Wigerius et al. identify the polarity protein AMOT-130 as vital for dendritic spine morphogenesis. They show that reduced Lats1 kinase activity in the neonatal brain is required for the recruitment of AMOT-130 to postsynaptic compartments to stabilize dendritic spines.
Abstract
The actin cytoskeleton is essential for the structural changes in dendritic spines that lead to the formation of new synapses. Although the molecular mechanisms underlying spine formation are well characterized, the events that drive spine maturation during development are largely unknown. In this study, we demonstrate that Angiomotin (AMOT-130) is necessary for spine stabilization. AMOT-130 is enriched in mature dendritic spines and functions to stabilize the actin cytoskeleton by coupling F-actin to postsynaptic protein scaffolds. These functions of AMOT are transiently restricted during postnatal development by phosphorylation imposed by the kinase Lats1. Our study proposes that AMOT-130 is essential for normal spine morphogenesis and identifies Lats1 as an upstream regulator in this process. Moreover, our findings may link AMOT-130 loss and the related spine defects to neurological disorders.
Introduction
Dendritic spines are critical for memory formation, and changes in spine structure contribute directly to neurological disorders (Brandon and Sawa, 2011; Verpelli and Sala, 2012; Yu and Lu, 2012). During development, neurons in the neonatal brain express immature filopodia-like protrusions that are replaced by mature spines. Scaffolding proteins and adhesion molecules are critical for this process as they recruit and stabilize glutamate receptors to form functional synapses (Sheng and Kim, 2011; Anggono and Huganir, 2012). These associations rely in turn on changes in the actin cytoskeleton, which power alterations in spine morphology and trigger rewiring of neural circuitry (Maletic-Savatic et al., 1999; Saneyoshi and Hayashi, 2012). However, it is unclear how mature spines develop from transient filopodia and how synaptic molecules that modulate actin filaments influence these events.
The Hippo signal transduction pathway is conserved throughout metazoa and plays an important role in limiting epithelial tissue growth by controlling the balance between proliferation and apoptosis (Barry and Camargo, 2013; Piccolo et al., 2014; Varelas, 2014; Moroishi et al., 2015). Central in this pathway are the mammalian Ste20-like kinases (MST1/2) and the nuclear dbf2-related (NDR) family kinases large tumor suppressor 1/2 (Lats1/2) that restrict the activity of the transcription coactivators Yes-associated protein (YAP)/TAZ (Yu and Guan, 2013). Yet how these signaling components operate in postmitotic neurons is less well understood. Studies in Drosophila melanogaster show that deregulation of the Hippo signaling cascade alters brain size and restricts neuronal differentiation (Jukam et al., 2016; Poon et al., 2016). In mammalian neurons, Hippo kinase activity has been linked to actin remodeling (Ultanir et al., 2012, 2014) and, conversely, actin cytoskeleton changes increase Lats1/2 activity (Piccolo et al., 2014). Collectively, this suggests that Hippo kinases both target and respond to the signaling networks that impact on neuronal structure.
The Angiomotin (AMOT) proteins are expressed within various tissues as alternative splice variants with distinct and redundant functions in cell morphology and migration (Moleirinho et al., 2014). The major isoforms AMOT-80 and AMOT-130 share a central coiled-coil domain and a carboxyl terminal PSD-95/Dlg-1/ZO-1 (PDZ) motif but are distinguished by an N-terminal region that contains an F-actin–binding domain encoded by AMOT-130 (Ernkvist et al., 2006). Both proteins can act as scaffolds for cell polarity proteins, including Rho family GTPases. However, AMOT-130, unlike AMOT-80, has been implicated in the regulation of Hippo signaling (Wells et al., 2006; Zhao et al., 2011). Previous work has established that AMOT-130 and AMOT-80 are expressed in the central nervous system (CNS; Wells et al., 2006), and recent genetic research has implicated a role for AMOT-130 in autism (Schanzenbächer et al., 2016). Despite this, there is little understanding of the role of AMOT proteins in the brain.
In this study, we examined the function of AMOT-130 in the developing CNS. We found that AMOT-130 accumulates in the postsynaptic density (PSD) and is essential for spine formation by controlling actin turnover and PSD integrity through interactions with the scaffolding proteins MUPP1 and PSD-95. Our results furthermore identify Lats1-mediated serine 175 (S-175) phosphorylation of AMOT-130 as a critical regulatory step for its role in developing spines. Collectively, these findings provide insight into the understanding of how AMOT-130 loss-of-function influences synapse structure and may link to congenital spine defects observed in autism-spectrum disorder pathology.
Results
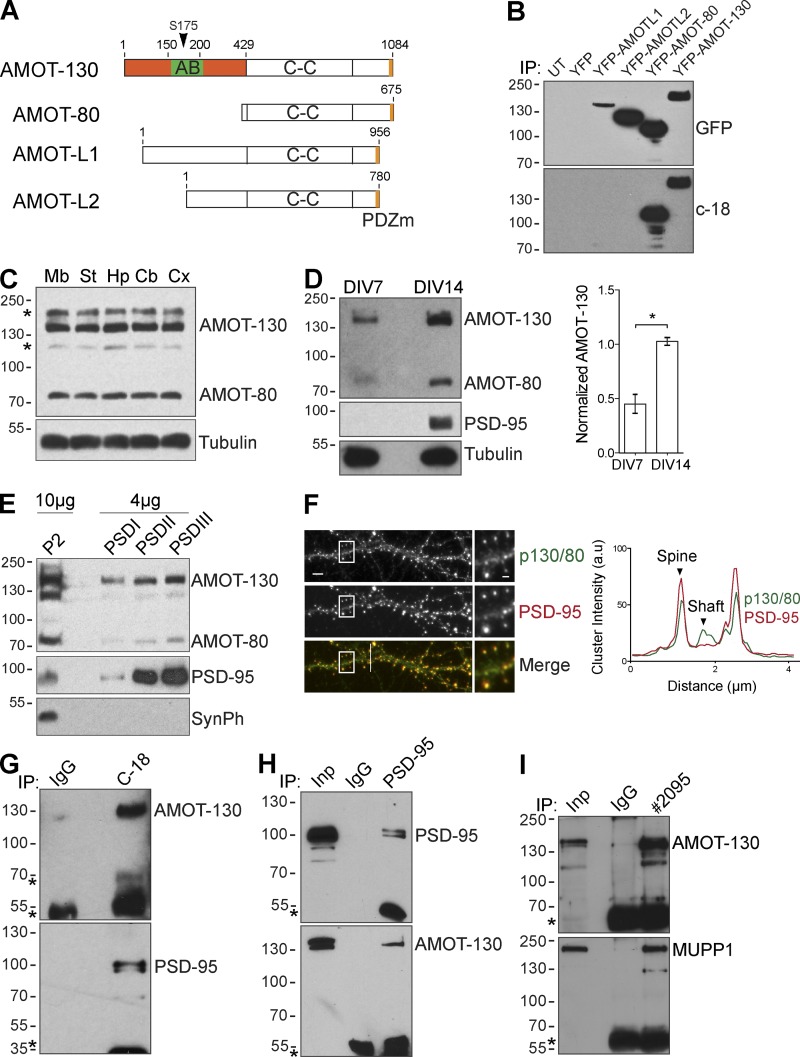
Expression of AMOT-130 and AMOT-80 in the CNS
The AMOT protein family impacts cellular polarity and embryonic development by acting as scaffolds for signaling complexes that participate in these processes (Wells et al., 2006; Hirate et al., 2013). However, it is not known whether AMOTs share similar functions in the CNS despite indications of abundant distribution in the brain (Lein et al., 2007). Thus, we examined the roles of the core splicing isoforms AMOT-130 and AMOT-80 during development. We evaluated a commercial antibody (C-18) for its specificity to detect AMOT but not the related gene products AMOT-L1 or AMOT-L2 by transfecting HEK293T cells with cDNAs encoding YFP–AMOT-L1, AMOT-L2, AMOT-80, and AMOT-130 (Fig. 1 A). All members were detected with anti-GFP antibody, whereas the C-18 antibody specifically recognized AMOT-130 and AMOT-80 (Fig. 1 B). To examine the distribution of AMOT-130/80 in the CNS, we analyzed lysates from whole rat brain regions or cultures of dissociated rat hippocampal neurons. Both AMOT-130 and AMOT-80 were abundantly expressed in various brain regions (Fig. 1 C) and in neuron cultures, with approximately twofold higher expression levels detected at 14 d in vitro (DIV) before onset of synaptogenesis, compared with 7-DIV cultures (Fig. 1 D; *, P < 0.05; two-tailed Student’s t test). However, variations in AMOT-80 expression between brain and primary hippocampal cultures might be an effect from contributions of other cell types including glia and vasculature, which are known to express AMOT-80 (Aase et al., 2007; Yi et al., 2011), and enhanced AMOT-130 levels at 14 DIV are consistent with a role in neuron maturation.
Figure 1.
Distribution of AMOT isoforms in the CNS. (A) Schematic of structural domains in the four major AMOT isoforms. The location of the actin-binding (AB) region, the N-terminal region (orange), coiled-coil (C-C) domain, and the PDZ-binding motif (PDZm) are indicated. The arrowhead denotes the conserved serine (S-175) within the actin-binding domain. (B) Lysates from HEK293T cells transfected as indicated were immunoprecipitated (IP) and probed with anti-GFP antibody (top). Membranes were reprobed with C-18 antibody (bottom). UT, untranslated. (C) Expression of AMOT in the indicated brain regions. Mb, midbrain; St, striatum; Hp, hippocampus; Cb, cerebellum; Cx, cortex. Asterisks indicate unspecific bands. (D) Clarified lysates from 7- and 14-DIV hippocampal neuron cultures analyzed by immunoblotting with indicated antibodies. The graph to the right shows quantification of AMOT-130 protein normalized to tubulin. Error bars represent means ± SD. *, P < 0.05; Student’s t test. (E) Protein isolated from crude synaptosomal fraction (P2) and enriched PSD fractions were analyzed by immunoblotting. SynPh, synaptophysin. (F) Double immunostaining of PSD-95 (red) and AMOT-130/80 (green) in hippocampal neurons. Bars: (main images) 5 µm; (insets) 1.25 µm. Right: intensity profile along white line across the dendrite. (G–I) Coimmunoprecipitation with IgG or antibody recognizing endogenous AMOT-130/80 (C-18), PSD-95 (H), or AMOT-130 (2095; I), which does not recognize AMOT-80, from hippocampal lysates. Bound complexes were analyzed by immunoblotting (top) and reprobed (bottom) with the indicated antibodies. Asterisks denote IgG heavy chains. Molecular masses are given in kilodaltons.
We next investigated the subcellular distribution of AMOT-130/80 by performing fractionation of whole rat brain homogenates (Fig. S1 A). AMOT-130 was detected in membrane/microsomal fractions but was largely absent in crude cytosolic fractions (Fig. S1 B). A similar subcellular distribution was noted for AMOT-80 (Fig. S1 B). In sucrose gradient fractions of HEK293T lysate, endogenous AMOT-130 and AMOT-80 had distinct but overlapping localization profiles (Fig. S2 C), consistent with the previous finding that all AMOT family members can self-associate (Moleirinho et al., 2014). To analyze their subcellular distribution in the synapse, we isolated synaptosomes by differential centrifugation. Both AMOT-130 and AMOT-80 were found in the crude synaptosomal (P2) fractions (Figs. 1 E and S1 B), whereas AMOT-130, like the PSD marker PSD-95, was detected in PSD fractions resistant to treatment with Triton X-100 (PSDI/II) and sarkosyl (PSDIII) detergent (Fig. 1 E). This suggests that both AMOT proteins are associated with the postsynaptic density. We next stained neurons for endogenous AMOT-130/80, which demonstrated a punctate pattern along dendritic shafts and in spine heads that overlapped with PSD-95 (Fig. 1 F). To examine whether AMOT proteins interact with PSD-95, we immunoprecipitated AMOT-130/80 from neuron lysates. PSD-95 was detected in AMOT-130/80 coprecipitates (Fig. 1 G). Conversely, AMOT-130 was detected in PSD-95 coprecipitates (Fig. 1 H), confirming that both proteins form a complex in neurons. Because PSD-95 has been shown to associate with MUPP1, an AMOT-interacting protein (Wells et al., 2006; Sugihara-Mizuno et al., 2007), we also tested whether endogenous AMOT-130 precipitated MUPP1 from neuronal tissues. An antibody generated against the N-terminal region (2095) unique to AMOT-130 precipitated MUPP1 from CNS tissues (Fig. 1 I). Collectively, these data demonstrate that AMOT-130 is present in a complex together with PSD scaffolds. They also suggest that both splicing isoforms are abundantly expressed in CNS tissues.
