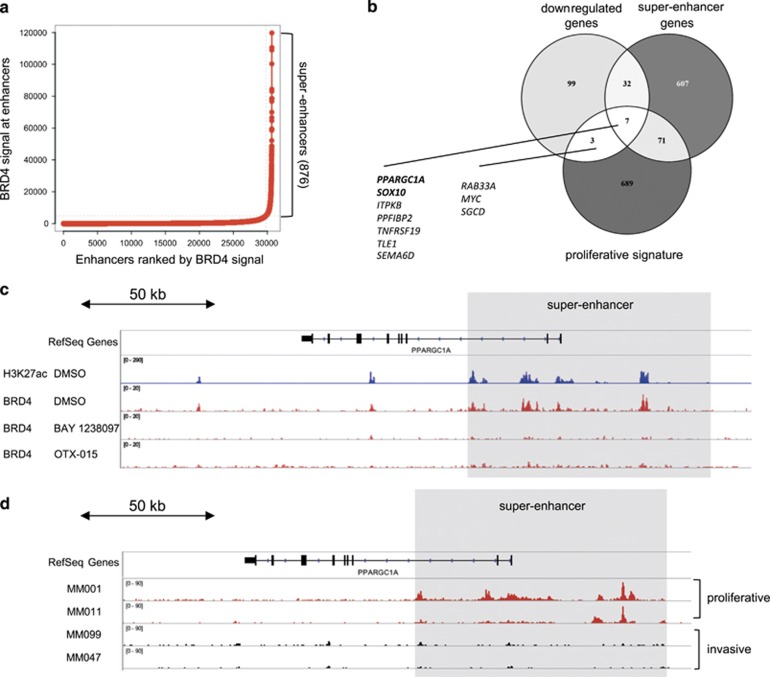
We next characterized the epigenetic profile of the BET inhibitor-sensitive CHL-1 cells to identify super-enhancers that can contribute to cell identity.28 High BRD4 binding densities, together with elevated H3K27 acetylation (H3K27ac) levels, have recently been identified as regulatory hallmarks of genes encoding cell type-specific transcription factors.28, 29 We performed chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) using antibodies against H3K27ac and BRD4. We found high BRD4 occupancy at H3K27ac-positive enhancers, which was reverted by the BET inhibitors BAY 1238097 and OTX-015, a BET inhibitor currently being evaluated in clinical trials (Supplementary Figure 2a).30, 31 Genome-wide analysis of putative enhancers and corresponding BRD4 signals allowed the identification of 876 super-enhancers in CHL-1 cells (Figure 2a). Of note, BRD4 occupancy at super-enhancers was recently suggested not to be solely predictive of gene-specific transcriptional activation.32 This led us to compare the overlap of CHL-1 super-enhancer assigned genes with the early response genes (significantly downregulated after 4 h of BET inhibition) and the melanoma proliferative gene signature. We confirmed that PGC-1α and SOX10 were strong candidates for BRD4-dependent genes (Figure 2b). A comparison of ChIP-seq profiles revealed a strong enrichment of H3K27ac peaks and BRD4 binding in the PGC-1α gene upstream and super-enhancer regions in untreated CHL-1 cells, but not after application of 500 nM BAY 1238097 or OTX-015 for 4 h (Figure 2c). BET inhibitor treatment was also accompanied by reduction of H3K27ac at the PGC-1α super-enhancer (Supplementary Figure 2b). Similarly, ChIP-seq profiles defined a BRD4-bound super-enhancer upstream of the SOX10 gene (Supplementary Figure 2c). Analysis of primary melanoma samples (GSE60666)25 revealed that the presence of the super-enhancer upstream of the SOX10 gene correlated with its expression, which defines the proliferative or invasive phenotype of the cell (Supplementary Figure 2d).25 Most importantly, the presence of a super-enhancer upstream of the PGC-1α gene was also predictive of its expression in a subset of proliferative, but not in invasive models (Figure 2d and Supplementary Figure 2e).
Figure 2.
