Abstract
In mammals, FOXO transcriptional factors form a family of four members (FOXO1, 3, 4, and 6) involved in the modulation proliferation, apoptosis, and carcinogenesis. The role of the FOXO family in breast cancer remains poorly elucidated. According to the cellular context and the stage of the disease, FOXOs can have opposite effects on carcinogenesis. To study the role of FOXOs in breast carcinogenesis in more detail, we examined their expression in normal tissues, breast cell lines, and a large series of breast tumours of human origin. We found a very low physiological level of FOXO6 expression in normal adult tissues and high levels of expression in foetal brain. FOXO gene expressions fluctuate specifically in breast cancer cells compared to normal cells, suggesting that these genes may have different roles in breast carcinogenesis. For the first time, we have shown that, among the various FOXO genes, only FOXO6 was frequently highly overexpressed in breast cell lines and tumours. We also found that inhibition of the endogenous expression of FOXO6 by a specific siRNA inhibited the growth of the human breast cell lines MDA-MB-468 and HCC-38. FACS and Western blot analysis showed that inhibition of endogenous expression of FOXO6 induced accumulation of cells in G0/G1 phase of the cell cycle, but not apoptosis. These results tend to demonstrate that the overexpression of the human FOXO6 gene that we highlighted in the breast tumors stimulates breast carcinogenesis by activating breast cancer cell proliferation.
Keywords: gynecological cancers, cervical squamous cell carcinoma, endometrial adenocarcinoma, uc.189, prognosis
INTRODUCTION
The FOXO genes encode the proteins of the O-subfamily belonging to the large family of forkhead transcription factors that share a highly conserved DNA-binding domain, the forkhead domain or winged-helix domain [1, 2]. In mammals, the O-subfamily is composed of four genes: FOXO1, FOXO3, FOXO4, and FOXO6, involved in the regulation of various cellular processes, such as cell cycle progression, apoptosis, metabolism, and DNA-repair. FOXO genes are ubiquitously expressed in varying degrees in all mouse tissues examined [3–5]. It has been shown that FOXO6 is also expressed in mouse embryo, mainly in the brain [6]. The transcriptional activity of FOXO proteins is regulated by posttranslational modifications, such as acetylation, ubiquitination, and phosphorylation [1]. Notably, FOXOs are negatively modulated by growth factors, such as insulin, via activation of the PI3K-AKT pathway. Activation of this pathway induces phosphorylation of FOXO proteins by AKT, leading to their exclusion from the nucleus, thereby terminating their ability to induce target genes [6, 7]. Human tumours frequently harbour activating mutations in PIK3CA (or p110α, the catalytic subunit of PI3K) or inactivating mutations in PTEN (negative regulator of the PI3K-AKT pathway), leading to over-stimulation of PI3K-AKT pathway activity [8].
Because of their anti-proliferative and pro-apoptotic functions, and the fact that conditional deletion of FOXO1/2/4 alleles in adult mouse tissues leads to the appearance of lymphoblastic thymic lymphomas and haemangiomas, FOXOs have been considered to be tumour suppressors [2]. However, various studies have described unexpected functions of FOXOs in resistance to cancer treatment and cancer promotion, suggesting a complex role of FOXOs in this disease. Overexpression of FOXO1 and FOXO3 has been shown to inhibit the growth of breast cancer cells [9–12]. IκB kinase and ERk promote breast carcinogenesis via inhibition of FOXO3 [9, 11]. Moreover, cytoplasmic FOXO3 staining is positively correlated with poor patient survival [9]. These results strongly suggest that FOXOs act as tumour suppressors in breast cancer. However, FOXO1 and 3 have also been implicated in the promotion of breast tumour cell invasion [13, 14]. The results reported by Sisci et al. suggest that the role of FOXO3 in breast cancer is linked to the oestrogen receptor α (ERα) status: in ERα-positive cells, FOXO3 inhibits breast carcinogenesis, while in ERα-negative cells, FOXO3 tend to promote breast carcinogenesis [15]. The role of FOXOs in breast carcinogenesis therefore appears to depend on the cellular context and the stage of disease. The possible role of other FOXOs proteins (FOXO4 and 6) in breast cancer remains unknown.
To more clearly define the role of the FOXO family in breast cancer, we studied their expression in normal tissues, breast cell lines, and tumours of human origin. Surprisingly, we found that FOXO6, but not FOXO1, 3, and 4, was frequently overexpressed in breast cell lines and tumours compared to normal cells, suggesting that this FOXO gene could act as an oncogene in human breast carcinogenesis. To further examine this possibility, we studied the effect of inhibition of endogenous FOXO6 expression on cell growth of two different human breast cell lines expressing high levels of FOXO6.
RESULTS
Expression analysis of FOXO genes in a variety of human normal tissues and cancer
To study the involvement of the four FOXO genes in cancer, FOXO gene expression was first determined in 24 normal human tissues by qRT-PCR (Supplementary Table 1). These genes were ubiquitously expressed in all normal tissues examined. The highest FOXO1 expression was detected in the uterus, skeletal muscle and ovary, the highest expression FOXO3 was detected in bone narrow and skeletal muscle, and the highest FOXO4 expression was detected in the placenta, adrenal gland, ovary and skeletal muscle. Moderate to low FOXO6 expression was observed in normal adult tissues and was lower than the expression of other FOXO genes. This FOXO gene was highly expressed in foetal brain (mRNA expression level = 502) but not in foetal liver (mRNA expression level = 9) in keeping with earlier report [6]. The human FOXO6 gene is therefore expressed in a specific temporal and spatial pattern.
We then compared the expression of these genes in four types of cancer and corresponding normal tissues: breast, brain, bladder, and colon. Five normal tissues and 10 cancer tissues were analysed for each cancer type. FOXO1, FOXO3, and FOXO4 were significantly underexpressed in tumour samples: in breast, bladder and colon tumours for FOXO1, in breast tumours for FOXO3, and in breast, bladder, and colon tumours for FOXO4 (Supplementary Figure 1). Interestingly, in contrast with the other tumors, slight significant overexpression of FOXO1 was observed in brain tumours, and the expression of FOXO3 was found to be overexpressed in some of these tumors.
