Abstract
The light, oxygen, voltage (LOV) domain proteins are blue light photoreceptors that utilize a non-covalently bound flavin mononucleotide (FMN) cofactor as the chromophore. The modular nature of these proteins has led to their wide adoption in the emerging fields of optogenetics and optobiology, where the LOV domain has been fused to a variety of output domains leading to novel light-controlled applications. In the present work, we extend our studies of the sub-picosecond to several hundred microsecond transient infrared spectroscopy of the isolated LOV domain AsLOV2 to three full-length photoreceptors in which the LOV domain is fused to an output domain: the LOV-STAS protein, YtvA, the LOV-HTH transcription factor, EL222, and the LOV-histidine kinase, LovK. Despite differences in tertiary structure, the overall pathway leading to cysteine adduct formation from the FMN triplet state is highly conserved, although there are slight variations in rate. However significant differences are observed in the vibrational spectra and kinetics after adduct formation, which are directly linked to the specific output function of the LOV domain. While the rate of adduct formation varies by only 3.6-fold amongst the proteins, the subsequent large-scale structural changes in the full-length LOV photoreceptors occur over the micro- to sub-millisecond timescales and vary by orders of magnitude depending on the different output function of each LOV domain.
Keywords: LOV domain, Photoreceptor, Ultrafast infrared, Flavoprotein, AsLOV2, YtvA, EL222, LovK
Graphical Abstract
Introduction
The light, oxygen, voltage (LOV) domain is part of the PAS domain superfamily and functions in a variety of light activated biological activities including phototropism, gene regulation, and stress response.1 The LOV domain binds oxidized flavin mononucleotide (FMN) in a highly conserved hydrogen bonding network composed of two Asn and two Gln residues.2 Changes in the LOV domain upon blue light illumination take place through formation of a covalent adduct between C4a of FMN and a conserved Cys concomitant with protonation of FMN-N5 (Figure S1).3 Adduct formation leads to rotation of a conserved Gln residue (Gln513 in AsLOV2), which is thought to initiate large scale structural changes by perturbing interactions with the LOV β-sheet (Figure 1A).3,4 Together these events lead to activation of an effector domain, initiating biological activity.5 Despite over a decade of intense research on LOV photoactivation, the mechanistic link between the ultrafast structural changes around the chromophore and the structural dynamics at longer timescales remains to be fully elucidated.6
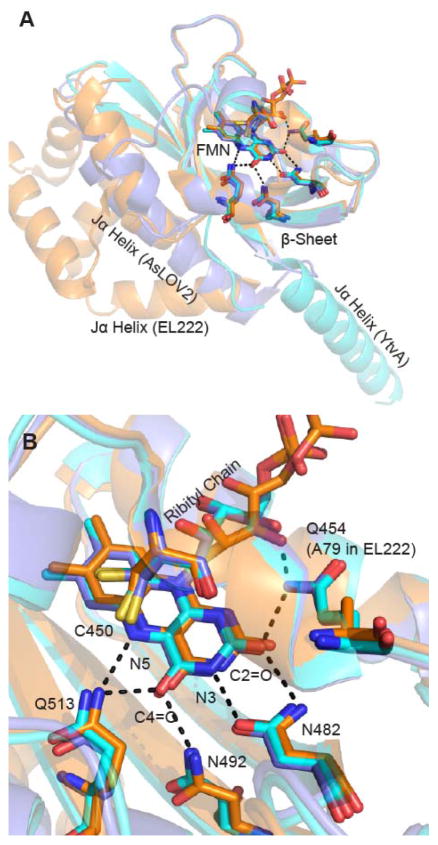
Figure 1. Superposition of LOV photoreceptor structures.
The X-ray structures of AsLOV2 (2V0U, slate blue), YtvA (2PR5, cyan) and EL222 (3P7N, orange) have been superimposed using PyMOL.7 No structure is currently available for LovK. (A) The Jα helix adopts different positions relative to the FMN binding pocket in each photoreceptor. In AsLOV2, the Jα helix is docked to the β-sheet, while this surface is occupied by the helix-turn-helix (HTH) domain in EL222 in the dark state and acts as a dimerization site in the EL222 light state. In YtvA the β-sheet acts as the obligate dimerization site in both light and dark states with the Jα helix extending out to the STAS domain (which is not present in this structure). (B) The flavin binding pocket is comprised of several conserved residues that interact with the flavin through hydrogen bonds. This includes Q513, N492, N482, and Q454 in AsLOV2; Q123, N104, N94, and Q66 in YtvA; Q138, N117, N107, and A79 in EL222. In the Figure, residues are numbered based on the sequence of AsLOV2.
The modularity of the LOV domain enables blue-light control of a variety of biological functions. For example, in Avena sativa, light activation of the LOV domain (AsLOV2) is coupled to unfolding of the Jα helix which results in activation of a Ser/Thr kinase in the full length phototropin.8 AsLOV2 has also been used as a tool in the emerging field of optobiology. The AsLOV2 domain has been fused to the GTPase Rac1 where it functions as a photocage for the enzyme,9 and also as the phototrigger for the light-inducible dimerization system iLID.10 The ps-ns time-resolved infrared spectra (TRIR) of AsLOV2 in H2O buffer were reported by both Alexandre et al. and Pfeifer et al. in which the spectrum of 3FMN* was resolved using transient IR and step-scan IR, respectively.11,12 Recently, we and others have measured the ps-ms TRIR spectra of AsLOV2 and we have fully assigned the spectra using isotope editing by labelling the protein and the FMN cofactor.13,14 In addition, a global fit of the data enabled the resolution of dispersive kinetics in triplet state decay to adduct formation.13
In the present work, we now extend our analysis to three photoreceptors in which the LOV domain is fused to an output partner via the Jα helix: YtvA from Bacillus subtilis which includes a sulfate transporter anti-sigma factor agonist (STAS) domain, LovK from Caulobacter crescentus which includes a histidine kinase (HK) domain, and EL222 from Erythrobacter litoralis which includes a helix-turn-helix (HTH) DNA binding domain. In YtvA and LovK, photoactivation is thought to involve a tilt and rotation around a dimerization site without large-scale secondary structure changes.5,15,16 In contrast, in EL222 photoactivation results in undocking of the HTH domain from the LOV β-sheet and formation of a head-to-head DNA-binding homodimer.17,18 These four proteins show very different steady state recovery lifetimes at 25 °C: 27s for AsLOV2,19 75 min for YtvA,20 2 hr for LovK,16 and 25 s for EL222.21
The flavin binding pocket is composed of several conserved residues that have non-covalent interactions with the isoalloxazine ring of FMN: Q513 and N492 are hydrogen bonded to the FMN C4=O and N5 groups, while Q454 and N482 form a network around the FMN N3 and C2=O groups (Figure 1B). Previous studies have shown that mutation of the Gln to Ala supresses large scale structural changes in the protein,3,22 while mutation of the Asn residues primarily results in lower adduct yield and modulation of triplet decay kinetics.23 Additionally Q454 also forms a hydrogen bond with the C2=O and the ribityl chain of FMN in AsLOV2 and YtvA, however this residue is an Ala in EL222. An α-helix, located above the FMN chromophore, contains the conserved Cys residue that is required for LOV photocycling and forms an adduct with the photoexcited FMN that is characterized by an absorption band at 390 nm in the light state (A390, Figure S1).
