Abstract
Post-translational modifications of the transcription elongation complex provide mechanisms to fine-tune gene expression, yet their specific impacts on RNA polymerase II regulation remain difficult to ascertain. Here, in Schizosaccharomyces pombe, we examine the role of Cdk9, and related Mcs6/Cdk7 and Lsk1/Cdk12 kinases, on transcription at base-pair resolution with Precision Run-On sequencing (PRO-seq). Within a minute of Cdk9 inhibition, phosphorylation of Pol II-associated factor, Spt5 is undetectable. The effects of Cdk9 inhibition are more severe than inhibition of Cdk7 and Cdk12, resulting in a shift of Pol II toward the transcription start site (TSS). A time course of Cdk9 inhibition reveals that early transcribing Pol II can escape promoter-proximal regions, but with a severely reduced elongation rate of only ~400 bp/min. Our results in fission yeast suggest the existence of a conserved global regulatory checkpoint that requires Cdk9 kinase activity.
While post-translational modifications of the RNA polymerase II complex regulate gene expression, the exact mechanism remains unknown. Here, the authors use altered-specificity kinase mutations to examine the role of Cdk9, Mcs6/Cdk7, and Lsk1/Cdk12 on transcription at base-pair resolution in yeast.
Introduction
Multiple kinases modify RNA polymerase II (Pol II) and associated pausing and elongation factors to regulate Pol II transcription and pre-mRNA processing1,2. For example, the concerted action of Cdk7, Cdk9, and Cdk12 are required for the early transition of Pol II from an initiating to a productively elongating and RNA processing complex3–6. Cdk7, the kinase subunit of the TFIIH general transcription factor complex (Mcs6 in Schizosaccharomyces pombe, Cdk7 in humans, Kin28 in S. cerevisiae), phosphorylates the largest subunit of Pol II within the C-terminal domain (CTD) at the ser5 and ser7 positions of heptad amino acid sequence, YSPTSPS, which is repeated 29 times in S. pombe7–9. The modification of Pol II by Mcs6 is critical for recruitment of the 5′ RNA capping machinery and release of the mediator complex at the earliest stages of gene transcription10–13. Ser2 of the CTD heptad repeat is also phosphorylated as Pol II elongates further into the gene body14, and both Cdk9 and Lsk1 (Cdk12 in humans, Ctk1 in S. cerevisiae) have been suggested to modify this residue, possibly to facilitate 3′ RNA processing15.
Similar to the Pol II CTD, targets within elongation factors can also mediate essential regulation of transcription elongation. Spt4 and Spt5 comprise a highly conserved elongation factor (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB)-sensitivity inducing factor, or DSIF in metazoans)16. Spt5 possesses an unstructured C-terminal repeat (CTR) domain, with specific residues actively targeted by Cdk917. Moreover, the structure of eukaryotic Spt4/5 bound to transcribing RNA Pol II reveals contacts with upstream DNA and nascent RNA exiting Pol II18, and depletions of Spt5 in fission yeast globally reduce levels of elongating Pol II19. Cdk9 (Bur1 in S. cerevisiae) positively impacts transcription elongation20, and has been suggested to convert Spt5 from a negative to positive elongation factor in humans21. Such observations may reflect regulated recruitment of elongation-coupled factors, as association of capping enzymes and polymerase-associated factor 1 (Paf1) with the transcription complex are influenced by the phosphorylation status of Spt522–25.
In most metazoan systems, the phosphorylation of Spt5 and negative elongation factor (NELF) by positive transcription elongation factor b (P-TEFb, the Cdk9/cyclin T1 complex), is thought to be required for the release of elongating Pol II from a promoter-proximal pause site26,27. This Cdk9-regulated process (referred to hereafter as pausing) is now known to occur on the vast majority of genes in mammals28, serving to regulate gene expression, or to ensure appropriate processing of RNA and/or maturation of the elongation complex29. In budding yeast, which lack homologs of all NELF subunits, such pausing has not been observed, yet perturbation of Cdk9 and Spt5 impair normal transcription by Pol II20,30,31. In contrast, pause-like distributions of Pol II have been found in the fission yeast S. pombe, where they depend on Spt432. In addition, S. pombe Cdk9 interacts with the triphosphatase component of the 5′ RNA capping apparatus (Pct1), possibly to alleviate a Spt4-5-induced checkpoint that ensures proper pre-mRNA processing17. Still, such a checkpoint in yeast remains largely speculative, and the relation to Cdk9-regulated pausing in metazoans is unknown.
Investigations of the direct influence of kinase activity on the dynamics and regulation of transcription in vivo requires highly selective inhibitors, which can be difficult to obtain. Mutant kinases have been designed with exquisite specificity for bulky ATP analogs that selectively inhibit their activity33. The use of such analog-sensitive (AS) strains in fission yeast revealed correlated changes in global mRNA levels upon inhibition of Mcs6 or Cdk9 within minutes of inhibition34. Inhibitions of an AS TFIIH kinase resulted in a shift of Pol II levels toward the 5′ ends of genes in budding yeast after 1 h35 and impaired pausing and termination in humans after a day of inhibition3,36. However, even within just 1 h, the primary effects of kinase inhibition on Pol II dynamics may be masked, especially in yeast where genes are short and transcribed in a matter of minutes37.
In this study, to gain an insight into the impact of transcription complex-targeting kinases on Pol II elongation, we use AS kinase mutants in the fission yeast S. pombe to rapidly and selectively inhibit the activities of Cdk9, Mcs6, and Lsk1 individually or in combination. Using precision run-on sequencing analysis, we observe global changes in transcriptional dynamics within minutes of treatment, indicative of the general influence of these kinases on transcription by Pol II. While almost no instantaneous changes in Pol II distribution result from the loss of lsk1as activity, inhibition of cdk9as and/or mcs6as dramatically alter the transcription landscape within 5 min, despite non-overlapping substrates. Spt5 phosphorylation becomes undetectable after just 1 min of cdk9as inhibition; however, unlike metazoans, elongating Pol II is not trapped at the promoter-proximal pause site28. Instead, a fine-scale time course of cdk9as inhibition reveals that promoter-proximal Pol II has a severely compromised elongation rate, while downstream Pol II appears less affected. Our results support the existence of a Cdk9-dependent early elongation checkpoint in fission yeast, which may reflect a primitive form of Cdk9-regulated promoter-proximal pausing in metazoans.
Results
Cdk9 is not required for promoter-proximal pause escape
Recently, we reported promoter-proximal pause-like distributions of Pol II on many genes in fission yeast32, yet whether escape from such pausing is regulated through kinase activity is not known. In metazoans, kinase inhibitors, including DRB and flavopiridol (FP), which primarily inhibit Cdk9, have been instrumental in the discovery of promoter-proximal pausing as a major regulatory hurdle for most genes4,26–28. However, off-target effects of chemical inhibitors make selective ablation of Cdk9 activity in vivo nearly impossible38.