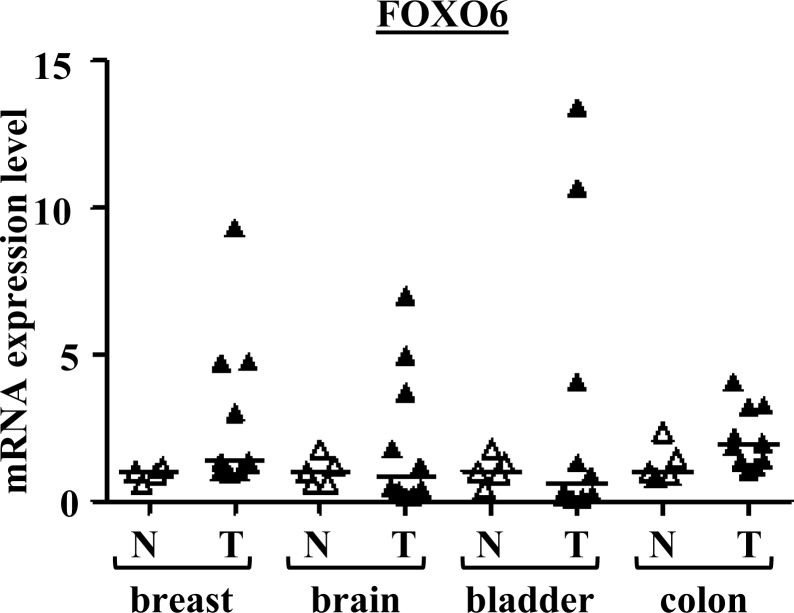
Surprisingly, very heterogeneous FOXO6 expression was observed in the various cancer tissues examined (Figure 1). Most importantly, this gene was found to be highly overexpressed in several bladder, brain and breast tumours. Therefore, FOXO6 would play an important role in carcinogenesis, notably in breast cancer. These observations have led us to study further the involvement of this gene in breast carcinogenesis.
Figure 1. FOXO6 mRNA expressions in various cancers and normal tissues.
Scatter dot plot with median of qRT-PCR data for FOXO6 in the series of breast, brain, bladder and colon tissues (n = 5 normal tissues and n = 10 tumour tissues for each). p-values (Mann-Whitney U Test) are indicated: *, 0.01 < p-value < 0.05; **, p-value < 0.01.
Expression analysis of FOXO genes in human breast cell lines and tumours
To study further the role of FOXO6 in breast cancer, first we used qRT-PCR to examine the expression of FOXO genes in a series of 39 human breast cell lines (including seven normal breast cell lines (N) and thirty-two tumorigenic breast cell lines (T), Supplementary Table 2) and in a large series of 527 human breast tumours (clinical parameters presented in Supplementary Table 3). We confirmed that FOXO6, but not FOXO1, 3 or 4, was frequently overexpressed in breast cell lines (25%) and in breast tumours (26.9%) (Table 1 and Supplementary Table 2).
Table 1. mRNA expressions of FOXO in breast cell lines and tumors.
| Cell line/ Tumor | Gene | FOXO1 | FOXO3 | FOXO4 | FOXO6 |
|---|---|---|---|---|---|
| Expression | Number (%) | ||||
| Tumorigenic cell lines (n = 32) | Overexpression Normal expression |
0 (0) 32 (100) |
0 (0) 32 (100) |
0 (0) 32 (100) |
8 (25) 24 (75) |
| Tumors (n = 527) |
Overexpression Normal expression |
1 (0.2) 526 (99.8) |
23 (4.4) 504 (95.6) |
6 (1.1) 521 (98.9) |
142 (26.9) 385 (73.1) |
FOXO6-expression values of the breast cell lines were normalized so that the ‘basal FOXO6 mRNA level’ (smallest quantifiable amount of mRNA (Ct = 35)) was equal to 1. Overexpression was defined as Ct values under 30 (values above 32 (2ΔCt = 235–30 = 32)).
FOXO6-expression values of the breast tumors were normalized so that the median FOXO6-expression value of normal breast tissues was equal to 1. Overexpression was defined as threefold variations of expression relative to the median expression of normal samples.
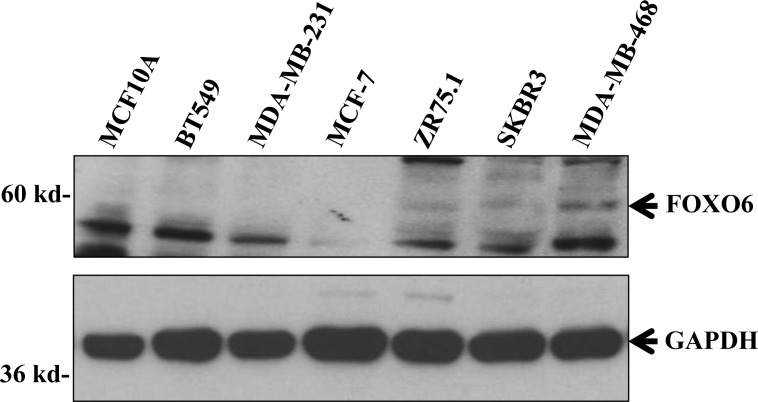
Western blot analysis demonstrated FOXO6 protein expression in breast cell lines expressing a high level of FOXO6 mRNA, but not in breast cell lines expressing low levels of this mRNA or in the normal mammary cell line MCF10A, confirming FOXO6 protein overexpression in breast cancer cells (Figure 2 and Supplementary Table 2).
Figure 2. FOXO6 protein expression in various breast cell lines.
Cellular extracts of various breast cell lines were analysed by immunoblotting for their expressions of FOXO6 and GAPDH (loading control).
Altogether, these results indicate that, among the four members of the FOXO gene family, only FOXO6 is overexpressed in breast cancer.
FOXO6 expression is negatively correlated with expression of an active form of AKT1 and positively correlated with PTEN expression in human breast tumours
The PI3K-AKT pathway has been shown to inhibit FOXO1, 3 and 4 expression [16], suggesting that FOXO6 overexpression in breast cancers could be due to impaired activity of this pathway. To test this hypothesis, we investigated the correlations between FOXO6 mRNA expression level and various proteins involved in the PI3K-AKT pathway.
