Abstract
During aging, decreased efficiency of nuclear factor E2-related factor 2 (Nrf2) activation and autophagic processes in the brain may be a contributing factor in the pathogenesis of neurodegenerative diseases such as Alzheimer’s disease. Therefore, we analyzed the expression of BAG3, a co-chaperone that mediates autophagy, and the autophagy adaptors NBR1, NDP52 and SQSTM1/p62 in the brains of 4, 8 and 12 month old wild-type and Nrf2 knockout (−/−) mice. We also analyzed the levels of total tau, as well as phospho-tau species. There were minimal differences in the expression of autophagy-related genes or tau species in 4 month old animals, however by 12 months all of these autophagy-associated genes were expressed at significantly lower levels in the Nrf2(−/−) mice. The decreases in the autophagy associated genes was accompanied by significantly elevated levels of phospho-tau species in the 12 month Nrf2(−/−) brains. These findings indicate that Nrf2 regulation of autophagy-related genes likely plays a greater role in mediating the clearance of tau as an organism ages.
Keywords: Nrf2, tau, autophagy adaptors, BAG3
1. Introduction
Nuclear Factor (erythroid-derived 2)-like 2 (Nrf2) is a transcription factor that plays a crucial role in the cytoprotective response to stress. Nrf2 was first described as a regulator of antioxidant and detoxifying proteins (Alam, et al., 1999, Ishii, et al., 2000, Ishii, et al., 2002, Itoh, et al., 1997, Wild, et al., 2011), but it is now evident that Nrf2 regulates numerous genes involved in cytoprotective processes, including protein quality control (Pajares, et al., 2017, Pajares, et al., 2016). Nrf2 plays a central role during aging as the cumulative effects of environmental stressors increase (Lewis, et al., 2010, Zhang, et al., 2015). In a study of 10 rodent species with varying maximal lifespan potential (MLSP), Nrf2 signaling positively correlated with MLSP (Lewis, et al., 2015). Because neurons are terminally differentiated they cannot dilute toxic protein species during cell division. Instead, they must rely on efficient cytoprotective responses, such as protein quality control processes, to maintain cell health. Given the dependence of neurons on these processes, Nrf2 has been proposed to play a pivotal role in maintaining neuronal health.
In neurons, macroautophagy (autophagy) plays a significant role in protein quality control (Boland, et al., 2008). In particular, selective autophagy, i.e., the recognition and targeting of client proteins to the developing autophagosome and subsequent degradation in the autophagic vacuoles, is essential for effective protein quality control (Gestwicki and Garza, 2012, Vidal, et al., 2014). Selective autophagy is dependent on chaperone/co-chaperone complexes, which include autophagy cargo adaptor proteins, to engage specific client proteins and target them to the autophagy system (Gamerdinger, et al., 2009, Gamerdinger, et al., 2011, Rapino, et al., 2014, Ulbricht, et al., 2013). Nrf2 directly regulates the expression of two autophagy cargo adaptor proteins, SQSTM1/p62 and NDP52 (Jain, et al., 2010, Jo, et al., 2014, Pajares, et al., 2016). Further, Nrf2 also regulates genes involved in induction, autophagosome formation and elongation as well as autolysosome clearance (Pajares, et al., 2016). Therefore, Nrf2 plays a key role in allowing neurons to respond to stressors by increasing the machinery needed to remove damaged or misfolded proteins.
As organisms age oxidative stress increases along with the accumulation of dysfunctional or aggregate prone proteins (David, 2012), increasing the need for efficient autophagy. Indeed, interventions that increase autophagy result in extended lifespans (Madeo, et al., 2015). Conversely, deletion of the autophagy cargo adaptor protein p62, which also functions as an upregulator of Nrf2 signaling, results in reduced lifespan and indicators of premature aging (Kwon, et al., 2012). Neurons also exhibit increased reliance on autophagy and less on proteasome-dependent clearance mechanisms during aging (Gamerdinger, et al., 2009). In addition, the levels of the co-chaperone BAG3, which facilitates selective autophagy, increase during aging (Gamerdinger, et al., 2009), and knockdown of BAG3 in neurons attenuates autophagic clearance of tau (Lei, et al., 2014). Overall these and other data indicate that Nrf2 plays a key role in maintaining a healthy proteome during aging, in part by regulating the expression of genes involved in the selective autophagy process.
Given the fact that Nrf2 and selective autophagy processes play crucial roles in the maintenance of the neuronal proteome, especially in the context of aging, this study focused on determining the impact of Nrf2 deletion on autophagy cargo adaptor proteins and BAG3 in 4, 8 and 12 month old mice. mRNA and protein levels of BAG3 and the autophagy cargo adaptor proteins NBR1, NDP52 and p62 were measured in the hippocampus of wild type and Nrf2(−/−) mice at each age. Both hippocampal and cerebral cortical tissue were used for protein expression. Protein expression of LC3II was also measured as an indication of autophagy. Additionally, we quantitatively analyzed the expression of total tau as well as phosphorylated tau species (pT231, pS262 and pS396/404) in both the hippocampus and cortex at the 3 ages. Overall, our results indicate that deletion of Nrf2 does not play a significant role expression of these genes in young animals, but attenuates the age-dependent upregulation of BAG3 and the autophagy adaptor proteins concurrent with increased accumulation of phospho-tau species. These findings suggest that impaired Nrf2 signaling may be a contributing factor in development of aging-related neurodegenerative conditions that are characterized by the accumulation of aggregate prone proteins such as tau.
2. Materials and Methods
2.1. Mice
All mice were maintained on a 12 h light/dark cycle with food and water available ad libitum. All procedures were approved by University Committee on Animal Research of the University of Rochester. Wild-type (WT) C57BL/6J mice were originally purchased from Jackson laboratories. Nrf2-knockout (−/−) mice on a C57BL/6J background were generously supplied by Dr M Yamamoto (Itoh, et al., 1997) via the RIKEN BioResource Center, Tsukuba, Japan. Both male and female mice were used in the study. There were 5–6 animals in each group for all quantitative analyses. To prepare samples for quantitative RT-PCR analysis and immunoblotting, 4, 8 and 12 month old WT and Nrf2(−/−) mice were deeply anesthetized with isoflurane and perfused with phosphate-buffered saline (PBS). Brains were removed, sectioned along the sagittal axis and rapidly dissected. The cortex and hippocampus of a half hemisphere were collected and frozen for quantitative RT-PCR analysis and the other half hemisphere regions were frozen for immunoblotting analyses. All samples were stored at −80°C until use. To prepare brains for immunohistochemistry, PBS perfusion was followed by perfusion with 4% paraformaldehyde (PFA) for 15 min. The brains were removed and kept in 4% PFA for 2 h at 4°C, and then cryoprotected by transferring them to 15% sucrose and then 30% sucrose solution in PBS until they sank, respectively. The brains were then rapidly frozen in cooled 2-methylbutane for 30 s on dry ice, and then kept at −80°C for immunohistochemistry analysis.
2.2. Quantitative RT-PCR analysis
Total RNA was extracted from hippocampal samples using the E.Z.N.A. Total RNA kit (Omega Bio-tek). cDNA was generated using the Verso cDNA synthesis kit (Thermo Scientific). cDNA (10 ng) was used as a template, and quantitative PCR was performed in 20 μl reactions with perfeCTa SYBR Green FastMix (QuantaBio) according to the manufacturer’s protocol on a Bio-Rad CFX 96 Real-Time System. The primers were designed using NCBI/Primer-BLAST or were validated primers from the Primer Bank (https://pga.mgh.harvard.edu/primerbank/). The primer sequences for mouse Bcl-2 associated athanogene 3 (BAG3), NBR1, NDP52 (also known as calcium binding and coiled-coil domain 2 (Calcoco2)), sequestosome 1/p62 (SQSTM1/p62), microtubule-associated protein tau, and actin beta (ACTb) (reference gene) are shown in Table 1. Threshold cycle (Ct) values were used to calculate fold changes based on the 2−ΔΔCT method.
