Abstract
The global success of multidrug-resistant Acinetobacter baumannii has been associated with the dissemination of a high-risk clone designated clonal complex (CC) 92B (Bartual scheme)/CC2P (Pasteur scheme), which is the most frequent genetic lineage in European, Asian, and North American carbapenem-resistant Acinetobacter isolates. In these isolates, carbapenem resistance is mainly mediated by β-lactamases encoded by blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and/or blaOXA-58-like genes. In this study, we characterized the population genetics of 121 carbapenem-resistant A. baumannii complex isolates recovered from 14 hospitals in seven cities in Colombia (2008–2010). Multiplex PCR was used to detect blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like genes. Molecular typing was performed using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). PCR showed that 118 (97.5%) of the isolates were positive for both blaOXA-23-like and blaOXA-51-like genes, and three other isolates were only positive for blaOXA-51-like. PFGE identified 18 different pulsotypes, while MLST identified 11 different sequence types (STs), seven of which had not been previously described in Acinetobacter. None of the STs found in this study was associated with CC92B/CC2P. The most widespread STs in our isolates belonged to ST636 and their single-locus variants ST121/ST124/ST634 (CC636B) followed by STs belonging to CC110B. Our observations suggest a wide distribution of diverse A. baumannii complex clones containing blaOXA-23-like in Colombian hospitals (especially CC636B and CC110B) that differ from the high-risk clones commonly found in other regions of the world, indicating a distinct molecular epidemiology of carbapenem-resistant Acinetobacter spp. in Colombia.
Keywords: : Acinetobacter baumannii, antibiotic resistance, carbapenemase, OXA enzymes, multilocus sequence typing
Introduction
Acinetobacter baumannii is an opportunistic pathogen associated with hospital-acquired infections and outbreaks worldwide, especially in critically ill patients. A. baumannii is typically able to survive on inanimate and dry surfaces and to be multidrug resistant (MDR), even to carbapenems.1 Carbapenem resistance has been associated with the presence of carbapenemase-type β-lactamases, particularly those encoded by the blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like genes, and the presence of the insertion sequences ISAba1, ISAba3, or ISAba4.2,3
Multilocus sequence typing (MLST) has been used worldwide to study the population structures of A. baumannii.4 Two different MLST systems with their own methodology and databases have been developed: one described by Bartual et al. (http://pubmlst.org/abaumannii/4) and the other described by the Pasteur Institute (www.pasteur.fr/mlst).5 Although these schemes share three of the seven housekeeping genes they use, both tools are useful for characterization of A. baumannii in population-based studies.6 The sequence types (STs) belonging to clonal complex (CC) 92B (Bartual scheme)/CC2P (Pasteur scheme) (also designated international clonal lineage II) have been reported as the most widespread in Europe, Asia, and the United States.7 Given its epidemiological success and its frequent association with carbapenem resistance, this CC has been designated a high-risk clone of MDR A. baumannii.8,9
In South America, namely in Brazil and Argentina, CC79P/CC113B and CC15P/CC104B have been the predominant clones responsible for the dissemination of OXA-23-like-producing carbapenem-resistant A. baumannii (CRAB).10–20
We aimed to characterize the population structure of CRAB isolates recovered from hospitals in Colombia and to identify possible high-risk clones responsible for dissemination of these organisms.
Materials and Methods
Bacterial isolates
A total of 121 carbapenem-resistant A. calcoaceticus–A. baumannii complex isolates obtained from the same number of patients were recovered from the isolate collection of the Colombian Nosocomial Resistance Study Group housed at the International Center for Medical Research and Training (CIDEIM) in Cali, Colombia. Bacterial identification was performed by the VITEK 2® automated system (bioMérieux, Marcy-l'Étoile, France), and in vitro antimicrobial susceptibility testing was performed by broth microdilution (Sensititre panels; Trek Diagnostic Systems, Westlake, OH) according to the Clinical and Laboratory Standards Institute (CLSI).21 Clinical interpretation of susceptibility to tigecycline was based on breakpoints suggested previously by Jones et al.22
The isolates were collected between 2008 and 2010 from 14 tertiary care hospitals in seven cities (Bogotá, Medellín, Cali, Pereira, Bucaramanga, Barranquilla, and Ibagué). This collection period was chosen as part of the surveillance of MDR organisms made by CIDEIM in Colombia, and corresponds to the time in which emergence and widespread dissemination of carbapenemase-producing Enterobacteriaceae occurred. The most common sample source was the respiratory tract (n = 40, 33%), followed by blood (n = 35, 29%) and skin and soft tissues (n = 33, 27%). The majority of isolates were recovered from patients hospitalized in intensive care units (87, 72%).