BRD4-bound super-enhancers define gene regulatory regions of PGC-1α and SOX10 phenotype master regulators. (a) Dot plot showing putative enhancers ranked by normalized BRD4 signal (BRD4 signal—input signal in r.p.m./bp). (b) Venn diagram showing the overlap of downregulated, super-enhancer and proliferative signature genes. (c) ChIP-seq tracks of H3K27ac and BRD4 of DMSO- or BET inhibitor-treated CHL-1 cells (4 h, 1 μM) at the PGC-1α super-enhancer. Enrichment of signal is presented as log likelihood ratio (logLR) over background signal. (d) H3K27ac ChIP-seq tracks of proliferative or invasive primary melanoma cells at the PGC-1α super-enhancer (data from GSE60666).25
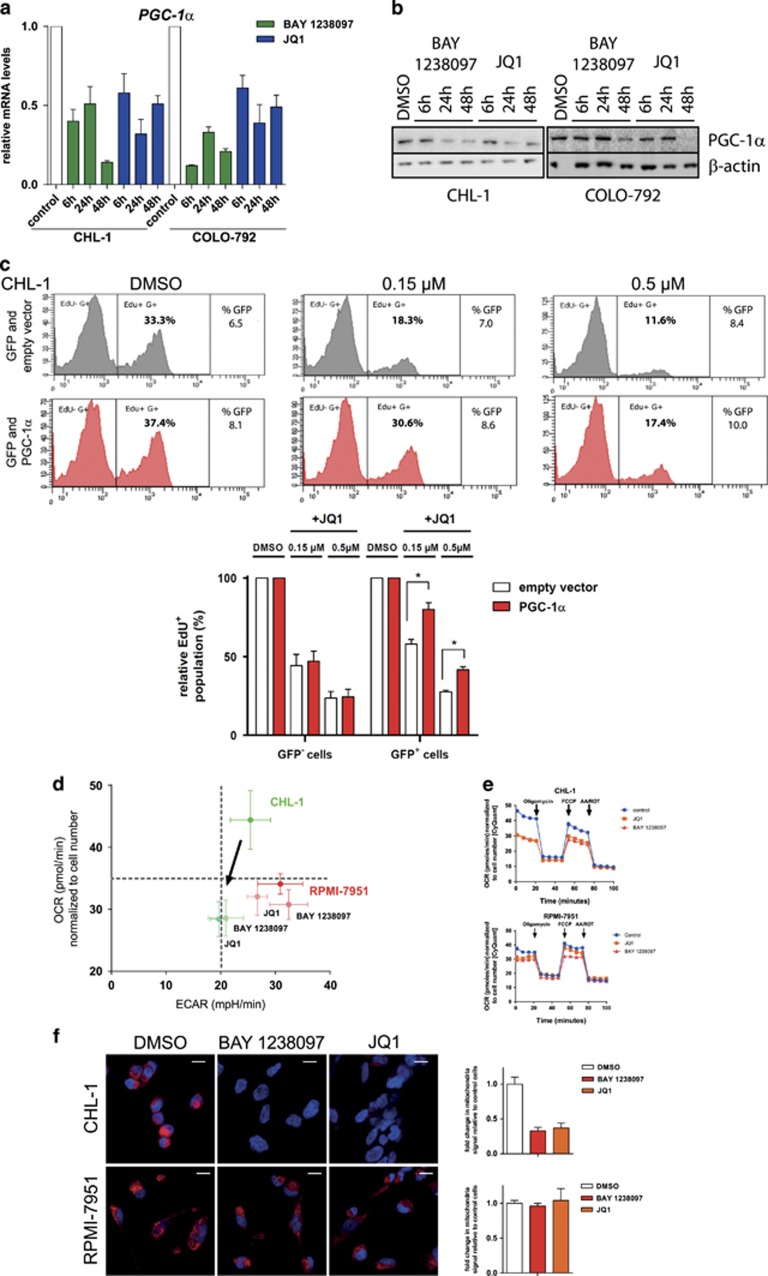
The impact of BET inhibition on PGC-1α expression was further confirmed by performing a time course study in the sensitive CHL-1 and COLO-792 melanoma models. PGC-1α expression was comparably downregulated by BAY 1238097 and JQ1 in both cell lines from 6 h onwards (Figure 3a). Also, a time-dependent reduction of PGC-1α protein was observed for both compounds (Figure 3b). Ectopic expression of PGC-1α partially rescued the anti-proliferative effects of JQ1, as evidenced by measuring the EdU+ cell population by flow cytometry in the CHL-1 and IPC-298 cell lines (Figure 3c and Supplementary Figure 3). The significant but partial rescue indicates that forced PGC-1α expression can indeed limit the cell response to BET inhibitors, but also that other factors are likely to be involved in the reduction of cell proliferation upon treatment.
Figure 3.
BET inhibitor-dependent downregulation of PGC-1α expression affects mitochondrial function, oxygen consumption rate and proliferation of OXPHOS melanoma cells. (a) Time-dependent response of PGC-1α gene expression to BET inhibition. After normalization to housekeeping genes, the control DMSO treatment group of each cell line was set to a value of 1. Gene expression at each time point was normalized to the control. Data are shown as the mean+s.e.m. of three replicates. (b) Time-dependent reduction of PGC-1α, as determined by western blot analysis. Neither protein was detectable at any time point in SK-MEL-2 or in RPMI-7951 cells, despite long exposure times (data not shown). Results are representative of triplicate experiments. (c) Overexpression rescue experiments. Transfected CHL-1 cells were treated with JQ1 for 24 h and subjected to 10 μM EdU 4–6 h prior to staining. The EdU-positive cell population was analyzed by flow cytometry. Representative results from a single experiment are shown as well as the mean values obtained in three independent experiments. *P<0.05% in t-test. Numerical results were normalized to the DMSO-treated control sample. (d) Determination of OCR and ECAR (an indirect measure of glycolysis) using a Seahorse analyzer to visualize the energy phenotype profile following BET inhibitor treatment (1 μM for 20 h). Data are represented as mean±s.d. of three separate experiments, each measured six times. (e) Determination of OCR using a Seahorse analyzer following consecutive injections of oligomycin (1 μM), FCCP (0.5 μM) and antimycin A (1 μM)/rotenone (1 μM). The plots are representative of two independent experiments, each measured six times. (f) Microscopy analysis and quantification of mitochondrial function. Red signal indicates Mitotracker Deep Red dye staining of functional mitochondria, while nuclei (blue) are visualized following DAPI staining. Images shown are representative of two independent replicates. The scale bar indicates 20 μm. Data are represented as the mean Mitotracker signal compared to DMSO-treated cells, +s.e.m. ECAR, extracellular acidification rate; OCR, oxygen consumption rate.