The levels of fourteen proteins (non-phosphorylated and/or phosphorylated) involved in the PI3K-AKT pathway were analysed using RPPA assays in 224 samples from our series of 527 human breast tumours (Table 2). A negative correlation was observed between the expressions of FOXO6 and AKT1 phosphorylated on serine 473, a marker of PI3K-AKT pathway activity [17] (Spearman test: r = –0.192, p = 0.0039). A positive correlation was also observed between the expressions of FOXO6 and PTEN, a negative regulator of the PI3K-AKT pathway [17] (Spearman test: r = +0.135, p = 0.044, confirmed at the mRNA level: r = +0.212, p = 0.000018).
Table 2. Relationship between levels of FOXO6 mRNA and a panel of proteins of the PI3K-AKT pathway in a series of 224 breast tumors.
| Proteins of the PI3K-AKT pathway | ra | p-valuea |
|---|---|---|
| PTEN | +0.135 | 0.044 |
| INPP4b | +0.036 | NS |
| Akt1 | +0.060 | NS |
| p-Akt1.ser473 | –0.192 | 0.0039 |
| Akt2 | –0.010 | NS |
| mTor | –0.100 | NS |
| p-mTor.ser2448 | –0.019 | NS |
| FOXO1 | –0.086 | NS |
| TSC2 | +0.038 | NS |
| p70.S6.Kinase | +0.076 | NS |
| p-p70.S6.Kinase.thr389 | +0.014 | NS |
| S6.Ribosomal.protein | +0.019 | NS |
| p-S6.Ribosomal.protein.ser235/ser236 | –0.058 | NS |
| p-S6.Ribosomal.protein.ser24 | –0.046 | NS |
a:Spearman rank correlation Test. In bold: p-values < 0.05.
These observations indicate that the FOXO6 overexpression observed in human breast cancers is correlated with a low PI3K-AKT pathway activity.
Relationship between FOXO6 mRNA expression in human breast tumours and classical clinicopathological parameters
We investigated the relationship between FOXO6 expression and clinicopathological parameters (Table 3). No correlation was observed between FOXO6 mRNA levels and age, grade, lymph node status, macroscopic tumour sizes, molecular subtypes and presence of metastases. However, a weakly positive correlation was observed between FOXO6 mRNA overexpression and PR-positive status (p = 0.035) and, more interestingly, with the proliferation marker MKI67 (p = 0.027).
Table 3. Relationship between FOXO6 transcript level and classical biological parameters in a series of 527 breast tumors.
| Clinical biological parameters | Number of patients (%) | p-valuea | ||
|---|---|---|---|---|
| Total population |
FOXO6 mRNA expression ˂ 3 relative to normal |
FOXO6 mRNA expression ≥ 3 relative to normal | ||
| Total | 527 (100) | 385 (73.1) | 142 (26.9) | |
| Age ≤50 >50 |
125 (23.7) 402 (76.3) |
99 (79.2) 286 (71.1) |
26 (20.8) 116 (28.9) |
0.076 (NS) |
| SBR histological gradeb,c I II III |
60 (11.7) 241 (47.1) 211 (41.2) |
42 (70) 187 (77.6) 145 (68.7) |
18 (30) 54 (22.4) 66 (31.3) |
0.088 (NS) |
| Lymph node statusd 0 1–3 >3 |
159 (30.5) 250 (47.9) 113 (21.6) |
114 (71.7) 179 (71.6) 88 (77.9) |
45 (28.3) 71 (28.4) 25 (22.1) |
0.42 (NS) |
| Macroscopic tumor sizee ≤25 mm >25 mm |
248 (48) 269 (52) |
181(73) 197 (73.2) |
67 (27) 72 (26.8) |
0.95 (NS) |
| ERα status Negative Positive |
181 (34.3) 346 (65.7) |
140 (77.3) 245 (70.8) |
41 (22.7) 101 (29.2) |
0.11 (NS) |
| PR status Negative Positive |
255 (48.4) 272 (51.6) |
197 (77.2) 188 (69.1) |
58 (22.7) 84 (30.9) |
0.035 |
| ERBB2 status Negative Positive |
397 (75.3) 130 (24.7) |
297 (74.8) 88 (67.7) |
100 (25.2) 42 (32.3) |
0.11 (NS) |
| Molecular subtypes HR– ERBB2– HR– ERBB2+ HR+ ERBB2– HR+ ERBB2+ |
102 (19.4) 72 (13.7) 295 (56) 58 (11) |
82 (80.4) 52 (72.2) 215 (72.9) 36 (62.1) |
20 (19.6) 20 (27.8) 80 (27.1) 22 (37.9) |
0.093 (NS) |
| Histological typesf Ductal Lobular Other |
398 (89.6) 28 (6.3) 18 (4.1) |
294 (73.9) 20 (71.4) 14 (77.8) |
104 (26.1) 8 (28.6) 4 (22.2) |
0.89 (NS) |
| MKI67 mRNA expressionh Median (range) |
12.5 (0.8–313) | 11.91 (0.8–313) | 14.05 (1.74–117.3) | 0.027 i |
| Metastasis No Yes |
317 (60.2) 210 (39.8) |
229 (72.2) 156 (74.3) |
88 (27.8) 54 (25.7) |
0.60 (NS) |
a: Chi-squared Test; b: Scarff Bloom Richardson classification; c: information available for 512 patients; d: information available for 522 patients; e: information available for 517 patients; f: information available for 444 patients; h: information available for 438 patients; i: Mann-Whitney’s U test. Abbreviations: NS: not significant; ERα: estrogen receptor alpha; PR: progesteron receptor; ERBB2: human epidermal growth factor receptor 2; HR: hormone receptor.