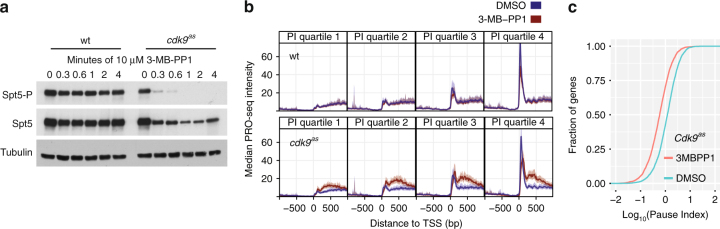
To isolate the immediate impact of loss of Cdk9 activity on Pol II pausing and elongation at single-nucleotide resolution in fission yeast, we performed PRO-seq in an AS mutant strain, cdk9as, which is vulnerable to inhibition by the addition of 3-MB-PP134,39. Phosphorylation of Spt5 became undetectable after 1 min of drug addition in cdk9as (Fig. 1a), while bulk measurements of phosphorylation on Pol II CTD residues were largely unaffected, within the timeframe tested (Supplementary Fig. 1a). The near-immediate loss of Spt5 phosphorylation upon cdk9as inhibition implies an extremely rapid action of the small molecule 3-MB-PP1 and supports a model of active, competing Spt5 dephosphorylation40. Taking advantage of the ability to rapidly inhibit AS kinase activity, PRO-seq libraries were prepared in two biological replicates (Supplementary Fig. 1b) from cdk9as cells treated for 5 min with 10 µM 3-MB-PP1 (treated) or an equivalent volume of DMSO (untreated). As a control, we prepared PRO-seq libraries from equivalently treated wild-type (wt) cells. Comparison of untreated cdk9as profiles with wt profiles revealed no obvious differences (Supplementary Fig. 2a) and addition of 3-MB-PP1 to wt cells produced almost no transcriptional changes (Supplementary Fig. 2b), indicating that this system has little to no basal phenotype of the AS mutation or off-target drug effects. To evaluate the effect of Cdk9 on promoter-proximal pause-like distributions, genes were divided into quartiles based on their pausing index (PI), which measures the enrichment of Pol II in the promoter-proximal region (TSS to +100 nt) relative to the gene body (“+” indicates downstream and “−” indicates upstream). Although we observed striking increases in early gene body PRO-seq signal (TSS to +1 kb) resulting from 5 min of cdk9as inhibition, pausing did not explain this effect (Fig. 1b).
Fig. 1.
Cdk9 is not required for promoter-proximal pause escape. a Western blot analysis using antibodies that specifically recognize pSpt5 compared with total Spt5 in wt and cdk9as over a time course of cultures treated with 10 μM 3-MB-PP1. Total Spt5 represents both phosphorylated and unphosphorylated forms. Tubulin serves as a loading control. b TSS-centered composite profiles of PRO-seq data before (blue) and after (red) 5 min of treatment with 10 μM 3-MB-PP1 for genes grouped by quartiles of increasing pausing index from left to right (calculated using untreated wild-type data from combined replicates). The top four panels represent wild-type data, while the bottom four panels represent cdk9as data. Each quartile contains 841 filtered genes. c Cumulative density functions for pausing index (log10) of all filtered genes in treated (red) and untreated (blue) samples for the cdk9as strain. Data used in b and c reflect the results of combined data from two biological replicates for each treatment
Inhibition of pause escape in mammals with drugs that target Cdk9 is known to result in an increase in engaged, promoter-proximal Pol II coinciding with loss of signal from the gene body28,41, yielding a greater PI for many genes. In contrast, we find few genes with significant increases in promoter-proximal Pol II upon Cdk9 inhibition (Supplementary Fig. 2c, d). Moreover, compared to wt, cdk9as cells exhibit a global decrease in PI as a result of 5 min of treatment with 3-MB-PP1 (Fig. 1c, Supplementary Fig. 2e). Together, our results reveal that Cdk9 activity in S. pombe is not required for release of promoter-proximal Pol II into elongation, but it is nonetheless critical for efficient elongation across gene bodies.
Mcs6 and Cdk9 impact Pol II at 5′ and 3′ ends of genes
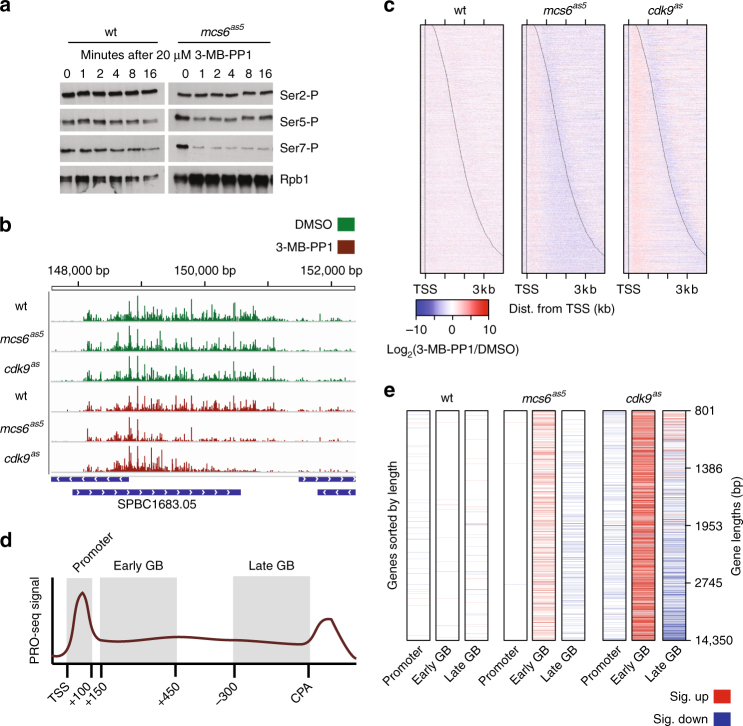
CTD modification by transcription-coupled kinases, Mcs6 and Lsk115, can modulate the association of auxiliary components with Pol II, to facilitate co-transcriptional RNA processing2 and elongation through chromatin42. Thus, in addition to Cdk9, we set out to measure the impact of Mcs6 and Lsk1 on global transcription. A novel AS variant of Mcs6 (here called mcs6as5) was as sensitive to treatment with 3-MB-PP1 as cdk9as (Supplementary Fig. 3a, b). Importantly, no gross transcriptional differences were observed in untreated mcs6as5 compared with wt (Supplementary Fig. 2a). Within minutes of treatment, we observed measurable losses in Ser5-P and Ser7-P in the mcs6as5 strain, while these marks in wt cells were unaffected (Fig. 2a). For consistency, PRO-seq experiments with mcs6as5 were performed using 10 μM 3-MB-PP1, which reduces Ser5-P levels to the same extent as 20 μM of the inhibitor (Supplementary Fig. 3b). The AS Lsk1 (lsk1as) was also inhibited by 10 μM 3-MB-PP1 (Supplementary Fig. 3c). However, despite the clear impact on Ser2 phosphorylation (Supplementary Fig. 4a), there was almost no visible change in transcription upon treatment of lsk1as with 10 μM 3-MB-PP1 for 5 min (Supplementary Fig. 4b–d). In contrast, loss of Mcs6 or Cdk9 activity produced abrupt changes at both ends of individual genes (Fig. 2b). Most notably in cdk9as, Pol II signal appeared to decrease with increasing distance from the transcription start sites (TSS) after inhibition. We also profiled distributions of Pol II in strains containing combinations of AS kinases. Combined inhibition of either lsk1as and cdk9as, or mcs6as5 and cdk9as produced transcriptional defects resembling those of cdk9as or mcs6as5 alone, although possible basal effects of the combined mutations limited further interpretation from these strains (Supplementary Fig. 5).
