The endoribonuclease DICER facilitates chromatin decondensation during lesion recognition following UV exposure. Chitale and Richly show that DICER mediates the recruitment of the methyltransferase MMSET, which catalyzes the dimethylation of histone H4 at lysine 20 and facilitates the recruitment of the nucleotide excision repair factor XPA.
Abstract
Ultraviolet (UV) irradiation triggers the recruitment of DNA repair factors to the lesion sites and the deposition of histone marks as part of the DNA damage response. The major DNA repair pathway removing DNA lesions caused by exposure to UV light is nucleotide excision repair (NER). We have previously demonstrated that the endoribonuclease DICER facilitates chromatin decondensation during lesion recognition in the global-genomic branch of NER. Here, we report that DICER mediates the recruitment of the methyltransferase MMSET to the DNA damage site. We show that MMSET is required for efficient NER and that it catalyzes the dimethylation of histone H4 at lysine 20 (H4K20me2). H4K20me2 at DNA damage sites facilitates the recruitment of the NER factor XPA. Our work thus provides evidence for an H4K20me2-dependent mechanism of XPA recruitment during lesion recognition in the global-genomic branch of NER.
Introduction
Nucleotide excision repair (NER) is one of the most versatile DNA repair pathways in the cell and handles several helix-distorting lesions caused by both intrinsic and extrinsic factors. The main types of lesions repaired by NER are photoproducts and pyrimidine dimers, which are caused by exposure to UV light (de Laat et al., 1999) Depending on the genomic location of the lesion, NER operates in two subpathways. Transcriptional coupled NER (TC-NER) recognizes lesions in transcriptionally active genes, whereas global genomic NER (GG-NER) deals with lesions in any chromatin environment. The recognition of the lesion is followed by lesion verification, unwinding of the DNA, excision of the lesion containing strand, and refilling of the DNA gap (de Laat et al., 1999; Fousteri and Mullenders, 2008; Marteijn et al., 2014). One of the critical factors linking lesion recognition to actual repair in both subpathways is the DNA-binding zinc-finger–containing protein XPA. Cells lacking XPA are completely deficient in both TC-NER and GG-NER (Kim et al., 1995). XPA patients are characterized by central nervous system disorders (Enokido et al., 1997; Kohji et al., 1998) and clinical skin defects and are very susceptible to UV light–induced skin tumors (Kraemer, 1994). In both the GG-NER and TC-NER pathways, XPA is recruited to chromatin by the transcription factor II H (TFIIH) complex (Yang et al., 2006; Feltes and Bonatto, 2015). This recruitment occurs together with the recruitment of replication protein A (RPA). RPA binds single-stranded DNA to stabilize the repair bubble, whereas XPA shows a high affinity for single-stranded DNA–double-stranded DNA junctions. Along with interacting with most of NER proteins, XPA also interacts with certain NER-regulating proteins such as PARP1 (King et al., 2012). Considering its interactions with both the NER repair bubble and various NER proteins, there is strong support for the idea that XPA functions as a scaffold protein. In addition, it may also be responsible for linking NER to other cellular processes such as cell cycle regulation (Wu et al., 2006).
One of the main constraints of GG-NER is the recognition and repair of a lesion in a chromatin context. A prominent histone mark involved in many DNA repair pathways is histone H2A ubiquitylation. With regard to NER, H2A ubiquitylation is catalyzed by the E3 ligase RNF8 and the UV–DDB-CUL4 and UV–RING1B complexes (Bergink et al., 2006; Kapetanaki et al., 2006; Guerrero-Santoro et al., 2008; Marteijn et al., 2009; Gracheva et al., 2016; Papadopoulou and Richly, 2016). We have recently demonstrated that the H2A-ubiquitin–binding protein ZRF1 is an essential factor in GG-NER that mediates the remodeling of E3 ligase multiprotein complexes (Gracheva et al., 2016) and contributes to the subnuclear localization of GG-NER (Chitale and Richly, 2017b). More recently, we have shown that ZRF1 operates in concert with the endoribonuclease DICER during GG-NER (Chitale and Richly, 2017a). DICER is most well known for its role in the RNAi pathway and has been shown to play a role in the establishment of heterochromatin (Wilson and Doudna, 2013; Holoch and Moazed, 2015; Chitale and Richly, 2017c). DICER is recruited to sites of DNA damage, and cells lacking DICER show impaired GG-NER. Importantly, we found that contrary to its function in heterochromatin formation, DICER is involved in chromatin decondensation during NER (Chitale and Richly, 2017a). This function of DICER is independent of its riboendonuclease activity and occurs upon the association of DICER with chromatin. This points toward a DICER function relatively early in the NER pathway, which probably enables the repair machinery to better access the lesion.
During DSB repair, H2A ubiquitylation is linked to the methylation of histone H4 (Fradet-Turcotte et al., 2013). The vast majority of H4K20me2 at chromatin is set by the di-/trimethylases SUV4-20H1 and SUV4-20H2 (Schotta et al., 2004, 2008). More recently, the enzymes SETD8 (Panier and Boulton, 2014; Milite et al., 2016) and MMSET/WHSC1 (Pei et al., 2011; Zimmermann and de Lange, 2014; Wang and Goldstein, 2016) were reported to affect the methylation status of H4K20 during DSB repair. SETD8 represents a member of the SET domain containing methyltransferases. It catalyzes the monomethylation of histone H4 at lysine 20 (H4K20), a modification that may be involved in modulating chromatin compaction (Lu et al., 2008). Moreover, methylation of H4K20 was reported to be essential for the recruitment of the signaling factor 53BP1 (Dulev et al., 2014). In particular, H4K20 dimethylation (H4K20me2) via MMSET and H2A ubiquitylation together were suggested to provide a binding platform for 53BP1 (Botuyan et al., 2006; Pei et al., 2011; Fradet-Turcotte et al., 2013). Interestingly, small RNAs produced by DICER were demonstrated to recruit MMSET to sites of DSBs suggesting a potential interplay of both factors in DNA repair pathways (Wang and Goldstein, 2016). Given our previous findings linking DICER to GG-NER, we sought to investigate a potential function for DICER and MMSET during GG-NER.
Here, we show a novel role for MMSET in GG-NER. MMSET is recruited to chromatin in a DICER-dependent manner, setting H4K20me2 in response to UV damage. We further provide evidence that H4K20me2 is responsible for the recruitment of XPA in the GG-NER pathway.
Results
DICER facilitates the generation of H4K20me2 during NER
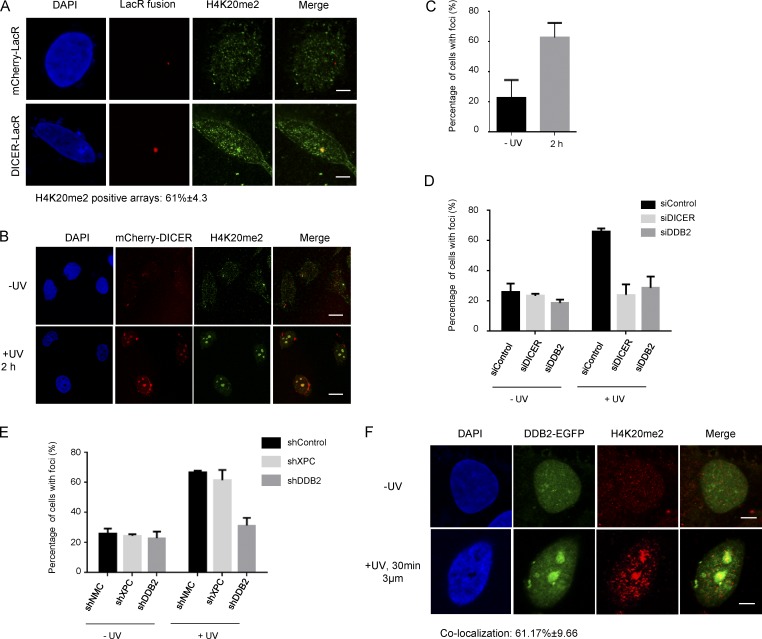
We have previously shown that DICER is responsible for decondensation of chromatin during NER (Chitale and Richly, 2017a). Modification of histones is an essential mechanism of establishing and maintaining chromatin states, as well as controlling the binding of chromatin associated proteins. Several histone modifications such as H2AX-phosphorylation, H2A ubiquitylation, and histone methylation are implicated in playing both structural and tethering roles in NER (Bergink et al., 2006; Li, 2012). To identify potential histone modifications that may mediate DICER-dependent decondensation, we microscopically screened selected histone marks. To this end, we used a LacO tethering system to tether LacR fusion proteins to a select chromatin locus in U2OS cells (Janicki et al., 2004). We found that tethering DICER did not result in local changes in H4K16 acetylation (H4K16ac), a mark linked to chromatin decondensation (Fig. S1 A). In contrast, when tethering DICER, we observed a prominent H4K20me2 mark at the chromatin array (Fig. 1 A). The control array, visualized by tethering of mCherry alone, did not show presence of the H4K20me2 mark (Fig. 1 A). Because we observed that chromatin-bound DICER was sufficient to generate a prominent H4K20me2 mark, we next wanted to address whether this held true in a system without forced tethering of DICER. We had previously observed that DICER is specifically recruited to chromatin after UV exposure and colocalizes with nuclear foci of ZRF1 (Chitale and Richly, 2017a). Therefore, we exposed U2OS cells to UV irradiation and monitored the nuclear distribution of H4K20me2 in relation to both DICER and ZRF1 foci. To this end, we preextracted the cells before fixation for subsequent immunofluorescence. This procedure washes off soluble proteins and facilitates the visualization of nonsoluble (chromatin-bound) factors. As previously observed, nuclear foci of chromatin-bound DICER appeared only after UV exposure. Interestingly, we also observed a prominent H4K20me2 mark at these foci (Fig. 1, B and C; and Fig. S1 B). Thus, recruitment of DICER to chromatin, either by forced tethering or after UV exposure, seems to be sufficient to result in a prominent H4K20me2 mark.