BET inhibition reduces mitochondrial function
In poorly metastatic melanoma models, knockdown of PGC-1α leads to upregulation of the WNT, TGF-β and integrin pathways, which are all implicated in metastasis.33 Importantly, this was not observed in proliferative cells after pharmacological BET inhibition (Supplementary Figure 4a). On the other hand PGC-1α is essential for mitochondrial function and thereby influences the metabolic and energetic states of melanoma tumors.11 It promotes mitochondrial respiration and resistance to oxidative stress in the predominantly OXPHOS subset of melanoma,11 whereas the subset that relies primarily on glycolysis for energy production generally does not express PGC-1α.10 In view of the dramatic effects of BET inhibition on PGC-1α expression, we determined whether changes in the cellular energy phenotype ensued. Indeed, a shift towards reduced basal oxygen consumption rate was observed for the sensitive models CHL-1 and G-361, but not for the insensitive models RPMI-7951 and SK-MEL-2 (Figure 3d and Supplementary Figure 4b). Further investigation using the mitochondrial uncoupler carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP) showed that BET inhibition also resulted in diminished maximal respiratory capacity of the mitochondria in sensitive, but not in insensitive cells (Figure 3e and Supplementary Figure 4c).
Mitochondrial function was then examined after 3 days of treatment using the Mitotracker Deep Red dye for detection. Significant signal loss was observed in the BET inhibitor-sensitive CHL-1 and G-361 cells, but there was only a trend in COLO-792 cells. Conversely, no signal loss was seen in the insensitive RPMI-7951 or SK-MEL-2 cells (Figure 3f and Supplementary Figure 4d). However, the CHL-1 cells, which lost around two-thirds of the mitotracker signal, maintained staining with a specific mitochondrial outer membrane antibody, confirming that mitochondria were still present in the presence of inhibitor (Supplementary Figure 4e).
BAY 1238097 shows in vivo efficacy in melanoma models with high PGC-1α levels
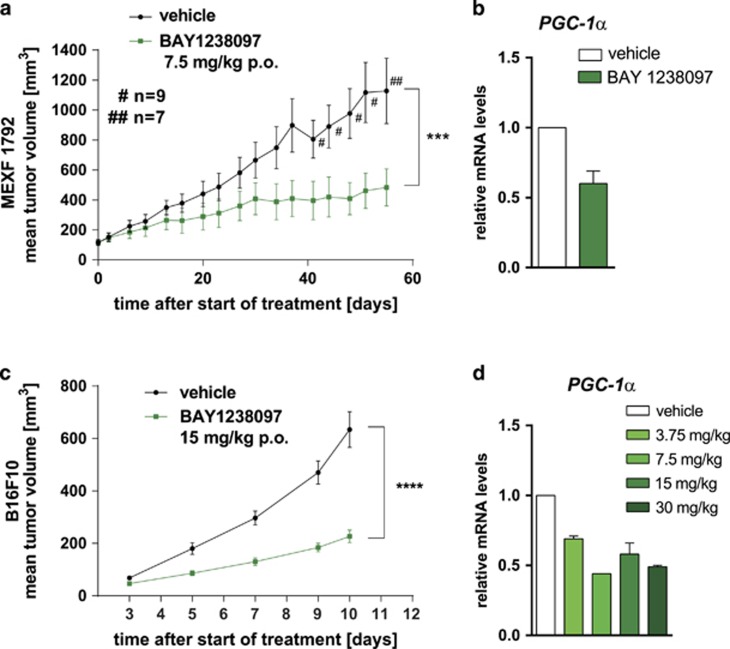
Pharmacokinetic profiling of BAY 1238097 revealed a low clearance and high oral bioavailability (Supplementary Figures 5a and b), thereby supporting oral dosing for in vivo evaluation. We first selected a patient-derived xenograft (PDX) melanoma model with elevated PGC-1α expression (Supplementary Figure 6a). This model, MEXF 1792, was transplanted onto mice, which were then treated daily, orally with 7.5 mg/kg BAY 1238097 (maximal tolerated dose, MTD). Significantly reduced tumor growth was observed (35% treated/control (T/C) tumor volume, P=0.03) on day 51 post-start of treatment (Figure 4a). A marked reduction of PGC-1α in the tumor tissue was confirmed 3 h post treatment (Figure 4b). We tested an additional PDX model, MEXF 1341, with low PGC-1α expression (Supplementary Figure 6a), using the same treatment schedule. Here, no biologically significant anti-tumor activity was observed (Supplementary Figure 6b).
Figure 4.
PDX and syngeneic mouse melanoma models exhibit sensitivity to BAY 1238097 related to downregulation of PGC-1α expression. (a) Mean tumor volume (±s.e.m.) of PDX model MEXF 1792 during treatment with vehicle or BAY 1238097 (7.5 mg/kg p.o.). Vehicle-treated mice with high tumor burden were killed before the final day of the study, as shown by the ‘n’ value over the vehicle group curve. (b) PGC-1α gene expression in MEXF 1792 samples from a, reported as the mean+s.e.m. relative to the vehicle control group (treated samples n=9, control samples n=7). Tumor samples were harvested 6 h post treatment on day 56. (c) B16F10 mean tumor volume (±s.e.m.) during treatment with vehicle or BAY 1238097 (15 mg/kg p.o.). (d) Dose-dependent reduction of PGC-1α gene expression in BAY 1238097-treated B16F10 tumors, 3 h post treatment. Gene expression results are shown relative to the vehicle-treated control group as the mean+s.e.m. of three tumor samples per dose.
