Abstract
Background
Identification of differentially methylated regions (DMRs) is the initial step towards the study of DNA methylation-mediated gene regulation. Previous approaches to call DMRs suffer from false prediction, use extreme resources, and/or require library installation and input conversion.
Results
We developed a new approach called Defiant to identify DMRs. Employing Weighted Welch Expansion (WWE), Defiant showed superior performance to other predictors in the series of benchmarking tests on artificial and real data. Defiant was subsequently used to investigate DNA methylation changes in iron-deficient rat hippocampus. Defiant identified DMRs close to genes associated with neuronal development and plasticity, which were not identified by its competitor. Importantly, Defiant runs between 5 to 479 times faster than currently available software packages. Also, Defiant accepts 10 different input formats widely used for DNA methylation data.
Conclusions
Defiant effectively identifies DMRs for whole-genome bisulfite sequencing (WGBS), reduced-representation bisulfite sequencing (RRBS), Tet-assisted bisulfite sequencing (TAB-seq), and HpaII tiny fragment enrichment by ligation-mediated PCR-tag (HELP) assays.
Electronic supplementary material
The online version of this article (10.1186/s12859-018-2037-1) contains supplementary material, which is available to authorized users.
Keywords: Epigenetics, DNA Methylation, WGBS, Differentially Methylated regions (DMR), RRBS, Bisulfite sequencing
Background
DNA methylation plays a critical role in gene regulation [1]. In human somatic cells, 70–80% of all CpG dinucleotides in the genome are methylated [2]. DNA methylation represents one type of epigenetic modification which has been shown to control transcription in mammals [3], interacting sometimes with DNA binding proteins [4]. DNA methylation regulates many diverse biological functions, such as embryonic stem cell differentiation [5], aging [6], gene imprinting [7, 8], and X-chromosome inactivation [9]. DNA methylation is conserved and somatically heritable mark that is generally associated with transcriptional repression [10]. Aberrant methylation has been found in multiple diseases such as cancer [11], imprinting defects [12] and mental disorders such as schizophrenia [13, 14]. Environmental exposures such as uteroplacental insufficiency [15, 16] or cigarette smoking [17, 18] have also been observed to alter DNA methylation.
Recent developments in sequencing technology enabled genome-wide characterization of DNA methylation. Whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS) have been widely used to measure DNA methylation at a single CpG resolution [19]. DMRs are the contiguous genomic regions whose DNA methylation status differs between two groups of samples. DMRs have been used to characterize cell-type or condition specific DNA methylation [20–22].
Computational approaches have been developed based on statistical frameworks to identify DMRs. BSmooth [23] identifies DMRs using a local-likelihood approach to estimate a sample-specific methylation profile. For a statistical test between samples, BSmooth uses a Welch’s t-test [24]. MethylKit identifies DMRs based on logistical regression if multiple replicates are available, or a Fisher’s exact test [25] if only one sample is available. MethylSig [26] uses a beta-binomial approach to identify DMRs based on read coverage and biological variability, which showed high sensitivity in comparison with MethylKit, BSmooth, a standard t-test and the Wilcoxon rank test. Metilene [27] uses a binary segmentation algorithm combined with a two-dimensional Kolmogorov-Smirnov test. Genomic regions are pre-segmented, and gradually reduced in size until the region contains less than a defined minimum number of CpGs or statistical significance is not improved. RADMeth [28] employs a beta-binomial regression and a Stouffer-Liptak test. And RnBeads [29] calculates p-values for each CpG in the data set using hierarchical linear models and M-values [30]. BiSeq uses a smooth-based approach while considering coverage to call DMRs [31].
With the exception of Metilene, many well-known DMR callers require knowledge of the R programming language [32] or even a specific version of R (MethylKit & BiSeq). In addition, each DMR caller requires specific input formats to run it properly. Furthermore, many of them use extreme computing resources for genome-scale analysis. Table 1 summarizes the characteristics and algorithms of the widely used DMR callers we investigated.
Table 1.
Comparison of DMR calling software
| Program | DMR Identification | Execution |
|---|---|---|
| Defiant | Weighted Welch Expansion | Binary |
| BSmooth [23] | Local-likelihood smoothing with binomial test | R |
| MethylKit [25] | Fisher’s exact test [37] or logistic regression with tiling | R |
| MethylSig [26] | beta-binomial [64] | R |
| Metilene [27] | p-value by beta binomial | Binary |
| MOABS [65] | beta-binomial | Binary |
| RADMeth [28] | beta-binomial regression | Binary |
The numerous disadvantages of these programs/statistical methods prompted the development of a new DMR-identification program. Defiant is a standalone program following GNU99 standard, which reduces the issues of portability. Defiant automatically detects ten different input formats widely used for DNA methylation. It does not require installation of any libraries and runs with a single command line. We evaluate Defiant’s performance with comprehensive benchmarking tests using both artificial and real WGBS data. We applied Defiant to analyzing the DNA methylation changes induced by iron deficiency during the critical neuro-developmental period (fetus and newborn) in the rat hippocampus.
Implementation
Animals and Hippocampal dissection
G2 pregnant CD1 Sprague-Dawley rats were purchased from Charles Rivers Laboratories. The experimental conditions for induction of fetal-neonatal iron deficiency were following the previously described protocol [33]. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Minnesota. Hippocampal dissection and storage from PND15 rats was performed as previously described [33]. Genomic DNA from PND15 rat hippocampi was isolated using Allprep DNA/RNA mini kit (Qiagen).
