Abstract
Long-range chromatin interactions between enhancers and promoters are essential for transcription of many developmentally controlled genes in mammals and other metazoans. Currently, the exact mechanisms that connect distal enhancers to their specific target promoters remain to be fully elucidated. Here, we show that the enhancer-specific histone H3 lysine 4 monomethylation (H3K4me1) and the histone methyltransferases MLL3 and MLL4 (MLL3/4) play an active role in this process. We demonstrate that in differentiating mouse embryonic stem cells, MLL3/4-dependent deposition of H3K4me1 at enhancers correlates with increased levels of chromatin interactions, whereas loss of this histone modification leads to reduced levels of chromatin interactions and defects in gene activation during differentiation. H3K4me1 facilitates recruitment of the Cohesin complex, a known regulator of chromatin organization, to chromatin in vitro and in vivo, providing a potential mechanism for MLL3/4 to promote chromatin interactions between enhancers and promoters. Taken together, our results support a role for MLL3/4-dependent H3K4me1 in orchestrating long-range chromatin interactions at enhancers in mammalian cells.
Keywords: enhancer, H3K4me1, MLL3, MLL4, chromatin loop
Introduction
Enhancers play a critical role in regulating spatiotemporal gene expression programs in animals1,2,3,4,5. These cis-regulatory elements can regulate target gene transcription in specific cell lineages and developmental stages by recruiting sequence-specific transcription factors (TFs)2,6 and chromatin remodeling complexes1,3,7,8,9,10. Active enhancers exhibit characteristic histone modifications such as H3K4me1 and H3K27ac, DNase I hypersensitivity, occupancy by H3.3 and H2A.Z histone variants, and production of short-lived RNA known as eRNAs11,12,13,14,15,16,17. Based on these biochemical features, millions of candidate enhancers have been annotated in the human genome18,19.
Lineage-specific enhancers undergo step-wise activation during development, beginning with the binding of sequence-specific TFs, which recruit various chromatin-remodeling complexes to promote chromatin modification and nucleosome dynamics. The chromatin modification H3K4me1 is a hallmark of the initial stage of enhancer activation, while the appearance of active chromatin marks such as H3K27ac defines an active state12,14,20,21,22. Mll3 and Mll4 encode the histone H3 lysine 4 (H3K4) monomethyltransferases with partially redundant functions23,24,25. Recruitment of MLL3 and MLL4 by TFs is necessary for enhancer-mediated activation of target genes during cellular differentiation24. It has been reported that MLL3/4 regulate enhancer activation through the recruitment of the co-activator protein p300, a histone acetyltransferase that mediates H3K27 acetylation and transcriptional activation of target genes25,26. H3K4 methyltransferase activity of MLL3/4 (and the Drosophila homolog TRR1) appears to be dispensable for gene expression in mouse embryonic stem (ES) cells and during Drosophila development, but may be involved in fine-tuning of enhancer function27,28.
In addition to trans-acting factors, activation of enhancers is also accompanied by formation of long-range chromatin interactions between these distal cis-regulatory elements and target gene promoters8,29. The Mediator and Cohesin complexes have been shown to play an important role in enhancer/promoter interactions30. Depletion of Cohesin resulted in genome-wide loss of chromatin domains and local chromatin interactions31,32,33. Interestingly, previous observations show that changes in local chromatin interactions correlate with H3K4me1 dynamics during differentiation of human ES cells34. However, it is unclear whether deposition of H3K4me1 by MLL3/4 contributes to chromatin interactions at enhancers. We hypothesized that MLL3/4, and potentially H3K4me1, may be involved in regulating chromatin interactions at enhancers. To test this hypothesis, we examined chromatin interactions in mouse ES cells (mESCs) either lacking both histone lysine methyltransferases MLL3/4 (DKO) or harboring point mutations in the two genes that render them catalytically inactive (dCD). We confirmed that MLL3/4 and H3K4me1 are indeed required to maintain proper chromatin interactions between enhancer and promoters. Through genome-wide analysis of chromatin contacts, we demonstrated that local chromatin contacts at distal regulatory elements are dependent on MLL3/4 in mouse ES cells. We obtained evidence that MLL3/4 modulate local chromatin organization through recruitment of the Cohesin complex. In addition, we show that H3K4 methyltransferase activities of MLL3/4 are needed for maximal Cohesin recruitment and chromatin interactions at enhancers. Altogether, these results provide new molecular details on the dynamic chromatin structure at enhancers.
Results
MLL3/4 are required for enhancer/promoter chromatin interactions at Sox2
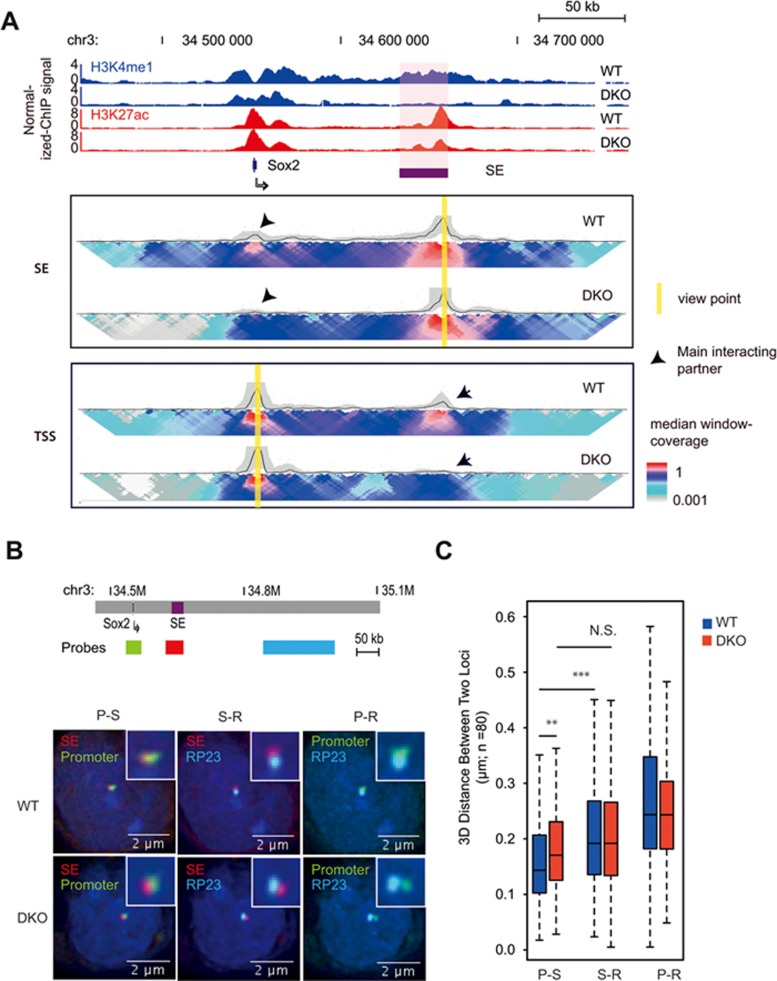
To determine the role of MLL3/4 on chromatin organization at enhancers, we first focused on a previously identified super-enhancer (SE), located 130 kb downstream of the Sox2 gene (Figure 1A and Supplementary information, Figure S1A)35,36. Deletion of this SE caused a drastic, allele-specific reduction of SOX2 expression in cis35,36. In addition, this SE was shown to interact with the Sox2 gene in mESCs35,36,37. H3K4me1 was present at the SE in wild-type cells (WT), but was largely lost in DKO cells (Figure 1A and Supplementary information, Table S5), confirming an essential role for MLL3/4 in deposition of active chromatin marks at the Sox2 distal SE (Sox2 SE).
Figure 1.