Analysis of bla genes
Detection of OXA-type enzymes was performed by a multiplex PCR assay targeting blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, and blaOXA-58-like genes, following protocols previously reported.23 In addition, identification of ISAba1 and ISAba4 associated with blaOXA-23-like gene was performed using PCR and sequencing in the isolates analyzed through MLST.23
Molecular typing
Pulsed-field gel electrophoresis (PFGE) was performed in all 121 isolates studied, following the method described by Gautom24 and using the SmaI restriction enzyme. Analysis of pulsotypes and construction of dendrograms were performed using Fingerprinting II software (Bio-Rad Laboratories, Hercules, CA) and the Dice coefficient for comparison. Isolates were considered as clonal if percentages of similarity were higher than 75%. Based on these results, at least one isolate from each PFGE pulsotype was selected for MLST analysis using the Bartual scheme,4 following the protocol and conditions described in http://pubmlst.org/abaumannii/. The goeBURST algorithm of PHYLOViZ software (version 1.1) was used to establish relationships between the STs found in our study and those reported worldwide.
Results and Discussion
PCR results showed the presence of both blaOXA-23-like and blaOXA-51-like genes occurring simultaneously in 97.5% (118 out of 121) of the A. baumannii complex isolates analyzed. None of the isolates was positive for blaOXA-24-like or blaOXA-58-like genes, which is in agreement with the infrequent reports of these OXA-type carbapenemases known to date in Colombia.20 Of note, three isolates were negative for blaOXA-23-like but were positive for blaOXA-51-like. This carbapenem-resistant gene distribution is similar to other reports in Latin America and around the world, in which the prevalent enzyme in CRAB is OXA-23.25–27 In addition, we found the blaOXA-23 gene accompanied by the insertion sequence ISAba1 (part of the widely disseminated transposon Tn2006) in 19 of the 21 isolates selected for MLST. This mobile genetic element has been previously reported in CRAB isolates.28 The other two isolates were negative for blaOXA-23-like gene but positive for blaOXA-51-like, and the insertion sequence ISAba1 accompanied one of them. These isolates exhibited an MDR phenotype that might be explained by mechanisms not examined in this study such as the loss or modification of the carbapenem-associated outer membrane protein CarO, or modification of penicillin-binding proteins as previously reported elsewhere.29,30 blaOXA-51-like has only been found to lead to carbapenem resistance if ISAba1 is located upstream of the gene.2
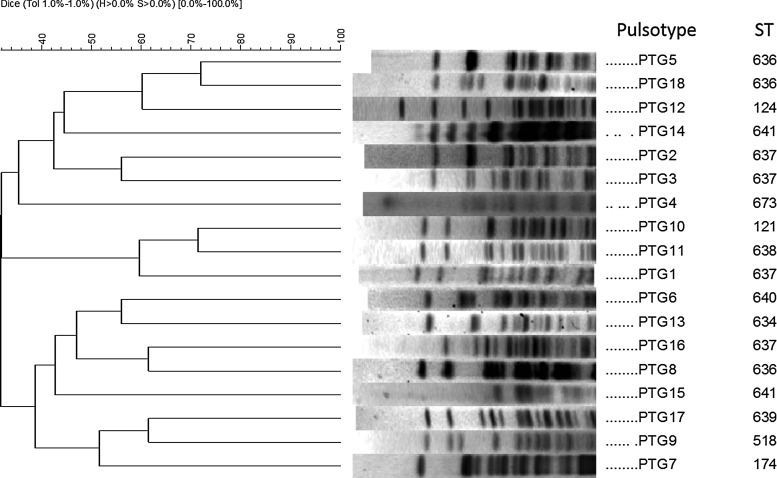
Analysis by PFGE showed the presence of 18 different band profiles (pulsotypes; Fig. 1); five of them represented by polyclonal isolates. Based on these results, 21 isolates (at least one isolate per pulsotype) were selected for MLST (Table 1). Results showed 11 different STs, seven of which had not been previously reported in A. baumannii (ST636, ST634, ST637, ST638, ST639, ST640, and ST641). These STs have been submitted in the MLST database (http://pubmlst.org/abaumannii/). The diversity of STs in A. baumannii has been associated with the variability found in gyrB and gpi genes in the MLST scheme.31 However, in our study, the cpn60 gene was the one that exhibited high sequence diversity among the Colombian A. baumannii complex isolates.
