Abstract
The rapid pace of conversion of natural areas to agricultural systems is highly concerning, and the consequences for conservation and pest management are not yet fully understood. We examined mitochondrial (COI and Cytb) and nuclear (ITS1) gene regions of 21 populations of the stink bug Euschistus heros, to investigate the genetic diversity, genetic structure, and demographic history of this emerging soybean pest in South America. Two deep lineages that diverged in the Pliocene (4.5 Myr) occur over wide areas of Brazil. Historical changes during the Plio-Pleistocene led to significant genetic differences between E. heros populations, which differentiated further in several biomes. The northern lineage is older, more diverse, and prevalent in the Amazon and Caatinga, while the southern lineage is younger, less diverse, and prevalent in the Atlantic Forest and Chaco biomes. Euschistus heros populations are expanding in size and range but at different rates, strongly affected by environmental variables. Secondary contact between the main lineages is now occurring, mainly in areas of intensive farming and particularly in the Cerrado, an important agricultural frontier. Individuals adapted to different environmental conditions and to large monocultures might currently be combining into a panmictic and hard-to-control pest population.
Introduction
The drastic climate changes during the Plio-Pleistocene have been considered the main cause of high levels of diversification in many areas in Brazil1–3. The four major Brazilian biomes, the Atlantic Forest (AF), Cerrado (central savannas), Amazon and Caatinga (seasonally dry tropical forest) have undergone profound changes during glaciation and interglacial cycles4. Due to this complexity and landscape composition, the processes of diversification by vicariance and habitat refugia are frequently invoked to explain the high levels of species endemism and diversity in this part of the planet5–8. Recent findings suggest that forest expansion and contraction and ‘historically stable areas’ may also have played a major role in the differentiation of lineages9.
Today, the Amazon and Atlantic forests are separated by a unique mosaic composed of savannas and woodlands that covers a large area between the two forest biomes, from Argentina and Paraguay (the Argentinean and Paraguayan Chaco) and continues through the central Brazilian Cerrado to the Caatinga in northeastern Brazil. This belt of mostly sparse dry vegetation is known as the ‘major South American disjunction’3,10,11 and forms a natural barrier preventing the movement of organisms between the northern Brazilian biomes and the Atlantic Forest12–14. Recent human impacts on these biomes, particularly the expansion of agricultural areas, have drastically changed the landscape and likely the connection between ecosystems, rearranging patterns that began to be formed millions of years ago.
The expansion of agricultural frontiers necessitates conversion of the native habitat to agriculture15. Fragments of native vegetation become embedded in a matrix of cropland and pasture that will eventually affect the species and ecosystem dynamics16. In some instances, habitat loss can limit the geographic range of endemic species, although certain species may thrive in their new surroundings. Cropland areas can provide exploitable resources, e.g. the expansion of soybean cultivation in Brazil in recent decades17. Soybean crops were formerly limited to southern Brazil (Atlantic Forest); in the early 1970s, advances in farming methods and new varieties allowed soybean farmers to expand into a new and important frontier, the Cerrado18–20.
Farming in the Cerrado has had both negative and positive impacts over the last 40 years. The expansion of soybean crops caused extensive environmental impacts such as fragmentation and the loss of natural areas, a matter of great concern for ecologists and conservationists2,21,22. On the other hand, Brazil is the second largest soybean producer after the USA, and soybeans account for an important share of its GDP23. For these reasons, it is important to understand both the impacts of soybean expansion on connecting natural populations, and the influx of insect pests from natural areas into soybean croplands.
The Neotropical brown stink bug, Euschistus heros (Fabr. 1798) (Hemiptera: Pentatomidae), is one of the most important pests of soybeans24. This Neotropical native is widespread in South America, living in markedly different environments including the Amazon Forest, Cerrado, Caatinga and Atlantic Forest25. The dispersal ability of E. heros is not well known, but is considered to be poor, perhaps because of its limited flight activity and diapause behavior26,27. This polyphagous pest feeds on members of Fabaceae, Solanaceae, Brassicaceae, Compositae, and Malvaceae, and often reaches high population densities on soybeans28–31. Rarely reported before the 1970s24,32,33, E. heros has since increased in abundance and is now found in all major soybean-producing regions24. Recently, this pest was recorded in Argentina34 and Paraguay24, raising concerns regarding a possible range expansion to other locations in South, Central and North America24.
Here we present a genealogy of mitochondrial and nuclear DNA sequences from E. heros. We addressed several questions regarding the genetic diversity, population structure, and demographic history of E. heros populations in Brazil and Paraguay, examining the potential role of past events in the differentiation of lineages, and the recent events (i.e. soybean expansion) promoting the admixture of ancient lineages. Our first objective was to determine the genetic distribution, studying population divergence and population structure. Second, we investigated the demographic history of E. heros in different biomes in South America. Finally, we used a modelling approach to explore how environmental variables and soybean expansion can explain the genetic pattern of current populations of E. heros in Brazil.
Results
Genealogical inferences
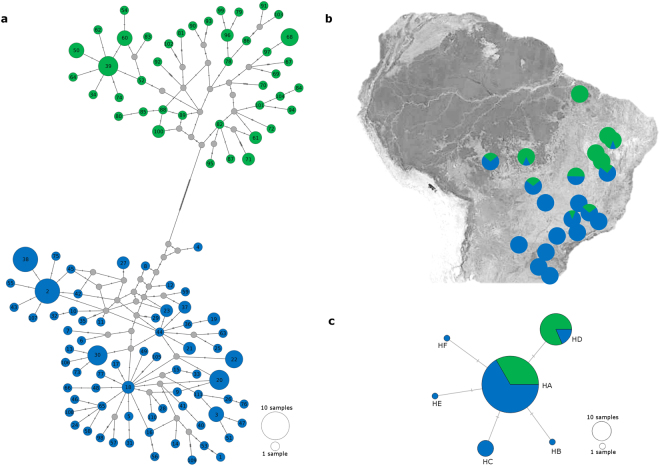
Genealogical relationships among 111 mitochondrial haplotypes indicate two well-supported E. heros lineages separated by 52 mutational steps, and an estimated genetic distance of D = 0.042 (Fig. 1a). The southern lineage (S) haplotypes occur mainly in southern regions of South America (Fig. 1b). Small percentages of lineage S haplotypes were also found in central and northeastern Brazil (Fig. 1b), in a wide range of habitats (Table 1). The northern lineage (N) occurs mainly in northern and northeastern South America and was not present in Paraguay or southern Brazil (Table 1). A total of 91 haplotypes were identified as private haplotypes; the most frequent variants were H2 and H38 (n = 8), both from lineage S (Table 1).
Figure 1.
Median-joining network and geographic distribution of Euschistus heros haplotypes in South America. (a) Network of 159 concatenated mitochondrial COI-Cytb sequences. Size of haplotype circles reflects sample size, and gray nodes represent missing haplotypes. Colors indicate the two mitochondrial lineages of haplotypes: lineage N (northern) in green, and lineage S (southern) in blue. Number of mutation steps is shown as hatch marks. (b) Geographic distributions of the mitochondrial haplotypes. Circles represent the proportion of each lineage. The map was obtained from Google Maps, Map ©2017 Google, INEGI. Pie charts were drawn with Microsoft Excel. Maps were modified with GIMP 2.8.22 (c) Network of 124 nuclear ITS1 region sequences. The haplotype color refers to the mitochondrial lineages of the individual from which the ITS1 sequence was obtained.
Table 1.