Fig. 2.
Mcs6 and Cdk9 impact Pol II at 5′ and 3′ ends of genes. a Western blot analysis of phosphorylated CTD residues (Ser2-P, Ser-5-P, Ser7-P) in relation to total CTD signal. Levels were measured over a time course of cultures treated with 20 μM 3-MB-PP1. b Browser track image from the SPBC1683.05 locus displaying normalized read counts from untreated (green) and treated data (red) for wild-type, mcs6as5, and cdk9as strains from top to bottom, respectively (plus strand only). All PRO-seq samples represent treatments for 5 min with 10 μM 3-MB-PP1 (treated) or equivalent volume of DMSO (untreated). c Heat maps depicting the log2 fold change in normalized PRO-seq signal (treated/untreated) within 10 bp bins from −250 bp to +4000 bp relative to the TSS (“+” indicates downstream and “−” indicates upstream) for wild-type, mcs6as5, and cdk9as strains from left to right, respectively. Data used in b and c reflect the results of combined data from two biological replicates for each treatment. Only one replicate was used for 3-MB-PP1-treated mcs6as5 (see Methods). Genes within each heat map are sorted by increasing length from top to bottom, with black lines representing observed TSS and CPS (n = 3383). d Hypothetical plot for the illustration of the definition of promoter (TSS to +100) early (+150 to +450 from TSS) and late (−300 to 0 from CPS) gene body regions used in e. e Heat maps depicting whether each gene exhibits a significant fold change (adjusted p < 0.01; treated/untreated; DESeq2: Wald test, Benjamini and Hochberg’s correction) in promoter, early, or late gene body regions. Genes were required to be longer than 800 nt (n = 3003) and are sorted from top to bottom by increasing gene length with length quartiles shown on the right. Significant increases and decreases in each region are shown as red and blue, respectively
Composite PRO-seq profiles around TSS and cleavage and polyadenylation sites (CPS) revealed global kinase-dependent effects at both ends of transcription units (Supplementary Fig. 5). Surprisingly, the loss of signal at 3′ ends of genes upon inhibition of Mcs6 or Cdk9 was only observed on longer genes (Fig. 2c, genes near the bottom of heatmaps). Differential expression analysis43 was used to verify differences in treated and untreated PRO-seq counts within three discrete regions of each gene: (1) the promoter-proximal region (TSS to +100), (2) the early gene body (+150 to +450 from the TSS), and (3) the late gene body (−300 to CPS) (Fig. 2d). Most strikingly in cdk9as, and to a lesser extent in mcs6as5, when genes were sorted by increasing length, distance from the TSS appeared to dictate the pattern of changes within the gene body regions. Although promoter-proximal regions were only subtly affected, early gene bodies were consistently found to show increases in signal in response to Cdk9 or Mcs6 inhibition. However, late gene bodies displayed differing results depending on the region’s distance from TSS, such that short genes exhibited increased signal and long genes had decreased signal within the last 300 bp of the transcription unit (Fig. 2e).
Several mechanisms might explain the observed gene length-dependent effects on transcription. Consistent with a loss of signal from the 3′ ends of longer genes (within bottom half of heatmaps in Fig. 2c), in the absence of cdk9as activity, Pol II may exhibit a compromised processivity, preventing transcription elongation beyond a certain distance from the TSS37. Alternatively, the altered transcription profiles may reflect a reduced rate of early transcribing ECs incapable, or delayed in, accelerating to the natural productive speed. Assuming an unchanged initiation rate, the increased density of elongating Pol II at the 5′ end would be compatible with a reduced elongation rate of Pol II, which within 5 min may not have allowed Pol II to transcribe fully across the longest genes.
A checkpoint during early elongation impacts Pol II rates
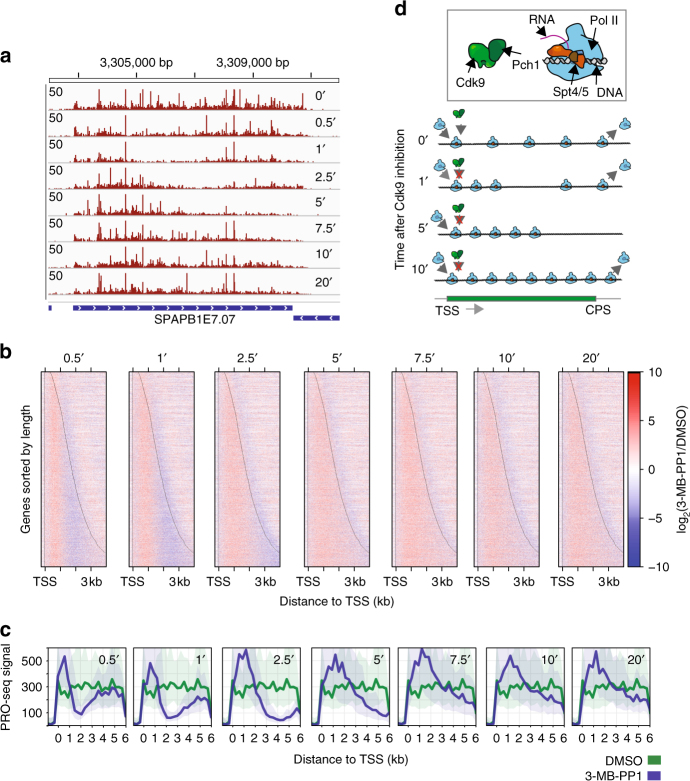
We reasoned that these proposed explanations—loss of processivity leading to premature termination vs. reduced elongation rate—could be resolved by examining a time course of Pol II distribution on genes following rapid Cdk9 inactivation. For instance, a defect in Pol II processivity would result in premature termination at roughly the same distance from the TSS, regardless of time after addition of 3-MB-PP1, whereas a rate defect might reveal a “wave front” shifting downstream with increasing time of inhibition. Thus, we performed a high-resolution time course of treatment with 3-MB-PP1 followed by PRO-seq, focusing specifically on the severe phenotype in cdk9as.