MLL3/4 and H3K4me1 are required for chromatin interactions at the Sox2 enhancer. (A) ChIP-seq and 4C-Seq analysis of Sox2 locus in wild type, MLL3/4 double-knockout mESCs. Top, genome browser snapshot of ChIP-seq data showing loss of H3K4me1 at Sox2 SE in MLL3/4 double-knockout mESCs. WT, wild type mouse ES cell line E14. DKO, MLL3/4 double-knockout mouse ES cell line. y-axis shows input normalized ChIP-seq RPKM. Sox2 SE is indicated in red shade. Sox2 gene locus is indicated by arrow. Bottom, 2D-heat map of 4C-seq analysis showing significant reduction in contact frequency between Sox2 TSS and Sox2-SE in DKO cells relative to WT cells. The same genomic position is aligned with genome browser snapshot for ChIP-seq analysis. Panel SE, 4C-seq with viewpoint at Sox2 SE locus, highlighted by yellow bar. Panel TSS, 4C-seq viewpoint at Sox2 TSS locus, highlighted by yellow bar. Black arrows emphasize the main interacting partner with the viewpoint. Heatmap shows the median genomic coverage using different sizes of sliding windows between 2 kb (top row) and 50 kb (bottom row). The gray shade above the heatmap shows the genomic coverage between 20th and 80th percentile values using a slide window of 5 kb. The black trend line indicates the median genomic coverage using a slide window of 5 kb. (B) 3D FISH microscopy images showing that physical distance between Sox2 SE and promoter becomes larger in DKO mESC, relative to WT. Top, schematic showing relative genomic positions for 3D FISH probes. Bottom, 3D FISH images showing overlay signals for probe set combinations in representative WT and DKO mESC. Nucleus was stained with DAPI. Insets show the zoom in of probe-detected foci for clearance. Red color, probes hybrid to SE locus, Green color, probes detecting promoter locus; Cyan color, probes detecting a region (RP23) that is located 100 kb downstream from SE. Note that Promoter is ∼100 kb upstream of SE. (C) Summary of FISH data from approximately 80 individual cells for both WT and DKO cell types. y-axis shows the distance between the centers of two foci represented by two colors. Scale bar is shown at the bottom right corner of each image. Note that distance between promoter and SE was significantly larger in DKO than WT. S-R, distance between RP23 and SE; P-R, distance between RP23 and Promoter; P-S, distance between Promoter and SE. Stars indicate statistical significance tested with Mann-Whitney U test (**P < 0.01; ***P < 0.005). RPKM, reads per kilobase pair per million total reads; SE, super enhancer. See also Supplementary information, Figures S1 and S2.
To test the hypothesis that loss of MLL3/4 would impair chromatin interactions at enhancers, we employed 4C-seq38 to examine the chromatin interactions centered at either the Sox2 promoter or the distal enhancers, in WT and DKO cells (Figure 1A and Supplementary information, Figure S1B). In WT cells, strong chromatin interactions could be detected between the Sox2 promoter and its enhancer located 130 kb downstream. Such interactions were dependent on the enhancer, since they were lost in homozygous Sox2 SE deletion mouse ES cells (DEL; Supplementary information, Figure S1C)35. Strikingly, the Sox2 promoter-enhancer interactions were dramatically reduced in DKO cells, suggesting that MLL3/4 are required for the formation of chromatin interactions between the Sox2 promoter and SE (Figure 1A and Supplementary information, Figure S1B).
We further employed three-dimensional fluorescent in situ hybridization technology (3D-FISH) as an orthogonal approach to validate the loss of enhancer-promoter interactions in vivo at single-cell resolution. Consistent with 4C-seq results, the spatial distance between Sox2 SE and Sox2 promoter significantly increased in both DKO and DEL cells (Figure 1B, 1C and Supplementary information, Figure S1D, S1E).
To exclude the possibility that loss of chromatin interactions in DKO cells is due to a reduced level of SOX2 expression, we carried out RNA-seq analysis of the WT and DKO cells. No apparent change in expression of other pluripotency TFs, such as OCT4 and NANOG, was detected, consistent with an earlier report25 (Supplementary information, Figure S1G). Therefore, the loss of chromatin interactions between the SE and target gene in the DKO cells is unlikely the result of secondary effects of reduced expression of other TFs.
Similar to the Sox2 locus, we also observed disruption of enhancer-promoter interactions and target gene expression at the Car2 gene locus in DKO cells (Supplementary information, Figure S1F), while an independent study identified a loss of promoter/enhancer interactions at the Lefty gene locus using the same MLL3/4 DKO cells25. Our results demonstrate that chromatin interactions between enhancers and target genes depend on H3K4me1 and the methyltransferases MLL3/4.
MLL3/4 loss leads to global reduction of chromatin interactions at enhancers in the ES cells
To determine whether MLL3/4 knockout also led to a general loss of chromatin interactions at enhancers, we first carried out ChIP-seq analysis in WT and DKO cells to examine the effects of MLL3/4 loss on genomic distribution of H3K4me1(Supplementary information, Table S6). Focusing on a set of enhancers previously determined in mouse ES cells39, we found that loss of MLL3/4 led to a significant decrease of H3K4me1 signals at these distal cis-regulatory elements genome-wide (Supplementary information, Figure S2A), whereas H3K4me1 level near transcriptional start sites (TSS) was unchanged or slightly increased, consistent with previous reports that MLL3/4 are primarily responsible for deposition of H3K4me1 to promoter-distal enhancers23 (Supplementary information, Figure S2B). We identified a total of 78 645 narrow H3K4me1 peaks in WT mESCs, and observed that H3K4me1 signals at 34 527 of them decreased by more than 50% in DKO cells. We referred to these as MLL3/4-dependent H3K4me1 regions (Supplementary information, Figure S2C and Table S1A). Little or no change was detected in the other 44 118 genomic regions, which we designated as MLL3/4-independent H3K4me1 regions (Supplementary information, Figure S2C and Table S1B). In addition, 9 228 genomic regions acquired H3K4me1 peaks upon MLL3/4 knockout (Other; Supplementary information, Figure S2C and Table S1C). Consistent with a previous report23, we found that more than 85% of MLL3/4-dependent H3K4me1 regions were promoter-distal (>2 kb), whereas MLL3/4-independent H3K4me1 regions were enriched at or near TSS. Most of the increased H3K4me1 peaks in DKO were located within TSS proximal regions (Supplementary information, Figure S2D). Motif analysis revealed strong enrichment of ES-specific TF-binding sites in MLL3/4-dependent H3K4me1 regions (Supplementary information, Figure S2E), consistent with a previously reported role for the sequence-specific TFs in recruiting MLL3/4 to establish H3K4me1 at distal enhancers24. Gene ontology analysis indicates that genes near the MLL3/4-dependent H3K4me1 regions are involved in pluripotency and stem cell maintenance (Supplementary information, Figure S2F), suggesting a role for MLL3/4-dependent H3K4me1 regions in cell type-specific gene expression.
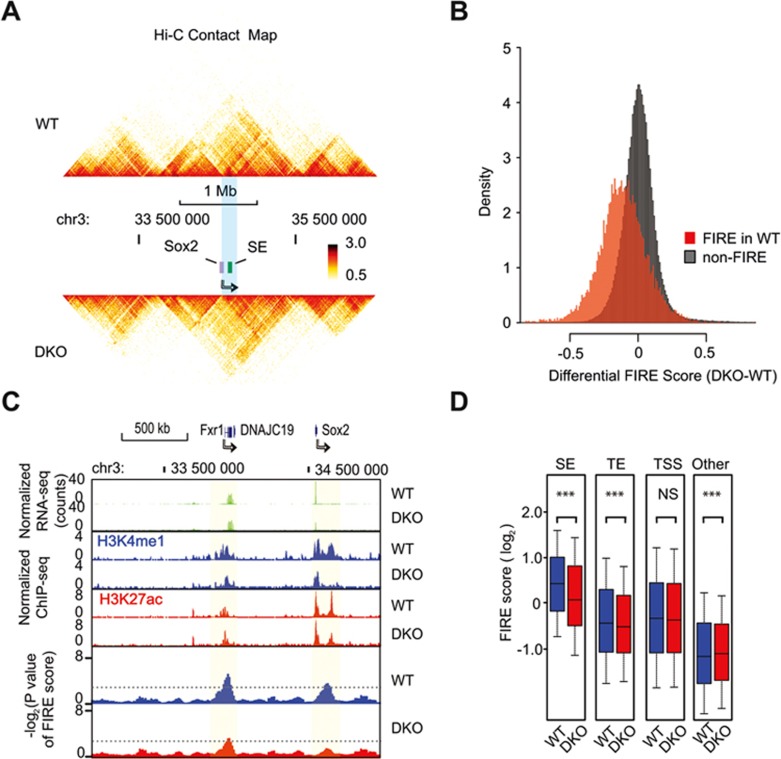
We next employed in situ Hi-C40 to investigate alterations in chromatin architecture in both wild type and DKO cells to determine whether loss of MLL3/4 would result in reduced chromatin interactions at enhancers genome-wide. We carried out Hi-C experiments in two biological replicates, and obtained ∼109 reads from each cell line. As shown in Figure 2A, global chromatin organization was well preserved upon MLL3/4 knockout (Figure 2A and Supplementary information, Figure S3A). The topologically associating domains (TADs) are largely unaffected9,41 (Supplementary information, Figure S3C and S3D). The strongest impact of MLL3/4 loss on contact frequency was observed at short-range chromatin interactions, especially between bins located shorter than 100 kb apart from each other (Supplementary information, Figure S3B). We observed that short-range chromatin contacts altered in DKO cells were not evenly distributed in the genome, and overlapped significantly with previously defined frequently interacting regions (FIREs)42, which were specifically enriched for SEs. We determined 14 190 FIREs in WT cells and 13 542 FIREs in DKO cells, covering ∼5% of total genome (Supplementary information, Figure S3E-S3H and Data S1). Roughly 70% of FIREs are shared between WT and DKO cells. Upon knockout of MLL3/4, the chromatin contacts in WT FIREs were significantly reduced compared to non-FIRE genomic regions (Figure 2B and Supplementary information, Figure S4A-S4D, Table S2). To illustrate how H3K4me1 signals are associated with chromatin interactions at individual loci, we used a 2 Mb-region containing Sox2 on chromosome 3 as an example. The FIRE score was significantly decreased near the Sox2 SE locus in DKO cells, coincident with depletion of H3K4me1, whereas the nearby Fxr1 locus was less affected (Figure 2C). We observed a significant decrease in average FIRE scores of typical enhancers (TE) and SE genome-wide, but not at the TSS (Figure 2D, Supplementary information, Figure S4G-S4I). The FIRE scores were tightly associated with MLL3/4-dependent H3K4me1 peaks and H3K27ac peaks. These peaks are mostly located at TSS-distal enhancer regions (Supplementary information, Figure S2). Our results further support that MLL3/4 play an active role in promoting chromatin interactions at the enhancers.