Sampling localities of Euschistus heros, with code, biomes, mitochondrial haplotype from two concatenated genes (COI-Cytb), haplotype nuclear ITS1 region, and geographic coordinates.
| ID | Locations (City, State) | Code | Biome | mtDNA haplotypes (n)* | ITS1 haplotypes (n) | Latitude (S) | Longitude (W) |
|---|---|---|---|---|---|---|---|
| 1 | Teutônia, RS | RST | Atlantic Forest | H1; H2(2); H3; H4; H5; H6; H7; H8 | HA(4); HB | 29°26′48.83″ | 51°48′50.44″ |
| 2 | Santa Bárbara do Sul, RS | RSSB | Atlantic Forest | H9; H10; H11; H12; H13; H14; H15; H16; H17 | HA(5) | 28°22′01.95″ | 53°15′06.23″ |
| 3 | Chopinzinho, PR | PRC | Atlantic Forest | H18; H19(2); H20(3); H21; H22(4); H23; H24; H25 | HA(6); HC(3) | 25°51′23.28″ | 52°32′14.01″ |
| 4 | Cornélio Procópio, PR | PRCP | Atlantic Forest | H2(3); H20; H26; H27; H28; H29 | HA(3) | 23°10′57.89″ | 50°38′44.37″ |
| 5 | Anhembi, SP | SPA | Atlantic Forest | H23; H30(2); H31; H32; H33 | HA(3) | 22°47′17.09″ | 48°07′52.29″ |
| 6 | Lavras, MG | MGL | Atlantic Forest | H27; H43; H44; H45 | HA(2); HC | 21°14′54.56″ | 45°00′04.95″ |
| 7 | General Higinio Morínigo, PY | PY | Chaco | H2; H20; H38; H105; H106; H107; H108; H109; H110; H111 | HA(3); HC(2) | 25°09′19.55″ | 55°29′59.24″ |
| 8 | Costa Rica, MS | MSCR | Cerrado | H3; H30; H37; H48; H49 | HA(2); HC | 18°32′37.15″ | 53°07′45.17″ |
| 9 | Jaboticabal, SP | SPJ | Cerrado | H2; H30; H34; H35; H36; H37; H38(2) | HA(8) | 21°15′09.05″ | 48°19′32.43″ |
| 10 | Capitólio, MG | MGC | Cerrado | H21; H30; H39(2); H40; H41; H42 | HA(6); HD | 20°36′50.88″ | 46°02′52.35″ |
| 11 | Santa Juliana, MG | MGSJ | Cerrado | H38(2); H46; H47 | — | 19°18′40.47″ | 47°31′57.69″ |
| 12 | Padre Bernardo, GO | GOPB | Cerrado | H18; H38; H39; H50; H51; H52 | HA(6); HD(3) | 15°09′39.38″ | 48°17′01.46″ |
| 13 | Rondonópolis, MT | MTR | Cerrado | H39; H53; H54; H55; H56; H57; H58 | HA(7); HD; HE | 16°27′55.71″ | 54°38′19.04″ |
| 14 | Sorriso, MT | MTS | Amazon Forest | H39; H50(2); H59; H60; H61; H62 | HA(7); HD(2); HF | 12°32′34.61″ | 55°43′17.53″ |
| 15 | Cerejeiras, RO | ROC | Amazon Forest | H2; H3; H60; H63; H64; H65; H66 | HA(5) | 13°11′14.64″ | 60°49′02.48″ |
| 16 | Paragominas, PA | PAP | Amazon Forest | H67; H68(4); H69; H70; H71(2); H72 | HA(4); HD(4) | 03°00′09.95″ | 47°21′11.19″ |
| 17 | Correntina, BA | BAC | Caatinga | H38(2); H60; H73; H74; H75; H76; H77 | HA(8); HD(2) | 13°20′33.19″ | 44°38′08.06″ |
| 18 | São Desidério, BA | BASD | Caatinga | H78; H79; H80; H81; H82 | HA(3); HD(5) | 12°21′27.47″ | 44°58′38.23″ |
| 19 | Luís Eduardo Magalhães, BA | BALE | Caatinga | H61; H83; H84; H85; H86; H87; H88; H89; H90 | HA(3); HD(6) | 12°05′25.63″ | 45°46′49.94″ |
| 20 | Bom Jesus, PI | PIB | Caatinga | H91; H92; H93; H94; H95; H96; H97; H98; H99 | — | 09°04′17.95″ | 44°21′33.65″ |
| 21 | Bom Jesus, PI | BJPI | Caatinga | H96; H100(2); H101; H102; H103; H104 | HA(2); HD(3) | 09°04′17.95″ | 44°21′33.65″ |
*Italics haplotype = Southern lineage; Bold haplotype = Northern lineage.
Analyses of the ITS1 region revealed a single nucleotide polymorphism variation separating the haplotypes. The six haplotypes were separated by a one-step mutation (Fig. 1c and Supplementary material S2). We created two alternative ITS1 sequences for all individuals that showed ambiguity in the polymorphic site, to recreate the heterozygotes. The ITS1 network had lower haplotype diversity than the mitochondrial network. Haplotype HA was the most frequent (70.16%) and was widely distributed across all regions (Table 1). Haplotype HC was found in southern and central South America, and associated individuals previously identified as mitochondrial lineage S (Table 1 and Supplementary Fig S2). Haplotype HD (21.77%) was found only in northern South America, and associated individuals previously identified as mitochondrial lineages N or S (Table 1). The single haplotypes HB, HE and HF were found in populations RS1, MT1, and MT2, respectively (Table 1). The sharing of nuclear haplotypes by specimens from both mitochondrial lineages may indicate that these lineages can interbreed (Fig. 1c).
Diversity statistics
These South American E. heros populations showed extensive mitochondrial diversity. Of 159 concatenated mitochondrial sequences of COI and Cytb analyzed, 111 haplotypes were found; most (82%) of these haplotypes were private and only 18% were shared among individuals. The overall haplotype diversity, nucleotide diversity and mean number of nucleotide differences were h = 0.991, π = 0.03312 and K = 32.892, respectively (Table 2). The haplotype diversity was similar in lineage N (h = 0.984) and lineage S (h = 0.982); however, lineage N (π = 0.0090) had higher nucleotide diversity than lineage S (π = 0.0062). For the different biomes, the haplotype diversity among biomes (groups) was low, ranging from 0.967 in the Amazon to 1.000 in the Chaco. The nucleotide diversity varied more widely among biomes, from 0.00627 in the Atlantic Forest to 0.03147 in the Amazon Forest (Table 2). Locations where both lineages were present had the highest levels of nucleotide diversity and mean numbers of nucleotide differences. Sequence analysis of the ITS1 region identified six haplotypes with a haplotype diversity of 0.461, nucleotide diversity of 0.0008 and mean number of nucleotide differences of 0.499 (Table 2). The haplotype diversity and nucleotide diversity were higher in lineage N (h = 0.503; π = 0.0008) than lineage S (h = 0.355; π = 0.0006), previously defined by the mitochondrial network. Among biomes, the highest diversity was observed in the Chaco (h = 0.600; π = 0.0009) and the lowest diversity in the Atlantic Forest (h = 0.315; π = 0.0005) (Table 2). The Amazon Forest, Caatinga and Cerrado have higher nucleotide diversity due to mixing of the two lineages in these areas.
Table 2.
Measures of genetic diversity for Euschistus heros based on two concatenated mitochondrial genes (COI-Cytb) and ITS1 region.
| Geographic regions | Sample size (N) | Haplotype number (H) | Haplotype diversity (h) | Nucleotide diversity (π) | Average # of nucleotide difference (K) |
|---|---|---|---|---|---|
| COI-Cytb | |||||
| Pooled | 159 | 111 | 0.991 | 0.0331 | 32.892 |
| Atlantic Forest | 50 | 36 | 0.979 | 0.0063 | 6.224 |
| Chaco | 10 | 10 | 1.000 | 0.0066 | 6.511 |
| Cerrado | 37 | 27 | 0.970 | 0.0256 | 25.396 |
| Amazon Forest | 24 | 18 | 0.967 | 0.0315 | 31.246 |
| Caatinga | 38 | 35 | 0.996 | 0.0252 | 25.007 |
| Lineage N* | 57 | 42 | 0.984 | 0.0090 | 8.921 |
| Lineage S* | 102 | 69 | 0.982 | 0.0062 | 6.198 |
| ITS1 region | |||||
| Pooled | 124 | 6 | 0.461 | 0.0008 | 0.499 |
| Atlantic Forest | 28 | 3 | 0.315 | 0.0005 | 0.325 |
| Chaco | 5 | 2 | 0.600 | 0.0009 | 0.600 |
| Cerrado | 36 | 4 | 0.340 | 0.0006 | 0.357 |
| Amazon Forest | 23 | 3 | 0.466 | 0.0008 | 0.490 |
| Caatinga | 32 | 2 | 0.516 | 0.0008 | 0.516 |
| Lineage N* | 50 | 2 | 0.503 | 0.0008 | 0.503 |
| Lineage S* | 74 | 6 | 0.355 | 0.0006 | 0.382 |
*Lineages were previously defined by mitochondrial network.