WGBS & data processing
WGBS was performed as a published protocol [34]. Briefly, 1 μg of genomic DNA was fragmented into 300 bp size using M220 Covaris Ultrasonicator. Sequencing libraries were generated using NEBNext genomic sequencing kit (New England Biolabs) and ligated with Illumina methylated paired end adaptors. Libraries were bisulfite-converted using Imprint DNA modification kit (Sigma), and the size of 300–600 bp was selected using Pippin Prep DNA size selection system (Sage Science). Libraries were then amplified using PfuTurbo Cx Hotstart DNA polymerase (Agilent Technologies). Samples were sequenced to 100 bps in either paired-end or single-read formation on an Illumina HiSeq 2000 with RTA version 1.13.48 and HiSeq control software version HiSeqCS:1.5.15.1. Adpaters were trimmed from the reads using a custom C language program. Trimmed reads were aligned against the rat genome (rn4) using bs seeker (v1) [35]. The methylation status was then tallied from the bs seeker output. When reads overlapped at a base, the methylation status from read 1 was used. Methylation data at the C and G in a CpG pair was merged to produce the estimate at that locus. The WGBS data have been deposited in the GEO repository (GSE98064).
Identification of DMRs using WWE
Defiant defines DMRs based on seven criteria. These can also be specified by the end-user.
All nucleotides in all samples are present and meet minimum coverage (default 10). The user can specify that some nucleotides can be missing from certain replicates, but we recommend against it as it can introduce false positives.
- Absolute value in the difference of the sum of the methylation percentages is above a given cutoff %. The default is 10%, but this will vary in every individual experiment based on the chemistry [36]. The mean methylation percentage, , is weighted based on coverage, i.e.
where Cr is the coverage for replicate r, mCr is the number of 5mC for replicate r, and R is the number of replicates.1 A 2-tailed p-value, default 0.05
If there is only 1 replicate in either sample, a p-value between groups A and B is calculated by Fisher’s exact test [37]
| 2 |
where mC = number of 5-methyl Cystosine and C = number of Cytosine.
If there are multiple replicates in both groups, the p-value is based on Welch’s t-test [24] in sum of methylation percentages is below a given cutoff.
If both samples have multiple replicates, the t-test between groups A and B is calculated thus:
| 3 |
where the unbiased sample variance s for any group A is also weighted based on coverage:
| 4 |
The Benjamini-Hochberg [38] approach is applied to the identified DMRs to adjust p-value for multiple testing.
a minimum number of CpN constituting differentially methylated region (default CpN = 5),
a minimum range of the differentially methylated nucleotides (default 0)
a maximum range between CpN (default 20,000 nucleotides).
a maximum similar, i.e. non-differentially methylated, CpG count (default 5). If a DMR is currently expanding, the DMR expands until one criterion of differential methylation stops. By allowing a similar CpG count, DMRs can contain similarly methylated CpG inside the DMR. Once the similar nucleotide is broken, the DMR shrinks to the point where differential methylation stopped.
All criteria are easily set by command-line options. Defiant is designed to test multiple parameters in parallel, to make the final decision on each parameter up to the end user. Defiant has a companion program in Perl, plot_results.pl, which plots the number of DMRs as a function of different parameters using GNUPlot if a user has chosen to test multiple parameters. The effects of the variation of each parameter on the number of DMRs found in the rat hippocampus data can be seen in (Additional file 1: Figures S2-S16).
The number of DMR is sensitive to CpN, differentially methylated CpN (d), and p-value, but less sensitive to minimum percent change, minimum coverage. Minimum CpN = 5 is a point where the number of DMR is large enough and less sensitive to minimum coverage (Additional file 1: Figure S7). Minimum coverage determines the number of DMRs. The default (10) is the point where the number of DMR is robust to the “Differentially methylated CpN” (Additional file 1: Figure S8). We set a large number for “Maximum range for CpN” (default = 20,000). The difference in the methylation percentage (default 10%) can further be used to identify subtle but significant changes. A minimum range between CpN is not used for the rat data (default 0) but provide a user with flexibility in defining DMRs.
Comparison with other methods
DMR Overlay is the percent of nucleotides predicted by the program inside of the artificial DMR. DMR overlay is measured in two directions, once with respect to the benchmark DMRs and once with respect to the predicted DMRs (Fig. 5). For example, in Fig. 1, the overlay of predicted DMR 1 with respect to Benchmark DMR “A” is calculated as
while for the reverse, DMRs A and B would each have their own DMR overlay equal to 100%. This is similar, but more informative than a Jaccard index, which is symmetric while DMR overlay depends on direction of comparison.
Defiant is run with default minimum coverage of 10, p = 0.05, minimum percent change of 10%, and five CpN (CpG) in each DMR.
MethylKit [25] scans for DMRs using a tiling window of 1000 nucleotides and a step size of 1000 nucleotides. Consecutive windows that score q < 0.05 are considered as a single DMR. Tiles can report q = 0 with p ≈ 1, so tiles with q = 0 are treated as non-DMRs. MethylSig [26] also uses a tiling approach, with a window and step size of 25 nucleotides. As for MethylKit, consecutive windows with q < 0.05 are considered as a single DMR.
Metilene [27] is a command-line program, and was run according to defaults. Metilene was run with a minimum CpG count of 5 with the rat data to make results comparable to Defiant.
RADMeth [28] is run as a series of three commands to the Linux shell. RADMeth showed itself superior to ComMet [39] and DSS [40] so we do not compare these methods here.
RnBeads [29] differentially methylated regions are defined by 500 nucleotide tiles. Similarly with MethylKit and MethylSig, consecutive tiles with q < 0.05 are considered as a single DMR.
Fig. 5.
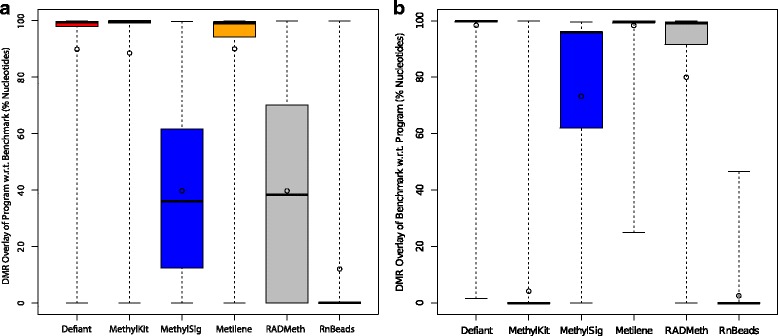
Comparison of DMR overlay of program with respect to benchmark in panel (a), and benchmark with respect to program in panel (b) for the combined RRBS and WGBS data sets
Fig. 1.