Figure 2.
MLL3/4-dependent H3K4me1 shows reduced chromatin interactions in DKO mESC. (A) Heatmap showing the chromosomal contacts near the Sox2 locus, as determined in in situ Hi-C experiments. The upper panel shows the WT cells and lower panel shows the DKO cells. Sox2 gene is indicated by the violet box and an arrow. Color key shows the normalized contact frequency. (B) Histogram showing the distribution of differential FIRE scores across 10 kb bins (DKO-WT). Red, bins classified as FIRE regions in WT cells; Grey, non-FIRE bins in WT cells. (C) Genome browser track shows correlation between the change of FIRE scores (bottom) and changes of H3K4me1 ChIP-seq RPKM (middle) upon MLL3/4 knockout. Partial loss of SOX2 expression is also shown (top). For comparison, FXR1 or DNAJC19 expression is stable, consistent with the stable FIRE structure. The light green shades indicate the FIRE regions. Dashed lines label the cutoff for FIRE. (D) Boxplots comparing log2 value of FIRE scores in WT (blue) and DKO (red) for bins containing different classes of cis-regulatory elements (TE, SE and TSS). Other, bins with increased FIRE scores in DKO relative to WT. The P value below each category was computed by two-tail paired Welch t-test. Asterisk indicates that the difference is statistically significant. Note that SE displays the highest FIRE score among all elements tested here and SE, TE and TSS generally have higher FIRE score than the average genome. SE, super enhancer; TE, typical enhancer; TSS, transcription-starting site. See also Supplementary information, Table S1, Figures S3 and S4.
Histone methyltransferase activities of MLL3/4 are necessary for enhancer/promoter interactions at the Sox2 locus
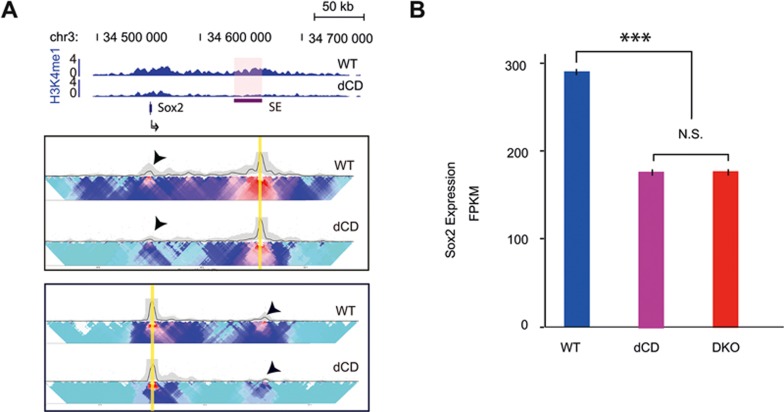
The above results showed that the histone methyltransferases MLL3/4 play an active role in establishment or maintenance of enhancer/promoter interactions at the Sox2 locus, and in promoting local chromatin interactions at active enhancers in mouse ES cells. To determine if the histone methyltransferase activities of the MLL3/4 are necessary for their role in chromatin structure at enhancers, we again carried out 4C-seq experiments using dCD cell line, focusing on the Sox2 locus. Consistent with previous reports that MLL3/4 primarily mediate mono-methylation at distal enhancers27,28, H3K4me1 was absent from the Sox2 enhancer in dCD cells. Compared to the WT mESC, the chromatin interactions between Sox2 and the downstream enhancer was greatly reduced in the dCD mESC line (Figure 3A), suggesting that the histone methyltransferase activities of MLL3/4 indeed are required for enhancer/promoter interactions at Sox2 gene.
Figure 3.
MLL3/4 methyltransferase activity is required for chromatin interactions and gene expression at the Sox2 enhancer. (A) ChIP-seq and 4C-Seq analysis of Sox2 locus in wild type and MLL3/4 catalytic mutant mESCs. Similar to DKO cells (Figure 1A), reduction of chromatin interactions between Sox2 SE and TSS was detected in dCD cells. (B) RNA-seq data shows that expression of Sox2 decreased in dCD and DKO cells compared with WT. FPKM is shown in y-axis. Error bars are derived from three biological replicates. See also Supplementary information, Figure S5.
In accordance with previous observations linking Sox2 promoter-enhancer interactions to Sox2 transcription35,36, a significant decrease in Sox2 mRNA expression was observed (∼45%) in both dCD and DKO cells, in the mRNA-seq data obtained from WT, DKO or dCD cell lines27 (Figure 3B). Previously, it was shown that removal of Sox2 SE led to more than 90% reduction of Sox2 expression35. The relative mild effect of MLL3/4 knockout on Sox2 expression indicates that additional mechanisms, for example proximal enhancers of Sox2, may be involved in regulating Sox2 expression that compensate for the reduction of MLL3/4 at the distal enhancer and subsequent loss of long-range interactions.
Cohesin acts downstream of MLL3/4 to promote chromatin interactions at enhancers
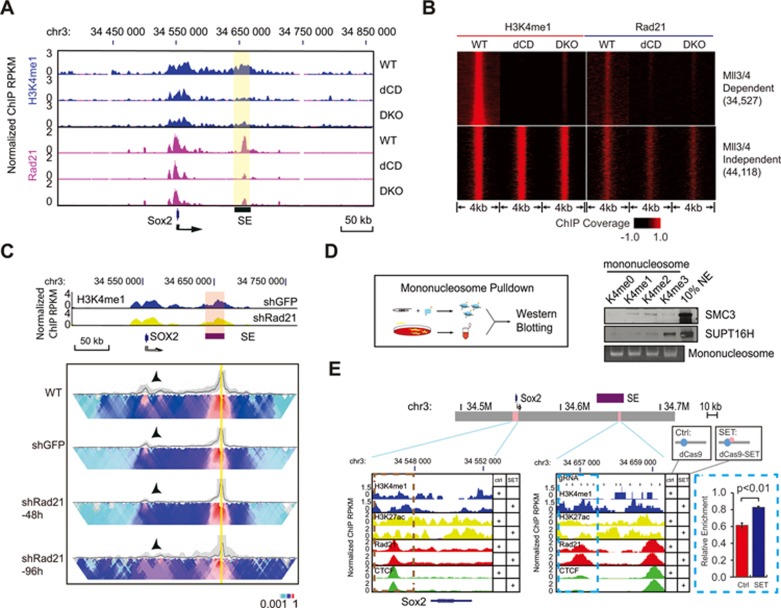
To gain insight into the molecular mechanisms by which MLL3/4 promote chromatin interactions at enhancers, we next examined the relationship between MLL3/4 and Cohesin complex at enhancers30. The Cohesin complex, consisting of SMC1, SMC3, RAD21 and SA1/2, has been proposed to mediate chromatin contacts between enhancers and promoters30,32,43,44,45,46,47. To test whether MLL3/4 facilitate the recruitment of Cohesin complex to enhancers, we first asked whether MLL3/4 and/or H3K4me1 are required for the recruitment of Cohesin at the Sox2 SE. We performed ChIP-seq experiments in WT, dCD and DKO cells using antibodies against the Cohesin subunit Rad21. Consistent with previous reports30,47, the Cohesin complex co-localized with H3K4me1 peaks at the Sox2 SE in WT mESC (Figure 4A). In the dCD and DKO cells, occupancy of Cohesin at Sox2 SE was dramatically reduced as compared to that in the WT cells (Figure 4A), suggesting that Cohesin occupancy at enhancers is H3K4me1-dependent. Genome-wide analysis further showed that binding of Cohesin complex to MLL3/4-dependent H3K4me1 peaks was drastically reduced in dCD and DKO cells, whereas the occupancy of RAD21 remained unchanged near the Mll3/4-independent H3K4me1 peaks (Figure 4B).