Several studies have focused on elucidating the structural changes in LOV photoreceptors on the μs-ms timescale. Initial characterization of Arabidopsis thaliana Phot1 LOV2 by the thermal grating method revealed a 2 ms time constant for the unfolding of the Jα helix,24 which was further extended to include a two-step mechanism.25 In YtvA it was proposed that the structural changes resolvable by circular dichroism are minimal and that smaller scale changes in hydrogen bonding or salt bridge interactions are occurring in the activation mechanism.26,27 More recently, the thermal grating technique has been applied to YtvA constructs lacking the effector domain and it was found that structural changes are confined to the Jα helices.28
In this study, the femtosecond to millisecond dynamics of LOV photoreceptor activation have been probed using time resolved IR with the femtosecond to millisecond Time-Resolved Multiple Probe Spectroscopy (TRMPS) method and Fourier Transform Infrared Spectroscopy (FTIR). This has enabled the interrogation of the structural dynamics that link the initial photochemistry to generation of the signalling state in the three full-length photoreceptors.
Materials and Methods
Protein Constructs
The truncated gene containing residues 404–546 of A. sativa Phototropin (herein referred to as AsLOV2) and the genes for full length YtvA and LovK were cloned into pET15b (Novagen) in frame with an N-terminal 6x-His-Tag. The gene containing full length EL222 was codon optimized, synthesized, and subcloned into pET15b by Genscript, Inc. (Piscataway, NJ).
Expression and Purification of AsLOV2, YtvA, and LovK
BL21 (DE3) (Novagen) competent E. coli cells were transformed with the respective plasmids by heat shock and plated on LB-Agar (Diffco) supplemented with 200 μg/mL ampicillin (Gold Biosciences). After incubation overnight at 37 °C, a single colony was used to inoculate 10 mL of 2X-YT media containing 200 μg/mL ampicillin which was then incubated in an orbital shaker (250 rpm) at 37 °C for 2–3 hr. This starter culture was then used to inoculate 1L of 2X-YT (IBI Sci) containing 200 μg/mL ampicillin, which was then incubated under the same conditions as the starter culture until the OD600 reached 1.0. Protein expression was then induced by the addition of 1 mM IPTG (Gold Biosciences), and the culture was shaken at 20 °C for 16 h. Subsequently, the cells were harvested by centrifugation at 12,000 g and then frozen at −20 °C until needed.
The frozen cell pellets were resuspended in 40 mL of 20 mM Tris buffer pH 8.0 containing 150 mM NaCl (resuspension buffer) and the cells were then disrupted using a French Press (Constant Systems Cell Disruptor) at a pressure of 27 kPSI using a cell that was maintained at 4 °C. After adding 100 μM phenylmethylsulfonylfluoride (PMSF) and 20 μL of β-mercaptoethanol, the lysate was clarified by ultracentrifugation at 250,000 g for 30 min. A 3 mL Ni-NTA (Novagen) column was prepared by equilibrating the resin with resuspension buffer and the clarified lysate was loaded onto the column. The column was washed with 100 mL of resuspension buffer and eluted using a step-gradient of resuspension buffer containing 10, 20, 30, and 500 mM imidazole. Fractions containing the purified protein were pooled and dialyzed against 20 mM Tris buffer pH 8.0 containing 150 mM NaCl. In each case, the proteins were shown to be ≥ 95% pure by SDS-PAGE. Purified and desalted protein fractions were lyophilized and dissolved in D2O (Cambridge Isotope Labs) prior to measurement.
Expression and Purification of EL222
EL222 was expressed and purified using the method described above with the following modifications. After loading the clarified cell extract onto the Ni-NTA column, the bound protein was first washed with resuspension buffer and then eluted using resuspension buffer containing 5 mM imidazole. Fractions containing the protein were pooled and dialyzed overnight into 20 mM Tris buffer pH 7.6 containing 30 mM NaCl. The dialyzed protein was then loaded onto a 5 mL Q-Sepharose column that had been equilibrated with the same buffer. The eluent was reloaded onto the column to maximize the amount of EL222 that bound to the resin. The protein was eluted using a gradient of 30 mM to 500 mM NaCl in 20 mM Tris buffer pH 7.6. Fractions containing EL222 were pooled and the protein was shown to be ≥ 95% pure by SDS-PAGE. Purified and desalted EL222 was lyophilized and suspended in D2O prior to measurement.
Time Resolved Multiple Probe Spectroscopy (TRMPS)
TRMPS measurements were conducted at the Central Laser Facility of the Rutherford Appleton Laboratory using the apparatus previously described.29 Briefly, 800nJ/pulses of the 450 nm pump excitation wavelength were provided by a Ti:Sapphire laser pumped OPA at a rate of 1 kHz and a pulse duration of 100 fs. A broadband mid-IR probe was generated using a 10 kHz Ti:Sapphire laser pumping an OPA with difference frequency generation (DFG) stage. The signal and idler outputs of the OPA were mixed to form the mid-IR broadband probe pulse with a duration of <100 fs. Two MCT detectors were used for data collection and gave ~400 cm−1 spectral bandwidth with 3 cm−1 resolution. The spectra were calibrated using transmission of a polystyrene film. To avoid photodegradation and conversion to the light state, the sample was flowed through a Harrick cell with CaF2 windows and a 50 μm spacer. The flow rate was set to 1 mL/min and the cell was rastered in the beam to minimize unwanted secondary photochemistry. Data were analyzed by global analysis using the Glotaran Software Package which globally fits a selected kinetic model to the entire time-resolved spectral data set; in the present case we fit a sequential first order model to the data.30
Fourier Transform Infrared Spectroscopy (FTIR)
FTIR spectra were recorded on a Bruker Vertex80 modified to accommodate an LED and a temperature controlled sample holder to maintain the sample temperature at 20°C. Dark state spectra were recorded with 1 cm−1 resolution and 128 scans were collected and averaged. The light state was generated after 2 min of irradiation at 450 nm using the LED, and the dark state spectrum was subtracted from the light state spectrum to generate the difference spectrum.
Results
In this study we have characterized the activation of the LOV domain from the initial dark state structure to the final light state structure. Below we compare time resolved infrared difference spectra (TRIR) tracking the conversion from singlet excited state (1FMN*, ns timescale) to triplet state (3FMN*, μs timescale), and then adduct formation (A390) and finally the subsequent protein structural evolution (μs-ms timescale).