Strikingly, individual genes reproducibly exhibited a shifting distribution of transcribing Pol II, which appeared to propagate from the TSS toward the 3′ ends of genes over time (Fig. 3a, Supplementary Fig. 6a–c). Heat maps of the fold change after each duration of 3-MB-PP1 exposure, relative to untreated (DMSO), revealed an immediate global impact of Cdk9 inhibition on transcription that progressed as a spreading of increased signal across genes over time. Meanwhile, an early drop in signal at gene 3′ ends was gradually recovered with the advancing Pol II density (Fig. 3b).
Fig. 3.
A checkpoint during early elongation impacts Pol II rates. a Browser track image displaying the normalized PRO-seq read count signal at the SPBAPB1E7.07 locus for cdk9as cells treated with 10 μM 3-MB-PP1 for an increasing duration, from top to bottom. b Heat maps of log2(treated/untreated) normalized PRO-seq signal within 10 bp windows from −250 bp to +4000 bp around the TSS for all filtered genes (n = 3383) ordered by increasing gene length from top to bottom. Panels show PRO-seq data from each time point of drug treatment (relative to the 0-min sample), with increasing duration from left to right. c Composite PRO-seq signal for all filtered genes at least 6 kb in length (n = 42) before and after treatment. Panels from left to right show profiles after increasing duration of treatment compared with DMSO-treated cells. Data used in a–c reflect the results of combined data from two biological replicates for each treatment. d Illustration of two populations of transcribing Pol II, which have rates differentially affected by Cdk9 inhibition
To better understand the propagation of increased Pol II density following the initial loss in downstream signal, we prepared composite profiles at each time-point considering only the longest genes in our filtered set (at least 6 kb, n = 42). Intriguingly, average profiles for these genes reveal a rapid recession of Pol II density toward the CPS (Fig. 3c)—a phenomenon also obvious within individual genes (Fig. 3a; see 1-min and 2.5-min time points).
We reason that these dynamic profiles may represent two distinct populations of transcribing Pol II captured at the moment of cdk9as inhibition. One population had yet to transit a 5′ checkpoint dependent on Cdk9 activity and is thus impaired in elongation when Cdk9 is inhibited. The other population, already beyond this checkpoint, may no longer require Cdk9 to maintain its rate of transcription (Fig. 3d), which has previously been estimated in budding yeast to be ~2 kb/min37. Accordingly, these observations are indicative of a Cdk9-dependent regulatory step during the early stages of elongation in fission yeast (Fig. 3d), albeit not one fixed at a specific pause position, as is the case in mammals29.
Distinct effects of Cdk9 inhibition on Pol II rates
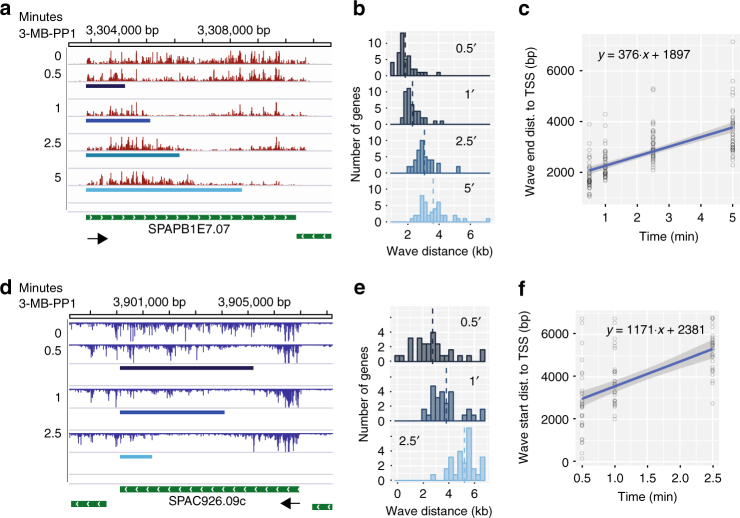
Gene activation and repression have been used to derive transcription kinetics in mammals by following changes in Pol II distributions over time28,44,45. By adapting a previously described model46, the high temporal and spatial resolution of our data allowed us to determine the distance covered by the advancing waves accurately, despite the comparatively short lengths of S. pombe genes (Fig. 4a). For 43 filtered genes longer than 4 kb we were able to estimate advancing wave distance at every time point from 30 s to 5 min based on the differences between treated and untreated signals within tiled windows across each gene (Supplementary Fig. 7a). Consistent with a forward moving population of Pol II, distance estimates increased with time of Cdk9 inhibition (Fig. 4b). Moreover, by fitting a linear regression to distance traveled over time elapsed, we determined that the average rate of transcription in this population was 376 bp/min (Fig. 4c, Supplementary Fig. 8a), much lower than previous estimates in yeast37.
Fig. 4.
Distinct effects of Cdk9 inhibition on Pol II rates. a Browser image displaying normalized PRO-seq signal reflecting combined data from two biological replicates for increasing lengths of cdk9as inhibition (top to bottom) over the SPBAPB1E7.07 locus with advancing wave estimates (blue rectangles) for each time point (0.5, 1, 2.5, and 5 min) plotted below the corresponding data. b Histograms of estimated advancing wave end distances (relative to TSS) for filtered genes (n = 43) at each time point after addition of 10 μM 3-MB-PP1 to cdk9as cells. Dotted, vertical lines represent the mean distance for each distribution. c Linear regression of distances traveled against time elapsed gives the estimated rate of advancing wave progression (slope). d Browser image showing clearing wave measurements (blue rectangles) for each time point plotted below the corresponding data at the SPAC926.09c locus. e Histograms of estimated clearing wave tail end distances (relative to TSS) for filtered genes (n = 32) at time points (0.5, 1, and 2.5 min) after cdk9as inhibition. f Linear regression of clearing wave distances traveled vs. time elapsed gives the estimated rate of clearing wave progression (slope)
Slower moving Pol II is thought to be an easier target for exonuclease-mediated termination, as the window within which Pol II terminates downstream of the CPS is directly related to transcription rates47. We therefore expected that on average, the slow population of polymerase, once downstream of the CPS would terminate within a shorter distance from the CPS. Indeed, analysis of global, post-CPS elongation after 20 min of Cdk9 inhibition shows a narrowed zone of termination (Supplementary Fig. 8b) and significantly reduced PRO-seq signal within this region when compared with untreated cells (Supplementary Fig. 8c, d).