Figure 1.
UV exposure results in formation of H4K20me2 foci. (A) DICER leads to H4K20me2 at foci. Immunofluorescence images showing distribution of H4K20me2 in 2–6-3 cells, containing a chromatin array tethered with either mCherry-LacR (Control) or mCherry-LacR-DICER. H4K20me2 is observed at ≈61% of DICER-LacR arrays and 0% of mCherry-LacR arrays. Bars, 5 µm. (B) UV exposure results in colocalization of H4K20me2 with DICER foci. Immunofluorescence images showing nuclear distribution of H4K20me2 in UV unexposed and UV exposed U2OS cells expressing mCherry-DICER. Bars, 25 µm. (C) The graph shows the percentage of U2OS cells showing foci of H4K20me2 overlapping with DICER in unexposed cells and after UV exposure (mean ± SEM). Cells were counted from three independent experiments, with 200 cells counted per experiment. (D and E) Formation of H4K20me2 foci is dependent on NER. The graph shows the percentage of cells showing foci of H4K20me2 in unexposed U2OS cells (−UV) and 2 h after UV exposure (+UV) in the indicated knockdown cell lines (mean ± SEM). Cells were counted from three independent experiments, with 200 cells counted per experiment. (F) H4K20me2 is observed at the site of UV damage. Immunofluorescence images showing distribution of DDB2-EGFP and H4K20me2 in U2OS cells unexposed to UV and subjected to damage through a 3-µm micropore membrane. The damage sites are marked by DDB2-EGFP. H4K20me2 is observed at ≈61% of lesions 30 min after UV exposure. Bars, 5 µm.
To confirm that the formation of the H4K20me2 foci is indeed a consequence of DICER recruitment to chromatin, we performed an siRNA-mediated knockdown of DICER, irradiated the cells, and subsequently quantified the number of cells with H4K20me2 foci (Figs. 1 D and S1 E). We found that knockdown of DICER significantly reduced the number of H4K20me2 foci, further pointing to a link between DICER and H4K20me2 in UV-exposed cells. Next, we wanted to determine whether the H4K20me2 foci are formed as a result of NER and not as a general response to UV exposure. Because we had previously linked DICER to the GG-NER pathway (Chitale and Richly, 2017a), we analyzed the formation of H4K20me2 foci in cells after knocking down DDB2 or XPC, the two main GG-NER lesion recognition proteins (Fig. 1 E). We found that DDB2 knockdown cells (shDDB2) showed impaired formation of H4K20me2 foci after UV damage. In contrast, foci formation occurred normally in XPC knockdown cells (shXPC).
To determine the extent of H4K20me2 after UV irradiation, we monitored the total H4K20me2 levels in cells at various time points after UV exposure. Surprisingly, we did not observe a significant change in global H4K20me2 levels after UV irradiation (Fig. S1 C). We further assessed the H4K20me2 levels upon irradiating cells with different UV doses (Fig. S1 D). Upon increasing the UV dose, we observed a significant increase in the H4K20me2 levels. Finally, we directly assessed the role of H4K20me2 in NER by monitoring H4K20me2 levels in cells subjected to localized damage through a micropore. We observed the presence of H4K20me2 at ∼60% of lesions marked by DDB2 (Fig. 1 F). Collectively, these data point to a role for DICER-mediated dimethylation of H4K20 as an important early step during NER.
MMSET is required for setting of H4K20me2 after UV exposure
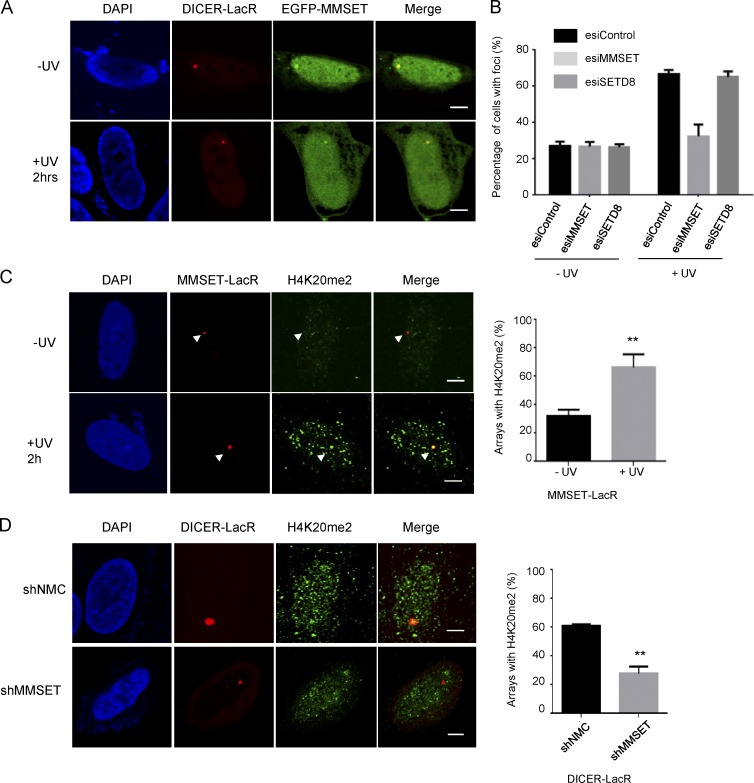
Having established a role for H4K20me2 during NER, we next wanted to determine whether H4K20me2 is set by a particular histone methyltransferase in response to UV exposure. H4K20me2 is a histone modification known to play a role in double-strand break (DSB) repair, where it provides a tethering platform for binding of 53BP1. During DSB repair, SETD8 and MMSET have been implicated in setting the H4K20me2 mark. Thus, we addressed a potential role for SETD8 and MMSET in setting H4K20me2 during NER. Because we had observed that tethering of DICER alone was sufficient to set the H4K20me2 mark at chromatin, we analyzed the interaction of DICER with both SETD8 and MMSET. We tethered DICER-LacR to the LacO array while coexpressing either EGFP-MMSET or EGFP-SETD8. We observed that chromatin-bound DICER was specifically able to strongly recruit MMSET to the array in 100% of both UV-exposed and unexposed cells (Figs. 2 A and S1 F). On the contrary, SETD8 showed only a very mild recruitment to tethered DICER, and recruitment was only observed in ∼20% cells in both UV-unexposed and exposed conditions (Fig. S1 G). Next, we assessed the appearance of H4K20me2 foci in UV-irradiated cells after depletion of SETD8 or MMSET, respectively. We found that endoribonuclease-prepared siRNA (esiRNA)–mediated knockdown of SETD8 did not affect the appearance of H4K20me2 foci formation, whereas knockdown of MMSET resulted in a dramatic loss of the foci (Fig. 2 B and Fig. S1, I and J). To substantiate our finding, we tethered an mCherry-LacR-MMSET fusion protein to the LacO array and examined the presence of H4K20me2 at the array by immunofluorescence (Fig. 2 C). Interestingly, we found that in unexposed cells MMSET was able to set the H4K20me2 mark in only ∼20% of tethered arrays, consistent with the enzymatic function of MMSET. However, after exposure to UV irradiation, we observed a prominent H4K20me2 mark in ∼70% of the arrays. In contrast, tethering of SETD8-LacR coincided with H4K20me2 in only ∼30% of the analyzed cells after exposure to UV light (Fig. S1 H). To further address the functional interplay of DICER, MMSET, and SETD8, we analyzed H4K20 methylation when tethering DICER-LacR to the array while simultaneously knocking down either MMSET or SETD8. Indeed, we found that upon knockdown of MMSET, DICER-tethered arrays showed H4K20me2 in ∼20% of cells compared with 60% observed in the control cells (Fig. 2 D). In contrast, knockdown of SETD8 did not affect the setting of H4K20me2 at the DICER tethered array (Fig. S1 K). Collectively, these data suggest that MMSET is likely the primary enzyme involved in setting H4K20me2 at chromatin in response to UV light. However, tethering MMSET to chromatin is insufficient to catalyze H4K20 dimethylation, suggesting that an additional UV-dependent component is probably required.
Figure 2.
