Abstract Abstract
The Ophiostomatales is an Ascomycete order of fungi that accommodates several tree pathogens and many species that degrade wood. These fungi are commonly vectored by Scolytine bark and ambrosia beetles. In recent years it has also been shown that hyperphoretic mites on these beetles can vector some Ophiostomatales. Little is known regarding the Ophiostomatales in China and we have consequently explored the diversity of these fungi associated with conifer-infesting beetles and mites in Yunnan province. Galleries and beetles were collected for 17 beetle species, while 13 mite species were obtained from six of these beetle species. Collectively, 340 fungal isolates were obtained, 45 from beetles, 184 from mites, 56 from galleries and 55 isolates where the specific niche was not clear. DNA sequences for five gene regions (ITS, LSU, BT, EF, and CAL) were determined for fungal isolates representing different morphological groups. Phylogenetic analyses confirmed the presence of 19 fungal taxa, including five novel species described here as Ophiostoma acarorum sp. nov., Ophiostoma brevipilosi sp. nov., Graphilbum kesiyae sp. nov., Graphilbum puerense sp. nov., and Leptographium ningerense sp. nov. Ophiostoma ips was the most frequently isolated species, representing approximately 31% of all isolates. Six of 19 taxa were present on mites, beetles and in the galleries of the beetles, while three species were found on mites and galleries. Two species were found only on mites and one species only on a beetle. Although the numbers of beetles and mites were insufficient to provide statistical inferences, this study confirmed that mites are important vectors of the Ophiostomatales in China. We hypothesize that these mites are most likely responsible for horizontal transfer of fungal species between galleries of different beetle species. The fact that half of the fungal species found were new to science, suggests that the forests of east Asia include many undescribed Ophiostomatales yet to be discovered.
Keywords: Symbiont, species diversity, fungal vector, Ophiostoma quercus
Introduction
The ophiostomatoid fungi represent a polyphyletic group of tree- or wood-infecting fungi, most often staining freshly exposed sapwood and thus lowering the value of timber (Seifert 1993). Some of the species are important tree pathogens that have dramatically impacted natural forests and caused major losses to forestry companies during the course of the last century (Harrington 1993, Wingfield et al. 2017). The sexual and often asexual spore-bearing structures of the ophiostomatoid fungi are specifically adapted for dispersal by arthropods, producing spores in sticky droplets that attach to the bodies of their vectors (Malloch and Blackwell 1993). However, these morphological and biological traits have evolved more than once in the Ascomycota and phylogenetic analyses have shown that these fungi reside in two different orders, the Ophiostomatales and Microascales (De Beer et al. 2013). The Ophiostomatales currently accommodates a single family, the Ophiostomataceae, that includes ten genera (Bateman et al. 2017, De Beer et al. 2016a, De Beer et al. 2016b, Van der Linde et al. 2016), while the Microascales comprises of five families, of which the Ceratocystidaceae, Gondwanamycetaceae and Graphiaceae include 14 genera of ophiostomatoid fungi (De Beer et al. 2014, 2017, Mayers et al. 2015, Nel et al. 2017).
Bark beetles (Coleoptera: Scolytinae) are well known vectors of ophiostomatoid fungi (Harrington 2005, Kirisits 2004, Six 2003). Other than a few bark beetle species that are considered to be primary pests causing significant economic losses, most species of bark beetles are secondary invaders, which colonize injured or stressed trees (Harrington 2005, Six and Wingfield 2011). Some bark beetle species have specialized structures known as mycangia that carry fungal conidia or ascospores, but most of these insects carry fungi on their exoskeletons (Six 2003). They construct their galleries, where the ophiostomatoid fungi grow, in the phloem and cambial layers under the bark of trees (Paine et al. 1997). The nature of the relationships between fungi and bark beetles is recognized as being very diverse. Some beetle-fungus symbioses appear to be obligate, while others may be facultative (Kirisits 2004). The role of the fungi can vary from their being nutritional symbionts of the beetles to saprophytes in the galleries, although in most cases the exact nature of the symbiosis is not well understood (Six and Wingfield 2011).
Along with the fungi, mites are also common symbionts of bark beetles (Hofstetter and Moser 2014, Roets et al. 2009). Due to their small body size and low mobility, many mite species rely on beetles or other animals for dispersal (Mitchell 1970). The relationship between mites and their vectors is usually commensal, but in some cases these are antagonistic or even mutualistic (Pfammatter et al. 2013). The phoretic mites of only a limited number of bark beetles have been studied in detail, including mostly North American species of Scolytus, Dendroctonus and Ips (Hofstetter et al. 2013).
As is the case with bark beetles, their mite associates can also vector ophiostomatoid fungi (Hofstetter and Moser 2014). Some mites such as Tarsonemus spp. possess sporothecae that are specialized structures of the integument that serve to carry fungal spores (Hofstetter et al. 2015). Ophiostomatoid fungi associated with mites include genera such as Ceratocystis, Knoxdaviesia, Ceratocystiopsis, Ophiostoma, Leptographium, Grosmannia and Sporothrix (Hofstetter et al. 2013). The interactions between mites and fungi are dynamic and vary with tree host condition and climate (Klepzig and Hofstetter 2011). It is for example known that mites can promote some fungal growth by increasing fungal transmission, reproduction, and survival, or by reducing the abundance of antagonistic fungi (Hofstetter and Moser 2014). Mites can also negatively affect some fungi because of exploitative/interference competition, predation, or encouragement of antagonistic fungi (Hofstetter et al. 2006).
Research on the interactions between ophiostomatoid fungi and bark beetles have been ongoing for more than a century in North America (Harrington 2005, Hedgcock 1906), Europe (Kirisits 2004, Münch 1907), and Japan (Masuya et al. 2013, Nisikado and Yamauti 1933). The associations of ophiostomatoid fungi with mites have been studied only in North America and Europe (Hofstetter and Moser 2014, Hofstetter et al. 2013), with a single report from Japan (Moser et al. 1997) and with ongoing studies on Protea spp. in Southern Africa (Roets et al. 2009, Roets et al. 2007). It is only more recently that the fungal associates of bark beetles have been explored in China, despite the fact that more than 160 bark beetle species have been recorded from that country (Yin et al. 1984).
The first new beetle associated ophiostomatoid species described from China for which the description was supported by DNA sequence data, was Grosmannia yunnanensis (Zhou et al. 2000, Yamaoka et al. 2008). Additional new species described during the past decade from a variety of conifer-infesting beetles in China, include one Graphium (Paciura et al. 2010a), 12 Leptographium (Liu et al. 2017, Lu et al. 2008, Paciura et al. 2010b, Yin et al. 2015), and five Ophiostoma (Yin et al. 2016) species. The most comprehensive study on the fungal associates of a specific beetle species in China has been on Dendroctonus valens, an economically important invasive bark beetle that was introduced to China from the USA and is attacking native pine species (Lu et al. 2008, 2009a, b, Taerum et al. 2013). Together with the fungal associates of D. valens, almost 40 ophiostomatoid species have been recorded and described from conifers in China (Liu et al. 2017, Paciura et al. 2010a, b, Yin et al. 2015, 2016, Zhou et al. 2013). Most of these are from bark beetles or their galleries. The fungal associates of mites have not been studied in this country (Hofstetter et al. 2013, Zhou et al. 2013).
Yunnan province forms part of in the southwestern forest zone, the second largest forest area in China (http://www.china.org.cn/english/shuzi-en/en-shuzi/gq/htm/zrzy-land-sl.htm). This province has a unique geography where three climatic regions meet: the eastern Asia monsoon region, the Tibetan plateau region, and the tropical monsoon region of southern Asia and Indo-China. As a result , species diversity in Yunnan province is high when compared to other parts of China. For example, more than 18000 plant species and 1836 vertebrate species are found here which represent 51.6% and 54.8% respectively of total species numbers in China (Yang et al. 2004). However, only 13 ophiostomatoid species have been reported from Yunnan (Zhou et al. 2013). Based on the high diversity of trees and vertebrates, but the relatively low number of ophiostomatoid species reported from Yunnan to date, we hypothesize that there are many undiscovered species from this group of fungi associated with bark beetles and mites in this province. The aim of this study was thus to further explore the diversity of ophiostomatoid fungi associated with conifer-infesting beetles and mites in Yunnan.
Material and methods
Collection of bark beetles and mites
Three surveys were conducted in Yunnan during the flight period of bark beetles in July 2001, July 2002, and between June and September 2010 (Table 1, Suppl. material 2: Figure S1). Surveys were conducted both in sawmill log-yards and forest areas. During the 2001 and 2002 surveys, only galleries and beetles were considered from pine species in conifer and mixed forests in central and northwestern Yunnan (Table 1, Suppl. material 2: Figure S1). During the 2010 survey, some collections were conducted using Pinus kesiya log traps in the pine forests around Pu’er city in southwestern Yunnan. Both adult bark beetles and their galleries were collected and stored in Eppendorf tubes and re-sealable bags, respectively. Mites were carefully removed from these beetles and galleries in the laboratory and individually placed in Eppendorf tubes. After the isolation of fungi, beetles and mites were stored in 75% ethanol for later identification. Beetles and mites were identified based on the morphology by Dr R. Beaver (Chiang Mai University, Thailand) and Dr E.A. Ueckermann (Plant Protection Research Institute, Agricultural Research Council, South Africa) respectively.
Table 1.
Conifer-infesting bark and ambrosia beetles (Scolytinae), weevils (Molytinae), true weevils (Cossoninae), cylindrical bark beetles (Colydiinae), and mites (Acari) collected from Yunnan in this study. Numbers in the table refer to number of mite individuals collected.
| Beetle species | Family | Subfamily | Tree host | Origin | Collection dates | Mite species | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | d | e | f | g | h | i | j | k | l | m | Total | |||||||
| A | Coccotrypes cyperi | Curculionidae | Scolytinae | Pinus kesiya | Ning’er | Sep. 2010 | 16 | 3 | 9 | 0 | 0 | 0 | 2 | 4 | 0 | 0 | 0 | 0 | 3 | 37 |
| B | Cyrtogenius luteus | Curculionidae | Scolytinae | P. kesiya | Ning’er | Sep. 2010 | 0 | 2 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
| C | Hylurgops major | Curculionidae | Scolytinae | Pinus yunnanensis | Zixishan | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D | Ips acuminatus | Curculionidae | Scolytinae | P. kesiya | Ning’er, Puer | Jun. 2010 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 8 |
| E | Lasconotus sp. | Zopheridae | Colydiinae | P. kesiya | Ning’er | Jun. 2010 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| F | Orthotomicus angulatus | Curculionidae | Scolytinae | P. kesiya | Ning’er | Sep. 2010 | 0 | 0 | 4 | 13 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 19 |
| G | Pissodes sp. | Curculionidae | Molytinae | Pinus armandii | Lijiang, Midu | Jul. 2001 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H | Polygraphus aterrimus | Curculionidae | Scolytinae | P. kesiya | Ning’er | Jun. 2010 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| I | Polygraphus sp. | Curculionidae | Scolytinae | P. kesiya | Ning’er | Jun. 2010 | 0 | 0 | 0 | 6 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| J | P. szemaoensis | Curculionidae | Scolytinae | P. kesiya | Ning’er, Simao | Jun. 2010 | 0 | 0 | 0 | 14 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 1 | 20 |
| K | P. verrucifrons | Curculionidae | Scolytinae | P. yunnanensis | Lufeng | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L | Stenoscelis sp. | Curculionidae | Cossoninae | P. kesiya | Ning’er | Jun. 2010 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| M | Tomicus minor | Curculionidae | Scolytinae | P. yunnanensis | Zixishan | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N | T. piniperda | Curculionidae | Scolytinae | P. yunnanensis | Lufeng, Zixishan, Changhu | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| O | T. brevipilosus | Curculionidae | Scolytinae | P. kesiya | Ning’er | Jun. 2010 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P | Unknown sp.1 | Curculionidae | Scolytinae | P. semaonensis | Chuxiong | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q | Unknown sp.2 | Curculionidae | Molytinae | Tsuga sp. | Dali | Jul. 2002 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 16 | 5 | 23 | 39 | 1 | 4 | 2 | 4 | 1 | 2 | 2 | 1 | 6 | 106 | ||||||
Mite species: a = Dendrolaelaps sp. 1 (Mesostigmata, Digamasellidae); b = Dendrolaelaps sp. 2 (Mesostigmata, Digamasellidae); c = Histiostoma cf. sapromyzarum (Sarcoptiformes, Histiostomatidae); d = Insectolaelaps sp. 1 (Mesostigmata, Digamasellidae); e = Lasioseius sp. 1 (Mesostigmata, Blattisociidae); f = Proctolaelaps nr. hystrix (Mesostigmata, Melicharidae); g = Rhizoglyphus sp. 1 (Sarcoptiformes, Acaridae); h = Schwiebea taiwanensis (Sarcoptiformes, Acaridae); I = Unknown sp.1 (Mesostigmata); j = Unknown sp.2 (Oribatei); k = Unknown sp.3 (Mesostigmata, Uropodoidea); l = Unknown sp.4 (Mesostigmata); m = Unknown sp.5.
Isolation of fungi
Fungi were isolated from the bark beetles and their galleries following the methods described by Linnakoski et al. (2008). At the time of collection, living mites collected from the galleries were placed on the surface of 2% MEA medium (20 g Difco agar, 20 g Difco BactoTM malt extract [Becton, Dickinson & Company], 1 L deionized water) and allowed to crawl over the plate for 24 hours. The mites were then removed and stored in Eppendorf tubes for identification. MEA plates were monitored daily for fungal growth and hyphal tips of emerging colonies were transferred to fresh MEA plates in order to purify the fungi. Pure cultures were grouped based on culture morphology, and representatives from each group were selected for DNA sequencing. All isolates used in this study were deposited in the Culture Collection (CMW) of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, Republic of South Africa. Isolates representing types of new species were also deposited in the culture collection (CBS) of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands.
DNA sequencing and phylogenetic analyses
Isolates were grown on 2% MEA in Petri dishes. DNA was extracted using PrepMan Ultra Sample Preparation reagent (Applied Biosystems, Foster City, CA) following the manufacturer’s instructions. The primer pairs ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990) were used to amplify internal transcribed spacer regions (ITS), the primary barcode of fungi (Schoch et al. 2012). LR0R and LR5 (Vilgalys and Hester 1990) were used to amplify nuclear large subunit (LSU). For ITS2-LSU used in Leptographium, ITS and LSU sequences were combined. EF2F (Marincowitz et al. 2015) and EF2R (Jacobs et al. 2004) were used to amplify the elongation factor 1-α (EF) gene, the secondary barcode of fungi (Stielow et al. 2015). BT2A and BT2B (Glass and Donaldson 1995) were used to amplify part of the β-tubulin (BT) gene, and CL2F and CL2R (Duong et al. 2012) for the calmodulin (CAL) gene. PCR and sequencing were conducted using the methods described by Duong et al. (2012).
The sequences obtained using the forward and reverse primers were aligned in Geneious Pro v. 7.1.4 (Biomatters, Auckland, New Zealand). ITS sequences were submitted to BLAST searches in NCBI Genbank for preliminary identifications. Based on these results, the ITS data were separated into different data sets according to genus. ITS2-LSU data were used for analyses of taxa residing in Leptographium s.l., while ITS was used for analyses of taxa belonging to the other genera. The BT, EF and CAL data were separated into smaller data sets based on the species complexes as defined by De Beer and Wingfield (2013). All sequences obtained in this study were submitted to GenBank (Table 2). Alignments were done online with MAFFT v. 7 (Katoh and Standley 2013). Maximum likelihood (ML), maximum parsimony (MP) and Bayesian Inference (BI) were conducted on the datasets as described in Duong et al. (2012). For haplotype analyses of O. quercus and O. tsotsi, the online server ElimDupes was used (http://www.hiv.lanl.gov/content/sequence/ELIMDUPES/elimdupes.html).