AMOT-130 clusters with PSD scaffolds in the PSD
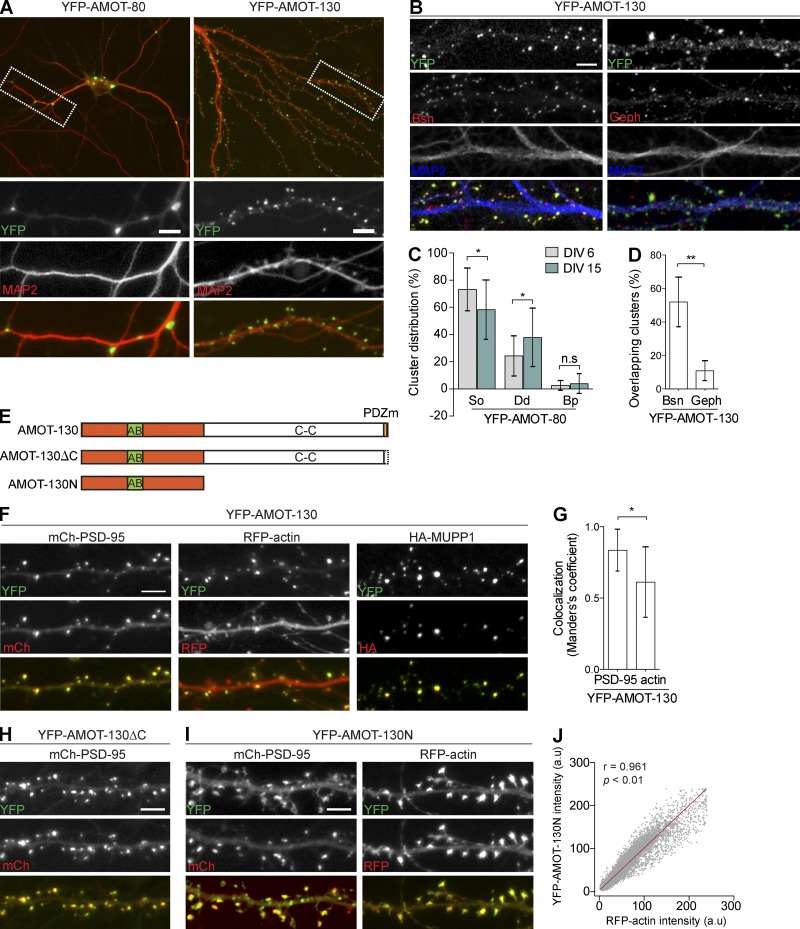
Our biochemical and staining analysis is consistent with a differential localization of AMOT-130 and AMOT-80, with AMOT-130 distributed in the PSD of mature synapses. To confirm this finding, we examined the localization of exogenous AMOT-130 and AMOT-80. In cultures, YFP–AMOT-130 localized in dendritic spines (Fig. 2 A, right), and immunostaining of MAP2 together with presynaptic Bassoon or postsynaptic gephyrin, markers for excitatory and inhibitory synapses, indicated that a majority of excitatory synapses are positive for AMOT-130 (Fig. 2, B and D; **, P < 0.01; two-way ANOVA with Sidak’s post hoc multiple comparisons test). These results show that a majority of excitatory synapses contain AMOT-130. In contrast, YFP–AMOT-80 accumulated in somatodendritic compartments of MAP2-labeled neurons (Fig. 2 A, left), indicating that AMOT-80 is not enriched in the synapse by itself. AMOT-80 puncta were highest in the cell soma in young cultures at 6 DIV (73 ± 0.03% clusters versus dendrite 58 ± 0.05% clusters; n = 22 neurons) and changed their distribution toward distal compartments in more mature cultures at 14 DIV (soma, 24 ± 0.03%, versus dendrite, 37 ± 0.05% punctate; n = 22; Fig. 2 C; *, P < 0.05; one-way ANOVA with Tukey’s post hoc test). We also noted that AMOT-80 puncta showed some accumulation at dendrite branch points (Fig. 2 A, bottom left), a pattern previously reported for neural Septin7 (Tada et al., 2007).
Figure 2.
Subcellular distribution of AMOT-130 and -80 in hippocampal neurons. (A) Neurons transfected with cDNAs encoding YFP–AMOT-80 and YFP–AMOT-130 (green) were immunostained for endogenous MAP2 (red). (B) Coimmunostaining of endogenous MAP2 (blue) together with Bassoon (Bsn, red; left) or gephyrin (Geph, red; right) in neurons expressing YFP–AMOT-130. (C) Quantification of YFP–AMOT-80 punctate as a fraction of total punctate numbers in soma (So), dendrites (Dd), and branch points (Bp) in culture at indicated stages. *, P < 0.05; one-way ANOVA with Tukey’s post hoc test. (D) Percentage of Bassoon and gephyrin clusters that overlap with YFP–AMOT-130. **, P < 0.01; two-way ANOVA with post hoc Sidak’s multiple comparisons test. (E) Schematic of AMOT-130 mutant constructs used in this study. AB, actin-binding domain. (F) Clusters of YFP–AMOT-130 distributed with mCherry–PSD-95 (left), RFP-actin (middle), or HA-MUPP1 (right). (G) Manders’s colocalization coefficient for clusters of YFP–AMOT-130 that overlap with mCherry–PSD-95 or RFP-actin. Error bars represent means ± SD. *, P < 0.05; Student’s t test. (H) Neurons coexpressing YFP–AMOT-130–ΔC and mCherry–PSD-95. (I) Neurons expressing YFP–AMOT-130N together with mCherry–PSD-95 or RFP-actin. Bars, 5 µm. (J) Overlap of YFP–AMOT-130N and RFP-actin clusters in spines. Pearson’s correlation coefficient = 0.961; P < 0.01.
To identify the domains that are responsible for AMOT-130 synapse localization, we expressed cDNAs encoding various AMOT-130 mutants: an AMOT-130–ΔC mutant lacking the last six amino acids comprising the PDZ-binding motif or an AMOT-130N construct containing the unique AMOT-130 N-terminal region (Fig. 2 E). The mutants were coexpressed with mCherry–PSD-95, RFP-actin, or HA-MUPP1, a known AMOT-130–interacting protein (Sugihara-Mizuno et al., 2007) and PSD component (Fig. 2 F; Krapivinsky et al., 2004). Accumulation of YFP–AMOT-130 was apparent with all PSD scaffolding members (Fig. 2, F and G). The association between AMOT-130 and MUPP1 (Fig. 1 I) suggested that the PDZ-binding motif of AMOT-130 was perhaps necessary for the enrichment of AMOT-130 in dendritic spines; however, exogenously expressed AMOT-80, which also contains the PDZ-binding motif, failed to enrich in spines (see Fig. 2 A). To exclude that the PDZ-binding motif directs AMOT-130 to spines, we examined the ability of AMOT-130–ΔC to distribute in spines. AMOT-130–ΔC localized in dendritic spines (Fig. 2 H) similar to WT AMOT-130, supporting the idea that the PDZ-binding motif is dispensable for AMOT-130 spine distribution. This suggests that other domains in AMOT-130 are involved in spine targeting. Because the main structural component of spines is actin and a conserved motif in the N-terminal region of AMOT-130 has been shown to directly associate with F-actin (Ernkvist et al., 2006), we transfected neurons with YFP–AMOT-130N. Indeed, YFP–AMOT-130N was highly enriched in spines, with a majority of punctates codistributing with mCherry–PSD-95 clusters (Fig. 2 I, left) and particularly with RFP–actin clusters (Fig. 2 I, right). Quantification of the relative AMOT-130N and RFP-actin fluorescence intensities revealed a near-complete linear correlation in overlapping actin clusters (Fig. 2 J; AMOT-130N and RFP-actin AU; Pearson correlation = 0.961; P < 0.01). As the AMOT-80 isoform that lacks the N-terminal domain is restricted from spines, our findings ascertain that the N-terminal domain is both necessary and sufficient for the localization of AMOT-130 into dendritic spines.
AMOT-130 silencing alters dendritic spine morphology
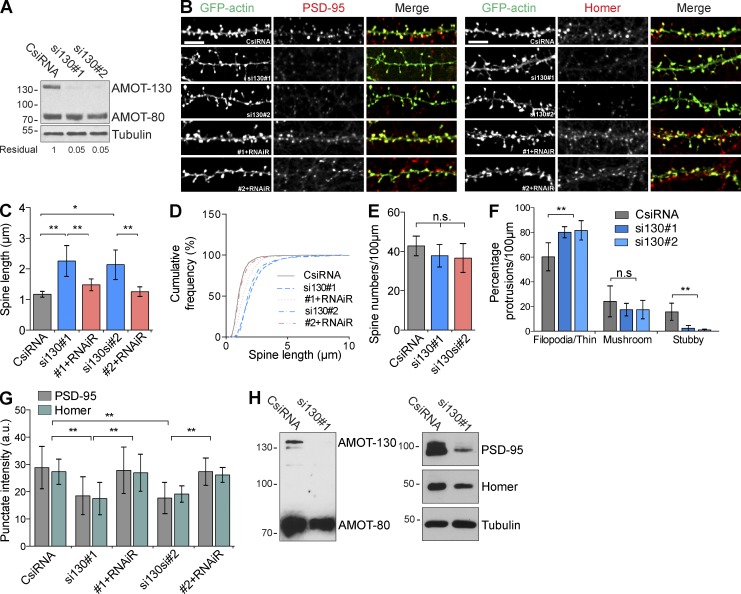
The synaptic distribution of AMOT-130 through the N-terminal domain that contains the F-actin–binding motif together with previous work showing that AMOT functions to regulate Rho-GTPase signaling (Wells et al., 2006; Ernkvist et al., 2009) suggests that AMOT-130 might be involved in spine remodeling. To address this possibility, we designed two AMOT-130 isoform-specific siRNAs (si130#1 and si130#2) that did not target other AMOT isoforms. The efficacy of the siRNAs was evaluated together with a negative control (CsiRNA) in rat IEC-18 cells for their high transfection efficiency. Treatment with si130#1 or si130#2 reduced the levels of endogenous AMOT-130 to ∼95% compared with cells treated with control siRNA (Fig. 3 A). We next introduced the siRNA in 11-DIV hippocampal neurons together with GFP-actin to mark dendritic spines and imaged cells 4 d later. As shown in Fig. 3 (B and C), AMOT-130 knockdown caused an elongated filopodia-like phenotype marked by protrusions that were 83–93% longer than those in control cells (Fig. 3 C; **, P < 0.01; ANOVA with Tukey’s post hoc test). In 2–3-wk-old cultures, most spines are assumed to have reached spine lengths between 1–1.5 µm (Boyer et al., 1998); however, the mean length of protrusions in AMOT-depleted cells was 2.25 ± 0.16 µm (si130#1) and 2.13 ± 0.15 µm (si130#2) compared with 1.17 ± 0.03 µm (Fig. 3 C). The shift toward longer protrusions was also apparent in a cumulative frequency distribution analysis (Fig. 3 D). However, AMOT-130–depleted neurons showed no difference in spine density compared with control cells (Fig. 3 E). To verify that the defect in length is caused by loss of AMOT-130, we performed rescue experiments by expressing a cDNA encoding human Flag–AMOT-130 resistant to rat siRNAs (RNAiR). Coexpression of the RNAiR construct together with siRNAs efficiently reversed the elongation phenotype (Fig. 3, B–D). Given the increase in immature protrusions, we also analyzed the distribution of filopodia-like/thin, mushroom, and stubby spines in knockdown neurons. Although the percentage of filopodia/thin spines was significantly increased by 33–35% compared with control (Fig. 3 F; **, P < 0.01; ANOVA with Tukey’s post hoc test), other spine types were reduced. However, only stubby spines reached significance (Fig. 3 F; **, P < 0.01; ANOVA with Tukey’s post hoc test), consistent with the unaltered total spine density (Fig. 3 E). The change in spine structure to an immature filopodia-like morphology is consistent with a role for AMOT-130 in stabilizing spines. To determine whether AMOT-130 depletion alters protein composition, we examined the immunoreactivity of PSD-95 and Homer. Loss of AMOT-130 provoked a significant reduction in total fluorescence of both scaffolding proteins, reducing intensity by ∼35% of PSD-95 (Fig. 3, B and G; CsiRNA vs. si130#1 or si130#2; **, P < 0.01) and by 30% of Homer clusters (Fig. 3, B and G; CsiRNA vs. si130#1 or si130#2; **, P < 0.01), correlating with the fractional increase of immature filopodia. This reduction was rescued by RNAiR expression (Fig. 3 G; CsiRNA versus si130#1 or si130#2; *, P < 0.05), suggesting that AMOT-130 levels stabilize PSD-95 and Homer in the spine. Lastly, to examine the effect of AMOT-130 depletion on the cytosolic levels of PSD-95 and Homer, we electroporated dissociated hippocampal neurons before plating (0 DIV) with control or si130#1. Lysates were collected 9 d later and analyzed by immunoblotting. Reduction of endogenous AMOT-130 diminished the levels of Homer and markedly of PSD-95 (Fig. 3 H), thus concluding that AMOT-130 plays a pivotal role for PSD scaffold stability.