The syngeneic PGC-1α-expressing B16F10 mouse melanoma model was then evaluated, due to its strong response to BET inhibition in cell culture (Figure 1b). Here also, daily oral treatment with 15 mg/kg BAY 1238097 (maximal tolerated dose) led to a significant reduction of tumor growth (36% T/C tumor volume on day 10 post start of treatment P<0.001; Figure 4c). In a separate experiment, up to two-fold downregulation of PGC-1α gene expression was observed with increasing doses of the inhibitor, when measured 6 h post application (Figure 4d). Altogether these in vivo results further underscore the strong activity of BET inhibitors in PGC-1α-positive melanoma models.
Discussion
Here we provide evidence that elevated PGC-1α expression in melanoma correlates to increased sensitivity to BET inhibitor treatment. Extensive characterization of the responsive models with regard to their epigenetic and bioenergetic status revealed a mechanism by which BRD4-bound super-enhancer elements drove the expression of PGC-1α. Pharmacological inhibition of BRD4 subsequently led to loss of BRD4 from these epigenetic regulatory elements, decreased the expression of PGC-1α, and ultimately resulted in reduced mitochondrial function together with cell cycle arrest. These results were further supported by in vivo studies in PDX and syngeneic models of melanoma.
Initial studies with I-BET151 showed that BET inhibition blocked melanoma proliferation and induced apoptosis at high compound concentration.16, 18 We show here that submicromolar BET inhibitor treatment impaired melanoma proliferation preferentially in models expressing PGC-1α. This cofactor interacts with chromatin remodeling and histone acetyltransferase complexes,34 and is a master regulator of mitochondria biogenesis.35 We now provide evidence for regulation of cellular bioenergetics dependent on the epigenetic reader BRD4 in melanoma. The transcription factor MITF is overexpressed in melanoma19, 36 and induces PGC-1α expression, thus leading to a shift towards OXPHOS energy production and increased survival of BRAF-driven melanoma.10 However, we found that neither MITF expression nor BRAF mutation status were associated with BET inhibitor sensitivity (Supplementary Figure 1e and Supplementary Table 1b).
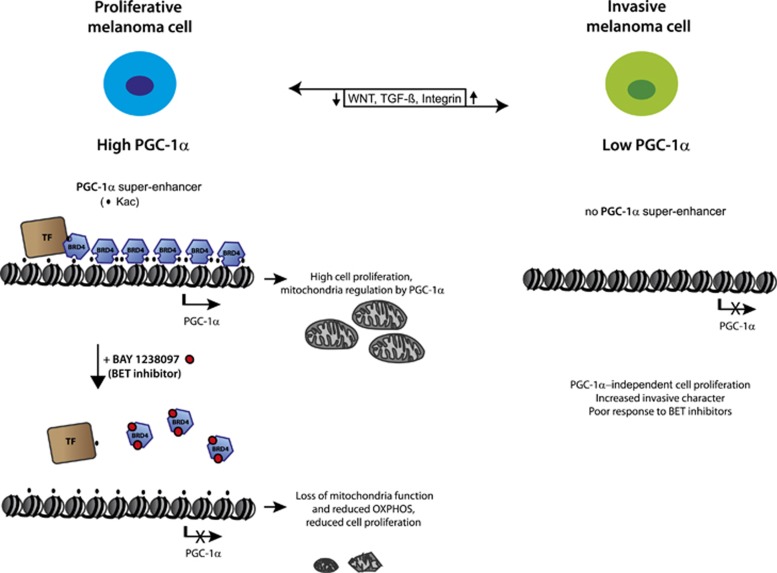
We propose a model where treatment with a BET inhibitor obviates BRD4 interaction with acetylated histones at super-enhancer regions, thus leading to the shutting-off of specific genes encoding factors with an essential role in melanoma proliferation such as PGC-1α and SOX10 (Figure 5). This is followed by loss of mitochondrial function, reduced oxygen consumption and presumably oxidative damage, leading to inhibition of tumor cell proliferation. While this is not the only pathway influenced by BET inhibitors, our results suggest that a strong link exists between metabolic response and the epigenetic reader BRD4 in melanoma.
Figure 5.
Model of BRD4-bound super-enhancer defining a ‘PGC-1α high’ subgroup of proliferative melanoma cells. PGC-1α suppresses the metastatic character of melanoma cells (PGC-1α high vs PGC-1α low), while promoting mitochondria biogenesis and protecting from oxidative stress. Pharmacological blockade of PGC-1α expression by BET inhibition leads to downstream loss of mitochondrial function and reduced OXPHOS, ultimately resulting in impaired cell proliferation. This does not induce WNT, TGF-β and integrin pathway activity,33 which could otherwise promote a switch from a proliferative to an invasive phenotype.
The sensitivity of the OXPHOS subgroup to BET inhibition may be a unique feature linked to the BRD4-dependent epigenetic regulation of the metabolic state in melanoma cancer. A classification of diffuse large B-cell lymphoma based on metabolic signatures has been proposed,37 but we did not observe an increased response of the OXPHOS subset to BET inhibition (unpublished data). Altogether, these findings further support the essential roles of epigenetics and metabolism crosstalk in various diseases and tumor types.
The present data add an important novel facet to the role of BRD4-dependent super-enhancers in driving master regulators of the bioenergetic pathways and thereby define a subgroup of melanoma cells. Altogether these findings suggest a novel strategy to target metabolic dependencies of melanoma by pharmacological inhibition of BRD4 in PGC-1α-expressing melanoma. Further clinical studies with BET inhibitors12 should help clarify whether this can indeed be translated to the benefit of melanoma patients.