Table 2.
Isolates of ophiostomatoid fungi obtained from different beetles, their galleries and mites in Yunnan.
| Taxon | Species | Isolate number1,2 | Host | Location | Beetle | G/B/M3 | GenBank number4 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CMW | CBS | ITS/ITS2-LSU5 | BT | EF | CAL | ||||||
| Ophiostomatales | |||||||||||
| 1 | Sporothrix sp. A | 41787 | Pinus kesiya | Ning’er | Coccotrypes cyperi | M | MG205645 | MG205681 | – | – | |
| 2 | S. nebularis | 11791 | Pinus yunnanensis | Zixishan | Tomicus piniperda | * | MG205646 | MG205682 | – | – | |
| 41762 | P. kesiya | Ning’er | Co. cyperi | M | MG205647 | MG205683 | – | – | |||
| 41776 | P. kesiya | Ning’er | Co. cyperi | M | MG205648 | MG205684 | – | – | |||
| 41779 | P. kesiya | Ning’er | Co. cyperi | M | MG205649 | =MG205684 | – | – | |||
| 41782 | P. kesiya | Ning’er | Co. cyperi | M | MG205650 | =MG205684 | – | – | |||
| 41816 | P. kesiya | Ning’er | Cyrtogenius luteus | M | MG205651 | MG205685 | – | – | |||
| 41819 | P. kesiya | Ning’er | Orthotomicus angulatus | M | MG205652 | =MG205685 | – | – | |||
| 41835 | P. kesiya | Ning’er | Polygraphus sp. | M | MG205653 | =MG205684 | – | – | |||
| 3 | Ophiostoma acarorum sp. nov. | 41630 | P. kesiya | Ning’er | Ips acuminatus | G | =MG205656 | MG205686 | – | – | |
| 41641 | P. kesiya | Ning’er | Polygraphus szemaoensis | G | =MG205656 | =MG205686 | – | – | |||
| 41642 | P. kesiya | Ning’er | P. szemaoensis | G | =MG205656 | =MG205686 | – | – | |||
| 41647 | P. kesiya | Pu’er | I. acuminatus | M | =MG205656 | =MG205686 | – | – | |||
| 41789 | P. kesiya | Ning’er | Cy. luteus | M | =MG205657 | =MG205686 | – | – | |||
| 41791 | P. kesiya | Ning’er | Cy. luteus | M | =MG205657 | =MG205686 | – | – | |||
| 41795 | P. kesiya | Ning’er | Cy. luteus | M | =MG205657 | =MG205686 | – | – | |||
| 41798 | 139643 | P. kesiya | Ning’er | Cy. luteus | M | =MG205657 | =MG205686 | – | – | ||
| 41812 | 139658 | P. kesiya | Ning’er | Cy. luteus | M | MG205656 | =MG205686 | – | – | ||
| 41850H | 139748 | P. kesiya | Ning’er | O. angulatus | M | MG205657 | =MG205686 | – | – | ||
| 41852 | P. kesiya | Ning’er | Co. cyperi | M | =MG205657 | =MG205686 | – | – | |||
| 41987 | P. kesiya | Ning’er | I. acuminatus | G | =MG205656 | =MG205686 | – | – | |||
| 4 | O. ips | 41620 | P. kesiya | Ning’er | I. acuminatus | G | MG205658 | MG205687 | – | – | |
| 41644 | P. kesiya | Pu’er | I. acuminatus | B | =MG205658 | =MG205687 | – | – | |||
| 4 | O. ips | 41653 | P. kesiya | Ning’er | P. szemaoensis | G | =MG205658 | MG205688 | – | – | |
| 41695 | P. kesiya | Simao | P. szemaoensis | M | =MG205658 | =MG205688 | – | – | |||
| 41697 | P. kesiya | Simao | P. szemaoensis | B | =MG205658 | =MG205688 | – | – | |||
| 41709 | P. kesiya | Simao | P. szemaoensis | B | =MG205658 | =MG205688 | – | – | |||
| 41745 | P. kesiya | Ning’er | Polygraphus sp. | M | =MG205658 | =MG205688 | – | – | |||
| 41916 | P. kesiya | Ning’er | P. szemaoensis | G | =MG205658 | =MG205688 | – | – | |||
| 41993 | P. kesiya | Ning’er | I. acuminatus | M | =MG205658 | =MG205687 | – | – | |||
| 5 | Ophiostoma sp. B | 12032 | Pinus semaonensis | Chuxiong | Unknown sp.1 | * | MG205659 | MG205689 | MG205731 | – | |
| 6 | O. brevipilosi sp. nov. | 41624 | 139661 | P. kesiya | Ning’er | Tomicus brevipilosus | B | =MG205660 | MG205690 | MG205732 | – |
| 41662H | 139659 | P. kesiya | Ning’er | T. brevipilosus | B | MG205660 | =MG205690 | =MG205732 | – | ||
| 41760 | P. kesiya | Ning’er | T. brevipilosus | B | =MG205660 | =MG205690 | =MG205732 | – | |||
| 41873 | 139660 | P. kesiya | Ning’er | T. brevipilosus | B | =MG205660 | =MG205690 | =MG205732 | – | ||
| 41932 | P. kesiya | Ning’er | T. brevipilosus | B | =MG205660 | =MG205690 | =MG205732 | – | |||
| 41995 | P. kesiya | Ning’er | T. brevipilosus | B | =MG205660 | =MG205690 | =MG205732 | – | |||
| 7 | O. setosum | 12152 | Tsuga sp. | Dali | Unknown sp.2 | * | MG205661 | MG205691 | MG205733 | – | |
| 12192 | Tsuga sp. | Dali | Unknown sp.2 | * | =MG205661 | =MG205691 | MG205734 | – | |||
| 12337 | Tsuga sp. | Dali | Unknown sp.2 | * | =MG205661 | =MG205691 | MG205735 | – | |||
| 8 | O. quercus | 11747 | P. yunnanensis | Lufeng | Polygraphus verrucifrons | * | =AF198238 | =FJ455565 | MG205736 | – | |
| 11748 | P. yunnanensis | Lufeng | P. verrucifrons | * | =AF198238 | =FJ455565 | =MG205736 | – | |||
| 11756 | P. yunnanensis | Lufeng | P. verrucifrons | * | MG205662 | MG205692 | MG205737 | – | |||
| 11806 | P. yunnanensis | Changhu | T. piniperda | * | =FJ434947 | =FJ455565 | MG205738 | – | |||
| 11807 | P. yunnanensis | Changhu | T. piniperda | * | =FJ434947 | =AY466647 | =MG205738 | – | |||
| 11981 | Abies sp. | Chuxiong | Unknown sp.1 | * | =FJ434947 | =FJ455570 | MG205739 | – | |||
| 12015 | P. semaonensis | Chuxiong | Unknown sp.1 | * | =AY466624 | MG205693 | MG205740 | – | |||
| 12037 | Unknown | Chuxiong | Unknown sp.1 | * | =AY466624 | MG205694 | MG205741 | – | |||
| 12039 | Unknown | Chuxiong | Unknown sp.1 | * | =FJ434947 | MG205695 | MG205742 | – | |||
| 8 | O. quercus | 12122 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | MG205696 | MG205743 | – | |
| 12146 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | =MG205696 | =MG205743 | – | |||
| 12185 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | MG205697 | =MG205740 | – | |||
| 12195 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | =MG205697 | =MG205740 | – | |||
| 12286 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | =FJ455570 | MG205744 | – | |||
| 12350 | Tsuga sp. | Dali | Unknown sp.2 | * | =FJ434947 | MG205698 | MG205745 | – | |||
| 12359 | Tsuga sp. | Dali | Unknown sp.2 | * | =AF198238 | =FJ455570 | MG205746 | – | |||
| 12364 | Tsuga sp. | Dali | Unknown sp.2 | * | =AF198238 | MG205699 | MG214780 | – | |||
| 12370 | Tsuga sp. | Dali | Unknown sp.2 | * | =AF198238 | =MG205698 | MG205747 | – | |||
| 12382 | Tsuga sp. | Dali | Unknown sp.2 | * | =AF198238 | =FJ455570 | MG205748 | – | |||
| 41659 | P. kesiya | Pu’er | I. acuminatus | G | MG205664 | MG205700 | MG205749 | – | |||
| 41693 | P. kesiya | Simao | P. szemaoensis | G | =MG205664 | =MG205700 | =MG205749 | – | |||
| 41715 | P. kesiya | Simao | P. szemaoensis | B | =MG205664 | =MG205700 | =MG205749 | – | |||
| 41718 | P. kesiya | Simao | P. szemaoensis | B | =MG205664 | =MG205700 | =MG205749 | – | |||
| 41724 | P. kesiya | Ning’er | P. szemaoensis | B | MG205665 | MG205701 | MG205750 | – | |||
| 41732 | P. kesiya | Ning’er | Cy. luteus | G | =MG205664 | =MG205700 | =MG205749 | – | |||
| 9 | O. tsotsi | 41730 | P. kesiya | Ning’er | Co. cyperi | G | =FJ441284 | MG205704 | MG205755 | – | |
| 41731 | P. kesiya | Ning’er | Co. cyperi | G | =FJ441284 | MG205705 | MG205756 | – | |||
| 41733 | P. kesiya | Ning’er | Cy. luteus | G | =FJ441284 | =MG205704 | =MG205755 | – | |||
| 41734 | P. kesiya | Ning’er | Cy. luteus | G | =FJ441284 | =MG205704 | =MG205755 | – | |||
| 41735 | P. kesiya | Ning’er | Co. cyperi | M | =FJ441284 | =MG205704 | =MG205755 | – | |||
| 41742 | P. kesiya | Ning’er | Co. cyperi | M | =FJ441284 | =MG205705 | =MG205756 | – | |||
| 41746 | P. kesiya | Ning’er | Co. cyperi | M | =FJ441284 | =MG205704 | =MG205755 | – | |||
| 41758 | P. kesiya | Ning’er | Co. cyperi | M | =FJ441284 | =MG205705 | =MG205756 | – | |||
| 10 | Ophiostoma sp. C | 12150 | Tsuga sp. | Dali | Unknown sp.2 | * | MG205666 | MG205709 | MG205762 | – | |
| 11 | Graphilbum fragrans | 11778 | P. yunnanensis | Zixishan | Tomicus minor | * | MG205667 | MG205710 | – | – | |
| 12 | Gra. kesiyae sp. nov. | 41626 | P. kesiya | Ning’er | P. szemaoensis | M | =MG205669 | – | – | – | |
| 41657 | 139639 | P. kesiya | Ning’er | Polygraphus sp. | M | MG205668 | =MG205714 | – | – | ||
| 12 | Gra. kesiyae sp. nov. | 41686 | 139641 | P. kesiya | Simao | P. szemaoensis | M | =MG205668 | MG205711 | – | – |
| 41691 | 139642 | P. kesiya | Simao | P. szemaoensis | M | =MG205669 | =MG205713 | – | – | ||
| 41703 | P. kesiya | Simao | P. szemaoensis | G | – | MG205712 | – | – | |||
| 41716 | 139657 | P. kesiya | Simao | P. szemaoensis | M | =MG205669 | =MG205712 | – | – | ||
| 41729H | 139652 | P. kesiya | Ning’er | P. szemaoensis | G | MG205669 | MG205713 | – | – | ||
| 41774 | 139653 | P. kesiya | Ning’er | Polygraphus aterrimus | B | MG205668 | MG205714 | – | – | ||
| 46468 | P. kesiya | Ning’er | P. aterrimus | G | MG205668 | =MG205714 | – | – | |||
| 46469 | P. kesiya | Ning’er | P. aterrimus | B | MG205668 | =MG205714 | – | – | |||
| 13 | Gra. puerense sp. nov. | 41619 | P. kesiya | Ning’er | P. szemaoensis | B | =MG205670 | MG205715 | – | – | |
| 41667 | 139651 | P. kesiya | Pu’er | I. acuminatus | G | MG205670 | MG205716 | – | – | ||
| 41670 | P. kesiya | Ning’er | I. acuminatus | B | =MG205670 | – | – | – | |||
| 41671 | P. kesiya | Ning’er | I. acuminatus | B | =MG205670 | MG205717 | – | – | |||
| 41673 | 139640 | P. kesiya | Ning’er | I. acuminatus | B | =MG205670 | MG205718 | – | – | ||
| 41942H | 139650 | P. kesiya | Ning’er | P. szemaoensis | G | MG205671 | MG205719 | – | – | ||
| 41971 | P. kesiya | Ning’er | P. szemaoensis | M | =MG205671 | MG205720 | – | – | |||
| 41996 | P. kesiya | Ning’er | I. acuminatus | M | =MG205671 | MG205721 | – | – | |||
| 41998 | P. kesiya | Ning’er | I. acuminatus | G | =MG205670 | =MG205715 | – | – | |||
| 14 | Leptographium gracile | 12305 | Pinus armandii | Lijiang | Pissodes sp. | * | MG205672 | MG205722 | MG205763 | MG205782 | |
| 12397 | P. armandii | Midu | Pissodes sp. | * | =MG205672 | =MG205722 | =MG205763 | =MG205782 | |||
| 12399 | P. armandii | Midu | Pissodes sp. | * | =MG205672 | =MG205722 | =MG205763 | =MG205782 | |||
| 12404 | P. armandii | Midu | Pissodes sp. | * | =MG205672 | =MG205722 | =MG205763 | =MG205782 | |||
| 12407 | P. armandii | Midu | Pissodes sp. | * | =MG205672 | =MG205722 | =MG205763 | =MG205782 | |||
| 12412 | P. armandii | Midu | Pissodes sp. | * | =MG205672 | =MG205722 | =MG205763 | =MG205782 | |||
| 15 | Grosmannia radiaticola | 12323 | Tsuga sp. | Dali | Unknown sp.2 | * | MG205673 | MG205723 | MG205764 | – | |
| 16 | L. ningerense sp. nov. | 41773 | P. kesiya | Ning’er | Co. cyperi | M | – | =MG205724 | MG205765 | =MG205783 | |
| 41786H | 139663 | P. kesiya | Ning’er | Co. cyperi | M | MG205674 | MG205724 | =MG205765 | MG205783 | ||
| 41831 | 139664 | P. kesiya | Ning’er | O. angulatus | M | MG205675 | =MG205724 | =MG205765 | =MG205783 | ||
| 17 | G. yunnanensis | 41622 | P. kesiya | Ning’er | P. szemaoensis | B | MG205676 | MG205725 | MG205766 | – | |
| 41627 | P. kesiya | Ning’er | P. szemaoensis or I. acuminatus | M | – | =MG205725 | – | – | |||
| 41633 | P. kesiya | Ning’er | P. szemaoensis or I. acuminatus | M | – | MG205726 | – | – | |||
| 41635 | P. kesiya | Ning’er | Lasconotus sp. | B | – | =MG205726 | MG205767 | – | |||
| 41636 | P. kesiya | Ning’er | Lasconotus sp. | B | – | =MG205726 | MG205768 | – | |||
| 41666 | P. kesiya | Ning’er | Polygraphus sp. | M | – | =MG205725 | – | – | |||
| 41687 | P. kesiya | Simao | P. szemaoensis | M | – | =MG205725 | MG205769 | – | |||
| 41694 | P. kesiya | Simao | P. szemaoensis | M | – | =MG205725 | – | – | |||
| 41707 | P. kesiya | Simao | P. szemaoensis | B | – | =MG205725 | MG205770 | – | |||
| 41720 | P. kesiya | Simao | P. szemaoensis | G | – | =MG205725 | MG205771 | – | |||
| 41721 | P. kesiya | Ning’er | P. szemaoensis | M | – | =MG205725 | MG205772 | – | |||
| 41726 | P. kesiya | Ning’er | P. szemaoensis | M | – | =MG205725 | – | – | |||
| 41728 | P. kesiya | Ning’er | P. szemaoensis | M | – | =MG205726 | MG205773 | – | |||
| 41777 | P. kesiya | Ning’er | Co. cyperi | M | – | MG205727 | MG205774 | – | |||
| 41778 | P. kesiya | Ning’er | Co. cyperi | M | – | MG205728 | =MG205774 | – | |||
| 41781 | P. kesiya | Ning’er | Co. cyperi | M | – | =MG205727 | =MG205774 | – | |||
| 41783 | P. kesiya | Ning’er | Co. cyperi | M | – | =MG205725 | MG205775 | – | |||
| 41805 | P. kesiya | Ning’er | Polygraphus sp. | M | – | =MG205725 | – | – | |||
| 41814 | P. kesiya | Ning’er | Polygraphus sp. | M | – | =MG205725 | – | – | |||
| 41858 | P. kesiya | Ning’er | P. szemaoensis | G | – | =MG205725 | MG205776 | – | |||
| 41863 | P. kesiya | Ning’er | P. szemaoensis | G | MG205677 | =MG205725 | MG205777 | – | |||
| 41945 | P. kesiya | Ning’er | P. szemaoensis | M | – | =MG205725 | – | – | |||
| 41963 | P. kesiya | Ning’er | P. szemaoensis | M | – | =MG205725 | – | – | |||
| 41990 | P. kesiya | Ning’er | I. acuminatus | G | – | =MG205725 | =MG205776 | – | |||
| 17 | G. yunnanensis | 41992 | P. kesiya | Ning’er | I. acuminatus | G | – | =MG205725 | MG205778 | – | |
| 41999 | P. kesiya | Ning’er | P. szemaoensis | B | – | =MG205725 | – | – | |||
| 42000 | P. kesiya | Ning’er | P. szemaoensis | B | – | – | =MG205778 | – | |||
| 18 | L. conjunctum | 11782 | P. yunnanensis | Zixishan | Hylurgops major | * | MG205678 | MG205729 | MG205779 | – | |
| 41761 | P. kesiya | Ning’er | Polygraphus sp. | M | MG205679 | MG205730 | MG205780 | – | |||
| Microascales | |||||||||||
| 19 | Graphium pseudormiticum | 41665 | P. kesiya | Pu’er | I. acuminatus | M | MG205680 | – | MG205781 | – | |
1 The culture collection (CBS) of Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; CMW Culture Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, Pretoria, South Africa 2 H = ex–holotype isolate 3 G = Gallery; B = Beetle; M = Mite; * = Unknown 4 ITS = internal transcribed spacer regions 1 and 2 of the nuclear ribosomal DNA operon, including the 5.8S region; ITS2–LSU = the internal transcribed spacer 2 region and partial large subunit of the nrDNA operon; ΒT = beta–tubulin; EF = translation elongation factor 1–alpha; CAL = Calmodulin
Growth studies
Growth studies were conducted on three isolates of each novel taxon. Mycelium-covered agar plugs were transferred from the actively growing margins of one-week-old cultures and placed at the centers of 90 mm Petri dishes containing 2% MEA. Cultures were incubated in the dark at temperatures ranging from 5–35 °C at 5 °C intervals. Three replicates were used for each isolate at each temperature. Colony diameters were measured every day until hyphae reached the edges of the Petri dishes. Optimum and maximum growth temperatures were calculated for each species.