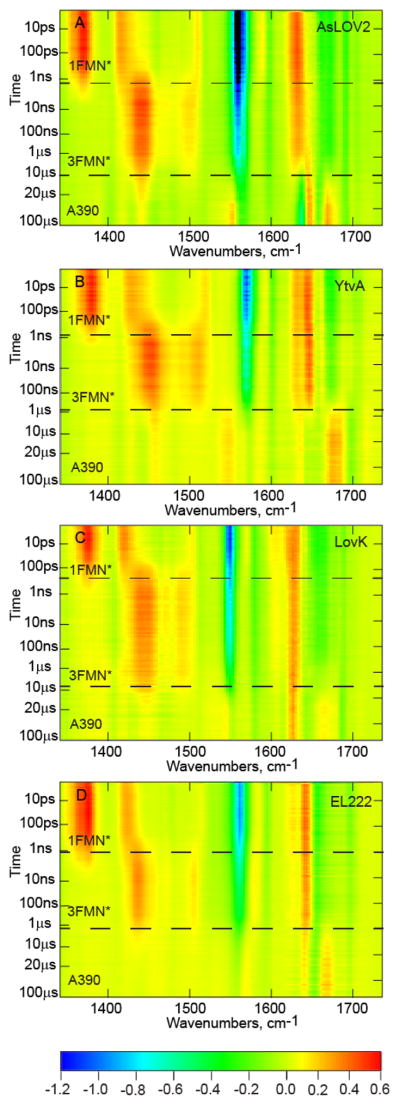
TRIR of YtvA, LovK and EL222, together with AsLOV2, are shown in Figure 2. Negative signals (bleaches) correspond to depopulated ground state modes and positive transients are vibrational modes arising from the excited state or new ground state populations. Evolution associated spectra (EAS) of the four LOV proteins were subsequently determined from a sequential decay model globally fit to the experimental TRIR data (Figure 3); the quality of the fit is shown in Figure S2. The data for each of the proteins is shown as a heat map with time on the y-axis and wavenumber on the x-axis with a color bar showing approximate intensity values. Horizontal lines denote the three major phases of the LOV photocycle that are resolvable in the TRMPS experiment: 1FMN*, 3FMN*, and A390. Below we compare the EAS for the three full-length LOV proteins with the EAS for AsLOV2 which we recently published.13 We first briefly review the time resolved vibrational spectrum of AsLOV2 as a foundation for the subsequent discussion.
Figure 2. TRIR of LOV photoreceptors.
(A) AsLOV2, (B) YtvA, (C) LovK, and (D) EL222 are displayed as two-dimensional heat maps with time on the y-axis and frequency (cm−1) on the x-axis. A color bar legend is shown below with approximate ΔA values for each of the normalized heat maps.
Figure 3. EAS Analysis of LOV Photoreceptors.
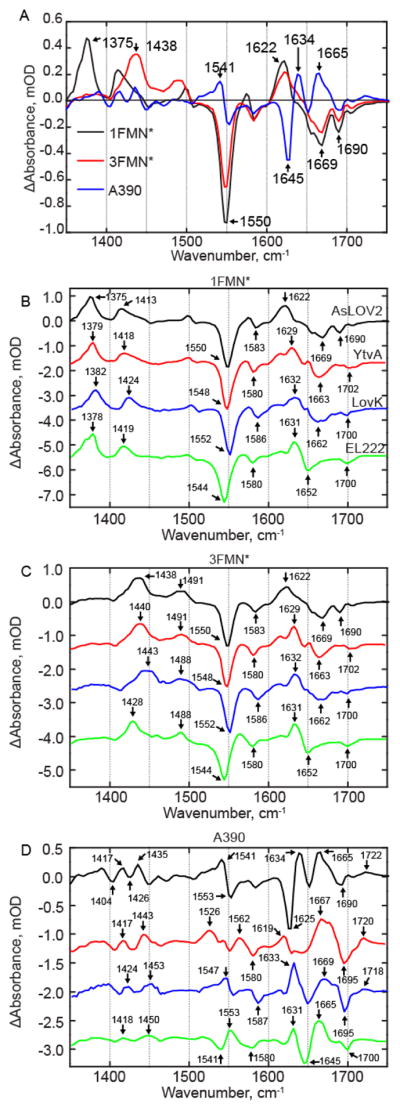
(A) EAS of AsLOV2 is shown overlayed. EAS corresponding to (B) 1FMN*, (C) 3FMN*, and (D) A390 are shown on a stacked y-axis plot. Small differences are observed in the singlet and triplet excited states while more significant differences are reflected in the A390 spectrum and show the most variation between the four LOV proteins. AsLOV2 is shown in black, YtvA in red, LovK in blue, and EL222 in green.
The first EAS (Figure 3A, black line) shows the instantaneously formed singlet excited state of FMN (1FMN*) in AsLOV2. This excited state is characterized by positive transients at ~1375 cm−1 and ~1413 cm−1 and ground state bleaches at ~1550 cm−1 and ~1580 cm−1 assigned to the C10a-N1 and C4a-N5 FMN modes, respectively.13,31,32 In addition, signals in the 1600–1660 cm−1 region can be assigned to overlapping flavin (C2=O) and protein modes, while the bleaches at 1690 and 1705 cm−1 are assigned to Q513 and the FMN C4=O, respectively.13 The second EAS forms with an ~2 ns time constant (Figure 3A, red line) and shows the triplet excited state (3FMN*) which is evident by the disappearance of excited state transients at 1375 cm−1 and 1413 cm−1 and the concomitant rise of transients at 1438 cm−1 and 1491 cm−1 associated with vibrations involving the isoalloxazine moiety. 3FMN* decays in 9.5 μs under formation of an adduct between Cys450 and FMN-C4a (A390), which is characterized by a difference spectrum in which large changes in protein modes are observed (Figure 3A blue line).13
EAS Comparison of Picosecond to Sub-Millisecond Structural Dynamics
EAS were plotted on a stacked y-axis to facilitate direct and convenient comparison between the four wild-type proteins (Figure 3 B–D). The EAS for YtvA and LovK most closely resemble the EAS for AsLOV2 (Figure 3B and C, red and blue lines, respectively). The first EAS for YtvA (Figure 3B, red line) shows the 1FMN* excited state. Transients assigned to ring modes are observed at 1379 cm−1 and 1418 cm−1, ~4 cm−1 blue-shifted compared to AsLOV2, while the strong bleach at 1548 cm−1 is, within the 3 cm−1 resolution of the experiment except for EL222, in which the strong bleach is observed at 1544 cm−1.
The most significant variation between the 1FMN* state of AsLOV2 and the other three proteins are found in the 1600–1670 cm−1 region, where the composite transient at 1622 cm−1 in AsLOV2 is split into at least three distinct transients in YtvA, with the most intense absorption shifted by 7 cm−1 to 1629 cm−1 with shoulders at 1615 cm−1 and 1650 cm−1. In LovK, a similar doublet peak to YtvA is observed with frequencies of 1610 cm−1 and 1632 cm−1 and in EL222 at 1615 cm−1 and 1631 cm−1. The bleach in this region of the YtvA spectrum is shifted by 6 cm−1 to 1663 cm−1 compared to AsLOV2, and the modes associated with Gln123 and the FMN C4=O are merged to form a single bleach at 1702 cm−1. In LovK a bleach is observed at 1669 cm−1, similar to that found in AsLOV2, however a lower frequency shoulder is present at 1656 cm−1 while in LovK this shoulder is observed at 1663 cm−1. The EAS of 3FMN* in YtvA (Figure 3C, red line) is similar to that of AsLOV2 and shows the disappearance of 1FMN* transients and the appearance of 3FMN* transients at 1440 and 1491 cm−1, with all other protein/flavin modes changing only in intensity.
The A390 EAS shows the largest differences between the four proteins. Perhaps most interesting is the C4-C10a ring stretching vibration which is found at 1553 cm−1 (-)/1541 cm−1 (+) in AsLOV2 and YtvA. This vibrational mode was previously assigned by 13C labelling of the chromophore, for which numbering in shown in the SI.13 In YtvA, the transient at 1553 cm−1 is greatly diminished in intensity due to the presence of a new transient at 1526 cm−1, which is not found in the other three proteins. The 1526 cm−1 transient may represent either an N-C stretch or N-D bend vibration based on FTIR spectra of [apoprotein-15N]-YtvA (Supplemental Figure S3), and could be due to dimerization, as a similar but weaker transient is also found in LovK. In LovK, the corresponding transient is shifted by 6 cm−1 to 1547 cm−1, while in EL222, this bleach/transient pair appears to be reversed, with the transient at 1550 cm−1 and the bleach at a lower frequency (1535 cm−1).