Figure 3.
AMOT-130 knockdown destabilizes PSD scaffolds and triggers spine elongation. (A) Rat IEC-18 lysate transfected with control (CsiRNA) or AMOT-130 siRNA (si130#1 and si130#2) was analyzed by immunoblotting with the indicated antibodies. Numbers indicate residual amounts of AMOT-130 relative to control. (B) Hippocampal neurons transfected with siRNA together with GFP-actin or with RNAi resistant (RNAiR) AMOT-130 constructs were immunostained for endogenous PSD-95 and Homer. Bars, 5 µm. (C) Quantification of dendritic spine length in neurons transfected with siRNA. (D) Cumulative frequency plot of spine lengths. (E) Quantification of dendritic spine density. P = 0.55. (F) Characterization of spine shapes classified as filopodia/thin, mushroom, or stubby spines on neurons transfected with siRNA. (G) Quantification of PSD-95 and Homer cluster intensity in AMOT-130–depleted neurons. Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; one-way ANOVA followed by Tukey’s post hoc test. (H) CsiRNA or si130#1 electroporated into dissociated hippocampal neurons at 0 DIV before plating. Lysates were analyzed by immunoblotting with AMOT antibody and reprobed against PSD-95, Homer, and tubulin. Molecular masses are given in kilodaltons.
Kinetic properties of AMOT-130 in spines are determined by actin binding
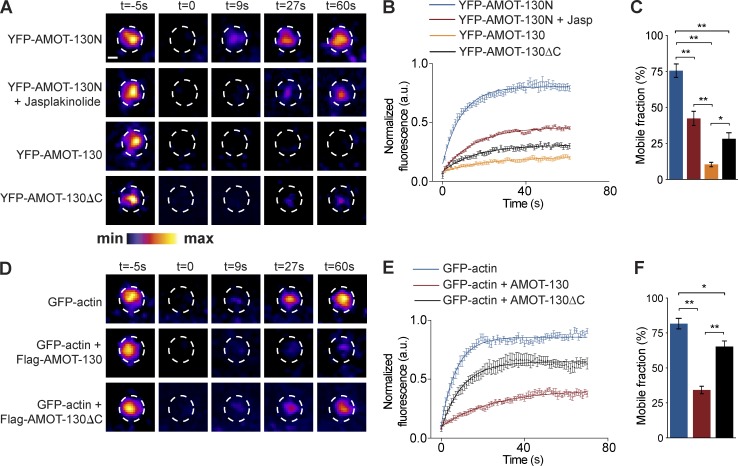
Because reducing AMOT-130 levels affects spine development, we next set out to more directly determine the role for AMOT-130 in spine stabilization. We first focused on the turnover of AMOT-130 protein in dendritic spines by performing FRAP experiments. Given the high correlation in cluster overlap of YFP–AMOT-130N and RFP-actin (Fig. 2 J), we first tested the mobility of the N-terminal region in neurons 2 d after transfection. YFP–AMOT-130N showed complete recovery within 60 s of photobleaching, similar to GFP-actin (Fig. 4, A, B, and D). The recovery was suppressed by treatment with the F-actin–stabilizing compound jasplakinolide (10 nM) linking the turnover of YFP–AMOT-130N to actin dynamics (Fig. 4, A and B). The mobile fraction of YFP–AMOT-130N was 75.5 ± 6.0% but was reduced to 42.3 ± 11% with jasplakinolide (Fig. 4 C; **, P < 0.01; ANOVA with Tukey’s post hoc test). These results show that the rapid exchange of AMOT-130N molecules is similar to that of actin (Star et al., 2002) and is reduced when filaments are pharmacologically stabilized, supporting the idea that in spines, AMOT-130 is coupled to the actin cytoskeleton. Furthermore, these results are consistent with previous findings reporting that the N-terminal region of AMOT-130 directly associates with F-actin (Chan et al., 2013; Dai et al., 2013).
Figure 4.
AMOT-130 stabilizes the actin cytoskeleton in dendritic spines. (A) FRAP results of YFP–AMOT-130 and derivative AMOT constructs in neurons treated with DMSO or 10 nM jasplakinolide (Jasp) as indicated. Time-lapse images show spine turnover of the indicated constructs before and at 0, 9, 27, and 60 s after photobleaching. The pseudo color scale is indicated at the bottom. Bar, 2 µm. (B) Normalized data fit in single exponential decay curves of fluorescence recovery (y-axis) plotted against time after bleaching (x-axis). (C) Quantification of AMOT recovery in dendritic spines. (D) FRAP results of GFP-actin alone or expressed together with Flag–AMOT-130 or Flag–AMOT-130–ΔC. Time-lapse images show spine protein turnover before and at 0, 9, 27, and 60 s after photobleaching. Dashed circles indicate photobleached spines. (E) Normalized data fit in single exponential decay curves of fluorescence recovery (y-axis) plotted against time after bleaching (x-axis). (F) Quantification of GFP-actin recovery in dendritic spines. Error bars represent means ± SD of the mobile fraction after photobleaching. *, P < 0.05; **, P < 0.01; one-way ANOVA followed by Tukey’s post hoc test.
In contrast with YFP–AMOT-130N, photobleaching YFP–AMOT-130 demonstrated strikingly different kinetics; only 10 ± 1.52% YFP–AMOT-130 recovered within the same time frame monitored (Fig. 4, A–C; AMOT-130 vs. AMOT-130N; AMOT-130N + Jasp; **, P < 0.01). This represented an 86% increase in the stable fraction, indicating that AMOT-130 is almost unaffected by changes in actin mobility. Given that AMOT-130 is highly stable in contrast with the AMOT-130N mutant, we reasoned that the C-terminal region that includes the PDZ-binding motif likely participates in the stabilization of AMOT-130. To examine this possibility, we expressed YFP–AMOT-130–ΔC in spines and monitored its recovery. The mobile fraction of YFP–AMOT-130–ΔC was approximately threefold larger than that of YFP–AMOT-130, but it was still reduced compared with YFP–AMOT-130N (Fig. 4, A–C; *, P < 0.05) or with jasplakinolide treatment (Fig. 4 C; AMOT-130–ΔC vs. AMOT-130N; **, P < 0.01). These data indicate that the PDZ domain–binding motif as well as a currently uncharacterized site in the coiled-coil domain of AMOT-130 influences its turnover rate.
Given that the spine morphology is altered by depletion of AMOT-130 and that actin stabilization reduces the turnover of YFP–AMOT-130N, we asked whether AMOT-130 directly affects the exchange of spine actin. To address this, we monitored the recovery of photobleached GFP-actin when coexpressed together with Flag-tagged AMOT-130. The mobile GFP-actin pool was unaffected by Flag–AMOT-130N expression (not depicted). However, in spines expressing Flag–AMOT-130, GFP-actin mobility was reduced to 34 ± 2.72% (Fig. 4, D–F; **, P < 0.01). Moreover, coexpression of Flag–AMOT-130–ΔC reduced the mobility of GFP-actin to 65 ± 3.94% relative to 82 ± 3.73% with GFP-actin alone (Fig. 4 D–F; *, P < 0.05). Thus, although PDZ domain interactions are not necessary for the synaptic targeting of AMOT-130, they do play a significant role in regulating spine actin turnover. Collectively, these results suggest that AMOT-130 controls spine morphogenesis though stabilization of actin filaments, consistent with its role as core organizer of the PSD.
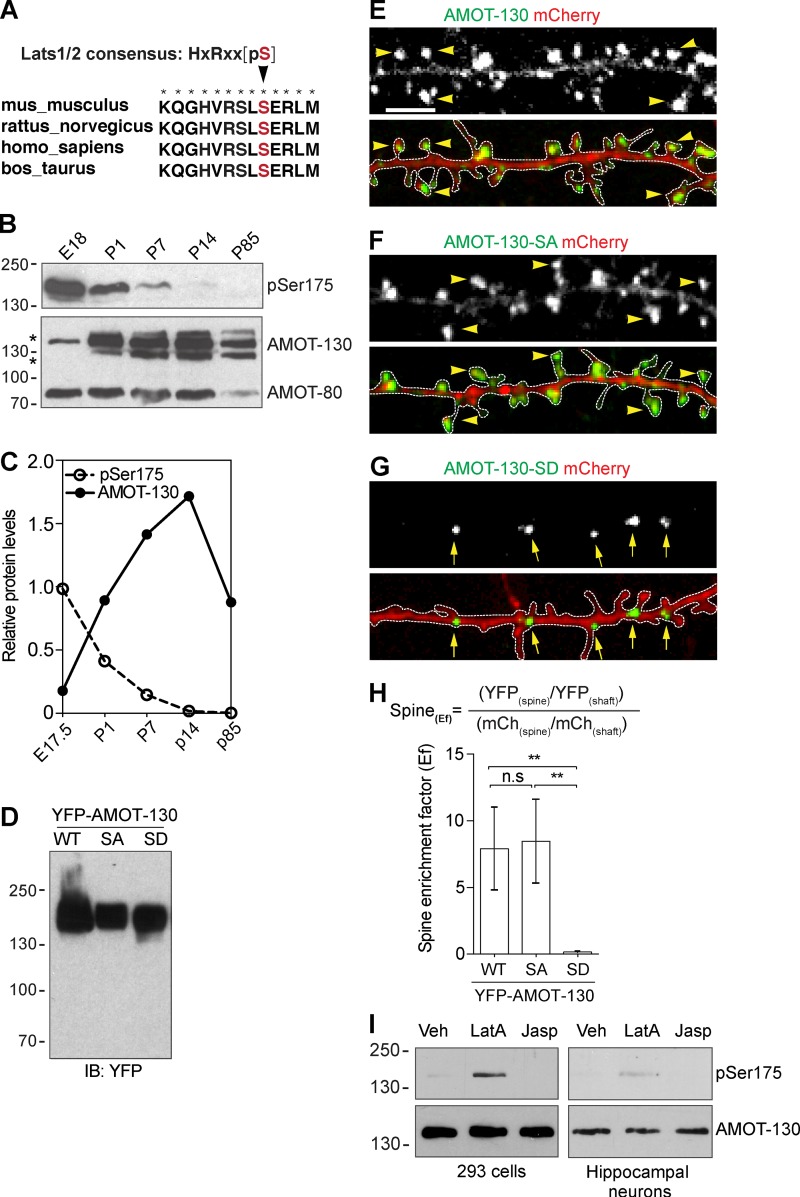
S-175 phosphorylation excludes AMOT-130 from spines
Combined, our data are consistent with AMOT-130 contributing to spine stability by controlling actin dynamics, in part, by directly coupling F-actin to the PSD. We next asked what regulates this association. Previously, a conserved serine residue (S-175), which is located in the AMOT-130 N-terminal region (Fig. 5 A), has been reported to disengage AMOT-130 from the actin cytoskeleton when phosphorylated (Adler et al., 2013; Chan et al., 2013; Dai et al., 2013; Leung and Zernicka-Goetz, 2013). If AMOT-130 contributes to spine actin stabilization, we reasoned that this association might be under control by phosphorylation during normal brain development. To determine whether the phosphorylation status of S-175 in AMOT-130 is affected during development when spines mature in vivo, we probed equal amounts of brain lysates prepared at different developmental stages with a phospho site-specific antibody (pSer175). We detected high levels of phosphorylated AMOT-130 in the late embryonic brain with the expression decreasing sharply until P14 (Fig. 5 B). This pattern contrasted with the expression of total AMOT-130 protein that was lower in the embryonic brain but increased during development, with peak expression at P14 (Fig. 5, B and C). Notably, AMOT-80 protein levels remained stable throughout early development. These results show that Ser175 in AMOT-130 is specifically phosphorylated before birth but is largely unphosphorylated in the postnatal brain.