Table 1.
| Gene name | primers sequence |
|---|---|
| BAG3 | REV 5′-GCTGAAGATGCAGTGTCCTTAG-3′ FOR 5′-CTGGGAGATCAAAATCGACCC-3′ |
| NBR1 | REV 5′-TCCCAAGACTCTCACCAGTG-3′ FOR 5′-GGAAATCAGCTACAGATGCAAGT-3′ |
| SQSTM1/p62 | REV 5′-TGTTCCCGCCGGCACTCCTT-3′ FOR 5′-GGCGCACTACCGCGATGAGGA-3′ |
| NDP52 | REV 5′-TCCTTGCGTCGAGGGATGAACTTT-3′ FOR 5′-TGGCAACTTCTCTCAGGTCCTGTT-3′ |
| TAU | REV 5′-CTCTTACTAGCTGATGGTGAC-3′ FOR 5′-GAACCACCAAAATCCGGAGA-3′ |
| β-ACTIN | FOR 5′-CTAAGGCCAACCGTGAAAAG-3′ REV 5′-ACCAGAGAGGCATACAGGGACA-3′ |
2.3. Immunoblotting
Four to five volumes of cold RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EGTA, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS) containing 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF and 10 μg/ml each of aprotinin, leupeptin and pepstatin (Jo, et al., 2014) was added to pre-chilled Dounce homogenizers containing the cortical or hippocampal tissue. Proteins were extracted using 10–15 strokes of the pestle while holding the tube in the ice. The homogenates were transferred to pre-chilled Eppendorf tubes, centrifuged at 16,000 × g for 10 min at 4 °C, the supernatant was collected, and protein concentration was determined using the BCA assay. For analysis, samples (10–50 μg protein) were separated on 10% or 12% SDS-polyacrylamide gels followed by transfer to nitrocellulose membranes. After blocking in Tris-buffered saline Tween-20 (TBST) containing 5% non-fat dry milk at room temperature for 1 h, the membranes were incubated in the same solution with antibodies against pS396/404 tau (PHF1) (1:5000, a gift from Dr. P. Davies), pS262 tau (12E8) (1:1000, a gift from Dr. P. Seubert), pT231 tau (AT180) (1:1000, Thermo Scientific), BAG3 (1:5000, Proteintech), mouse NDP52 (1:1000, Millipore), p62/SQSTM1 (1:2500, Cell Signaling Technology), NBR1 (1:2000, Boster Biological Technology), LC3 (1:5000, Novus Biologicals) or beta actin (1:10000, Thermo Scientific), at 4 °C, overnight. The membranes were rinsed three times with TBST, incubated with either goat anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase (1:5000, Bio-Rad) at room temperature for 1 h, and then developed with chemiluminescence. The protein levels were normalized using beta actin. Data were analyzed by densitometry using Un-Scan-It (Silk Scientific Corp.) with background subtraction and normalized to loading controls. Analyses of WT and Nrf2 (−/−) samples at each age and for each protein were carried using data from a single immunoblot/exposure.
2.4. Immunohistochemical staining
Brain slices (30 μm) were prepared from 4 and 12 month WT and Nrf2 (−/−) mice brains fixed with 4% PFA. The brain sections were washed three times using 0.15 M phosphate buffer (0.05M NaH2PO4, 0.1M Na2HPO4) to remove the cryoprotectant. After transfer to polysine microscope adhesion slides (Thermo Scientific), antigen retrieval was carried out. For p62 and BAG3 staining, the slides were incubated at 94°C for 10 min in 10 mM sodium citrate buffer pH 6.0, then cooled at room temperature for 30 min. For AT180 staining, the slides were incubated at 60°C for 15 min in L.A.B solution (Polysciences), and then washed twice with PBS. After antigen retrieval, the slides were blocked in PBS containing 5% BSA and 0.5% Triton X-100 for 1 h at room temperature. The sections were incubated with AT180 (1:100), BAG3 (1:200) or p62 (1:100) antibody, LC3 (1:100, Cell Signaling Technology) and biotin-conjugated NeuN (1:3000, Millipore) in 5% BSA and 0.1% Triton X-100 in PBS overnight at 4°C. The next day, slices were incubated for 1 h at room temperature with Alexa Fluor 488 donkey-anti-rabbit (BAG3, p62) or Alexa Fluor 488 donkey-anti-mouse (AT180) and streptavidin Alexa Fluor 594 conjugate (NeuN) secondary antibody (1:1000, Invitrogen) followed by three washes with PBS. The coverslips were mounted on the glass slides with ProLong® diamond antifade reagent (Thermo Scientific) following three washes with PBS. The slides were imaged using a laser scanning confocal microscope (Olympus FV1000). Images from one slide containing brain sections from mice of each age and genotype were collected under identical settings for each region. Three mice at each age and for each genotype were imaged. The brain sections were imaged at level V of the cortex, and CA1, CA3 and dentate gyrus (DG) area of hippocampus. However, only images from the CA1 region are shown. Fiji ImageJ software was used for analyzing these images. Average fluorescence intensity was measured from two areas of each brain section; areas at approximately same location of three brain sections were analyzed.
2.5. Statistical analysis
Statistical analyses were conducted using Student’s t-test, and statistical significance was defined as *p < 0.05, #p < 0.01 and ^p < 0.001. Data were analyzed using GraphPad.
3. Results
3.1. BAG3, NBR1, NDP52 and p62 mRNA levels are lower in older Nrf2 (−/−) mice
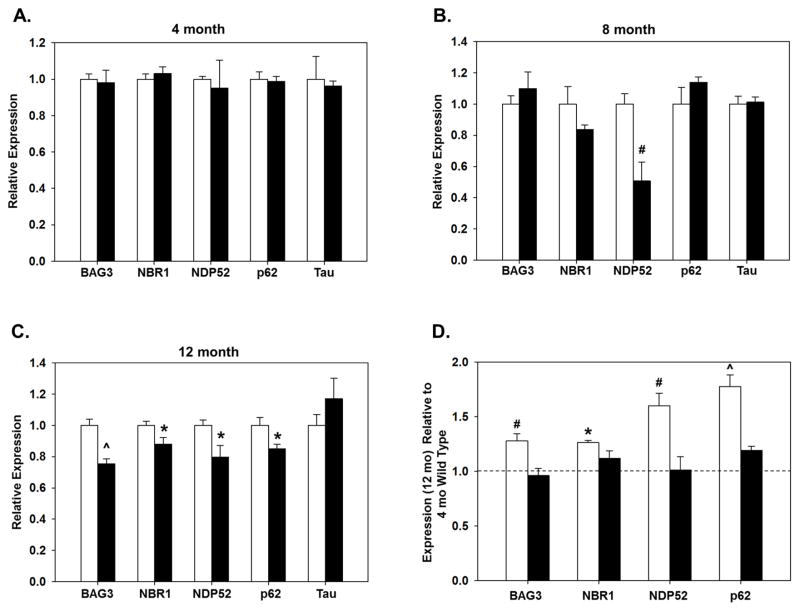
The expression of BAG3, NBR1, NDP52, p62 and tau was measured in hippocampal samples from 4, 8 and 12 month old WT and Nrf2(−/−) mice using quantitative RT-PCR. There were no differences in tau mRNA expression between WT and Nrf2 (−/−) mice at any of the ages (Fig. 1A–C). In 4 month old mice no differences were observed between WT and Nrf2 (−/−) for any of these genes (Fig. 1A). However, in 8 month old Nrf2 (−/−) mice NDP52 expression was significantly lower than in WT, and by 12 months the expression of BAG3, NBR1, NDP52, and p62 were also significantly lower in the Nrf2 (−/−) mice compared to WT at the same age (Fig. 1B,C). Interestingly, the expression of BAG3, NBR1, NDP52, and p62 were significantly elevated in 12 month old animals compared to 4 month old animals in the WT mice but not in the Nrf2 (−/−) mice (Fig. 1D). These data indicate that Nrf2 plays a significant role in facilitating the upregulation of these genes as the animal ages.
Figure 1. Gene expression of BAG3 and autophagy adaptors are not upregulated in older Nrf2(−/−) mice.