Mitochondrial divergence dating
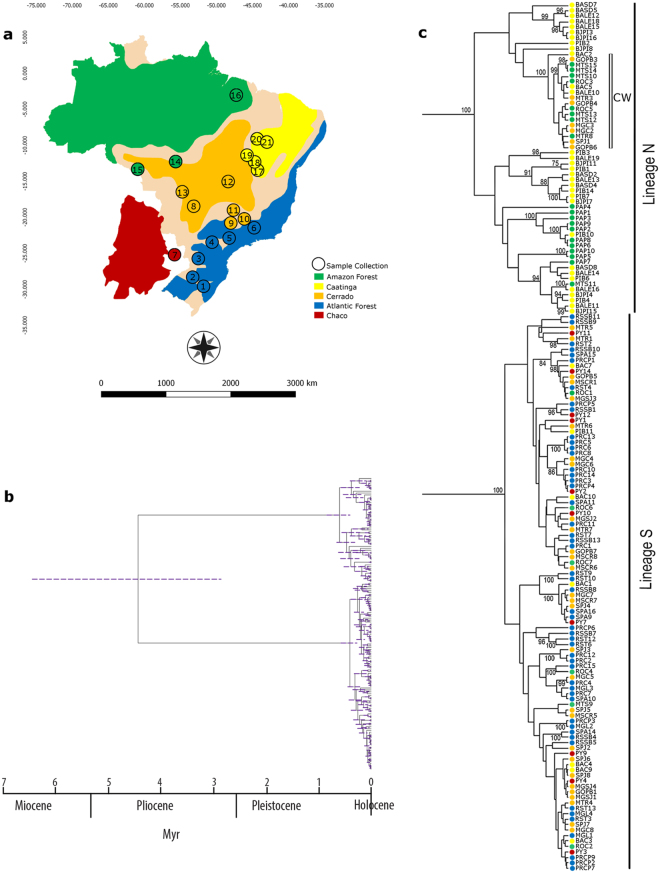
The estimated age of origin of the lineage S (southern lineage) clade was 4.5 Myr (95% C.I 2.801–6.453 Myr), during the Pliocene, with intense diversification in the Pleistocene and Holocene (Fig. 2b).
Figure 2.
Bayesian coalescent tree for Euschistus heros. (a) Geographic and biome distribution of each population of E. heros sampled (see Table 1) (map based on Ab’Saber77 and modified with GIMP 2.8.22); (b) Bayesian phylogeny tree of 159 concatenated mitochondrial sequences (COI-Cytb). Gray bars at nodes indicate 95% highest probability density intervals (HPD) confidence intervals for nodal age. (c) Bayesian phylogeny tree showing posterior probability values (>75) and biome where individuals were collected (taxon names provided in Table 1).
Population Structure
Lineage N haplotypes were associated mainly with the Amazon Forest and Caatinga, with one, more recent clade (CW) associated with central Brazil, a transitional region among the Cerrado, Caatinga and Amazon Forest (Fig. 2a,c). Lineage S predominates in the Atlantic Forest and Chaco, with a lower frequency in the Cerrado, Caatinga and Amazon Forest (Fig. 2a,c).
At the regional scale, the E. heros populations showed high genetic structure, as assessed by the Analysis of Molecular Variance (AMOVA). Differences among populations accounted for most of the genetic variances in mtDNA (56.57%, P < 0.001) and a high and significant value in ITS1 regions (20.18%, P < 0.001) (Table 3). A test of the hypothesis that the genetic variation is structured by biomes showed that 45.07% of the mtDNA total variance was distributed among biomes (ΦCT = 0.450, P < 0.001). Furthermore, the larger portion of genetic variation within populations (39.61%, ΦST = 0.603) indicates overall genetic differentiation in these populations (Table 3). Analysis of the ITS1 region supported the mitochondrial data, showing a significant structuring by biome (ΦCT = 0.173, P < 0.001), in which most of the genetic variation was within populations (Table 3).
Table 3.
Analysis of molecular variance (AMOVA) for genetic structure of Euschistus heros based on two concatenated mitochondrial genes (COI-Cytb) and ITS1 region.
| Source of variation | d.f. | Variance components | Percentage variance | Fixation indices (P-value) |
|---|---|---|---|---|
| (a) COI-CytB | ||||
| Among populations | 20 | 9.550 | 56.57 | ΦST = 0.566 (P = 0.00) |
| Within populations | 138 | 7.333 | 43.43 | |
| Total | 158 | 16.883 | ||
| Among biomes | 4 | 8.342 | 45.07 | ΦCT = 0.450 (P = 0.00) |
| Among populations within biomes | 16 | 2.836 | 15.32 | ΦSC = 0.278 (P = 0.00) |
| Within populations | 138 | 7.333 | 39.61 | ΦST = 0.603 (P = 0.00) |
| Total | 158 | 18.511 | ||
| (b) ITS1 region | ||||
| Among populations | 18 | 0.050 | 20.18 | ΦST = 0.201(P = 0.00) |
| Within populations | 105 | 0.201 | 79.82 | |
| Total | 123 | 0.252 | ||
| Among biomes | 4 | 0.045 | 17.30 | ΦCT = 0.173 (P = 0.00) |
| Among populations within biomes | 14 | 0.014 | 5.49 | ΦSC = 0.066 (P = 0.07) |
| Within populations | 105 | 0.201 | 77.21 | ΦST = 0.227 (P = 0.00) |
| Total | 123 | 0.260 | ||
Demographic statistics inferred for mitochondrial genes
Considering the two lineages, significant negative values were found in both the Tajima’s D and Fu’s Fs neutrality tests, indicating population expansion or purifying selection. Considering the biomes, the neutrality test statistics did not fully agree with one another. Fu’s Fs statistic was significantly negative for all biomes, but only the Atlantic Forest biome had a significant negative Tajima’s D value (Table 4).
Table 4.
Neutrality test statistics and mismatch distribution analysis for Euschistus heros based on two concatenated mitochondrial genes (COI-Cytb).
| Geographic regions | Sample size (n) | Tajima’s D | Fs de Fu | τ (SD95%) | SSD (P-value) | r (P-value) |
|---|---|---|---|---|---|---|
| COI-Cytb | ||||||
| Pooled | 159 | 0.445 | −23.799* | 56.2 (5.30 – 89.24) | 0.0268 (P = 0.08) | 0.0041 (P = 0.98) |
| Atlantic Forest | 50 | −1.920* | −25.224* | 6.4 (4.25–7.48) | 0.0016 (P = 0.66) | 0.0136 (P = 0.55) |
| Chaco | 10 | −1.253 | −4.883* | 6.0 (2.81–7.01) | 0.0325 (P = 0.12) | 0.0563 (P = 0.34) |
| Cerrado | 37 | 0.858 | −18.632* | 57.5 (3.66–79.53) | 0.0394 (P = 0.09) | 0.0160 (P = 0.80) |
| Amazon Forest | 24 | 0.805 | −7.198* | 58.7 (3.98–196.99) | 0.0363 (P = 0.10) | 0.0419 (P = 0.26) |
| Caatinga | 38 | −0.067 | −19.748* | 7.3 (5.36–11.13) | 0.0199 (P = 0.03*) | 0.0071 (P = 0.84) |
| Lineage N | 57 | −1.779** | −24.741** | 10.0 (7.25–11.03) | 0.0112 (P = 0.04*) | 0.0145 (P = 0.17) |
| Lineage S | 102 | −2.271** | −25.120** | 6.5 (4.50–7.27) | 0.0033 (P = 0.11) | 0.0144 (P = 0.20) |
Lineage were previously defined by mitochondrial network. τ = Expansion parameter; SSD = Sum of Squared Deviation; r = Harpending’s Raggedness Index.