Figure 5.
Phosphorylation at S-175 excludes AMOT-130 from spines. (A) Sequence alignment of AMOT-130. The arrowhead pointing to Ser175 (red) within the actin-binding domain of AMOT-130 is the consensus phosphogroup acceptor residue for the Lats1/2 kinases. Asterisks indicate fully conserved residues. (B) AMOT expression in the rat brain at the indicated stages analyzed by immunoblotting with antibodies against Ser175-phosphorylated AMOT-130 (pSer175) and total AMOT-130/80 protein. (C) Quantification of the relative expression of pSer175 and AMOT-130 protein in the rat brain. (D) Expression levels of YFP–AMOT-130–WT, YFP–AMOT-130–SA, and YFP–AMOT-130–SD in HEK293T cells. IB, immunoblot. (E–G) Neurons expressing mCherry together with YFP–AMOT-130–WT, YFP–AMOT-130–SA, or YFP–AMOT-130–SD. Panels show dendritic sections outlined by dashed lines with punctates in spines (arrowheads) or along dendritic shafts (arrows). Bar, 5 µm. (H) Spine enrichment of AMOT-130 by calculating the Ef. Error bars represent means ± SD. **, P < 0.01; one-way ANOVA with Tukey’s post hoc test. (I) HEK293T cells or 10-DIV hippocampal neurons were treated with Vehicle (Veh), LatA (2 µM), or jasplakinolide (2 µM) for 1 h and analyzed by immunoblotting as indicated. Molecular masses are given in kilodaltons.
To investigate the functional importance of the Ser175 phosphorylation, we generated YFP-tagged constructs encoding a phosphomimetic mutant, AMOT-130–SD, and a mutant that cannot be phosphorylated, AMOT-130–SA. The level of mutant expression was same as WT AMOT-130 when transfected in heterologous cells (Fig. 5 D). We then examined the subcellular distribution of the mutants relative to AMOT-130 in neurons. Similar to AMOT-130, the AMOT-130–SA mutant clustered in spine heads, whereas some expression was seen in the spine neck and along dendrites (Fig. 5, E and F). In contrast, AMOT-130–SD clusters were largely excluded from the spine but accumulated in dendritic shafts (Fig. 5 G). To quantify the accumulation of AMOT at synapses, we calculated the spine enrichment factor (Ef) by normalizing YFP spine fluorescence to mCherry in dendritic shafts (Fig. 5 H) as described (see the Confocal image acquisition and analysis section of Materials and methods). This revealed a drastic reduction of AMOT-130–SD expression in the spine head. The mean fluorescence intensity of AMOT-130–SD was 0.153 ± 0.016 compared with 7.91 ± 0.987 for AMOT-130 and 8.46 ± 0.996 for AMOT-130–SA (Fig. 5 H; **, P < 0.01; ANOVA with Tukey’s post hoc test), indicating that the phosphomimetic mutant less efficiently accumulates in spines compared with WT AMOT-130 or AMOT-130–SA. Because the AMOT-130 interaction with F-actin is controlled by phosphorylation, we asked whether changes in actin dynamics might influence the phosphorylation of AMOT-130. To test for this possibility, we treated cells with actin-modulating compounds that impair (latrunculin A; LatA) or increase (jasplakinolide) actin polymerization. Treatment with LatA triggered phosphorylation in both HEK293T cells and in neurons, suggesting that the integrity of the actin network affects AMOT-130 localization and phosphorylation (Fig. 5 I). Altogether, these results show that phosphorylation of Ser175 negatively controls enrichment of AMOT-130 in the synapse by disrupting actin filament binding.
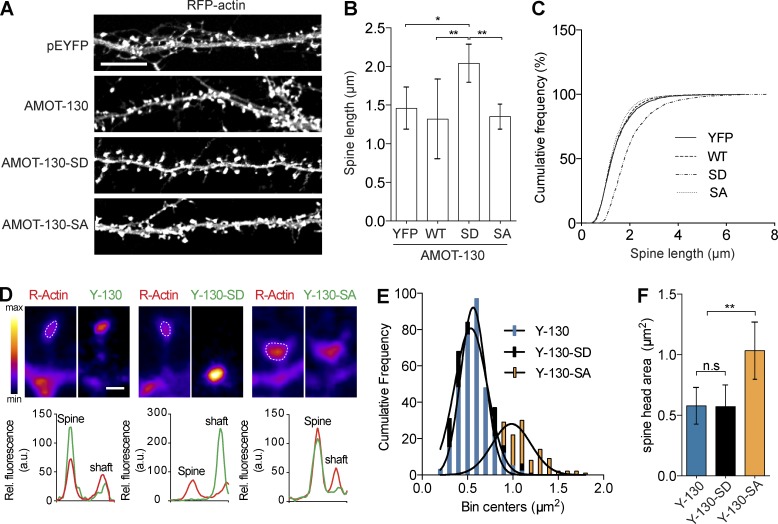
Effects of AMOT-130 point mutants on spine growth and morphology
Given the altered spine morphology by RNAi-mediated depletion of AMOT-130, we hypothesized that expression of AMOT-130–SD, which is unable to accumulate in spines, and AMOT-130–SA, which associates with F-actin, might impact spine morphology. Indeed, expression of AMOT-130–SD in neurons coexpressing RFP-actin to label spines induced a significant increase in spine length by 40% compared with cells expressing AMOT-130 or AMOT-130–SA (Fig. 6 A). The mean length in AMOT-130–SD neurons was 2.0 ± 0.1 µm compared with 1.3–1.5 µm for AMOT-130, AMOT-130–SA, or GFP-actin alone (Fig. 6, B and C; *, P < 0.05; **, P < 0.01; ANOVA with Tukey’s post hoc test). Thus, the loss-of-function effect on spine growth suggests that AMOT-130–SD affects disassembly of components that target, directly or indirectly, actin filaments, consistent with a common role of AMOT-130 as signaling platform (Yi et al., 2011; Mana-Capelli et al., 2014). Furthermore, we found in AMOT-130–SA–expressing neurons that the spine head area was consistently enlarged (Fig. 6 D). Quantification demonstrated a 40% increase in spine head area that was not observed with the other constructs (Fig. 6, E and F; **, P < 0.01; ANOVA with Tukey’s post hoc test). One possibility to account for the increase in spine volume might lie in the ability of the nonphosphorylated AMOT-130 to bundle actin. To test this, we expressed N-terminal mutants (Fig. S2 A; 130N-SD and 130N-SA) in COS-7 cells and analyzed their effects on actin filament formation. The 130N-SA construct colocalized to cortical RFP-actin and triggered significant amounts of actin stress fibers, which formed thick bundles that were absent in cells expressing 130N-SD or YFP (Fig. S2, B and C; *, P < 0.05; **, P < 0.01; ANOVA with Tukey’s post hoc test). Moreover, this effect was synergistically enhanced in cells coexpressing mCherry-Vasp (Fig. S2, C and D; *, P < 0.05; **, P < 0.01; ANOVA with Tukey’s post hoc test), which is known to promote bundling of actin filaments (Bachmann et al., 1999; Walders-Harbeck et al., 2002). Collectively, these results propose that AMOT-130 has bundling activity that contributes to actin stabilization, which can impact spine head size.
Figure 6.
AMOT-130 mutants trigger distinct effects on spine morphology. (A) Neurons expressing RFP-actin together with pEYFP, YFP–AMOT-130, YFP-AMOT-SD, or YFP-AMOT-SA. Panels show dendritic sections with RFP-actin expression. Bar, 10 µm. (B) Quantification of spine lengths. (C) Cumulative frequency plots of spine lengths. (D) Magnification of spines expressing the indicated constructs (top) and corresponding relative intensity profiles (bottom). Outlined spine heads with actin and pseudo color scale are indicated. Bar, 5 µm. (E) Bin distribution of spine head area. (F) Quantification of spine head area outlined by RFP-actin illustrated in D. Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; one-way ANOVA with Tukey’s post hoc test.
Lats1 specifically phosphorylates AMOT-130 on S-175 in neurons
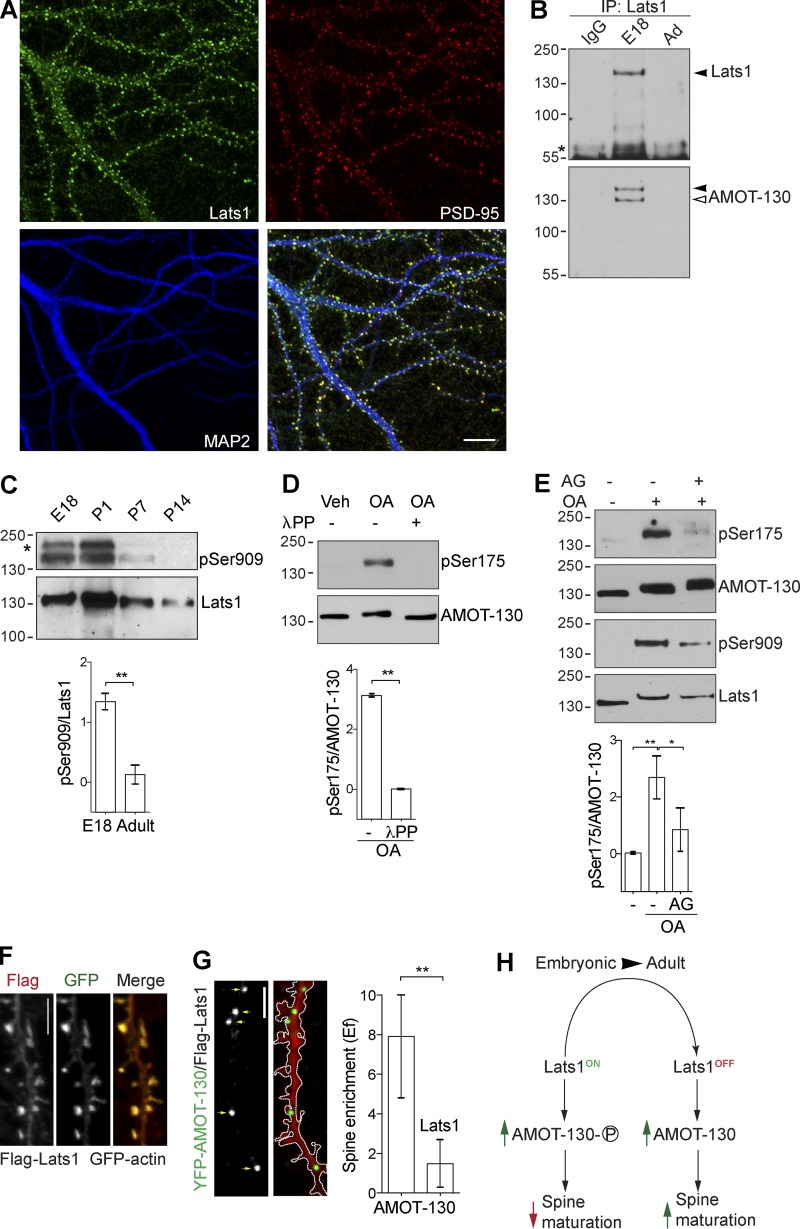
Given the differential distribution of AMOT-130 regulated by Ser175 phosphorylation, we next sought to determine the kinase responsible for phosphorylation. We focused on the NDR family kinase Lats1, which has been shown to directly phosphorylate AMOT-130 on Ser175 in heterologous cells (Adler et al., 2013; Dai et al., 2013; Mana-Capelli et al., 2014; Ultanir et al., 2014). To determine whether Lats1 targets AMOT-130 in the CNS, we first assessed its subcellular distribution in neurons. At 18 DIV, endogenous Lats1 was enriched in spines and overlapped with PSD-95 punctate (Fig. 7 A). No antibody cross-reactivity with exogenously expressed Lats2 was detected (Fig. 3 A), further affirming that Lats1 is specifically accumulated in spines. We next examined whether AMOT-130 and Lats1 form a complex by immunoprecipitating endogenous Lats1 from adult rat brain lysates collected from E18 or adult (>7 wk) animals. Precipitating equal amounts of Lats1 from E18 and adult brain tissue revealed that AMOT-130 significantly interacted in embryonic but not adult tissues (Fig. 7 B). Given the critical impact of AMOT-130 Ser175 phosphorylation on its function in spines, we next examined the kinase activity of Lats1 in vivo during development. Using an antibody that specifically recognizes phosphorylation of serine 909 in Lats1 (pSer909) as a readout for elevated kinase activity (Chan et al., 2005), we detected major phosphorylation in the embryonic and P1 stages with drastically lower amounts at P7 and no detectable signal in adult samples (Fig. 7 C; **, P < 0.01; two-tailed Student’s t test). The expression of total Lats1 levels peaked at P1 and then gradually decreased during brain development (Fig. 7 C), coinciding with the observed phosphorylation pattern of AMOT-130 in the brain (Fig. 5 B). These results indicated that Lats1 is a neuronal regulator of AMOT-130 during a developmental phase that coincides with the onset of synaptogenesis.