FIG. 1.
Dendrogram of genomic comparison for the 18 representative PTGs identified by pulsed-field gel electrophoresis in Acinetobacter baumannii complex isolates using the UPGMA based on Dice similarity. PTGs, pulsotype groups; ST, sequence type (Bartual scheme); UPGMA, unweighted pair group method with arithmetic mean.
Table 1.
Molecular Characterization and Antimicrobial Susceptibility Testing of Carbapenem-Resistant Acinetobacter baumannii Complex from Colombian Hospitals
| Antibiotic MIC (μg/ml) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | City | PTG | STB | blaOXA-23-like | blaOXA-51-like | ISAba1 | Sample | AMK | CEP | CEF/SUL | CIP | IMI | MEM | PIP/TAZ | POLB | TIG |
| 1 | CLO | PTG1 | 637 | POS | POS | POS | Respiratory tract | >64 | 32 | 128/64 | >8 | 64 | 64 | >256/4 | 1 | 0.5 |
| 2 | BOG/CLO/MED/PEI | PTG2 | 637 | POS | POS | POS | Respiratory tract | >64 | 16 | 64/32 | >8 | 32 | 64 | >256/4 | 2 | 0.25 |
| 3 | BOG/CLO/MED/PEI | PTG2 | 637 | POS | POS | POS | Skin and soft tissues | >64 | 32 | 64/32 | >8 | 32 | 64 | >256/4 | 1 | 0.5 |
| 4 | BGA/CLO | PTG3 | 637 | NEG | POS | POS | Respiratory tract | >64 | 32 | 128/64 | >8 | 32 | 64 | >128/4 | 1 | 0.5 |
| 5 | BGA/BOG/CLO | PTG4 | 637 | POS | POS | POS | Respiratory tract | >64 | 32 | 64/32 | >8 | 64 | 64 | >256/4 | 1 | 0.5 |
| 6 | BGA/BOG/CLO | PTG4 | 637 | POS | POS | POS | Gastrointestinal tract | >64 | 64 | 64/32 | >8 | 32 | 64 | >256/4 | 2 | 0.25 |
| 7 | BGA/BOG/CLO | PTG4 | 641 | POS | POS | POS | Skin and soft tissues | >64 | 64 | 64/32 | >8 | 32 | 64 | >256/4 | 2 | 0.25 |
| 8 | CLO/MED | PTG5 | 636 | POS | POS | POS | Respiratory tract | >32 | >16 | 64/32 | >2 | 32 | 64 | >64/4 | 1 | 0.25 |
| 9 | BOG/CLO/IBE/PEI | PTG6 | 640 | POS | POS | POS | Respiratory tract | >64 | 32 | 32/16 | >8 | 64 | 64 | 256/4 | 2 | 0.5 |
| 10 | CLO/PEI | PTG7 | 174 | POS | POS | POS | Respiratory tract | 64 | 128 | 128/64 | >8 | 32 | 64 | 256/4 | 2 | 4 |
| 11 | BOG/CLO/IBE | PTG8 | 636 | POS | POS | POS | Blood | >64 | 32 | 128/64 | >8 | 64 | 64 | >256/4 | 2 | 0.5 |
| 12 | BGA/BOG/BQA/IBE/MED | PTG9 | 518 | POS | POS | POS | Blood | >64 | >32 | 128/64 | >8 | 128 | 128 | >256/4 | 4 | 2 |
| 13 | BGA/BOG/BQA/IBE/MED | PTG10 | 121 | POS | POS | POS | Blood | >64 | 64 | 64/32 | >8 | 32 | 64 | >256/4 | 2 | 0.25 |
| 14 | BOG/BQA | PTG11 | 638 | POS | POS | POS | Urine | >64 | 32 | 32/16 | >8 | 32 | 64 | >128/4 | <0.5 | 0.5 |
| 15 | BOG | PTG12 | 124 | POS | POS | POS | Urine | 64 | >64 | >128/64 | >8 | >64 | >64 | >128/4 | 1 | 0.5 |
| 16 | BOG | PTG13 | 634 | POS | POS | POS | Respiratory tract | 32 | 16 | 64/32 | >8 | 32 | 64 | >256/4 | 1 | 0.5 |
| 17 | BGA/BQA | PTG14 | 641 | POS | POS | POS | Respiratory tract | >64 | 64 | 64/32 | >8 | 64 | 128 | >256/4 | 1 | 0.5 |
| 18 | BQA | PTG15 | 641 | POS | POS | POS | Blood | 64 | 32 | 64/32 | 8 | 32 | 64 | 256/4 | <0.5 | 1 |
| 19 | BGA | PTG16 | 637 | NEG | POS | NEG | Gastrointestinal tract | >64 | 64 | 64/32 | >8 | 32 | 64 | >256/4 | 2 | 0.25 |
| 20 | BOG | PTG17 | 639 | POS | NEG | POS | Blood | >64 | 32 | 64/32 | <1 | 128 | 128 | >256/4 | 1 | 1 |
| 21 | CLO | PTG18 | 636 | POS | POS | POS | Blood | >64 | 32 | 64/32 | <1 | 128 | 128 | >256/4 | 1 | 1 |
Isolates selected for multilocus sequence typing are presented.