Some aspects of the adduct spectra are common to the LOV proteins, such as the protein/FMN transient centered at ~1665 cm−1; however, there is some variation in the shape of the band, suggesting that a portion of this band arises from a protein mode that is not conserved. The bleach at 1625 cm−1 in AsLOV2 was previously assigned to β-sheet and the adjacent transient at 1634 cm−1 to α-helix.14 This β-sheet mode appears to be blue-shifted in EL222 to 1645 cm−1 and to 1650 cm−1 in LovK. In YtvA, there is no defined bleach in this region, rather there is a broad feature from ~1630 cm−1 to 1650 cm−1 that could reflect the lack of changes in the Jα helix upon signaling state formation. Application of a 70 μs component to the kinetic analysis of YtvA based on Choi et al.28 revealed the presence of a small bleach at 1642 cm−1 (Supplementary Figure S3). While this bleach can tentatively be assigned to the Jα helix, we note that the entire spectrum changes in intensity on this timescale, complicating a full analysis of the data. Finally, a transient assigned to C4=O is observed after adduct formation and is found at 1722 cm−1 (AsLOV2), 1720 cm−1 (YtvA), 1718 cm−1 (LovK), and 1719 cm−1 (EL222).
Structural differences in EL222 are evident from excited state spectra
The EAS of EL222 shows the most changes when compared to the EAS of the other LOV domain proteins (Figure 3D). In the first EAS (black line), the frequency of the ground state bleach is red-shifted to 1544 cm−1 compared to 1550±2 cm−1 in the other LOV domains. Notably, this is not accompanied by any major changes in the excited state transients at 1378 cm−1 and 1419 cm−1 which are within 3 cm−1 of the position observed in AsLOV2, and thus within the resolution of the experiment. The 1600–1690 cm−1 region is much less complex in EL222 compared to the other LOV proteins. A single strong transient/bleach pair is observed at 1631(+)/1652(-) cm−1 whereas multiple bands are observed in the EAS of the other proteins. In addition, the bleach at 1700 cm−1 is more similar to that found in YtvA and LovK in which there is only one bleach in this region assigned to the conserved Gln (Gln513 in AsLOV2).
In EL222, the transients at 1378 cm−1 and 1419 cm−1 arising from the 1FMN* state are found in essentially the same position as in AsLOV2, however the lower frequency 3FMN* transient is shifted by 10 cm−1 to 1429 cm−1 while the higher frequency transient at 1488 cm−1 is within 3 cm−1 of the position observed in the other proteins. Upon adduct formation, the C4-C10a vibrational mode is perturbed and a bleach/transient pair is formed at 1541(-)/1553(+) cm−1. However, unlike the other proteins, this bleach/transient is reversed in EL222 with the transient at a lower frequency than the bleach (i.e. 1541(+)/1553(-) cm−1). Transients in the 1600–1690 cm−1 region are also observed at 1631 cm−1 and 1665 cm−1 and are assigned to protein modes, while bleaches at 1645 cm−1 and 1700 cm−1 are assigned to modes arising from β-sheet and Gln138, respectively.
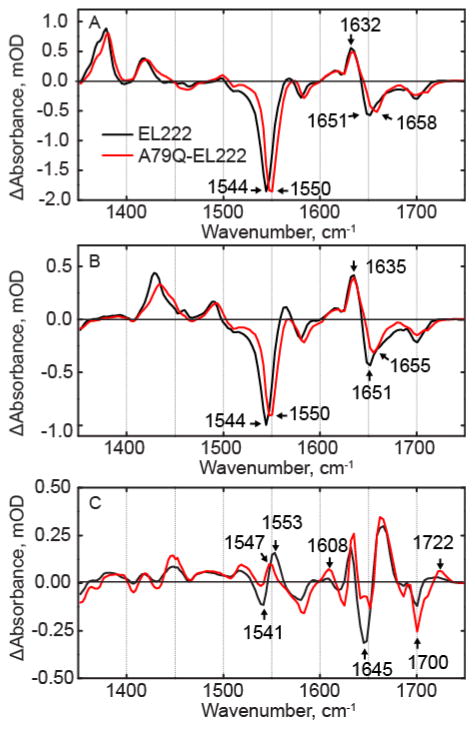
A79Q-EL222
To resolve a possible cause for the differences between EL222 and the other LOV proteins, we prepared the A79Q-EL222 mutant to restore the Gln known to interact with C2=O and the FMN ribityl chain in AsLOV2, LovK and EL222 (Figure 1). The EAS of A79Q-EL222 and wild-type EL222 are compared in Figure 4. In the 1FMN* EAS (Figure 4A), the major bleach at 1544 cm−1 in wild-type EL222 is blue shifted to 1550 cm−1, which is closer to the frequency of this bleach in the other LOV proteins which are at 1550 cm−1, 1548 cm−1, and 1552 cm−1 in AsLOV2, YtvA, and LovK respectively. The band at ~1550 cm−1 is a delocalized ring stretching vibration, and previous 13C labelling of the chromophore revealed that FMN-C2 contributes to this vibrational mode. In addition, there is a 7 cm−1 blue-shift from 1651 cm−1 to 1658 cm−1 in a bleach corresponding to a protein mode. The transients at ~1635cm−1 can be assigned to N117, while the lower frequency transient is likely N107 which is hydrogen bonded to FMN C2=O. Similar to the other LOV proteins, the EAS for 3FMN* in A79Q-EL222 is characterized by the decay of ES transients at 1381 cm−1 and 1421 cm−1 and the rise of transients at 1433 cm−1 and 1494 cm−1 with minimal changes in the 1600–1700 cm−1 region.
Figure 4. EAS of A79Q-EL222.
from global fitting of the data showing (A) 1FMN*, (B) 3FMN* and (C) A390 of EL222 and A79Q-EL222. Changes in ring modes at 1544 cm−1, 1380 cm−1, and 1425 cm−1 indicate that hydrogen bonding to FMN is restored compared to wild-type EL222.
Interestingly, there are several differences in the A390 EAS of A79Q compared to that of the wild-type protein. There is a 6 cm−1 red-shift of the C4-C10a transient associated with Cys adduct formation from 1553 cm−1 to 1547 cm−1. This causes a decrease in amplitude of the transient/bleach pair, while the bleach is now observed at 1541 cm−1 in A79Q-EL222. The transient at 1722 cm−1 in the mutant is assigned to FMN-C4=O which is shifted ~10 cm−1 from the position found in the wild-type protein and thus more similar to the position of this transient in the other LOV domain proteins, whereas in wild-type EL222 this transient is much broader. In summary, the changes in the bands assigned to the FMN ring modes between wild-type and A79Q-EL222 indicate that introduction of Q79 results in a FMN environment that more closely matches that found in the other proteins. However, protein modes in the 1600 cm−1 to 1700 cm−1 region still resemble wild-type EL222, despite the changes in the flavin modes.