The transient expansion of the region of Pol II clearance seen on the longest genes (Fig. 3c) suggests that Pol II further into the gene body is moving faster than the advancing wave upon loss of Cdk9 activity. To determine the rate of this “clearing” population of Pol II, we further adapted the Danko et al.46 model to identify the region between the advancing and clearing waves, which thus contains the start of the clearing wave (Supplementary Fig. 7b). Because the clearing population is fleeting and only present on the longest genes, we were forced to omit the 5-min time point and further restrict our analysis to 32 genes longer than 6 kb. Again, clearing wave positions predicted by the model agreed well with PRO-seq profiles (Fig. 4d) and receded away from the TSS on average (Fig. 4e). Moreover, the average transcription rate of clearing complexes was estimated to be 1171 bp/min (Fig. 4f), significantly faster than the advancing wave (Supplementary Fig. 8a). This rapid clearing of gene bodies is reminiscent of the effects of FP in mammals, where late transcribing polymerases continue rapidly to clear off genes, apparently unaffected by the drug28. Additionally, our results demonstrate disparate effects of Cdk9 inhibition on the elongation complex, dictated by its position relative to the TSS. Similar to the inability of metazoan Pol II to escape promoter proximal pausing in the absence of Cdk9 activity4, we observe a general requirement for Cdk9 at the 5′ ends of genes in S. pombe. These observations suggest a general, spatially localized requirement for Cdk9 activity on the elongation complex within the promoter-proximal region that is conserved in fission yeast. In the absence of this modification, most Pol II is able to transcribe the full length of even the longest genes, but suffers a significantly reduced elongation rate.
Discussion
Discerning how post-translational modifications of the elongation complex influence transcription is essential for understanding gene regulation, as these mechanisms are capable of tuning the output of gene expression29. The work presented here advances our understanding of three transcription-coupled kinases, Cdk9, Mcs6, and Lsk1, and their conserved roles in transcription elongation.
Unlike Cdk9 and Mcs6, Lsk1 is non-essential in S. pombe48, and loss of lsk1as activity had little impact on transcription elongation within 5 min, despite clearly affecting Ser2-P levels. Nonetheless, subtle reductions in elongating Pol II seen within the 3′ post-CPS termination region (Supplementary Fig. 4b) might be a symptom of inefficient recruitment of 3′ processing machinery15. On the other hand, 5-min inhibition of Cdk9, Mcs6, or both, produced highly related shifts in genome-wide Pol II distributions toward the 5′ ends of genes. Interestingly, modification of Pol II by Mcs6 has previously been suggested to enhance recruitment of Cdk9 in S. pombe34. Consequently, the inhibition of Mcs6 might indirectly inhibit Cdk9, producing a cdk9as-like effect on transcription. However, experiments in budding yeast using an irreversible AS Kin28 mutant produced altered transcription profiles similar to what we observe after 5 min in S. pombe, but a full hour after inhibition35.
As previously reported, we find that the most striking effect of inhibiting Cdk9 is a rapid reduction of Spt5 phosphorylation17. This near complete loss of Spt5 phosphorylation within minutes of cdk9as inhibition is consistent with the identification of a phosphatase opposing the activity of Cdk9 throughout the transcription cycle40. Genome-wide analysis of elongating Pol II within this timeframe of Cdk9 inhibition communicates several novel mechanistic insights into the possible role for this activity in transcription elongation. Cdk9 activity is not essential for escape of transcribing Pol II from the promoter-proximal region in S. pombe, but without Cdk9 activity these transcription complexes exhibit a severely impaired elongation rate. The progression of increased PRO-seq signal (compared with untreated cells) across the entire length of genes with increasing time of Cdk9 inhibition excludes premature termination as the sole explanation for effects on Pol II elongation. However, we cannot rule out a combination of defects in the elongation rate and processivity of affected Pol II, which might account for non-uniform increases in Pol II density seen at later time points after cdk9as inhibition (see Fig. 3c).
Surprisingly, our Pol II distributions upon loss of Spt5 phosphorylation differ notably from the depletions of total Spt5 in S. pombe19. Shetty et al.19 reported general reduction of transcribing Pol II within gene bodies after 2 h of Spt5 depletion. In contrast, continued inhibition of cdk9as (10–20 min) resulted in higher density across genes due to reduced elongation rate, similar to the phenotype of strains lacking the Spt5 binding partner Spt432. Importantly, we cannot rule out effects being a consequence of another known or unknown Cdk9 target. Considering that Cdk9 is capable of targeting additional transcription-coupled substrates, including the Pol II largest subunit CTD49, it is possible that the effects we observe are attributable to other targets, or combinations thereof. Alternatively, the discrepancy of effects on Pol II transcription may represent separable functions of Spt5. The depletion of Spt5 might directly influence transcription elongation and processivity through loss of known interactions with Pol II, template DNA, and the nascent RNA18,37,50, whereas inhibiting-specific phosphorylation of the unstructured, CTR of Spt5 might only affect transcription rate, perhaps by impairing recruitment of additional factors51.
The loss of Spt5 phosphorylation at the early stages of elongation may reduce association of factors, which help to negotiate nucleosomes52. Transcription elongation is known to both, modulate and be influenced by the chromatin environment23,53,54. For example, inability to recruit Paf1 and the facilitates chromatin transcription (FACT) complex could reduce the ability of the elongation complex to progress through nucleosomes55. Alternatively, Pol II slowing may occur to ensure recruitment of 5′ capping components, similar to pre-mRNA processing steps at gene 3′ ends22,56. Inhibiting phosphorylation of Spt5 might prevent or delay the association of capping enzymes, potentially reducing elongation rates across entire transcription units. Nonetheless, further work comparing the importance of Spt5 phosphorylation with other Cdk9 substrates will be crucial for understanding the mechanisms by which Cdk9 exerts influence over the kinetics of transcription elongation in fission yeast.
The observation that Pol II farther downstream of the TSS (~1 kb) was less, or possibly unaffected by Cdk9 inhibition appears directly comparable to clearing waves observed in metazoan systems following the inhibition of P-TEFb28. But whereas most other transcribing Pol II is trapped at the promoter-proximal pause site in flies and mammals4,28, these early gene body elongation complexes continue through the gene with severely impaired rates in S. pombe. We propose that NELF, which is a critical pausing factor in Drosophila and mammals57–59, may serve to exploit an early elongation checkpoint, observed here in fission yeast, creating a rate limiting, Cdk9-dependent, mechanism to control gene expression. Indeed, inhibiting the ability of NELF to associate with Pol II in flies has been found to reduce the effects of Cdk9 inhibition on pause intensity60. Further work will be required to determine if the four-subunit NELF complex is sufficient to induce Cdk9-regulated pausing in other eukaryotes, and whether reduced Pol II elongation rates would result from pause escape in the absence of both NELF and Cdk9.
Methods
Yeast strains
Yeast strains are listed in Supplementary Table 1.