Hippocampal BAG3, NBR1, NDP52, p62 and tau gene expression in WT (open bars) and in Nrf2 (−/−) mice (black bars) at 4 (A), 8 (B) and 12 (C) months. Relative mRNA level of each gene is compared with that of WT mice. Age-dependent changes in gene expression were analyzed in 12 month old mice relative to 4 month old WT mice (D). N=5–6 in each group, mean ± SEM, (*p<0.05, #p < 0.01, ^p < 0.001).
3.2. Protein expression levels of BAG3, NBR1, NDP52 and p62 are lower in older Nrf2(−/−) mice
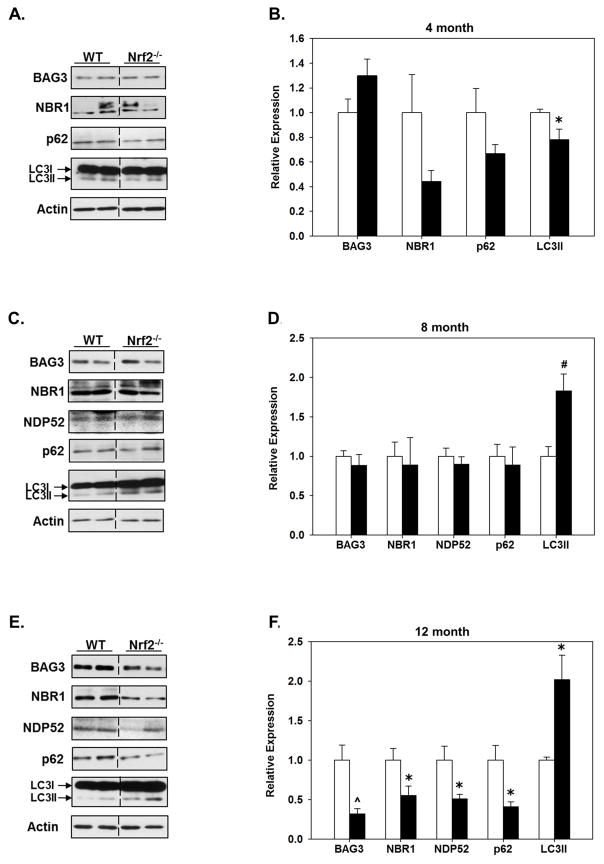
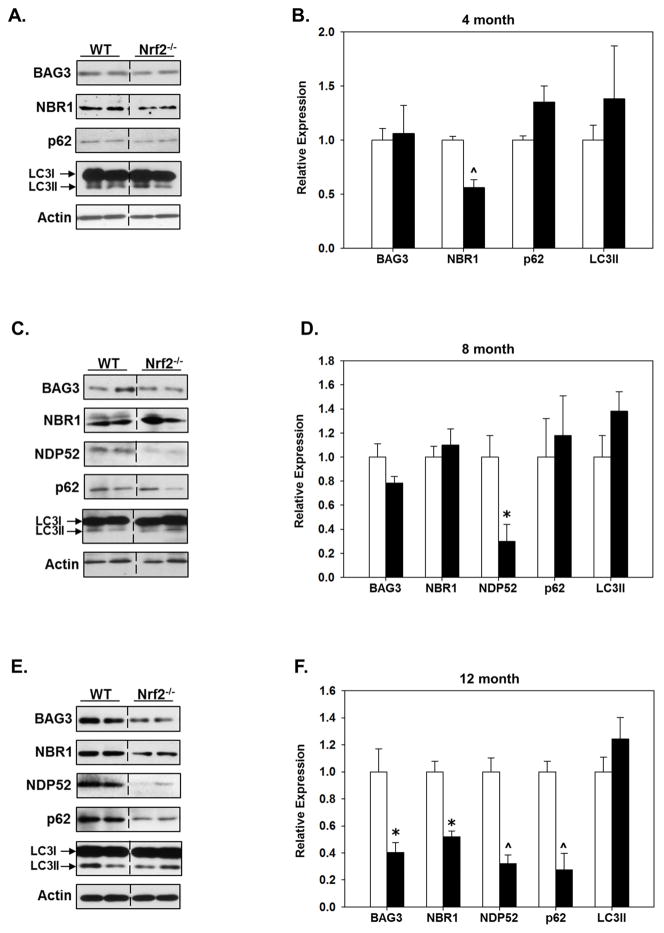
The protein expression of BAG3, NBR1, NDP52, p62 and LC3II in the cortex and hippocampus of 4, 8 and 12 month old WT and Nrf2 (−/−) mice was quantitatively assessed. In 4 month old animals NDP52 expression was below the level of detection on immunoblots in both WT and Nrf2 (−/−) mice, however NDP52 expression was detectable in both genotypes and regions at 8 and 12 months of age (Figs. 2&3). In 4 month old animals there was a statistically significant difference in LC3II in the hippocampus of Nrf2 (−/−) mice (Fig. 2A, B). Interestingly, at 8 months there was an increase in LC3II in the hippocampus in Nrf2 (−/−) mice, which remained significantly elevated in 12 month old Nrf2 (−/−) mice (Fig. 2C–F). Further we found that NDP52 protein expression in the hippocampus was similar in 8 month old WT and Nrf2 (−/−) mice, but was downregulated in the hippocampus of Nrf2 (−/−) mice at 12 months. NDP52 protein expression was dramatically decreased in the cortex of both 8 and 12 month old Nrf2 (−/−) mice (Fig. 3). In the hippocampus, no differences were observed in the expression of BAG3, NBR1, or p62 between WT and Nrf2 (−/−) mice at 8 months (Fig. 2C, D), but expression of these proteins was significantly lower in 12 month old Nrf2 (−/−) mice (Fig. 2E, F). A similar expression profile of these proteins was observed in the cortex (Fig. 3), although LC3II levels were not significantly changed in the cortex at any age (Fig. 3). Interestingly, the levels of NBR1 were significantly decreased in the cortex of 4 month old Nrf2 (−/−) mice (Fig. 3A, B) with a trend towards being decreased in the hippocampus as well (Fig. 2A, B), but these latter data did not reach significance. However, as noted above, there was no difference in NBR1 expression at 8 months between Nrf2(−/−) and WT mice, although by 12 months NBR1 levels were significantly lower in Nrf2(−/−) mice compared to WT in both regions (Fig. 2E,F; Fig. 3E,F). The reason why NBR1 is decreased in Nrf2(−/−) mice at 4 months and 12 months, but not 8 months is not known, but may be due to the complexity of its regulation through a bidirectional promoter (Dimitrov, et al., 2001, Whitehouse, et al., 2004) (see discussion). Nonetheless, taken together these data suggested that Nrf2 plays a pivotal role in maintaining the appropriate expression levels of BAG3, NBR1, NDP52 and p62 during aging, and thus selective autophagy processes.
Figure 2. Protein levels of BAG3 and autophagy adaptors are significantly lower in the hippocampus of older Nrf2 (−/−) mice compared to WT mice.
Hippocampal tissue was collected from 4 (A, B), 8 (C, D) and 12 (E, F) month old WT and Nrf2(−/−) mice and immunoblotted for BAG3, NBR1, NDP52, p62 and LC3. Actin served as loading control. Representative immunoblots are shown in (A, C, E) and quantitated data in (B, D, F). At 12 month the levels of BAG3 and all the autophagy adaptors are significantly lower in the Nrf2(−/−) (black bars) compared to the WT mice (open bars), the levels of LC3II are significantly higher. Relative data are expressed as function of WT levels at each age. N=5–6 in each group, mean ± SEM, (*p<0.05, #p < 0.01, ^p < 0.001).
Figure 3. Protein levels of BAG3 and autophagy adaptors are significantly lower in the cortex of older Nrf2 (−/−) mice compared to WT mice.
Cerebral cortical tissue was collected from 4 (A, B), 8 (C, D) and 12 (E, F) month old WT and Nrf2(−/−) mice and immunoblotted for BAG3, NBR1, NDP52, p62 and LC3. Actin served as loading control. Representative immunoblots are shown in (A, C, E) and quantitated data in (B, D, F). At 12 month the levels of BAG3 and all the autophagy adaptors are significantly lower in the Nrf2(−/−) (black bars) compared to the WT mice (open bars). Relative data are expressed as function of WT levels at each age. N=5–6 in each group, mean ± SEM, (*p<0.05, #p < 0.01, ^p < 0.001).