Morphological studies
To facilitate morphological descriptions of new taxa, asexual and sexual structures (where present) were mounted in lactophenol on glass slides, covered with a coverslip and examined with a Zeiss Axioskop2 Plus compound microscope or a Zeiss Discovery V12 dissection microscope with an Axiocam digital camera (Axiovision 3.1) (München-Hallbergmoos, Germany). Measurements were made for each taxonomically characteristic structure. The measurements are presented in the format (minimum–) mean minus standard deviation–mean plus standard deviation (–maximum). For reference to asexual states that resemble morphological features of well-known asexual genera, we followed the reference style (e.g. hyalorhinocladiella-like) suggested by Hawksworth (2011). Descriptions of morphological features of Leptographium species were based on the style of Jacobs and Wingfield (2001).
Frequency of isolation
Frequencies of isolation of the ophiostomatoid species in all samples were calculated as follows: F = (NF/NT) × 100, where F represents the frequency of isolation (%), NT represents the total number of isolates collected, and NF represents the number of isolates of a particular fungal species.
Results
Bark beetles and mites
Collectively, 17 beetle species belonging to four sub-families were collected from conifer hosts at 10 sites (Table 1). Seven of the species were collected in 2001 and 2002 and ten species in 2010. In total, 106 mites were collected from galleries of six Scolytine bark beetle species during the 2010 survey. One hundred of these mites represented 12 mite species belonging to eight families (Table 1). The remaining six mites (Table 1, Column m) could not be identified. The abundance of each mite species collected varied considerably, with Insectolaelaps sp. 1, Histiostoma cf. sapromyzarum and Dendrolaelaps sp. 1 being the most abundant overall. The other species were present in very low numbers. Among the most abundant mites, Insectolaelaps sp. 1 was found on four beetle species, and Histiostoma cf. sapromyzarum was found on three beetle species, while Dendrolaelaps sp. 1 was associated with only one beetle species. Coccotrypes cyperi was the most common beetle vector as it was phoreticed with 34 mites representing eight species, including three unknown species. Nineteen mites representing five species (including one unknown) came from Polygraphus szemaoensis, and Orthotomicus angulatus vectored 19 mites representing four species.
Fungal isolation
A total of 340 fungal isolates were obtained, 54 from beetles in 2001 and 2002, and 286 isolated from beetles, galleries and mites in 2010 (Tables 3 and Suppl. material 1: Table S1). Bark beetle niches in this study include the beetles themselves, their galleries or the mites associated with a particular bark beetle or its galleries. The numbers of fungal isolates collected from the different bark beetle niches varied substantially. For example, 85 isolates were collected from the P. szemaoensis niche, 60 isolates from that of C. cyperi, but only two and four isolates from the niches of Lasconotus sp. and P. aterrimus respectively.
Table 3.
Numbers of fungal isolates per species obtained from beetles (B), their galleries (G) or mite (M) associates
| Year of survey → | 2010 | 2001 | 2002 | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Host tree species1 → | I | II | III | IV | V | Total | ||||||||||||||||||||||||||||||||
| Beetle species2 → | A | B | D | E | F | H | I | J | L | O | G | C | K | M | N | P | Q | |||||||||||||||||||||
| B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | B | G | M | * | * | * | * | * | * | * | ||
| Taxon. Fungus species↓ | ||||||||||||||||||||||||||||||||||||||
| 1. Sporothrix sp. A | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 2. S. nebularis | 0 | 0 | 4 | 0 | 0 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 0 | 0 | 0 | 5 | 1 | 3 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 45 |
| 3. Ophiostoma acarorum sp. nov. | 0 | 0 | 21 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 29 |
| 4. O. ips | 0 | 0 | 1 | 0 | 1 | 17 | 6 | 11 | 14 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 1 | 0 | 0 | 0 | 2 | 9 | 15 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 4 | 4 | 0 | 106 |
| 5. Ophiostoma sp. B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 6. O. brevipilosi sp. nov. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| 7. O. setosum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 5 |
| 8. O. quercus | 0 | 1 | 0 | 0 | 1 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2 | 4 | 11 | 43 |
| 9. O. tsotsi | 0 | 2 | 0 | 0 | 2 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 |
| 10. Ophiostoma sp. C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 11. Graphilbum fragrans | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| 12. Gra. kesiyae sp. nov. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| 13. Gra. puerense sp. nov. | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 |
| 14. Leptographium gracile | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 10 |
| 15. G. radiaticola | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 16. L. ningerense sp. nov. | 0 | 0 | 2 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 |
| 17. G. yunnanensis | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 4 | 3 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 39 |
| 18. L. conjunctum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| 19. Graphium pseudormiticum | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 0 | 3 | 28 | 0 | 4 | 56 | 10 | 19 | 18 | 2 | 0 | 0 | 0 | 0 | 34 | 2 | 2 | 0 | 0 | 0 | 12 | 21 | 28 | 36 | 1 | 0 | 0 | 9 | 0 | 0 | 10 | 1 | 6 | 1 | 8 | 12 | 17 | 340 |
1 Tree host species: I = Pinus kesiya; II = P. armandii; III = P. yunnanensis; IV = P. semaonensis; V = Tsuga sp. 2 Beetle species: A = Coccotrypes cyperi; B = Cyrtogenius luteus; C = Hylurgops major; D = Ips acuminatus; E = Lasconotus sp.; F = Orthotomicus angulatus; G = Pissodes sp.; H = Polygraphus aterrimus; I = Polygraphus sp.; J = P. szemaoensis; K = P. verrucifrons; L = Stenoscelis sp.; M = Tomicus minor; N = T. piniperda; O = T. brevipilosus; P = Unknown sp.1; Q = Unknown sp.2.* = information about specific niche not available.
DNA sequencing and phylogenetic analyses
From the total of 340 isolates obtained in the study, DNA sequences were generated for 134 isolates (Table 2), representing all the morphological groups. Based on preliminary BLAST results, ITS data generated in the present study were separated into two data sets, the first including Ophiostoma, Sporothrix and Graphilbum in the Ophiostomatales, and the second including Graphium spp. in the Microascales. Because amplification of the ITS1 and 2 regions is often problematic for Leptographium s.l. species (De Beer and Wingfield 2013), a reference data set consisting of ITS2-LSU data was compiled to determine the species complexes in Leptographium s.l. to which the isolates from China belonged.
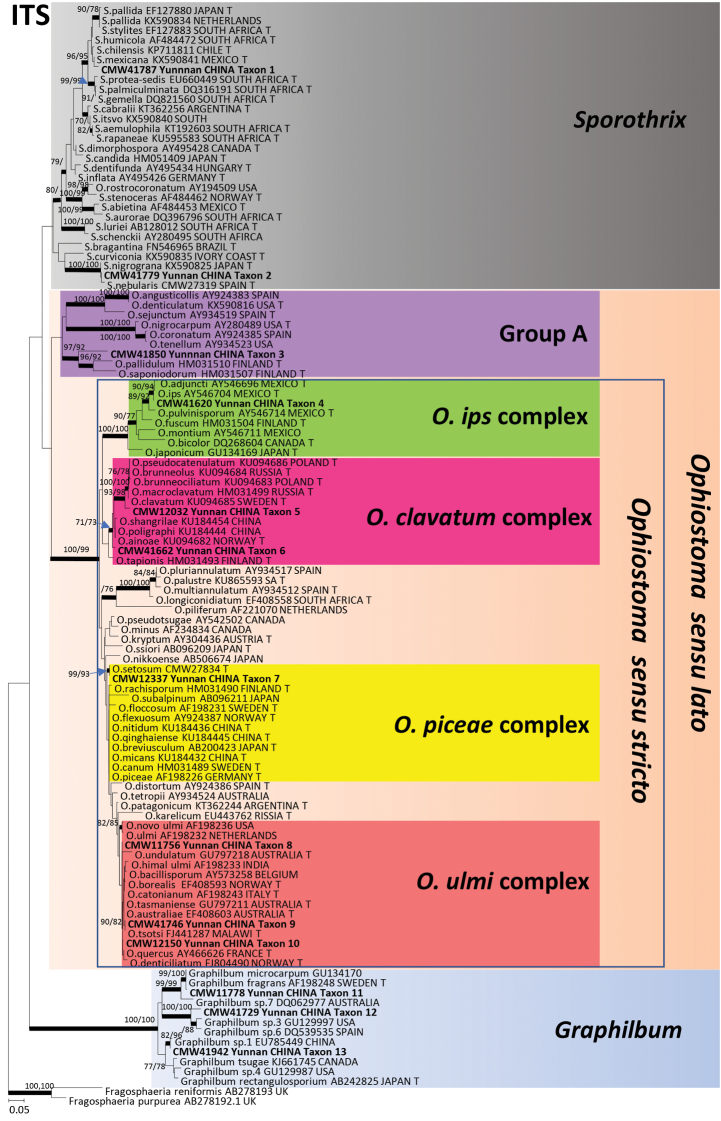
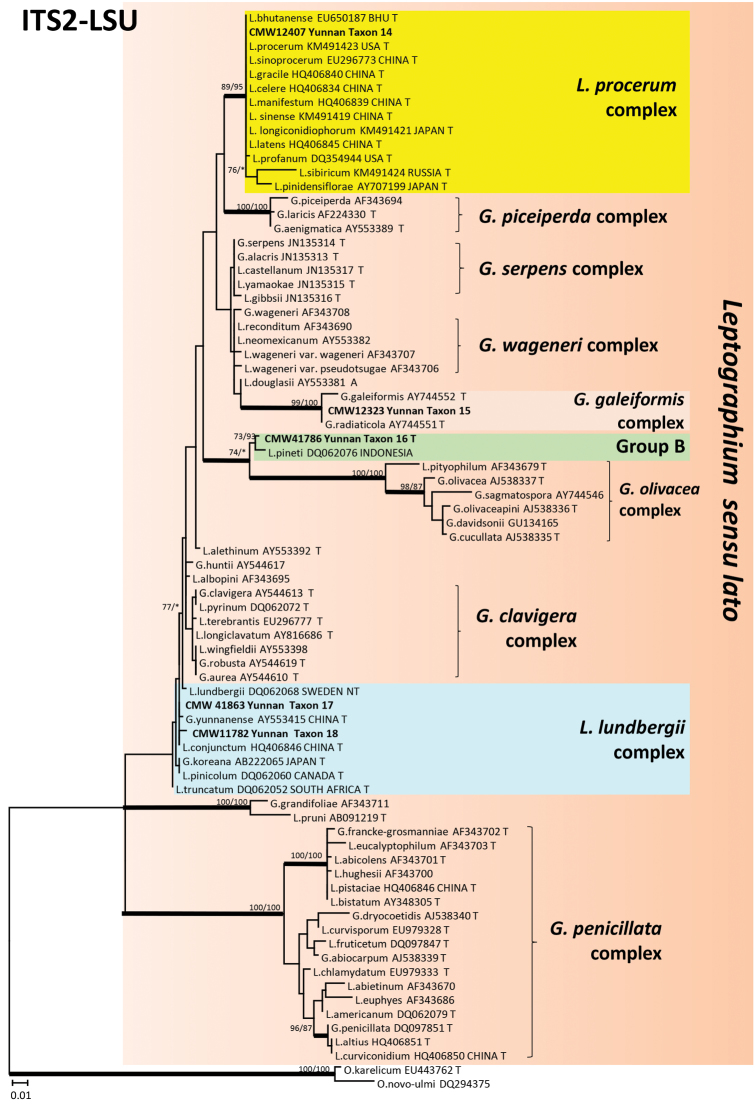
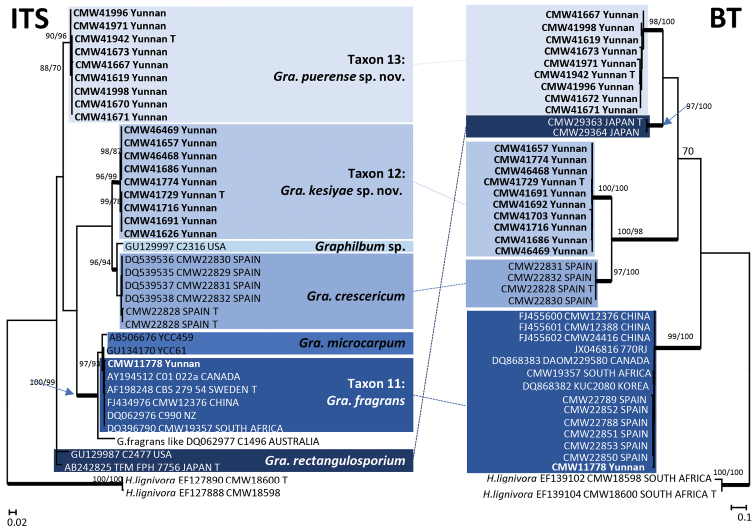
The 340 isolates were separated in 19 taxa (Taxa 1 to 19) based on DNA sequences (Table 2) and culture morphology. Taxa 1 and 2 (Fig. 1) represented species of Sporothrix, Taxa 3 to 10 resided in five species complexes in Ophiostoma s.l., and Taxa 11 to 13 belonged to Graphilbum. Taxa 14 to 18 belonged to four species complexes in Leptographium s.l. (Fig. 2). One taxon (Taxon 19) resided in the genus Graphium (Suppl. material 2: Figure S1).