MMSET plays a role in setting of H4K20me2. (A) DICER recruits MMSET to chromatin. Immunofluorescence images showing distribution of EGFP-MMSET in cells with DICER-LacR–tethered arrays in both UV-unexposed and exposed cells. Recruitment was observed in 20 out of 20 cells in both conditions. Bars, 5 µm. (B) H4K20me2 foci are dependent on MMSET. The graph shows the percentage of cells showing foci of H4K20me2 in unexposed cells (−UV) and 2 h after UV exposure (+UV) in U2OS cells transfected by the indicated esiRNA (mean ± SEM). Cells were counted from three independent experiments, with 200 cells counted per experiment. (C) MMSET tethered to a chromatin array can set the H4K20me2 mark after UV exposure. Immunofluorescence images on the left show H4K20me2 distribution in U2OS 2–6-3 cells, expressing mCherry-LacR-MMSET (MMSET-LacR). The LacO array is visualized by binding of MMSET-LacR. Bars, 5 µm. The graph on the right shows a quantification of the number of arrays showing an H4K20me2 mark in unexposed cells (−UV) and 2 h after UV exposure (+UV; mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001. (D) MMSET is required for DICER-dependent setting of H4K20me2. Immunofluorescence images on the left show the distribution of H4K20me2 in cells with DICER-LacR–tethered arrays and the indicated knockdowns. Bars, 5 µm. The graph on the right shows a quantification of the number of DICER-LacR arrays showing an H4K20me2 mark in undamaged cells (−UV; mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001.
MMSET catalyzes the UV-dependent formation of H4K20me2 foci
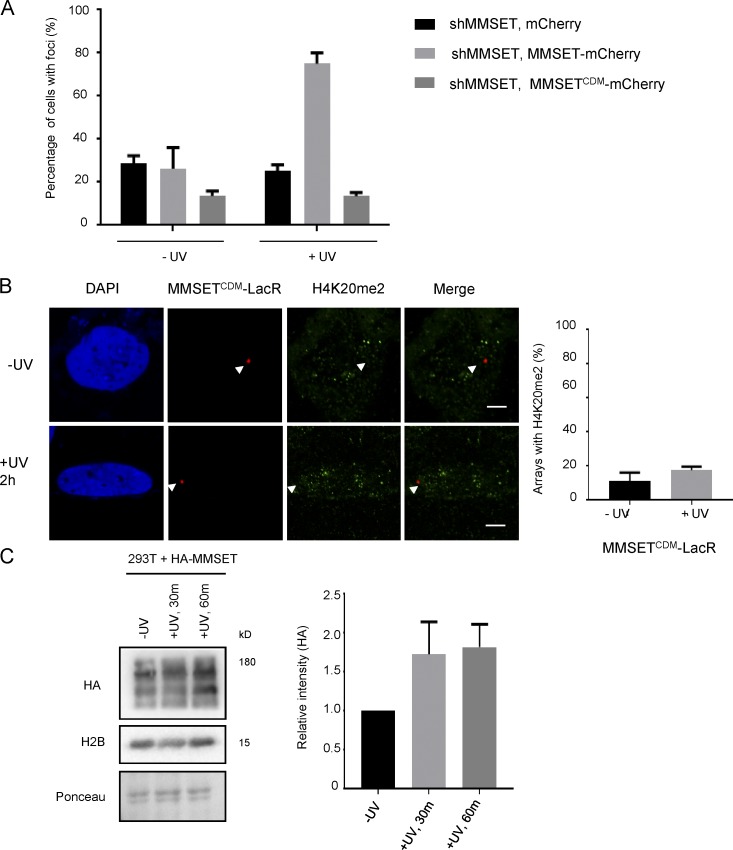
Our previous data indicated that MMSET is required to set H4K20me2 in response to UV irradiation. To further confirm this observation, we created a stable shRNA-mediated MMSET knockdown in U2OS cells (Fig. S2 A). We observed that shMMSET cells did not form H4K20me2 foci in response to UV irradiation (Fig. 3 A), reiterating our previous observations. Next, we wanted to determine whether this phenotype was rescued by reintroduction of MMSET into the shMMSET cells. Hence, we expressed MMSET-mCherry in the MMSET knockdown cells and monitored H4K20me2 foci formation. We observed that reintroduction of MMSET restored the formation of H4K20me2 foci (Figs. 3 A and S2 B). Notably, expression of a catalytically dead mutant (CDM) of MMSET (R1138A, C1144A; Huang et al., 2013) in shMMSET cells did not restore H4K20me2 foci formation (Figs. 3 A and S2 B). Likewise, the MMSETCDM mutant did not set H4K20me2 at the LacR-tethered array, even upon UV exposure (Fig. 3 B). These data confirm that the catalytic activity of MMSET is required for setting the H4K20me2 mark in response to UV irradiation.
Figure 3.
MMSET enzymatic activity is required to set the H4K20me2 mark. (A) MMSET expression can restore formation of H4K20me2 foci. The graph shows the percentage of cells showing foci of H4K20me2 in unexposed cells (−UV) and 2 h after UV exposure (+UV) in shMMSET U2OS cells transfected with either mCherry, MMSET-mCherry or MMSETCDM-mCherry (mean ± SEM). Cells were counted from three independent experiments, with 200 cells counted per experiment. (B) MMSETCDM tethered to a chromatin array is unable to set the H4K20me2 mark after UV exposure. (Left) Immunofluorescence images showing H4K20me2 distribution in U2OS 2–6-3 cells, expressing mCherry-LacR-MMSETCDM (MMSETCDM-LacR). Bars, 5 µm. Arrowhead marks the position of the array. The graph on the right shows a quantification of the number of arrays showing an H4K20me2 mark in unexposed cells (−UV) and 2 h after UV exposure (+UV; mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. (C) MMSET is recruited to chromatin upon UV damage. The blot on the left shows levels of HA-MMSET in the chromatin fraction at the indicated time points after UV exposure. H2B is used as a loading control. The graph on the right shows the quantification of the relative band intensity of HA-MMSET in the indicated samples. Intensity was measured from three independent experiments.
NER factors, and also DICER, are specifically recruited to chromatin after UV damage. MMSET, however, has functions outside of NER that also presumably require its presence at chromatin. Thus, we analyzed whether UV irradiation caused an increase in chromatin-bound MMSET. To assess whether MMSET is recruited to chromatin upon UV irradiation, we expressed HA-tagged MMSET in HEK 293T cells and monitored its levels in the chromatin fraction after UV exposure. We found that MMSET is specifically recruited to chromatin after UV exposure (Fig. 3 C). Our earlier data indicated that total H4K20me2 levels do not increase significantly after UV exposure (Fig. S1 C); however, increasing UV doses (i.e., creating more lesions per unit stretch of DNA) can lead to an increase in H4K20me2 levels. This likely suggests a specific local setting of H4K20me2, whereas the total cellular H4K20me2 levels do not change in a measurable way. We additionally addressed whether we could boost the H4K20me2 levels by overexpression of MMSET. We found that upon overexpression of MMSET, the overall levels of H4K20me2, as well as H3K36me2 (another histone mark catalyzed by MMSET), increased (Fig. S2 C). Collectively, these data suggest that MMSET is recruited to chromatin and that it catalyzes the H4K20me2 mark in response to UV damage.
MMSET is required for efficient NER
Because our previous data linked the appearance of H4K20me2 foci to lesion recognition via DDB2 (Fig. 1, D and E), we additionally determined whether MMSET is an essential factor of GG-NER and TC-NER. To this end, we first analyzed the levels of unscheduled DNA synthesis (UDS) in control and MMSET (esiMMSET) knockdown fibroblasts (Fig. 4 A). Knockdown of MMSET resulted in an ∼50% decrease of 5-ethynyl-2′-deoxyuridine (EdU) incorporation as compared with control cells. XPA fibroblasts showed a drastic reduction of EdU incorporation, whereas CSA fibroblasts only showed a minor reduction. Next, we determined whether MMSET also affects recovery of RNA synthesis (RRS) after exposure to UV irradiation as a measure for TC-NER. We found that knockdown of MMSET had a mild effect on RRS as measured by 5-ethynyl uridine (EU) incorporation after UV exposure (Fig. 4 B). To further assess the overall effect of MMSET knockdown on NER, we monitored the removal of CPDs in control and MMSET knockdown cells. We found that cells lacking MMSET showed an impaired removal of CPDs, with 50% of the CPDs retained even after 48 h (Fig. S2 D). Finally, we assessed the effect of MMSET depletion on cell survival. We found that MMSET-deficient cells showed impaired survival after UV exposure compared with control cells (Fig. 4 C). Collectively, our data suggest that MMSET plays a role in GG-NER and, to a minor extent, TC-NER.
Figure 4.
MMSET is required for efficient NER. (A) MMSET is required for efficient NER. The graph shows relative EdU incorporation as a result of UDS in MRC5 cells transfected with the indicated esiRNAs, XPA-deficient fibroblasts, and CSA-deficient fibroblasts (mean ± SEM). EdU intensity was measured in three independent experiments, with 200 cells counted per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001; ***, P ≤ 0.0001. (B) MMSET is required for efficient NER. RRS assay showing levels of EU incorporation in the indicated cells in unexposed cells and cells exposed to UV damage. The EU intensity is normalized to the intensity of the −UV condition for each cell type. EU intensity was measured for 200–300 cells per experiment (n = 3). (C) Knockdown of MMSET impairs cell survival of UV-exposed cells. The graph shows the relative cell viability (as measured by MTT assay at 72 h after UV) of U2OS cells with stable shRNA knockdowns of XPC and MMSET and control cells (shNMC) at the indicated UV dose (n = 3).