AMK, amikacin; BGA, Bucaramanga; BOG, Bogotá; BQA, Barranquilla; CEF/SUL, cefoperazone/sulbactam; CEP, cefepime; CIP, ciprofloxacin; CLO, Cali; IBE, Ibagué; IMI, imipenem; MED, Medellín; MEM, meropenem; MIC, minimum inhibitory concentration; NEG, negative; PEI, Pereira; PIP/TAZ, piperacillin/tazobactam; POLB, polymyxin B; POS, positive; PTG, pulsotype group; STB, sequence type (Bartual scheme); TIG, tigecycline.
Overall, carbapenem resistance in A. baumannii complex from Colombian hospitals in this study was linked to the widespread dissemination of blaOXA-23-like β-lactamase. Although the isolates were analyzed between 2008 and 2010, the resistance profile of A. baumannii in Colombia has likely not changed (based on our latest surveillance data), and as a result, the resistance mechanisms are expected to be similar to date.32–35
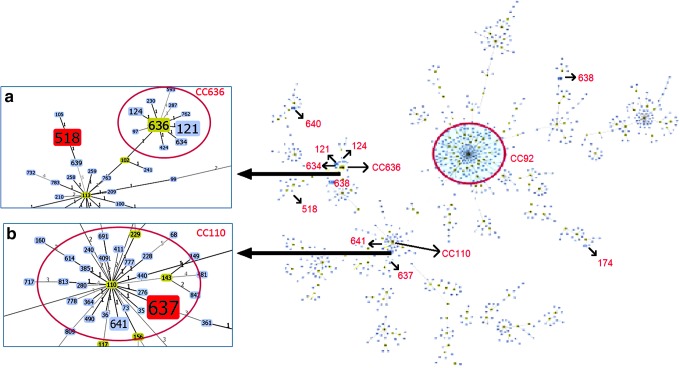
The most common STs found within the isolate collection were the novel ST636 and its single-locus variants (SLVs) ST121 (Spain),7 ST124 (USA),7 and ST634 (novel in this study), all of these belonging to CC636B (designated in this study) (Fig. 2). Other STs included ST637 and ST641 (SLVs of CC110B), as well as ST518. CC636B has been predominantly related to the spread of CRAB in Mexico.36
FIG. 2.
Distribution of the different STs found in carbapenem-resistant A. baumannii complex in Colombia, using goeBURST (PHYLOViZ). The red arrows indicate the ST location. CC92B, shown with a red circle, was not found in our study but is included for reference since it is one of the clonal complexes associated with the global spread of blaOXA-23. (a) Closer look at CC636B. (b) Closer look at CC110B. CC, clonal complex.
Interestingly, ST637 and ST641 are SLVs of ST110, which is the founder of the CC110B previously reported in Argentina, Korea, and the United States.7 The only other study of its kind in Colombia, recently reported the presence of ST229 (CC110B) and ST758.35 However, this study included 32 A. baumannii complex isolates collected from five hospitals in only one city (Medellín).
Of note, none of the STs found in this study was associated with CC92B/CC2P, which has been reported as the most prevalent Acinetobacter MDR lineage in 34 countries of the world.7 In Latin America, MLST allelic profiles for CRAB have seldom been reported related to CC92B/CC2P in Brazil and Mexico.7,14,36 It is also important to note that almost all STs described in this study were present in Bogotá, the capital and largest city of Colombia (Table 1). This phenomenon is likely explained by the fact that third-level hospitals in Bogotá are referral centers for the rest of the country and have high levels of complexity.