Figure 1.
ML tree of the ITS region of Ophiostoma, Sporothrix, Graphilbum. Novel sequences obtained in this study are printed in bold type. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure 2.
ML tree of the ITS2-LSU region of Leptographium. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Two species of Sporothrix were collected (Suppl. material 2: Figure S3). Taxon 1 was represented by a single isolate and both ITS and BT data suggested that this was of an undescribed species. Taxon 2 included 45 isolates, eight for which sequence data were included in our analyses (Suppl. material 2: Figure S3). In the ITS tree, these isolates grouped in a monophyletic clade that included the ex-type isolates of S. nebularis and S. nigrograna. In the BT tree these isolates again grouped with S. nebularis.
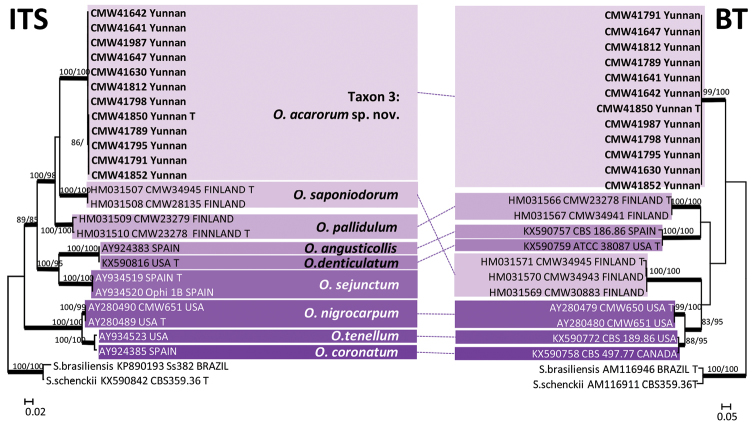
Taxon 3 included 29 isolates (Table 3), 12 of which (Table 2) were included in the analyses. These isolates grouped with a number of Ophiostoma spp. peripheral to the well-defined complexes in Ophiostoma s.str., and they were treated as Group A in Ophiostoma s.l. (Fig. 1). In both the ITS and BT trees (Fig. 3), the Yunnan isolates formed a well-supported lineage, closest to but clearly distinct from O. pallidulum and O. saponiodorum.
Figure 3.
ML trees of Group A generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Taxon 4 formed part of the O. ips complex (Fig. 1). This included 106 isolates (Table 3), nine of which were included in the analyses (Table 2). ITS and BT data (Suppl. material 2: Figure S4) confirmed that these isolates all represented O. ips.
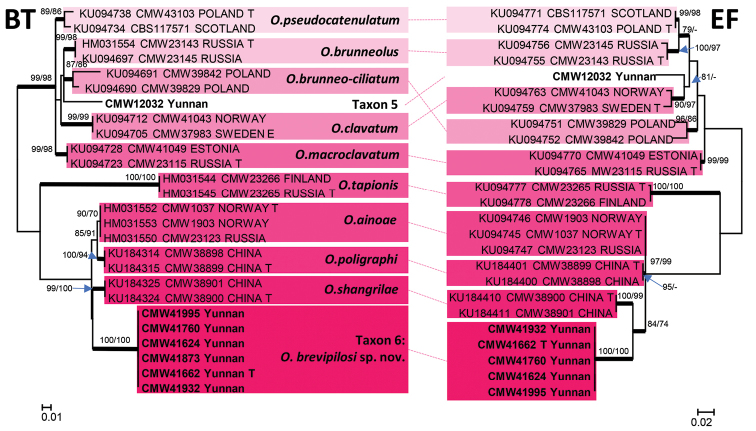
Taxon 5, including only one isolate, together with Taxon 6 that included eight isolates (Table 3), six for which sequences were produced, formed part of the O. clavatum complex (Fig. 1). BT and EF data confirmed that both these taxa grouped distinct from all known species in the complex (Fig. 4).
Figure 4.
ML trees of the O. clavatum complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Five isolates comprised Taxon 7 (Tables 2 and 3), which formed part of the O. piceae complex (Fig. 1). Both BT and EF analyses confirmed that these isolates grouped with O. setosum isolates previously identified from Canada, Korea and China (Suppl. material 2: Figure S5).
There were 55 isolates (Table 3) belonging to the O. ulmi complex (Fig. 1). The sequences for these isolates were quite variable. Thus more detailed analyses, including all available related sequences from Genbank generated in previous studies dealing with the variable haplotypes of these taxa (Grobbelaar et al. 2009, Kamgan Nkuekam et al. 2010), were required. Taxon 8 represented O. quercus and Taxon 9 O. tsotsi based on ITS (Suppl. material 2: Figure S6), BT (Suppl. material 2: Figure S7), and EF (Suppl. material 2: Figure S8). Sequences of the 36 putative O. quercus isolates were very variable and respectively represented 13, 23 and 28 haplotypes for the ITS, BT and EF gene regions (Suppl. material 1: Table S2). When combined, 51 unique haplotypes were found (Suppl. material 1: Table S2). The 18 isolates representing Taxon 9 grouped with isolates of O. tsotsi (Suppl. material 2: Figures S6, S7, S8). The ITS data for O. tsotsi was less variable than those of O. quercus (Suppl. material 2: Figure S6), including only one haplotype. However, the BT (Suppl. material 2: Figure S7) and EF (Suppl. material 2: Figure S8) of O. tsotsi had 14 and 12 unique haplotypes respectively. The combined ITS, BT and EF data, represented 20 unique haplotypes (Suppl. material 1: Table S3). Based on BT (Suppl. material 2: Figure S7) and EF (Suppl. material 2: Figure S8) data, the isolate representing Taxon 10 grouped distinct from all other lineages and could represent an undescribed species.
For Graphilbum, results from the analyses of the ITS and BT sequences (Fig. 5) suggested that the Yunnan isolates resided in three taxa. These included one isolate of Gra. fragrans (Taxon 11) and two distinct, well-supported lineages (Taxa 12 and 13) each including 11 isolates (Table 3), representing undescribed species.
Figure 5.
ML trees of the Graphilbum generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
In Leptographium s.l. (Fig. 2), ten isolates (Taxon 14) grouped in the L. procerum complex (Table 3). Although both BT and EF sequences (Suppl. material 2: Figure S9) of this taxon differed in 1 bp from those of L. gracile, CAL sequences were identical to those of L. gracile and we conclude that Taxon 14 represents the latter species. However, species delineation in this complex requires reconsideration because sequences of several species in the complex are very similar.
A single isolate (Taxon 15) grouped in the Grosmannia galeiformis complex (Fig. 2, Table 2). Both the BT and EF sequences placed this isolate among those of G. radiaticola (Suppl. material 2: Figure S10).
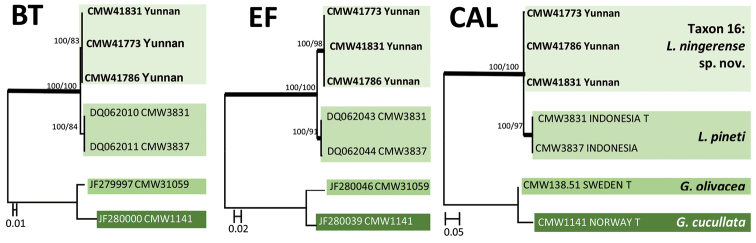
Taxon 16 (Fig. 2) included 13 isolates (Table 3) that grouped closest to L. pineti, peripheral to the G. olivacea complex. BT, EF and CAL analyses (Fig. 6) showed that Taxon 16 was distinct from L. pineti and represented a novel species.
Figure 6.
ML trees of the L. pineti generated from DNA sequences of BT, EF and CAL regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Forty one isolates (Tables 2 and 3) belonged to the L. lundbergii complex (Fig. 2). The analyses of BT and EF sequences (Suppl. material 2: Figure S11) showed that these isolates separated in two groups, respectively aligning with G. yunnanensis (Taxon 17) and L. conjunctum (Taxon 18).
A single isolate represented Taxon 19 (Table 2) in the genus of Graphium. Both the ITS and EF sequences of this isolate grouped with Graphium pseudormiticum isolates (Suppl. material 2: Figure S1).
Frequencies of isolation
The origin and sources of the 340 isolates representing 19 taxa are presented in Tables 2, 3 and Suppl. material 1: S1. The 54 isolates collected from beetles and their galleries in 2001 and 2002 belonged to 11 taxa (Table 3). During the 2010 survey, 286 isolates belonging to 12 taxa were collected (Table 3). The 45 isolates collected directly from six bark beetle species belonged to seven taxa, the 184 isolates from mites to 11 taxa, and the 57 isolates from galleries represented nine taxa. Six taxa were present on beetles, galleries and mites, three taxa on galleries and mites, one taxon only on beetles, and two taxa only on mites. No taxa were found only in galleries that were not also found on beetles and/or mites. Four taxa were collected from both the 2001–2002 survey and the 2010 survey.
Grosmannia yunnanensis, O. ips, O. quercus, S. nebularis and Taxon 3 were the most frequently isolated species, representing 12.5%, 31.2%, 12.6%, 13.2% and 8.5% of the isolated fungi respectively. The remaining species were isolated only occasionally. O. ips was isolated from the niches (beetles, galleries, mites) of ten different bark beetles species. However, most of the O. ips were found associated with niches of I. acuminatus, P. szemaoensis and C. cyperi representing approximately 9.7%, 8.5% and 5.3%, respectively. S. nebularis was also isolated from the niches of ten beetle species and the highest frequency of isolation was 5.5%, associated with the niche of O. angulatus.
The number of fungal species isolated from different beetle or weevil niches varied between different species. There were ten fungal species, representing about 27.9% of total fungal isolates associated with the niche of P. szemaoensis. Among them, O. ips and L. yunnanense were the most frequently isolated, representing about 8.5% and 7.1%, respectively. There were eight fungal species, representing about 15.3%, associated with Co. cyperi and of these, the most frequently isolated fungus was O. ips with the frequency of 5.3%.
Only 13.2% of the total number of isolates were obtained directly from beetles (Table 3). Of these, 6.2% was obtained from P. szemaoensis. Most of the isolates from beetles were identified as O. ips (2.6%) and O. quercus (1.8%), while 2.9% and 2.6% of isolates were collected from I. acuminatus and T. brevipilosus respectively. Among these Taxon 6, representing 2.4%, was isolated only from T. brevipilosus. The other beetles vectored very low numbers of fungi. O. ips was the fungus most frequently isolated from the beetles, representing 4.4%, followed by Taxon 6 represting 2.4 % of the isolates.
Approximately 54% of all isolates were collected from mites (Table 3, Suppl. material 1: Table S1). The fungi associated with mites varied between the different mite species. Ten fungal species were isolated from Insectolaelaps sp. 1, representing 21.2% of the total. From some mite species, such as Lasioseius sp. 1, Dendrolaelaps sp. 2 and the unknown species in the families of Mesostigmata and Oribatei, only one fungal species was found, representing a frequency of isolation of 0.3%, 1.8%, 1.2% and 1.5%, respectively. O. ips, S. nebularis and G. yunnanensis were the three most frequently isolated species associated with mites, representing 15.3%, 10.6% and 7.9%, respectively.
Taxonomy
Eight of the 19 taxa obtained in the present study represented undescribed species. For three of these, only a single isolate was obtained and we have chosen not to formally describe these. The remaining five taxa including two Ophiostoma, two Graphilbum, and one Leptographium species, are described as follows:
Taxon 3
Ophiostoma acarorum
R.Chang & Z.W.de Beer sp. nov.
823693
Figure 7.
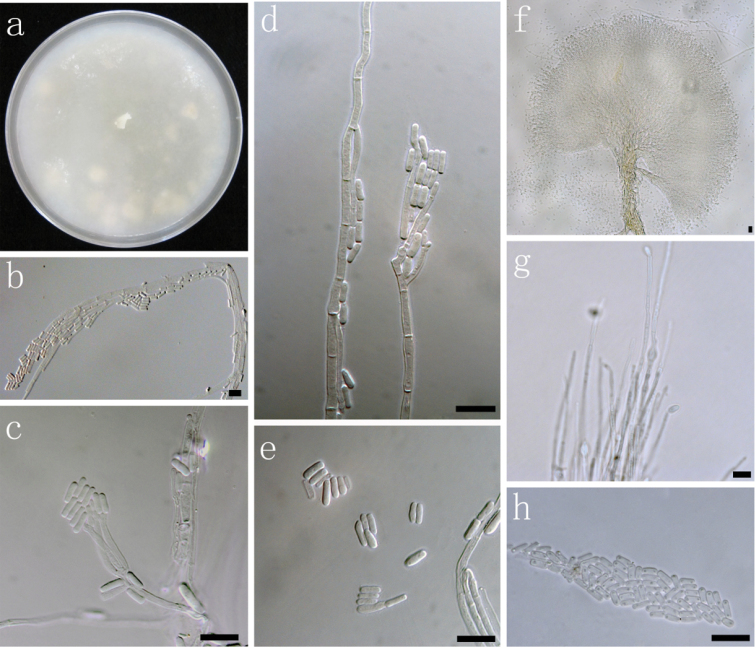
Morphological characters of asexual structures of Ophiostoma acarorum sp. nov. a fourteen days old culture on OA b–c Hyalorhinocladiella-like asexual state d conidia. Scale bars: a–d = 10 μm.
Etymology.
The epithet acarorum refers to the subclass Acari in the Arachnida to which all mite species belong from which 25 of the 29 isolates of this species were isolated.
Description.
Sexual state not observed. Hyalorhinocladiella-like asexual state: conidiophores (7–) 18–76.5 (–140) μm long; cells arising directly from the hyphae, (10.5–) 13.5–24.5 (–31) × (1–) 1.5–2 (–2) μm; conidia hyaline, smooth, oblong, (3–) 3.5–5 (–6.5) × (0.7–) 1–1.5 (–2.5) μm.
Culture characteristics.
Colonies hyaline at the beginning, becoming white to dark brown with age. Mycelium superficial on the 3% OA. Colony margin smooth. Colonies on 2% MEA flat, reaching 69 mm diam in 13 d at 30 °C. No growth observed at 5 °C. Optimal temperature for growth 25 °C.
Type material.
CHINA, Yunnan Province, Puer City, from Insectolaelaps sp. 1 in Orthotomicus angulatus gallery on Pinus kesiya bark, 17 Sep. 2010, S.J.Taerum, herbarium specimen of dried culture, PREM 61539 (holotype), CMW41850 = CBS139748 (ex-holotype culture).
Additional specimens examined.
CHINA, Yunnan Province, Puer City, from Histiostoma cf. sapromyzarum in Cyrtogenius luteus gallery on Pinus kesiya bark, 16 Sep. 2010, S.J.Taerum, PREM 61540, CMW41812 = CBS139658; from Histiostoma cf. sapromyzarum in Cyrtogenius luteus gallery on Pinus kesiya bark, 16 Sep. 2010, S.J.Taerum, CMW41798 = CBS139643.
Host.
Pinus kesiya.
Beetle vectors.
Ips acuminatus, Polygraphus szemaoensis.
Mite vectors.
Histiostoma cf. sapromyzarum (phoretic on Cyrtogenius luteus), Insectolaelaps sp. 1 (phoretic on Ips acuminatus and Orthotomicus angulatus).