MMSET localizes to UV inflicted DNA damage sites
Next, we wanted to determine whether MMSET plays a direct role at the sites of DNA damage. Thus, we induced localized DNA damage in U2OS cells using a 3-µm micropore membrane and analyzed the recruitment of mCherry-tagged MMSET to the sites of damage. We found recruitment of MMSET to ∼53% of CPD lesions 30 min after UV irradiation (Fig. 5 A). Similarly, MMSET colocalized with DDB2-EGFP in ∼40% of UV irradiated U2OS cells (Fig. S3 A). We had observed earlier that chromatin-tethered MMSET sets the H4K20me2 mark only upon UV irradiation (Fig. 2 C) and that chromatin-tethered DICER is sufficient to recruit MMSET and to facilitate H4K20 methylation (Fig. 2 A). To better understand how UV irradiation mediates H4K20 methylation by MMSET, we examined the interplay of MMSET with other NER factors. Hence, we analyzed whether chromatin-tethered MMSET recruited any of the upstream GG-NER factors linked with damage recognition (Fig. S3 F). To this end, we tethered MMSET-LacR to the LacO array and examined the recruitment of DICER as well as DDB2, XPC, and RING1B, which are recruited upstream of DICER (Chitale and Richly, 2017a). We found that MMSET alone was unable to recruit any of the upstream factors to chromatin in both UV conditions (Fig. S3, B–F). Next, we monitored the presence of H4K20me2 at the LacO array upon tethering MMSET-LacR after knocking down various NER factors and after exposure to UV light. We observed, in line with our previous data (Fig. 1 E), that knockdown of DDB2 resulted in a decreased formation of H4K20me2 at the array, whereas knockdown of XPC had no effect (Fig. 5 B). Additionally, siRNA-mediated knockdown of DICER also decreased the levels of H4K20 methylation (Fig. 5 C). Therefore, we surmise that UV triggered recruitment of upstream NER factors to damaged DNA is essential to enhance the catalytic activity of MMSET at the array.
Figure 5.
MMSET is required for efficient NER and interacts with the NER machinery. (A) MMSET is recruited to the site of UV damage. Immunofluorescence images showing distribution of CPDs and mCherry-MMSET in U2OS cells subjected to damage through a 3-µm micropore membrane. MMSET is recruited to ≈53% of lesions 30 min after UV exposure. Bars, 5 µm. (B) DDB2 is required for UV-dependent setting of H4K20me2 by MMSET. The graph shows a quantification of the number of MMSET-LacR arrays showing H4K20me2 in the indicated knockdown cell lines cells 2 h after UV exposure (mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. *, P ≤ 0.01. (C) DICER is required for UV-dependent setting of H4K20me2 by MMSET. The graph shows a quantification of the number of MMSET-LacR arrays showing H4K20me2 in cells transfected with the indicated siRNAs 2 h after UV exposure (mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001. (D) MMSET interacts with ZRF1 and DICER. Western blot showing HA immunoprecipitations (IP) from cells transfected with either empty plasmid or HA-MMSET.
We next performed immunoprecipitations after expressing HA-tagged MMSET in HEK293T cells (Fig. 5 D). Notably, we found that MMSET interacts robustly with DICER and ZRF1, but not with XPA or XPB, respectively (Fig. 5 D). Reverse immunoprecipitation experiments using FLAG-ZRF1 or FLAG-DICER in combination with HA-MMSET (Fig. S3 G) also recapitulated the interaction of MMSET with ZRF1 and DICER. These data together suggest that MMSET localizes to UV light–inflicted DNA damage sites and that it physically interacts with DICER and ZRF1. Additionally, MMSET requires the presence of DICER and DDB2 at chromatin to set the H4K20me2 mark.
H4K20me2 provides a tethering platform for XPA recruitment
We next wanted to elucidate a possible function of H4K20 methylation during NER. During DSB repair, H4K20me2 provides a tethering signal for binding of 53BP1. Thus, we reasoned that H4K20me2 in NER could potentially provide a tethering platform for downstream operating repair factors. To this end, we tethered MMSET-LacR to chromatin and exposed the cells to UV irradiation to have maximum arrays bearing an H4K20me2 mark. We had previously noticed that MMSET was unable to recruit selected factors of the upstream NER machinery to chromatin (Fig. S4, B–F). Therefore, we examined the recruitment of NER factors presumably acting downstream of DICER (Fig. S4 F). Interestingly, we found that XPA was strongly recruited to the MMSET tethered array in UV-exposed cells. In contrast, we observed only minimal XPA recruitment in UV-unexposed cells (Fig. 6 A). This, along with our previous data showing that XPA does not directly interact with MMSET (Fig. 5 D), led us to hypothesize that XPA might be recruited to chromatin by the H4K20me2 mark rather than by MMSET itself. Additionally, we assayed the recruitment of XPF, a protein operating further downstream of XPA according to the sequential NER recruitment model (Fig. S3 F). We found a very mild recruitment of XPF compared with that of XPA in UV-exposed cells (Fig. S4 B), suggesting that XPA may be the primary protein recruited by H4K20me2.
Figure 6.
MMSET mediated H4K20me2 is required for recruitment of XPA. (A) MMSET-mediated H4K20me2 recruits XPA to chromatin. Immunofluorescence images showing distribution of XPA-EGFP and H4K20me2 in cells with MMSET-LacR tethered arrays in both UV-unexposed and exposed cells (left). The graph on the right shows a quantification of the number of arrays showing XPA-EGFP recruitment in unexposed cells (−UV) and 2 h after UV exposure (+UV; mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Bars, 5 µm. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001. (B) DICER can also recruit XPA to chromatin. Immunofluorescence images showing distribution of XPA-EGFP in cells with DICER-LacR tethered arrays, in both UV unexposed and exposed cells. Bars, 5 µm. (C) MMSET is required for DICER mediated XPA recruitment. The graph shows a quantification of the number of DICER-LacR arrays showing XPA-EGFP recruitment in the indicated knockdown cell lines (mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. ***, P ≤ 0.0001. (D) MMSET is required for XPA recruitment to localized sites of DNA damage. Immunofluorescence images showing distribution of CPDs and XPA in U2OS cells subjected to damage through a 3-µm micropore membrane. XPA recruitment to CPDs was quantified in both control (shNMC) and MMSET knockdown (shMMSET) cell lines. Bar, 10 µm. (E) MMSET is required for XPA recruitment to chromatin. Western blot showing chromatin association experiment in unexposed cells (−UV) and cells after 2 h UV exposure (+UV) in the indicated knockdown cell lines (right). Quantification of the relative intensity of XPA in the indicated conditions (left). H2B was used as a loading control (mean± SEM; n = 3).
XPA is known to be recruited to chromatin via TFIIH (Yang et al., 2006; Feltes and Bonatto, 2015). Hence, we additionally assessed the recruitment of XPB, a TFIIH subunit, to the MMSET-LacR–tethered array (Fig. S4 A). Surprisingly, we found no recruitment of XPB to the array in either UV-exposed or unexposed cells. We had previously observed that DICER recruits MMSET to chromatin in a UV-independent fashion (Fig. 2 A). Therefore, we assessed whether DICER was able to recruit XPA in a similar experimental setting. In line with our previous findings (Fig. 1 A), tethered DICER recruited XPA to chromatin in both unexposed and exposed cells (Fig. 6 B). DICER does not directly interact with XPA (Fig. S3 G), again suggesting the observed recruitment of XPA probably originated from a DICER-interacting protein or a histone modification linked with DICER function at chromatin (Fig. 1, A and B). Interestingly, DICER-mediated recruitment of XPA was significantly diminished in MMSET knockdown cells, further linking XPA recruitment to the H4K20me2 mark (Fig. 6 C). To confirm that H4K20me2 is required for XPA recruitment, we next tethered a catalytically dead MMSET mutant (MMSETCDM-LacR) to the array. We found that the MMSET mutant does not set the H4K20me2 mark in UV-exposed cells (Fig. 3 B) and that it was unable to recruit XPA even in UV-exposed cells (Fig. S4 C). We next addressed whether MMSET, and thus H4K20me2 are also necessary for the recruitment of XPA to DNA damage sites. We assessed XPA recruitment to localized sites of UV damage in control and MMSET knockdown U2OS cells (Fig. 6 D). Depletion of MMSET led to a significant reduction in XPA recruitment to the damage site (Fig. 6 D). Further, depletion of MMSET in HEK 293T cells exhibited a drastic reduction of XPA levels at chromatin after UV irradiation (Figs. 6 E). In summation, our data suggest that MMSET-catalyzed H4K20me2 facilitates the recruitment of XPA to chromatin after UV damage.