For the lineages, the mismatch distribution analysis resulted in a nonsignificant SSD (P > 0.05), indicating a recent demographic expansion of lineage S but not lineage N (P = 0.04). For the biomes, a nonsignificant SSD (P > 0.05) was also found for the E. heros populations in all biomes but the Caatinga (P = 0.03) (Table 4). The nonsignificant raggedness index (P > 0.17) supports the spatial-expansion model of populations of lineages, biomes, and the entire group (all populations combined) (Table 4). The τ values were higher in the Cerrado and the Amazon Forest, τ = 57.5 and τ = 58.7, respectively, compared to the other three biomes, Atlantic Forest (τ = 6.4), Chaco (τ = 6.0) and Caatinga (τ = 7.3) (Table 4).
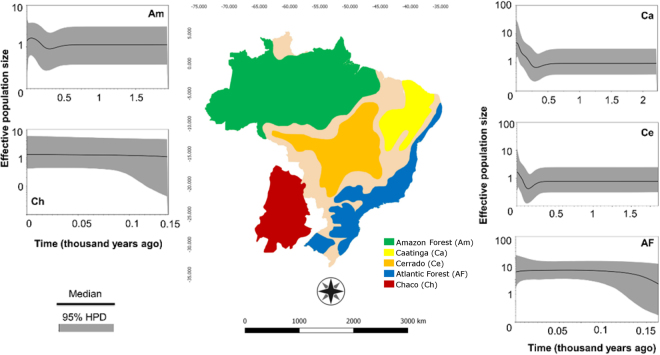
The expansion of populations in the Amazon Forest, Chaco, Caatinga, Cerrado and Atlantic Forest occurred within the last 500 years, corresponding to a recent expansion during the Quaternary according to the Bayesian skyline plot analysis (Fig. 3). The Chaco and Atlantic Forest populations remained stable during the past 100 years, while the Caatinga and Cerrado populations are still expanding. According to the effective population size (Ne), the Atlantic Forest population is the largest, followed by the Caatinga. The Cerrado, Amazon Forest and Chaco populations have similar sizes, but the Cerrado population is still expanding very rapidly, while the Chaco population is expanding slowly and the Amazon population is now contracting (Fig. 3).
Figure 3.
Bayesian skyline plot (BSP) showing population size dynamics for Euschistus heros in five biomes. The y-axis indicates effective population size (Ne) scaled by mutation rate (µ) as a function of time. Black horizontal line shows median BSP estimate, and gray area shows upper and lower 95% highest posterior density limits. The map was based on Ab’Saber77 and modified with GIMP 2.8.22.
Environmental features and soybean expansion modelling the current mitochondrial lineage distribution
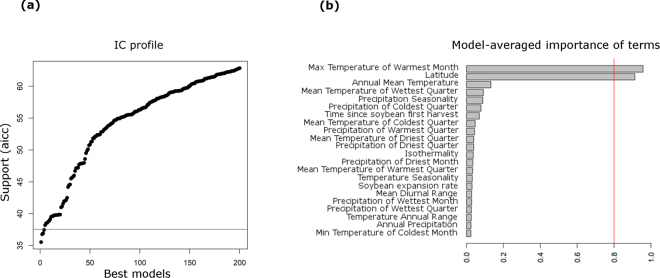
Three models passed the cutoff (i.e. models that were less than 2 units away from the “best” model) to explain the presence (%) of the southern lineage at a given location (i.e. the probability of finding an individual from the southern lineage). The best predictors were the ‘max temperature of the warmest month’, ‘latitude’, and ‘annual mean temperature’ (Fig. 4 and Table 5). The most important variables were the ‘max temperature of the warmest month’ and ‘latitude’, which received the highest score in all top models. The model performance improved when ‘annual mean temperature’ was excluded, i.e. AICc (36.22 and 35.35) and wi (0.09 and 0.14) (Table 5). The best model selected (AICc = 35.35) was 22.29 units away from the null model (AICc = 57.64). The variable ‘latitude’ (0.50) was more important than ‘max temperature of the warmest month’ (0.43) in the best model according to the z-scored beta (null deviance = 18 on 18 d.f., residual deviance = 4.04 on 16 d.f.) (Table 5). Latitude (0.9) was also the most important variable in the second-best model compared to the ‘max temperature of the warmest month’ (0.7) and ‘annual mean temperature’ (0.65) (null deviance 18 on 18 d.f., residual deviance = 3.469 on 15 d.f.) and in the third-best model (0.66) compared to ‘mean temperature of wettest quarter’ (0.35) and ‘max temperature of the warmest month’ (0.56) (null deviance 18 on 18 d.f., residual deviance = 3.531 on 15 d.f.). None of the three best models included soybean variables. The two soybean variables, ‘time since first soybean harvest’ and ‘soybean expansion rate’, ranked 7th and 16th in overall importance. The ‘time since first soybean harvest’ was strongly correlated with ‘latitude’ (r = 0.90, d.f. = 17, P = 0.000), ‘max temperature of the warmest month’ (r = 0.61, d.f. = 17, P = 0.005), and ‘annual mean temperature’ (r = 0.76, d.f. = 17, P = 0.000).
Figure 4.
Model selection and variable importance. (a) AICc values for the 200 top models for percentage of the southern lineage. Horizontal red line separates models with AICc less than 2 units away from the “best” model. (b) Relative importance of predictors, considering all models. Relative importance is the sum of weights for the models in which the variable appears. Vertical red line indicates the 0.8 cutoff used to separate important variables.
Table 5.
Model-selection results showing the top three models for the response variable ‘presence of the southern lineage’.
| Competing models | AIC c | w i (AIC) | |
|---|---|---|---|
| Presence of southern lineage (%) | |||
| (1) | +0.50 Latitude −0.43 Max Temperature of Warmest Month |
35.35 | 0.14 |
| (2) | +0.90 Latitude −0.70 Max Temperature of Warmest Month +0.65 Annual Mean Temperature |
36.22 | 0.09 |
| (3) | +0.66 Latitude +0.35 Mean Temperature of Wettest Quarter −0.56 Max Temperature of Warmest Month |
36.56 | 0.07 |
Selected models are those with lowest AICc values within two units from that of the best model. Best models are those with lower AICc values. The Akaike weights (wi) represent the relative likelihood of a model and can be used to compare the strength of evidence of alternative models.
Discussion
Our results revealed two divergent deep lineages of E. heros in South America. The two COI-Cytb haplotype groups are separated by 52 mutational steps and have an estimated genetic distance of 4.2% (K2P). The number of mutation steps separating the two E. heros lineages is exceptionally high, raising the question of the possible presence of cryptic species. Rolston35 described a species in Middle America that is morphologically similar to E. heros, Euschistus atrox (Westwood) 1837, distributed in Colombia, Guiana, and Panama. However, examination of the external morphology, the sharing of ITS1 alleles by both lineages, and the admixture of lineages in the laboratory support the hypothesis of a single species that encompasses the two divergent lineages. Our results reveal the need for a thorough systematic review of the genus Euschistus.
The two E. heros lineages are geographically separated from one another, with one clade clustering the northernmost populations (i.e. northern and northeastern Brazil), and a second clade clustering the southernmost populations (i.e. southern and southeastern regions). Both mitochondrial lineages expanded to form a mixed zone upon secondary contact in the central and southwestern regions. It is not clear when the reunion occurred, but the formerly isolated populations seem to have come into contact before reproductive isolation was complete36,37. A related point to consider is that all but one of the northern haplotypes found in the Cerrado (CW) were phylogenetically grouped together in one clade, indicating a subgroup differentiation. The central-western (CW) subgroup likely occupied the region much earlier than the southern lineage arrived and before the first soybean fields were established. This is strong evidence against the hypothesis that the E. heros expansion was purely associated with the expansion of soybean cultivation during the 1970s.