Figure 4.
MLL3/4-dependent H3K4me1 facilitates Cohesin binding to enhancers. (A) Genome browser tracks showing loss of Cohesin (Rad21) binding at the Sox2 gene and Sox2 SE (indicated in violet shade) in MLL3/4 DKO and dCD cells. Top, relative genomic positions of the Sox2 gene and Sox2 SE; Middle, RNA-seq tracks show Sox2 expression for reference, quantified in Figure 1D; Bottom, normalized ChIP-seq signals of H3K4me1, Rad21 and Med12 at the Sox2 locus. (B) ChIP-seq analysis showing binding of Cohesin to MLL3/4-dependent H3K4me1 peaks in DKO and dCD cells. ChIP-seq signals are centered around H3K4me1 peaks and extended 2 kb upstream and downstream along the genome. x-axis indicates relative coordinates to peaks center. Heatmap shows ChIP-seq coverage value. Note that Cohesin (Rad21) is affected at MLL3/4-dependent H3K4me1 regions in both dCD and DKO cells, coinciding with the loss of H3K4me1. (C) Genome browser tracks showing knockdown of Cohesin complex (shRad21) does not cause overt change of H3K4me1 signals and 4C interactions at the Sox2 locus. Top, normalized H3K4me1 ChIP-seq tracks for control and Rad21 knockdown cells. shGFP, control using shRNA against GFP sequences. SE, super enhancer, indicated also by violet shade. Bottom, 4C-seq analysis showing that chromatin interactions between Sox2 SE and promoter are reduced upon Rad21 depletion by shRNA. shRad21-48h, shRNA knockdown targeting RAD21 mRNA 48 h after lentivirus infection. shRad21-96h, shRNA knockdown targeting RAD21 mRNA 96 h after lentivirus infection. Black arrows emphasize the main interacting partner with the viewpoint. Heatmap shows the median genomic coverage of the indicated position using different sizes of sliding windows between 2 kb (top row) and 50 kb (bottom row). The gray shade above the heatmap shows the genomic coverage between 20th and 80th quantile values using a slide window of 5 kb. The black trend line indicates the median genomic coverage using a slide window of 5 kb. Note that the interaction between Sox2 SE and gene body is dramatically lost at 96 h post knock-down via lentivirus. (D) The in vitro pull-down assay showing that Cohesin (Smc3) preferentially associate to H3K4me1 and H3K4me2 mononucleosomes. Top, schematic showing assay workflow. Modified H3 histones are assembled into biotinylated DNA-bound nucleosomes in vitro. Nucleosomes were then incubated with HeLa nuclear lysate and the Streptavidin pull-down fractions were assayed for binding factors with Western blotting. Bottom, Western blots showing binding of SMC3 and SUPTH binding to H3K4 unmodified (K4me0), mono- (K4me1), di- (K4me2) and tri- (K4me3) methylated nucleosomes. The FACT complex subunit SUPT16H is used as a control to show that FACT binds preferentially to H3K4me3 mono-nucleosomes. Agarose gel staining for 601λ DNA was used as loading control for mononucleosomes. (E) Genome browser tracks showing increased H3K4me1 and Cohesin (RAD21 subunit) binding in the Sox2 SE locus in DKO cells expressing dCas9-MLL3SET. DKO cells were transfected with vectors co-expressing tiling CRIPSR guides targeting Sox2 SE and dCas9 proteins with or without MLL3SET domain fusion. Top, schematic of the Sox2 loci assayed. Pink boxes indicate genomic regions of interest. SE (purple), Sox2 SE. Bottom panels, genome browser tracks showing normalized ChIP-seq coverage in RPKM for H3K4me1, H3K27ac, RAD21 and CTCF at control region (Sox2 promoter, encircled by brown dashed box) and the guide RNA targeted region of Sox2 SE (encircled by blue dashed box). y-axis, relative RPKM coverage was calculated by normalizing to the constant, CTCF-overlapping major peak right to the dashed box. The bar plot shows the quantification of RAD21 ChIP-qPCR enrichment at the blue dash box indicated region in control cells (red) and cells transfected with dCas9-MLL3SET (blue). Ctrl, dCas9 without MLL3SET domain. SET, dCas9-MLL3SET fusion protein. SE, super enhancer. See also Supplementary information, Figure S5.
To determine whether Cohesin complex acts downstream of MLL3/4 to regulate chromatin interactions, we depleted the expression of Cohesin complex component RAD21 using shRNA-mediated knockdown (Supplementary information, Figure S5A). No obvious change of H3K4me1 at either Sox2 locus or at genome-wide level was detected (Figure 4C and Supplementary information, Figure S5B, Table S5), in spite of loss of chromatin interactions between Sox2 SE and the promoter (Figure 4C, lower panel). Sox2 expression decreased significantly upon Rad21 knockdown, to a similar degree as in DKO and dCD mESC lines, supporting the model that Cohesin, MLL3/4 and H3K4me1 act together to modulate enhancer-promoter interactions and gene activation (Supplementary information, Figure S5C). This result further suggests that MLL3/4 likely act upstream of the Cohesin complex to mediate chromatin interactions.
H3K4me1 may facilitate the recruitment of Cohesin complex to enhancers
The observation that enhancer/promoter interactions at Sox2 were strongly reduced in the catalytically dead MLL3/4 mutant cell line suggests that one potential mechanism for MLL3/4 to facilitate recruitment of Cohesin complex at enhancers is via H3K4me1. Consistent with this hypothesis, the Cohesin complex is generally co-localized with H3K4me1 peaks in vivo (Figure 4A, B and Supplementary information, Figure S5D). Change of H3K4me1 due to MLL3/4 depletion also caused decreased Cohesin binding around MLL3/4-dependent H3K4me1 peaks but not at MLL3/4-independent H3K4me1 peaks (Figure 4B and Supplementary information, Figure S5E). Meanwhile, we observed that FIRE score around the decreased Cohesin peaks was also significantly attenuated, further indicating Cohesin's role in chromatin interaction (Supplementary information, Figure S5F).
To further test this hypothesis, we performed in vitro pull down assays using nuclear extracts from HeLa cells and reconstituted mononucleosomes bearing either unmodified H3 histones or H3 with chemically-modified epitopes mimicking lysine 4, including mono-, di- and tri-methylation. We found that the Cohesin complex bound more strongly to the mononucleosomes with H3K4me1 and H3K4me2 modifications than unmodified nucleosomes. As a control, the FACT complex showed stronger preference to H3K4me3 than H3K4me148,49 (Figure 4D and Supplementary information, Figure S5G).
Our ChIP-seq and in vitro pull-down assays strongly suggest that H3K4me1 may either directly or indirectly facilitate the binding of the Cohesin complex to enhancers to mediate chromatin interactions. To further determine whether H3K4me1 deposition is sufficient for Cohesin recruitment, we repurposed the catalytically dead Cas9 protein (dCas9) to induce ectopic H3K4me1 at targeted locus by fusing it to the MLL3 SET domain (MLL3SET), which catalyzes monomethylation of H3K423. We showed that transfection of dCas9-MLL3SET and a set of guide RNAs targeting the Sox2 SE to DKO cells significantly increased local H3K4me1 levels at the targeted region (Figure 4E) as well as Cohesin occupancy at Sox2 SE, indicating that localization of MLL3/4 to H3K4me1 nucleosomes may facilitate Cohesin recruitment in vivo. As a control, H3K4me1 and Cohesin occupancy at the non-targeted Sox2 promoter was not significantly altered (Figure 4E).