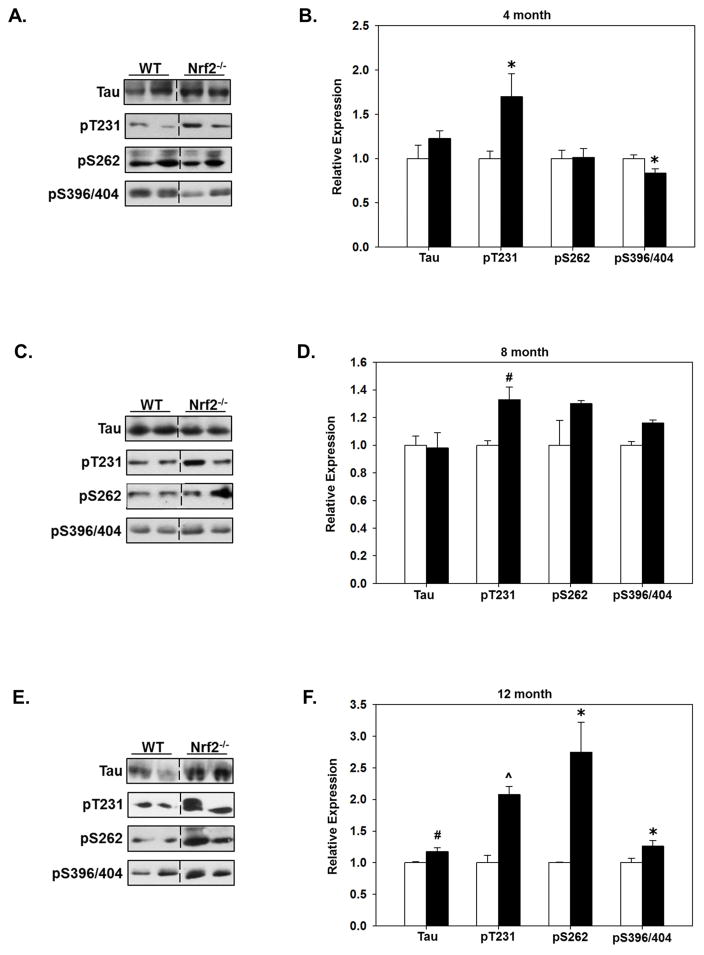
3.3. The levels of phosphorylated tau species are increased in older Nrf2(−/−) mice
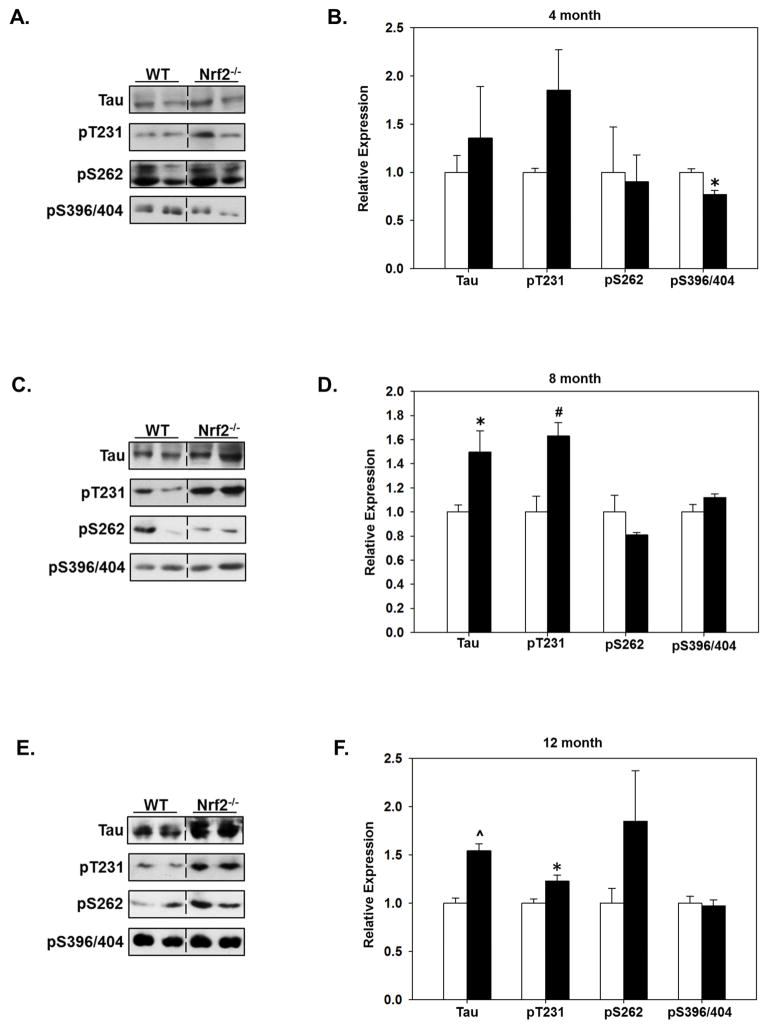
To determine the effects of Nrf2 in mediating the clearance of phosphorylated tau species, we quantitated the expression levels of total tau and phosphorylated tau in the cortex and hippocampus of WT and Nrf2 (−/−) mice at each age. In a previous study our data indicated that deletion of Nrf2 resulted in increased levels of phosphorylated tau species, however in these studies we did not control for the age of the animals (Jo, et al., 2014). As shown in Fig. 4, in the hippocampus, tau and pT231-tau (AT180) levels were significantly higher in 8 and 12 months old Nrf2 (−/−) mice, while the levels of tau phosphorylated at S262 (12E8) and at S396/S404 (PHF1) was not increased at any of the ages compared with WT mice (Fig. 4). An interesting observation was that at 4 months the levels of PHF1 reactivity were significantly decreased in both the hippocampus (Fig. 4A, B) and cortex (Fig. 5A, B). The reason for this initial decrease is not known, however given that soluble fractions were immunoblotted and tau phosphorylated at S396/S404 is more insoluble (Abraha, et al., 2000), it can be suggested that at this early time point a portion of tau phosphorylated at this epitope was relatively less soluble and thus was decreased in the fraction we immunoblotted. In the cortex, the levels of tau phosphorylated at T231 (AT180) were significantly elevated at all ages in Nrf2 (−/−) mice (Fig. 5). At 12 months of age all tau species examined were significantly higher in Nrf2 (−/−) cortex compared to WT (Fig. 5). However, it should be noted that the increases in tau phosphorylated at S396/S404 at 12 months in the cortex were not as robust as what was observed for the other phospho-tau species, and the increase did not reach significance in the hippocampus. This could be due to the tau solubility issue described above. However these results, which support our previous findings (Jo, et al., 2014), suggest that Nrf2 regulates the processes that mediate tau clearance, and increases in Nrf2 responsive genes during aging is likely to contribute to the maintenance of physiological levels of tau.
Figure 4. The levels of tau phosphorylated at T231 increase in the hippocampus of Nrf2 (−/−) mice.
Hippocampal tissue was collected from 4 (A, B), 8 (C, D) and 12 (E, F) month old WT and Nrf2(−/−) mice and immunoblotted for tau phosphorylated at T231 (AT180), S262 (12E8) or S398/404 (PHF1), or total tau. Representative immunoblots are shown in (A, C, E) and quantitated data in (B, D, F). Data were normalized to actin levels and quantitated as a function of WT levels at each age. At 8 and 12 months, total tau and pT231 tau levels were significantly higher in in the Nrf2(−/−) (black bars) compared to the WT mice (open bars). N=5–6 in each group, mean ± SEM, (*p<0.05, #p < 0.01, ^p < 0.001).
Figure 5. Tau and phosphorylated tau levels are significantly higher in the cortex of 12 month old Nrf2 (−/−) mice.