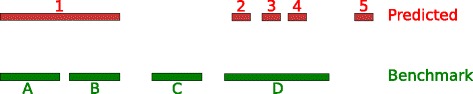
Illustration of how different DMR results are compared. Artificial (or “Benchmark”) DMRs are depicted in green, while predicted DMRs are shown in red
Results
DMR identification by weighted Welch expansion
Defiant calculates a p-value using a Welch’s t-test for the weighted means and variance (Method). A weight is to give more credence to the replicate with a high coverage. If there is only one replicate in either set, Welch’s t-test cannot be used, and thus Defiant uses a Fisher’s exact test. For accurate detection of the boundary of a DMR, Defiant detects the start point based on the following factors: differences in methylation levels, coverage, p-value, and a minimum number of CpGs. If all of these criteria are satisfied, the next nucleotide is checked for constituting differential methylation, i.e. the DMR expands. It terminates when there is a number of consecutive CpGs that do not pass the criteria. We call this algorithm weighted Welch expansion (WWE).
Figure 2 demonstrates how Defiant identifies DMRs using WWE. Defiant calculates the weighted mean based on the coverage. The weighted mean for the two datasets (“B” and “F”) are shown on the top two panels. The p-values were calculated using a Welch’s t-test (on the third panel). The DMR on the left side starts at 120,124 when the difference in methylation levels is − 88% and p = 7.7 × 10− 20. The DMR expands as long as it sees CpGs that pass the criteria. At 120,052, Defiant stops its expansion when it observes at least 5 consecutive CpGs that fail the criteria. Defiant does not require setting the size of a window or “tiles” as is done in MethylKit [25], so the data determines the size of the differentially methylated regions. This approach gives much greater flexibility and power in analyzing the data, as small differentially methylated regions can be easily missed when using a tiling approach.
Fig. 2.
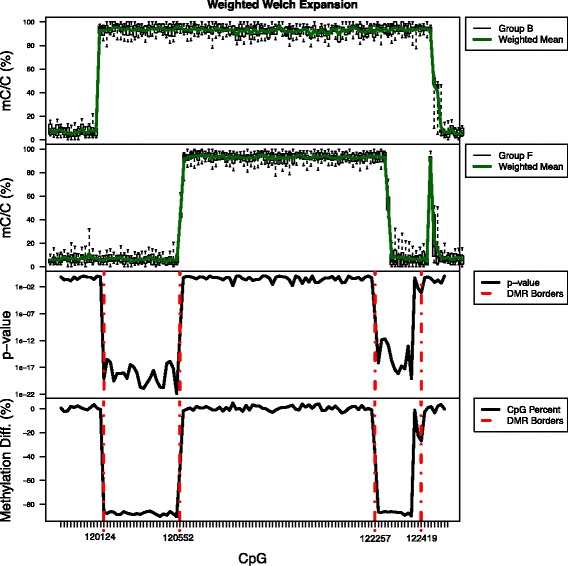
Defiant uses WWE for DMR identification. We used artificial data designed for Metilene [27]. Each group is composed of ten replicate DNA methylation samples. The top two panels show the level of DNA methylation for each CpG (box-and-whisker plots). The mean is weighted based on coverage. The third panel from the top shows the weighted Welch p-value between the sets for individual CpG. The bottom panel shows differences between the weighted mean. Defiant calls a DMR when it finds consecutive CpGs with 1) differences in methylation levels, 2) minimum coverage, 3) p-value. When Defiant finds consecutive CpGs that do not match the above criteria, the expansion of a DMR stops. The third and fourth panels show the DMR start and end points in red
Datasets
We used two datasets to evaluate Defiant’s performance.
Artificial benchmarking datasets used for Metilene [27]
Metilene [27] simulated RRBS and WGBS datasets using beta binomial distribution. The dataset are composed of two different backgrounds, each with four subsets. These are named 1.1 (strongest methylation differences) through 1.4 (weakest differences) for the first background, and 2.1 through 2.4 for the second (Additional file 1: Figure S1). In total, there are 16 subsets (eight for RRBS and eight for WGBS) to test DMR calling for various data configurations. Each subset has ten replicates. We downloaded them from http://www.bioinf.uni-leipzig.de/Software/metilene/Downloads/.
WGBS data from postnatal day 15 iron deficient and iron sufficient rat hippocampi
WGBS data were generated to analyze changes in hippocampal DNA methylation due to iron-deficiency during the fetal and neonatal periods. Pregnant/nursing rat dams were fed an iron-deficient diet (4 ppm iron) from gestational day 2 through postnatal day (P) 7, at which time they were given an iron-sufficient control diet (200 ppm iron). Iron sufficient control rats were fed an iron sufficient diet through the entire experimental duration. At P15, rat pups from both groups were euthanized and hippocampi were isolated. This diet manipulation induced a 60% iron deficiency in the P15 rat hippocampus [41, 42]. Three biological replicates of WGBS data were generated from hippocampi of both groups.
Performance evaluation
We evaluate the DMR-identification programs Defiant, Metilene, MethylKit, MethylSig, RadMeth, and RnBeads. We chose not to use BSmooth and MOABS as they have already shown inferior performance as compared to Metilene [27] using the same test set we use in this experiment. Using the DMRs in the artificial datasets as the gold standard, we defined true positive (TP) when a predicted DMR overlapped with a DMR in the benchmark, otherwise it was defined as a false positive (FP). False negative (FN) was defined when a DMR in the benchmark dataset was not predicted. FN, FP, and TP values for all DMR callers for each artificial dataset are listed in Additional file 1: Table S1.