Dynamic chromatin organization at lineage-specific enhancers during mouse stem cell differentiation
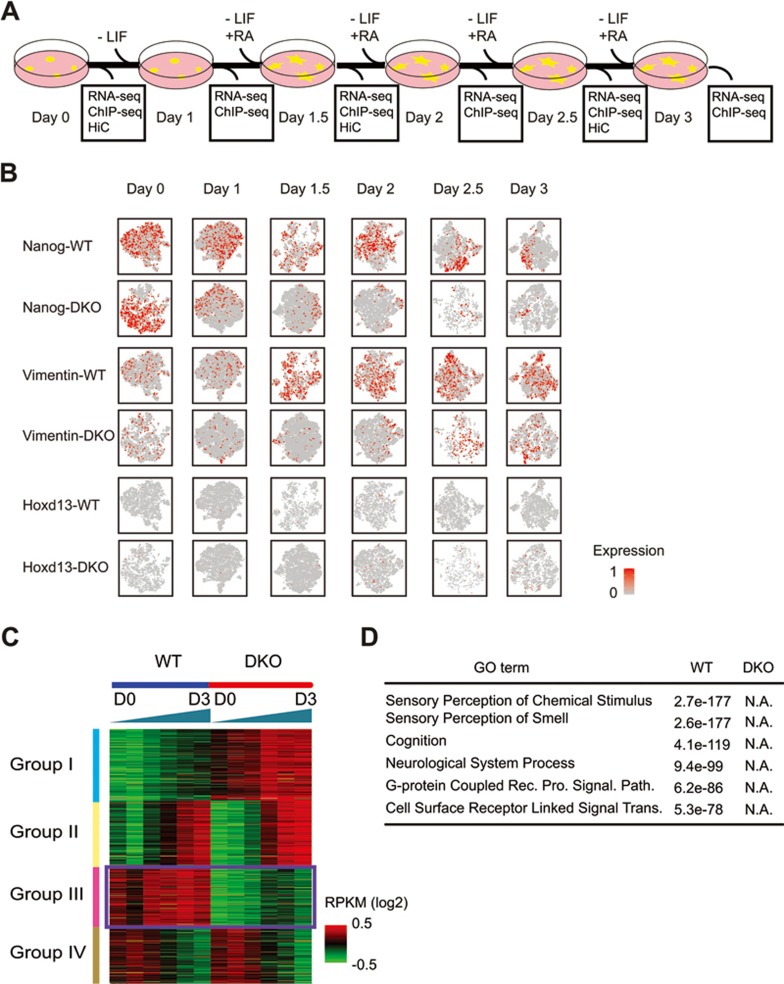
We next interrogated the role of MLL3/4 in the temporal dynamics of enhancer-promoter interactions and dynamic gene expression during cellular differentiation. If MLL3/4 are indeed critical for establishing chromatin interactions at enhancers, as the above results would suggest, we would expect chromatin interactions at lineage-specific enhancers to be MLL3/4-dependent during mESC differentiation. To test this prediction, we treated WT and DKO cells with retinoic acid (RA) to induce the differentiation of these cells towards the neural progenitor cell (NPC) lineage. We collected the cells every 12 h over a 60-h period (Figure 5A). Single-cell RNA-seq analyses showed that both WT and DKO cells lost Nanog expression after RA treatment. However, while the WT cells successfully differentiated into the NPC lineage, as evidenced by the expression of NPC-specific markers, such as Vimentin, the DKO cells failed to completely differentiate into the NPC lineage (Figure 5B). Bulk RNA-seq analysis showed broad defects in the induction of genes involved in neuronal function (Figure 5C, D and Supplementary information, Table S8). In particular, group III genes were induced in WT but not in DKO cells (Figure 5C and Supplementary information, Figure S5H, Table S8). Lack of induction of these genes in DKO cells indicated a failure of differentiation towards NPC in the absence of MLL3/4, supported by Gene Ontology analysis of group III genes (Figure 5D). This result strongly supports the model that MLL3/4 are critical for stem cell differentiation50,51.
Figure 5.
Characterization of gene expression during NPC differentiation in WT and DKO mESC. (A) Scheme of differentiation protocol. Data was collected for RNA-seq and ChIP-seq at every 12-h time points and Hi-C at every 24-h time points. (B) Single-cell RNA analysis of NPC differentiation. Each panel represents 2-D t-SNE (stochastic neighbor embedding) projection of the cells colored by the total UMI counts per cell. x-axis represents t-SNE-1 and y-axis shows t-SNE-2. Red color indicates high expression of the gene that is noted on the left of each row. Each column represents a time point indicated by A. Expression patterns of Nanog and Vimentin showed that the cell population was well synchronized and no overt subgroup was observed. Nanog, pluripotency marker in embryonic stem cells. Hoxd13 had slight increase in expression in DKO cells. Vimentin, marker for neural progenitor. Hoxd13, posterior HOX transcription factor normally expressed in the posterior part of the body plan during development. (C) Clustering analysis of bulk RNA-seq from WT and DKO cells showing that genes could be clustered to four different groups depending on the panel of change along differentiation. Note that Group I and Group III genes are mostly MLL3/4-dependent and they behave differentially between WT and DKO cells. Group II and Group IV are MLL3/4-independent genes and they are expressed in the same pattern in two cell types. (D) Gene ontology analysis showing that genes induced only in WT cells are mostly related to neuron function. Note that no GO terms could be detected in genes that are induced only in DKO cells. See also Supplementary information, Figure S5 and Table S7.
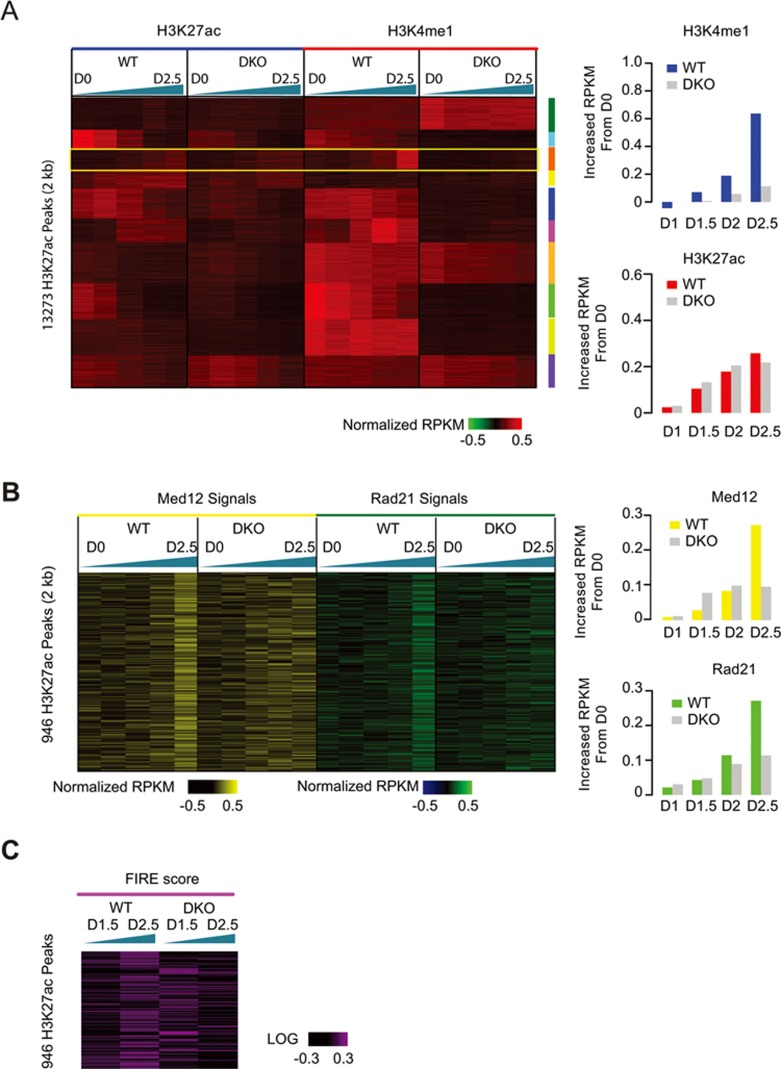
We also examined the dynamic H3K27ac and H3K4me1 profiles in WT and DKO cells at various time points during NPC differentiation using ChIP-seq (Figure 6A and Supplementary information, Table S3). Strikingly, at the majority of the distal H3K27ac peaks, H3K4me1 signals were depleted in DKO cells, and this resulted in a failure to induce cell type-specific gene expression (Supplementary information, Figure S5I). This observation provides further support for a role for MLL3/4 in regulating the chromatin epigenetic landscape at distal enhancers.
Figure 6.