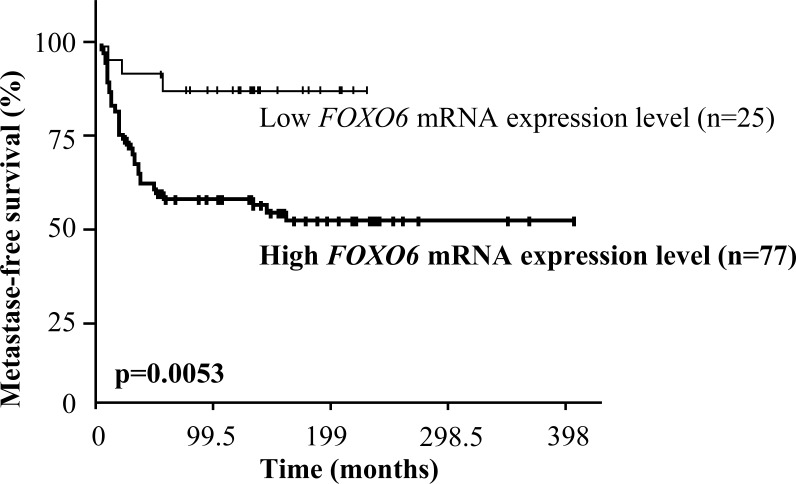
Most importantly, a log rank test was used to identify relationships between metastase-free survival (MFS) and FOXO6 mRNA levels. FOXO6 overexpression was not a prognostic marker in our series of 527 breast tumors (data not shown). We therefore investigated if FOXO6 overexpression could be a prognostic marker in a subpopulation of breast cancer (see Table 3). The high expression of FOXO6 was not a marker of poor prognostic in HR- ERBB2+, HR+ ERBB2-, HR+ ERBB2+, lobular, or ductal breast cancer. However, this approach allowed us to highlight that the high expression of FOXO6 was a marker of poor prognostic in our subpopulation of breast tumors HR- and ERBB2- (triple negative breast tumors) (p = 0.0053, Figure 3). The classical biological parameters: age, SBR histological grade, lymph node status, macroscopic tumor size, and PIK3CA mutation status, were not prognostic markers in this series of triple negative breast tumors (data not shown). The prognostic value of the FOXO6 expression in this subpopulation of breast tumors is therefore independent of biological parameters studied.
Figure 3. Survival curves of two groups of patients according to FOXO6 mRNA expression level in the cohort of 102 triple negative breast tumors.
AUC analysis was used to divide the population into two relevant FOXO6 expression subgroups.
Altogether, our results raise the hypothesis that FOXO6 overexpression may play an important role in the development of triple negative breast tumor.
Effect of FOXO6 siRNA on the growth of two different human breast cell lines
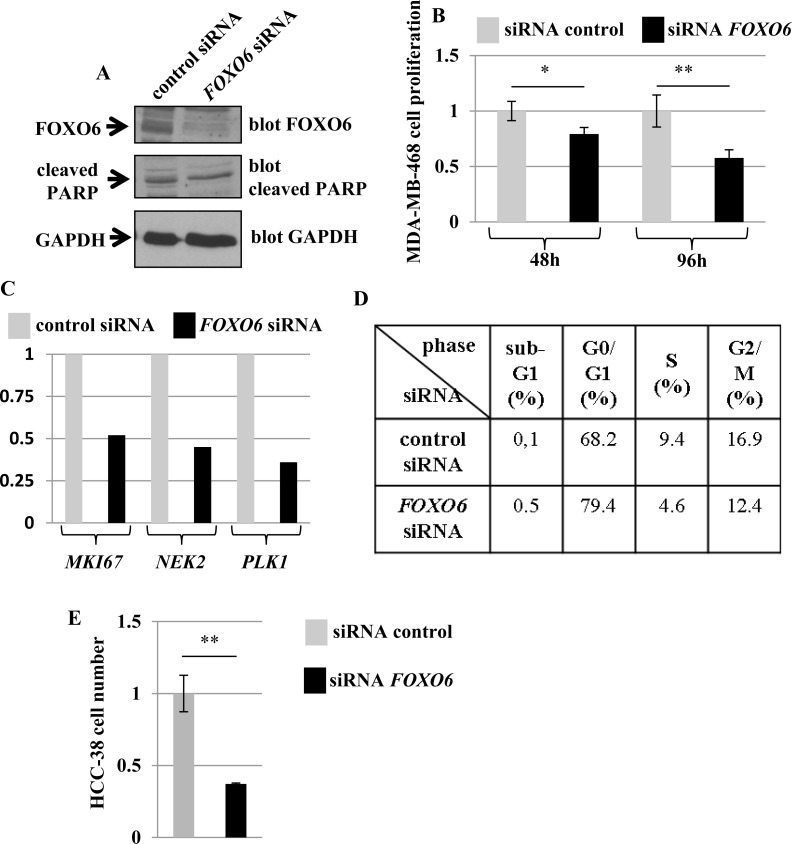
To further investigate the possibility that FOXO6 might modulate the proliferation of breast cancer cells, we examined the effect of inhibiting the expression of endogenous FOXO6 on proliferation of the breast cell line MDA-MB-468 expressing high levels of this FOXO gene (Supplementary Table 2 and Figure 2). A specific FOXO6 siRNA with confirmed efficacy and specificity was used to inhibit FOXO6 expression in our cellular model (Figure 4A and Supplementary Figure 2).
Figure 4.
Effect of a specific FOXO6 siRNA on the growth of the human breast cell lines MDA-MB-468 and HCC-38 (A) Effect of FOXO6 siRNA on the expressions of FOXO6 and cleaved PARP (marker of apoptosis). MDA-MB-468 cells were transfected with control siRNA or FOXO6 siRNA as indicated. Six days after transfection, the expressions of FOXO6, cleaved PARP, and GAPDH (loading control) were determined by Western blot. (B–D) Effect of FOXO6 siRNA on cell growth. MDA-MB-468 cells were transfected with control siRNA or FOXO6 siRNA as indicated. The viable cell count was determined two and four days after transfection (B). Six days after transfection, the expressions of MKI67, NEK2, and PLK1 were evaluated by qRT-PCR (expressions were normalised to that detected in cells transfected with control siRNA) (C) and FACS analysis was performed (D). (E) Effect of FOXO6 siRNA on cell growth of HCC-38 cell line. Cells were transfected with control siRNA or FOXO6 siRNA as indicated. The viable cell count was determined 7 days after transfection.
Inhibition of FOXO6 expression in MDA-MB-468 cells resulted in inhibition of cell proliferation (Figure 4B). In order to confirm this observation, three genes involved in proliferation (MKI67, NEK2, and PLK1 [18]) were quantified by qRT-PCR in MDA-MB-468 cells treated with control siRNA or FOXO6 siRNA. MKI67, NEK2 and PLK1 expressions were 1.9-, 2.2-, and 2.8-fold lower, respectively, in MDA-MB-468 cells treated with FOXO6 siRNA compared to control cells (Figure 4C). The FOXO6 siRNA also induced a positive effect on expression of the PPARGC1A gene, an established target gene of FOXO6 [4], confirming the specificity of this siRNA.