For comprehensive evaluation, we compared precision (TP/(TP + FP)) against FN as well as against recall (TP/(TP + FN)) for 8 sets of RRBS and WGBS data (Fig. 3). In these tests, both Defiant and Metilene showed excellent precision and recall with very low FNs compared with other DMR callers. They scored perfect precision because their FP is 0 for all 16 tests. RnBeads also showed FP equal or close to zero. However, it suffered a high FN (Fig. 3). MethylKit showed the worst performance in these tests mainly due to excessive number of FPs. MethylSig showed high number of FPs for WGBS datasets (Additional file 1: Table S1). Moreover, MethylSig did not predict any DMRs for RRBS datasets (TPs and FPs were 0). RADMeth scored moderately well, but scored behind both Defiant and Metilene.
Fig. 3.
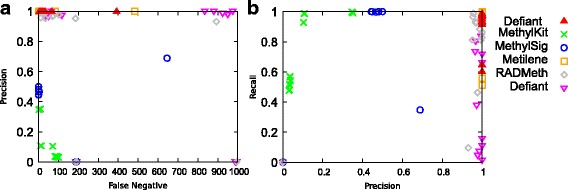
Performance comparison using all 16 Metilene’s benchmark datasets composed of RRBS and WGBS. Each point represents a data set. Precision is compared against FN (a) and recall against precision (b). In plot a, 15 points from Defiant, 15 from Metilene, 10 from RADMeth, and 5 from RnBeads overlap in the upper-left corner. Both Defiant and Metilene outperformed other predictors
To obtain the overall performance we calculated the F1 score (=. The F1 score is the harmonic mean of precision and recall. Both Defiant and Metilene showed outperforming F1 scores compared with other predictors (Fig. 4). The performances between Defiant and Metilene were comparable when we investigated the statistical differences between the predictions using a Welch’s t-test [24] (p = 0.81, Additional file 1: Table S2). Considering that the artificial datasets were generated using a beta binomial distribution, providing a favorable environment for Metilene, the comparable performance of Defiant is notable. Examining the results more closely, we found that Defiant outperformed Metilene especially for background 2, dataset 4 where there were subtle but significant differences in DNA methylation levels (Fig. 3 and Additional file 1: Table S1). To examine the DMR calling boundary more carefully, we calculated the overlapping ratios of DMRs, quantified as “DMR overlay” (Methods). Compared with Metilene, Defiant showed better overlay with respect to the predicted DMRs, suggesting that DMRs that Defiant calls capture the robust portion of DMRs (Fig. 5).
Fig. 4.
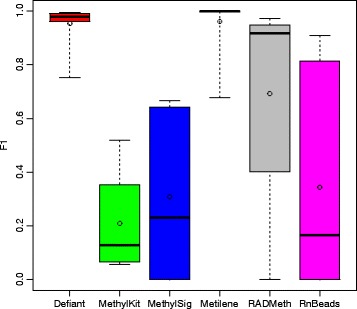
F1 values of 16 different artificial data sets. Circles indicate mean values. Both Defiant and Metilene outperformed other predictors
Comparison using real WGBS data
We applied Defiant to identify DMRs in the iron-deficient rat hippocampus using the same parameters as were used with the artificial data. Fetal-neonatal iron deficiency, which is one of the most common nutritional deficiencies in the world [43], affecting as many as 2 billion people and approximately 30% of pregnancies [44, 45], causing long-term cognitive deficits despite iron treatment [46, 47]. Because these changes persist into adulthood even in the face of normal iron levels, and the known link between fetal exposures and long-term outcomes after birth, we investigated whether DNA methylation is altered in the developing rat hippocampus induced by fetal-neonatal iron-deficiency using WGBS datasets. The methods used to induce fetal-neonatal iron-deficiency have been previously described [48]. We investigated if DNA methylation is affected by iron-deficiency during fetal-neonatal periods using WGBS datasets with three iron-deficient rats and three iron sufficient rats.
We compared performance of Defiant to that of Metilene because they showed comparable performances based on the benchmarking test. Between the iron-deficient and iron-sufficient groups, Defiant identified 229 DMRs, while Metilene identified only 80 DMRs, with ten regions showing overlap between the two approaches. Figure 6 showed the DNA methylation status of the DMRs identified both by Defiant and Metilene.
Fig. 6.