Characterization of H3K4me1, Cohesin and chromatin interactions during NPC differentiation of WT and DKO mESC. (A) Clustering analysis showing that H3K27ac peaks could be classified to 10 different groups per change in both H3K27ac and H3K4me1 signals in WT and DKO cells during differentiation. The color key shows the log2 value of transformed input normalized RPKM value of each peak. Green triangle shows the differentiation time with the thicker side representing later time points. Yellow box emphasizes the group that shows induced H3K27ac and H3K4me1 signal in WT cells along differentiation. The right bar charts show the averaged signal of H3K4me1 (top) and H3K27ac (bottom) of group III at D0, Day1, D2 and D2.5. (B) Heatmap showing the change of input normalized Mediator subunit Med12 (left) and Cohesin subunit Rad21 RPKM (right) at Group III H3K4me1 peaks emphasized in A. Note that Cohesin and Mediator binding also tends to increase in WT cells but not in DKO cells. The right bar charts show the averaged signal of H3K4me1 (top) and H3K27ac (bottom) of group III. (C) Heatmap showing the change of FIRE score at Group III H3K4me1 peaks emphasized in A. Note that FIRE score tends to increase in WT cells but not in DKO cells. See also Supplementary information, Figure S6 and Table S3.
To dissect the temporal relationships between H3K4me1 deposition, Cohesin occupancy and chromatin organization, we examined Cohesin and Mediator occupancy and FIRE scores in the regions where accumulating H3K4me1 and H3K27ac signals were detected along differentiation in WT cells but not in DKO cells (Figure 6A). If H3K4me1 is necessary for Cohesin recruitment, gradual induction of H3K4me1 and H3K27ac at enhancers during mESC differentiation would lead to binding of Cohesin. As expected, these loci showed increased Cohesin binding during differentiation only in WT cells, along with increased H3K4me1 signals (Figure 6B). More interestingly, regions with induced H3K4me1 but not H3K27ac also displayed increased binding of Cohesin, suggesting that H3K4me1 played a major role in Cohesin recruitment than H3K27ac (Supplementary information, Figure S6). The FIRE score of these regions were also gradually elevated in WT cells, in accordance with the role of MLL3/4 in regulation of chromatin interactions (Figure 6C).
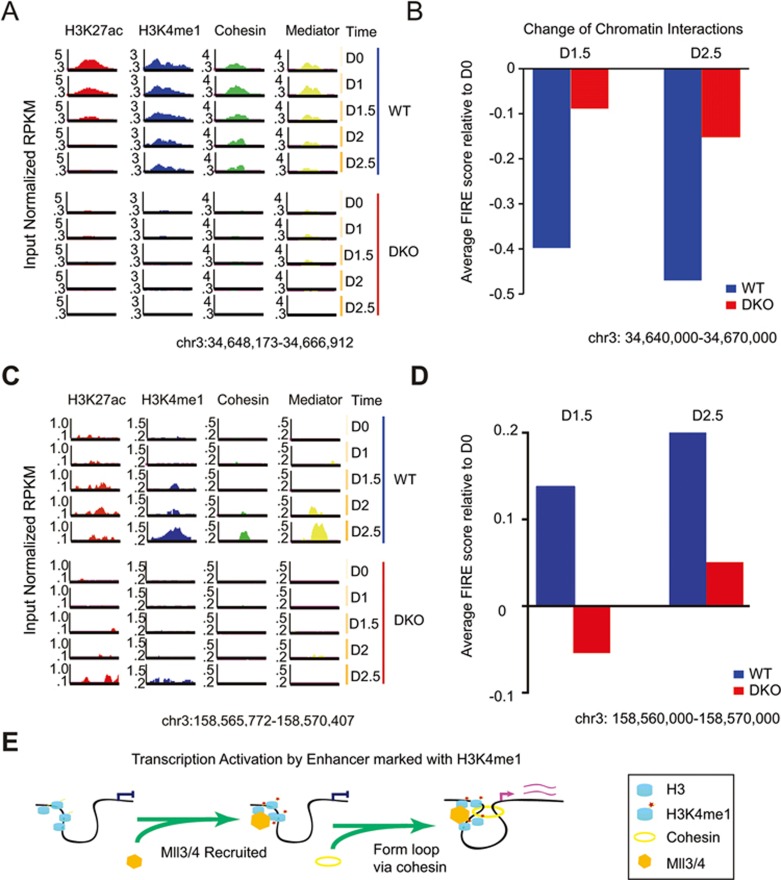
In order to more clearly reveal the concordant change of H3K4me1 signal, Cohesin binding and FIRE score, we focused our analysis on the Sox2 SE. We observed a gradual decrease in Cohesin binding along differentiation, concurrently with decreased H3K4me1 signals (Figure 7A). The FIRE score spanning this region also gradually decreased, in accordance with the role of H3K4me1 in regulation of chromatin interactions mediated by Cohesin (Figure 7B). At another representative locus, we observed accumulation of H3K4me1 but not H3K27ac signals at Day 1.5, while Cohesin- and Mediator-binding signals were detected at Day 2, 12 h later than the histone marks (Figure 7C). As expected, FIRE score of this region increased in WT cells, consistent with increased Cohesin occupancy (Figure 7D). In DKO cells, neither Cohesin nor Mediator could be detected in both loci at any time point, likely due to lack of H3K4me1 at enhancers. This striking example demonstrated that changes of Cohesin binding preceded or coincided with H3K4me1, suggesting that the histone modification could potentially stabilize Cohesin loading at enhancers, as well as chromatin organization.
Figure 7.
Dynamic histone modification, Cohesin occupancy and changes in chromatin interactions at representative loci. (A) Genome browser track showing that at Sox2 SE locus, H3K27ac and H3K4me1 signals were enriched in WT cells but not in DKO cells on Day 0. Along cellular differentiation, H3K27ac and H3K4me1 signals gradually decreased, while Mediator and Cohesin binding were also alleviated. Note that Cohesin and Mediator started decreasing at Day 1.5 while both H3K27ac and H3K4me1 started decreasing at Day 1. By contrast, none of the factors could be detected in DKO. y-axis shows input normalized ChIP-seq RPKM. (B) Bar chart showing change of chromatin interactions (FIRE score). Blue bars indicate FIRE score at each time point relative to Day 0. The 30 kb regions are enclosing the entire Sox2 SE in A. The change of FIRE score in DKO (red bars) was mild compared to WT. (C) Genome browser track showing at a representative locus, H3K27ac and H3K4me1 signals gradually increased along cellular differentiation, starting from Day 1.5. Meanwhile, Mediator and Cohesin are also detected to be initiated at Day 2. By contrast, in DKO cells, none of the factors could be detected until Day 2.5. y-axis shows input normalized ChIP-seq RPKM. (D) Bar chart showing change of chromatin interactions enclosed in C (FIRE score). Blue bars indicate FIRE score at each time point relative to Day 0. The 10 kb regions are enclosing the entire 5 kb region shown in panel A. Less change in FIRE score was observed in DKO (red bars) than in WT. (E) A model depicting MLL3/4 and H3K4me1's role in establishment of chromatin interactions at enhancers. When H3K4 is not methylated at distal enhancer, the promoter is physically separated in different nuclear territory (left). MLL3/4 are recruited to distal enhancers, leading to monmethylation of lysine 4 of histone H3 (middle). Subsequently, the Cohesin complex is recruited to mediate the looping interaction between gene promoter and enhancer. RPKM, reads per kilobase pair per million total reads; SE, super enhancer. See also Supplementary information, Figure S7 and Table S4
Dynamic chromatin contacts at super enhancers during ES cell differentiation depend on MLL3/4
To more clearly demonstrate the role of MLL3/4 in mediating chromatin interactions, we focused on SE, which are regions with highly clustered sites of TF- and co-factor-binding sites, and involved in activating transcription of cell identity genes through long-range chromatin interactions52. We identified SEs in WT and DKO cells at different time points, based on their unusually high levels of H3K27ac signals39,53,54 (Supplementary information, Figure S7A and Table S4). Consistent with a recent report42, over half of the SEs identified above were within FIREs in WT mESC, and the H3K27ac signals at these SEs were gradually lost during differentiation. In DKO cells, FIRE scores at the SEs were significantly reduced, providing strong support for a role of MLL3/4 in chromatin interactions at enhancers (Supplementary information, Figure S7B). For example, at the Sox2 locus, the SE gradually lost H3K27ac signal until fully depleted by Day 1.5 (Supplementary information, Figure S7C). FIRE score at this enhancer also decreased along differentiation (Supplementary information, Figure S7D). In DKO cells, the SE was not properly formed due to lack of MLL3/4 and the chromatin interaction frequency was also low at this region.