Distribution.
At present known only from Yunnan, China.
Notes.
The hyalorhinocladiella-like asexual state of O. acarorum resembles that of O. pallidulum (Linnakoski et al. 2010), one of its two closest relatives based on phylogeny (Fig. 3). Ophiostoma saponiodorum, the other close relative has a similar hyalorhinocladiella-like state, but can be distinguished based on the presence of a second, synnematous asexual state (Linnakoski et al. 2010).
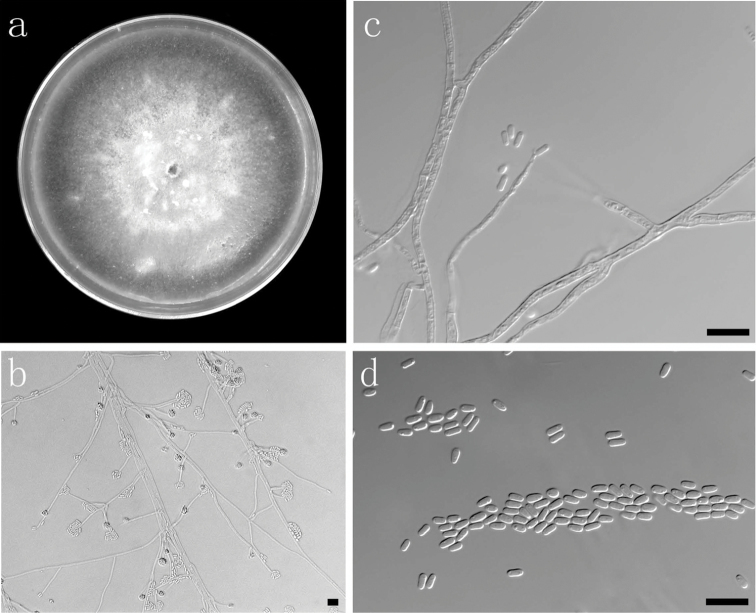
Taxon 6
Ophiostoma brevipilosi
R.Chang & Z.W.de Beer sp. nov.
823694
Fig. 8.
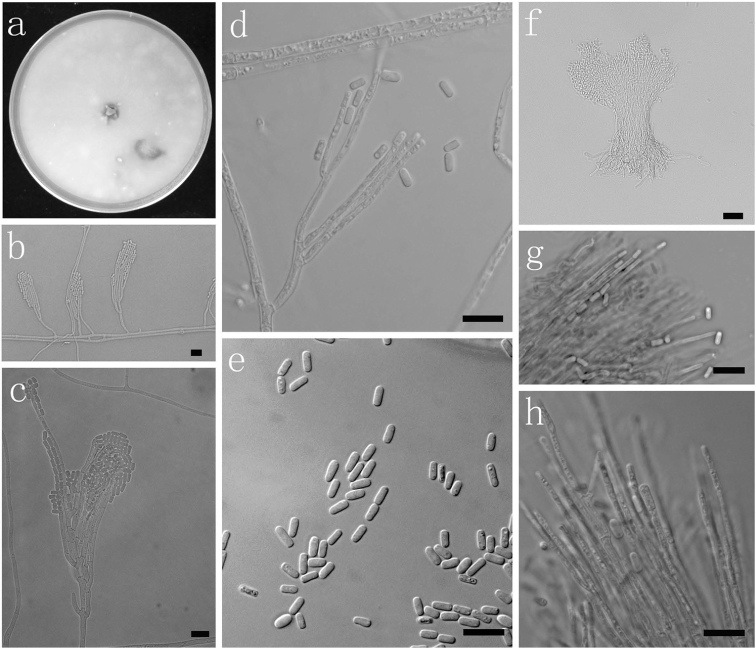
Morphological characters of asexual structures of Ophiostoma brevipilosi sp. nov. a fourteen days old culture on OA b–d Hyalorhinocladiella-like asexual state and condia e Pesotum-like macronematal asexual state f cells of Pesotum-like macronematal asexual state g conidia. Scale bars: a–g = 10 μm.
Etymology.
The epithet brevipilosi refers to the bark beetle vector Tomicus brevipilosus from which all eight isolates of this taxon were obtained.
Description.
Sexual state not observed. Pesotum-like macronematal asexual state predominant. Synnemata simple, dark brown at the base, (179.5–) 227–468 (–667) μm long including conidiogenous apparatus, (22–) 32.5–58 (–69) μm wide at base; cells (13–) 16–26 (–32.5) μm long, conidia hyaline, 1-celled, smooth, oblong, (3–) 3–4.5 (–5.5) × (1.5–) 1.5–2.5 (–3) μm. Hyalorhinocladiella-like asexual state: conidiophores (14.5–) 33–115 (–145) μm long; cells arising directly from the hyphae, (12–) 15–38 (–47) × (1.1–) 1.5–2 (–2.5) μm; conidia hyaline, smooth, obovoid, (2.5–) 3–5.5 (–8) × (1.5–) 2–2.5 (–3) μm.
Culture characteristics.
Colonies hyaline at the beginning, then becoming white to dark. Mycelium superficial on the 3% OA. Colony margin smooth. Colonies on 2% MEA flat, reaching 67 mm diam in 11 d at 25 °C. No growth observed at 5 and above 30 °C. Optimal temperature for growth 20 and 25 °C.
Type material.
CHINA, Yunnan Province, Puer City, from Tomicus brevipilosus on Pinus kesiya bark, 27 Jun. 2010, S.J.Taerum, herbarium specimen of dried culture, PREM 61537 (holotype), CMW41873 = CBS139660 (ex-holotype culture).
Additional specimens examined.
CHINA, Yunnan Province, Puer City, from Tomicus brevipilosus on Pinus kesiya bark, 27 Jun. 2010, S.J.Taerum, PREM 61538, CMW41624 = CBS139661; CHINA, Yunnan Province, Puer City, from Tomicus brevipilosus on Pinus kesiya bark, 27 Jun. 2010, S.J.Taerum, CMW41662 = CBS139659.
Host.
Pinus kesiya.
Beetle vector.
Tomicus brevipilosus.
Distribution.
At present known only from Yunnan, China.
Notes.
The synnematous asexual state of O. brevipilosi corresponds with similar structures of O. brunneo-ciliatum as described by Linnakoski et al. (2016). The hyalorhinocladiella state resembles those of O. brunneolum, O. macroclavatum, O. pseudocatenulatum (Linnakoski et al. 2016) and O. poligraphi (Yin et al. 2016). However, the morphology of these structures is not sufficient to distinguish between the species in the complex, and DNA sequences of the BT and EF gene regions are recommended for accurate species identification.
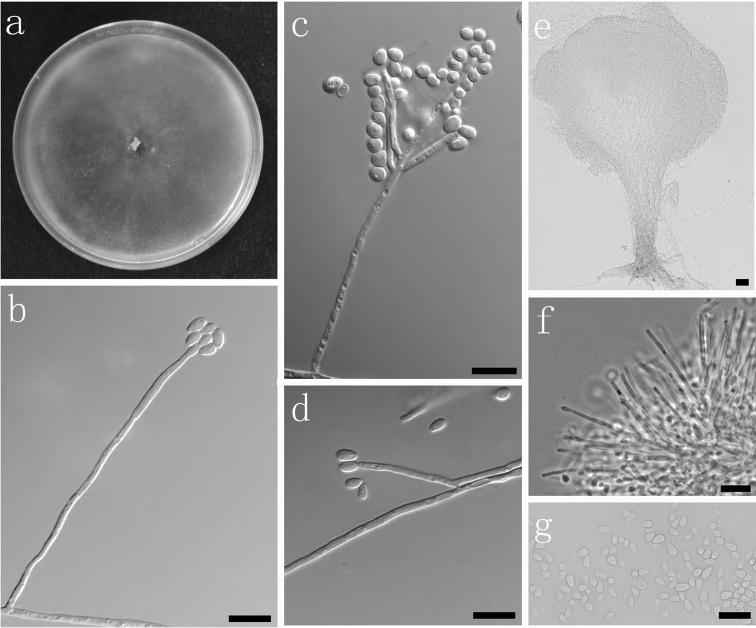
Taxon 12
Graphilbum kesiyae
R.Chang & Z.W.de Beer sp. nov.
823695
Fig. 9.
Morphological characters of asexual structures of Graphilbum kesiyae sp. nov. a fourteen days old culture on OA b–e Hyalorhinocladiella-like asexual state and conidia f Pesotum-like macronematal asexual state g cells of Pesotum-like macronematal asexual state h conidia. Scale bars: a–h = 10 μm.
Etymology.
The epithet kesiyae refers to the tree host of all beetles and mites from which the 12 isolates of this species were collected.
Description.
Sexual state not observed. Pesotum-like macronematal asexual states predominant. Synnemata simple, dark brown at the base, (85.5–) 112.5–173 (–203) μm long including conidiogenous apparatus, (9–) 14–45.5 (–65.5) μm wide at base; cells (8.5–) 10–18.5 (–25.5) μm long; conidia hyaline, 1-celled, smooth, oblong, (3.5–) 4–5 (–5.5) × (1.5–) 1.5–2 (–2.5) μm. Hyalorhinocladiella-like asexual state: conidiophores (22–) 38–101.5 (–166) μm long; cells arising directly from the hyphae, (10–) 12–27(–40) × (1.2–) 1.5–2 (–2.5) μm; conidia hyaline, smooth, obovoid, (3.5–) 4–5.5 (–8.5) × (1–) 1.5–2 (–3) μm.
Culture characteristics.
Colonies hyaline. Mycelium superficial on the 3% OA. Colony margin smooth. Colonies on 2% MEA flat, reaching 85 mm diam in 10 d at 25 °C. No growth observed at 5 and 35 °C. Optimal temperature for growth 25 °C.
Type material.
CHINA, Yunnan Province, Puer City, from Polygraphus szemaoensis gallery on Pinus kesiya bark, 12 Aug. 2010, S.J.Taerum, herbarium specimen of dried culture, PREM 61541 (holotype), CMW41729 = CBS139652 (ex-holotype culture).
Additional specimens examined.
CHINA, Yunnan Province, Puer City, from Insectolaelaps sp. 1 in Polygraphus szemaoensis gallery on Pinus kesiya bark, 10 Aug. 2010, S.J.Taerum, CMW41691 = CBS139642; CHINA, Yunnan Province, Puer City, from Proctolaelaps nr. hystrix in Polygraphus szemaoensis gallery on Pinus kesiya bark, 11 Aug. 2010, S.J.Taerum, PREM 61542, CMW41716 = CBS139657.
Host.
Pinus kesiya.
Beetle vectors.
Polygraphus aterrimus, Polygraphus szemaoensis.
Mite vectors. Proctolaelaps nr. hystrix (phoretic on Polygraphus szemaoensis), Insectolaelaps sp. 1 (phoretic on Polygraphus szemaoensis).
Distribution.
At present known only from Yunnan, China.
Notes.
Graphilbum kesiyae and Gra. puerense can be distinguished from Gra. crescericum by the presence of both synnematous and hyalorhinocladiella-like asexual states in culture. Gra. crescericum produces only the hyalorhinocladiella-like asexual state. The optimal temperature for growth of Gra. puerense is 30 °C while that for Gra. kesiyae is 25 °C, and synnemata of Gra. puerense reach double the length of those of Gra. kesiyae.
Taxon 13
Graphilbum puerense
R.Chang & Z.W.de Beer sp. nov.
823696
Fig. 10.
Morphological characters of asexual structures of Graphilbum puerense sp. nov. a fourteen days old culture on OA b–d Hyalorhinocladiella-like asexual state e conidia f Pesotum-like macronematal asexual state g–h cells of Pesotum-like macronematal asexual state and conidia. Scale bars: a–h = 10 μm.
Etymology.
The epithet puerense refers to the city from which this species was collected.
Description.
Sexual state not observed. Pesotum-like macronematal asexual states predominant. Synnemata simple, dark brown at the base, (187.5–) 206– 357(–437.5) μm long including conidiogenous apparatus, (12–) 15.5–45 (–61) μm wide at base; conidiogenous cells (15.5–) 18.5–30.5 (–34) μm long, conidia hyaline, 1-celled, smooth, oblong, (4–) 4–5 (–5.5) × (1–) 1.5–2 (–2.5) μm. Hyalorhinocladiella-like asexual state: conidiophores (17–) 3–140 (–232.5) μm long; cells arising directly from the hyphae, (6.5–) 10–25.5 (–42.5) × (1–) 1–2 (–3) μm; conidia hyaline, smooth, obovoid to oblong, (3.5–) 4–8 (–12) × (1–) 1.5–2.5 (–3) μm.
Culture characteristics.
Colonies hyaline. Mycelium superficial on the 3% OA. Colony margin smooth. Colonies on 2% MEA flat, reaching 76 mm diam in 5 d at 30 °C. No growth observed at 5 °C. Optimal temperature for growth 30 °C.
Type material.
CHINA, Yunnan Province, Puer City, from Polygraphus szemaoensis gallery on Pinus kesiya bark, 29 Jun. 2010, S.J.Taerum, herbarium specimen of dried culture, PREM 61543 (holotype), CMW41673 = CBS139640 (ex-holotype culture).
Additional specimens examined.
CHINA, Yunnan Province, Puer City, from Ips acuminatus gallery on Pinus kesiya bark, 4 Jul. 2010, S.J.Taerum, PREM 61544, CMW41667 = CBS139651; CHINA, Yunnan Province, Puer City, from Ips acuminatus gallery on Pinus kesiya bark, Jul 2010, S.J.Taerum, CMW41942 = CBS139650.
Host.
Pinus kesiya.
Beetle vectors.
Ips acuminatus, Polygraphus szemaoensis.
Mite vectors.
Proctolaelaps nr. hystrix (phoretic on Ips acuminatus), Insectolaelaps sp. 1 (phoretic on Ips acuminatus), and Uropodoidea sp. 2 (phoretic on Polygraphus szemaoensis).
Distribution.
At present known only from Yunnan, China.
Notes.
See comparison between Gra. kesiyae and Gra. puerense above under notes of Gra. kesiyae.
Taxon 16
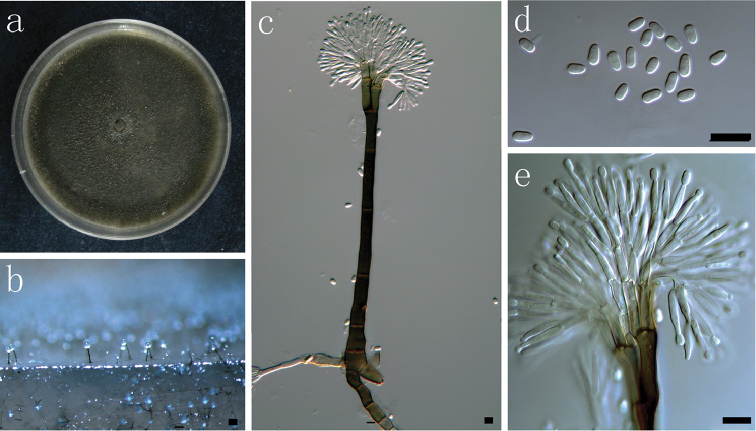
Leptographium ningerense
R.Chang & Z.W.de Beer sp. nov.
823697
Fig. 11.
Morphological characters of asexual structures of Leptographium ningerense sp. nov. a fourteen days old culture on OA b mononematous asexual morph on wood tissue on WA c conidiophore d conidia e conidiogenous apparatus. Scale bars: a–e = 10 μm.
Etymology.
The epithet ningerense refers to the Ning’er county where all isolates of this taxon were collected.
Description.