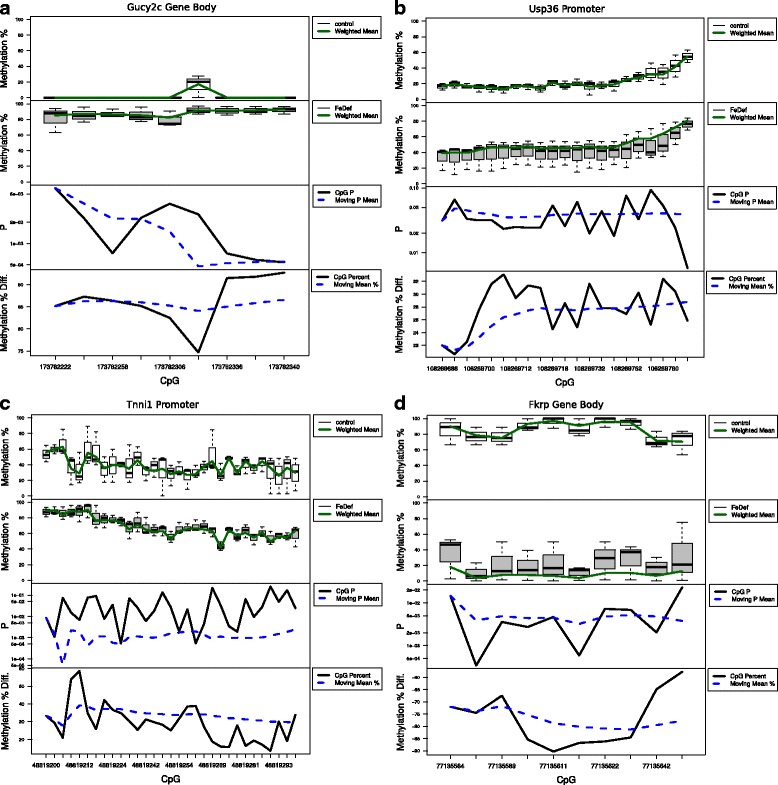
An iron-deficient diet induces strong changes in methylation for many genes, the strongest of which are shown here. a Gucy2c displays strongly increased methylation from an Fe-deficient diet. b Usp36 is hypermethylated in Fe-deficient pups. c Tnni1 shows moderate, but consistently significant hypermethylation. d Fkrp shows strong and consistent hypomethylation
We found that the DMRs identified by Metilene but not by Defiant were mostly in low coverage areas. When methylation levels were weighted by the coverage, they showed non-significant p-values (Fig. 7a and b). Indeed, compared with the artificial datasets which have the distribution of coverage in a short range, the WGBS data in rat hippocampus were with a wider range of coverage (Fig. 7c). These results indicate a superior performance of Defiant when applied to real WGBS data. Figure 8 shows the examples of DMRs detected by Defiant but not by Metilene. The DNA methylation profiles showed clear differences in DNA methylation between the iron sufficient and the iron deficient groups for regions near genes such as Pde6c, Chd2, Mobp, and Pck1. The differences between Defiant’s and Metilene’s predictions are due to Defiant’s use of coverage-weighted means. The coverage-weighted means used in WWE allow Defiant to avoid artifacts such as Simpson’s paradox [49].
Fig. 7.
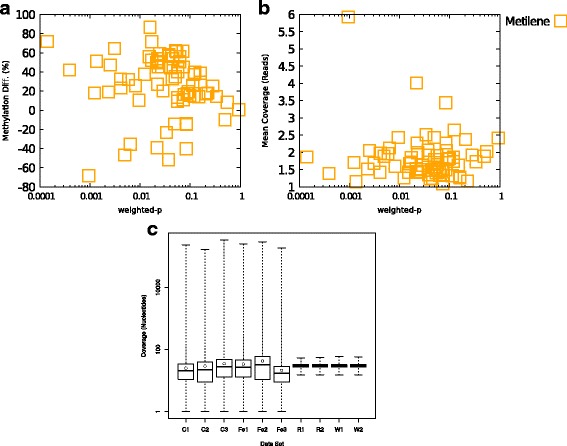
Statistics on CpG regions predicted uniquely by Metilene. While, these genes show differences in the methylation levels, their p-values were unacceptably high when coverage information is considered. a The differences in the methylation levels over weighted Welch’s p-values. b The mean coverage against the weighted p-value c distribution of coverage. Real WGBS shows much wider distribution of coverage than the artificial data set. “C” = iron sufficient rat samples, “Fe” = Iron-deficient rat samples, “R” = artificial RRBS data, and “W” = artificial WGBS data
Fig. 8.
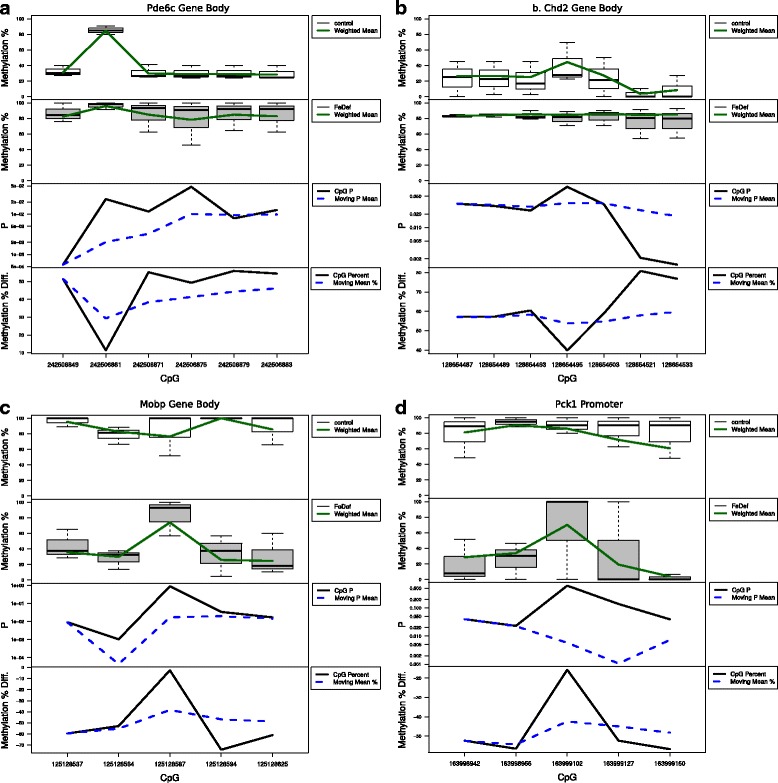
The DMRs identified by Defiant but missed by Metilene. DMRs on a. Pde6c gene body b. Cdh2 gene body, c. Mobp gene gody, and d. Pck1 promoter region
Defiant identifies DMRs close to genes potentially affected by iron-deficiency in fetal-neonatal periods
The 229 DMRs that Defiant identified mapped within 15 Kbps of 108 genes (Additional file 1: Table S3). Among the 108 genes, 45 showed hypomethylated and 63 showed hypermethylated regions (Additional file 1: Tables S4 and S5, respectively). We identified that considerable portion of them (37 out of 43 hypomethylated and 31 out of 62 hypermethylated DMRs) were associated with neuronal function or development (Additional file 1: Tables S4 and S5), corroborating a previous finding [50]. Gene ontology (GO) analysis using Enrichr [51, 52] identified “abnormal nervous system” in Mouse Genome Informatics [53] mammalian phenotype level 3 (p-value = 0.005). Genes associated with this term were Camk2b, Fkrp, Ncf1, St8sia1, Itsn1, Cacna1c, Usf2, Mib1, Fig4, Jph3, Mobp, Ush1g, Prkar1b, Tal1, and Pde6c. We also observed terms “Rho GTPase cycle” (p-value: 0.0005), and “Axon guidance” (p-value = 0.001). In total, 7 out of 43 and 12 out of 62 genes were associated with GTPase activity for gain and loss of DNA methylation levels, respectively. The rho family of GTPases [54, 55], one of the G-protein coupled receptors, regulates neuronal morphogenesis [56], dendritogenesis [57], and spinogenesis [58]. We also found terms “synapse part” (p-value = 0.017; Arf1, Tenm2, Pde2a, Cacna1c, Srgap2, and Mib1). Collectively, Defiant identified DMRs close to the genes critical for neuronal synaptogenesis and plasticity.