Discussion
Great strides have been made in the identification of cis-regulatory sequences in the human genome18,19. Since most candidate cis-regulatory sequences reside far from the transcription start sites, it is generally believed that 3D chromatin architecture plays an essential role in activation of target genes by enhancers. However, the mechanisms by which long-range chromatin interactions are established at lineage-specific enhancers during development are still incompletely understood34,37. In particular, it has yet to be shown whether chromatin remodeling complexes play an active role in chromatin organization at enhancers, despite the data showing close correlation between dynamic histone modifications and chromatin organization during human ES cell differentiation34,41,55,56,57. Here, we provide multiple lines of evidence supporting an active role for the histone methyltransferases MLL3/4 and their enzymatic product H3K4me1 in establishment of long-range chromatin interactions at enhancers. First, deletion or catalytic-inactivating point mutations of MLL3/4 resulted in loss of chromatin interactions between the enhancer and Sox2 promoter in the mouse ES cells; second, loss of MLL3/4 also led to a decrease in local chromatin interactions at regions bearing MLL3/4-dependent H3K4me1; third, deletion or catalytic-dead mutations of MLL3/4 resulted in reduced occupancy by the Cohesin complex at MLL3/4-dependent H3K4me1 sites. Furthermore, MLL3/4 are required for depositing H3K4me1 and establishing local chromatin interactions at a large number of distal enhancers during mouse ES cell differentiation. Finally, pull-down experiment using nuclear extracts showed that the Cohesin complex favored H3K4me1-modified nucleosomes over other forms of nucleosomes, and targeted deposition of H3K4me1 led to recruitment of Cohesin complex in the mouse ES cells. Taken together, our results support a model that MLL3/4 and H3K4me1 modulate local chromatin interactions at enhancers by facilitating the recruitment of Cohesin complex (Figure 7E).
The present study revealed a novel mechanism of enhancer/promoter interactions in mammalian cells. Cohesin has been shown to mediate chromatin interactions and chromatin domains in metazoan27,30,32,33,43,44,45. Our results suggest that MLL3/4 could facilitate or stabilize Cohesin recruitment at enhancers to promote DNA interactions with other regions. We observed that Cohesin would not bind to the SEs at Sox2 and Car2 loci and thus no chromatin interactions were formed in the absence of MLL3/4 (Figure 1 and Supplementary information, Figure S1). In a time course experiment along NPC differentiation, we also observed that H3K4me1 signals coincided or preceded Cohesin loading. While our results strongly support a role for H3K4me1 in the recruitment of Cohesin complexes to enhancers, two questions remain. First, how does this histone mark promote Cohesin binding to enhancers? SMC1, SA1/2, SMC3 and RAD21 are known as the core components of the Cohesin complex, and none of them contains a known H3K4me1-binding domain. Thus, the Cohesin likely indirectly associates with the H3K4me1 mononucleosome. Second, depletion of Cohesin only affected a small number of genes, and had only modest effects on gene expression32,33. Clearly, additional studies are needed to determine how Cohesin complex's binding to enhancer is facilitated by H3K4me1, and how Cohesin recruitment at enhancers leads to enhancer/promoter interactions and activation of target genes.
Since we observe partial loss of H3K27ac at Sox2 and Car2 SE in DKO cells, it is possible that H3K27ac could also help recruitment of Cohesin to enhancers. H3K27ac is a histone modification that often coincides with H3K4me1 at active enhancers, and might act downstream of H3K4me1 to recruit Cohesin. In a previous study, knockdown of EP300/CBP histone acetyltransferases resulted in loss of pluripotency and long-range chromatin interactions at several loci in mouse ES cells58. However, we consider that H3K4me1 has a more prominent role in recruitment of Cohesin and establishing chromatin interactions at enhancers than H3K27ac, because many genomic regions with induced H3K4me1 but not H3K27ac during mESC differentiation also displayed increased binding of Cohesin.
Materials and Methods
Cell culture
Mouse ES cell lines were derived from E14 strain and strains reported in other studies25,27. All three mouse ES cells were cultured in mouse ES cell media: DMEM 85%, 15% fetal bovine serum (Hyclone), penicillin/streptomycin, 1× non-essential amino acids (Gibco), 1× GlutaMax, 1000 U/mL LIF (Millipore), 0.4 mM β-mercaptoethanol. Mouse ES cells were initially cultured on 0.1% gelatin-coated petri-dish with CF-1-irradiated mouse embryonic fibroblasts (GlobalStem) and were passaged twice on 0.1% gelatin-coated feeder-free plates before harvesting. Lenti-X 293 cells (Clontech) were cultured in DMEM containing 10% Tet-approved fetal bovine serum (Clontech), penicillin/streptomycin and GlutaMax (Gibco). Alkaline phosphatase staining was performed using the Alkaline Phosphatase Staining kit (STEMGENT) in the presence of MEF feeder cells.
In situ Hi-C
The in situ Hi-C experiments were conducted according to a previous study40. Briefly, 2 million cells were cross-linked with 1% formaldehyde for 10 min at RT and the reaction was quenched using 125 mM of Glycine for 5 min at RT. Nuclei were isolated and directly applied for digestion using 4 cutter restriction enzyme MboI (NEB) at 37 °C o/n. The single strand overhang was filled with biotinylated-14-ATP (Life Tech.) using Klenow DNA polymerase (NEB). Different from traditional Hi-C, with in situ protocol the ligation was performed when the nuclear membrane was still intact. DNA was ligated for 4 h at 16 °C using T4 ligase (NEB). Protein was degraded by proteinase K (NEB) treatment at 55 °C for 30 min. The crosslinking was reversed with 500 mM of NaCl and heat at 68 °C o/n. DNA was purified and sonicated to 300-700 bp small fragments. Biotinylated DNA was selected with Dynabeads MyOne T1 Streptavidin beads (Life Tech.). Sequencing library was prepared on beads and intensive wash was performed between different reactions. Libraries were checked with Agilent Bioanalyzer 2100 and quantified using Qubit (Life Tech.). Libraries were sequenced with Illumina Hiseq 2500 or Hiseq 4000 with 50 or 100 cycles of paired-end reads.
Data processing and FIRE analysis
Hi-C pre-processing pipeline was applied to all the raw Hi-C data as previously described34. All intra-chromosome reads within 15 kb were removed from the downstream analyses. The filtered intra-chromosome reads connecting two genomic regions located greater than 15 kb were selected and binned into 10 kb resolution to build the Hi-C contact matrices. For each 10 kb bin, the raw FIRE score was defined as the total number of chromatin interactions within 200 kb. A custom normalization pipeline, 'HiCNormCis', was modified from the HiCNorm59 and applied to normalize raw FIRE score. Briefly, a Poisson regression model was fitted for each 10 kb bin, with the raw FIRE score as the outcome variable, and three local genomic features, such as effective fragment length, GC content and mappability score, as the covariates. The residuals from the Poisson regression model were used as the normalized FIRE score, which were comparable among different 10 kb bins and different Hi-C data sets. Next, for each 10 kb bin, z-score was calculated based on the normalized FIRE score. 10 kb bins with z-score > 1.64 (P < 0.05) were defined as FIRE bins. More details could be found in Supplementary information, Data S1.
3D-FISH
WT or DKO cells were cultured on laminin-coated coverslips (Neuvitro) for 1 h at 37 °C, and then rinsed with PBS and fixed with 4% PFA in PBS for 10 min. The fixation was quenched with 0.1 Tris-HCl, pH 7.5, for 10 min, rinsed and stored in PBS.
To generate probes, fosmid clone spanning Sox2 SE locus or promoter locus (BACPAC), was labeled with Alexa-fluor 568-5-dUTP or Alexa-fluor 488-5 dUTP (Life Technologies) using Nick Translation Kit (Roche) and incubated at 15 °C for 4 h. The reaction was stopped by 1 μL 0.5 M EDTA, pH 8 and heat-inactivated at 65 °C for 10 min. Unbound dyes were removed using Illustra ProbeQuant G-50 Micro Columns (GE Healthcare) following the manufacturer's instructions. For each hybridization, 100 ng of FISH probes were ethanol precipitated with 10 μg of sonicated salmon sperm DNA, 4 μg of mouse Cot-1 DNA, 1/10th volume of 3 M sodium acetate, pH 5.2, and 2.5 volume of 100% ethanol. Each probe was precipitated and dissolved in 5 μL of formamide and 5 μl of 2× hybridization mix (8× SSC/40% dextran sulfate) at 55 °C for 20 min. The probes were denatured at 75 °C for 5 min before applying to slides.