Kinetics of the LOV Photoreceptors
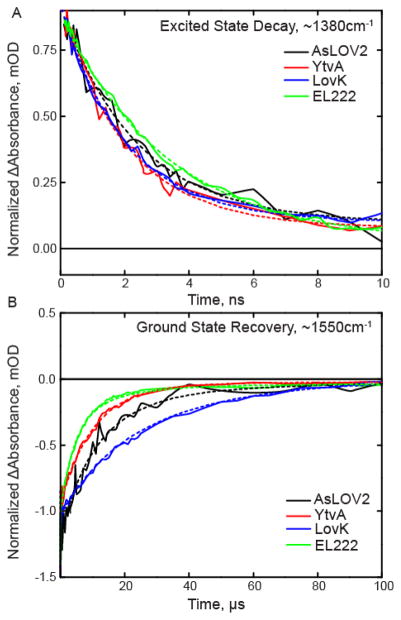
Kinetic parameters were determined from the same global fit of the entire dataset used for EAS analysis (Table 1, Figure 5). AsLOV2, YtvA, and LovK show the most similarity in excited singlet state decay (~2.2 ns), whereas EL222 has a slower decay of 3.3 ns. Triplet state decay rates for AsLOV2 and YtvA are nearly identical at ~10 μs while LovK is slower at 15 μs and EL222 is faster with a rate of 4.2 μs. Previous visible transient absorption studies performed on YtvA in H2O show triplet state decay times on the order of 1–2 μs.23,33 Since the EAS kinetics were determined in D2O, we can conclude that the rate of triplet state decay and adduct formation occurs with a kinetic isotope effect of ~5–10 for YtvA.
Table 1.
Kinetic parameters from global fitting of each dataset
| AsLOV2 | YtvA | LovK | EL222 | A79Q-EL222 | |
|---|---|---|---|---|---|
| 1FMN* | 2.4ns | 2.1ns | 2.0ns | 3.3ns | 2.7ns |
| 3FMN* | 9.5μs | 10.2μs | 15.3μs | 4.2μs | 1.9μs |
| A390 | Long | Long | Long | Long | Long |
Figure 5. Excited state decay and ground state recovery kinetics of the LOV proteins fit to the sequential model.
Raw data are shown as a solid line and results of the global fit are shown as dashed lines. (A) 1FMN* decay is shown on the ns timescale and is extracted from the transient absorption at 1380 cm−1. (B) Ground state recovery is shown on the μs timescale and is extracted from the main bleach at 1550 cm−1. Global fits for each photoreceptor are shown as dashed lines.
As previously reported for AsLOV2,13 formation of the A390 state occurs through dispersive kinetics, reflecting the dynamic and global changes in protein structure that occur upon adduct formation. This leads to variable structural changes in the subsequent steps leading to light state formation at later times in the photocycle. When A79 is mutated to Gln in EL222, the decay of the singlet excited state remains the same, while the decay of the triplet state is accelerated 2.2-fold.
Continued structural evolution is observed on the microsecond timescale
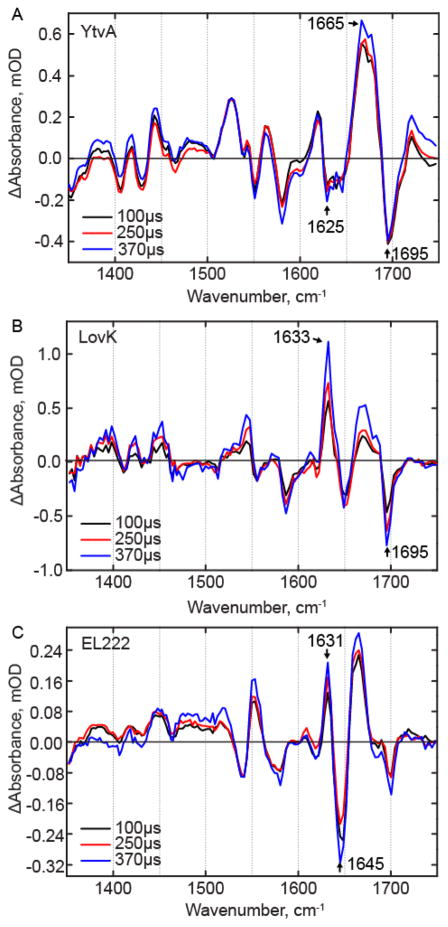
Thus far, we have described the early events up to adduct formation in the LOV photocycle, however structural evolution is also observed on the hundreds of μs timescale in the TRMPS experiment with the most dramatic changes being observed in LovK. In AsLOV2 it was shown previously that when TRMPS spectra from 100–400 μs are normalized to the adduct C4-C10a bleach at 1553 cm−1, a bleach at 1625 cm−1 continues to increase with a sub-ms time constant, indicating a decrease in β-sheet content.34 The transient in the ~1660 cm−1 region also increases on this longer time scale. A similar evolution in a bleach at 1625 cm−1 is observed in the YtvA TRMPS spectrum (Figure 6A), together with an increase in a bleach at 1642 cm−1 which is assigned to uncoiling of the coiled-coil helices linking the LOV domain to the STAS domain of YtvA based on studies with model coiled-coils.35 In contrast to AsLOV2 and YtvA, the TRMPS data for LovK (Figure 6B) and EL222 support an overall increase in β-sheet content over this longer timescale, based on an increase in transients at 1633 cm−1and 1631 cm−1, respectively. The TRMPS data thus suggest significant variation in the structural dynamics of the LOV domains at longer timescales where AsLOV2 and YtvA show a loss of β-sheet content (bleach upon light state formation), whereas β-sheet content increases in LovK and EL222 (transient).
Figure 6. TRMPS spectra of the LOV photoreceptors on the microsecond timescale.
Spectra after 100 μs were normalized to the adduct bleach mode at ~1550 cm−1 for (A) YtvA, (B) LovK, and (C) EL222.
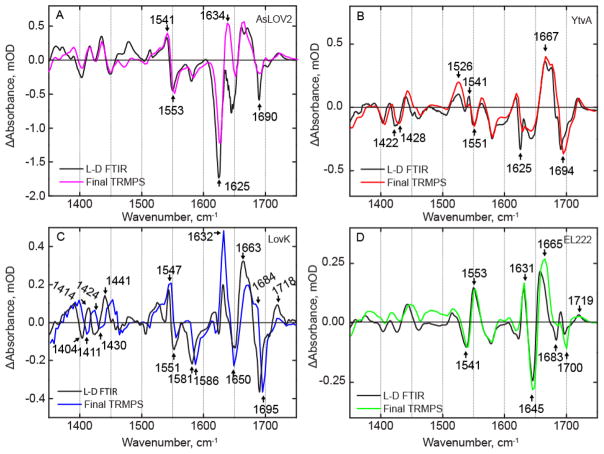
The TRMPS spectra provide insight into the structural evolution of the photoreceptors out to ~400 μs. However, comparison of the final TRMPS spectrum with the steady state FTIR difference spectrum (Figure 7) reveals that structural dynamics of the proteins must occur on even longer timescales. In AsLOV2, evolution of bleaches at 1625 cm−1 and 1690 cm−1 and a transient at 1634 cm−1 is observed to occur between the final TRMPS spectrum and the light – dark (L-D) FTIR spectrum, which were previously assigned to β-sheet, Q513, and α-helix, respectively.36,37 In the case of YtvA (Figure 7B), a bleach at 1625 cm−1 clearly evolves between the 370 μs TRMPS spectrum and the steady state spectrum as shown in the light – dark FTIR spectrum (Figure 7B). In EL222 (Figure 7D), the only modes that are evolving on these longer timescales are a bleach at 1683 cm−1 and the large transient at 1665 cm−1 which red-shifts to 1657 cm−1.
Figure 7. Comparison of latest TRMPS and steady state FTIR spectra reveal structural evolution post 400 μs.