Cerebral cortical tissue was collected from 4 (A, B), 8 (C, D) and 12 (E, F) month old WT and Nrf2(−/−) mice and immunoblotted for tau phosphorylated at T231 (AT180), S262 (12E8) or S398/404 (PHF1), or total tau. Representative immunoblots are shown in (A, C, E) and quantitated data in (B, D, F). Data were normalized to actin levels and quantitated as a function of WT levels at each age. At 12 months, total tau and phospho-tau levels were significantly higher in in the Nrf2(−/−) mice (black bars) compared to the WT mice (open bars). N=5–6 in each group, mean ± SEM, (*p<0.05, #p < 0.01, ^p < 0.001).
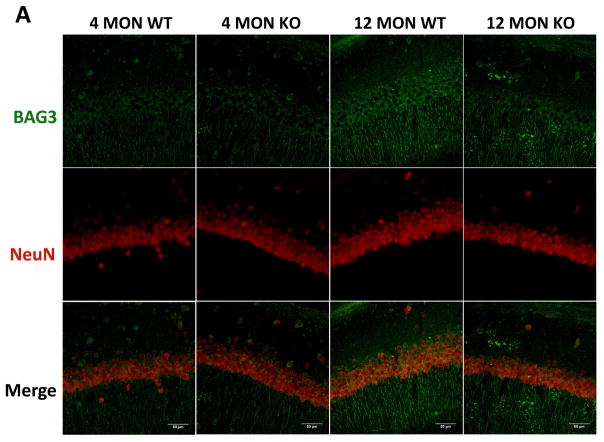
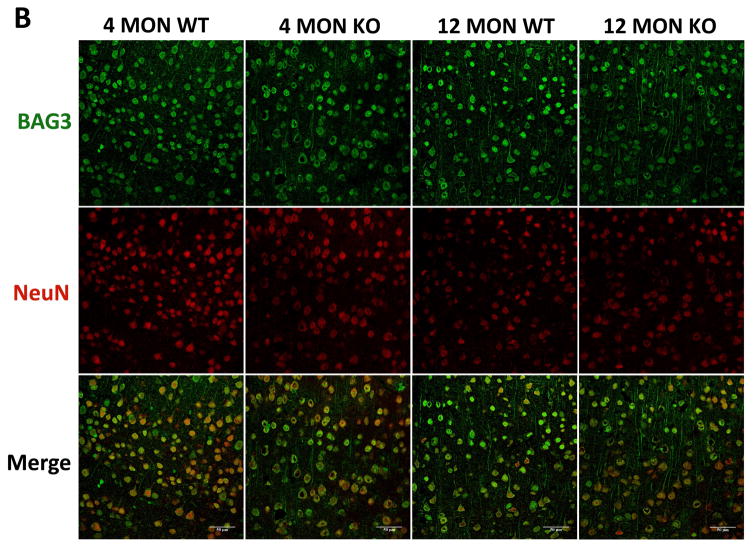
3.4 Immunohistochemical analyses of BAG3, p62 and pT231-tau in the cortex and hippocampus of WT and Nrf2 (−/−) mice
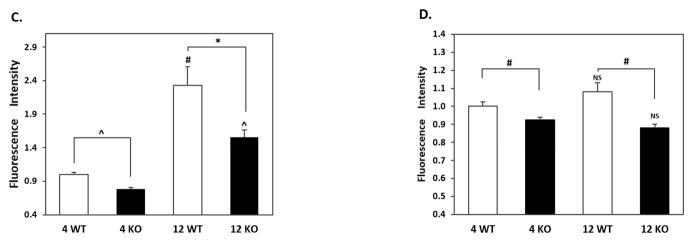
Our quantitative RT-PCR and immunoblotting data showed significant age-dependent changes in BAG3, p62 and tau phosphorylated at T231 (AT180) in the cortex and hippocampus. To further investigate these changes, we carried out immunohistochemical analyses by comparing WT and Nrf2 (−/−) mice at 4 and 12 months. In agreement with the quantitative RT-PCR and immunoblotting data, the BAG3 fluorescence in neurons of the cortex and CA1 region was greater in WT mice at 12 months compared to Nrf2(−/−) mice. Interestingly this difference in BAG3 fluorescence was also apparent in the 4 month old mice (Fig. 6). In the CA1 region there was an age-dependent increase in BAG3 fluorescence for both genotypes, although more pronounced for the WT mice (Fig. 6). Similarly, the levels of p62 were significantly lower in the hippocampus and cortex of 12 months old Nrf2(−/−) mice compared to WT mice of the same age (Fig. 7). We also noticed that the NeuN staining of the CA1 region from 12 months old Nrf2(−/−) mice appeared somewhat lighter (Fig. 6 & 7). Therefore we counted the number of NeuN positive cells in four brain sections in each of 3 Nrf2(−/−) and 3 WT mice at 12 months of age. The areas that were used for counting were the same for both genotypes. These data showed that there was no significant difference in neuronal number between WT and Nrf2(−/−) mice (data not shown). Overall the immunohistochemical results support the quantitative RT-PCR and immunoblot data indicating that Nrf2 plays a role in modulating the expression of these genes during aging.
Figure 6. BAG3 immunostaining in brains of 4 and 12 month WT and Nrf2 (−/−) mice.
(A) Representative immunohistochemical staining for BAG3 (green) and NeuN (red) in the CA1 of the hippocampus. (B) Representative immunohistochemical staining for BAG3 (green) and NeuN (red) in the frontal cortex layer V. NeuN was used as a neuron marker. WT: wild type, KO: Nrf2 (−/−). Scale bars: 50μm. Quantitation of BAG3 immunofluorescence in the CA1 region of the hippocampus (C) and frontal cortex layer V (D). BAG3 fluorescence intensity is presented as mean ± SD (*p < 0.05, #p < 0.01, ^p < 0.001, NS: Not Significant). The symbols over the 12 month bars are comparisons to 4 month data for each genotype.
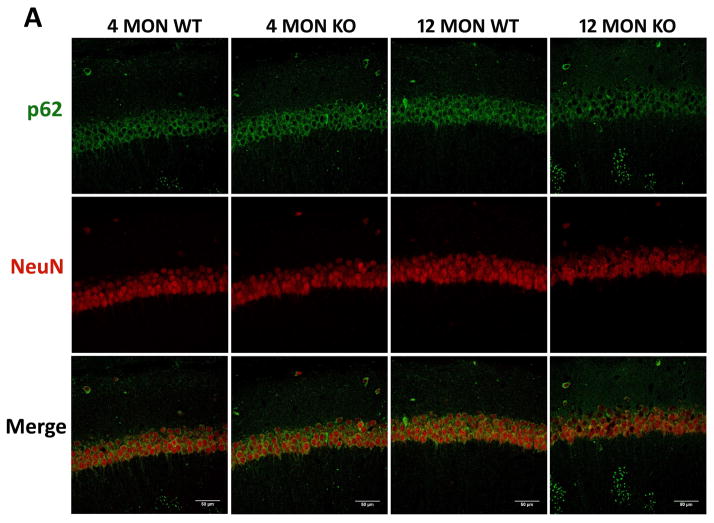
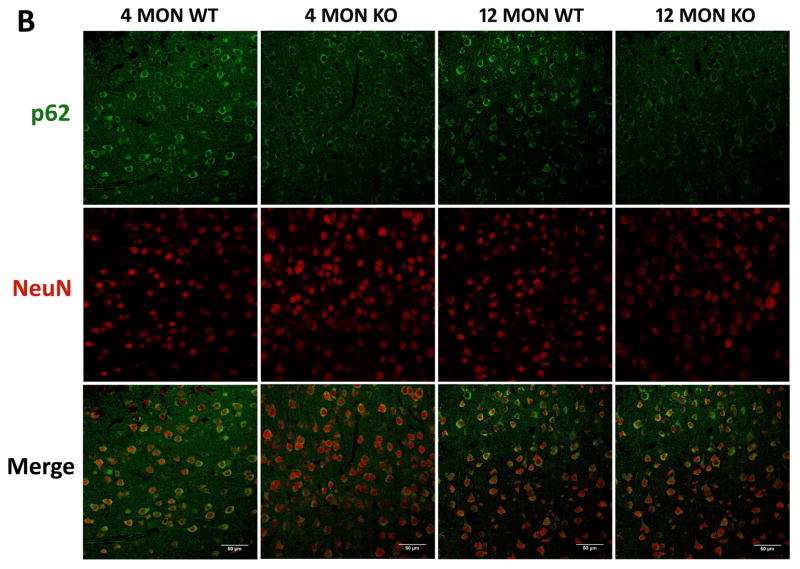
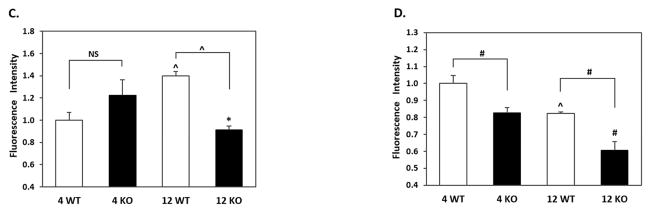
Figure 7. p62 immunostaining in brains of 4 and 12 month WT and Nrf2 (−/−) mice.