The divergence time of the two main clades is estimated as occurring during the Pliocene (i.e. 4.5 Myr). This divergence seems to be associated with a cooling and drying of the global environment, which caused the separation of the Amazon Forest from the southern part of the Atlantic Forest and the consequent expansion of grasslands and savannas38. Temperature cycles were also associated with more recent diversification events during the Pleistocene (i.e. differentiation of the CW group). Deep sequence divergence dating to the Pliocene is also reported for other organisms in the Neotropics39,40, and phylogeographic structure has been found in amphibians in the Atlantic Forest3, reptiles in the Cerrado41, and plants4,42.
Spatial genetic structuring by biomes was also found among subpopulations of E. heros. Separation into the Amazon Forest, Caatinga, Cerrado, Atlantic Forest and Chaco biomes seems to be the best way to explain the genetic variance hierarchically. Thus, separating insects by biomes can help us to understand the pattern of lineage mixing, diversity and demographic history. The haplotype diversity of E. heros was high and similar among biomes and lineages. This pattern is the result of the high number of private haplotypes found in E. heros populations in all biomes. The higher nucleotide diversity of lineage N compared to lineage S can be explained under the ‘historical climate’ stability models, where a stable environment such as the Amazon Forest can offer conditions for a population to persist, resulting in elevated intraspecific genetic diversity43,44. Unstable regions, on the other hand, would be associated with recent or multiple-event colonization, resulting in lower intraspecific genetic diversity and signatures of expansion4. Therefore, the northern biomes (Amazon Forest and Caatinga) were the most stable environment, while the Atlantic Forest was the least stable environment. Another consideration is that lineage S is associated with areas that have undergone intense transformation due to agricultural practices, and has experienced population dynamics linked with farming cycles and control tactics.
Although E. heros’ limited dispersal capacity likely helped to preserve the pattern formed during the late Tertiary and Quaternary as an outcome of the climate changes, the last 100 years were an important turning point for E. heros populations (i.e. soybean introduction and expansion of farming starting at the end of the 19th century). It is plausible that farming and trade routes have increased the admixture process in certain areas, especially the Cerrado and connecting areas, even though there are still large areas where the two lineages have not yet encountered each other, showing that the pattern is still well preserved.
Recent signals of expansion were detected for E. heros lineages and in all biomes sampled. The inferences regarding population growth were supported by the neutrality tests, the unimodal mismatch distribution and the demographic expansion parameters (τ). Spatial expansion is also occurring, given that no significant Raggedness values were found. Apart from differences in test sensitivity, the lack of full agreement between tests for E. heros in each biome might indicate a more complex scenario. Multiple processes affecting local diversity and the noise from human intervention causing population reduction, population subdivision, bottlenecks, and facilitation of dispersal resulting in the secondary contact might affect the precise demographic estimates for a species45–47.
We also conducted a Bayesian Skyline analysis to test the hypothesis of recent expansion in all biomes and to determine how the effective sizes of the populations behaved over time. The period of E. heros population growth in all areas overlaps with the period of intense changes caused by the increase of urban occupation and agricultural area in South America48. It may be that the resulting habitat loss not only did not affect E. heros populations negatively, but has even been advantageous. One possible hypothesis to explain the success of E. heros is shifting hosts from natural areas to agricultural fields, especially soybeans but also cotton and bean fields31. A second hypothesis is that one or more traits occur in a latitudinal cline49–51. The species’ association with environmental gradients should also be considered, given the possibility of differences in traits and adaptations such as reproductive diapause27,52,53.
We used environmental and soybean variables to make phylogeographic inferences to predict the predominance of lineage S over lineage N at a given location. Selected models had similar AICc scores and considerably reduced the number of variables, down to 4 for the percentage of lineage S models. The two most important variables were the maximum temperature of the warmest month and the latitude. Temperature and photoperiod both affect this species, and might induce quiescence behavior and other possible differences in physiological responses. Latitude, on the other hand, can be correlated with geographic distance, environmental gradients, and agricultural gradients, as in the case of the soybean expansion. Our data support the predictions of the latitudinal-gradient hypothesis, even though distinct demographic scenarios can be expected at different times of E. heros’ evolutionary history. The time since the first soybean harvest correlates with latitude and other bioclimate variables, which likely decreased the importance of this variable in the model.
The reunion of the two long-separated lineages might have unforeseen consequences for one of the largest soybean-producing regions in the world. The two lineages are united again in central Brazil, where an agricultural revolution started in the 1970s and continues today, pushing soybean fields northward54. It is possible that the northern and southern populations of E. heros are exchanging adaptations in admixture zones. However, knowledge of the differences between the two lineages is limited, because their presence was unknown until this point55,56. The changing status of E. heros from a secondary to a primary pest in soybean crops and the reasons for this are poorly understood. In recent years, the increase of population densities in soybean fields, the shorter quiescence period, larger host range (i.e. damage in cotton crops) and pesticide tolerance/resistance have been frequently reported in E. heros populations30,31,57. These concerns increase in a scenario of GM soybean introduction, no-till management, and expansion to diversity-hotspot areas.
Material and Methods
Sample collection and DNA extraction
One hundred fifty-nine specimens of E. heros were collected between 12/2015 and 07/2016 from 21 different localities across five South American biomes. Twenty sampling sites were in Brazil and one site in Paraguay. Specimens were collected as adults, from the canopy of soybeans, using a beating cloth under the plants. Individuals were preserved in ethanol (>95%) at –20 °C until laboratory manipulation, after which the remaining tissue from all specimens was stored at –80 °C. DNA was extracted from the head tissue of an adult specimen, using the modified CTAB protocol58.
PCR amplification and DNA sequencing
Fragments of two mitochondrial and one nuclear region were amplified by polymerase chain reaction (PCR), using specific mitochondrial primers developed for this project and previously developed ITS1 primers59. The Cytochrome c Oxidase Subunit 1 (COI) fragment was amplified using the forward primer (5′-ACCGCACATGCATTTGTAATAA-3′) and the reverse primer (5′-GTGGCTGATGTGAAGTATGCTC-3′), and the Cytochrome b (Cytb) fragment was amplified using the forward primer (5′-GGATATGTTTTACCTTGAGGACA-3′) and the reverse primer (5′-GGAATTGATCGTAAGATTGCGTA-3′). To amplify the ITS1 rnDNA region (18 S partial – ITS1 complete – 5.8 S partial) we used the forward primer CAS18SF1 (5′- TACACACCGCCCGTCGTACTA-3′) and the reverse primer CAS5p8sB1d (5′- ATGTGCGTTCRAAATGTCGATGTTCA-3′). The PCR reactions were performed in a total volume of 25 μL containing 80 ng total DNA, 1.5 mM/μL MgCl2, 0.1 mM/μL dNTPs, 0.4 pmol/μL of each primer, 1 U of Taq DNA Polymerase (Synapse Inc.) and Buffer (10 × Taq DNA Buffer). PCR cycles consisted of denaturation at 95 °C for 3 min, followed by 35 cycles with denaturation at 95 °C for 30 s, annealing at 54 °C for 40 s, polymerization at 72 °C for 1.5 min and final extension at 72 °C for 10 min. Subsequently, the PCR products were separated on agarose gel (1.5% w/v) and observed under ultraviolet light. The amplicons were purified using 0.33 μL EXO I, 0.33 μL FastAp and 0.34 μL of ultra-pure water together with 10 μL of each PCR product, held at 37 °C for 30 min, then at 80 °C for 15 min. The PCR product Sanger sequencing was performed by the Animal Biotechnology Laboratory at ESALQ, University of São Paulo.