Defiant showed best performance based on the running time
Running time is important for genome-scale analysis. We evaluated the performance based on resource use. Using the benchmarking datasets, all pertinent DMR callers were run on the same system (Intel Xeon CPU 2.1GHz with 32GB RAM). For fair comparison, we converted the format of the benchmarking datasets into the format preferred by each DMR caller before evaluation. Therefore, mapping and input format conversion were not needed for this benchmarking. We observed dynamic range of running time and memory use. Defiant showed much faster performance compared with other methods: 5× faster than Metilene, 10× faster than MethylKit and MethylSig, and > 300× faster than RADMeth and RnBeads. When we applied Defiant to our WGBS rat datasets on the same system, it took less than 2 min to obtain the DMR results for the entire genome. Defiant’s memory usage is light, about 1GB for genome-wide WGBS data. Compared to the memory usages of the DMR callers, Defiant used slightly more memory than Metilene because Defiant is designed to run in a single step.
Conclusions
We developed Defiant a new method to identify DMRs. Defiant is designed to provide easy and fast implementation of DMR calling while guaranteeing the prediction performance. We also put more credence to the CpGs with high coverage. For a rigorous test while weighing DNA methylation based on coverage, we used a Welch’s t-test. A Welch’s t-test does not assume equal variation between the sets and simplifies the incorporation of the weight information.
One of the widely used approaches for modeling DNA methylation is a beta binomial distribution. In our benchmarking tests, Defiant showed superior performance to other beta binomial distribution based predictors such MethylSig, MOABS, and RADMeth. It is noteworthy that the artificial data we used were generated by the developers of Metilene using a beta binomial distribution. Despite the potential bias against it, Defiant showed comparable performance with Metilene. Our results indicate that using a Welch’s t-test is appropriate for DMR identification. Close examination found that Defiant performed better then Metilene when modest but significant differences were observed (Fig. 3). More importantly, Defiant identified more DMRs in the rat hippocampus datasets which showed clear DMRs (Fig. 8). The DMRs uniquely observed by Metilene were in very low coverage areas (Fig. 7). Together, these suggest that Defiant performs better than Metilene for analysis of real data.
For genome-scale analysis, reduced running time is highly desirable. In our test, Defiant ran remarkably faster than other competitors using memory less than 1GB (Fig. 9). When applied to the whole-genome data in rats, it took less than 2 min for DMR calling for the entire genome. Defiant accepts diverse formats for DNA methylation including the format for Bismark coverage, and Bismark cytosine [59], BisSNP [60], MethylKit & MethylSig input, UCSC ENCODE, and EPP [61]. Defiant also runs as a standalone software.
Fig. 9.
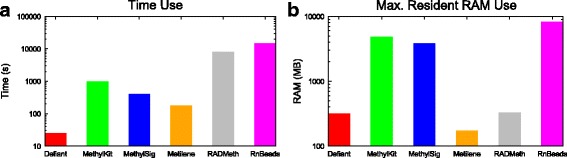
Resources used by DMR callers in time used (panel a) and RAM (panel b). We used eight artificial WGBS data sets. All evaluation was done on a computer with Intel(R) Xeon(R) CPU E5–2620 v2 @ 2.10GHz with 32 GB RAM
For convenient analysis, Defiant provides annotation about the genes located around DMRs. Additional file 1: Table S3 shows the example of output of Defiant. Defiant is applicable to bisulfite-sequencing data, RRBS [19], HpaII tiny fragment enrichment by ligation-mediated PCR-tag (HELP-Tag) data [62], and Tet-assisted bisulfite sequencing (TAB-Seq) [63]. The source code is available on http://github.com/hhg7/defiant.
Acknowledgements
Not applicable.
Funding
We thank the Functional Genomics Core of the Penn Diabetes Research Center (P30- DK19525). This work is supported by NIH grant (R01 DK106027 to KJW and R01 HD029421 to MG).
Availability and requirements
Input files for all programs compared with benchmark artificial data, Defiant source code, and installation instructions are available on http://github.com/hhg7/defiant. Data is publicly available on the Gene Expression Omnibus under GSE98064.
Abbreviations
- DMR
Differentially methylated region
- GO
Gene ontology
- RRBS
Reduced Representation Bisulfite Sequencing
- WGBS
Whole Genome Bisulfite Sequencing
- WWE
Weighted Welch Expansion
Additional file
Supplementary document. (PDF 475 kb)
Authors’ contributions
DC developed the Defiant method, wrote the source code, and compared performance. PT and YL performed bisulfite sequencing for the rat hippocampus. RS and KW conceived the project. YL, DC, RS and KW designed the experiment. JS prepared the preprocessing for the DNA methylation data and helped implementation of various DMR callers. DC, PT, RS, JS, YL, and KW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All experimental procedures involving the use of live animals were approved by the Institutional Animal Care and Use Committee review boards of the University of Minnesota, the Children’s Hospital of Philadelphia, and the University of Pennsylvania.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12859-018-2037-1) contains supplementary material, which is available to authorized users.