FACS analysis showed that FOXO6 siRNA treatment decreased the number of cells located in the S and G2/M phases of the cell cycle, induced an accumulation of cells in the G0/G1 phase, and did not increase the number of cells with sub-G1 DNA content (apoptotic cells) (Figure 4D). Moreover, FOXO6 siRNA had no effect on the expression of cleaved PARP, a marker of apoptosis (Figure 4A).
These results strongly suggest that inhibition of FOXO6 expression affects the growth of MDA-MB-468 cells mainly by inducing cell cycle arrest in the G0/G1 phase.
To validate our results, we also tested the effect of the siRNA FOXO6 on proliferation of the breast cell line HCC-38, another breast cell line expressing high levels of FOXO6 gene (Supplementary Table 2). The siRNA FOXO6 inhibited, as for the MDA-MB-468 cells, the cell proliferation (Figure 4E).
DISCUSSION
This study demonstrates specific fluctuations of FOXO gene expressions in human breast cancer, suggesting that these genes play different roles in this cancer. We showed that FOXO1, 3 and 4 are frequently underexpressed these genes could therefore act as tumour suppressor genes. This hypothesis is supported by the work of Guttilla and White (2009) showing that FOXO1 mRNA is down-regulated in breast cancers compared to normal breast tissue, and that overexpression of FOXO1 induces MCF-7 cell death [12]. Many observations indicate that FOXO3 also exerts tumour suppressor activity in breast cancer [2]. However, alteration of FOXO3 expression does not appear to be a major mechanism of inhibition of the biological function of FOXO3 in this cancer type, as no significant variation of the expression of this gene was observed in our large series of breast cancers.
Surprisingly, we found that FOXO6 was often highly overexpressed in human breast tumours and cell lines at both the mRNA and protein levels. In contrast with FOXO1, FOXO3, and FOXO4, FOXO6 could therefore be an oncogene in breast cancer. This finding is consistent with the fact that FOXO6 is the most distant member of the FOXO family [6]. Indeed, FOXO6 exhibits major structural differences compared to the other three family members, and, unlike FOXO1 and 3, activation of the PI3K-AKT pathway by growth factors inhibits FOXO6 transcriptional activity mainly via a mechanism independent of shuttling to the cytosol [6, 19]. FOXO1, 3, and 4 contain an N- and C-terminal AKT motif and a third AKT motif located in the forkhead domain. FOXO6 lacks the conserved C-terminal AKT motif, which is the cause of the shuttling impairment [1, 19]. However, the reason why FOXO6 plays a different role in breast cancer compared to the other FOXO members has yet to be elucidated.
To our knowledge, this is the first time that FOXO6 overexpression has been demonstrated in breast cancer. In particular, no data concerning FOXO6 expression in breast cancer were found in the cbioportal (www.cbioportal.org) or TCGA databases due to the absence of a specific probe for FOXO6. Low to moderate FOXO6 expression was observed in all human adult tissues examined in this study, but high FOXO6 expression was detected in foetal brain tissue in keeping with earlier report [20]. It is noteworthy that we observed overexpression of this FOXO gene also in several brain, bladder and colon tumours. FOXO6 gene would therefore be involved in various cancer types.
The molecular mechanisms responsible for altered FOXO6 expression in breast cancer are unknown. FOXO6, located at 1p34.2, is amplified in only 2% of breast cancers (www.cbioportal.org). The FOXO6 overexpression observed in human breast tumours and cell lines would therefore not be due to this molecular mechanism. The study by Guttilla et al. indicated that FOXO1 expression is modulated by several microRNAs in breast cancer cells [12]. However, the involvement of microRNAs in the regulation of FOXO6 expression has not been described [2]. We found a negative correlation between the expressions of FOXO6 and AKT phosphorylated on serine 473, a marker of the PI3K-AKT pathway activity, and a positive correlation between the expressions of FOXO6 and PTEN at the mRNA and protein level, a negative regulator of the PI3K-AKT pathway [17]. FOXO6 overexpression is therefore associated with low activity of the PI3K/AKT pathway in breast cancers. Essaghir et al. showed that activation of the PI3K/AKT pathway by various growth factors inhibited the expression of FOXO1, FOXO3, and FOXO4 in human fibroblasts [16]. These observations and our results therefore suggest that the FOXO6 overexpression that we have highlighted in human breast cancers could be at least partly due to low activity of the PI3K/AKT pathway. Other studies are required to more clearly define the molecular mechanisms responsible for FOXO6 overexpression in breast cancer.
We found that a specific siRNA FOXO6 inhibited growth of two different breast cell lines expressing high levels of FOXO6 gene. FACS and Western blot analysis showed that inhibition of endogenous FOXO6 expression induced accumulation of MDA-MB-468 cells in G0/G1 phase of the cell cycle, but did not induce apoptosis. Our results therefore strongly suggest that FOXO6 might be an oncogene in human breast cancer, which positively regulates cell proliferation by activating the progression of cancer cells through the G0/G1 phase. This hypothesis is supported by the results of various studies. Li Qinyu et al. (2013) showed that FOXO6 mRNA and protein levels are upregulated in gastric cancer tissues, and this overexpression promotes gastric cancer cell tumorigenicity via upregulation of Myc [21]. However, we have not found any correlation between the expressions of FOXO6 and c-Myc in breast tumors (data not shown), strongly suggesting that the overexpression of FOXO6 would act on breast carcinogenesis via a specific mechanism independent of the transcriptional factor Myc. Moreover, Chen et al. demonstrated that FOXO6 is overexpressed in hepatocellular cancer and that FOXO6 siRNA increases the percentage of cells in G0/G1 phase [22]. However, FOXO6 has also been shown to be downregulated in lung cancer compared to adjacent normal tissue [23]. Moreover, FOXO6 overexpression inhibits the proliferation of A549 human lung cancer cells, whereas knockdown of endogenous FOXO6 expression enhances cell proliferation [23]. FOXO6 therefore behaves like a tumour suppressor gene in lung cancer. These findings suggest that FOXO6 may have opposite roles in cancer depending on the cancer type.
In conclusion, we provide evidences strongly suggesting that the overexpression of FOXO6 promotes breast carcinogenesis by stimulating cellular proliferation. FOXO6, which is intensely expressed in breast cancers, but expressed at very low levels in normal adult tissues, may therefore be a potential candidate as a target for breast cancer therapy.