Conclusions
This study shows that blaOXA-23 was the most prevalent determinant of carbapenem resistance in A. baumannii complex strains collected from 14 Colombian hospitals between 2008 and 2010. The blaOXA-23-harboring A. baumannii complex isolates belonged to the MLST-defined CC110B, previously reported in Argentina, Korea, and the United States, and the novel CC636B, containing diverse STs, some of them previously found in Spain and the United States. Genetic lineages of CRAB reported to be widely distributed in all the continents, such as the high-risk clone CC92B/CC2P, were not found in our study.
Our observations serve as an initial characterization of the population structure of CRAB in Colombia during the same time in which dissemination of carbapenemases in Enterobacteriaceae occurred in our country and became epidemic. Molecular studies contribute to the understanding of the population genetic diversity and mechanisms of carbapenem resistance in A. baumannii.
Acknowledgments
We thank the participating institutions that form the Colombian Nosocomial Resistance Study Group led by the International Center for Medical Research and Training (CIDEIM) in Cali, Colombia: Hospital Central de la Policía, Hospital Militar Central, Fundación Santa Fe de Bogotá, Hospital Universitario San Ignacio, Hospital Universitario del Valle, Clínica Fundación Valle del Lili, Clínica de las Américas, Hospital Pablo Tobón Uribe, Hospital Universitario San Jorge, Fundación Cardiovascular de Colombia, Hospital Universitario de Santander, Clínica FOSCAL, Clínica General del Norte, and Hospital Federico Lleras Acosta.
Funding
This work was supported by Merck Sharp & Dohme, Janssen-Cilag, Pfizer, AstraZeneca, Merck, Novartis, Amarey Nova Medical, Merck, and bioMérieux Colombia, which helped funding the Colombian Nosocomial Resistance Study Group.
Disclosure Statement
A.C. has received speaker honoraria from Merck Sharp & Dohme and bioMérieux. C.A.A. has received grant support form Pfizer, Forest Pharmaceuticals, and Theravance, Inc., has served on the speaker bureaus of Cubist, Forest Pharmaceuticals, Pfizer, Novartis, and AstraZeneca, and has performed consulting activities for Cubist, Bayer, and AstraZeneca. He is supported by grant K24-AI114818. R.C.'s research at the Servicio de Microbiología del Hospital Universitario Ramón y Cajal was funded by the European Commission (grants R-GNOSIS-FP7-HEALTH-F3-2011-282512 and FP7-HEALTH-F3-2013-MON4STRAT-602906-2). M.V.V. has received consulting fees and research grants from Merck Sharp & Dohme, Pfizer, Janssen-Cilag, Novartis, Merck, and AstraZeneca. The other authors declare no competing interests.
References
- 1.Runnegar N., Sidjabat H., Goh H.M.S., Nimmo G.R., Schembri M.A., and Paterson D.L. 2010. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a single institution over a 10-year period. J. Clin. Microbiol. 48:4051–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turton J.F., Ward M.E., Woodford N., Kaufmann M.E., Pike R., Livermore D.M., and Pitt T.L. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72–77 [DOI] [PubMed] [Google Scholar]
- 3.Evans B.A., and Amyes S.G.B. 2014. OXA β-lactamases. Clin. Microbiol. Rev. 27:241–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartual S.G., Seifert H., Hippler C., Luzon M.A.D., Wisplinghoff H., and Rodríguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diancourt L., Passet V., Nemec A., Dijkshoorn L., and Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomaschek F., Higgins P.G., Stefanik D., Wisplinghoff H., and Seifert H. 2016. Head-to-head comparison of two multi-locus squence typing (MLST) schemes for characterization of Acinetobacter baumannii outbreak and sporadic isolates. PLoS One 11:e0153014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karah N., Sundsfjord A., Towner K., and Samuelsen Ø. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247 [DOI] [PubMed] [Google Scholar]