Contributor Information
David E. Condon, Email: dec986@gmail.com
Phu V. Tran, Email: tranx271@umn.edu
Yu-Chin Lien, Email: ylien@mail.med.upenn.edu.
Jonathan Schug, Email: jschug@mail.med.upenn.edu.
Michael K. Georgieff, Email: georg001@umn.edu
Rebecca A. Simmons, Email: rsimmons@upenn.edu
Kyoung-Jae Won, Email: kyoung.won@bric.ku.dk.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Ehrlich M, Gama-Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 1982;10(8):2709. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy M-K, Movassagh M, Goh H-G, Bennett MR, Down TA, Foo RS. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Gen. 2010;11(1):1–10. doi: 10.1186/1471-2164-11-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, Schubeler D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 5.Altun G, Loring JF, Laurent LC. DNA methylation in embryonic stem cells. J Cell Biochem. 2010;109(1):1–6. doi: 10.1002/jcb.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson V, Jones P. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 7.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. TIG. 1991;7(2):45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 9.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9(19):2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 10.Ushijima T, Watanabe N, Okochi E, Kaneda A, Sugimura T, Miyamoto K. Fidelity of the Methylation pattern and its variation in the genome. Genes Res. 2003;13(5):868–874. doi: 10.1101/gr.969603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6 10.1038/nrg1655. [DOI] [PubMed]
- 13.Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, Yoshikawa T, Kato T. DNA Methylation status of SOX10 correlates with its Downregulation and Oligodendrocyte dysfunction in schizophrenia. J Neurosci. 2005;25(22):5376–5381. doi: 10.1523/JNEUROSCI.0766-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H-S, Akbarian S. GAD1 mRNA expression and DNA Methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2(8):1–6. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham TD, MacLennan NK, Chiu CT, Laksana GS, Hsu JL, Lane RH. Uteroplacental insufficiency increases apoptosis and alters p53 gene methylation in the full-term IUGR rat kidney. Am J Physiol Regul Integr Comp Physiol. 2003;285(5):962–970. doi: 10.1152/ajpregu.00201.2003. [DOI] [PubMed] [Google Scholar]
- 16.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, Slieker RC, Stok AP, Thijssen PE, Müller F, van Zwet EW, Bock C, Meissner A, Lumey LH, Eline Slagboom P, Heijmans BT. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592. doi: 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, Strauch K, Waldenberger M, Illig T. Tobacco smoking leads to extensive genome-wide changes in DNA Methylation. PLoS One. 2013;8(5):1–14. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, Guan W, Xu T, Elks CE, Aslibekyan S, Moreno-Macias H, Smith JA, Brody JA, Dhingra R, Yousefi P, Pankow JS, Kunze S, Shah SH, McRae AF, Lohman K, Sha J, Absher DM, Ferrucci L, Zhao W, Demerath EW, Bressler J, Grove ML, Huan T, Liu C, Mendelson MM, Yao C, Kiel DP, Peters A, Wang-Sattler R, Visscher PM, Wray NR, Starr JM, Ding J, Rodriguez CJ, Wareham NJ, Irvin MR, Zhi D, Barrdahl M, Vineis P, Ambatipudi S, Uitterlinden AG, Hofman A, Schwartz J, Colicino E, Hou L, Vokonas PS, Hernandez DG, Singleton AB, Bandinelli S, Turner ST, Ware EB, Smith AK, Klengel T, Binder EB, Psaty BM, Taylor KD, Gharib SA, Swenson BR, Liang L, DeMeo DL, O’Connor GT, Herceg Z, Ressler KJ, Conneely KN, Sotoodehnia N, Kardia SLR, Melzer D, Baccarelli AA, van Meurs JBJ, Romieu I, Arnett DK, Ong KK, Liu Y, Waldenberger M, Deary IJ, Fornage M, Levy D, London SJ. Epigenetic signatures of cigarette SmokingCLINICAL PERSPECTIVE. Circ Cardiovasc Genet. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33(18):5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak P, Stampfer MR, Munoz-Rodriguez JL, Garbe JC, Ehrich M, Futscher BW, Jensen TJ. Cell-type specific DNA Methylation patterns define human breast cellular identity. PLoS One. 2012;7(12):1–9. doi: 10.1371/journal.pone.0052299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gervin K, Page CM, Aass HCD, Jansen MA, Fjeldstad HE, Andreassen BK, Duijts L, van Meurs JB, van Zelm MC, Jaddoe VW, Nordeng H, Knudsen GP, Magnus P, Nystad W, Staff AC, Felix JF, Lyle R. Cell type specific DNA methylation in cord blood: a 450K-reference data set and cell count-based validation of estimated cell type composition. Epigenetics. 2016;11(9):690–698. doi: 10.1080/15592294.2016.1214782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakatsu T, Stuart T, Valdes M, Breakfield N, Schmitz RJ, Nery JR, Urich MA, Han X, Lister R, Benfey PN, Ecker JR. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nature Plants. 2016;2 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed]
- 23.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13(10):83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch BL. The generalization of Student’s problem when several different population variances are involved. Biometrika. 1947;34(1–2):28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 25.Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13:87. 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed]
- 26.Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30(17):2414–2422. doi: 10.1093/bioinformatics/btu339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jühling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF, Hoffmann S. Metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2015; 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed]
- 28.Dolzhenko E, Smith AD. Using beta-binomial regression for high-precision differential methylation analysis in multifactor whole-genome bisulfite sequencing experiments. BMC Bioinformatics. 2014;15(1):215. doi: 10.1186/1471-2105-15-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assenov Y, Mueller F, Lutsik P, Walter J, Lengauer T, Bock C. Compehensive analysis of DNA Methylation data with RnBeads. Nat Methods. 2014;11(11):1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11(1):1–9. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebestreit K, Dugas M, Klein H-U. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics. 2013;29(13):1647–1653. doi: 10.1093/bioinformatics/btt263. [DOI] [PubMed] [Google Scholar]