Sexual state not observed. Asexual state, conidiophores occurring singly or in groups of up to 3, macronematous, mononematous, erect, arising directly from the mycelium, (93.5–) 141.5–195.5 (–210.5) μm long. Rhizoids present. Stipes dark olivaceous, 4–6 septa, not constricted at septa, (66–) 119.5–142 (–159) μm long. Apical cells not swollen at apex, (3–) 5–6.5 (–7) μm wide. Basal cells occasionally swollen at apex, (5.5–) 7–10 (–11.5) μm wide. Conidiogenous apparatus (28–) 35–58 (–70) μm long, excluding the conidial mass, with multiple series of cylindrical branches. Primary branches olivaceous, smooth, cylindrical, not swollen at apex, aseptate, arrangement of primary branches was Type B—two or more branches, (12.5–) 14.5–18 (–19.5) × (3.5–) 4–5.5(–6.5) μm. Secondary branches light olivaceous, frequently swollen at apex, aseptate, (6.5–) 9–13(–15) × (3.5–) 4–5 (–5.5) μm. Tertiary branches light olivaceous, aseptate, (7–) 8–10 (–12) × (3–) 3.5–4.5 (–5) μm. Conidiogenous cells discrete, hyaline, 2–3 per branch, aseptate, cylindrical, tapering slightly at the apex, (10.5–) 12–17.5 (–20.5) × (2–) 2–2.5 (–3) μm. Conidia hyaline, aseptate, elliptical, (3–) 3.5–5.5 (–6.5) × (1.5–) 2–3 (–4) μm.
Culture characteristics.
Colonies on 3% OA flat, hyaline at the beginning, then becoming light olivaceous to dark olivaceous. Colonies hyaline at the beginning, then becoming dark olivaceous. Mycelium superficial on the 3% OA with olivaceous aerial mycelium. Colony margin smooth. Conidiophores forms abundantly in clusters on OA. Colonies on 2% MEA flat, reaching 76 mm diam in 10 d at 25 °C. No growth observed at 5 and 35 °C. Optimal temperature for growth 25 °C, reaching 30.5 mm in diam. in 7 days.
Type material.
CHINA, Yunnan Province, Puer City, from Schwiebea (Jacotietta) taiwanensis hyperphoretic on Coccotrypes cyperi on Pinus kesiya bark, 16 Sep. 2010, S.J.Taerum, herbarium specimen of dried culture, PREM 61545 (holotype), CMW41786 = CBS139663 (ex-holotype culture).
Additional specimens examined.
CHINA, Yunnan province, Puer City, from Insectolaelaps sp. 1 in Orthotomicus angulatus on Pinus kesiya bark, 17 Sep. 2010, S.J.Taerum, PREM 61546, CMW41831 = CBS139664.
Host.
Pinus kesiya.
Beetle vectors.
Polygraphus szemaoensis.
Mite vectors.
Dendrolaelaps sp. 1 (phoretic on Coccotrypes cyperi), Dendrolaelaps sp. 2 (phoretic on Coccotrypes cyperi and Cyrtogenius luteus), Schwiebea (Jacotietta) taiwanensis (phoretic on Coccotrypes cyperi), Insectolaelaps sp. 1 (Orthotomicus angulatus).
Distribution.
At present known only from Yunnan, China.
Notes.
L. ningerense is morphologically similar to L. pineti, but grows much more rapidly, reaching 30 mm in 7 d on 2% MEA at 25 °C while L. pineti reaches a diameter of only 15 mm in 6 d. However, the two species are best distinguished with BT, EF and CAL sequences.
Discussion
This study resulted in a total of 340 fungal isolates of ophiostomatoid fungi obtained from the beetles and weevils, their galleries and phoretic mites in a province of China where these fungi are poorly known. The fungi resided in the two phylogenetically unrelated Microascales and Ophiostomatales and included a species of Graphium (Microascales) as well as species of Sporothrix, Graphilbum, Leptographium and Ophiostoma (Ophiostomatales). Analysis showed that these isolates belonged to 19 distinct taxa, eight of which represented undescribed species of which five were provided with names. Of the remaining 11 species, 10 had previously been reported from China, with only S. nebularis representing a new report for this country. This study also includes the first records of fungi associated with the Scolytine beetle species Coccotrypes cyperi, Cyrtogenius luteus, and Tomicus brevilopus and it is the first time that fungal associates are reported from beetle-associated mites in China.
Surveys in this study aimed to explore fungal diversity in conifer-infesting beetle ecosystems in Yunnan. This is in contrast to determining the specificity of these interactions where a much more focused and systematic sampling would have been required. The exact nature of the interactions between the beetles and mites, mites and fungi, beetles and fungi, and all of these with their host trees, thus remains largely unknown. The discussion below therefore focuses on the fungi collected in these surveys, with limited notes on the hosts, beetles and mites and some general observations.
The two Sporothrix species collected in this study were primarily from mites. Sporothrix nebularis was the second most abundant species and 36 of the 45 isolates came from five different mite species phoretic on five different beetle species. The fungus was first described from Hylastes attenuatus infesting Pinus radiata in Spain (Romón et al. 2014b) and was later also found on Hylastes ater and Hylurgops palliatus from the same host and country (Romón et al. 2014a). Our results suggest that S. nebularis has a much wider Eurasian distribution and host range than previously realized. It is also possible that in Europe, many of the previous reports of S. stenoceras from nine bark beetle species (Kirisits 2004), actually represent S. nebularis. This is particularly because the two taxa are morphologically almost indistinguishable, and S. nebularis might have been misidentified as S. stenoceras where only morphology was considered (Romón et al. 2014b). The single isolate of Sporothrix sp. A was from a mite on Cyrtogenius luteus. Overall, the results support the suggestion by De Beer et al. (2016a) that the majority of Sporothrix species are mite associates.
Ophiostoma acarorum grouped peripheral to the well-known species complexes in Ophiostoma s.str., and closest to, but distinct from O. pallidulum and O. saponiodorum. O. pallidulum is known from ten different bark beetle and one weevil species infesting Pinus sylvestris in Finland (Linnakoski et al. 2010), Poland (Jankowiak and Bilański 2013a, b, c), and the Ukraine (Davydenko et al. 2017). Interestingly, O. pallidulum was also isolated from dead P. sylvestris roots in Poland in the absence of beetle attack (Jankowiak et al. 2012). O. saponiodorum is known from two spruce-infesting beetles in Finland and Russia (Linnakoski et al. 2010), pine-infesting Hylastes and Pissodes spp. in Poland and Spain (Jankowiak and Bilański 2013a, c, Romón et al. 2014a), and Orthotomicus erosus caught in traps in Italy (Malacrinò et al. 2017). Similar to O. pallidulum and O. saponiodorum, O. acarorum does not appear to be beetle-specific as it was isolated from three mite species phoretic on four beetle species, and from the galleries of two beetle species. However, the fact that 25 of 29 isolates in the present study were from mites, suggests that O. acarorum is a symbiont of mites on conifer-infesting beetles, rather than the beetles themselves. The wide range of beetle species from which O. pallidulum and O. saponiodorum have been isolated in Europe, always in relatively low abundance, also suggests that these two species are mite rather than beetle symbionts.
Ophiostoma ips was the species most frequently isolated in the study, constituting 31% of the 340 isolates. It was also the fungus found in association with the largest number of beetle species i.e. 10 of 17, and with three of the five host trees in central and southern Yunnan. This is consistent with current knowledge of the fungus that is known to have a global distribution (Zhou et al. 2007) and is associated with a wide range of conifer-infesting bark beetles in various genera (Kirisits 2004, Rumbold 1931, Upadhyay 1981). Shortly after the description of O. ips (Rumbold 1931), Leach et al. (1934) had already shown that ascospores of the fungus were vectored by mites associated with two Ips spp. Furthermore, Stone and Simpson (1990) observed that mycetophagous mites associated with Ips grandicollis fed directly on O. ips in the galleries of the beetle. About half of the 106 O. ips isolates from Yunnan came from mites associated with six of the bark beetle species. The results thus provide additional weight to the hypothesis that O. ips is primarily a mite associate, rather than a bark beetle associate.
Two taxa from Yunnan formed part of the O. clavatum complex as recently defined by Linnakoski et al. (2016). These authors showed that most of the species in the complex appear to be specific to one or two closely related Ips spp. on either a pine or spruce host from Europe. The only exception is O. macroclavatum, that is known from on three Ips spp. and Pityogenes chalcographus on both pine and spruce. Yin et al. (2016) subsequently described two additional species in the complex from Polygraphus poligraphus and Ips shangrila respectively, on spruce in Qinghai Province. Both fungi were also isolated from Dendroctonus micans in the same area. One of the species that we collected, O. brevipilosi, was from Tomicus brevipilosus beetles infesting P. kesiya. It is thus the first species in the complex associated with a Tomicus species. The other species from Yunnan, Ophiostoma sp. B (Taxon 5) was represented by only one isolate from the gallery of an unknown beetle on P. semaonensis.
Only a single species of the O. piceae complex, O. setosum, was obtained during the surveys. Five isolates of this species were from Pissodes galleries on Tsuga dumosa, confirming a previous report from the same host in China (Paciura et al. 2010b). The fungus was originally described from Tsuga heterophylla in Canada (Uzunovic et al. 2000). It was also reported from Tsuga and Pseudotsuga in the USA (Harrington et al. 2001), from Hylastes ater on pine in New Zealand (Reay et al. 2005) and pine logs imported from New Zealand to Korea (Kim et al. 2005a). In Bhutan, this species is associated with Ips schmutzenhoferi on Picea spinulosa and Pinus wallichiana (Kirisits et al. 2012). It thus seems as if O. setosum is a generalist associate of beetles infesting conifers in native Northern Hemisphere forests, and was most likely introduced into New Zealand.
The third most abundant species collected in this study was O. quercus that forms part of the O. ulmi species complex (De Beer and Wingfield 2013). This fungus is generally considered as hardwood-infecting with many reports from wounds and stained wood of well-known tree genera such as Quercus, Betula, Populus, Eucalyptus, Acacia, and Nothofagus from North and South America, Europe, Africa, Australasia, and Central and East Asia (De Beer et al. 2003, De Errasti et al. 2016, Grobbelaar et al. 2009, Harrington et al. 2001, Paciura et al. 2010b). Little is known regarding the insect vectors of O. quercus apart from reports that the fungus is vectored by nitidulid beetles on oak (Juzwik et al. 1998) and eucalypts (Kamgan Nkuekam et al. 2012). Unlike most other species in the O. ulmi complex, there is a growing body of evidence suggesting that O. quercus is also a successful colonist of conifer wood. It has been reported from stained pine wood in the USA, Sweden, Australia, New Zealand and South Africa (De Beer et al. 2003, Harrington et al. 2001), but also in association with pine bark beetles such as Tomicus yunnanensis in China (Paciura et al. 2010b), Hylastes plumbeus in Japan (Masuya et al. 2009), as well as Ips sexdentatus and Hylobius abietus in Poland (Jankowiak 2012, Jankowiak and Bilański 2013b). Furthermore, it has been found with Pissodes weevils on Tsuga in China (Paciura et al. 2010b) and pine in Poland (Jankowiak and Bilański 2013c).
The 43 O. quercus isolates obtained in this study came from four of the host trees distributed in eight of the ten study sites in both central and southern Yunnan. The O. quercus isolates were found in association with eight beetle species, with 13 of the isolates coming from three mite species. To the best of our knowledge, this represents the first records of this fungus from mites. However, spores of the closely related and morphologically similar O. novo-ulmi have been observed in the guts and sporothecae of mites phoretic on Scolytus beetle vectors of the Dutch Elm Disease fungi (Brasier 1978, Moser et al. 2010). The difference between the latter group of fungi and O. quercus is that they are specific to elm trees and elm-infesting beetles. In contrast, O. quercus is probably of all ophiostomatoid species the one with the widest array of hosts and the plasticity to utilize as a vector, any arthropod passing a wound or gallery in which it is growing. It is thus not surprizing that O. quercus is also one of the most genetically diverse ophiostomatoid species (Grobbelaar et al. 2009, Kamgan Nkuekam et al. 2010), which is supported by the large variety of haplotypes revealed in analyses of the BT and EF gene regions in this study.
Ophiostoma tsotsi was the second species in the O. ulmi complex obtained in this study. This species, which is morphologically indistinguishable from O. quercus, is known from wounds on planted Eucalyptus and Acacia mearnsii trees in Africa (Grobbelaar et al. 2010), Australia (Kamgan Nkuekam et al. 2011) and China (Grobbelaar et al. 2011), as well as native mangroves in South Africa (Osorio et al. 2016). The only insects from which the fungus has previously been reported are nitidulid beetles (Kamgan Nkuekam et al. 2012). We isolated O. tsotsi from galleries of two pine bark beetles, as well as from two mite species associated with one of these beetles. This was an unexpected result because, much like O. quercus, the assumption has been that O. tsotsi is an exclusively hardwood-infesting member of the O. ulmi complex. This complex was initially referred to as the ‘hardwood lineage’ of the O. piceae complex (De Beer et al. 2003, Harrington et al. 2001) before De Beer and Wingfield (2013) distinguished between the O. piceae and O. ulmi complexes. Although O. tsotsi is known from fewer hosts and locations than O. quercus, it also displays a relatively high level of genetic diversity as was evident from our BT and EF sequence analyses.
A single isolate referred to as Ophiostoma sp. C (Taxon 10) came from an unknown beetle gallery on Tsuga. This isolate was clearly distinct from all other species in the O. ulmi complex. Although we have chosen not to describe it as new, together with O. quercus and O. tsotsi, this is only the third of the 18 species currently recognized in the complex to be reported from conifer hosts.
Three of the taxa collected in the surveys resided in the genus Graphilbum that includes 10 known species and several undescribed taxa (De Beer and Wingfield 2013, Lu et al. 2009a, Reid and Hausner 2015, Romón et al. 2014b). All the known species are from conifers (Abies, Picea, Pinus, Tsuga) and mostly from bark beetles or their galleries, and in some cases stained wood. Only two undescribed species are from hardwoods (Geldenhuis et al. 2004, Kamgan Nkuekam et al. 2008). Graphilbum fragrans is by far the most common species with one report from bark beetles on spruce and fir in Canada (Jacobs et al. 2003). The remainder of the species have been collected from a variety of pine bark beetles and weevils from Europe (Mathiesen-Käärik 1953, Romón et al. 2007), the USA, New Zealand and Australia (Harrington et al. 2001), South Africa (Zhou et al. 2006), Japan (Masuya et al. 2009) and Korea (Kim et al. 2007).
Graphilbum fragrans found in this study had previously been reported from T. yunnanensis on Pinus yunnanensis, and Pissodes spp. on Tsuga dumosa and P. armandii in China (Paciura et al. 2010b, Zhou et al. 2013). In the present study, the fungus was isolated from galleries of Tomicus minor on P. kesiya. We collected 11 isolates each for Gra. kesiyae (Taxon 12) and Gra. puerense (Taxon 13). Gra. kesiyae was found in association with three beetle species with seven isolates from mites. Gra. puerense was from two beetle species with three isolates from mites. These are the first reports of any Graphilbum species isolated from mites.
Five species of fungi residing in Leptographium sensu lato were collected in this study, including one novel species. Ten isolates representing Taxon 14 were from Pissodes sp. on P. armandii and were shown to represent L. gracile. This species was described from the same insects and tree host as the isolates in the present study, as well as from Ips typographus on Picea koraiensis (Paciura et al. 2010a). Four other species grouped very closely to L. gracile in the L. procerum complex. Leptographium sinoprocerum and L. sinense were both described from China where they were associated with the introduced bark beetle Dendroctonus valens that infests and kills native P. tabuliformis (Lu et al. 2008), and native Hylobitelus xiaoi on non-native P. elliottii (Yin et al. 2015). Leptographium longiconidiophorum was originally isolated from P. densiflora in Japan (Yin et al. 2015) and L. bhutanense from Hylobitelus chenkupdorjii on Pinus wallichiana in Bhutan (Zhou et al. 2008).