- 8.Dai W., Huang S., Sun S., Cao J., and Zhang L. 2013. Nosocomial spread of carbapenem-resistant Acinetobacter baumannii (types ST75 and ST137) carrying blaOXA-23-like gene with an upstream ISAba1 in a Chinese hospital. Infect. Genet. Evol. 14:98–101 [DOI] [PubMed] [Google Scholar]
- 9.Ruan Z., Chen Y., Jiang Y., Zhou H., Zhou Z., Fu Y., Wang H., Wang Y., and Yu Y. 2013. Wide distribution of CC92 carbapenem-resistant and OXA-23-producing Acinetobacter baumannii in multiple provinces of China. Int. J. Antimicrob. Agents 42:322–328 [DOI] [PubMed] [Google Scholar]
- 10.Grosso F., Carvalho K.R., Quinteira S., Ramos A., Carvalho-Assef A.P.D., Asensi M.D., and Peixe L. 2011. OXA-23-producing Acinetobacter baumannii: a new hotspot of diversity in Rio de Janeiro? J. Antimicrob. Chemother. 66:62–65 [DOI] [PubMed] [Google Scholar]
- 11.Martins N., Martins I.S., de Freitas W.V., de Matos J.A., Girão V.B.D.C., Coelho-Souza T., Maralhães A.C.D.G., Cacci L.C., de Figueiredo M.P., Dias R.C.S., Costa-Lourenço A.P.R., Ferreira A.L.P., Dalla-Costa L., Nouér S.A., Santoro-Lopes G., Riley L.W., and Moreira B.M. 2013. Imported and intensive care unit-born Acinetobacter baumannii clonal complexes: one-year prospective cohort study in intensive care patients. Microb. Drug Resist. 19:216–223 [DOI] [PubMed] [Google Scholar]
- 12.Clímaco E.C., de Oliveira M.L., Pitondo-Silva A., Oliveira M.G., Medeiros M., Lincopan N., and da Costa Darini A.L. 2013. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect. Genet. Evol. 19:127–133 [DOI] [PubMed] [Google Scholar]
- 13.Coelho-Souza T., Reis J.N., Martins N., Martins I.S., Menezes A.O., Reis M.G., Silva N.O., Dias R.C.S., Riley L.W., and Moreira B.M. 2013. Longitudinal surveillance for meningitis by Acinetobacter in a large urban setting in Brazil. Clin. Microbiol. Infect. 19:E241–E244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martins N., Dalla-Costa L., Uehara A.A., Riley L.W., and Moreira B.M. 2013. Emergence of Acinetobacter baumannii international clone II in Brazil: reflection of a global expansion. Infect. Genet. Evol. 20:378–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chagas T.P.G., Carvalho K.R., de Oliveira Santos I.C., Carvalho-Assef A.P.D., and Asensi M.D. 2014. Characterization of carbapenem-resistant Acinetobacter baumannii in Brazil (2008–2011): countrywide spread of OXA-23-producing clones (CC15 and CC79). Diagn. Microbiol. Infect. Dis. 79:468–472 [DOI] [PubMed] [Google Scholar]
- 16.Vasconcelos A.T.R., Barth A.L., Zavascki A.P., Gales A.C., Levin A.S., Lucarevschi B.R., et al. . 2015. The changing epidemiology of Acinetobacter spp. producing OXA carbapenemases causing bloodstream infections in Brazil: a BrasNet report. Diagn. Microbiol. Infect. Dis. 83:382–385 [DOI] [PubMed] [Google Scholar]
- 17.Cardoso J.P., Cayô R., Girardello R., and Gales A.C. 2016. Diversity of mechanisms conferring resistance to β-lactams among OXA-23-producing Acinetobacter baumannii clones. Diagn. Microbiol. Infect. Dis. 85:90–97 [DOI] [PubMed] [Google Scholar]
- 18.Camargo C.H., Tiba M.R., Saes M.R., de Vasconcellos F.M., Dos Santos L.F., Romero E.C., and Garcia D.de O. 2016. Population structure analysis of carbapenem-resistant Acinetobacter baumannii clinical isolates from Brazil reveals predominance of clonal complexes 1, 15, and 79. Antimicrob. Agents Chemother. 60:2545–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stietz M.S., Ramírez M.S., Vilacoba E., Merkier A.K., Limansky A.S., Centrón D., and Catalano M. 2013. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect. Genet. Evol. 14:294–301 [DOI] [PubMed] [Google Scholar]
- 20.Escandón-Vargas K., Reyes S., Gutiérrez S., and Villegas M.V. 2017. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev. Anti. Infect. Ther. 15:277–297 [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th edition. CLSI; document M100-S24. Wayne, PA [Google Scholar]