Assembly of sequence datasets
All sequences were aligned and edited manually using the software Sequencher 4.0.1 (Gene Codes Corp., Ann Harbor, MI, USA). To eliminate missing data, sequences were interrupted at 607 bp for the COI gene, 386 bp for the Cytb gene and 638 bp (18 S partial – 52 bp; ITS1 complete – 416 bp; 5.8 S partial – 170 bp) for the ITS1 region. There were no insertions or deletions in the sequences obtained. All sequences (datasets) obtained in this study were deposited in NCBI-GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession numbers MG651970 - MG652128 (COI), MG652129 - MG652287 (Cytb), and MG654513 - MG654636 (ITS1).
The presence of nuclear paralogs of mitochondrial origin (termed numts)60 was inspected in the mitochondrial gene fragments, using the software MEGA v.5.261. Three signatures of numts were searched: (i) indels that introduce frameshifts, (ii) out-of-place inframe stop codons that lead to premature termination of protein translation, and (iii) lack of codon position substitution bias toward the 3rd position, that lead to a higher rate of non-synonymous mutations. The presence of signatures (i) and (ii) is enough to consider a given sequence a numt. No numts were detected in the COI or CtyB sequences; therefore, we included all mitochondrial sequences in our analysis. The posterior analyses were performed using concatenated mitochondrial genes (COI-Cytb).
Genealogical inferences
The genealogical relationships between haplotypes of the mitochondrial and ITS1 regions were reconstructed by a network of median-joining haplotypes, using the PopArt software62. Preliminary analysis revealed two putative mitochondrial lineages associated with E. heros populations in Brazil and Paraguay. The genetic distance (D) between the two mitochondrial lineages was inferred by dividing the haplotypes in two groups and calculating the 2-parameter Kimura method (K2P) in MEGA v.5.2 software61.
Diversity statistics
The diversity analysis was performed by dividing individuals into two groups according to the mitochondrial lineages, or into five groups according to the biome to which the individuals belonged: Amazon Forest, Cerrado, Caatinga, Atlantic Forest or Chaco. Number of haplotypes, haplotype diversity (h), nucleotide diversity (π) and mean number of nucleotide differences (S) were estimated using the DNAsp v.563.
Divergence dating
We estimated the relative age of divergence between the two mitochondrial lineages using the Bayesian relaxed phylogenetic approach implemented in BEAST v.1.8.464, based on the combined mitochondrial genes. The substitution model was determined using the software PARTITIONFINDER version 1.1.1.65 that selected the GTR + G + I model. A strict molecular-clock model to estimate the substitution rate and coalescent tree priors set to the constant size model were implemented. We used the insect molecular clock (mean = 0.0177, SD = 0.001)66 that corresponds to 3.54% pairwise divergence per Myr. Three independent runs were performed for 150 million generations, sampling every 1000 steps and discarding 20% as burn-in. TRACER v.1.6 was used to determine convergence, measure the effective sample size (ESS), and calculate the mean and 95% highest posterior density interval (HPD) for divergence times. Effective sample size (ESS) for all parameters exceeded 200, and the three runs converged to similar distributions. Runs were then combined with LogCombiner v.1.4.764.
Population Structure
Variance Analysis (AMOVA) was performed in Arlequin with parametric bootstrap (1000 replicates) using a 5% significance level67. The analyses were conducted to examine the presence of genetic structure among individuals, considering all sites sampled (non-hierarchical), among populations according to the sampling location (populations) and among biomes in three hierarchical levels.
Demographic statistics inferred for mitochondrial data
Tajima’s D and Fu’s Fs neutrality tests were calculated using Arlequin v.3.567. Both tests used 1,000 permutations using coalescing simulations. Fu’s Fs statistic was considered significant at the 95% confidence level when the P-value was less than 0.02. For each biome, we also estimated tau (τ) with its 95% confidence intervals, using a generalized least-squares approach and 1,000 coalescent simulations in Arlequin v.3.5. The parameter τ denotes the age of the expansion (t), so that t = τ/2 u; u = μLg68. The parameter μ represents the estimated mutation rate, L is the length of the sequence, and g is the generation time. For E. heros, we did not estimate t directly, because the number of generations per year cannot be estimated straightforwardly. Thus, if we assume that the estimated mutation rate has not changed in E. heros (substitution rate = 1.345%), then u will be constant and we can consider that a smaller τ value indicates a newly established population, and a larger τ value an older one.
We conducted a mismatch distribution analysis using a spatial expansion model. The sum of square of deviations (SSD), raggedness index (r) statistics, and their associated P-value were calculated using Arlequin v.3.5. A nonsignificant SSD value means that the hypothesis of population expansion cannot be rejected, and a nonsignificant raggedness index indicates a good fit of the data to the spatial expansion model. We also used a Bayesian Skyline Plot (BSP) in Beast to reconstruct the demographic history, using TRACER v.1.6, based on the COI-Cytb data using 10 groups. We used the same substitution model and molecular-clock model that were used to estimate the divergence time.
Environmental features and soybean expansion modelling the current mitochondrial lineage distribution
We used a model selection approach to identify and select variables that could be influencing the presence of a lineage at a given location69,70. Therefore, our response variable was the proportion of the southern lineage S calculated at each location as a percentage of the total composition. As predictor variables, we used ‘latitude’, 19 WorldClim variables based on all pixels of a CFR at 30 arc-second image71, and two soybean variables. The soybean variables consisted of the estimated time since the first harvest and the rate of increase of soybean production, given the cultivated area. We used linear regression to compile data from different sources and to estimate the two soybean variables, using the regression slope and the predicted year when the cultivated area was 100 hectares (Supplementary Table S1 and Fig. S1)54,72,73.
We evaluated the fit and plausibility of possible candidate models using glmulti74. We used a selection considering only the main effect, keeping the 200 best models. The criterion for selection was the corrected Akaike Information Criterion (AICc)75. We selected models with AICc less than two units away from the best model. We also evaluated the Akaike weight of the best models, to assess the probability that a model is the best76. All variables were standardized by z-score, and the significance of each predictor was assessed by a GLM. We also assessed the importance of each variable by summing the Akaike weight for the models in which the variable appeared. Variables that appear many times in the top models, tend to be more important. We used the cutoff of 0.8 to separate the most important variables under the weight criterion69,70.
Electronic supplementary material
Acknowledgements
We thank all collaborators who helped with insect sampling (Arias, O.; Azevedo, T.; Baptistella, M.G.; Bordignon, A.; Brogin, R.L.; Farinha, J.F.; Ferreira, V.E.N.; Gomes, J.; Gontijo, P.; Leite, N.A.; Lerner, M.A.; Machado, R.; Netto, J.; Omoto, C.; Pavan, G.B.; Rondon, M.; Rossi, G.D.; Silva, E.H.; Silva, L.B.; Tonelli, M.; Trebien, E.; Vidotti, M.S.; Vivan, L.M.; Zachrisson, B.; Zenati, L.). Our thanks to Julia Barbosa de Paiva for valuable assistance with the maps. We also thank the following entities for financial support: Fundação de Amparo à Pesquisa do Estado de São Paulo – FAPESP (grant #2014/11495-3 and fellowship #2017/02393-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico - CNPq (grant #423040/2016-2 and fellowship #309167/2015-9) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - CAPES for fellowships.