The RPA32 domain of XPA facilitates its localization to H4K20me2 sites
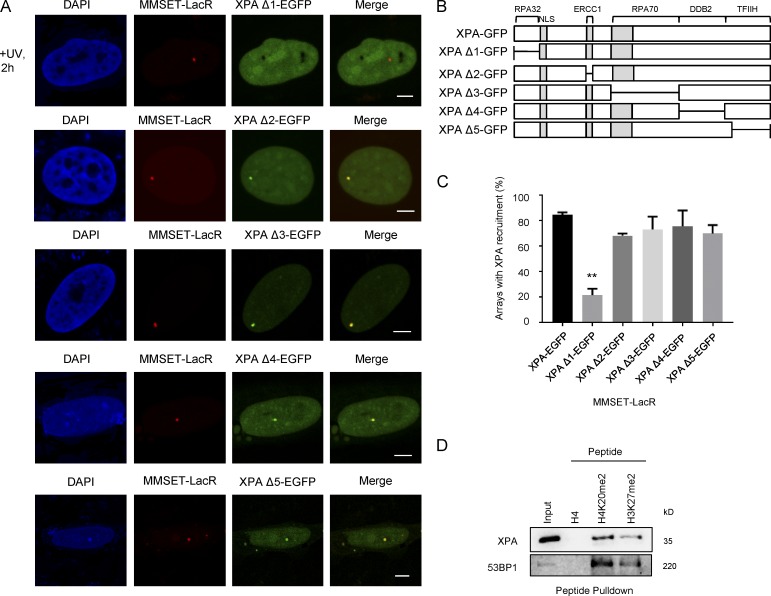
Our data so far pointed to a role for the MMSET-catalyzed H4K20me2 mark in the recruitment of XPA. To find out which XPA domain is essential for its recruitment to H4K20me2 sites, we generated various previously characterized deletion mutants of GFP-XPA fusion proteins lacking either the binding domains for RPA32 (XPA-Δ1), ERRC1 (XPA-Δ2), RPA70 (XPA-Δ3), DDB2 (XPA-Δ4), or TFIIH (XPA-Δ5), respectively (Fig. 7, A and B). Next, we expressed these mutants together with MMSET-LacR and analyzed their colocalization at the MMSET-marked array in UV-exposed cells (Fig. 7 A). We observed a significant reduction in recruitment of the XPA mutant devoid of the RPA32-binding domain (XPA-Δ1; Fig. 7 C). Interestingly, deletion of the other interaction domains (XPA-Δ2-5) did not affect the recruitment to the MMSET-LacR array. Thus, either interaction with RPA32 or other interactions mediated by this domain are essential for XPA binding to H4K20me2.
Figure 7.
XPA requires the RPA32-binding domain for interaction with H4K20me2. (A) Fluorescence microscopy images showing distribution of XPA deletions (Δ1–Δ5) in cells with MMSET-LacR tethered arrays 2 h after UV exposure. (B) Schematic showing the location of the generated XPA deletions. Bars, 5 µm. (C) RPA32 binding domain is required for XPA association with H4K20me2. The graph shows a quantification of the number of MMSET-LacR arrays showing recruitment of the indicated GFP tagged XPA deletion, 2 h after UV exposure (mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001. (D) H4K20me2 associates with XPA and 53BP1. Peptide pull-downs with biotinylated H4, H3K27me2, and H4K20me2 peptides and nuclear extracts expressing GFP-XPA. Specific interactions were analyzed by immunoblotting using the indicated antibodies.
Our previous data had shown that XPA does not interact with MMSET (Fig. 5 D). XPA also does not contain any known methyl-binding domain. This suggests that the observed recruitment (Fig. 6, A and B) is facilitated by other factors. To further support these findings, we examined whether XPA or a multiprotein complex harboring XPA shows an affinity for H4K20me2. To this end we performed peptide pull-downs from nuclear extracts of HEK293T cells expressing GFP-XPA using H4, H4K20me2 and H3K27me2 peptides (Fig. 7 D). We observed highly specific binding of 53BP1 to H4K20me2 in good agreement with previous studies (Botuyan et al., 2006; Tuzon et al., 2014). Importantly, we noticed a specific binding of XPA to H4K20me2 and, to a lesser extent, H3K27me2. We further repeated the experiment comparing the binding of wild-type XPA, as well as the XPA-Δ1 deletion that was observed to lose the specific recruitment in vivo. Similar to our previous results, we found that the XPA-Δ1 deletion was unable to bind H4K20me2 in vitro (Fig. S5 A).
Collectively, our data suggest that XPA interacts with the H4K20me2 mark and that the RPA32 interaction domain is essential for this interaction.
XPA recruitment to H4K20me2 requires 53BP1
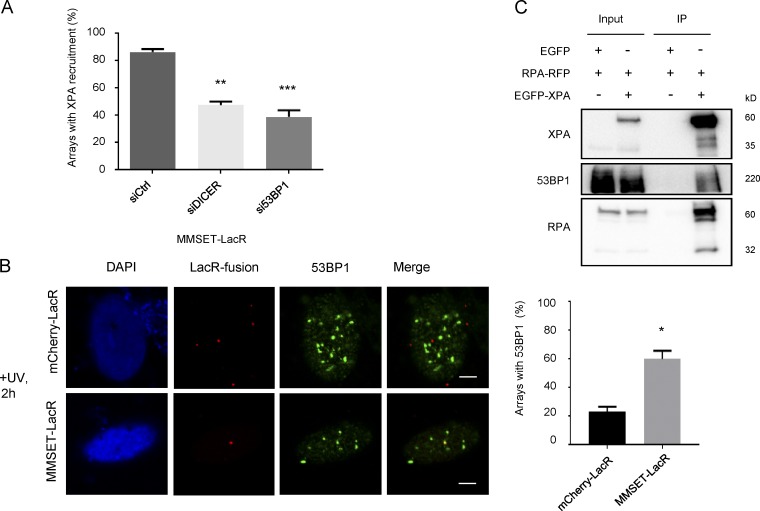
53BP1 is one of the known H4K20me2-binding proteins and is implicated in DSB repair (Botuyan et al., 2006; Tuzon et al., 2014). We reasoned that 53BP1 might also be recruited to H4K20me2 in GG-NER and potentially bridge the interaction of H4K20me2 with XPA. To test this possibility, we examined the localization of GFP-XPA to the MMSET-tethered array in cells depleted for either 53BP1 or DICER as a control (Fig. 8 A). Interestingly, siRNA-mediated knockdown of 53BP1 (Figs. 8 A and S5 B) significantly reduced the recruitment of XPA.
Figure 8.
53BP1 mediates XPA recruitment to H4K20me2. (A) 53BP1 is required for XPA recruitment to H4K20me2. The graph shows a quantification of the number of MMSET-LacR arrays showing XPA-EGFP recruitment in the indicated knockdown cell lines 2 h after UV exposure (mean ± SEM). Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. **, P ≤ 0.001; ***, P ≤ 0.0001. (B) 53BP1 can selectively bind an MMSET-LacR array. Immunofluorescence images showing distribution of 53BP1 in UV-exposed cells with either a mCherry-LacR– or MMSET-LacR–tethered array (left). Bars, 5 µm. The graph on the right shows a quantification of the number of arrays showing colocalization with 53BP1 in UV-exposed cells for the indicated tethered protein. Colocalization was measured from three independent experiments, with 30 cells per experiment. Statistical significance was assessed by an unpaired t test. *, P ≤ 0.01. (C) Western blot showing interaction of XPA and 53BP1. EGFP-XPA was immunoprecipitated using GFP-Trap beads from HEK293T cells expressing the indicated proteins. The blot shows levels of the indicated proteins in the input and immunoprecipitation (IP) samples.
To further assess the involvement of 53BP1 in tethering XPA to H4K20me2, we analyzed its recruitment to the MMSET-tethered array (Fig. 8 B). Using specific antibodies, we noted that 53BP1 localized to a great number of discrete foci in UV-damaged cells. A complete overlap of these foci with the MMSET-tethered array occurred in ∼60% of cells. In contrast, in the control (mCherry-LacR) the foci overlapped with the array in only ∼20% of cells, indicating that 53BP1 is specifically binding the MMSET-tethered array. Similarly, a DICER-tethered array colocalized with 53BP1 even in UV-unexposed cells in ∼85% of cells (Fig. S5 C). We next wanted to determine whether XPA and 53BP1 interact directly. However, we could not find strong evidence for this interaction, either in endogenous immunoprecipitations or in immunoprecipitations with either overexpressed EGFP-XPA or HA-MMSET (unpublished data), suggesting that this interaction is fairly weak or transient. Because our previous experiments had indicated that the RPA2-binding domain is important for XPA recruitment to H4K20me2 (Fig. 7 A), we next performed an immunoprecipitation of EGFP-XPA in HEK293T cells also expressing RPA2-RFP. Notably, we observed an interaction of XPA and 53BP1 upon simultaneous expression of RPA2-RFP (Fig. 8 C), likely indicating that RPA2 stabilizes the XPA–53BP1 interaction. Collectively, our data suggest that XPA is recruited to the DNA damage site by MMSET-mediated H4K20 dimethylation in a 53BP1- and RPA2-dependent manner.