- 32.Team, R.C.: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2015). R Foundation for Statistical Computing. https://www.R-project.org
- 33.Tran PV, Kennedy BC, Pisansky MT, Won K-J, Gewirtz JC, Simmons RA, Georgieff MK. Prenatal choline supplementation diminishes early-life iron deficiency induced reprogramming of molecular networks associated with behavioral abnormalities in the adult rat hippocampus. J Nutr. 2016;146(3):484–493. doi: 10.3945/jn.115.227561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schbeler D, Kaestner KH. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28(6):652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P-Y, Cokus SJ, Pellegrini M. BS seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11(1):203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marabita F, Almgren M, Lindholm ME, Ruhrmann S, Fagerström-Billai F, Jagodic M, Sundberg CJ, Ekström TJ, Teschendorff AE, Tegner J, Gomez-Cabrero D. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013;8(3):333–346. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;85(1):87–94. doi: 10.2307/2340521. [DOI] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 39.Saito Y, Tsuji J, Mituyama T. Bisulfighter: accurate detection of methylated Cytosines and differentially methylated regions. Nucleic Acids Res. 2014;42(6):45. doi: 10.1093/nar/gkt1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng H, Conneely KN, Wu H. A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res. 2014;42(8):69. doi: 10.1093/nar/gku154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgenson LA, Wobken JD, Georgieff MK. Perinatal iron deficiency alters apical Dendritic growth in Hippocampal CA1 pyramidal neurons. Dev Neurosci. 2003;25 10.1159/000075667. [DOI] [PubMed]
- 42.Jorgenson LA, Sun M, O’Connor M, Georgieff MK. Fetal iron deficiency disrupts the maturation of synaptic function and efficacy in area ca1 of the developing rat hippocampus. Hippocampus. 2005;15(8):1094–1102. doi: 10.1002/hipo.20128. [DOI] [PubMed] [Google Scholar]
- 43.Carlson ES, Tkac I, Magid R, O’Connor MB, Andrews NC, Schallert T, Gunshin H, Georgieff MK, Petryk A. Iron is essential for neuron development and memory function in mouse hippocampus. J Nutr. 2009;139(4):672–679. doi: 10.3945/jn.108.096354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao R, Georgieff MK. Iron in fetal and neonatal nutrition. Semin Fetal Neonat M. 2007;12(1):54–63. doi: 10.1016/j.siny.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLean E, Cogswell M, Egli I, Wojdyla D, de Benoist B. Worldwide prevalence of anaemia, WHO vitamin and mineral nutrition information system, 1993-2005. Public Health Nutr. 2009;12(4):444–454. doi: 10.1017/S1368980008002401. [DOI] [PubMed] [Google Scholar]
- 46.Lozoff B. Iron deficiency and child development. Food Nutr Bull. 2007;28(4):suppl4), 560–suppl4), 571. doi: 10.1177/15648265070284S409. [DOI] [PubMed] [Google Scholar]
- 47.Lukowski AF, Koss M, Burden MJ, Jonides J, Nelson CA, Kaciroti N, Jimenez E, Lozoff B. Iron deficiency in infancy and neurocognitive functioning at 19 years: evidence of long-term deficits in executive function and recognition memory. Nutr Neurosci. 2010;13(2):54–70. doi: 10.1179/147683010X12611460763689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran PV, Kennedy BC, Lien Y-C, Simmons RA, Georgieff MK. Fetal iron deficiency induces chromatin remodeling at the Bdnf locus in adult rat hippocampus. Am J Physiol Regul Integr Comp Physiol. 2015;308(4):276–282. doi: 10.1152/ajpregu.00429.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson EH. The interpretation of interaction in contingency tables. J R Stat Soc Series B. 1951;13:238–241. [Google Scholar]
- 50.Brunette KE, Tran PV, Wobken JD, Carlson ES, Georgieff MK. Gestational and neonatal iron deficiency alters apical dendrite structure of CA1 pyramidal neurons in adult rat hippocampus. Dev Neurosci. 2010;32 10.1159/000314341. [DOI] [PMC free article] [PubMed]
- 51.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):90. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ. Mouse genome database (MGD)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45(D1):723. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boettner B, Aelst LV. The role of rho GTPases in disease development. Gene. 2002;286(2):155–174. doi: 10.1016/S0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- 55.Auer M, Hausott B, Klimaschewski L. Rho GTPases as regulators of morphological neuroplasticity. Annals of Anatomy - Anatomischer Anzeiger. 2011;193(4):259–266. doi: 10.1016/j.aanat.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luo L. RHO GTPASES in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 57.Nakayama AY, Harms MB, Luo L. Small GTPases Rac and rho in the maintenance of Dendritic spines and branches in Hippocampal pyramidal neurons. J Neurosci. 2000;20(14):5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by rho GTPase regulatory proteins. Prog Neurobiol. 2011;94(2):133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed]
- 60.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13(7):1–14. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, Gnirke A, Meissner A. Genomic distribution and inter-sample variation of non-CpG Methylation across human cell types. PLoS Genet. 2011;7(12):1–15. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, Hatchwell E, Selzer RR, Richmond TA, Green RD, Melnick A, Greally JM. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crowder, M.J.: Beta-binomial Anova for proportions. Journal of the Royal Statistical Society. Series C (Applied Statistics) 27(1), 34–37 (1978). doi:10.2307/2346223.
- 65.Sun D, Xi Y, Rodriguez B, Park HJ, Tong P, Meong M, Goodell MA, Li W. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 2014;15(2):38. doi: 10.1186/gb-2014-15-2-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]