Author Contributions
A.S.C., C.O. and P.L.S. conceived and designed the study. E.M.G.C., F.N.S. and P.L.S. collected the data. A.S.C. and C.O. provided reagents and analytical tools. A.S.C., E.M.C.G. and PLS analyzed the data. A.S.C., E.M.C.G. and P.L.S. wrote the manuscript. All authors read, corrected, and approved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20187-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mittermeier RA, et al. Hotspots Revisited: Earth’s biologically richest and most endangered terrestrial ecoregions. Washington. Cemex. 2005;1:392. [Google Scholar]
- 2.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 3.Thomé MTC, et al. Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Molecular Phylogenetics and Evolution. 2010;55:1018–1031. doi: 10.1016/j.ympev.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Werneck FP. The diversification of eastern South American open vegetation biomes: Historical biogeography and perspectives. Quaternary Science Reviews. 2011;30:1630–1648. doi: 10.1016/j.quascirev.2011.03.009. [DOI] [Google Scholar]
- 5.Haffer J. Speciation in Amazonian forest birds. Science. 1969;165:131–137. doi: 10.1126/science.165.3889.131. [DOI] [PubMed] [Google Scholar]
- 6.Bennett KD, Provan J. What do we mean by ‘refugia’? Quaternary Science Reviews. 2008;27:2449–2455. doi: 10.1016/j.quascirev.2008.08.019. [DOI] [Google Scholar]
- 7.Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 8.Carnaval AC, Moritz C. Historical climate modeling predicts patterns of current biodiversity in the Brazilian Atlantic forest. Journal of Biogeography. 2008;35:1187–1201. doi: 10.1111/j.1365-2699.2007.01870.x. [DOI] [Google Scholar]
- 9.Leite YLR, et al. Neotropical forest expansion during the last glacial period challenges refuge hypothesis. PNAS. 2016;113:1008–1013. doi: 10.1073/pnas.1513062113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa L. The historical bridge between the Amazon and the Atlantic Forest of Brazil: a study of molecular phylogeography with small mammals. Journal of Biogeography. 2003;30:71–86. doi: 10.1046/j.1365-2699.2003.00792.x. [DOI] [Google Scholar]
- 11.Brieger, F. G. Contribuição à fitogeografia do Brasil com referência especial as orquídeas. Anais do XX Congresso Nacional de Botânica, 41–44 (1969).
- 12.Moojen J. Speciation in the Brazilian Spiny Rats (Genus Proechimys, Family Echimyidae) University of Kansas Publications, Museum of Natural History. 1948;1:301–406. [Google Scholar]
- 13.Mori SA, Boom BM, Prance GT. Distributional patterns and conservation of eastern Brazilian coastal forest tree species. Brittonia. 1981;33:233–245. doi: 10.2307/2806330. [DOI] [Google Scholar]
- 14.Por, D. F. S, The Atlantic rain forest of Brazil. SPB Academic Publishing (1992).
- 15.Schiesari L, Waichman A, Brock T, Adams C, Grillitsch B. Pesticide use and biodiversity conservation in the Amazonian agricultural frontier. Philosophical Transactions of the Royal Society B. 2013;368:20120378. doi: 10.1098/rstb.2012.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabarelli M, Da Silva JMC, Gascon C. Forest fragmentation, synergisms and the impoverishment of neotropical forests. Biodiversity & Conservation. 2004;13:1419–1425. doi: 10.1023/B:BIOC.0000019398.36045.1b. [DOI] [Google Scholar]
- 17.Paterniani E, Malavolta E. La conquista del ‘cerrado’ en el Brasil. Victoria de la investigación científica. Interciencia. 1999;24:173–181. [Google Scholar]
- 18.Klink, C.A. De Grão em Grão: O Cerrado Perde Espaço. Brasilia, Brazil: World Wide Fund for Nature (WWF-Brasil): 66 (1995).
- 19.Klink, C. A., Macedo, R. H. & Mueller, C. C. Cerrado: Processo de ocupação e implicações para a conservação e utilização da sua diversidade biológica. Brasilia, Brazil: World Wide Fund for Nature (WWF-Brasil): 104 (1994).
- 20.Spehar CR. Impact of strategic genes in soybean on agricultural development in the Brazilian tropical savannahs. Field Crops Research. 1995;1:141–146. doi: 10.1016/0378-4290(95)00007-D. [DOI] [Google Scholar]
- 21.Klink, C. A., Moreira, A. G. & Solbrig, O. T. Ecological impacts of agricultural development in the Brazilian Cerrados. The World’s savannas: Economic driving forces, ecological constraints and policy options for sustainable land in the Biosphere 12, (eds Young, M. D. & Solbrig O. T.) 259–283 (Parthenon Publishing, London. 1993).
- 22.Fearnside PM. Soybean cultivation as a threat to the environment in Brazil. Environmental Conservation. 2001;28:23–38. [Google Scholar]
- 23.Brazilian Institute of Geography and Statistics – IBGE. http://www.ibge.gov.br/home/ (2017).
- 24.Panizzi AR. Growing problems with stink bugs (Hemiptera: Heteroptera: Pentatomidae): species invasive to the US and potential Neotropical invaders. American Entomologist. 2015;61:223–233. doi: 10.1093/ae/tmv068. [DOI] [Google Scholar]
- 25.Panizzi AR, Slansky F., Jr. Review of phytophagous pentatomids (Hemiptera: Pentatomidae) associated with soybean in the Americas. Florida Entomologist. 1985;68:184–214. doi: 10.2307/3494344. [DOI] [Google Scholar]
- 26.Aldrich JR. Dispersal of the southern green stink bug, Nezara viridula (L.) (Heteroptera: Pentatomidae), by hurricane Hugo. Proceedings of the Entomological Society of Washington. 1990;92:757–759. [Google Scholar]
- 27.Mourão APM, Panizzi AR. Photophase influence on the reproductive diapause, seasonal morphs, and feeding activity of Euschistus heros (Fabr., 1798) (Hemiptera: Pentatomidae) Brazilian Journal of Biology. 2002;62:231–238. doi: 10.1590/S1519-69842002000200006. [DOI] [PubMed] [Google Scholar]
- 28.Link D, Grazia J. Pentatomídeos da região central do Rio Grande do Sul (Heteroptera) Anais da Sociedade Entomológica do Brasil. 1987;16:115–129. [Google Scholar]
- 29.Panizzi AR. Stink bugs on soybean in northeastern Brazil and a new record on the southern green stink bug, Nezara viridula (L.) (Heteroptera: Pentatomidae) Neotropical Entomology. 2002;31:331–332. doi: 10.1590/S1519-566X2002000200023. [DOI] [Google Scholar]
- 30.Soria MF, Degrande PE, Panizzi AR. Algodoeiro invadido. Revista Cultivar. 2010;131:18–20. [Google Scholar]
- 31.Smaniotto LF, Panizzi AR. Interactions of selected species of stink bugs (Hemiptera: Heteroptera: Pentatomidae) from leguminous crops with plants in the Neotropics. Florida Entomologist. 2015;98:7–17. doi: 10.1653/024.098.0103. [DOI] [Google Scholar]
- 32.Rolston LH. Revision of the genus Euschistus in Middle America (Hemiptera, Pentatomidae, Pentatomini). Revisión del género Euschistus en Centroamérica (Hemiptera, Pentatomidae, Pentatomini) Entomologica Americana. 1974;48:1–102. [Google Scholar]
- 33.Panizzi, A. R. et al. Insetos da soja no Brasil. EMBRAPA Soja – Boletim de Pesquisa e Desenvolvimento (INFOTECA-E) (1977).
- 34.Saluso A, Xavier L, Silva FAC, Panizzi AR. An invasive pentatomid pest in Argentina: Neotropical brown stink bug, Euschistus heros (F.) (Hemiptera, Pentatomidae, Pentatomini) Neotropical Entomology. 2011;40:704–705. [PubMed] [Google Scholar]
- 35.Rolston LH. Revision of the genus Euschistus (Hemiptera: Pentatomidae) in Middle America. Entomologica Americana. 1974;48:1–102. [Google Scholar]
- 36.Coyne, J. A. & Orr, H. A. Speciation. Sinauer, (Sunderland, MA, 2004).