Construction of mcs6as5 mutant fission yeast
We previously described mcs6 and cdk9 AS mutant strains, each with single substitutions of the gatekeeper residues, Leu87 and Thr120, respectively, with Gly34. Although growth of both strains was sensitive to 3-MB-PP1, the mcs6L87G strain was ~5-fold less sensitive than was the cdk9T120G strain34. By standard methods for gene replacement in S. pombe61, we introduced structure-guided second site mutations to attempt to optimize performance of mcs6as alleles62 and obtained a double-point mutant, mcs6N84T/L87G (referred to hereafter as mcs6as5), which had a growth rate indistinguishable from that of wild-type cells in the absence of 3-MB-PP1, and was sensitive to growth inhibition by 3-MB-PP1 with an IC50 ≈ 5 µM, which was nearly identical to that of cdk9as (Supplementary Fig. 3a).
Monitoring loss of target phosphorylation upon kinase inhibition
For measuring loss of phosphorylation on Spt5 and Rpb1 upon CDK inhibition, cell lysates were rapidly prepared using the trichloroacetic acid (TCA) lysis method63. Briefly, cultures were grown to a density of ~1.2 × 107 cells/mL in YES media at 30 °C prior to treatment. After the specified duration of treatment with of 3-MB-PP1 or DMSO, ~ 6 × 107 cells were transferred into 500 μl 100% (w/v) TCA and collected by centrifugation. Pellets were resuspended in 20% (w/v) TCA and protein extracts were prepared using a bead beater to mechanically lyse cells with glass beads. Antibodies used in this study recognized Spt5-P (used at 1:2000) or total Spt5 (used at 1:500)64, total Pol II (BioLegend, MMS-126R; used at 1:1000), Pol II Ser2-P (Abcam, ab5095; used at 1:1000), Pol II Ser5-P (BETHYL A304-408A; used at 1:1000), Pol II Ser7-P (EMD Millipore 04-1570; used at 1:1000), and α-tubulin (Sigma, T-5168; used at 1:5000). Raw images of all scanned western blots used in this work are provided (Supplementary Figs. 9–11).
Treatment of AS strains for PRO-seq
All samples were grown and treated in YES medium at 30 °C. Biological replicates were prepared by picking distinct colonies of a particular strain and growing them in separate liquid cultures. For each experiment, cultures were grown overnight and diluted to an optical density OD600 = 0.2. Diluted cultures were grown to mid-log phase (OD600 = 0.5–0.6), before treatments. Prior to treatment, all culture volumes were adjusted (based on OD600) to have an equal number of S. pombe cells (in 10 mL volume). After normalization for cell number, a fixed amount of thawed S. cerevisiae (50 µL OD600 = 0.68) culture was spiked in to each sample. Treatments were performed by adding either 10 µM 3-MB-PP1 (stock concentration = 40 mM) or an equivalent volume of DMSO. Sample treatments were terminated by pouring into 30 mL ice-cold water, treated with diethyl pyrocarbonate DEPC. Samples were immediately spun down and processed for cell permeabilization as described below (see PRO-seq section).
Time course experiments
The time course experiment was performed in two biological replicates, where all time points for a given replicate was performed on cells from the same large culture. Each time point treatment was performed as described above. In order to limit differences in time on ice before permeabilization, treatment time points were carried out in reverse order. The 2.5, 1, and 0.5-min treatments were performed separately to ensure accuracy of timing. The 0-min time point was treated with DMSO for 20 min to account for any possible effects of adding the solvent for the maximum duration.
PRO-seq
After experimental treatments and spiking in S. cerevisiae, cells were spun down and washed in cold H20. Cells were then permeabilized in 10 mL 0.5% sarkosyl at 4 °C, and incubated on ice for 20 min, then spun at a reduced RCF (400 × g) for 5 min at 4 °C. For run-on reactions, yeast pellets were resuspended in 120 μL of 2.5× run-on reaction buffer [50 mM Tris-HCl, pH 7.7, 500 mM KCL, 12.5 mM MgCl2] with 6 μL 0.1 M DTT and 3.75 μL of each 1 mM biotin-NTP added immediately before use. The volume of the run-on reaction mix was brought to 285 μL with DEPC-treated H20. 15 μL 10% sarkosyl was then added to the reaction and the run-on was allowed to run on for 5 min at 30 °C. RNA was extracted using a hot phenol approach65; after the run-on reaction cells were pelleted at 400 × g for 5 min at 4 °C and quickly resuspended in 500 μL acid phenol. An equal volume of AES buffer [50 mM NaAc, pH 5.3, 10 mM EDTA, 1% SDS] was added and placed at 65 °C for 5 min with periodic vortexing, followed by 5 min on ice. 200 μL chloroform was added and mixed followed by centrifugation at 14,000 × g for 5 min (4 °C). 3 M NaOAc was added to the aqueous layer (200 mM) followed by ethanol precipitation with 3× volume of 100% ethanol. The RNA pellet was air dried before being resuspended in 20 μL DEPC-treated water. The standard PRO-seq protocol66 was then followed, beginning with base hydrolysis, through to sequencing. For base hydrolysis of RNA, 5 μL 1 N NaOH was added to the resuspended RNA and left on ice for 10 min before being neutralized with 25 μL 1 M Tris-HCL pH 6.8. After passing hydrolyzed RNA through a P-30 column (Bio-Rad), RNA was incubated with streptavidin-conjugated magnetic beads (M280, Invitrogen) for 20 min at room temperature, then washed with 500 μL of each of the following: high salt solution (50 mM Tris-Cl, pH 7.4, 2 M NaCl, 0.5% Triton X-100), bead binding solution (10 mM Tris-Cl, pH 7.4, 300 mM NaCl, 0.1% Triton X-100), low salt solution (5 mM Tris-Cl, pH 7.4, 0.1% Triton X-100). Note that prior to incubation with sample RNA, M280 beads were washed once with a solution of 50 mM NaCl and 0.1 N NaOH, and then twice with 100 mM NaCl. After bead binding and washing, enriched nascent RNA was extracted using TRIzol (Ambion) and ethanol precipitated. An RNA adapter was then ligated to the 3′ ends of enriched RNAs (see below) using T4 RNA ligase 1 (NEB), followed by another round of bead binding, washing, and TRIzol extraction, as before. RNA 5′ ends were then biochemically prepared for adapter ligation by first removing any cap structures with RNA 5′ pyrophosphohydrolase (RppH, NEB), then restoring mono-phosphates to base hydrolyzed RNA 5′ ends with T4 polynucleotide kinase (T4 PNK, NEB). T4 RNA ligase 1 was then used to ligate the 5′ RNA adapter (5′-CCUUGGCACCCGAGAAUUCCA-3′) to enriched RNAs. Following another round of bead binding and washing, as before, cDNA was prepared by reverse transcription using superscript reverse transcriptase III (SSRTIII, Invitrogen) and the primer, RP1 (Illumina, TruSeq Small RNA Sample Prep oligos). Libraries were then amplified via PCR using Phusion (NEB) polymerase and Illumina oligos, RP1 and one indexed RNA PCR Primer (RPI-x; Illumina, TruSeq Small RNA Sample Prep oligos). PCR products were run on a native polyacrylamide gel, size selected (~130–400 bp), extracted, quantified, and submitted to sequencing. More details regarding preparing PRO-seq libraries can be found in the published protocol for precision run-on and sequencing66.