Materials and Methods
Reagents, plasmids and cell lines
The synthesis of BAY 1238097 is described in patent application WO 2014/026997 as example 127. JQ1 and OTX-015 were synthesized in-house. Plasmids were constructed using standard methods (LGC Genomics GmbH, Berlin, Germany). The FLAG-tagged PGC-1α plasmid was constructed by Life Technologies (Ober-Olm, Germany) in the pcDNA 3.1 backbone. Melanoma cell lines were from the American Type Culture Collection (ATCC, Manassas, VA, USA) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany), and were routinely grown in the recommended medium in the presence of 10% (v/v) fetal calf serum (Life Technologies) at 37 °C and in 5% CO2 atmosphere. Authentication was performed by the DSMZ using short tandem repeat DNA typing analysis. Cell lines were confirmed to be free of mycoplasma (MycoAlert Mycoplasma Detection Assay, Lonza, Cologne, Germany).
BET binding profile
Competition binding assays were performed at DiscoverX (Fremont, CA, USA) with purified DNA-tagged BET bromodomains and a universal, immobilized ligand. Details are available under https://www.discoverx.com/technologies-platforms/competitive-binding-technology/bromoscan-technology-platform.
Cell viability and colony formation assays
The cells were seeded in triplicate in 96-well plates. For the viability assay, they were plated at a density of 3000–5000 cells/well and incubated in culture medium. The next day the culture medium was exchanged to assay medium containing various compound concentrations. Cell viability was measured during the exponential growth phase 3–6 days later using the Alamar Blue assay (Thermo Fisher Scientific, Pinneberg, Germany). Mean GI50 values (Supplementary Table 1b) were calculated from duplicate or triplicate experiments with GraphPad Prism (GraphPad Software, San Diego, CA, USA). For the colony formation assay, 7500–10 000 cells in their respective medium were diluted 1–8.3 in soft agar and then added to wells pre-coated with 0.6% soft agar. Once the mixture had solidified at room temperature, 50 μl growth medium was added. The medium was exchanged to assay medium containing various compound concentrations the following day. Colony formation was measured after 7 days using the CellTiter 96 Aqueous assay system (Promega, Mannheim, Germany).
Gene expression analysis
Cells were grown to 80% confluence in six-well culture dishes. RNA was purified after cell lysis using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) before preparing complimentary DNA using the Superscript III reagents (Thermo Fisher Scientific) or the RT2 First Strand kit (Qiagen). Samples were then analyzed by quantitative PCR, either with TaqMan probes (Life Technologies, BRD4, Hs04188087_m1; PGC-1α, Hs01016719_m1; HPRT1, 4326321E) or RT2 Custom Profiler Arrays (Qiagen, BRD4, PPH15694; PGC-1α, PPH00461; HPRT1, PPH01018; RPLP0, PPH21138; B2M, PPH01094). Ct values and fold-changes were determined from triplicate experiments using standard protocols. Relative quantification was performed after normalization to housekeeping genes.
For time course assays, CHL-1 and COLO-792 cells were treated with BAY 1238097 (1 μM) or JQ1 (1 μM) prior to harvest. For gene expression analysis of tumor samples, slices were homogenized and total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen). Samples were analyzed using the TaqMan probes above, except for the murine B16F10 samples, which were analyzed with mouse-specific TaqMan probes: PGC-1α, Mm01208835_m1; HPRT1, Mm03024075_m1. Gene expression profiles of PDX models were determined by Affymetrix Inc. Technology (Lahr, Germany).
Western blot analysis
Cells were grown to 80% confluence in six-well culture dishes, and lysed in 100 μl RIPA lysis buffer supplemented with complete protease inhibitor cocktail (Roche, Mannheim, Germany). Lysates were clarified by centrifugation (13 200 g, 15 min, 4 °C). The supernatant was transferred to a new tube, and protein levels were quantitated with the BCA method (Thermo Fisher Scientific). About 30–60 μg total protein was analyzed using SDS–PAGE (Nu-PAGE 4–12% Bis-Tris protein gels) and western blotting with anti-PGC-1α (ST1202, Millipore, Darmstadt, Germany) and anti-β-actin (Sigma, St Louis, MO, USA). The anti-FLAG M2 monoclonal antibody F3165-5MG (Sigma) was used to monitor protein levels after transfection. A goat-anti-mouse (IRDye800CW, LI-COR Biosciences, Bad Homburg, Germany) or goat-anti-rabbit (IRDye680LT, LI-COR Biosciences) secondary antibody was used for detection with the help of an Odyssey Fc system (LI-COR Biosciences).
Microscopy
For detection of functional mitochondria, cells were seeded into eight-well chamber slides (Millipore) at 2000–4000 cells/well in 500 μl media and incubated overnight at 37 °C and 5% CO2. The cells were treated with BAY 1238097 (1 μM) or JQ1 (1 μM) for 72 h. Mitotracker Deep Red (100 nM, Invitrogen, Karlsruhe, Germany) was added for 20 min before fixing with 3.7% formaldehyde for 15 min at 37 °C. Fixed cells were washed with phosphate-buffered saline (PBS) and permeabilized with cold acetone for 5 min. After washing four times with PBS, the slides were incubated with 300 nM DAPI for 5 min. The cells were then washed and sealed with coverslips and Vectashield mounting medium (Vector Labs, Burlingame, CA, USA). Mitotracker and DAPI staining were visualized using an LSM700 confocal microscope (Zeiss, Berlin, Germany), with a × 20 objective utilizing separate 405 and 647 nm laser excitation wavelengths. Quantitation was performed using ImageJ software (National Institute of Health, Bethesda, MD, USA). Total mitochondria staining intensity in a specified area was divided by the total count of nuclei within the same area. Data are represented as the mean signal in two replicates relative to DMSO-treated cells, ±s.e.m., with 90–200 cells quantitated per treatment group.