- 37.Sedghifar A, Brandvain Y, Ralph P. Beyond clines: lineages and haplotype blocks in hybrid zones. Molecular Ecology. 2016;25:2559–2576. doi: 10.1111/mec.13677. [DOI] [PubMed] [Google Scholar]
- 38.Haywood AM, Valdes PJ. Vegetation cover in a warmer world simulated using a dynamic global vegetation model for the Mid-Pliocene. Palaeogeography, Palaeoclimatology, Palaeoecology. 2006;237:412–427. doi: 10.1016/j.palaeo.2005.12.012. [DOI] [Google Scholar]
- 39.Bermingham E, Martin AP. Comparative mtDNA phylogeography of neotropical freshwater fishes: testing shared history to infer the evolutionary landscape of lower Central America. Molecular Ecology. 1998;7:499–517. doi: 10.1046/j.1365-294x.1998.00358.x. [DOI] [PubMed] [Google Scholar]
- 40.Da Silva MNF, Patton JL. Molecular phylogeography and the evolution and conservation of Amazonian mammals. Molecular Ecology. 1998;7:475–486. doi: 10.1046/j.1365-294x.1998.00276.x. [DOI] [PubMed] [Google Scholar]
- 41.Santos MG, Nogueira C, Giugliano LG, Colli GR. Landscape evolution and phylogeography of Micrablepharus atticolus (Squamata, Gymnophthalmidae), an endemic lizard of the Brazilian Cerrado. Journal of Biogeography. 2014;41:1506–1519. doi: 10.1111/jbi.12291. [DOI] [Google Scholar]
- 42.Garcia MG, et al. Molecular evidence of cryptic speciation, historical range expansion, and recent intraspecific hybridization in the Neotropical seasonal forest tree Cedrela fissilis (Meliaceae) Molecular Phylogenetics and Evolution. 2011;61:639–649. doi: 10.1016/j.ympev.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 43.Carnaval AC, Moritz C. Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. Journal of Biogeography. 2008;35:1187–1201. doi: 10.1111/j.1365-2699.2007.01870.x. [DOI] [Google Scholar]
- 44.Carnaval AC, Hickerson MJ, Haddad CF, Rodrigues MT, Moritz C. Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science. 2009;323:785–789. doi: 10.1126/science.1166955. [DOI] [PubMed] [Google Scholar]
- 45.Bremer JRA, Viñas J, Mejuto J, Ely B, Pla C. Comparative phylogeography of Atlantic bluefin tuna and swordfish: the combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Molecular Phylogenetics and Evolution. 2005;36:169–187. doi: 10.1016/j.ympev.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 46.Marí-Mena N, Lopez-Vaamonde C, Naveira H, Auger-Rozenberg MA, Vila M. Phylogeography of the Spanish moon moth Graellsia isabellae (Lepidoptera: Saturniidae) BMC Evolutionary Biology. 2016;16:139. doi: 10.1186/s12862-016-0708-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corrêa AS, Vinson CC, Braga LS, Guedes RNC, de Oliveira LO. Ancient origin and recent range expansion of the maize weevil Sitophilus zeamais, and its genealogical relationship to the rice weevil S. oryzae. Bulletin of Entomological Research. 2017;107:9–20. doi: 10.1017/S0007485316000687. [DOI] [PubMed] [Google Scholar]
- 48.Laurance WF, Sayer J, Cassman KG. Agricultural expansion and its impacts on tropical nature. Trends in Ecology & Evolution. 2014;29:107–116. doi: 10.1016/j.tree.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Tyukmaeva VI, Salminen TS, Kankare M, Knott KE, Hoikkala A. Adaptation to a seasonally varying environment: a strong latitudinal cline in reproductive diapause combined with high gene flow in Drosophila montana. Ecology and Evolution. 2011;1:160–168. doi: 10.1002/ece3.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hut RA, Paolucci S, Dor R, Kyriacou CP, Daan S. Latitudinal clines: an evolutionary view on biological rhythms. Proceedings of the Royal Society of London B: Biological Sciences. 2013;280:20130433. doi: 10.1098/rspb.2013.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paolucci S, Zande L, Beukeboom LW. Adaptive latitudinal cline of photoperiodic diapause induction in the parasitoid Nasonia vitripennis in Europe. Journal of Evolutionary Biology. 2013;26:705–718. doi: 10.1111/jeb.12113. [DOI] [PubMed] [Google Scholar]
- 52.Ruberson JR, Yeargan KV, Newton BL. Variation in diapause responses between geographic populations of the predator Geocoris punctipes (Heteroptera: Geocoridae) Annals of the Entomological Society of America. 2001;94:116–122. doi: 10.1603/0013-8746(2001)094[0116:VIDRBG]2.0.CO;2. [DOI] [Google Scholar]
- 53.Pegoraro M, et al. Geographical analysis of diapause inducibility in European Drosophila melanogaster populations. Journal of Insect Physiology. 2017;98:238–244. doi: 10.1016/j.jinsphys.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 54.C N de Abastecimento – CONAB. S H de Á Plantada, Produtividade e Produção, Relativas às Safras 1976/77 a 2015/16 de soja http://www.conab.gov.br/conteudos.php?a=1252 (2017).
- 55.Lombaert E, et al. Bridgehead effect in the worldwide invasion of the biocontrol harlequin ladybird. PloS one. 2010;5:e9743. doi: 10.1371/journal.pone.0009743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rius M, Darling JA. How important is intraspecific genetic admixture to the success of colonising populations? Trends in Ecology & Evolution. 2014;29:233–242. doi: 10.1016/j.tree.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Sosa-Gómez DR, et al. Insecticide susceptibility of Euschistus heros (Heteroptera: Pentatomidae) in Brazil. Journal of Economic Entomology. 2009;102:1209–1216. doi: 10.1603/029.102.0346. [DOI] [PubMed] [Google Scholar]
- 58.Clark TL, Meinke LJ, Foster JE. Molecular phylogeny of Diabrotica beetles (Coleoptera: Chrysomelidae) inferred from analysis of combined mitochondrial and nuclear DNA sequences. Insect Molecular Biology. 2001;10:303–314. doi: 10.1046/j.0962-1075.2001.00269.x. [DOI] [PubMed] [Google Scholar]
- 59.Ji YJ, Zhang D’X, He LJ. Evolutionary conservation and versatility of a new set of primers for amplifying the ribosomal internal transcribed spacer regions in insects and other invertebrates. Molecular Ecology Resources. 2003;3:581–585. doi: 10.1046/j.1471-8286.2003.00519.x. [DOI] [Google Scholar]
- 60.Lopez JV, Yuhki N, Masuda R, Modi W, O’Brien SJ. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. Journal of Molecular Evolution. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leigh JW, Bryant D. POPART: full‐feature software for haplotype network construction. Methods in Ecology and Evolution. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 63.Librado P, Rozas J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 64.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lanfear R, Calcott B, Ho SY, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 66.Papadopoulou A, Anastasiou I, Vogler AP. Revisiting the insect mitochondrial molecular clock: the mid-Aegean trench calibration. Molecular Biology and Evolution. 2010;27:1659–1672. doi: 10.1093/molbev/msq051. [DOI] [PubMed] [Google Scholar]
- 67.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 68.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology & Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 69.Anderson, D. R. Information Theory and Entropy. Model Based Inference in the Life Sciences: A Primer on Evidence, 51–82 (2008).
- 70.Burnham, K. P. & Anderson, D. R. Model selection and multimodel inference: a practical information-theoretic approach. Springer Science & Business Media (2003).
- 71.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 72.Conceição, A. O. A expansão da soja no Rio Grande do Sul 1975-75. (Dissertation). Porto Alegre. Universidade Federal do Rio Grande do Sul. FEE (1986).
- 73.Pissinato, B. A cultura de cana-de-açúcar no Estado de São Paulo entre 1950 e 2010: evolução histórica da área e da produtividade. (Dissertation) Universidade de São Paulo (2013).
- 74.Calcagno V, de Mazancourt C. glmulti: an R package for easy automated model selection with (generalized) linear models. Journal of Statistical Software. 2010;34:1–29. doi: 10.18637/jss.v034.i12. [DOI] [Google Scholar]
- 75.Johnson JB, Omland KS. Model selection in ecology and evolution. Trends in Ecology & Evolution. 2004;19:101–108. doi: 10.1016/j.tree.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 76.Wagenmakers EJ, Farrell S. AIC model selection using Akaike weights. Psychonomic Bulletin & Review. 2004;11:192–196. doi: 10.3758/BF03206482. [DOI] [PubMed] [Google Scholar]
- 77.Ab’Saber AN. Domínios morfoclimáticos e províncias fitogeográficas do Brasil. Orientação. 1967;3:45–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.