MATERIALS AND METHODS
Patients and samples
Samples of 527 primary unilateral invasive breast tumours (composed of 89.6% of ductal breast cancer, 6.3% of lobular breast cancer, and 4.1% for the other subtypes from information available for 444 patients) excised from women managed at Institut Curie-René Huguenin Hospital (Saint-Cloud, France) from 1978 to 2008 were analysed. Samples were immediately stored in liquid nitrogen until mRNA and protein extraction. Tumour samples were considered suitable for our study when the proportion of tumour cells exceeded 70%. All patients (mean age: 62 years, range: 29–91 years) met the following criteria: primary unilateral non metastatic breast carcinoma, for which complete clinical, histological and laboratory data were available; no neoadjuvant radiotherapy or chemotherapy; and complete follow-up at Institut Curie-René Huguenin Hospital. Treatment consisted of modified radical mastectomy in 320 cases (61.1%) and breast-conserving surgery plus locoregional radiotherapy in 204 cases (38.9%) (information available for 524 patients). Patients underwent physical examination and routine chest radiography every 3 months for 2 years, then annually. Mammograms were performed annually. Adjuvant therapy was administered to 415 patients, consisting of chemotherapy alone in 129 cases, hormone therapy alone in 178 cases and both treatments in 108 cases. The histological type and the number of positive axillary nodes were established at the time of surgery. The malignancy of infiltrating carcinomas was scored according to Scarff-Bloom-Richardson’s (SBR) histo-prognostic system. Hormone receptor (HR) (estrogen receptor α (ERα), progesterone receptor (PR)) and human epidermal growth factor receptor 2 (ERBB2) status were determined by protein assay using biochemical methods (dextran-coated charcoal method, enzyme immunoassay or immunohistochemistry) and confirmed by real-time quantitative RT-PCR assays [24, 25]. The population was divided into four groups according to HR (ERα and PR) and ERBB2 status, as follows: HR+/ERBB2+ (n = 58), HR+/ERBB2- (n = 295), HR-/ERBB2+ (n = 72) and HR-/ERBB2- (n = 102). The median follow-up was 9.1 years (range: 5 months to 33 years); 210 patients developed metastatic disease. Sixteen specimens of adjacent normal breast tissue from breast cancer patients or normal breast tissue from women undergoing cosmetic breast surgery were used as sources of normal mRNA.
Samples of 39 breast tissue-derived cell lines were analysed. These cell lines, obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) or the German Resource Centre for Biological Material (DSMZ, Braunschweig, Germany), were cultured under the conditions recommended by the suppliers, and authenticated in our laboratory by using the GenePrint 10 System kit (Promega, Madison, WI, USA) just before the extraction.
We also analysed mRNA samples from 24 normal adult tissues [26, 27] and four types of normal and cancer tissues: breast, brain, bladder, and colon (5 normal tissues and 10 cancer tissues were examined for each cancer).
Ethic approval and consent to participate
All patients who entered our institution before 2007 were informed that their tumor samples might be used for scientific purposes and had the opportunity to decline. Since 2007, patients entering our institution have given their approval also by signed informed consent. This study was approved by the local ethics committee (Breast Group of René Huguenin Hospital).
Cell culture
MDA-MB-468 and HCC-38 cells, purchased from ATCC, were maintained in DMEM or RPMI medium respectively containing 10% foetal bovine serum (Invitrogen, Carlsbad, CA) and 1% antibiotics (50 μg/mL penicillin, 50 μg/mL streptomycin, 100 μg/mL neomycin), and grown at 37°C in a humidified atmosphere of 5% (v/v) CO2 in air. This cell line was authenticated in our laboratory by using the GenePrint 10 System kit (Promega, Madison, WI, USA). We perform authentication of our cell lines each 20 passages.
RNA extraction
Total RNA was extracted from normal human tissues, and breast cell lines and tumours by using acid-phenol guanidium, as previously described [28]. RNA quality was determined by electrophoresis on agarose gels, staining with SYBR® Safe (ThermoFisher Scientific, San Jose, CA, USA) and visualization of the 18S and 28S RNA bands under blue light.
Real-time qRT–PCR
Quantitative values were obtained from the cycle number (Ct value) at which the increase in the fluorescence signal associated with exponential growth of PCR products started to be detected by the laser detector of the ABI Prism 7900 sequence detection system (Perkin Elmer Applied Biosystems, Foster City, CA, USA), using the PE Biosystems analysis software (Perkin Elmer Applied Biosystems) according to the manufacturer’s manuals. As the precise amount of total mRNA added to each reaction mix (based on optical density) and its quality (i.e., lack of extensive degradation) are both difficult to assess, we therefore also quantified TBP gene transcripts (Genbank accession NM_003194) encoding the TATA box-binding protein (a component of the DNA-binding protein complex TFIID) as an endogenous RNA control and normalised each sample on the basis of its TBP content. TBP was selected as endogenous control due to the moderate prevalence of its transcripts and the absence of any known TBP retro-pseudogenes (retro-pseudogenes lead to co-amplification of contaminating genomic DNA and consequently interfere with qRT–PCR, despite the use of primers in separate exons) [24]. Results expressed as N-fold differences in FOXO target gene expression relative to TBP gene expression and termed ‘NFOXO’ were determined as NFOXO = 2ΔCtsample, where the ΔCt value of the sample was determined by subtracting the Ct value of the FOXO gene from the Ct value of the TBP gene. TBP was then used as endogenous control. NFOXO values of the samples were also subsequently normalised so that the median NFOXO values for normal breast tissues (N) was equal to 1 (Tables 1 and 3, and, Figure 1, and Supplementary Figure 1), and/or the ‘basal mRNA level’ (smallest quantifiable amount of mRNA (Ct = 35)) was equal to 1 (Table 1 and Supplementary Tables 1 and 2). Primers for TBP and FOXO genes were chosen with the assistance of Oligo 6.0 software (National Biosciences, Plymouth, MN, USA) (Supplementary Table 4). We scanned the dbEST and nr databases to confirm the total gene specificity of the nucleotide sequences chosen for the primers. To avoid amplification of contaminating genomic DNA, one of the two primers was placed at the junction between two exons. Agarose gel electrophoresis was used to verify the specificity of PCR amplicons. Total RNA extraction, cDNA synthesis and PCR were performed under previously described conditions [28]. Over- and under-expressions were defined as threefold variations of expression relative to the median expression of normal samples, or as Ct values under 30 (values above 32 (2ΔCt = 235–30 = 32)) for normalization relative to ‘the basal mRNA level’.
siRNA experiments
Control siRNA and FOXO6 siRNA (1027281 and SI05195617, respectively, Qiagen, Santa Clarita, CA) were transfected using HiPerfect transfection Reagent (Qiagen, Santa Clarita, CA) according to the manufacturer’s instructions. 48 hours (Figure 2B) or 72 hours (Supplementary Figure 3, and Figure 2A, 2C, and 2D) after the transfection, cells were transfected once again.