To evaluate the presence of mitochondria, CHL-1 cells were seeded and treated as above. Cells were incubated with 3.7% formaldehyde in PBS for 15 min, washed with cold PBS and permeabilized with 0.5% Triton X-100 in PBS for 10 min. After washing three times with PBS, cells were blocked in 1% BSA in PBS with 0.02% Tween 20 (BSA–PBST) for 30 min before incubating them with anti-TOMM 20 antibody (ab56783, Abcam, Cambridge, UK) in BSA–PBST for 1 h. They were washed three times with PBST, followed by incubation with Alexa Fluor 555-conjugated anti-mouse secondary antibody (Invitrogen) for 1 h. After three washes with PBST, slides were prepared and visualized as above, utilizing separate 405 and 555 nm laser excitation wavelengths.
Cellular bioenergetics analysis
Extracellular flux analyses were performed using the Seahorse XF96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). Tissue culture plates were seeded at 20 000–30 000 cells/well. When fully attached, the cells were treated with BAY 1238097 (1 μM) or JQ1 (1 μM). After 20 h, mitochondrial function and mitochondrial stress in response to BAY 1238097 were determined using the XF Mito Stress Test Kit (Seahorse Bioscience), which measures key parameters of mitochondrial function including basal respiration, ATP production and respiratory capacity. To this end, cell medium was changed for non-buffered Dulbecco’s modified Eagle’s medium (DMEM) containing 8.3 g/l DMEM (Sigma), 2 mM GlutaMAX (Invitrogen), 5 mM glucose (Sigma), 63.3 mM NaCl (Sigma) adjusted to pH 7.4 with NaOH. The drugs were injected at a final concentration of 1 μM oligomycin, 0.5 μM FCCP, 1 μM rotenone and 1 μM antimycin A. All treatment conditions were analyzed with six replicates. oxygen consumption rate and ECAR values were normalized to cell numbers, which were determined following staining with Cyquant (Life Technologies).
Chromatin immunoprecipitation
CHL-1 cells were treated for 4 h with 500 nM BAY 1238097 or OTX-015 prior to cross-linking and harvest. One 15 cm cell culture dish with 80% confluent cells was used per immunoprecipitation. ChIP DNA preparation for sequencing was carried out as described previously38 with the following changes: cross-linked chromatin was sheared to 200–800 base pair fragments using sonication (30 s on/30 s off, high power, 2 × 7.5 min with a water bath change between the two cycles) in a Bioruptor sonicator UCD-200 (Diagenode, Liège, Belgium). About 1.5 μg antibodies (anti-BRD4 (A301-985A100, Bethyl Labs, Montgomery, TX, USA); anti-H3K27ac (Diagenode, C15410174)) were used per immunoprecipitation. DNA was purified using a PCR purification kit (Qiagen), followed by library preparation and genome-wide sequencing. In brief, samples were analyzed on a 2100 bioanalyzer using the DNA high sensitivity assay. Concentration was determined using a High Sensitivity DNA Kit. End repair and phosphorylation, and ligation of TrueSeq index adapter and amplification before subjection to sequencing (HiSeq Sequencing System) were done according to the TrueSeq ChIP Sample Preparation Guide (Illumina, Munich, Germany). Sequencing data are available in the GEO database with the accession number GSE95585.
Fluorescence-activated cell sorting and rescue analysis
CHL-1 or IPC-298 cells were seeded 24 h prior to transfection. Plasmid DNA was transfected using Lipofectamine LTX with Plus reagent (Life Technologies). Cells were co-transfected using a human FLAG-tagged PGC-1α or empty vector, and TagGFP vector (pTagGFP-C, Xia 2000, Evrogen, Heidelberg, Germany). Transfection conditions were optimized by monitoring fluorescence by microscopic analysis and FLAG-tagged PGC-1α levels by western blot analysis. For the flow cytometry analysis, transfected cells were treated with JQ1 for 24 h and stained using the EdU-staining kit (Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit). Cells were subjected to 10 μM EdU 4–6 h before staining, washed with cold PBS, detached using Trypsin/EDTA solution (Biochrom, Berlin, Germany), re-suspended and filtered through a 70-μm mesh (Falcon, Sigma-Aldrich, Taufkirchen, Germany). The EdU+ population was determined using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Life Technologies) and stained with DAPI at 1 μg/ml final concentration for 1 h at 4 °C in the dark, before performing flow cytometry analysis (FACS Canto 2, BD Biosciences, Heidelberg, Germany).
Gene expression profiling of CHL-1 melanoma cells
Cells were treated with GI50 or GI90 concentrations of BAY 1238079 before RNA extraction and profiling with Affymetrix HuGene-2.1ST arrays.23 Data are available in the GEO database under GSE92736. Probe set intensities were condensed to meta-probe set levels in Genedata Expressionist 9.0 using robust multi-array (RMA) algorithm followed by LOWESS normalization. A 2-group t-test was used to compare the treatment and control groups. The resulting P-values were corrected for multiple testing using the Benjamini–Hochberg correction (BH-q). Genes with a BH-q value <0.05 and a log2 expression fold change of ±0.5 were considered to be significantly altered by the treatment.