(A) Representative immunohistochemical staining for p62 (green) and NeuN (red) in the CA1 of the hippocampus. (B) Representative immunohistochemical staining for p62 (green) and NeuN (red) in the frontal cortex layer V. NeuN was used as a neuron marker. WT: wild type, KO: Nrf2 (−/−). Scale bars: 50μm. Quantitation of p62 immunofluorescence in the CA1 region of the hippocampus (C) and frontal cortex layer V (D). p62 fluorescence intensity is presented as mean ± SD (*p < 0.05, #p < 0.01, ^p < 0.001, NS: Not Significant). The symbols over the 12 month bars are comparisons to 4 month data for each genotype.
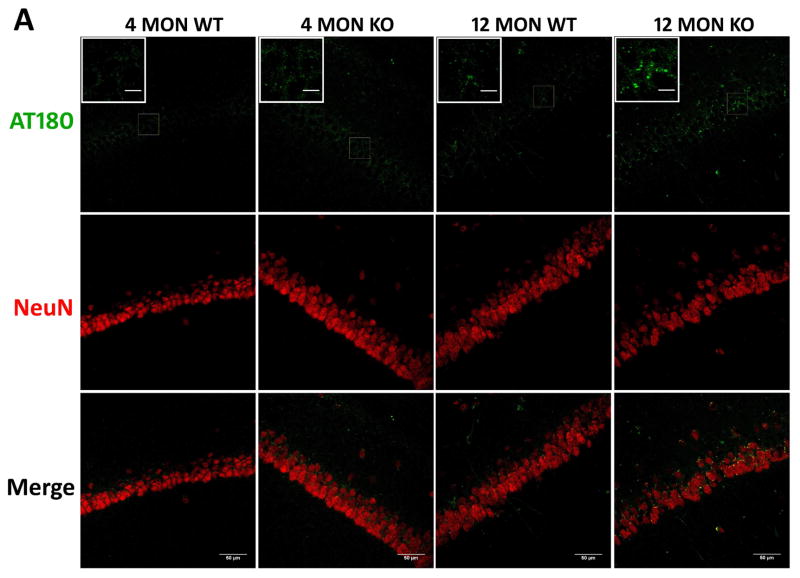
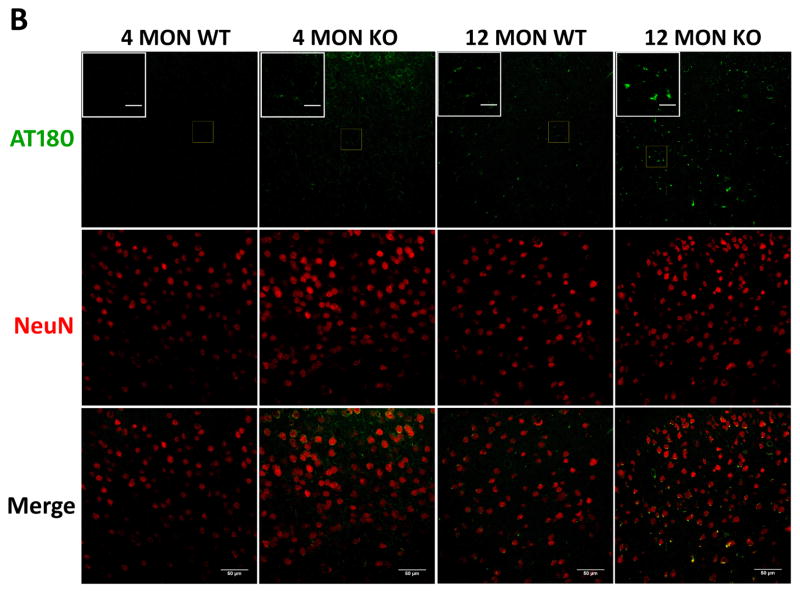
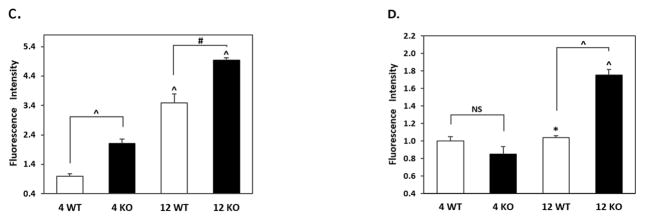
The presence of tau phosphorylated at T231 (AT180) was also immunohistochemically examined in the cortex and CA1 region of 4 and 12 month old WT and Nrf2 (−/−) mice. Increased AT180 staining was observed in 12 month old mice, and this increase was most apparent in the Nrf2 (−/−) mice, where the staining pattern appeared more punctate (Fig. 8). These data suggest that Nrf2 likely modulates the processes that facilitate the clearance of this phospho-tau species.
Figure 8. Phospho-T231 tau (AT180) immunostaining in brains of 4 and 12 month WT and Nrf2 (−/−) mice.
(A) Representative immunohistochemical staining for pT231 tau (AT180) (green) and NeuN (red) in the CA1 of the hippocampus. (B) Representative immunohistochemical staining for pT231 tau (AT180) (green) and NeuN (red) in the frontal cortex layer V. In the hippocampus and cortex of 12 month Nrf2 (−/−) mice, pT231 tau (AT180) staining appears to be increased and more punctate compared to WT mice. NeuN was used as a neuron marker Scale bars: 50μm. Higher magnification images are shown in insets, scale bars: 10 μm. Quantitation of AT180 immunofluorescence in the CA1 region of the hippocampus (C) and frontal cortex layer V (D). AT180 fluorescence intensity is presented as mean ± SD (*p < 0.05, #p < 0.01, ^p < 0.001, NS: Not Significant). The symbols over the 12 month bars are comparisons to 4 month data for each genotype.
4. Discussion
Nrf2 signaling plays a pivotal role in mediating cytoprotective responses which become increasingly important with increasing age. These cytoprotective responses include not only an upregulation of antioxidant genes but also genes involved in mediating autophagy (Pajares, et al., 2017, Pajares, et al., 2016, Zhang, et al., 2015). During aging, the expression of Nrf2 has been reported to increase in the brain (Zhang, et al., 2015, Zhang, et al., 2012), and the levels of Nrf2 and Nrf2 responsive genes are significantly higher in longer-lived rodents compared to those with shorter life spans (Lewis, et al., 2015). Given the crucial role of Nrf2 in autophagy (Bitto, et al., 2014, Pajares, et al., 2017, Pajares, et al., 2016), and the fact that autophagy is constitutively active in neurons and essential for maintaining the proteome (Boland, et al., 2008), we examined the expression of the autophagy adaptors p62, NDP52 and NBR1 and the autophagic co-chaperone BAG3 in the hippocampus and cerebral cortex of young (4 month), mature (8 month) and middle-aged (12 month) (Flurkey, et al., 2007) WT and Nrf2(−/−) mice. Further, given the importance of selective autophagy in regulating the turnover of tau in neurons (Jo, et al., 2014, Kruger, et al., 2012, Lei, et al., 2014), we also examined the levels of tau and phospho-tau species. Our results show that in young WT and Nrf2(−/−) mice the only significant difference in autophagy-related gene expression was a significant decrease in the protein level of NBR1 in the cortex of Nrf2(−/−) mice. However, while autophagy-related genes p62, NDP52, NBR1 and BAG3 expression increased in middle-aged WT mice, the same was not observed in middle-aged Nrf2(−/−) mice, which also exhibited, increased levels of phospho-tau species. No changes were observed in tau mRNA. These novel and important findings demonstrate the importance of Nrf2 in maintaining protein quality control processes as animals age.