Discussion
Timed recruitment of DNA repair factors is an essential determinant of damage recognition and necessary for the transition to downstream events of repair. Although we have learned much about how the NER machinery operates, it is still largely unknown how the chromatin environment, and in particular histone marks, contribute to the different steps of NER. We had previously observed a novel function for DICER in chromatin decondensation during NER. To gain a better understanding of the implications of histone marks during NER, we examined whether selected histone modifications are linked to DICER function during the DNA damage response. We found that chromatin-bound DICER can result in setting of the H4K20me2 mark (Fig. 1 A). DICER is recruited to chromatin after UV irradiation, and UV-dependent binding of DICER to chromatin also results in the formation of H4K20me2 foci (Fig. 1 B). Interestingly, generation of these H4K20me2 foci requires the presence of the NER factors DDB2 and DICER, thus linking it specifically to NER (Fig. 1, D and E). We additionally observed setting of H4K20me2 at sites of micropore-mediated UV damage (Fig. 1 F), suggesting that this chromatin mark is set during the UV damage response.
H4K20me2 has previously been shown to play a role in DSB repair, and the two enzymes implicated in H4K20 methylation are SETD8 and MMSET (Pei et al., 2011; Milite et al., 2016). To identify the H4K20 methylase that operates during NER, we analyzed whether SETD8 or MMSET interacts with DICER and whether each has an impact on H4K20me2 foci formation after UV exposure. We only observed a strong interaction of DICER and MMSET (Fig. 2 A), raising the possibility that the interplay of DICER and MMSET forms the basis for H4K20 methylation at the lesion site. In agreement with this hypothesis, knockdown of MMSET (Fig. 2 B) nearly abolished the generation of H4K20me2 foci, whereas knockdown of SETD8 did not significantly alter the nuclear H4K20me2 pattern. Additionally, by using the LacO tethering system, we determined that MMSET specifically sets the H4K20me2 mark at chromatin only after UV exposure (Fig. 2 C). To confirm that the catalytic activity of MMSET contributes to H4K20me2 foci formation, we performed rescue experiments with both wild-type and catalytically dead MMSET in shMMSET cells. We found that only the catalytically active protein could rescue H4K20me2 foci formation (Fig. 3 A). Further, MMSETCDM was also unable to set the H4K20me2 mark in the LacO tethering system (Fig. 3 B). This indicates that the setting of the H4K20me2 mark at the array is not a stochastic effect and is directly linked to the presence of active MMSET.
Our data further suggest that a UV light–dependent factor enhances the catalytic activity of MMSET (Fig. 2 C). To identify potential UV-dependent components, we analyzed the protein interactions of MMSET. We found that MMSET interacts strongly with ZRF1 and DICER (Fig. 5 D). This finding reiterates the DICER–MMSET interaction observed with the LacO tethering system (Fig. 2 A) and is in agreement with the previously reported association of DICER with ZRF1 (Chitale and Richly, 2017a). However, although DICER tethered to the LacO array recruits MMSET (Fig. 2 A), tethering of MMSET did not cause recruitment of DICER (Fig. S3 E). This finding suggests that the recruitment of MMSET to the lesion site might depend on ancillary factors. Thus, we hypothesized that UV-inflicted DNA damage is essential to target DICER to the damage site, which then in turn causes the recruitment of MMSET. The size of the LacO array is ∼4 Mb, and multiple lesions likely exist within the array given the UV dosage applied. Thus, although chromatin-tethered MMSET cannot recruit the tested factors of the NER machinery, UV damage probably recruits the NER machinery to chromatin-bearing tethered MMSET. To test this hypothesis, we analyzed MMSET-catalyzed H4K20 methylation in response to UV irradiation in DICER-depleted cells. Depletion of DICER diminished MMSET-mediated H4K20 methylation (Fig. 5 C), supporting our hypothesis. We speculate that DICER recruitment to chromatin might be accompanied by MMSET and that DICER enhances MMSET activity and hence the setting of the H4K20me2 mark.
It has been reported that MMSET and TIP60 are recruited to sites of DSBs through DICER-dependent small RNAs, which in turn facilitate the generation of H4K20me2 and H4K16ac at damaged chromatin and the decondensation of chromatin (Wang and Goldstein, 2016). However, the function of DICER in chromatin decondensation during NER is independent of its riboendonuclease activity (Chitale and Richly, 2017a,c). Thus, we additionally tested whether MMSET recruitment to chromatin via DICER is tied to its riboendonuclease activity. We observed that a catalytically defective DICER mutant still recruited MMSET and XPA to chromatin (Fig. S5, D and E). This suggests that the direct interaction between DICER and MMSET and their recruitment to chromatin seems to be a unique feature of NER. Interestingly, our findings are contrary to the observations in DSB repair, where the catalytic activity of DICER and hence the generation of small RNAs are essential. We also did not find a link between DICER and the setting of H4K16ac (Fig. S1 A). Thus, our observations suggest that the recruitment and function of MMSET in NER and DSB repair occurs by independent mechanisms.
H4K20me2 was previously shown to provide a binding platform for 53BP1 during DSB repair (Botuyan et al., 2006; Tuzon et al., 2014). DICER tethered to the LacO array strongly recruited XPA in both unexposed and UV-exposed cells (Fig. 6 B). In contrast, tethering of MMSET-LacR resulted in recruitment of XPA only in UV-exposed cells (Fig. 6 A). UV-triggered recruitment of DICER, and potentially other NER factors, seems to represent a critical step in the NER signaling cascade. Thus, when artificially tethering DICER to chromatin, downstream factors are recruited independent of UV irradiation. Additionally, neither DICER nor MMSET shows an interaction with XPA, further pointing toward an H4K20me2-dependent recruitment mechanism. This idea is supported by a lack of XPA recruitment when tethering the MMSETCDM mutant (Fig. S4 C). We also observed that MMSET was required for direct recruitment of XPA to CPDs, thereby recapitulating the data obtained with an artificial tethering system in a system with endogenous distribution of proteins (Fig. 6 D). Our peptide pull-down experiments further indicated that XPA shows an affinity for the H4K20me2 mark (Fig. 7 D). To characterize XPA binding to H4K20me2, we generated several deletions of XPA lacking specific protein interaction domains (Fig. 7 A). We found that the RPA2/32 interaction domain of XPA was essential for its interaction with H4K20me2. Deletion of this domain abolished binding of XPA to H4K20me2 both in vivo and in vitro (Figs. 7 A and S5 A). XPA does not contain any characterized methyl-binding domain, and in agreement, we observed that purified recombinant XPA was unable to bind H4K20me2 (unpublished data). Thus, it is likely that XPA associates with chromatin as part of a multiprotein complex containing factors capable of reading the H4K20me2 mark.
A known reader of the H4K20me2 mark that plays a role in DSB repair is 53BP1. 53BP1 has also been shown to play a role in NER. It is recruited to sites of UV damage and might potentially play a role in linking damage repair to cell cycle regulation as part of a universal DNA damage response (Marteijn et al., 2009). We found that knocking down 53BP1 reduced XPA recruitment to H4K20me2-decorated chromatin, suggesting that 53BP1 operates as a histone mark reader during NER as well. Additionally, we found that XPA and 53BP1 interact likely via RPA2 (Fig. 8 C), further linking XPA recruitment to 53BP1. Additionally, although we observed a strong recruitment of XPA to the H4K20me2 mark at the LacO array, we did not observe any recruitment of XPB to the array (Fig. S4 A). Furthermore, XPB also did not interact with either MMSET or DICER. This may suggest that besides recruitment via TFIIH or XPB, XPA may be recruited to the damage site by an additional H4K20me2-mediated mechanism. However, based on our data, we cannot conclude that TFIIH- and H4K20me2-based recruitment are independent mechanisms.
XPA is a critical factor of NER that serves in both subpathways, TC-NER and GG-NER. We found that knockdown of MMSET affected UDS in UV-exposed cells. Furthermore, knockdown of MMSET impaired cell survival and direct CPD removal in UV exposed cells. Thus, MMSET is an essential factor of NER. Because DICER seems to operate only in the GG-NER pathway (Chitale and Richly, 2017a), we assume that MMSET might contribute more significantly to GG-NER. In agreement, we found that MMSET knockdown affected RRS only to a mild extent. It is thus unclear what role, if any, MMSET plays in TC-NER.
The recruitment of XPA to the DNA damage site has been discussed controversially over the past few years, because XPA was demonstrated to have a function during lesion recognition and in the damage verification step. Given the strong phenotypes of XPA-deficient mice and the high susceptibility for cancer upon mutation of XPA alleles (de Vries and van Steeg, 1996; van Steeg et al., 2001), the molecular mechanisms of XPA recruitment should be of paramount interest. Here, we have described a chromatin-based mechanism for XPA recruitment to the DNA damage site during GG-NER (Fig. 9) that may either complement or be part of TFIIH-mediated XPA recruitment. Collectively, our data underline the importance of specific chromatin marks for recruiting DNA repair factors to the DNA damage site during NER.
Figure 9.
A model of H4K20me2-mediated recruitment of XPA during NER. The figure shows a proposed model for DICER- and MMSET-mediated setting of H4K20me2 and subsequent recruitment of XPA.