- 22.Jones R.N., Ferraro M.J., Reller L.B., Schreckenberger P.C., Swenson J.M., and Sader H.S. 2007. Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J. Clin. Microbiol. 45:227–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodford N., Ellington M.J., Coelho J.M., Turton J.F., Ward M.E., Brown S., Amyes S.G.B., and Livermore D.M. 2006. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 27:351–353 [DOI] [PubMed] [Google Scholar]
- 24.Gautom R.K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castanheira M., Costello S.E., Woosley L.N., Deshpande L.M., Davies T.A., and Jones R.N. 2014. Evaluation of clonality and carbapenem resistance mechanisms among Acinetobacter baumannii-Acinetobacter calcoaceticus complex and Enterobacteriaceae isolates collected in European and Mediterranean countries and detection of two novel β-lactamases, GES-22 and VIM-35. Antimicrob. Agents Chemother. 58:7358–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortivo G.D., Gutberlet A., Ferreira J.A., Ferreira L.E., Deglmann R.C., Westphal G.A., and de França P.H.C. 2015. Antimicrobial resistance profiles and oxacillinase genes in carbapenem-resistant Acinetobacter baumannii isolated from hospitalized patients in Santa Catarina, Brazil. Rev. Soc. Bras. Med. Trop. 48:699–705 [DOI] [PubMed] [Google Scholar]
- 27.Zowawi H.M., Sartor A.L., Sidjabat H.E., Balkhy H.H., Walsh T.R., Al Johani S.M., AlJindan R.Y., Alfaresi M., Ibrahim E., Al-Jardani A., Al Salman J., Dashti A.A., Johani K., and Paterson D.L. 2015. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J. Clin. Microbiol. 53:896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mugnier P.D., Poirel L., Naas T., and Nordmann P. 2010. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 16:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catel-Ferreira M., Coadou G., Molle V., Mugnier P., Nordmann P., Siroy A., Jouenne T., and Dé E. 2011. Structure-function relationships of CarO, the carbapenem resistance-associated outer membrane protein of Acinetobacter baumannii. J. Antimicrob. Chemother. 66:2053–2056 [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Cuenca F., Martínez-Martínez L., Conejo M.C., Ayala J.A., Perea E.J., and Pascual A. 2003. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565–574 [DOI] [PubMed] [Google Scholar]
- 31.Hamouda A., Evans B.A., Towner K.J., and Amyes S.G.B. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48:2476–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martínez P., and Mattar S. 2012. Imipenem-resistant Acinetobacter baumannii carrying the ISAba1-blaOXA-23,51 and ISAba1-blaADC-7 genes in Monteria, Colombia. Braz. J. Microbiol. 43:1274–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reguero M.T., Medina O.E., Hernández M.A., Flórez D.V., Valenzuela E.M., and Mantilla J.R. 2013. Antibiotic resistance patterns of Acinetobacter calcoaceticus-A. baumannii complex species from Colombian hospitals. Enferm. Infect. Microbiol. Clin. 31:142–146 [DOI] [PubMed] [Google Scholar]
- 34.Hernández-Gómez C., Blanco V.M., Motoa G., Correa A., Maya J.J., de la Cadena E., Perengüez M., Rojas L., Hernández A., Vallejo M., Villegas M.V., and de R G.en Colombia B.N. 2014. Evolution of antimicrobial resistance in Gram negative bacilli from intensive care units in Colombia. Biomedica 34:91–100 [DOI] [PubMed] [Google Scholar]
- 35.Vanegas J.M., Higuita L.F., Vargas C.A., Cienfuegos A.V., Rodríguez É.A., Roncancio G.E., and Jiménez J.N. 2015. Carbapenem-resistant Acinetobacter baumannii causing osteomyelitis and infections of skin and soft tissues in hospitals of Medellín, Colombia. Biomedica 35:522–530 [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Villoria A.M., Tamayo-Legorreta E., Garza-Ramos U., Barrios H., Sanchez-Pérez A., Rodríguez-Medina N., Uribe-Aviña N., Cevallos M.A., CRAB Study Group, and Silva-Sanchez J. 2016. A multicenter study in Mexico finds Acinetobacter baumannii clinical isolates belonging to clonal complexes 636B (113B) and 92B harboring OXA-72, OXA-239, and OXA-469. Antimicrob. Agents Chemother. 60:2587–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]