p62/SQSTM1, NDP52 and NBR1 are well established autophagy adaptors that are expressed in brain, and specifically in neurons. They bind to misfolded, damaged or ubiquitylated substrates and bring them to autophagosomes by interacting with an autophagosomal membrane-anchored member of the ATG8 family (LC3 and GABARAP family members in mammals) via an LC3 interacting region (LIR) (Codogno, et al., 2012, Lamark, et al., 2009, Randow and Youle, 2014). In the present study, a significant decrease in the mRNA and protein levels for p62/SQSTM1, NDP52 and NBR1 was observed in the hippocampus of 12 month Nrf2 (−/−) mice compared to WT mice, but showed no difference in 4 month and 8 month mice. The data strongly indicated that Nrf2 played a role in regulating p62/SQSTM1, NDP52 and NBR1 in the context of aging. The promoter regions for the SQSTM1/p62 and NDP52 genes contain antioxidant-response elements (AREs) where Nrf2 binds and activates the transcription of the gene, promoting the expression of p62 and NDP52 (Jain, et al., 2010, Jo, et al., 2014). Both p62 and NDP52 have previously been implicated in mediating the clearance of phosphorylated tau species as accumulation of hyperphosphorylated tau was observed in the brains of p62/SQSTM1 (−/−) mice (Ramesh Babu, et al., 2008), and in an Alzheimer disease mouse model, NDP52 and phosphorylated tau were associated (Kim, et al., 2014).
NBR1 (neighbor of BRCA1 gene 1), which is another important autophagy adaptor (Johansen and Lamark, 2011), is also expressed at significantly lower levels in 12 month old Nrf2(−/−) brain compared to controls, both in the hippocampus and cortex. Although a functional ARE has not been unequivocally identified in the NBR1 promoter, data suggest that it is regulated by Nrf2. In mouse, rat and human the NBR1 and BRCA1 genes lie head-to-head, and the expression of these two genes is under complex regulation through a bidirectional promoter (Dimitrov, et al., 2001, Whitehouse, et al., 2004). Analysis of this BRCA1/NBR1 intergenic region in mice and humans has shown that this region is highly conserved and a putative transcription factor binding site for Nrf2 has been identified (Dimitrov, et al., 2001). Further, it has been reported that Nrf2 played an important role in increasing BRCA1 transcription (Wang, et al., 2013). Given the fact that NBR1 expression in 12 month Nrf2(−/−) mice parallels that of SQSTM1/p62 and NDP52, which have established Nrf2 binding sites in their promoters, it is highly likely that NBR1 expression is also directly controlled by Nrf2.
SQSTM1/p62 plays several key roles in the Nrf2 signaling pathway and autophagy. SQSTM1/p62 expression is not only regulated by Nrf2, it also modulates Nrf2 signaling. SQSTM1/p62 can bind to the Nrf2 inhibitory protein Keap1 preventing Nrf2 binding and degradation (Bitto, et al., 2014, Kwon, et al., 2012). Our quantitative RT-PCR data showed that p62 expression in the hippocampus was significantly up-regulated in 12 month old WT mice compared with 4 month old WT mice, but not Nrf2(−/−) mice. This suggests that p62 may play a role in mediating aging-related changes. These differences were also observed in the immunohistochemical data. Further, data indicate that p62 may promote mammalian longevity through both its adaptor function for selective autophagy and by facilitating Nrf2 activation in basal conditions (Kwon, et al., 2012, Zhang, et al., 2015).
BAG3 is a co-chaperone that is essential for proteostasis (Klimek, et al., 2017) and plays a role in mediating clearance of tau by selective autophagy (Lei, et al., 2014). Previous studies have suggested that expression of BAG3 increases in the brain during aging, and that this increase is due to increased dependence on the autophagic system for degradation of defective proteins (Gamerdinger, et al., 2009). Our results concur and extend these previous data. BAG3 expression increased in 12 month old WT mice compared with 4 month old mice, however no increases were observed with age in the Nrf2(−/−) mice. These data suggest that the expression of BAG3 is regulated by Nrf2. In support of this, we have found that BAG3 expression is upregulated in primary neurons by the Nrf2 activators, sulforaphane and tert-butylhydroquinone (tBHQ) (data not shown). It has also been demonstrated that treatment with MG132, a proteasome inhibitor, protected cells against oxidative damage by activating the Nrf2-ARE signaling pathway (Duan, et al., 2011), and BAG3 is also upregulated in neurons by MG132 treatment (Lei, et al., 2014). Finally, promoter analysis (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) of the BAG3 promoter shows the presence of a putative Nrf2 binding site.
In our previous studies, we have shown that Nrf2 activation or increased expression of exogenous BAG3 results in decreases in phospho-tau in primary neurons (Chesser, et al., 2016, Jo, et al., 2014, Lei, et al., 2014). Here, quantitative RT-PCR data confirmed that tau mRNA levels did not differ between Nrf2 (−/−) and WT mice at 4 or 12 months, while immunoblotting data indicated that tau phosphorylated at T231 increased in the Nrf2 (−/−) mice, most prominently in the hippocampus and cortex of 12 month old mice. Further immunohistochemistry showed that in the 12 month old Nrf2 (−/−) mice pT231 tau (AT180) staining was more prominent and punctate in appearance than in 12 month old WT mice. In a previous study we found a similar trend of the levels of phospho-tau species being higher in Nrf2(−/−) mice, and the levels of the autophagy adaptor NDP52 being lower (Jo, et al., 2014). However, these previous studies were not carried out with mice that were matched for age, and thus the expression levels showed some variance in comparison to this present study. For example, although in the previous study a significant decrease in the mRNA for p62 was observed in Nrf2(−/−) mice, NBR1 levels were not significantly different (Jo, et al., 2014). In addition, in this previous study we did not consider the aging variable. In this study, we carefully matched the ages of the WT and Nrf2(−/−) and thus these data suggest that deletion of Nrf2 results in an aging dependent impairment in the expression of autophagy related genes, including BAG3, which contributes to increased levels of phosphorylated tau species.
LCII levels in the cerebral cortex were not significantly different in Nrf2(−/−) compared to WT mice at any age. In contrast, LC3II levels were slightly but significantly decreased in 4 month old Nrf2(−/−) mice but increased significantly at 8 and 12 months compared to WT mice. The reason for these regional differences are unknown, however caution needs to be exercised in interpreting the data as increases in LC3II can be associated with increased autophagosome formation or reduced autophagic flux (Barth, et al., 2010). Given the fact that both the mRNA and protein levels of the autophagy adaptors were decreased in 12 month old Nrf2(−/−) mice and pT231 tau protein levels were increased it is more likely the increase in LC3II in the older Nrf2(−/−) cortices represents reduced autophagic flux. Indeed previous studies have shown that with impaired autophagy LC3II levels increase in brain (Maetzel, et al., 2014, Sarkar, et al., 2014). Co-staining of 12 month old brains for LC3 and pT231 tau indicated that there appears to be more LC3 puncta present in Nrf2(−/−) mice which showed col-localization with pT231-tau. These data support the suggestion that deletion of Nrf2(−/−) may result in a reduction of autophagic flux. Interestingly, in some neuron of 12 month Nrf2 (−/−) mice, there were pT231-tau positive puncta that did not co-localize with LC3 mice (see supplementary figure). This may indicate inefficient targeting of aggregated tau species to the autophagy system in Nrf2 (−/−) mice.