Light minus dark (L – D) FTIR difference spectra of (A) AsLOV2, (B) YtvA, (C) LovK, and (D) EL222 were measured to evaluate structural evolution from the microsecond timescale (390 μs spectrum from TRMPS) to the final steady-state.
Several differences are observed in the LovK L – D FTIR spectrum (Figure 7C) compared to the 390 μs TRMPS spectrum. Most prominent in this regard is the shoulder at 1684 cm−1 and the shift of the protein/flavin transient at 1663 cm−1, causing the shift of the 1695 cm−1 bleach. In addition, both bleaches and transients in the 1400–1500 cm−1 region appear to blue-shift by ~10 cm−1 in LovK. A similar effect is observed for YtvA where a bleach at 1422 cm−1 is shifted to 1428 cm−1. These modes are assigned to proline residues that are perturbed due to changes in the central β-sheet.38 The L – D FTIR spectra of YtvA and LovK have been measured previously in H2O buffer and the data shown here agree with the previous measurements despite a few differences mainly due to experimental conditions that will be addressed in the discussion section.26,39
Discussion
LOV activation proceeds through four major phases: a singlet excited state (i) intersystem-crossing on the ~2 ns timescale to a triplet state (ii), from which a cysteine adduct is formed in ~20 μs (iii) leading to large scale changes in protein structure on the microsecond timescale and longer. The TRMPS spectra and kinetics for each of these phases were measured and compared for four LOV photoreceptors. Further evolution beyond 400 μs is observed for each photoreceptor using FTIR difference spectroscopy. Transients at ~1380 cm−1 and the ground state bleaches at ~1550 cm−1 for all four proteins vary no more than 10 cm−1 of each other, as expected for flavin modes. Each protein shares a conserved β-sheet motif, and changes in the vibrational modes assigned to β-sheet are observed on the hundreds of microseconds timescale. The L – D FTIR data in the present work were obtained in D2O buffer at a concentration of 1 mM. While these spectra are similar to those published previously,26,40 there are a few differences. For example, the L – D spectrum in Bednarz et al.26 has a well resolved transient at ~1680 cm−1 that appears as a large red shifted transient in our data. In addition, we observe a band at 1632 cm−1 that is not resolved in the spectrum presented by Alexandre et al.40 These differences could be due to an H/D effect on the amide I region of the spectrum and/or the fact that the other studies were performed on dehydrated samples.
Differences between the excited state spectra are observed among the four proteins. AsLOV2, YtvA, and LovK show very similar excited state spectra and kinetics except for the protein transients in the 1600–1670 cm−1 region. In AsLOV2 and EL222, the transient at 1622 cm−1 appears as a symmetrical peak while in YtvA and LovK there are well-resolved shoulders on the transient. When the spectra of these three proteins are compared to that of EL222, the differences are more significant with a ~6 cm−1 shift in the main bleach from ~1550 cm−1 to 1544 cm−1 which is a result of the Gln to Ala replacement in EL222, as confirmed by studies of the A79Q mutation in which the main bleach shifts from 1544 cm−1 in WT-EL222 to 1550 cm−1 in A79Q-EL222. In addition, the bleach at 1652 cm−1 in EL222 is ~10 cm−1 red shifted compared to the other LOV domains which is assigned to stronger hydrogen bonding around FMN.
The singlet excited state of FMN forms within the instrument response time for each of the proteins and decays to the triplet state with a rate of ~2 ns for AsLOV2, YtvA, and LovK while a 2-fold decrease in decay rate to 3.5 ns is observed in EL222. The reduced rate of excited state decay for EL222 has not been previously described, however Raffelberg and coworkers reported modulation of the fluorescence quantum yield through mutation of residues interacting with the C2=O hydrogen bonding network in YtvA.23 The triplet state decays in ~10 μs for AsLOV2 and YtvA, 15 μs for LovK, and 4.2 μs for EL222. Triplet decay rates for AsLOV2 and YtvA were previously reported in the range of 1–3 μs.33,41 The latter measurements were performed in H2O, whereas our samples were exchanged into D2O prior to data collection. The observation of a large (5–10-fold) kinetic isotope effect for decay of the AsLOV2 and YtvA triplet states, suggests that proton transfer to FMN-N5 occurs in the rate limiting step leading to adduct formation, consistent with the kinetic isotope effect studies reported by Corchnoy et al.41 In contrast, no kinetic isotope effect is observed on the decay rates for the singlet excited state, as the values obtained in our measurements are comparable to those obtained in H2O.11,39 Finally, the structural dynamics that occur in the μs-ms regime have not been measured in H2O and thus it is not possible to assess whether these longer time-scale events are subject to a kinetic isotope effect. Using protein folding as a paradigm for the structural changes that occur between the LOV domain dark and light states, we note that while deuterium substitution has a well-accepted, albeit usually small, impact on the thermodynamics of protein folding, the effect on the rates of protein folding is complex and difficult to predict owing to the multiple effects caused by the impact of deuteration on protein backbone hydrogen bonds, on protein solvent hydrogen bonds and on the hydrophobic effect.42–47
The X-ray crystal structures of AsLOV2 (2V0U), YtvA (2PR5), and EL222 (3P7N) were superimposed to identify key differences in LOV domain structure including the orientation of the C-terminal effector domain (Figure 1). In addition, while the X-ray structure of LovK is currently unavailable, the kinetic and spectral parameters have been analyzed in the context of a sequence alignment of the four LOV domain sequences (Figure S4). The hydrogen bond network is highly conserved among the three proteins shown in Figure 1 except for EL222: while a Gln residue is hydrogen bonded to the FMN ribityl chain and C2=O in AsLOV2 and YtvA, in EL222 this Gln residues is replaced by an Ala residue. Based on sequence alignment, we propose that the homologous residue in LovK is a Gln (Figure S4). In addition to the Gln to Ala substitution in EL222, one other difference in the flavin binding pocket is the variation of Phe494, Leu106 or Val119 in AsLOV2, YtvA and EL222, respectively. Sequence alignment (Figure S4) suggests that this residue is a Leu in LovK. F494 is notable due to its distance and conformation relative to the isoalloxazine ring, which allows for π-stacking (Figure S5).
We have rationalized some of the differences in the TRIR spectra between EL222 and the other LOV proteins by studying the A79Q-EL222 mutant which slows the dark state recovery rate of EL222.18 Despite restoring an interaction to FMN as evident by the shift of flavin ring C=N modes from 1544 cm−1 to 1550 cm−1, which is found at 1548 cm−1, 1552 cm−1, and 1555 cm−1 in AsLOV2, YtvA, and LovK, this mutant does not fully restore the LOV hydrogen bonding network. Our data suggests that Q79 does not occupy the same conformation in the flavin binding pocket compared to the other LOV proteins and that a hydrogen bond is disturbed rather than restored in A79Q-EL222 due to the shift in frequency from 1651 cm−1 in wild-type EL222 to 1658 cm−1 in A79Q-EL222, which we assign to Gln or Asn derived protein C=O modes that comprise the hydrogen bonding network. Another difference is the kinetics of triplet decay/adduct formation in A79Q-EL222, which are faster compared to WT-EL222 and it has been shown previously that mutations around the C2=O hydrogen bonding network can modulate triplet decay kinetics.23
The global structural changes in EL222 that lead to signaling state formation must occur faster than those that occur in YtvA and LovK, since only minor changes in IR difference spectra occur after hundreds of μs. Bands at 1633 cm−1 in LovK and 1631 cm−1 in EL222 assigned to the β-sheet grow in on the hundreds of μs timescale while the corresponding band in YtvA (1625 cm−1) does not change on this timescale but is observed in L-D FTIR. On the μs to steady-state timescale, evolution of vibrational modes assigned to the β-sheet are observed. This suggests that changes due to adduct formation propagate slowly as distance from the conserved Cys increases. The appearance of a 1625 cm−1 bleach in AsLOV2 and YtvA suggests a loss of β-sheet content while the appearance of transient bands at 1631 cm−1 and 1633 cm−1 in EL222 and LovK, respectively, suggests an increase in β-sheet content. Significant evolution is observed in the 1400–1450 cm−1 region in YtvA and LovK, which we assigned based on 15N labelling to proline N-D modes, specifically P110 in YtvA and P118 in LovK, which are located on the β-sheet adjacent to a disordered loop (Figure S6).