Figure 7.
AMOT-130 is a substrate for Lats1 in hippocampal neurons. (A) Neurons immunostained for Lats1 (green), PSD-95 (red), and MAP2 (blue) were imaged at 18 DIV. Bar, 10 µm. (B) Immunoprecipitation (IP) from 500 µg of E18 or adult rat brain lysate with the indicated antibodies. Resulting immunoblots were probed with Lats1 antibody (top, filled arrowhead) and reprobed with AMOT-130 2095 antibody (bottom, open arrowhead). The asterisk indicates the IgG heavy chain. (C) Analysis of phosphorylated (top) and Lats1 (middle) expression in rat brain at the indicated developmental stages using antibodies as indicated. Quantification of pSer-909 levels normalized to Lats1 protein in E18 and adult samples (bottom). **, P < 0.01; Student’s t test. (D) Neurons (10 DIV) stimulated with vehicle (Veh) or 100 nM Okadaic acid (OA) for 30 min followed by posttreatment with λ-protein phosphatase (λPP) were analyzed with the indicated antibodies (top and middle). Quantification of pSer175 levels normalized to AMOT-130 (bottom). **, P < 0.01; Student’s t test. (E) Neurons were exposed to 100 nM 17-AAG (AG) for 60 min before OA stimulation followed by immunoblotting with the indicated antibodies. Quantification of pSer175 levels normalized to AMOT-130. *, P < 0.05; **, P < 0.01; one-way ANOVA followed by Tukey’s post hoc test. Molecular masses are given in kilodaltons. (F) Immunostaining of Flag (red) in neurons coexpressing Flag-Lats1 together with GFP-actin. (G) Neurons expressing mCherry together with YFP–AMOT-130 and Flag-Lats1. Panels show dendritic sections of mCherry and YFP–AMOT-130 as outlined with punctate clusters along dendritic shafts indicated by arrows. Quantification of YFP–AMOT-130 spine enrichment (Ef). Error bars represent means ± SD. **, P < 0.01; Student’s t test. Bars, 5 µm. (H) Summary of AMOT-130 functions during synapse development (see Discussion).
To more directly assess whether Lats1 regulates AMOT-130 in vivo, we next asked whether neuronal AMOT-130 is also subject to modulation by serine/threonine phosphatases. As we reported previously, AMOT can associate with the protein phosphatase PP2A (Wells et al., 2006), which together with the structurally related PP-1 are important for synaptic plasticity. Importantly, both phosphatases have been shown to inhibit NDR/Lats kinases reversibly in response to the PP2A/PP-1 antagonist okadaic acid (OA; Kilili and Kyriakis, 2010; Couzens et al., 2013; Lv et al., 2015). Thus, we examined the effects of OA on the phosphorylation of Lats1 and AMOT-130. In neurons at 10 DIV, acute treatment with OA at 100 nM induced robust phosphorylation over 30 min of Ser175 (Fig. 7 D). We also noted an upward mobility shift on the immunoblots of total AMOT-130 protein levels, recapitulating the increased S-175 phosphorylation, which was abolished with phosphatase treatment (Fig. 7 D; **, P < 0.01; two-tailed Student’s t test). These results indicate that either PP2A, PP-1, or both are involved in regulating AMOT-130 phosphorylation. To confirm that Ser175 phosphorylation is dependent specifically on Lats1, we treated neurons with OA together with 17-(allylamino) geldamycin (AG), an inhibitor of Lats1 catalytic activity (Huntoon et al., 2010). Pretreatment with 100 nM AG for 1 h before OA exposure for 30 min caused a reduction in phosphorylation of both Lats1 and AMOT-130 (Fig. 7 E; vehicle vs. pSer175 with OA, **, P < 0.01; OA + 17-AAG, *, P < 0.05; ANOVA with Tukey’s post hoc test), supporting the notion that AMOT-130 is a target of Lats1 in hippocampal neurons. We note that despite loss of Ser175 phosphorylation with OA/17-AAG, the Lats1 inhibition did not impose a mobility shift in total AMOT-130 levels, suggesting that OA treatment is inducing other posttranslational effects (Wang et al., 2017). Nonetheless, these results provide direct evidence that Lats1 is a principal upstream kinase that phosphorylates AMOT-130 on Ser175 during brain development and in hippocampal neurons.
If Lats1 phosphorylation can affect AMOT-130 spine distribution as indicated by expression of the phosphomimetic AMOT-130–SD mutant (Fig. 5 G), one might expect a similar change in distribution upon phosphorylation of WT AMOT-130. To test this idea, we expressed cDNAs encoding Flag-Lats1 together with YFP–AMOT-130 or the phosphoresistant YFP–AMOT-130–SA mutant in neurons and analyzed the subcellular localization. Neurons stained for Flag showed Lats1 localization in GFP-actin–labeled spines (Fig. 7 F) in agreement with the endogenous staining (Fig. 7 A). Moreover, Flag-Lats1 coexpression displaced YFP–AMOT-130 clusters from the spine head (Fig. 7 G) but not YFP–AMOT-130–SA (Fig. S3, B and C); instead, increased fluorescence intensity was detected along the dendritic shaft (Fig. 7 G). The YFP–AMOT-130 clusters were significantly reduced in spines and favored in dendritic shafts (Fig. 7 G; YFP–AMOT-130 vs. YFP–AMOT-130 + Flag-Lats1; P < 0.01; two-tailed Student’s t test). Collectively, these results demonstrate that AMOT-130 and Lats1 act in a common pathway during the early development of synapse formation.
Discussion
A fundamental structural component of the spine is the cytoskeleton, which undergoes rapid remodeling that is supported by actin-binding proteins. Although some of the steps in this process are generally understood, our knowledge of how individual molecules influence the emergence of new spines remains unclear. In this study, we present evidence to suggest that AMOT-130 has a pivotal function in the early development of synapses by controlling actin cytoskeleton dynamics and spine remodeling.
Our findings suggest a model (Fig. 7 H) wherein AMOT-130 is spatially restricted from nascent filopodia/spine extensions at a time when levels of Lats1 expression and activity are high (Fig. 7 C). During later stages of development, Lats1 activity and/or expression decreases, allowing AMOT-130 to accumulate in spines, where it associates with F-actin and provides a positive modulator driving actin bundling via its N-terminal region. Once engaged in the spine, AMOT-130 then associates with the PSD and clusters with structural proteins including MUPP1 and PSD-95. These intermolecular interactions are likely to hold AMOT-130 anchored in the synapse in order to stabilize actin filaments and spine morphology.
Silencing of AMOT-130 leads to an increase in spine length that produces elongated filopodia-like protrusions, which show a reduction in PSD-95 and Homer fluorescence intensity (Fig. 3). It is thus likely that the effect of AMOT-130 silencing on spine growth arises from changes in the organization of the actomyosin cytoskeleton. Absence of AMOT-130 in spines also affected the stability of protein scaffolds in the PSD because AMOT-130 formed clusters with structural components including MUPP1 and PSD-95 (Fig. 2). These associations suggest that AMOT-130 functions to connect postsynaptic components to the actin cytoskeleton. Moreover, in cells depleted of AMOT-130, the ability of these scaffolds to form clusters was compromised, indicating that a critical level of AMOT-130 is needed for their stability. Indeed, exogenous AMOT-130 that is resistant to RNAi and contains an intact PDZ motif can reverse the loss of the PSD-95/Homer clusters as well as the spine growth defect in AMOT-depleted cells (Fig. 3). Restoration of AMOT-130 levels might therefore counterbalance loss in crowding in the normally dense postsynaptic membrane, which is necessary for the organization of scaffolding proteins (Frost et al., 2010).
This packing mechanism could also provide an explanation for the immobility of AMOT-130 in our FRAP analysis. Once AMOT-130 accumulates in the synapse, it becomes attached to stable structures that are unaffected by actin turnover. This is consistent with the observation that the amino terminal domain is highly dynamic, whereas the C-terminal region including the PDZ motif adds stability through intermolecular interactions (Fig. 4). Does AMOT-130 directly influence actin turnover? We observed little effect on GFP-actin mobility when the amino terminal region (Flag–AMOT-130N) was coexpressed (not depicted). However, expression of full-length AMOT-130 caused a significant stabilization of actin filaments that was reversed to some extent by depletion of the PDZ motif (Fig. 4). Still, these results suggest that the central coiled-coil domain might carry targeting information for AMOT-130 in the synapse. At this time it is unclear whether AMOT-130 stabilizes the total actin pool or whether only actin filaments in contact with the PSD are uncoupled. Actin filaments have been shown to undergo rapid turnover and are the main cytoskeletal structures responsible for modulation of spine shape (Okamoto et al., 2004). Given that the amino terminal region shows turnover rates similar to that of GFP-actin, which can assemble into stable filaments (Star et al., 2002), a possibility is that AMOT-130 prevents the growth of filaments.
In addition to the importance of direct coupling of the N-terminal region to F-actin, our morphological changes suggest that AMOT-130 signals through the actin cytoskeleton to control actin filament turnover. One likely mechanism is that AMOT-130 influences Rho-GTPase activity through its ability to spatially restrict the Rho-GAP Rich1/nadrin (Wells et al., 2006; Yi et al., 2011), which, at least in epithelial systems, has a pivotal role in regulating the actin cytoskeleton. However, whether the same signaling pathway accounts for AMOT-130 functions in the spine remains to be determined. In addition to a role for AMOT-130 on Rho-GAP activity, inhibition of the Rho-associated kinase (ROCK) has been reported to disrupt cellular polarity by mislocalizing AMOT-130 in a feedback-dependent pathway, leading to reduced F-actin formation (Newell-Litwa et al., 2015; Mihajlović and Bruce, 2016). This suggests that AMOT-130 strikes a critical balance between actin assembly and disassembly by bundling filaments, thereby reshaping the cytoskeleton. Whether AMOT-80 affects these events remains to be tested. Nonetheless, our data are consistent with previous work in endothelial cells showing that AMOT-130 is important for stabilizing cell–cell contact (Ernkvist et al., 2008).
Our data support an important role for the serine/threonine kinase Lats1 in phosphorylating AMOT-130 in the brain. The Lats1/2 kinases are core components of the Hippo pathway that are best known for regulating tissue growth by turning off the proproliferative activity of the transcriptional activators YAP/TAZ (Piccolo et al., 2014). Like AMOT-130, however, the role of the Lats1/2 kinases in the CNS has been poorly investigated. Our data show that Lats1 is found in dendritic spines and that AMOT-130 associates with Lats1 during the peak kinase activity in development (Fig. 7). Lats1 shares substrate recognition with Lats2, which can also phosphorylate AMOT-130 (Mana-Capelli et al., 2014). It is therefore possible that Lats2 is part of an AMOT-130–Lats1 complex, consistent with proteomic data showing that Lats2 is highly expressed in the hippocampus (Uhlén et al., 2015). In the brain, increase in the levels of postsynaptic proteins during spine maturation is a critical step for the stabilization of developing synapses (Sans et al., 2000; Petralia et al., 2005). Likewise, the rise in AMOT-130 levels appears to be an important feature to promote spine growth, which in turn is dependent on the reduction of Lats1 activity (Figs. 5 and 7). It is still, however, unclear whether Lats1 can influence synapse formation directly, although Lats1 can itself directly modulate actin polymerization and has altered kinase activity during actin remodeling (Visser-Grieve et al., 2011). A possibility, therefore, is that a high level of AMOT-130 in developing synapses acts to outcompete Lats1 for binding to actin, which could impact actin stability and spine growth.