A single isolate from the gallery of an unknown bark beetle on Tsuga grouped with Grosmannia radiaticola. This species was originally described from stained Pinus radiata wood imported from New Zealand to Korea (Kim et al. 2005b). However, it has been reported from a variety of pine root-infesting bark beetles from South Africa, Chile, Sweden (Linnakoski et al. 2012), California (Kim et al. 2011) and Poland (Jankowiak and Bilański 2013a, b). In China, it has been reported from a gallery of the invasive D. valens on P. tabuliformis in Shanxi, which might have resulted in the in the impression that the fungus was introduced with the beetle into China (Lu et al. 2009b). However, the present report from another conifer host in a distant province suggests that the fungus could be native across Eurasia.
Of the 13 isolates of L. ningerense collected in this study one isolate was from the gallery of Polygraphus szemaoensis on Pinus kesiya, while the remaining 12 isolates were from four mite species associated with Coccotrypes cyperi, Cyrtogenius luteus and Orthotomicus angulatus. This species grouped closest to L. pineti, which was described from the gallery of an Ips sp. under the bark of Pinus merkusii growing on the island of Sumatra, Indonesia (Jacobs et al. 2000). The fact that no isolates were obtained from beetles might indicate that this species is a preferential symbiont of mites.
The fourth most prevalent species collected in this study was G. yunnanensis including 39 isolates, 27 of which were vectored by seven mite species. G. yunnanensis was originally described from Tomicus yunnanensis on P. yunnanensis in China (Zhou et al. 2000), and was subsequently reported from Polygraphus major on Pinus kesiya in Thailand (Yamaoka et al. 2007), and four other pine bark beetle species on P. densiflora in Japan (Masuya et al. 2009). Results of the present study have added an additional three beetle species to this list including Cryphales luteus, Polygraphus szemaoensis and an unidentified Polygraphus sp. This seemingly promiscuous relationship of G. yunnanensis with nine different beetle species in East Asia, together with the fact that more than two thirds of the isolates in our study were from mites, suggests an association of this fungus with mites, rather than beetles.
Two isolates of L. conjunctum, including one from a mite, were collected in the present study. L. conjunctum has not previously been reported from any location other than in the case of the original study in which it was described from Hylurgops major on Pinus yunnanensis in China (Paciura et al. 2010a). Our one isolate was from H. major, while the mite from which the second isolate came, was collected from the unknown Polygraphus sp.
The single Graphium isolate arising from this study was from a mite on Ips acuminatus, and had identical sequences to Gr. pseudormiticum. This fungus was first described from South Africa in association with Orthotomicus erosus on pine bait logs (Mouton et al. 1994). It was subsequently found in Sweden associated with Ips typographus on an unknown host tree, in Austria with Tomicus minor on Pinus sylvestris (Lackner and de Hoog 2011), and in China associated with Pissodes sp. on Tsuga dumosa (Paciura et al. 2010b). This is the first report of a Graphium species associated with a mite. The low frequency with which it was encountered could result from the fact that Graphium species are unable to grow on media including the antibiotic cycloheximide, which is commonly used as in a selective medium for the Ophiostomatales in surveys of bark beetle fungi.
Conclusions
Three surveys of pine-infesting bark beetles in Yunnan revealed several new fungal species and new beetle-fungus associations. This supports the view that the diversity of fungi associated with bark beetles in China is high and that it deserves further exploration. The results of the 2010 survey that included isolations from mites, revealed that many of the ophiostomatoid species often considered as beetle associates can also be isolated frequently from mites. It has been suggested that mycetophagous mites are often generalists with the ability to feed on and vector several fungal species (Hofstetter et al. 2013). It is also known that phoretic mites are promiscuous in terms of their beetle vectors, and that they will infest virtually any beetle species in order to reach their tree hosts (Hofstetter et al. 2013). Our results underscore these views although structured surveys supported by statistical analyses should be undertaken to better understand these relationships.
One of the most surprising and intriguing outcomes of the study was that fungal species such as O. quercus and O. tsotsi, which were considered to be primarily hardwood-infecting species, are prevalent on several of the pine-specific beetles and their associated mites. Bark beetles commonly have broad host ranges on either the hardwoods or conifers, but not across these groups. The question thus arises how these hardwood-infecting fungi can move between these host groups. One possibility is that they simply move with mites from one tree in a forest to another, crossing the forest floor and carrying the fungi with them. However, it is also known that most bark beetle-associated mites are not monospecific and may be common on other non-bark beetle hosts as well (e.g. Tenebrionidae, Cleridae, Histeridae, Elateridae) (Hofstetter et al. 2013). Some of these often predatory beetles, could thus vector the mites and their fungi across the otherwise hardwood-conifer barrier. The determining factor would then be whether the fungus has the ability to survive in the new environment. The results of this study suggest that O. quercus and O. tsotsi are the rare exceptions among the ophiostomatoid fungi that are equally fit in both the angiosperm and gymnosperm environments.
None of the known fungal species collected in the present study are considered pathogens based on current published knowledge, and we have not found any evidence that any of the novel taxa are pathogens. However, the study underscores the possibility that many ophiostomatoid fungi that have been considered beetle associates in the past, might actually have a closer association with mites, or that mites at least have the ability to vector them. Generally, bark beetles and their fungal associates are considered serious threats with invasive potential when they are introduced into new environments through the movement of wood and wood products (Wingfield et al. 2017). However, most pest risk analyses consider only the beetles and the fungi. Our results suggest that even in cases where beetles do not successfully establish and reproduce in a new environment, mites could vector potential fungal pathogens and introduce them into established beetle ecosystems. The role of mites in mixing fungal assemblages of different beetle species on a single tree or log, especially in the case of invasive versus native beetle species, deservers futher exploration.
Supplementary Material
Acknowledgements
We acknowledge members of Tree Protection and Cooperation Programme (TPCP), Center of Excellence in Tree Health Biotechnology (CTHB), the National Research Foundation (NRF), the Department of Science and Technology (DST)/NRF and the University of Pretoria, South Africa. We are deeply grateful to Prof. Zhao of the Yunnan Academy of Forestry for his support during the collection surveys. We also thank Prof. Yihe Zhao, Dr Mingliang Yin and Miss Dongxia Xu for assistance with the fieldwork, Mr Deer Konkarn for assistance with sequencing, and Drs Roger Beaver and Eddie Ueckermann for beetle and mite identifications.
Citation
Chang R, Duong TA, Taerum SJ, Wingfield MJ, Zhou X, de Beer ZW (2017) Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in Yunnan, China. MycoKeys 28: 19–64. https://doi.org/10.3897/mycokeys.28.21758
Supplementary materials
Table S1–S3
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Runlei Chang, Tuan A. Duong, Stephen J. Taerum, Michael J. Wingfield, Xudong Zhou, Z. Wilhelm de Beer
Data type: species data
Explanation note:
Table S1. Numbers of ophiostomatoid fungal isolates obtained from different mite species in this study.
Table S2. Haplotypes of Ophiostoma quercus.
Table S3. Haplotypes of Ophiostoma tsotsi.
Figure S1–S11
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Runlei Chang, Tuan A. Duong, Stephen J. Taerum, Michael J. Wingfield, Xudong Zhou, Z. Wilhelm de Beer
Data type: occurence
Explanation note:
Figure S1. Map of Yunnan Province in China showing the sites and tree hosts from which samples were collected during the three surveys in 2001, 2002 and 2010.
Figure S2. ML trees of the genus Graphium generated from DNA sequences of the ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S3. ML trees of the genus Sporothrix generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S4. ML trees of the O. ips complex generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S5. ML trees of the O. piceae complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S6. ML tree of the O. quercus complex generated from DNA sequences of ITS region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S7. ML tree of the O. quercus complex generated from DNA sequences of BT region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S8. ML tree of the O. quercus complex generated from DNA sequences of EF region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S9. ML trees of the L. procerum complex generated from DNA sequences of BT, EF and CAL regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S10. ML trees of the G. galeiformis complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S11. ML trees of the L. lundbergii complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
References
- Bateman C, Huang YT, Simmons DR, Kasson MT, Stanley EL, Hulcr J. (2017) Ambrosia beetle Premnobius cavipennis (Scolytinae: Ipini) carries highly divergent ascomycotan ambrosia fungus, Afroraffaelea ambrosiae gen. nov. et sp. nov. (Ophiostomatales). Fungal Ecology 25: 41–49. https://doi.org/10.1016/j.funeco.2016.10.008 [Google Scholar]
- Brasier CM. (1978) Mites and reproduction in Ceratocystis ulmi and other fungi. Transactions of the British Mycological Society 70: 81–89. https://doi.org/10.1016/S0007-1536(78)80175-2 [Google Scholar]
- Davydenko K, Vasaitis R, Menkis A. (2017) Fungi associated with Ips acuminatus (Coleoptera: Curculionidae) in Ukraine with a special emphasis on pathogenicity of ophiostomatoid species. European Journal of Entomology 114: 77–85. http://dx.doi.org/10.14411/eje.2017.011 [Google Scholar]
- De Beer ZW, Duong TA, Barnes I, Wingfield BD, Wingfield MJ. (2014) Redefining Ceratocystis and allied genera. Studies in Mycology 79: 187–219. https://doi.org/10.1016/j.simyco.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Duong TA, Wingfield MJ. (2016a) The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Studies in Mycology 83: 165–191. http://dx.doi.org/10.1016/j.simyco.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beer ZW, Marincowitz S, Duong TA, Kim JJ, Rodrigues A, Wingfield MJ. (2016b) Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biology 120: 1323–1340. http://dx.doi.org/10.1016/j.funbio.2016.07.004 [DOI] [PubMed] [Google Scholar]
- De Beer ZW, Marincowitz S, Duong TA, Wingfield MJ. (2017) Bretziella, a new genus to accommodate the oak wilt fungus, Ceratocystis fagacearum. MycoKeys 27: 1–19. https://doi.org/10.3897/mycokeys.27.20657 [Google Scholar]
- De Beer ZW, Seifert KA, Wingfield MJ. (2013) The ophiostomatoid fungi: their dual position in the Sordariomycetes. In: Seifert KA, De Beer ZW, Wingfield MJ. (Eds) The Ophiostomatoid Fungi: Expanding Frontiers. CBS, Utrecht, The Netherlands, 1–19.
- De Beer ZW, Wingfield BD, Wingfield MJ. (2003) The Ophiostoma piceae complex in the Southern Hemisphere: a phylogenetic study. Mycological Research 107: 469–476. https://doi.org/10.1017/S0953756203007445 [DOI] [PubMed] [Google Scholar]
- De Beer ZW, Wingfield MJ. (2013) Emerging lineages in the Ophiostomatales. In: Seifert KA, De Beer ZW, Wingfield MJ. (Eds) The Ophiostomatoid Fungi: Expanding Frontiers. CBS, Utrecht, The Netherlands, 21–46.
- De Errasti A, De Beer ZW, Coetzee MPA, Roux J, Rajchenberg M, Wingfield MJ. (2016) Three new species of Ophiostomatales from Nothofagus in Patagonia. Mycological Progress 15: 17. http://dx.doi.org/10.1007/s11557-016-1158-z
- Duong TA, De Beer ZW, Wingfield BD, Wingfield MJ. (2012) Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia 104: 715–732. http://dx.doi.org/10.3852/11-109 [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Geldenhuis MM, Roux J, Montenegro F, De Beer ZW, Wingfield MJ, Wingfield BD. (2004) Identification and pathogenicity of Graphium and Pesotum species from machete wounds on Schizolobium parahybum in Ecuador. Fungal Diversity 15: 137–151. https://goo.gl/4MVXew [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. http://aem.asm.org/content/61/4/1323.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobbelaar JW, Aghayeva DN, de Beer ZW, Bloomer P, Wingfield MJ, Wingfield BD. (2009) Delimitation of Ophiostoma quercus and its synonyms using multiple gene phylogenies. Mycological Progress 8: 221–236. https://doi.org/10.1007/s11557-009-0594-4 [Google Scholar]
- Grobbelaar JW, De Beer ZW, Bloomer P, Wingfield MJ, Wingfield BD. (2010) Ophiostoma tsotsi sp. nov., a wound-infesting fungus of hardwood trees in Africa. Mycopathologia 169: 413–423. https://doi.org/10.1007/s11046-009-9267-8 [DOI] [PubMed] [Google Scholar]
- Grobbelaar JW, De Beer ZW, Bloomer P, Wingfield MJ, Zhou XD, Wingfield BD. (2011) Discovery of Ophiostoma tsotsi on Eucalyptus wood chips in China. Mycoscience 52: 111–118. https://doi.org/10.1007/s10267-010-0081-4 [Google Scholar]
- Harrington TC. (1993) Diseases of conifers caused by species of Ophiostoma and Leptographium. In: Wingfield MJ, Seifert KA, Webber JF. (Eds) Ceratocystis and Ophiostoma: Taxonomy, Ecology and Pathogenicity. APS, St. Paul, Minnesota, 161–172.
- Harrington TC. (2005) Ecology and evolution of mycophagous bark beetles and their fungal partners. In: Vega FE, Blackwell M. (Eds) Insect-Fungal Associations: Ecology and Evolution. Oxford University Press, Oxford, UK, 257–291.
- Harrington TC, McNew D, Steimel J, Hofstra D, Farrell R. (2001) Phylogeny and taxonomy of the Ophiostoma piceae complex and the Dutch elm disease fungi. Mycologia 93: 111–136. https://doi.org/10.2307/3761610 [Google Scholar]
- Hawksworth DL. (2011) A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys 1: 7–20 https://doi.org/10.3897/mycokeys.1.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgcock GG. (1906) Studies upon some chromogenic fungi which discolor wood. Missouri Botanical Garden Annual Report 1906: 59–114. https://doi.org/10.2307/2400089 [Google Scholar]
- Hofstetter RW, Cronin JT, Klepzig KD, Moser JC, Ayres MP. (2006) Antagonisms, mutualisms and commensalisms affect outbreak dynamics of the southern pine beetle. Oecologia 147: 679–691. https://doi.org/10.1007/s00442-005-0312-0 [DOI] [PubMed] [Google Scholar]
- Hofstetter RW, Dinkins-Bookwalter J, Davis TS, Klepzig KD. (2015) Symbiotic associations of bark beetles. In: Vega FE, Hofstetter RW. (Eds) Bark Beetles: Biology and Ecology of Native and Invasive Species. Academic Press, San Diego, 209–245. https://doi.org/10.1016/B978-0-12-417156-5.00006-X
- Hofstetter RW, Moser JC. (2014) The role of mites in insect-fungus associations. Annual Review of Entomology 59: 537–557. https://doi.org/10.1146/annurev-ento-011613-162039 [DOI] [PubMed] [Google Scholar]
- Hofstetter RW, Moser JC, Blomquist S. (2013) Mites associated with bark beetles and their hyperphoretic ophiostomatoid fungi. In: Seifert KA, De Beer ZW, Wingfield MJ. (Eds) The Ophiostomatoid Fungi: Expanding Frontiers. CBS Biodiversity Series, Utrecht, The Netherlands, 165–176.
- Jacobs K, Bergdahl DR, Wingfield MJ, Halik S, Seifert KA, Bright DE, Wingfield BD. (2004) Leptographium wingfieldii introduced into North America and found associated with exotic Tomicus piniperda and native bark beetles. Mycological Research 108: 411–418. https://doi.org/10.1017/S0953756204009748 [DOI] [PubMed] [Google Scholar]
- Jacobs K, Seifert KA, Harrison KJ, Kirisits T. (2003) Identity and phylogenetic relationships of ophiostomatoid fungi associated with invasive and native Tetropium species (Coleoptera: Cerambycidae) in Atlantic Canada. Canadian Journal of Botany 81: 316–329. https://doi.org/10.1139/b03-025 [Google Scholar]
- Jacobs K, Wingfield MJ. (2001) Leptographium species: Tree Pathogens, Insect Associates, and Agents of Blue-stain. APS, St. Paul, Minnesota.