Based on analytical ultracentrifugation and SAXS data, it was previously concluded that large-scale structural changes such as a change in oligomerization or secondary structure do not occur during activation of YtvA.48 Our data are largely in agreement with this conclusion where the structural changes in YtvA are likely confined to alterations in hydrogen bonding or other interactions such as salt bridges that accompany local changes in secondary structure. In addition, the μs-ms structural dynamics reported in the present work agree with data from Choi et al. in which structural changes in the Jα helices occur on the order of ~100 μs and beyond.28
Conclusion
The LOV domain is a highly conserved flavin binding motif that couples light absorption to a variety of biological responses. In the present report, we describe the structural dynamics of three full-length LOV photoreceptors, from the instantaneous formation of the excited state to the structural changes that result in the biologically-relevant functional light state. Despite the high level of conservation, subtle differences are observed in excited state structure and the associated kinetics due to the specific flavin-protein interactions that are present in each photoreceptor. On the μs-s timescale, spectral evolution associated with large scale structural dynamics are observed and are distinct for each of the proteins measured. This work paves the way for future high-resolution spectroscopic and structural studies aimed at uncovering a pathway between FMN excitation and large scale structural changes associated with photoactivation.
Supplementary Material
Acknowledgments
Funding
This study was supported by the EPSRC (EP/G002916 to SRM) and NSF (CHE-1223819 to PJT). JI was supported by an NIH Chemistry-Biology Interface training grant (T32GM092714). AL is a Bolyai János Research Fellow and was supported by OTKA NN113090. JA was supported by the NSF REU program at Stony Brook University (NSF-CHE-1358959). SAHA was a Fulbright Scholar and gratefully acknowledges support from the Fulbright Program. JT was supported by the IMSD-MERGE Program at Stony Brook University (5R25GM103962-04).
We are grateful to STFC for access to the ULTRA laser facility. We are grateful to Professor Ray Owens and Anil Verma for assistance in protein preparation and access to the Oxford Protein Production Facility-UK.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.xxxxxxx.
Supporting information includes a table of band assignments for the four LOV photoreceptors (Table S1) and six figures. This includes the LOV photocycle and numbering of the isoalloxazine ring (Figure S1), kinetic traces for the excited state decay and ground state recovery together with a plot of the residuals to support the quality of data fitting (Figure S2), light minus dark FTIR spectra of unlabeled YtvA and 15N-labeled YtvA (Figure S3) to support band assignments, sequence alignment of the LOV domains to show sequence conservation between LovK, for which no X-ray structure is available, and the three other photoreceptors (Figure S4), and overlays of the AsLOV2, YtvA and EL222 structures showing the position of F494 in AsLOV2 (Figure S5) and a proline residue on the LOV β-sheet (P110 in YtvA) (Figure S6).
References
- 1.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crosson S, Moffat K. Photoexcited Structure of a Plant Photoreceptor Domain Reveals a Light-Driven Molecular Switch. Plant Cell Online. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 4.Harper SM. Structural Basis of a Phototropin Light Switch. Science (80) 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 5.Herrou J, Crosson S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol. 2011;9:713–723. doi: 10.1038/nrmicro2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol Plant. 2012;5:533–544. doi: 10.1093/mp/sss020. [DOI] [PubMed] [Google Scholar]
- 7.The PyMOL Molecular Graphics System, Version 1.8. Schrodinger, LLC; [Google Scholar]
- 8.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The Photocycle of a Flavin-binding Domain of the Blue Light Photoreceptor Phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 9.Winkler A, Barends TRM, Udvarhelyi A, Lenherr-Frey D, Lomb L, Menzel A, Schlichting I. Structural details of light activation of the LOV2-based photoswitch PA-Rac1. ACS Chem Biol. 2015;10:502–509. doi: 10.1021/cb500744m. [DOI] [PubMed] [Google Scholar]
- 10.Guntas G, Hallett Ra, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci. 2015;112:112–117. doi: 10.1073/pnas.1417910112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexandre MTA, Domratcheva T, Bonetti C, Van Wilderen LJGW, Van Grondelle R, Groot ML, Hellingwerf KJ, Kennis JTM, Van Wilderen LJGW, Van Grondelle R, Groot ML, Hellingwerf KJ, Kennis JTM. Primary reactions of the LOV2 domain of phototropin studied with ultrafast mid-infrared spectroscopy and quantum chemistry. Biophys J. 2009;97:227–237. doi: 10.1016/j.bpj.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeifer A, Majerus T, Zikihara K, Matsuoka D, Tokutomi S, Heberle J. Time-Resolved Fourier Transform Infrared Study on Photoadduct Formation and Secondary Structural Changes within the Phototropin LOV Domain. 2009;96:1462–1470. doi: 10.1016/j.bpj.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gil AA, Laptenok SP, French JB, Iuliano JN, Lukacs A, Hall CR, Sazanovich IV, Greetham GM, Bacher A, Illarionov B, Fischer M, Tonge PJ, Meech SR. Femtosecond To Millisecond Dynamics Of Light Induced Allostery In The Avena Sativa Lov Domain. J Phys Chem B. 2017;121:1010–1019. doi: 10.1021/acs.jpcb.7b00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konold PE, Mathes T, Weiβenborn J, Groot ML, Hegemann P, Kennis JTM. Unfolding of the C-Terminal Jα Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. Phys Chem Lett. 2016:3472–3476. doi: 10.1021/acs.jpclett.6b01484. [DOI] [PubMed] [Google Scholar]
- 15.Möglich A, Moffat K. Structural Basis for Light-dependent Signaling in the Dimeric LOV Domain of the Photosensor YtvA. J Mol Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell EB, McDonald CA, Palfey BA, Crosson S. An analysis of the solution structure and signaling mechanism of LovK, a sensor histidine kinase integrating light and redox signals. Biochemistry. 2010;49:6761–6770. doi: 10.1021/bi1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni Ra, Luecke H, Gardner KH. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci U S A. 2011;108:9449–54. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoltowski BD, Motta-mena LB, Gardner KH. Blue Light-Induced Dimerization of a Bacterial LOV – HTH DNA- Binding Protein. Biochemistry. 2013;52:6653–6661. doi: 10.1021/bi401040m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 20.Losi A, Polverini E, Quest B, Gärtner W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–2634. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash AI, McNulty R, Shillito ME, Swartz TE, Bogomolni Ra, Luecke H, Gardner KH. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc Natl Acad Sci U S A. 2011;108:9449–54. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zayner JP, Sosnick TR. Factors that control the chemistry of the LOV domain photocycle. PLoS One. 2014:9. doi: 10.1371/journal.pone.0087074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raffelberg S, Mansurova M, Gärtner W, Losi A. Modulation of the photocycle of a LOV domain photoreceptor by the hydrogen-bonding network. J Am Chem Soc. 2011;133:5346–5356. doi: 10.1021/ja1097379. [DOI] [PubMed] [Google Scholar]