Western blotting
Samples analysed in this study come from breast cell lines cultured under the conditions recommended by the suppliers, and authenticated in our laboratory by using the GenePrint 10 System kit (Promega, Madison, WI, USA). We perform authentication of our cell lines each 20 passages.
Methods are described in detail elsewhere [29]. Briefly, proteins were extracted from cell culture using TNMG buffer (20 mM Tris-HCl (pH 8), 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5% NP-40, pH 8) supplemented with protease inhibitors. For the siRNA experiments, cells were seeded in p60 plates (500,000 cells per well), transfected with control siRNA or FOXO6 siRNA and the cellular extracts were prepared six days after transfection. The following antibodies were used in this study: anti-GAPDH (sc-20357, Santa Cruz Biotechnology, Santa Cruz, CA), used as internal control, anti-FOXO6 (19122-1-AP, Proteintech, Chicago, USA), and anti-cleaved PARP (9541, Cell Signaling, Beverly, MA, USA). Proteins were detected by the ECL Western Blotting Analysis System procedure (GE Healthcare, Buckinghamshire, UK).
Cell proliferation assay
Cells were seeded in 96-well plates (10,000 cells per well) and transfected with control siRNA or FOXO6 siRNA. Cell proliferation was determined 48 hours and 96 hours after transfection using the Cell Titer kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Results were expressed as mean ± s.d. of triplicates from a representative experiment.
RPPA
RPPA was performed as previously described [30].
FACS analysis
Cells were seeded in p60 plates (500,000 cells per well) and transfected with control siRNA or FOXO6 siRNA. Six days after transfection, cells were harvested and DNA content was assessed by propidium iodide staining of methanol-fixed cells and monitoring by FACScan (LSRII).
Statistical analysis
The relative expression of each gene was characterized by the median and the range. Relationships between mRNA expression of genes and clinical parameters, and target mRNA and protein were assessed by nonparametric tests, Kruskal-Wallis H test (relationship between one quantitative parameter and one qualitative parameter) and Spearman’s rank correlation test (relationship between two quantitative parameters). To visualize the efficacy of a molecular marker to discriminate between two populations (patients that developed/or did not develop metastases) in the absence of an arbitrary cut-off value, data were summarized in a ROC (receiver operating characteristic) curve [31].The AUC (area under the curve) was calculated as a single measure to discriminate efficacy. Survival distributions were estimated by the Kaplan-Meier method, and the significance of differences between survival rates was ascertained with the log-rank test. Metastasis-free survival (MFS) was determined as the interval between initial diagnosis and detection of the first metastasis.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
We thank the staff of Institut Curie-René Huguenin Hospital for their assistance in specimen collection and patient care. This work was supported by the Association pour la Recherche en Cancérologie de Saint-Cloud (ARCS) and the French National Cancer Institute (INCa).
Author contributions
Conception and design: I. Bièche, R; Lidereau, B.S. Lopez, S. Zinn-Justin, N. Dalla-Venezia, S. M. Caputo; Development of methodology: I. Bièche, F. Lallemand, A. Petitalot; Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A. Petitalot, I. Bièche, F. Lallemand, L. de Koning, S. Vacher, W. Chemlali, A. Schnitzler, K. Taouis; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Petitalot, R. Lidereau, F. Lallemand; S. Vacher, S.M. Caputo, I. Bieche; Writing, review, and/or revision of the manuscript: F. Lallemand, S. M. Caputo, I. Bieche; Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. M. Caputo, A. Petitalot, F. Lallemand; Study supervision: S.M. Caputo, I. Bieche
CONFLICTS OF INTEREST
All the other authors declare to have no conflicts of interest.
FUNDING
Ambre Petitalot has obtained research grants from the “Institut National du Cancer” (INCA) (PRTK2011-046 and 2011-1-PL BIO-09-IC-1). Sandrine M. Caputo has a grant from INCA and Lopez’s team is “Ligue 2014” labelled.