For specific batches of experiments slight modifications to the library preparation were made. Samples prepared with wt, mcs6as5, or cdk9as, excluding the time course experiment, were prepared according to the standard procedure described above66. All libraries prepared from lsk1as mutant and mcs6as5 cdk9as double mutant strains received a novel 3′ RNA adapter during the first RNA ligation step. This 3′ adapter contains a random 6 nt unique molecular identifier at the 5′ end that, although not used here, can be used during read processing to remove PCR duplicates (5′-/5Phos/NNNNNNGAUCGUCGGACUGUAGAACUCUGAAC/Inverted dT/-3′). For all time course samples prepared in cdk9as, as well as lsk1as cdk9as samples, a different 3′ RNA adapter was used. Here, the RNA oligonucleotide possessed a known hexanucleotide sequence preceded by a guanine at the 5′ end, which differed for each library prepared (5′-/5Phos/GNNNNNNGAUCGUCGGACUGUAGAACUCUGAAC-/Inverted dT/-3′). The ligation of this adapter with distinct barcodes to each library permitted the pooling of all libraries after the 3′ end ligation step and thus facilitated handling. After pooling, the remainder of the library preparation was carried out as described above, but in a single tube. After sequencing, the inline barcode was used to parse reads based on their sample of origin.
Alignment and data processing
All sequencing was performed on an Illumina NextSeq 500 device. If samples were pooled during the library preparation using the novel 3′ RNA adapter described above, raw reads were parsed into their respective samples based on the inline barcode using fastx_barcode_splitter function from the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Raw reads from each sample were processed by removing any instances of partial or complete matches to the 5′ adapter sequence (5′-TGGAATTCTCGGGTGCCAAGG-3′) with the fastx_clipper function. Next, if a 3′ molecular, or sample barcode was included during 3′ adapter ligation, this length was removed from the beginning of each read. All reads were then trimmed to a maximum remaining length of 36 nt using the fastx_trimmer function. With the fastx_reverse_complement function we retrieved the reverse complement sequence of each read. All downstream alignments were performed using Bowtie (version 1.0.0)67. Reverse complemented reads derived from ribosomal RNA genes were then removed through alignment to a genome consisting of ribosomal RNA genes from Saccharomyces cereivisiae (spike-in) and then S. pombe. To parse reads based on organism of origin, while eliminating reads of ambiguous origin, non-ribosomal DNA reads were then aligned to a combined genome, containing each chromosome from S. cerevisiae (sacCer3 = S288C_reference_genome_R64-1-1_20110203) and S. pombe (version: ASM294v2). Only uniquely mapping reads were considered for downstream analysis. Ultimately, the normalization factor for each library was calculated as: total spike-in mapped reads/105. Unique read alignments to the fission yeast genome were processed using the Bedtools68 function, genomeCoverageBed, to generate bedgraph formatted files, reporting the number of read 3′ ends (the last base incorporated) at each position across the genome. Bedgraph files were further converted to bigwig format for downstream analysis. Sequencing, alignment, and batch information for each sample can be found in Supplementary Table 2.
Combined replicates
Biological replicates correlated very well with spike-in normalization centering gene-by-gene scatter around the line x = y (Supplementary Fig. 2), allowing us to combine the raw data from replicates (pre-alignment) for added read depth when useful. In one particular sample, mcs6as5 treated with 3-MB-PP1 for 5 min, we ultimately omitted biological replicate one (Supplementary Table 2) from all analysis in this work, including composite profiles, due to concerns with the library quality. Importantly, limiting differential expression analysis between treated and untreated samples to only one replicate in the treated condition reduces power to detect differences due to overestimates of variance43. Although the number of real changes in the tested gene regions after inhibition of mcs6as may be higher than listed in Fig. 2e, the reported global shifts in Pol II distribution can be seen in individual replicates.
Experimental batches
Three “batches” of experiments were performed to generate the data in this work (Supplementary Table 2). In order to minimize noise introduced from across-batch comparisons, all analyses presented herein were restricted to PRO-seq libraries prepared within the same batch.
Gene sets
Using previously published PRO-cap data32, we re-annotated TSS as the position with the maximum amount of background-subtracted PRO-cap signal within the region −250 to +250 around existing annotations (version: ASM294v1.16). The selected base was required to have >4 reads over background, while accounting for library size (annotations available at https://github.com/gregtbooth/Pombe_PROseq). All genes used in this work were required to have an observed TSS (n = 4654), and this observed TSS was used throughout. We then filtered genes for “activity” in our untreated, wild-type data. A gene was considered active if the read density within the gene body was significantly higher (p < 0.01) than a set of intergenic “background” regions, based on a Poisson distribution with λ = background density (λ = 0.0402; n active = 4576). Finally, to minimize the possible influence of read-through transcription from upstream genes, we filtered genes based on the relative amounts of PRO-seq signal directly upstream of the TSS. Thus, we only considered genes with more downstream (+250 to +550) relative to immediately upstream (−300 to observed TSS; n = 3383).
Differential expression analysis
Raw reads from the appropriate strand were counted within specified windows using custom scripts. Differential expression analysis was performed for desired regions using the DESeq2 R package43. Rather than using default between-sample normalization approaches, we supplied our own spike-in-based normalization factors (Supplementary Table 2) using the “sizeFactors” argument. Genes and regions were considered significantly changed (up or down) if they were computed to have an adjusted p-value < 0.01. To compute adjusted p-values for changes in counts DESeq2 uses the Wald test with Benjamini and Hochberg’s correction43.
Advancing and clearing wave analysis
Both advancing and clearing waves were identified using a three state Hidden Markov Model (HMM) that was previously developed and implemented on GRO-seq data from a human cell line46. To increase the number of time points for which we could identify advancing waves, we restricted our analysis to calling waves on filtered genes longer than 4 kb (n = 231) at each time point up to 5 min. Wave calling parameters were adjusted to accommodate the smaller genes of yeast. For calling advancing waves, the upstream region used was set to −500 to the observed TSS for each gene. Approximate wave distances, used to initialize the model, were set based on estimates derived from inspecting the fold-change heat maps (30 s = 1 kb; 1 min = 1.5 kb; 2.5 min = 2.5 kb; 5 min = 3.5 kb). Wave distances were determined based on the difference in signal within 50-bp windows between treated and untreated. The wave quality was determined based on the criteria used previously46. We further required that for each gene, a wave must be called for all used time points (maximum 5 min) using the combined replicate data, and that the calls must not recede toward the TSS with time (n = 43).