Fixed cells on coverslips were blocked in 5% BSA/0.1% Triton X-100/1× PBS for 30 min at 37 °C and washed with 0.1% Triton X-100 in PBS. Cover slips were then permeabilized in 0.1% saponin/0.1% Triton X-100/1× PBS for 10 min at room temperature, incubated in 20% glycerol in PBS for 20 min at room temperature, freeze-thawed three times in liquid nitrogen, incubated in 0.1 M HCl for 30 min at room temperature, blocked in 3% BSA and 100 μg/mL RNase A in PBS for 1 h at 37 °C, permeabilized again in 0.5% saponin/0.5% Triton X-100/1× PBS for 30 min at 37 °C and washed in 2× SSC. Cells were denatured with 70% formamide/2× SSC at 73 °C for 2.5 min and with 50% formamide/2× SSC at 73 °C for 1 min, after which denatured probes was applied to the slide. After overnight incubation at 37 °C, cells were washed twice with 50% formamide/2× SSC at 37 °C for 15 min and 2× SSC at 37 °C. The cover slips were stained with DAPI and rinsed in PBS before mounting on the slides with ProLong Gold Antifade Mountant and sealed with nail polish. Images were acquired at 100× magnification on DeltaVision RT Deconvolution microscope, controlled by SoftWorX software. DNA spots were identified and measured using TANGO v.0.93 software. Only cells detected with exactly two loci of each color probe in nucleus was included for analysis to avoid mitotic cell.
RNA-seq, single-cell RNA-seq and data analysis
Total RNA from ES cells was extracted with Trizol according to protocol (Thermo Scientific, 15596-026). PolyA+ RNA was purified with the Dynabeads mRNA purification kit (Life Tech). The mRNA libraries were prepared for strand-specific sequencing using Illumina TruSeq Stranded mRNA Library Prep Kit Set A (Illumina, RS-122-2101) or Set B (Illumina, RS-122-2102). Libraries were sequenced with Illumina Hiseq 2500 for 100 cycles single reads.
For single-cell RNA-seq, cells were collected after trypsin treatment and washed with PBS. For each time point, cell densities were estimated using a hemocytometer and 2 000-5 000 cells were collected for library preparation. The single-cell libraries were prepared with Chromium Single Cell 3′ v2 Library kit (10× Genomics). The libraries were sequenced using Illumina Hiseq 4000 and 16-20 million single-read reads were acquired for each libraries. The single-cell RNA-seq data were analyzed using Cellranger R kit that was provided by 10× Genomics with default parameters. The quality control statistics was listed in Supplementary information, Table S7.
Sequence reads were mapped to mouse mm9 reference genome with TopHat60. The differential expression was analyzed with Cuffdiff61. We plotted the differentially expressed genes if the fold change of adjusted FPKM value between E14 and DKO is larger than 2.
Gene Ontology analysis was carried out using DAVID release 6.7 with default parameters62.
Accession number
Sequencing data have been deposited in Gene Expression Omnibus (GEO) under accession number GSE74055.
Author Contributions
JY, SAC and BR conceived the project. JY, SAC, AL, TL, CMR, CW, KG, SP and ZY carried out experiments. JY, MH and YQ performed data analysis. JY, SAC and BR wrote the manuscript. KMD and JW provided dCD cells. All authors edited the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We would like to thank the Ren Lab members Drs David Gorkin, Tingting Du, Miao Yu, Jason Guoqiang Li, as well as Drs Shicai Fan (UCSD) and Xi Wang (DKFZ, Germany) for comments during manuscript preparation. We are also very grateful to Samantha Kuan and Dr Bin Li for technical assistance. We thank UCSD Neuroscience Microscopy Shared Facility (NS047101) for FISH imaging and the Murre Lab (UCSD) for sharing protocols and discussions for FISH assay. This work was supported by the Ludwig Institute for Cancer Research (BR), NIH (P50 GM085764-04 to BR, U54DK107977 to BR and MH, and R01 GM112720 to JW), an International Postdoc fellowship from the Swedish Vetenskapsrådet (537-2014-6796 to JY) and HHMI (JW).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Mll3/4 Deficient mESCs Reveal Lost Interaction between Sox2 Gene Body and SE, Related to Figure 1
Mll3/4 Are Required for Genome-wide Deposition of H3K4me1 at promoter-distal Enhancers, Related to Figure 1
H3K4me1 does not Affect Global Contact Frequency but is Required for FIREs, Related to Figure 2
H3K4me1 is Required for FIREs, Related to Figure 2
H3K4me1 Assisted Cohesin Binding to Chromatin, Related to Figure 3, 4
Change of H3K4me1 is Associated with Change of Cohesin Binding, Related to Figure 5
Super-enhancer Call Using H3K27ac ChIP-seq Data, Related to Figure 6
H3K4me1 peaks
FIRE score
H3K27ac peaks in different group I-IV
Super-enhancer
Primers used in 4C-seq and shRNA knockdown and qPCR
Antibodies
Overview statictics of single cell RNA-seq
Gene expression values (log2 transformed RPKM) in Wild type and DKO mESC during NPC differentiation, as shown in Figure 5C.
EXTENDED EXPERIMENTAL PROCEDURE
References
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell 2013; 152:1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol 2010; 20:R754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell 2014; 157:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makova KD, Hardison RC. The effects of chromatin organization on variation in mutation rates in the genome. Nat Rev Genet 2015; 16:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 2011; 12:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 1993; 72:73–84. [DOI] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet 2013; 14:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 2014; 14:762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 2012; 485:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol 2011; 357:450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet 2012; 28:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 2012; 13:469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- Rajagopal N, Xie W, Li Y, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS Comput Biol 2013; 9:e1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Wysocka J. Epigenomics of human embryonic stem cells and induced pluripotent stem cells: insights into pluripotency and implications for disease. Genome Med 2011; 3:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Yue F, Li Y, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015; 16:386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 2013; 33:4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2013; 2:e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee JE, Lai B, et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci USA 2016; 113:11871–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res 2017; 45:6388–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi KM, Swigut T, Henriques T, et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell 2017; 66:568–576 e564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, Herz HM, Sze CC, et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat Genet 2017; 49:1647–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012; 149:1233–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 2013; 32:3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci USA 2014; 111:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell 2017; 171:305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, et al. Chromatin architecture reorganization during stem cell differentiation. Nature 2015; 518:331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rivera CM, Ishii H, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One 2014; 9:e114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HY, Katsman Y, Dhaliwal NK, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev 2014; 28:2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013; 153:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Werken HJ, de Vree PJ, Splinter E, et al. 4C technology: protocols and data analysis. Methods Enzymol 2012; 513:89–112. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Jung I, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 2016; 17:2042–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009; 460:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing-Simmons E, Seitan VC, Faure AJ, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res 2015; 25:504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Fudenberg G, Mehta S, et al. Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature 2014; 516:432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 2008; 451:796–801. [DOI] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 2013; 154:801–813. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 1999; 400:284–288. [DOI] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 2009; 34:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 2010; 339:240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci 2013; 3:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 2014; 159:374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Li Y, Dixon JR, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013; 503:290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature 2012; 489:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012; 148:458–472. [DOI] [PubMed] [Google Scholar]
- Fang F, Xu Y, Chew KK, Chen X, Ng HH, Matsudaira P. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells 2014; 32:1805–1816. [DOI] [PubMed] [Google Scholar]
- Hu M, Deng K, Selvaraj S, Qin Z, Ren B, Liu JS. HiCNorm: removing biases in Hi-C data via Poisson regression. Bioinformatics 2012; 28:3131–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mll3/4 Deficient mESCs Reveal Lost Interaction between Sox2 Gene Body and SE, Related to Figure 1
Mll3/4 Are Required for Genome-wide Deposition of H3K4me1 at promoter-distal Enhancers, Related to Figure 1
H3K4me1 does not Affect Global Contact Frequency but is Required for FIREs, Related to Figure 2
H3K4me1 is Required for FIREs, Related to Figure 2
H3K4me1 Assisted Cohesin Binding to Chromatin, Related to Figure 3, 4
Change of H3K4me1 is Associated with Change of Cohesin Binding, Related to Figure 5
Super-enhancer Call Using H3K27ac ChIP-seq Data, Related to Figure 6
H3K4me1 peaks
FIRE score
H3K27ac peaks in different group I-IV
Super-enhancer
Primers used in 4C-seq and shRNA knockdown and qPCR
Antibodies
Overview statictics of single cell RNA-seq
Gene expression values (log2 transformed RPKM) in Wild type and DKO mESC during NPC differentiation, as shown in Figure 5C.
EXTENDED EXPERIMENTAL PROCEDURE