Bioinformatics
Microarray analysis and GSEA were carried out as described.23 Basal expression levels of genes from the proliferative and invasive melanoma signatures were extracted from GSE36133.26 Gene signatures of the proliferative and invasive state were derived from unsupervised clustering of clinical melanoma samples and showed a significant overlap with signatures identified previously in vitro.24 Signature gene expression levels for each cell line are shown as box-and-whisker plot in Supplementary Figure 1d.
Microarray expression profiling of BAY 1238097-treated CHL-1 cells was used to analyze the enrichment of the two proliferative gene signatures.24, 25 The resulting normalized enrichment score, adjusted P-value and false discovery rate were calculated by the GSEA software.39 Ranking metrics were generated by differences between median log2 normalized expression phenotypes (DMSO (n=4) vs treatment with IC90 concentration of BAY 1238097 for 4 h (n=5)). 1000 gene set permutations were used. The microarray data were deposited under GSE92736. A detailed description of the GSEA methodology and interpretation is provided at http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html.
For processing of the ChIP-seq data, paired-end genome-wide sequencing data were aligned to the human reference genome (assembly hg19) using Bowtie2 software,40 and sorted and indexed using SAMtools.41 Peak calling was done using the model-based analysis for ChIP-seq (MACS2) software. Broad peaks from H3K27ac were called using MACS2 broad mode. MACS2 was used to generate enrichment pileups (log likelihood ratio (logLR)) of treatment vs input control samples. A pseudocount of 0.00001 was added to avoid log10(0) during logLR calculation. ChIP-seq tracks were generated using the Integrative Genomics Viewer software.42, 43 Heatmaps and line plots were generated using the seqplots software (http://przemol.github.io/seqplots). The ChIP-seq data were deposited under GSE95585.
The rank ordering of super-enhancers (ROSE) script28, 44 was used to generate stitched enhancers and to separate super-enhancers from typical enhancers. Putative enhancers of close proximity (no further than 12.5 kb from one another) were stitched. Active putative enhancers were identified using the H3K27ac data set. The 2.5 kb region around the transcriptional start site of each gene was excluded from the calculation. BRD4 read density was normalized based on read density in the input control and sorted by signal. This resulted in the generation of two groups of enhancers, namely super-enhancers and typical enhancers. Super-enhancers were assigned to their closest coding gene using PeakAnalyzer v1.4.45 Gene annotation was done using Homo_sapiens.GRCH37.64 as reference. This method has been shown to be accurate for super-enhancer assignment.28
Animal studies
All in vivo experiments were discussed and approved by the internal animal welfare committee of the company, which included an ethical evaluation of the proposed procedures. Official permission was granted by the Governmental Animal Care and Use Office (Landesamt für Gesundheit und Soziales, Berlin, Germany). The studies on the PDX model were performed at Oncotest GmbH (Freiburg, Germany) and performed with patient consent. All experiments and protocols were approved by the Oncotest animal welfare body and the local authorities including the Governmental Animal Care and Use Office, and conducted according to all applicable international, national and local laws and guidelines. The PDX models MEXF 1792 and MEXF 1341 were derived from surgical specimens of melanoma patients and maintained in mice by serial passage. After removal from donor mice, tumors were cut into fragments (4–5 mm diameter) and implanted subcutaneously into the flank of NMRI nu/nu female mice (aged 5–7 weeks, Harlan Laboratories, Ittingen, Switzerland). Tumors were allowed to grow and treatment was initiated in animals distributed into groups, so that each randomized group contained a comparable group mean and median tumor volume of ~100–120 mm3 (n=10). The syngeneic mouse melanoma model B16F10 was inoculated in 0.1 ml DMEM/Ham’s F12 medium (0.5 × 106 cells) in C57BL/6 female mice (aged 10 weeks, Charles River, Worcester, MA, USA, n=12/group), and treatment was initiated on day 3 post tumor inoculation. No statistical methods were used to define the sample size of the in vivo studies. Depending on the studies, BAY 1238097 was dissolved in 0.9% NaCl in water, pH 4 or in 30% HPβ-CD in 0.2% NaCl in water, pH 7. It was administered orally, daily (10 ml/kg) at its maximal tolerated dose (7.5 or 15 mg/kg/day). In all studies, body weight was monitored as a measure for treatment-related toxicity. Tumors were measured by caliper at regular intervals by investigators who were not blinded to the treatment groups, and tumor volume was calculated according to the formula: (length × width × width)/2. The ratio treated/control tumor size was calculated at the end of the study according to the formula: %T/C=(mean tumor volume or weight of treatment group/mean tumor volume or weight of vehicle control group) × 100. For the studies in patient-derived models where treatment was started in mice bearing advanced tumors, individual relative tumor volumes were used to calculate the mean tumor volume, according to the following formula: relative tumor volume=(tumor volume on day of calculation/ tumor volume on day of start of treatment) × 100. Biologically significant activity was declared for %T/C<42% in agreement with the National Cancer Institute (NCI) criteria. All animals were included in the analysis, although some animals in the vehicle control groups were killed due to large tumor size before the final day of the study. In addition, tumor samples were collected for gene expression analysis at the end of the studies.
In the B16F10 study, where the effect of BAY 1238097 on tumor PGC-1α expression was evaluated in a dose response, animals bearing tumors of about 100 mm3 were distributed into five different groups (n=3 per group). BAY 1238097 (3.75, 7.5, 15 or 30 mg/kg) or vehicle was applied orally once and tumors were sampled 6 h post treatment.
Due to ethical considerations, no studies were initiated in mice using cell lines that showed a poor response to BET inhibitors in cell culture.