To identify regions containing the faster population of Pol II (i.e., the clearing wave), we adjusted the HMM to identify the region between the advancing and clearing waves. For this task, we used the first 1000 nt downstream of the observed TSS (within the advancing wave) to initialize the first state of the HMM. Approximate distances of the start of the clearing wave from the TSS were used to initialize the model and were set based on estimates derived from inspecting the fold-change heat maps (30 s = 3 kb; 1 min = 4 kb; 2.5 min = 5.5 kb). Unlike the model used to call the advancing wave, which assumed the upstream state to be normally distributed, all three states were presumed to behave according to distinct gamma distributions. Because of the fleeting nature of this population of polymerases observed in our time course, we were further limited to analysis of filtered genes longer than 6 kb (n = 42). Again, 50-nt windows were used to tile across genes; however, an additional smoothing parameter in the model arguments (TSmooth) was set to 5 as a way of restricting the effect of windows with outlying differences between treated and untreated. For each gene, we required that the model be able to identify a distinct clearing wave (Kullback–Leibler divergence > 1, between states 2 and 3) at each time point that did not move toward the TSS with time (n = 32).
We employed two methods to assess the confidence in measured average rates. For the first, bootstrapping approach, an average rate was calculated using 10% of genes for which we called waves. This process was repeated 1000 times (with replacement), giving a distribution of means. A more conservative, gene-by-gene estimate of variance was derived simply from the distribution of rates based on individual measurements for each gene.
Code availability
Custom scripts and alignment pipelines have been made publicly available through the following GitHub repository; https://github.com/gregtbooth/Pombe_PROseq.
Data availability
The raw and processed sequencing files have been submitted to the NCBI Gene Expression Omnibus under accession GSE102308.
Electronic supplementary material
Acknowledgements
Research reported in this publication was supported by NIH grant GM104291 to R.P.F. and GM25232 to J.T.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Cornell Genomics Facility for overseeing the sequencing of our libraries. We thank Chao Zhang (Department of Chemistry, University of Southern California), who originally advised us to make the N84T mutation for the generation of the mcs6as5 strain. We are grateful for valuable discussions with Charles Danko and Hojoong Kwak regarding transcription wave calling and other computational methods.
Author contributions
G.T.B. performed PRO-seq experiments and corresponding data analysis. P.K.P. conducted and analyzed biochemical assays. M.S. created the mcs6as5 strain. G.T.B., P.K.P., R.P.F., and J.T.L. designed experiments and prepared the manuscript.
Competing interests
The authors declare no competing financial interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-03006-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buratowski S. The CTD code. Nat. Struct. Mol. Biol. 2014;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 2.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larochelle S, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni Z, et al. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bösken CA, et al. The structure and substrate specificity of human Cdk12/Cyclin K. Nat. Commun. 2014;5:115. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glover-Cutter K, et al. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 2009;29:5455–5464. doi: 10.1128/MCB.00637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhtar MS, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azuma, Y., Yamagishi, M., Ueshima, R. & Ishihama, A. Cloning and sequence determination of the Schizosaccharomyces pombe rpb1 gene encoding the largest subunit of RNA polymerase II. Nucleic Acids Res. 19, 461–468 (1991). [DOI] [PMC free article] [PubMed]
- 10.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez CR, et al. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 2000;20:104–112. doi: 10.1128/MCB.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong KH, Jin Y, Struhl K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell. 2014;54:601–612. doi: 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson PJJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA. 2012;109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 15.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei Y, Schwer B, Shuman S. Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. J. Biol. Chem. 2003;278:7180–7188. doi: 10.1074/jbc.M211713200. [DOI] [PubMed] [Google Scholar]
- 18.Ehara H, et al. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- 19.Shetty A, et al. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol. Cell. 2017;66:77–88.e5. doi: 10.1016/j.molcel.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keogh MC, Podolny V, Buratowski S. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23:7005–7018. doi: 10.1128/MCB.23.19.7005-7018.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada T, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 23.Zhou K, Kuo WHW, Fillingham J, Greenblatt JF. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl Acad. Sci. USA. 2009;106:6956–6961. doi: 10.1073/pnas.0806302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, et al. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell. Biol. 2010;30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 27.Wada T. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription invitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 1991;11:3009–3019. doi: 10.1128/MCB.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booth GT, Wang IX, Cheung VG, Lis JT. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 2016;26:799–811. doi: 10.1101/gr.204578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 34.Viladevall L, et al. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol. Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Molina JB, Tseng SC, Simonett SP, Taunton J, Ansari AZ. Engineered covalent inactivation of TFIIH-kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Mol. Cell. 2016;63:433–444. doi: 10.1016/j.molcel.2016.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebmeier CC, et al. Human TFIIH kinase CDK7 regulates transcription-associated chromatin modifications. Cell Rep. 2017;20:1173–1186. doi: 10.1016/j.celrep.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 38.Bensaude O. Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cipak L, et al. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle. 2011;10:3527–3532. doi: 10.4161/cc.10.20.17792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parua, P. K. et al. A Cdk9-PP1 kinase-phosphatase switch regulates the elongation-termination transition of RNA polymerase II. Preprint at https://www.biorxiv.org/content/early/2017/09/20/190488 (2017). [DOI] [PMC free article] [PubMed]
- 41.Cheng B, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Love, M., Anders, S. & Huber, W. Differential analysis of count data—the DESeq2 package. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed]
- 44.Mahat DB, Salamanca HH, Duarte FM. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol. Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hah N, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danko CG, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fong N, et al. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell. 2015;60:256–267. doi: 10.1016/j.molcel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karagiannis J, Balasubramanian MK. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p sub-unit. PLoS ONE. 2007;2:e433. doi: 10.1371/journal.pone.0000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J. Biol. Chem. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wier AD, Mayekar MK, Héroux A, Arndt KM, VanDemark AP. Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin. Proc. Natl Acad. Sci. USA. 2013;110:17290–17295. doi: 10.1073/pnas.1314754110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindstrom DL, et al. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bintu L, et al. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat. Struct. Mol. Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ardehali MB, et al. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adelman K, et al. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/S0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 58.Wu CH, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujinaga K, et al. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson BA, et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353:45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 62.Zhang C, et al. A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods. 2005;2:435–441. doi: 10.1038/nmeth764. [DOI] [PubMed] [Google Scholar]
- 63.Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/S1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 64.Sansó M, et al. A positive feedback loop links opposing functions of P-TEFb/Cdk9 and histone H2B ubiquitylation to regulate transcript elongation in fission yeast. PLoS Genet. 2012;8:e1002822. doi: 10.1371/journal.pgen.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collart, M. A. & Oliviero, S. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. doi: 10.1002/0471142727 (2001). [DOI] [PubMed]
- 66.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat. Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed sequencing files have been submitted to the NCBI Gene Expression Omnibus under accession GSE102308.