- 24.Eitoku T, Nakasone Y, Matsuoka D, Tokutomi S, Terazima M. Conformational dynamics of phototropin 2 LOV2 domain with the linker upon photoexcitation. J Am Chem Soc. 2005;127:13238–13244. doi: 10.1021/ja052523i. [DOI] [PubMed] [Google Scholar]
- 25.Nakasone Y, Eitoku T, Matsuoka D, Tokutomi S, Terazima M. Dynamics of Conformational Changes of Arabidopsis Phototropin 1 LOV2 with the Linker Domain. J Mol Biol. 2007;367:432–442. doi: 10.1016/j.jmb.2006.12.074. [DOI] [PubMed] [Google Scholar]
- 26.Bednarz T, Losi A, Gärtner W, Hegemann P, Heberle J, Gartner W, Hegemann P, Heberle J. Functional variations among LOV domains as revealed by FT-IR difference spectroscopy. Photochem Photobiol Sci. 2004;3:575–579. doi: 10.1039/b400976b. [DOI] [PubMed] [Google Scholar]
- 27.Losi A, Ghiraldelli E, Jansen S, Gärtner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem Photobiol. 2005;81:1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
- 28.Choi S, Nakasone Y, Hellingwerf KJ, Terazima M. Photochemical Reactions of the LOV and LOV-Linker Domains of the Blue Light Sensor Protein YtvA. Biochemistry. 2016;55:3107–3115. doi: 10.1021/acs.biochem.6b00263. [DOI] [PubMed] [Google Scholar]
- 29.Greetham GM, Sole D, Clark IP, Parker AW, Pollard MR, Towrie M, Greetham GM, Sole D, Clark IP, Parker AW, Pollard MR, Towrie M. Time-resolved multiple probe spectroscopy Time-resolved multiple probe spectroscopy. 2015;103107:2–7. [Google Scholar]
- 30.Snellenburg JJ, Laptenok SP, Seger R, Mullen KM, Stokkum IHM van. Glotaran: A Java-Based Graphical User Interface for the R Package TIMP. J Stat Softw. 2012:49. [Google Scholar]
- 31.Kondo M, Nappa J, Ronayne KL, Stelling AL, Tonge PJ, Meech SR. Ultrafast vibrational spectroscopy of the flavin chromophore. J Phys Chem B. 2006;110:20107–20110. doi: 10.1021/jp0650735. [DOI] [PubMed] [Google Scholar]
- 32.Haigney A, Lukacs A, Zhao RK, Stelling AL, Brust R, Kim RR, Kondo M, Clark I, Towrie M, Greetham GM, Illarionov B, Bacher A, Römisch-Margl W, Fischer M, Meech SR, Tonge PJ. Ultrafast infrared spectroscopy of an isotope-labeled photoactivatable flavoprotein. Biochemistry. 2011;50:1321–1328. doi: 10.1021/bi101589a. [DOI] [PubMed] [Google Scholar]
- 33.Song SH, Madsen D, Van Der Steen JB, Pullman R, Freer LH, Hellingwerf KJ, Larsen DS. Primary photochemistry of the dark- and light-adapted states of the YtvA protein from bacillus subtilis. Biochemistry. 2013;52:7951–7963. doi: 10.1021/bi4012258. [DOI] [PubMed] [Google Scholar]
- 34.Buttani V, Losi A, Polverini E, Ga W. Blue news: NTP binding properties of the blue-light sensitive YtvA protein from Bacillus subtilis. 2006;580:3818–3822. doi: 10.1016/j.febslet.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Reisdorf WC, Krimm S. Infrared amide I′ band of the coiled coil. Biochemistry. 1996;35:1383–1386. doi: 10.1021/bi951589v. [DOI] [PubMed] [Google Scholar]
- 36.Herman E, Sachse M, Kroth PG, Kottke T. Blue-light-induced unfolding of the Jα helix allows for the dimerization of aureochrome-LOV from the diatom Phaeodactylum tricornutum. Biochemistry. 2013;52:3094–3101. doi: 10.1021/bi400197u. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee A, Herman E, Serif M, Maestre-Reyna M, Hepp S, Pokorny R, Kroth PG, Essen L-O, Kottke T. Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes. Nucleic Acids Res. 2016:1–14. doi: 10.1093/nar/gkw420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barth a. The infrared absorption of amino acid side chains. Prog Biophys Mol Biol. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 39.Alexandre MTA, Purcell EB, Van Grondelle R, Robert B, Kennis JTM, Crosson S. Electronic and protein structural dynamics of a photosensory histidine kinase. Biochemistry. 2010;49:4752–4759. doi: 10.1021/bi100527a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexandre MTA, Van Grondelle R, Hellingwerf KJ, Kennis JTM. Conformational heterogeneity and propagation of structural changes in the LOV2/Jα domain from Avena sativa phototropin 1 as recorded by temperature-dependent FTIR spectroscopy. Biophys J. 2009;97:238–247. doi: 10.1016/j.bpj.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J Biol Chem. 2003;278:724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- 42.Kuhlman B, Raleigh DP. Global analysis of the thermal and chemical denaturation of the N-terminal domain of the ribosomal protein L9 in H 2 O and D 2 O. Determination of the thermodynamic parameters, Δ H °, Δ S °, and Δ C °p. Protein Sci. 1998;7:2405–2412. doi: 10.1002/pro.5560071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krantz BA, Moran LB, Kentsis A, Sosnick TR. D/H amide kinetic isotope effects reveal when hydrogen bonds form during protein folding. Nat Struct Biol. 2000;7:62–71. doi: 10.1038/71265. [DOI] [PubMed] [Google Scholar]
- 44.Creswell CJ, Allred AL. The Strengths Of Hydrogen Bonds Formed By Protium and Deuterium. J Am Chem Soc. 1962;84:3966–3967. [Google Scholar]
- 45.Makhatadze GI, Clore GM, Gronenborn AM. Solvent isotope effect and protein stability. Nat Struct Biol. 1995;2:852–855. doi: 10.1038/nsb1095-852. [DOI] [PubMed] [Google Scholar]
- 46.Itzhaki LS, Evans PA. Solvent isotope effects on the refolding kinetics of hen egg-white lysozyme. Protein Sci. 1996;5:140–146. doi: 10.1002/pro.5560050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kresheck GC, Schneider H, Scheraga Ha. The effect of D2O on the thermal stability of proteins. Thermodynamic parameters for the transfer of model compounds from H2O to D2O. J Phys Chem. 1965;69:3132–3144. doi: 10.1021/j100893a054. [DOI] [PubMed] [Google Scholar]
- 48.Jurk M, Dorn M, Kikhney A, Svergun D, Gartner W, Schmieder P. The Switch that Does Not Flip: The Blue-Light Receptor YtvA from Bacillus subtilis Adopts an Elongated Dimer Conformation Independent of the Activation State as Revealed by a Combined AUC and SAXS Study. J Mol Biol. 2010;403:78–87. doi: 10.1016/j.jmb.2010.08.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.