Statistics
GraphPad Prism software was used for statistical analysis. A two-tailed t-test was performed to compare between two groups (two-group comparisons). In the cases where the variance was found to be unequal between the two groups, a Welch’s corrected unpaired t-test was applied. For these instances, the P-value remained ⩾0.05 with or without the correction. For the comparison of more than two groups, one-way Analysis of variance followed by Sidak’s multiple comparison test (pairwise comparisons) was used. For the in vivo studies, tumor weights were Log-transformed and a two-tailed t-test was performed between the BAY 1238097-treated group and the vehicle group. P<0.05 (*) was considered to indicate a statistically significant difference, P<0.0001 is reported as ****P 0.0001—0.001 is reported as ***P 0.001—0.01 is reported as **P 0.01—0.05 is reported as *P⩾0.05 is reported as (not significant) ‘NS’.
Acknowledgments
We thank Oncotest GmbH (Freiburg im Breisgau) for performing the in vivo studies with the PDX models. We are thankful to S Johnsen’s laboratory at the University of Göttingen for helpful ChIP-seq advice. We are indebted to H Weinmann and V Gekeler for their support. The dedicated help of N Dittmar, M Runge, T Sugawara, H Muckwar, F Knoth, K Reichert and B Hartmann is gratefully acknowledged.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
All authors are or were employees of Bayer AG.
Supplementary Material
References
- Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol 2014; 810: 120–140. [DOI] [PubMed] [Google Scholar]
- Lo JA, Fisher DE. The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science 2014; 346: 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP et al. A landscape of driver mutations in melanoma. Cell 2012; 150: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niezgoda A, Niezgoda P, Czajkowski R. Novel approaches to treatment of advanced melanoma: a review on targeted therapy and immunotherapy. Biomed Res Int 2015; 2015: 851387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. BRAF inhibitors: the current and the future. Curr Opin Pharmacol 2015; 23: 68–73. [DOI] [PubMed] [Google Scholar]
- Abildgaard C, Guldberg P. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol Med 2015; 21: 164–171. [DOI] [PubMed] [Google Scholar]
- Shakhova O, Cheng P, Mishra PJ, Zingg D, Schaefer SM, Debbache J et al. Antagonistic cross-regulation between Sox9 and Sox10 controls an anti-tumorigenic program in melanoma. PLoS Genet 2015; 11: e1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, de Moura MB, Lin Y, Vincent G, Thorne S, Duncan LM et al. Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer 2012; 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöckel L, Glasauer A, Basit F, Bitschar K, Truong H, Erdmann G et al. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer Metab 2015; 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 2013; 23: 302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013; 23: 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelato KA, Shaikhibrahim Z, Ocker M, Haendler B. Targeting epigenetic regulators for cancer therapy: modulation of bromodomain proteins, methyltransferases, demethylases, and microRNAs. Expert Opin Ther Targets 2016; 20: 783–799. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17: 330–339. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B. Targeting BET bromodomains for cancer treatment. Epigenomics 2015; 7: 487–501. [DOI] [PubMed] [Google Scholar]
- Gallagher SJ, Mijatov B, Gunatilake D, Tiffen JC, Gowrishankar K, Jin L et al. The epigenetic regulator I-BET151 induces BIM-dependent apoptosis and cell cycle arrest of human melanoma cells. J Invest Dermatol 2014; 134: 2795–2805. [DOI] [PubMed] [Google Scholar]
- Heinemann A, Cullinane C, De Paoli-Iseppi R, Wilmott JS, Gunatilake D, Madore J et al. Combining BET and HDAC inhibitors synergistically induces apoptosis of melanoma and suppresses AKT and YAP signaling. Oncotarget 2015; 6: 21507–21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SJ, Mijatov B, Gunatilake D, Gowrishankar K, Tiffen J, James W et al. Control of NF-kB activity in human melanoma by bromodomain and extra-terminal protein inhibitor I-BET151. Pigment Cell Melanoma Res 2014; 27: 1126–1137. [DOI] [PubMed] [Google Scholar]
- Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res 2013; 73: 6264–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi E, Gaudio E, Lejeune P, Tarantelli C, Cascione L, Kwee I et al. Preclinical evaluation of the BET bromodomain inhibitor BAY 1238097 for the treatment of lymphoma. Br J Haematol 2017; 178: 936–948. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O et al. Selective inhibition of BET bromodomains. Nature 2010; 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem 2008; 283: 9040–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil O, Lesche R, Gelato KA, Haendler B, Lejeune P. Inhibition of BET bromodomain-dependent XIAP and FLIP expression sensitizes KRAS-mutated NSCLC to pro-apoptotic agents. Cell Death Dis 2016; 7: e2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res 2006; 19: 290–302. [DOI] [PubMed] [Google Scholar]
- Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G et al. Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun 2015; 6: 6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011; 146: 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 2015; 58: 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 2016; 3: e196–e204. [DOI] [PubMed] [Google Scholar]
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol 2016; 3: e186–e195. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep 2016; 15: 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Lim JH, Lee Y, Granter SR, Thomas A, Vazquez F et al. A PGC1alpha-mediated transcriptional axis suppresses melanoma metastasis. Nature 2016; 537: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell 2003; 12: 1137–1149. [DOI] [PubMed] [Google Scholar]
- Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J 2015; 282: 647–672. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005; 436: 117–122. [DOI] [PubMed] [Google Scholar]
- Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012; 22: 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 2014; 8: 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013; 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 2010; 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.