Taken together, these data strongly indicate that Nrf2 plays an important role in maintaining a healthy nervous system proteome as animals age, in part by supporting the expression of proteins involved in mediating selective autophagy. Furthermore, interventions that facilitate Nrf2 signaling could promote the maintenance of proteostatic mechanisms which in turn may support healthier aging and increase longevity. However, further studies are needed to fully understand the role of Nrf2 signaling processes and how they are altered during aging.
Supplementary Material
Nrf2 deletion resulted in an age dependent decrease in p62, NBR1 and NDP52
BAG3 levels were significantly decreased in 12 month old Nrf2−/− brains
Deletion of Nrf2 resulted in an age-dependent increase in phospho-tau
Nrf2 may play a key role in selective autophagy and tau clearance
Acknowledgments
The authors thank the donors of the ADR, a program of BrightFocus Foundation, for support of this research. We also thank Dr. Robert Freeman for the use of his BioRad CFX96 Real Time System C1000 Thermo Cycler, and Dr. M. Kerry O’Banion for the use of his sliding microtome for preparing brain sections. The authors thanks Dr. Peter Davies for the PHF1 antibody and Dr. Peter Seubert for the 12E8 antibody.
Footnotes
Disclosure Statement: The authors have no actual or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraha A, Ghoshal N, Gamblin TC, Cryns V, Berry RW, Kuret J, Binder LI. C-terminal inhibition of tau assembly in vitro and in Alzheimer’s disease. Journal of cell science. 2000;113(Pt 21):3737–45. doi: 10.1242/jcs.113.21.3737. [DOI] [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. The Journal of biological chemistry. 1999;274(37):26071–8. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Bitto A, Lerner CA, Nacarelli T, Crowe E, Torres C, Sell C. P62/SQSTM1 at the interface of aging, autophagy, and disease. Age. 2014;36(3):9626. doi: 10.1007/s11357-014-9626-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28(27):6926–37. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesser AS, Ganeshan V, Yang J, Johnson GV. Epigallocatechin-3-gallate enhances clearance of phosphorylated tau in primary neurons. Nutritional neuroscience. 2016;19(1):21–31. doi: 10.1179/1476830515Y.0000000038. [DOI] [PubMed] [Google Scholar]
- Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13(1):7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- David DC. Aging and the aggregating proteome. Frontiers in genetics. 2012;3:247. doi: 10.3389/fgene.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Brennerova M, Forejt J. Expression profiles and intergenic structure of head-to-head oriented Brca1 and Nbr1 genes. Gene. 2001;262(1–2):89–98. doi: 10.1016/s0378-1119(00)00549-7. [DOI] [PubMed] [Google Scholar]
- Duan W, Guo Y, Jiang H, Yu X, Li C. MG132 enhances neurite outgrowth in neurons overexpressing mutant TAR DNA-binding protein-43 via increase of HO-1. Brain research. 2011;1397:1–9. doi: 10.1016/j.brainres.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Currer JM, Harrison DE. Mouse Models in Aging Research. In: Fox JG, Barthold S, Davisson M, Newcomer C, Quimby F, Smith A, editors. The Mouse in Biomedical Research. 2. Elsevier; Burlington, MA: 2007. pp. 637–72. [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO reports. 2011;12(2):149–56. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestwicki JE, Garza D. Protein quality control in neurodegenerative disease. Prog Mol Biol Transl Sci. 2012;107:327–53. doi: 10.1016/B978-0-12-385883-2.00003-5. [DOI] [PubMed] [Google Scholar]
- Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. The Journal of biological chemistry. 2000;275(21):16023–9. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- Ishii T, Itoh K, Yamamoto M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods in enzymology. 2002;348:182–90. doi: 10.1016/s0076-6879(02)48637-5. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and biophysical research communications. 1997;236(2):313–22. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. The Journal of biological chemistry. 2010;285(29):22576–91. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo C, Gundemir S, Pritchard S, Jin YN, Rahman I, Johnson GV. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat Commun. 2014;5:3496. doi: 10.1038/ncomms4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–96. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee D, Song JC, Cho SJ, Yun SM, Koh YH, Song J, Johnson GV, Jo C. NDP52 associates with phosphorylated tau in brains of an Alzheimer disease mouse model. Biochemical and biophysical research communications. 2014;454(1):196–201. doi: 10.1016/j.bbrc.2014.10.066. [DOI] [PubMed] [Google Scholar]
- Klimek C, Kathage B, Wordehoff J, Hohfeld J. BAG3-mediated proteostasis at a glance. Journal of cell science. 2017 doi: 10.1242/jcs.203679. [DOI] [PubMed] [Google Scholar]
- Kruger U, Wang Y, Kumar S, Mandelkow EM. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiology of aging. 2012;33(10):2291–305. doi: 10.1016/j.neurobiolaging.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Kwon J, Han E, Bui CB, Shin W, Lee J, Lee S, Choi YB, Lee AH, Lee KH, Park C, Obin MS, Park SK, Seo YJ, Oh GT, Lee HW, Shin J. Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO reports. 2012;13(2):150–6. doi: 10.1038/embor.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell cycle. 2009;8(13):1986–90. doi: 10.4161/Cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- Lei Z, Brizzee C, Johnson GV. BAG3 facilitates the clearance of endogenous tau in primary neurons. Neurobiology of aging. 2014;36:241–8. doi: 10.1016/j.neurobiolaging.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KN, Mele J, Hayes JD, Buffenstein R. Nrf2, a guardian of healthspan and gatekeeper of species longevity. Integrative and comparative biology. 2010;50(5):829–43. doi: 10.1093/icb/icq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KN, Wason E, Edrey YH, Kristan DM, Nevo E, Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(12):3722–7. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Zimmermann A, Maiuri MC, Kroemer G. Essential role for autophagy in life span extension. The Journal of clinical investigation. 2015;125(1):85–93. doi: 10.1172/JCI73946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Cuadrado A, Rojo AI. Modulation of proteostasis by transcription factor NRF2 and impact in neurodegenerative diseases. Redox biology. 2017;11:543–53. doi: 10.1016/j.redox.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, Escoll M, de Ceballos ML, Van Leuven F, Rabano A, Yamamoto M, Rojo AI, Cuadrado A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–16. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. Journal of neurochemistry. 2008;106(1):107–20. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell host & microbe. 2014;15(4):403–11. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F, Jung M, Fulda S. BAG3 induction is required to mitigate proteotoxicity via selective autophagy following inhibition of constitutive protein degradation pathways. Oncogene. 2014;33(13):1713–24. doi: 10.1038/onc.2013.110. [DOI] [PubMed] [Google Scholar]
- Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P, Behrends C, Furst DO, Volkmer R, Hoffmann B, Kolanus W, Hohfeld J. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013;23(5):430–5. doi: 10.1016/j.cub.2013.01.064. [DOI] [PubMed] [Google Scholar]
- Vidal RL, Matus S, Bargsted L, Hetz C. Targeting autophagy in neurodegenerative diseases. Trends in pharmacological sciences. 2014;35:583–91. doi: 10.1016/j.tips.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li J, Yang X, Sun H, Gao S, Zhu H, Wu J, Jin W. Nrf2 is associated with the regulation of basal transcription activity of the BRCA1 gene. Acta biochimica et biophysica Sinica. 2013;45(3):179–87. doi: 10.1093/abbs/gmt001. [DOI] [PubMed] [Google Scholar]
- Whitehouse C, Chambers J, Catteau A, Solomon E. Brca1 expression is regulated by a bidirectional promoter that is shared by the Nbr1 gene in mouse. Gene. 2004;326:87–96. doi: 10.1016/j.gene.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333(6039):228–33. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Davies KJA, Forman HJ. Oxidative stress response and Nrf2 signaling in aging. Free radical biology & medicine. 2015;88(Pt B):314–36. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu H, Davies KJ, Sioutas C, Finch CE, Morgan TE, Forman HJ. Nrf2-regulated phase II enzymes are induced by chronic ambient nanoparticle exposure in young mice with age-related impairments. Free radical biology & medicine. 2012;52(9):2038–46. doi: 10.1016/j.freeradbiomed.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.