Materials and methods
UDS
UDS experiments were performed as described previously (Jia et al., 2015). In brief, MRC5 fibroblasts were transfected with siRNAs, serum starved for 24 h, irradiated with 20 J/m2 UV light, and incubated with 10 µM EdU (Thermo Fisher) for 2 h. Alexa Fluor 488–azide (Thermo Fisher) was conjugated to EdU using the Click reaction. The coverslips were mounted in Vectashield with DAPI. Images were acquired with LAS AF software (Leica) using a AF-7000 widefield microscope (Leica) with a 63×/1.4 oil-immersion objective and an ORCA CCD camera (Hamamatsu). Images were analyzed using ImageJ. DAPI was used to define nuclei, and EdU intensity within nuclei was measured after background subtraction. 150–300 nuclei were analyzed per sample. Mean intensities of +UV and −UV conditions for all cells were calculated and used to estimate the DNA repair occurring in the particular sample.
RRS assay
The RRS assay was performed as described previously (Jia et al., 2015). In brief, MRC5 cells were transfected with siRNAs, irradiated with 11 J/m2 UV light, and incubated in DMEM containing 1% FBS for 12 h. This was followed by a 2-h incubation with EU (Sigma) and subsequent fixation. Alexa Fluor 488–azide (Thermo Fisher) was conjugated to EU using the Click reaction. The coverslips were mounted in Vectashield with DAPI. Images were acquired with the LAS AF software (Leica) using a AF-7000 widefield microscope (Leica) with a 63×/1.4 oil-immersion objective and an ORCA CCD camera. Images were analyzed using ImageJ as described (Jia et al., 2015). Images were analyzed using ImageJ. EU intensity within nuclei was measured after background subtraction and exclusion of nucleoli. 200–300 nuclei were analyzed per sample. Mean intensities of +UV and −UV conditions for all cells were calculated and used to estimate the RNA synthesis occurring in the particular sample.
CPD removal assay
CPD removal assay was performed as previously described (Gracheva et al., 2016). In brief, U2OS cells containing a stable integration of the indicated shRNA were plated on coverslips, exposed to 10 J/m2 UV light, and fixed at the indicated time points. Cells were stained with CPD antibody (Cosmo Bio) using the manufacturer’s protocol, followed by incubation with Alexa Fluor 488 fluorophore–conjugated secondary antibodies (Thermo Fisher Scientific). The cells were mounted in Vectashield with DAPI, and images were acquired with the LAS AF software (Leica) using an AF-7000 widefield microscope (Leica) with a 63×/1.4 oil-immersion objective and an ORCA CCD camera. Images were analyzed using ImageJ.
Cell survival assay (MTT)
Cell viability was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay as described in Sun et al. (2002). In brief, cells were plated in 96-well plates and exposed to UV. Cell survival was assessed 72 h after UV exposure by measuring conversion of MTT to formazan product.
Cell lines and transfections
HEK293T, U2OS, and U2OS 2–6-3 cells were cultured in DMEM supplemented with 10% FBS at 37°C and 5% CO2. The medium for U2OS 2–6-3 cells was additionally supplemented with 100 µg/ml Hygromycin to maintain stable insertion of the LacO cassette. Normal skin fibroblasts (GM15876) and MRC-5 fibroblasts (AG05965) were purchased from the Coriell Cell Repositories and cultured in DMEM, supplemented with 15% FBS.
Transfection of U2OS 2–6-3 and HEK293T cells was performed by Lipofectamine 2000 (Invitrogen) transfection according to manufacturer’s instructions. Plasmids used were mCherry-ZRF1, mCherry-LacR-DICER, mCherry-LacR-MMSETCDM, mCherry-LacR-SETD8, mCherry-LacR-MMSET, EGFP-MMSET, EGFP-SETD8, EGFP-XPA, HA-MMSET, FLAG-HA-DICER (19881; Addgene), and pCMV2-FLAG-ZRF1. Details on plasmids are provided in Table S1. Control nontargeting siGENOME siRNA pool 1 (D-001206-13-05) and DICER (M-003483-00-0005) siRNA was purchased from Dharmacon (siGENOME SMARTpool). esiRNAs for SETD8, MMSET, and 53BP1 were purchased from Sigma. Details regarding siRNAs are provided in Table S2.
UV irradiation and drug treatment
Cells were irradiated with 20 J/m2 UV-C using a CL-1000 UV cross-linker (UVP), unless stated otherwise. For micropore irradiation experiments, cells were exposed to 100 J/m2 localized UV damage using a micropore membrane with 3-µm pore size as described previously (Katsumi et al., 2001). In brief, cells were washed three times with PBS. Micropore membranes presoaked in PBS were then placed on the cell layer, superfluous PBS was aspirated, and cells were irradiated at 100 J/m2. This was followed by staining using the standard immunofluorescence protocol described.
Immunofluorescence
Cells were fixed in 4% PFA for 10 min at room temperature. When indicated, preextraction with CSK buffer (10 mM Pipes, pH 7.4, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2) containing 0.2% Triton X-100 was performed for 5 min on ice before fixation. Cells were incubated overnight with primary antibody at 4°C. Subsequently cells were incubated with Alexa fluorophore–conjugated secondary antibodies (Life Technologies). The mounting was performed in Vectashield with DAPI (Vector Laboratories).
Confocal microscopy
Images were acquired with the LAS AF software (Leica) using a TCS SP5 confocal microscope (Leica) with a 63×/1.4 oil-immersion objective at room temperature. The following lasers were used: 50-mW UV diode (405 nm), 65-mW argon, 20-mW DPSS (561 nm), and 10-mW HeNe (633 nm).
Antibodies
Antibodies used in this study were DICER (3363, rabbit; Cell Signaling), ZRF1 (NBP2-12802; Novus Biologicals). RING1B clone (D22F2, 5694, rabbit; Cell Signaling), XPA (GTX103168, mouse; Genetex), XPA (FL-273, rabbit; Santa Cruz) H2B (V119 8135, mouse; Cell Signaling), FLAG (mouse; Sigma), H4K20me2 (39173, rabbit; Active Motif), H4K16ac (39930, rabbit; Active Motif), H3K36me2 (39256, rabbit; Active Motif), CPD (mouse; CosmoBio), HA (C29F4, rabbit; Cell Signaling), XPB (S-19, rabbit; Santa Cruz), MMSET (39880, mouse; Active Motif), 53BP1 (NB100-304, rabbit; Novus), and SETD8 (C18B7, rabbit; Cell Signaling).
Chromatin association assays
HEK293T cells (unless stated otherwise) were irradiated with UV and cross-linked by formaldehyde at the indicated time points after UV irradiation. Assays were essentially performed as published (Richly et al., 2010). In brief, cells were cross-linked in 1% formaldehyde for 10 min, followed by quenching and washing. Cells were then lysed in buffer A, nuclei were spun down, and isolated nuclei were lysed in hypotonic buffer. The chromatin fraction was separated by centrifugation at maximum speed, and supernatant consisting of nucleoplasm was removed. Samples were prepared by adding SDS loading buffer to the chromatin fraction, followed by sonication and boiling.
FLAG/HA/GFP purifications
Cells were UV irradiated at 20 J/m2 and harvested 1 h after exposure (unless stated otherwise). FLAG affinity purifications were performed using FLAG-M2 agarose beads as described previously (Richly et al., 2010). HA affinity purifications were performed using HA agarose (Sigma) following an essentially similar protocol. GFP purifications were performed using GFP-Trap beads (Chromotek) following an essentially similar protocol. In brief, cells were harvested, washed with PBS, and then lysed for 10 min in buffer A. Nuclei were separated by centrifugation at 3,000 rpm for 4 min. Nuclei were then lysed in lysis buffer by sonication in a bioruptor for 20 min (30 s on/off). The lysate was spun at max speed to remove the nonsoluble fraction. The lysate was then incubated with the respective beads overnight; beads were washed and samples were analyzed on an SDS-PAGE gel.
Peptide pull-downs
Peptide pull-downs were performed essentially as described previously (Wysocka, 2006). In brief, biotinylated peptides (H4, H3K27me2, and H4K20me2) were immobilized on Streptavidin beads. Nuclear extracts from HEK293T cells expressing GFP-XPA were prepared and precleared as described previously (Wysocka, 2006). Protein extracts and peptide-bound beads were incubated overnight at 4°C on a turning wheel. After extensive washing, purified material was analyzed with immunoblotting.
Online supplemental material
Tables S1 and S2 give further information on the plasmids used in this study, as well as the sequences of all si/esiRNAs used in the study. Figs. S1–S5 contain all supporting data and additional controls.
Supplementary Material
Acknowledgments
We are grateful to the Institute of Molecular Biology microscopy facility for their expert help and assistance. We thank the members of the H. Richly laboratory for comments on the manuscript.
This work was supported by grants from the Boehringer Ingelheim Foundation and the Deutsche Forschungsgemeinschaft (RI-2413/1-1).
The authors declare no competing financial interests.
Author contributions: S. Chitale conducted and analyzed the experiments. S. Chitale and H. Richly designed the experiments and wrote the paper.