We also examined a role for PP2A/PP-1 in Lats1-dependent phosphorylation of AMOT-130, which can dephosphorylate Lats1/2 and is the main source of phosphatase activity in the synapse (Kilili and Kyriakis, 2010; Lohmann and Kessels, 2014; Lv et al., 2015). We find that treatment with the PP2A/PP-1 antagonist OA enhances Lats1 activity toward AMOT-130 with consequent robust phosphorylation in cultured neurons (Fig. 7). This effect can directly be ascribed Lats1 because attenuation of Lats1/2 activity by addition of a kinase inhibitor leads to a reduction in AMOT-130 phosphorylation (Fig. 7). These results suggest that the PP2A/PP-1 phosphatases are linked to the biochemical pathway in neurons that results in Lats-dependent phosphorylation of AMOT-130.
To date, there is limited information on the functions of the core Hippo components in the synapse. However, emerging evidence suggests that modulators of Hippo signaling can regulate synaptic integrity and are essential for cognition. For example, knockdown of Scribble or its interacting protein NOS1AP both affect synaptic morphology and have been coupled to memory defects and increased susceptibility to schizophrenia (Moreau et al., 2010; Richier et al., 2010; Cervantes-Sandoval et al., 2016; Hilal et al., 2017). Furthermore, the memory-related protein KIBRA has been shown to be critical for synaptic plasticity (Makuch et al., 2011; Zhang et al., 2014). In all of these cases, the principal target is the actin cytoskeleton, which suggests that several mechanisms exist for these factors to potentially intersect with the Hippo kinase cascade.
Collectively, we have demonstrated a role for AMOT-130 in the regulation of dendritic spine morphogenesis. The AMOT-130 knockdown phenotype rises from a disturbance in the molecular machinery that controls actin turnover, which triggers growth of immature filopodia-like protrusions. Similar structural abnormalities with underlying deficits in actin cytoskeleton organization are commonly reported in autism spectrum disorders (Ebert et al., 2013). Given that AMOT-130 has been linked to a protein network implicated in autism (Schanzenbächer et al., 2016), an important question for the future is whether AMOT-130 dysregulation contributes to the etiology of autism pathologies.
Materials and methods
Constructs and antibodies
Plasmids with cDNA encoding YFP–-AMOT-L1, YFP–AMOT-L2, YFP–AMOT-130, YFP–AMOT-80, YFP–AMOT-130–ΔC, Flag–AMOT-130, Flag–AMOT-130–ΔC, and HA-MUPP1 have been described previously (Wells et al., 2006). YFP–AMOT-130N was generated by PCR amplification (Expand High Fidelity PCR system; Roche), digested with MfeI and SalI, and cloned into the pEYFP-C1 (Takara Bio Inc.) vector. Point mutations (S175A and S175D) in YFP–AMOT-130 and YFP–AMOT-130N were generated by the QuikChange Site-Directed Mutagenesis approach (Agilent Technologies). The mCherry-tagged PSD-95 construct was generated by PCR amplification, digested with AgeI and EcoRI, and cloned into the corresponding sites in the mCherry (Takara Bio Inc.) plasmid. The following plasmids were obtained from Addgene: 2×Flag-Lats1 (18971; a gift from M. Sudol, National University of Singapore, Singapore), mCherry–VASP-5 (55151; a gift from M. Davidson; Florida State University, Tallahassee, Florida), and pCIneoMyc-Lats1 and pCIneoMyc-Lats2 (66851 and 66852; gifts from Y. Hata, Tokyo Medical and Dental University, Tokyo, Japan). The pEGFP–β-actin and RFP–β-actin plasmids were provided by R. Duncan (Dalhousie University, Halifax, Canada). Primary antibodies were as follows: AMOT-130/80 (1:200; C-18; Santa Cruz Biotechnology, Inc.), Rab11 (1:200; H-87; Santa Cruz Biotechnology, Inc.), AMOT-130 (1:2,000; ABS1024; EMD Millipore), and pS175-AMOT (1:100; ABS1045; EMD Millipore). AMOT-130 (1:1,000; 2095) was generated by subcloning the N-terminal 600 bp of the human AMOT-130 sequence as an EcoR1–EcoR1 fragment into pGEXT3. A GST-fusion protein was then generated and used to immunize rabbits. For staining or Western blotting, commercial antibodies to the following proteins were used: PSD-95 (1:400; ab2723; Abcam), Lats1 (1:2,000; NBP1-86860; Novus Biologicals), Lats2 (1:2,000; NB200-199; Novus Biologicals), pS909-Lats1 (1:5,000; 9157; Cell Signaling Technologies), Rab5 (1:1,000; 2143; Cell Signaling Technologies), gephyrin (1:500; 147-011; Synaptic Systems), Bassoon (1:400; 141-011; Synaptic Systems), Homer (1:1,000; 160-002; Synaptic Systems). For costaining with Bassoon or gephyrin, antibodies to MAP2 (1:500; 188-004; Synaptic Systems), synaptophysin (1:5,000; S-5768; Sigma-Aldrich), tubulin (1:500; T7451; Sigma-Aldrich), HA (1:2,000; H3663; Sigma-Aldrich), and GFP (1:5,000; ab290; Abcam) were used. Secondary antibodies were used as follows: IgG–horseradish peroxidase goat (1:4,000; sc-2768; Santa Cruz Biotechnology, Inc.), mouse (1:4,000; sc-2380; Santa Cruz Biotechnology, Inc.), and rabbit (1:4,000; 31466; Thermo Fisher Scientific). Alexa Flour 488 anti–goat, Alexa Fluor 488 anti–rabbit, Alexa Fluor 594 anti–mouse, Alexa Fluor 647 anti–guinea pig, and Alexa Fluor 647 anti–mouse (1:1,000; A-27012, A-11034, A-11032, A-21450, and A-21235, respectively; Thermo Fisher Scientific) were also used.
siRNA sequences and knockdown validation
Isoform-specific siRNA oligonucleotides that target the F-actin–binding domain in rat AMOT-130 were obtained from Ambion/Thermo Fisher Scientific and Sigma-Aldrich. The target sequences were: si130#1, 5′-GTCCGATCCTTGAGCGAAA-3′ (s152896; Ambion), and si130#2, 5′-CCAAGATGAAGGCCTTAGA-3′ (SASI_Rn02_00284990; Sigma-Aldrich). A CsiRNA whose sequence was not homologous to any vertebrate sequence was used as negative control (SIC001-1NMOL; Sigma-Aldrich). To evaluate the knockdown efficacy of endogenous AMOT-130, 20 µM siRNA were transfected to a final concentration of 10 nM in rat IEC-18 cells (passage 5–10) using Lipofectamine 2000 (LF2000; Invitrogen). Cells were harvested 4 d later, and the relative expression levels of endogenous AMOT-130 were analyzed by immunoblotting using anti–AMOT-130 antibody. Quantification of the Western blot membranes by densitometry estimated a reduction in the AMOT-130 levels to ∼90% in IEC-18 cells relative to controls (see Fig. 3 A). For knockdown experiments, neurons at 11 DIV were transfected with siRNA together with 5 µg pEGFP–β-actin plasmid using LF2000 and prepared for experiments at 15–16 DIV. Alternatively, 2.5 × 106 dissociated hippocampal neurons were electroporated with 1 µM si130#1 or CsiRNA at the day of plating (0 DIV) and analyzed by immunoblotting at 9 DIV.
Cell culture and transfection
Dissociated cultures of primary hippocampal neurons were prepared from E18 Sprague-Dawley rat embryos as described previously (Richier et al., 2010). Unless otherwise stated, experiments were performed in 15-DIV hippocampal cultures. HEK293T, COS-7, and IEC-18 cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% FBS (HEK293T and COS-7) or 5% FBS (IEC-18) and 10 U/ml penicillin and 10 µg/ml streptomycin. HEK293T cells were transfected with polyethylenimine (PEI) by mixing 2–4 µg DNA with PEI (1 mg/ml) at a ratio of 4:1 PEI to DNA. COS-7 cells were transfected with LF2000 with 2 µg of each plasmid according to the manufacturer’s recommendations. For transfection of neurons with various cDNA, the calcium phosphate method was used as previously described (Richier et al., 2010). Electroporation was performed with a GenePulserXcell (Bio-Rad Laboratories) using the following parameters: a square pulse with 220 V and a 180-µs pulse in a 0.2-mm cuvette.
Preparation of brain sections and immunoprecipitations
For preparation of brain sections, the midbrain, striatum, hippocampus, cerebellum, and cortex were dissected from adult (>P45) Sprague-Dawley rats and homogenized in ice-cold lysis buffer containing 150 mM NaCl, 75 mM Tris-HCl, pH 8.0, 1% NP-40, and protease inhibitors. For analysis of developmental AMOT-130/80 expression, primary hippocampal neurons and total rat brains were prepared at indicated time points. Protein concentrations were measured using the bicinchoninic assay method (Thermo Fisher Scientific), and 10 µg of protein per lane was loaded for SDS-PAGE. For rat brain immunoprecipitations, crude synaptosomal P2 fractions were solubilized in lysis buffer and centrifuged at 13,800 g for 30 min. For hippocampal neuron immunoprecipitations, cultures at 15 DIV were solubilized and centrifuged at 13,800 g for 30 min. Lysates were incubated with 1–2 µg of either isotype-matched IgG, anti–AMOT-130, anti–PSD-95, or anti-Lats1 antibody at 4°C overnight followed by incubation with protein A–Sepharose beads (GE Healthcare). The beads were washed three times in lysis buffer and eluted in 3× Laemmli sample buffer, and then bound proteins were subjected to SDS-PAGE. Proteins were analyzed by immunoblotting using indicated antibodies and visualized by chemiluminescence (Thermo Fisher Scientific). Relative protein intensities were quantified by densitometry using ImageJ (National Institutes of Health) software.
Isolation of PSD fractions from rat brain
Adult rat brains were homogenized (12 strokes with a Teflon homogenizer) in ice-cold sucrose buffer A (320 mM sucrose, 2 mM Hepes, and 2 mM EDTA, pH 7.4) supplemented with protease inhibitors. The homogenized brain extract was centrifuged at 900 g for 10 min, and the pellet (P1) was resuspended in buffer A and then homogenized (six strokes) and spun again at 900 g for 10 min. The resulting supernatants (S1) were pooled and centrifuged at 10,000 g for 10 min. The supernatant (S2) was centrifuged at 176,000 g for 1 h, which yielded the supernatant (S3) and the pellet (P3). The pellet (P2) was lysed in four volumes of ice-cold water, adjusted to 4 mM Hepes, pH 7.4, and mixed for 30 min at 4°C. The lysate was centrifuged at 25,000 g for 20 min, yielding the supernatant LS1 and LP1. LP1 was resuspended in Hepes-buffered sucrose, layered on top of a discontinuous sucrose gradient (1.2, 1, and 0.8 M sucrose dissolved in 4 mM Hepes, 2 mM EDTA, and protease inhibitors, pH 7.4) and centrifuged at 150,000 g for 2 h. The membrane fraction between 1 and 1.2 M sucrose was collected and adjusted to 320 mM Hepes-buffered sucrose. The suspension was centrifuged at 200,000 g for 30 min, yielding the synaptic plasma membrane (SPM) fraction. The SPM was resuspended (0.5% Triton X-100, 50 mM Hepes, and 2 mM EDTA) and mixed for 15 min at 4°C followed by centrifugation at 32,000 g for 20 min, yielding the PSD fraction. The pellet (PSD) was extracted with ice-cold buffer D (6 mM Tris-HCl, pH 7.4, 0.5% Triton X-100, and protease inhibitors) or with 3% sarkosyl and protease inhibitors and centrifuged again at 201,800 g for 1 h to obtain PSDII and PSDIII, respectively. All final pellets were resuspended in 40 mM Tris-HCl, pH 7.4, with 0.5% Triton X-100.