REFERENCES
- 1.Bullock M. FOXO factors and breast cancer: outfoxing endocrine resistance. Endocr Relat Cancer. 2016;23:R113–130. doi: 10.1530/ERC-15-0461. https://doi.org/10.1530/ERC-15-0461. [DOI] [PubMed] [Google Scholar]
- 2.Coomans de Brachène A, Demoulin JB. FOXO transcription factors in cancer development and therapy. Cell Mol Life Sci CMLS. 2016;73:1159–72. doi: 10.1007/s00018-015-2112-y. https://doi.org/10.1007/s00018-015-2112-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Perdomo G, Zhang T, Slusher S, Lee S, Phillips BE, Fan Y, Giannoukakis N, Gramignoli R, Strom S, Ringquist S, Dong HH. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes. 2011;60:2763–74. doi: 10.2337/db11-0548. https://doi.org/10.2337/db11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung SY, Huang WC, Su CW, Lee KW, Chi HC, Lin CT, Chen ST, Huang KM, Tsai MS, Yu HP, Chen SL. FoxO6 and PGC-1α form a regulatory loop in myogenic cells. Biosci Rep. 2013;33 doi: 10.1042/BSR20130031. https://doi.org/10.1042/BSR20130031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–99. doi: 10.1006/geno.1997.5122. https://doi.org/10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs FMJ, van der Heide LP, Wijchers PJ, Burbach JPH, Hoekman MFM, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–67. doi: 10.1074/jbc.M302804200. https://doi.org/10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 7.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 8.Shaw RJ, Cantley LC. Ras, PIK and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–30. doi: 10.1038/nature04869. https://doi.org/10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 9.Hu MCT, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 10.Zou Y, Tsai WB, Cheng CJ, Hsu C, Chung YM, Li PC, Lin SH, Hu MCT. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res BCR. 2008;10:R21. doi: 10.1186/bcr1872. https://doi.org/10.1186/bcr1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, Huang H, Kuo HP, Lee DF, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–48. doi: 10.1038/ncb1676. https://doi.org/10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284:23204–16. doi: 10.1074/jbc.M109.031427. https://doi.org/10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storz P, Döppler H, Copland JA, Simpson KJ, Toker A. FOXO3a promotes tumor cell invasion through the induction of matrix metalloproteinases. Mol Cell Biol. 2009;29:4906–17. doi: 10.1128/MCB.00077-09. https://doi.org/10.1128/MCB.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng X, Wu Z, Wu Y, Hankey W, Prior TW, Li L, Ganju RK, Shen R, Zou X. Cdc25A regulates matrix metalloprotease 1 through Foxo1 and mediates metastasis of breast cancer cells. Mol Cell Biol. 2011;31:3457–71. doi: 10.1128/MCB.05523-11. https://doi.org/10.1128/MCB.05523-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sisci D, Maris P, Cesario MG, Anselmo W, Coroniti R, Trombino GE, Romeo F, Ferraro A, Lanzino M, Aquila S, Maggiolini M, Mauro L, Morelli C, et al. The estrogen receptor α is the key regulator of the bifunctional role of FoxO3a transcription factor in breast cancer motility and invasiveness. Cell Cycle Georget Tex. 2013;12:3405–20. doi: 10.4161/cc.26421. https://doi.org/10.4161/cc.26421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Essaghir A, Dif N, Marbehant CY, Coffer PJ, Demoulin JB. The transcription of FOXO genes is stimulated by FOXO3 and repressed by growth factors. J Biol Chem. 2009;284:10334–42. doi: 10.1074/jbc.M808848200. https://doi.org/10.1074/jbc.M808848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29:233–42. doi: 10.1016/j.tibs.2004.03.006. https://doi.org/10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Bièche I, Vacher S, Lallemand F, Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E, Lerebours F, Ripoche H, Cizeron-Clairac G, Spyratos F, Lidereau R. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol Cancer. 2011;10:23. doi: 10.1186/1476-4598-10-23. https://doi.org/10.1186/1476-4598-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Heide LP, Jacobs FMJ, Burbach JPH, Hoekman MFM, Smidt MP. FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo-cytoplasmic shuttling. Biochem J. 2005;391:623–9. doi: 10.1042/BJ20050525. https://doi.org/10.1042/BJ20050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs FMJ, van der Heide LP, Wijchers PJ, Burbach JPH, Hoekman MFM, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–67. doi: 10.1074/jbc.M302804200. https://doi.org/10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 21.Qinyu L, Long C, Zhen-dong D, Min-min S, Wei-ze W, Wei-ping Y, Cheng-hong P. FOXO6 promotes gastric cancer cell tumorigenicity via upregulation of C-myc. FEBS Lett. 2013;587:2105–11. doi: 10.1016/j.febslet.2013.05.027. https://doi.org/10.1016/j.febslet.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Chen HY, Chen YM, Wu J, Yang FC, Lv Z, Xu XF, Zheng SS. Expression of FOXO6 is Associated With Oxidative Stress Level and Predicts the Prognosis in Hepatocellular Cancer: A Comparative Study. Medicine (Baltimore) 2016;95:e3708. doi: 10.1097/MD.0000000000003708. https://doi.org/10.1097/MD.0000000000003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu HJ, Zhang LG, Wang ZH, Guo XX. FoxO6 inhibits cell proliferation in lung carcinoma through up-regulation of USP7. Mol Med Rep. 2015;12:575–80. doi: 10.3892/mmr.2015.3362. https://doi.org/10.3892/mmr.2015.3362. [DOI] [PubMed] [Google Scholar]
- 24.Bièche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–56. [PubMed] [Google Scholar]
- 25.Bièche I, Parfait B, Laurendeau I, Girault I, Vidaud M, Lidereau R. Quantification of estrogen receptor alpha and beta expression in sporadic breast cancer. Oncogene. 2001;20:8109–15. doi: 10.1038/sj.onc.1204917. https://doi.org/10.1038/sj.onc.1204917. [DOI] [PubMed] [Google Scholar]
- 26.Bièche I, Maucuer A, Laurendeau I, Lachkar S, Spano AJ, Frankfurter A, Lévy P, Manceau V, Sobel A, Vidaud M, Curmi PA. Expression of stathmin family genes in human tissues: non-neural-restricted expression for SCLIP. Genomics. 2003;81:400–10. doi: 10.1016/s0888-7543(03)00031-4. [DOI] [PubMed] [Google Scholar]
- 27.Bièche I, Manceau V, Curmi PA, Laurendeau I, Lachkar S, Leroy K, Vidaud D, Sobel A, Maucuer A. Quantitative RT-PCR reveals a ubiquitous but preferentially neural expression of the KIS gene in rat and human. Brain Res Mol Brain Res. 2003;114:55–64. doi: 10.1016/s0169-328x(03)00132-3. [DOI] [PubMed] [Google Scholar]
- 28.Bieche I, Parfait B, Le Doussal V, Olivi M, Rio MC, Lidereau R, Vidaud M. Identification of CGA as a novel estrogen receptor-responsive gene in breast cancer: an outstanding candidate marker to predict the response to endocrine therapy. Cancer Res. 2001;61:1652–8. [PubMed] [Google Scholar]
- 29.Lallemand F, Seo SR, Ferrand N, Pessah M, L’Hoste S, Rawadi G, Roman-Roman S, Camonis J, Atfi A. AIP4 restricts transforming growth factor-beta signaling through a ubiquitination-independent mechanism. J Biol Chem. 2005;280:27645–53. doi: 10.1074/jbc.M500188200. https://doi.org/10.1074/jbc.M500188200. [DOI] [PubMed] [Google Scholar]
- 30.Rondeau S, Vacher S, De Koning L, Briaux A, Schnitzler A, Chemlali W, Callens C, Lidereau R, Bièche I. ATM has a major role in the double-strand break repair pathway dysregulation in sporadic breast carcinomas and is an independent prognostic marker at both mRNA and protein levels. Br J Cancer. 2015;112:1059–66. doi: 10.1038/bjc.2015.60. https://doi.org/10.1038/bjc.2015.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. https://doi.org/10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.