References
- Bergink S., Salomons F.A., Hoogstraten D., Groothuis T.A., de Waard H., Wu J., Yuan L., Citterio E., Houtsmuller A.B., Neefjes J., et al. 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 20:1343–1352. 10.1101/gad.373706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., and Mer G.. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127:1361–1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S., and Richly H.. 2017a DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 45:5901–5912. 10.1093/nar/gkx261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S., and Richly H.. 2017b Nuclear organization of nucleotide excision repair is mediated by RING1B dependent H2A-ubiquitylation. Oncotarget. 8:30870–30887. 10.18632/oncotarget.16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitale S., and Richly H.. 2017c Shaping chromatin with DICER. Oncotarget. 8:39937–39938. 10.18632/oncotarget.17773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W.L., Jaspers N.G., and Hoeijmakers J.H.. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768–785. 10.1101/gad.13.7.768 [DOI] [PubMed] [Google Scholar]
- de Vries A., and van Steeg H.. 1996. Xpa knockout mice. Semin. Cancer Biol. 7:229–240. 10.1006/scbi.1996.0031 [DOI] [PubMed] [Google Scholar]
- Dulev S., Tkach J., Lin S., and Batada N.N.. 2014. SET8 methyltransferase activity during the DNA double-strand break response is required for recruitment of 53BP1. EMBO Rep. 15:1163–1174. 10.15252/embr.201439434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokido Y., Inamura N., Araki T., Satoh T., Nakane H., Yoshino M., Nakatsu Y., Tanaka K., and Hatanaka H.. 1997. Loss of the xeroderma pigmentosum group A gene (XPA) enhances apoptosis of cultured cerebellar neurons induced by UV but not by low-K+ medium. J. Neurochem. 69:246–251. 10.1046/j.1471-4159.1997.69010246.x [DOI] [PubMed] [Google Scholar]
- Feltes B.C., and Bonatto D.. 2015. Overview of xeroderma pigmentosum proteins architecture, mutations and post-translational modifications. Mutat. Res. Rev. Mutat. Res. 763:306–320. 10.1016/j.mrrev.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Fousteri M., and Mullenders L.H.. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 18:73–84. 10.1038/cr.2008.6 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., and Durocher D.. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 499:50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva E., Chitale S., Wilhelm T., Rapp A., Byrne J., Stadler J., Medina R., Cardoso M.C., and Richly H.. 2016. ZRF1 mediates remodeling of E3 ligases at DNA lesion sites during nucleotide excision repair. J. Cell Biol. 213:185–200. 10.1083/jcb.201506099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Santoro J., Kapetanaki M.G., Hsieh C.L., Gorbachinsky I., Levine A.S., and Rapić-Otrin V.. 2008. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68:5014–5022. 10.1158/0008-5472.CAN-07-6162 [DOI] [PubMed] [Google Scholar]
- Holoch D., and Moazed D.. 2015. Small-RNA loading licenses Argonaute for assembly into a transcriptional silencing complex. Nat. Struct. Mol. Biol. 22:328–335. 10.1038/nsmb.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Wu H., Chuai S., Xu F., Yan F., Englund N., Wang Z., Zhang H., Fang M., Wang Y., et al. 2013. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 73:6277–6288. 10.1158/0008-5472.CAN-13-1000 [DOI] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., and Spector D.L.. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698. 10.1016/S0092-8674(04)00171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N., Nakazawa Y., Guo C., Shimada M., Sethi M., Takahashi Y., Ueda H., Nagayama Y., and Ogi T.. 2015. A rapid, comprehensive system for assaying DNA repair activity and cytotoxic effects of DNA-damaging reagents. Nat. Protoc. 10:12–24. 10.1038/nprot.2014.194 [DOI] [PubMed] [Google Scholar]
- Kapetanaki M.G., Guerrero-Santoro J., Bisi D.C., Hsieh C.L., Rapić-Otrin V., and Levine A.S.. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA. 103:2588–2593. 10.1073/pnas.0511160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumi S., Kobayashi N., Imoto K., Nakagawa A., Yamashina Y., Muramatsu T., Shirai T., Miyagawa S., Sugiura S., Hanaoka F., et al. 2001. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 117:1156–1161. 10.1046/j.0022-202x.2001.01540.x [DOI] [PubMed] [Google Scholar]
- Kim J.K., Patel D., and Choi B.S.. 1995. Contrasting structural impacts induced by cis-syn cyclobutane dimer and (6-4) adduct in DNA duplex decamers: implication in mutagenesis and repair activity. Photochem. Photobiol. 62:44–50. 10.1111/j.1751-1097.1995.tb05236.x [DOI] [PubMed] [Google Scholar]
- King B.S., Cooper K.L., Liu K.J., and Hudson L.G.. 2012. Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J. Biol. Chem. 287:39824–39833. 10.1074/jbc.M112.393504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohji T., Hayashi M., Shioda K., Minagawa M., Morimatsu Y., Tamagawa K., and Oda M.. 1998. Cerebellar neurodegeneration in human hereditary DNA repair disorders. Neurosci. Lett. 243:133–136. 10.1016/S0304-3940(98)00109-8 [DOI] [PubMed] [Google Scholar]
- Kraemer K.H. 1994. Nucleotide excision repair genes involved in xeroderma pigmentosum. Jpn. J. Cancer Res. 85 (inside front cover.). [PubMed] [Google Scholar]
- Li S. 2012. Implication of posttranslational histone modifications in nucleotide excision repair. Int. J. Mol. Sci. 13:12461–12486. 10.3390/ijms131012461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Simon M.D., Chodaparambil J.V., Hansen J.C., Shokat K.M., and Luger K.. 2008. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 15:1122–1124. 10.1038/nsmb.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn J.A., Bekker-Jensen S., Mailand N., Lans H., Schwertman P., Gourdin A.M., Dantuma N.P., Lukas J., and Vermeulen W.. 2009. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186:835–847. 10.1083/jcb.200902150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn J.A., Lans H., Vermeulen W., and Hoeijmakers J.H.. 2014. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 15:465–481. 10.1038/nrm3822 [DOI] [PubMed] [Google Scholar]
- Milite C., Feoli A., Viviano M., Rescigno D., Cianciulli A., Balzano A.L., Mai A., Castellano S., and Sbardella G.. 2016. The emerging role of lysine methyltransferase SETD8 in human diseases. Clin. Epigenetics. 8:102 10.1186/s13148-016-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., and Boulton S.J.. 2014. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15:7–18. 10.1038/nrm3719 [DOI] [PubMed] [Google Scholar]
- Papadopoulou T., and Richly H.. 2016. On-site remodeling at chromatin: How multiprotein complexes are rebuilt during DNA repair and transcriptional activation. BioEssays. 38:1130–1140. 10.1002/bies.201600094 [DOI] [PubMed] [Google Scholar]
- Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P.L., Wang L., You Z., and Lou Z.. 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 470:124–128. 10.1038/nature09658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H., Rocha-Viegas L., Ribeiro J.D., Demajo S., Gundem G., Lopez-Bigas N., Nakagawa T., Rospert S., Ito T., and Di Croce L.. 2010. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 468:1124–1128. 10.1038/nature09574 [DOI] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., and Jenuwein T.. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262. 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Sengupta R., Kubicek S., Malin S., Kauer M., Callén E., Celeste A., Pagani M., Opravil S., De La Rosa-Velazquez I.A., et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22:2048–2061. 10.1101/gad.476008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N.K., Kamarajan P., Huang H., and Chao C.C.. 2002. Restoration of UV sensitivity in UV-resistant HeLa cells by antisense-mediated depletion of damaged DNA-binding protein 2 (DDB2). FEBS Lett. 512:168–172. 10.1016/S0014-5793(02)02250-0 [DOI] [PubMed] [Google Scholar]
- Tuzon C.T., Spektor T., Kong X., Congdon L.M., Wu S., Schotta G., Yokomori K., and Rice J.C.. 2014. Concerted activities of distinct H4K20 methyltransferases at DNA double-strand breaks regulate 53BP1 nucleation and NHEJ-directed repair. Cell Reports. 8:430–438. 10.1016/j.celrep.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steeg H., de Vries A., van Benthem J., Beems R.B., and van Kreijl C.F.. 2001. DNA repair-deficient Xpa and Xpa/p53+/- knock-out mice: nature of the models. Toxicol. Pathol. 29(5 Suppl.):109–116. 10.1080/019262301753178519 [DOI] [PubMed] [Google Scholar]
- Wang Q., and Goldstein M.. 2016. Small RNAs Recruit Chromatin-Modifying Enzymes MMSET and Tip60 to Reconfigure Damaged DNA upon Double-Strand Break and Facilitate Repair. Cancer Res. 76:1904–1915. 10.1158/0008-5472.CAN-15-2334 [DOI] [PubMed] [Google Scholar]
- Wilson R.C., and Doudna J.A.. 2013. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42:217–239. 10.1146/annurev-biophys-083012-130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Shell S.M., Yang Z., and Zou Y.. 2006. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 66:2997–3005. 10.1158/0008-5472.CAN-05-3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J. 2006. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods. 40:339–343. 10.1016/j.ymeth.2006.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Roginskaya M., Colis L.C., Basu A.K., Shell S.M., Liu Y., Musich P.R., Harris C.M., Harris T.M., and Zou Y.. 2006. Specific and efficient binding of xeroderma pigmentosum complementation group A to double-strand/single-strand DNA junctions with 3′- and/or 5′-ssDNA branches. Biochemistry. 45:15921–15930. 10.1021/bi061626q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., and de Lange T.. 2014. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24:108–117. 10.1016/j.tcb.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.