Subcellular fractionation of HEK293T cells
Cells grown on 100-mm dishes were scraped in ice-cold buffer (250 mM sucrose, 3 mM imidazole, and 1 mM MgCl2, pH 7.4), homogenized by 10–15 strokes with a Dounce homogenizer, and centrifuged at 2,000 g for 20 min to yield the postnuclear supernatant (PNS). The PNS was adjusted to 7.5% sucrose, layered on top of a continuous (7.5–37.5% [wt/vol]) sucrose gradient, and centrifuged at 100,000 g for 4 h at 4°C. After ultracentrifugation, 10 fractions of equal volume were collected and analyzed by immunoblotting.
Cell stimulation and immunocytochemistry
Neurons at 8–10 DIV were treated with either vehicle (DMSO) or OA (100 nM; O8010; Sigma-Aldrich) and 17-AAG and 17-(Allylamino)-17-demethoxygeldanamycin (200 nM; sc-200461; Santa Cruz Biotechnology, Inc.). HEK293T cells and neurons were treated with jasplakinolide (EMD Millipore) or LatA (Cayman Chemical) at a final concentration of 2 µM. Lysate incubated with OA was posttreated with λ-protein phosphatase (5-10 U; P0753S; New England Biolabs, Inc.). During drug stimulations, the maintenance media was replaced with incubation solution containing 144 mM NaCl, 3 mM KCl, 2 mM MgCl2, and 10 mM Hepes, pH 6.7 and kept at 37°C in 5% CO2 for the indicated time periods. Neurons were fixed in 4% PFA with 4% sucrose in 1× PBS for 10–20 min at RT and permeabilized in 0.1% Triton X-100 for 10 min. For detection of Lats1, neurons were fixed for 3 min in 4% PFA/4% sucrose/1× PBS followed by fixation for another 10 min in ice-cold methanol. Fixed cells were washed extensively in 1× PBS and then blocked in 5% BSA in PBS with 0.1% Triton X-100 for 1 h. After blocking, cells were incubated with primary antibodies in 5% BSA overnight at RT. Then, cells were washed in 1× PBS three times for 2 min each, incubated with secondary antibodies in 5% BSA for 1 h at RT, washed in 1× PBS, and mounted on microscope slides with fluoromount (Sigma-Aldrich). COS-7 cells were fixed 2 d after transfection with 4% PFA followed by extensive washing in 1× PBS, and then they were mounted on microscope slides.
Confocal image acquisition and analysis
Images were acquired as z stack series taken at 0.3-µm step intervals with an LSM710 confocal scanning microscope using 40× 0.7 NA and 63× 1.4 NA oil immersion lenses and collected with a digital Axiocam camera controlled by ZEN software (ZEISS). The settings and acquisition parameters were the same across all experiments, and 15–20 neurons were analyzed for each experimental condition. Endogenous localization of AMOT-130/80 and PSD-95 in spines was measured by calculating the intensity profiles in the Alexa Fluor 488 and 594 channels across a line on dendrite images. For colocalization analysis, Manders’s overlap coefficient was calculated on background-subtracted images using the coloc2 plugin for Fiji (ImageJ). The correlation of YFP–AMOT-130N clusters overlapping with RFP-actin punctates was calculated by measuring fluorescence intensity of each channel. The corresponding overlap in pixel intensity was described by the Pearson’s correlation coefficient calculated in ImageJ. The AMOT-80 punctate was defined by calculating the ratio of punctate at a given compartment to the total numbers of punctate per neuron in equally thresholded images. To analyze spine enrichment of YFP–AMOT-130, YFP–AMOT-130–SD, and YFP–AMOT-130–SA, images were background subtracted and equally thresholded to include clusters twofold above the mCherry intensity in dendrites. Spines were outlined, and a mask of regions of interest (ROIs) was overlaid on 100-µm dendritic segments. The spine Ef was calculated by measuring the ratio of spine to shaft fluorescence intensity of YFP normalized to mCherry (mCh) according to the following equation:
where YFPspine and YFPshaft indicate peak green channel fluorescence intensities, and mChspine and mChshaft indicate peak red channel fluorescence intensities in the spine and shaft, respectively. The values are expressed as the total integrated fluorescence intensity. For analysis of PSD-95, Homer, or Bassoon spine clusters in AMOT-130–depleted neurons, z stacks of 10 images were collapsed by maximum-intensity projection. The threshold for different channels was identically adjusted and chosen such that all punctates were included in the analysis. Binary masks with pixel intensities were created, and punctates were highlighted by generating ROI masks that were applied along 100-µm dendritic segments that were quantified. Results were presented as the total integrated fluorescence intensity of each protein. All images were collected from at least two independent experiments per condition and analyzed with the experimenter blinded to the conditions.
FRAP
FRAP analysis was performed in a temperature- and CO2-controlled incubation chamber of a cell observer spinning-disk microscope (ZEISS) using the 488-nm line of a 100-mW argon laser with a 63× 1.4 NA oil immersion lens. During image acquisition, live neurons at 15 DIV were cultured in a glass-bottomed dish in Tyrode’s solution (25 mM Hepes, pH 7.4, 119 mM NaCl, 2.5 KCl, 2 mM CaCl2, 2 mM MgCl2, and 30 mM glucose) without or with jasplakinolide (J4580; Sigma-Aldrich) at a final concentration of 100 nM. To monitor the turnover of YFP-tagged AMOT or Pegfp–β-actin, a ROI with a diameter of ∼1 µm corresponding with individual spines of similar size was photobleached at full laser power (100% power and 100% transmission) for 2.5 s. Fluorescence recovery was monitored by automatic scanning of the whole cell in 0.2-s intervals at low (10–15%) laser power. No photobleaching was observed during recovery, whereas photodamage was accounted for by performing multiple bleach events within the same ROI. During image processing, the mean fluorescence of an untransfected area was measured as background and subtracted from the intensity of each frame to obtain the recovery. The fluorescence data were fit by nonlinear regression to an exponential one-phase association model in Prism (GraphPad Software). The mobile fraction (Mf) for analyzed proteins was approximated by photobleach correction of FRAP data using the following equation (Feder et al., 1996; Lippincott-Schwartz et al., 1999):
where Fprecell is the whole-cell prebleach intensity, Fpre is the bleach ROI prebleach intensity, F∞cell is the asymptote of fluorescence recovery of the whole cell, Fbackground is the mean background intensity, F∞ is the bleach ROI asymptote, and F0 is the bleach ROI immediately after bleach intensity. To obtain the Mf as percentage, the data were multiplied by 100, and results were presented by bar graphs made with Prism.
Data handling and statistical analysis
Statistical analysis was performed in Prism (6.0) software, and all datasets were checked for Gaussian normality distribution. Data testing of two groups was performed by a two-tailed unpaired Student’s t test. Multiple group analysis was performed using one-way ANOVA followed by the Tukey-Kramer post hoc test to account for multiple comparisons. Statistical significance was set at P < 0.05. In figures, significance levels were *, P < 0.05, and **, P < 0.01, as indicated in Prism. Quantitative analysis of spine density, spine length, and spine head area was performed manually in ImageJ by an observer blinded to the experimental conditions, and each experiment was reproduced at least two times with independent neuronal cultures. For spine density, the number of protrusions >100 µm dendritic segments was counted for each experimental condition. The length of a spine was measured from the base of the neck to the furthest point on the spine head. Spine head area was measured on spines at the best plane of view on z stack–collapsed images. All spines on primary, secondary, and tertiary dendrites located within 100 µm from the soma were included, and spine heads were manually traced in the actin channel. Dendritic spine numbers, lengths, and area on individual neurons were first averaged per neuron, and means from several neurons were then averaged to obtain a population mean. Spines visualized by GFP-actin were classified with regard to spine types as follows: stubby spines were ≤0.5 µm in length and lacked a clear head and neck, mushroom spines were ≥0.5 µm in length and had a short neck and large spine head, thin spines ranged between 1–2 µm in length and had elongated necks and small heads, and filopodial protrusions were ≥1.5–5 µm in length and lacked discernable spine heads. Collected data were presented as percentage frequency distributions of each spine type. Quantification of stress fiber bundles was performed on z-projected image stacks that were thresholded. The total number of clearly visible fibrillar structures in the resulting binary image was calculated in the actin and Vasp (red channel) in cells expressing AMOT-130 deletion mutants (green channel), and data from cells in each group were pooled. Measurements are given as means ± SD, and in figure legends, n refers to the number of cells counted for each condition. All figures were created in Illustrator CS5 (Adobe).
Online supplemental material
Fig. S1 A shows the fractionation protocol used in this study and subcellular distribution of AMOT-130/AMOT-80 in adult rat brain (Fig. S1 B) or in HEK293T cells (Fig. S1 C). Fig. S2 A shows a schematic of cDNA encoding the unique N-terminal region (AMOT-130N) in full-length AMOT-130, with mutations replacing S-175 with aspartic acid (AMOT-130N–SD) or alanine (AMOT-130N–SA). Fig. S2 B shows cDNA constructs expressing RFP-actin together with either control (pEYFP) or AMOT variants as outlined. Fig. S2 C shows cDNA constructs expressing mCherry-Vasp together with either control (pEYFP) or AMOT variants as outlined. Fig. S2 D shows the quantification of RFP-actin and mCherry-Vasp bundles. Fig. S3 A shows characterization of the Lats1 antibody used for immunostaining in Fig. 7 A, and clarified COS-7 lysates expressing cDNA as indicated were probed with antibodies against Lats1 (top), Lats 2 (middle), or myc (bottom). Fig. S3 B shows the neuronal distribution of the full-length YFP–AMOT-130–SA mutant in the abscence (left) or presence (right) of Lats1. Fig. S3 C shows quantification of YFP–AMOT-130–SA spine enrichment.
Supplementary Material
Acknowledgments
We thank Dr. Robert Brownstone and all members of the Fawcett and Krueger laboratories for critical evaluation and comments on the manuscript. We are grateful for the technical assistance from Stephen Whitefield.
This work was supported in part by funding from the Natural Sciences and Engineering Research Council of Canada to J.P. Fawcett (RGPIN-2017-05146) and S.R. Krueger (RGPIN-2011-326821), the Canadian Institutes of Health Research (MOP 341174), and a Research Scholar Award from the EJLB Foundation (to J.P. Fawcett). J.P. Fawcett was supported by the Canada Research Chairs Program through this work.
The authors declare no competing financial interests.
Author contributions: M. Wigerius designed experiments and performed biochemistry, imaging, and data analysis. L. Clattenburg performed antibody characterization. D. Quinn and A. Diab performed data analysis. A. Kolar and J. Qi performed animal handling and provided tissue cultures. S.R. Krueger provided advice and contributed to critical experimental work. M. Wigerius and J.P. Fawcett conceptualized and wrote the manuscript. J.P. Fawcett supervised the project.
References
- Aase K., Ernkvist M., Ebarasi L., Jakobsson L., Majumdar A., Yi C., Birot O., Ming Y., Kvanta A., Edholm D., et al. . 2007. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 21:2055–2068. 10.1101/gad.432007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J.J., Johnson D.E., Heller B.L., Bringman L.R., Ranahan W.P., Conwell M.D., Sun Y., Hudmon A., and Wells C.D.. 2013. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc. Natl. Acad. Sci. USA. 110:17368–17373. 10.1073/pnas.1308236110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anggono V., and Huganir R.L.. 2012. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 22:461–469. 10.1016/j.conb.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C., Fischer L., Walter U., and Reinhard M.. 1999. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J. Biol. Chem. 274:23549–23557. 10.1074/jbc.274.33.23549 [DOI] [PubMed] [Google Scholar]
- Barry E.R., and Camargo F.D.. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr. Opin. Cell Biol. 25:247–253. 10.1016/j.ceb.2012.12.006 [DOI] [PubMed] [Google Scholar]
- Boyer C., Schikorski T., and Stevens C.F.. 1998. Comparison of hippocampal dendritic spines in culture and in brain. J. Neurosci. 18:5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon N.J., and Sawa A.. 2011. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 12:707–722. 10.1038/nrn3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I., Chakraborty M., MacMullen C., and Davis R.L.. 2016. Scribble Scaffolds a Signalosome for Active Forgetting. Neuron. 90:1230–1242. 10.1016/j.neuron.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., and Silljé H.H.. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 24:2076–2086. 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Chan S.W., Lim C.J., Guo F., Tan I., Leung T., and Hong W.. 2013. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J. Biol. Chem. 288:37296–37307. 10.1074/jbc.M113.527598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzens A.L., Knight J.D., Kean M.J., Teo G., Weiss A., Dunham W.H., Lin Z.Y., Bagshaw R.D., Sicheri F., Pawson T., et al. . 2013. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 6:rs15 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- Dai X., She P., Chi F., Feng Y., Liu H., Jin D., Zhao Y., Guo X., Jiang D., Guan K.L., et al. . 2013. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 288:34041–34051. 10.1074/jbc.M113.518019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Gabel H.W., Robinson N.D., Kastan N.R., Hu L.S., Cohen S., Navarro A.J., Lyst M.J., Ekiert R., Bird A.P., and Greenberg M.E.. 2013. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 499:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernkvist M., Aase K., Ukomadu C., Wohlschlegel J., Blackman R., Veitonmäki N., Bratt A., Dutta A., and Holmgren L.. 2006. p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J. 273:2000–2011. 10.1111/j.1742-4658.2006.05216.x [DOI] [PubMed] [Google Scholar]
- Ernkvist M., Birot O., Sinha I., Veitonmaki N., Nyström S., Aase K., and Holmgren L.. 2008. Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim. Biophys. Acta. 1783:429–437. 10.1016/j.bbamcr.2007.11.018 [DOI] [PubMed] [Google Scholar]
- Ernkvist M., Luna Persson N., Audebert S., Lecine P., Sinha I., Liu M., Schlueter M., Horowitz A., Aase K., Weide T., et al. . 2009. The Amot/Patj/Syx signaling complex spatially controls RhoA GTPase activity in migrating endothelial cells. Blood. 113:244–253. 10.1182/blood-2008-04-153874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder T.J., Brust-Mascher I., Slattery J.P., Baird B., and Webb W.W.. 1996. Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70:2767–2773. 10.1016/S0006-3495(96)79846-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost N.A., Kerr J.M., Lu H.E., and Blanpied T.A.. 2010. A network of networks: cytoskeletal control of compartmentalized function within dendritic spines. Curr. Opin. Neurobiol. 20:578–587. 10.1016/j.conb.2010.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilal M.L., Moreau M.M., Racca C., Pinheiro V.L., Piguel N.H., Santoni M.J., Dos Santos Carvalho S., Blanc J.M., Abada Y.K., Peyroutou R., et al. . 2017. Activity-Dependent Neuroplasticity Induced by an Enriched Environment Reverses Cognitive Deficits in Scribble Deficient Mouse. Cereb. Cortex. 27:5635–5651. 10.1093/cercor/bhw333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y., Hirahara S., Inoue K., Suzuki A., Alarcon V.B., Akimoto K., Hirai T., Hara T., Adachi M., Chida K., et al. . 2013. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23:1181–1194. 10.1016/j.cub.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntoon C.J., Nye M.D., Geng L., Peterson K.L., Flatten K.S., Haluska P., Kaufmann S.H., and Karnitz L.M.. 2010. Heat shock protein 90 inhibition depletes LATS1 and LATS2, two regulators of the mammalian hippo tumor suppressor pathway. Cancer Res. 70:8642–8650. 10.1158/0008-5472.CAN-10-1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D., Viets K., Anderson C., Zhou C., DeFord P., Yan J., Cao J., and Johnston R.J. Jr. 2016. The insulator protein BEAF-32 is required for Hippo pathway activity in the terminal differentiation of neuronal subtypes. Development. 143:2389–2397. 10.1242/dev.134700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilili G.K., and Kyriakis J.M.. 2010. Mammalian Ste20-like kinase (Mst2) indirectly supports Raf-1/ERK pathway activity via maintenance of protein phosphatase-2A catalytic subunit levels and consequent suppression of inhibitory Raf-1 phosphorylation. J. Biol. Chem. 285:15076–15087. 10.1074/jbc.M109.078915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G., Medina I., Krapivinsky L., Gapon S., and Clapham D.E.. 2004. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 43:563–574. 10.1016/j.neuron.2004.08.003 [DOI] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. . 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Leung C.Y., and Zernicka-Goetz M.. 2013. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 4:2251 10.1038/ncomms3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Presley J.F., Zaal K.J., Hirschberg K., Miller C.D., and Ellenberg J.. 1999. Monitoring the dynamics and mobility of membrane proteins tagged with green fluorescent protein. Methods Cell Biol. 58:261–281. 10.1016/S0091-679X(08)61960-3 [DOI] [PubMed] [Google Scholar]
- Lohmann C., and Kessels H.W.. 2014. The developmental stages of synaptic plasticity. J. Physiol. 592:13–31. 10.1113/jphysiol.2012.235119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X.B., Liu C.Y., Wang Z., Sun Y.P., Xiong Y., Lei Q.Y., and Guan K.L.. 2015. PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep. 16:975–985. 10.15252/embr.201439951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makuch L., Volk L., Anggono V., Johnson R.C., Yu Y., Duning K., Kremerskothen J., Xia J., Takamiya K., and Huganir R.L.. 2011. Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron. 71:1022–1029. 10.1016/j.neuron.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maletic-Savatic M., Malinow R., and Svoboda K.. 1999. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 283:1923–1927. 10.1126/science.283.5409.1923 [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S., Paramasivam M., Dutta S., and McCollum D.. 2014. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell. 25:1676–1685. 10.1091/mbc.E13-11-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihajlović A.I., and Bruce A.W.. 2016. Rho-associated protein kinase regulates subcellular localisation of Angiomotin and Hippo-signalling during preimplantation mouse embryo development. Reprod. Biomed. Online. 33:381–390. 10.1016/j.rbmo.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Moleirinho S., Guerrant W., and Kissil J.L.. 2014. The Angiomotins--from discovery to function. FEBS Lett. 588:2693–2703. 10.1016/j.febslet.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M.M., Piguel N., Papouin T., Koehl M., Durand C.M., Rubio M.E., Loll F., Richard E.M., Mazzocco C., Racca C., et al. . 2010. The planar polarity protein Scribble1 is essential for neuronal plasticity and brain function. J. Neurosci. 30:9738–9752. 10.1523/JNEUROSCI.6007-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Hansen C.G., and Guan K.L.. 2015. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 15:73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K.A., Badoual M., Asmussen H., Patel H., Whitmore L., and Horwitz A.R.. 2015. ROCK1 and 2 differentially regulate actomyosin organization to drive cell and synaptic polarity. J. Cell Biol. 210:225–242. 10.1083/jcb.201504046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Nagai T., Miyawaki A., and Hayashi Y.. 2004. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7:1104–1112. 10.1038/nn1311 [DOI] [PubMed] [Google Scholar]
- Petralia R.S., Sans N., Wang Y.X., and Wenthold R.J.. 2005. Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol. Cell. Neurosci. 29:436–452. 10.1016/j.mcn.2005.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S., Dupont S., and Cordenonsi M.. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol. Rev. 94:1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Poon C.L., Mitchell K.A., Kondo S., Cheng L.Y., and Harvey K.F.. 2016. The Hippo Pathway Regulates Neuroblasts and Brain Size in Drosophila melanogaster. Curr. Biol. 26:1034–1042. 10.1016/j.cub.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Richier L., Williton K., Clattenburg L., Colwill K., O’Brien M., Tsang C., Kolar A., Zinck N., Metalnikov P., Trimble W.S., et al. . 2010. NOS1AP associates with Scribble and regulates dendritic spine development. J. Neurosci. 30:4796–4805. 10.1523/JNEUROSCI.3726-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saneyoshi T., and Hayashi Y.. 2012. The Ca2+ and Rho GTPase signaling pathways underlying activity-dependent actin remodeling at dendritic spines. Cytoskeleton (Hoboken). 69:545–554. 10.1002/cm.21037 [DOI] [PubMed] [Google Scholar]
- Sans N., Petralia R.S., Wang Y.X., Blahos J. II, Hell J.W., and Wenthold R.J.. 2000. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J. Neurosci. 20:1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanzenbächer C.T., Sambandan S., Langer J.D., and Schuman E.M.. 2016. Nascent Proteome Remodeling following Homeostatic Scaling at Hippocampal Synapses. Neuron. 92:358–371. 10.1016/j.neuron.2016.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M., and Kim E.. 2011. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 3:a005678 10.1101/cshperspect.a005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star E.N., Kwiatkowski D.J., and Murthy V.N.. 2002. Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 5:239–246. 10.1038/nn811 [DOI] [PubMed] [Google Scholar]
- Sugihara-Mizuno Y., Adachi M., Kobayashi Y., Hamazaki Y., Nishimura M., Imai T., Furuse M., and Tsukita S.. 2007. Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells. 12:473–486. 10.1111/j.1365-2443.2007.01066.x [DOI] [PubMed] [Google Scholar]
- Tada T., Simonetta A., Batterton M., Kinoshita M., Edbauer D., and Sheng M.. 2007. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 17:1752–1758. 10.1016/j.cub.2007.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. . 2015. Tissue-based map of the human proteome. Science. 347:1260419 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Ultanir S.K., Hertz N.T., Li G., Ge W.P., Burlingame A.L., Pleasure S.J., Shokat K.M., Jan L.Y., and Jan Y.N.. 2012. Chemical genetic identification of NDR1/2 kinase substrates AAK1 and Rabin8 Uncovers their roles in dendrite arborization and spine development. Neuron. 73:1127–1142. 10.1016/j.neuron.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultanir S.K., Yadav S., Hertz N.T., Oses-Prieto J.A., Claxton S., Burlingame A.L., Shokat K.M., Jan L.Y., and Jan Y.N.. 2014. MST3 kinase phosphorylates TAO1/2 to enable Myosin Va function in promoting spine synapse development. Neuron. 84:968–982. 10.1016/j.neuron.2014.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X. 2014. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 141:1614–1626. 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Verpelli C., and Sala C.. 2012. Molecular and synaptic defects in intellectual disability syndromes. Curr. Opin. Neurobiol. 22:530–536. 10.1016/j.conb.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Visser-Grieve S., Zhou Z., She Y.M., Huang H., Cyr T.D., Xu T., and Yang X.. 2011. LATS1 tumor suppressor is a novel actin-binding protein and negative regulator of actin polymerization. Cell Res. 21:1513–1516. 10.1038/cr.2011.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walders-Harbeck B., Khaitlina S.Y., Hinssen H., Jockusch B.M., and Illenberger S.. 2002. The vasodilator-stimulated phosphoprotein promotes actin polymerisation through direct binding to monomeric actin. FEBS Lett. 529:275–280. 10.1016/S0014-5793(02)03356-2 [DOI] [PubMed] [Google Scholar]
- Wang Y., Justilien V., Brennan K.I., Jamieson L., Murray N.R., and Fields A.P.. 2017. PKCι regulates nuclear YAP1 localization and ovarian cancer tumorigenesis. Oncogene. 36:534–545. 10.1038/onc.2016.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C.D., Fawcett J.P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., et al. . 2006. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 125:535–548. 10.1016/j.cell.2006.02.045 [DOI] [PubMed] [Google Scholar]
- Yi C., Troutman S., Fera D., Stemmer-Rachamimov A., Avila J.L., Christian N., Persson N.L., Shimono A., Speicher D.W., Marmorstein R., et al. . 2011. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 19:527–540. 10.1016/j.ccr.2011.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.X., and Guan K.L.. 2013. The Hippo pathway: regulators and regulations. Genes Dev. 27:355–371. 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W., and Lu B.. 2012. Synapses and dendritic spines as pathogenic targets in Alzheimer’s disease. Neural Plast. 2012:247150 10.1155/2012/247150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang S., Wennmann D.O., Chen Y., Kremerskothen J., and Dong J.. 2014. KIBRA: In the brain and beyond. Cell. Signal. 26:1392–1399. 10.1016/j.cellsig.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., Wang L.H., Liu C.Y., Lei Q., and Guan K.L.. 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25:51–63. 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.