- Jacobs K, Wingfield MJ, Bergdahl DR. (2000) New Leptographium species from Indonesia and Eastern North America. Mycoscience 41: 595–606. https://doi.org/10.1007/BF02460926 [Google Scholar]
- Jankowiak R. (2012) Ophiostomatoid fungi associated with Ips sexdentatus on Pinus sylvestris in Poland. Dendrobiology 68: 43–54. [Google Scholar]
- Jankowiak R, Bilański P. (2013a) Association of the pine-infesting Pissodes species with ophiostomatoid fungi in Poland. European Journal of Forest Research 132: 523–534. https://doi.org/10.1007/s10342-013-0693-2 [Google Scholar]
- Jankowiak R, Bilański P. (2013b) Diversity of ophiostomatoid fungi associated with the large pine weevil, Hylobius abietis, and infested Scots pine seedlings in Poland. Annals of Forest Science 70: 391–402. https://doi.org/10.1007/s13595-013-0266-z [Google Scholar]
- Jankowiak R, Bilański P. (2013c) Ophiostomatoid fungi associated with root-feeding bark beetles on Scots pine in Poland. Forest Pathology 43: 422–428. https://doi.org/10.1111/efp.12049 [Google Scholar]
- Jankowiak R, Bilański P, Kolařík M, Wasiuta D. (2012) Root-colonizing ophiostomatoid fungi associated with dying and dead young Scots pine in Poland. Forest Pathology 42: 492–500. https://doi.org/10.1111/j.1439-0329.2012.00783.x [Google Scholar]
- Juzwik J, Cease KR, Meyer JM. (1998) Acquisition of Ophiostoma quercus and Ceratocystis fagacearum by nitidulids from O. quercus-colonized oak wilt mats. Plant Disease 82: 239–243. http://dx.doi.org/10.1094/PDIS.1998.82.2.239 [DOI] [PubMed] [Google Scholar]
- Kamgan Nkuekam G, De Beer ZW, Wingfield MJ, Mohammed C, Carnegie AJ, Pegg GS, Roux J. (2011) Ophiostoma species (Ophiostomatales, Ascomycota), including two new taxa on eucalypts in Australia. Australian Journal of Botany 59: 283–297. http://hdl.handle.net/2263/16958 [Google Scholar]
- Kamgan Nkuekam G, De Beer ZW, Wingfield MJ, Roux J. (2012) A diverse assemblage of Ophiostoma species, including two new taxa on eucalypt trees in South Africa. Mycological Progress 11: 515–533. https://doi.org/10.1007/s11557-011-0767-9 [Google Scholar]
- Kamgan Nkuekam G, Jacobs K, De Beer ZW, Wingfield MJ, Roux J. (2008) Ceratocystis and Ophiostoma species including three new taxa, associated with wounds on native South African trees. Fungal Diversity 29: 37–59. https://goo.gl/FJF2KE [Google Scholar]
- Kamgan Nkuekam G, Solheim H, De Beer ZW, Grobbelaar JW, Jacobs K, Wingfield MJ, Roux J. (2010) Ophiostoma species, including Ophiostoma borealis sp. nov., infecting wounds of native broad-leaved trees in Norway. Cryptogamie, Mycologie 31: 285–303. http://hdl.handle.net/2263/16486 [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GH, Kim JJ, Breuil C. (2007) Sap-staining fungi from logs and boards of two commercially important pines in Korea. Holzforschung 61: 333–336. https://doi.org/10.1515/HF.2007.042 [Google Scholar]
- Kim GH, Kim JJ, Lim YW, Breuil C. (2005a) Ophiostomatoid fungi isolated from Pinus radiata logs imported from New Zealand to Korea. Canadian Journal of Botany 83: 272–278. https://doi.org/10.1139/b04-170 [Google Scholar]
- Kim JJ, Lim YW, Seifert KA, Kim SH, Breuil C, Kim GH. (2005b) Taxonomy of Ophiostoma radiaticola sp. nov. (Ophiostomatales, Ascomycetes), the teleomorph of Pesotum pini, isolated from logs of Pinus radiata. Mycotaxon 91: 481–496. [Google Scholar]
- Kim SJ, Harrington TC, Lee JC, Seybold SJ. (2011) Leptographium tereforme sp. nov. and other Ophiostomatales isolated from the root-feeding bark beetle Hylurgus ligniperda in California. Mycologia 103: 152–163. 10.3852/10-096 [DOI] [PubMed]
- Kirisits T. (2004) Fungal associates of european bark beetles with special emphasis on the ophiostomatoid fungi. In: Lieutier F, Day K, Battisti A, Grégoire JC, Evans H. (Eds) Bark and Wood Boring Insects in Living Trees in Europe, a Synthesis. Springer, Netherlands, 181–236. https://doi.org/10.1007/978-1-4020-2241-8_10
- Kirisits T, Konrad H, Wingfield MJ, Chhetri DB. (2012) Ophiostomatoid fungi associated with the Eastern Himalayan spruce bark beetle (Ips schmutzenhoferi) in Bhutan: Species assemblage and phytopathogenicity. Journal of Agricultural Extension and Rural Development 4: 266–268. https://doi.org/10.5897/JAERD12.075 [Google Scholar]
- Klepzig KD, Hofstetter RW. (2011) From attack to emergence: Interactions between southern pine beetle, mites, microbes, and trees. In: Coulson RN, Klepzig KD. (Eds) Southern Pine Beetle II. USDA Forest Service, 141–152.
- Lackner M, de Hoog GS. (2011) Parascedosporium and its relatives: phylogeny and ecological trends. IMA Fungus 2: 39–48. https://doi.org/10.5598/imafungus.2011.02.01.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JG, Orr LW, Christensen C. (1934) The interrelationship of bark beetles and blue staining fungi in felled Norway pine timber. Journal of Agricultural Research 49: 315–341. [Google Scholar]
- Linnakoski R, De Beer ZW, Ahtiainen J, Sidorov E, Niemela P, Pappinen A, Wingfield MJ. (2010) Ophiostoma spp. associated with pine- and spruce-infesting bark beetles in Finland and Russia. Persoonia 25: 72–93. https://doi.org/10.3767/003158510X550845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnakoski R, De Beer ZW, Duong TA, Niemela P, Pappinen A, Wingfield MJ. (2012) Grosmannia and Leptographium spp. associated with conifer-infesting bark beetles in Finland and Russia, including Leptographium taigense sp. nov. Antonie van Leeuwenhoek 102: 375–399. https://doi.org/10.1007/s10482-012-9747-6 [DOI] [PubMed] [Google Scholar]
- Linnakoski R, De Beer ZW, Rousi M, Niemela P, Pappinen A, Wingfield MJ. (2008) Fungi, including Ophiostoma karelicum sp. nov., associated with Scolytus ratzeburgi infesting birch in Finland and Russia. Mycological Research 112: 1475–1488. https://doi.org/10.1016/j.mycres.2008.06.007 [DOI] [PubMed] [Google Scholar]
- Linnakoski R, Jankowiak R, Villari C, Kirisits T, Solheim H, De Beer ZW, Wingfield MJ. (2016) The Ophiostoma clavatum species complex: a newly defined group in the Ophiostomatales including three novel taxa. Antonie van Leeuwenhoek 109: 987–1018. https://doi.org/10.1007/s10482-016-0700-y [DOI] [PubMed] [Google Scholar]
- Liu XW, Wang HM, Lu Q, Decock C, Li YX, Zhang XY. (2017) Taxonomy and pathogenicity of Leptographium species associated with Ips subelongatus infestations of Larix spp. in northern China, including two new species. Mycological Progress 16: 1–13. https://doi.org/10.1007/s11557-016-1245-1 [Google Scholar]
- Lu M, Zhou XD, De Beer ZW, Wingfield MJ, Sun JH. (2009a) Ophiostomatoid fungi associated with the invasive pine-infesting bark beetle, Dendroctonus valens, in China. Fungal Diversity 38: 133–145. http://hdl.handle.net/2263/13580 [Google Scholar]
- Lu Q, Decock C, Zhang XY, Maraite H. (2008) Leptographium sinoprocerum sp. nov., an undescribed species associated with Pinus tabuliformis-Dendroctonus valens in northern China. Mycologia 100: 275–290. http://dx.doi.org/10.1080/15572536.2008.11832482 [DOI] [PubMed] [Google Scholar]
- Lu Q, Decock C, Zhang XY, Maraite H. (2009b) Ophiostomatoid fungi (Ascomycota) associated with Pinus tabuliformis infested by Dendroctonus valens (Coleoptera) in northern China and an assessment of their pathogenicity on mature trees. Antonie Van Leeuwenhoek 96: 275–293. https://doi.org/10.1007/s10482-009-9343-6 [DOI] [PubMed] [Google Scholar]
- Malacrinò A, Rassati D, Schena L, Mehzabin R, Battisti A, Palmeri V. (2017) Fungal communities associated with bark and ambrosia beetles trapped at international harbours. Fungal Ecology 28: 44–52. https://doi.org/10.1016/j.funeco.2017.04.007 [Google Scholar]
- Malloch D, Blackwell M. (1993) Dispersal biology of the ophiostomatoid fungi. In: Wingfield MJ, Seifert KA, Webber JF. (Eds) Ceratocystis and Ophiostoma: Taxonomy, Ecology and Pathology. APS Press, St. Paul, 195–206.
- Marincowitz S, Duong TA, De Beer ZW, Wingfield MJ. (2015) Cornuvesica: A little known mycophilic genus with a unique biology and unexpected new species. Fungal Biology 119: 615–630. http://dx.doi.org/10.1016/j.funbio.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Masuya H, Yamaoka Y, Kaneko S, Yamaura Y. (2009) Ophiostomatoid fungi isolated from Japanese red pine and their relationships with bark beetles. Mycoscience 50: 212–223. https://doi.org/10.1007/s10267-008-0474-9 [Google Scholar]
- Masuya H, Yamaoka Y, Wingfield MJ. (2013) Ophiostomatoid fungi and their associations with bark beetles in Japan. In: Seifert KA, De Beer ZW, Wingfield MJ. (Eds) Ophiostomatoid Fungi: Expanding Frontiers. CBS, Utrecht, The Netherlands, 77–89.
- Mathiesen-Käärik A. (1953) Eine Übersicht über die gewöhnlichsten mit Borkenkäfern assoziierten Bläuepilze in Schweden und einige für Schweden neue Bläuepilze. Meddelanden från Statens Skogsforskningsinstitut 43: 1–74. https://goo.gl/NQTYoq [Google Scholar]
- Mayers CG, McNew DL, Harrington TC, Roeper RA, Fraedrich SW, Biedermann PHW, Castrillo LA, Reed SE. (2015) Three genera in the Ceratocystidaceae are the respective symbionts of three independent lineages of ambrosia beetles with large, complex mycangia. Fungal Biology 119: 1075–1092. https://doi.org/10.1016/j.funbio.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Mitchell R. (1970) An analysis of dispersal in mites. American Naturalist 104: 425–431. https://doi.org/10.1086/282677 [Google Scholar]
- Moser JC, Konrad H, Blomquist SR, Kirisits T. (2010) Do mites phoretic on elm bark beetles contribute to the transmission of Dutch elm disease? Naturwissenschaften 97: 219–227. https://doi.org/10.1007/s00114-009-0630-x [DOI] [PubMed]
- Moser JC, Perry TJ, Furuta K. (1997) Phoretic mites and their hyperphoretic fungi associated with flying Ips typographus japonicus Niijima (Col., Scolytidae) in Japan. Journal of Applied Entomology 121: 425–428. https://doi.org/10.1111/j.1439-0418.1997.tb01429.x [Google Scholar]
- Mouton M, Wingfield MJ, Van Wyk PS, Van Wyk PWJ. (1994) Graphium pseudormiticum sp. nov.: a new hyphomycete with unusual conidiogenesis. Mycological Research 98: 1272–1276. http://dx.doi.org/10.1016/S0953-7562(09)80298-0 [Google Scholar]
- Münch E. (1907) Die Blaufäule des Nadelholzes. I–II. Naturwissenschaftliche Zeitschrift für Forst- und Landwirtschaft 5: 531–573. [Google Scholar]
- Nel WJ, Duong TA, Wingfield BD, Wingfield MJ, De Beer ZW. (2017) A new genus and species for the globally important, multi-host root pathogen Thielaviopsis basicola Plant Pathology. https://doi.org/10.1111/ppa.12803
- Nisikado Y, Yamauti K. (1933) Contributions to the knowledge of the sap stains of wood in Japan. I. Studies on Ceratostomella ips Rumbold, the cause of blue stain of pine trees in Western Japan. Berichte des Ohara Instituts für Landwirtschaftliche Forschungen 5: 501–538. [Google Scholar]
- Osorio JA, De Beer ZW, Wingfield MJ, Roux J. (2016) Ophiostomatoid fungi associated with mangroves in South Africa, including Ophiostoma palustre sp. nov. Antonie van Leeuwenhoek 109: 1555–1571. https://doi.org/10.1007/s10482-016-0757-7 [DOI] [PubMed] [Google Scholar]
- Paciura D, De Beer ZW, Jacobs K, Zhou XD, Ye H, Wingfield MJ. (2010a) Eight new Leptographium species associated with tree-infesting bark beetles in China. Persoonia 25: 94–108. https://doi.org/10.3767/003158510X551097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciura D, Zhou XD, De Beer ZW, Jacobs K, Ye H, Wingfield MJ. (2010b) Characterisation of synnematous bark beetle-associated fungi from China, including Graphium carbonarium sp. nov. Fungal Diversity 40: 75–88. https://doi.org/10.1007/s13225-009-0004-x [Google Scholar]
- Paine TD, Raffa KF, Harrington TC. (1997) Interactions among scolytid bark beetles, their associated fungi, and live host conifers. Annual Review of Entomology 42: 179–206. https://doi.org/10.1146/annurev.ento.42.1.179 [DOI] [PubMed] [Google Scholar]
- Pfammatter JA, Moser JC, Raffa KF. (2013) Mites phoretic on Ips pini (Coleoptera: Curculionidae: Scolytinae) in Wisconsin red pine stands. Annals of the Entomological Society of America 106: 204–213. https://doi.org/10.1603/an12122 [Google Scholar]
- Reay S, Thwaites J, Farrell R. (2005) A survey of Ophiostoma species vectored by Hylastes ater to pine seedlings in New Zealand. Forest Pathology 35: 105–113. https://doi.org/10.1111/j.1439-0329.2004.00393.x [Google Scholar]
- Reid J, Hausner G. (2015) A new Graphilbum species from western hemlock (Tsuga heterophylla) in Canada. Mycotaxon 130: 399–419. https://doi.org/10.5248/130.399 [Google Scholar]
- Roets F, Crous PW, Wingfield MJ, Dreyer LL. (2009) Mite-mediated hyperphoretic dispersal of Ophiostoma spp. from the infructescences of South African Protea spp. Environmental Entomology 38: 143–152. https://doi.org/10.1603/022.038.0118 [DOI] [PubMed] [Google Scholar]
- Roets F, Wingfield MJ, Crous PW, Dreyer LL. (2007) Discovery of fungus-mite mutualism in a unique niche. Environmental Entomology 36: 1226–1237. https://doi.org/10.1093/ee/36.5.1226 [DOI] [PubMed] [Google Scholar]
- Romón P, De Beer ZW, Fernández M, Diez J, Wingfield BD, Wingfield MJ. (2014a) Ophiostomatoid fungi including two new fungal species associated with pine root-feeding beetles in northern Spain. Antonie van Leeuwenhoek 106: 1167–1184. https://doi.org/10.1007/s10482-014-0286-1 [DOI] [PubMed] [Google Scholar]
- Romón P, De Beer ZW, Zhou XD, Duong TA, Wingfield BD, Wingfield MJ. (2014b) Multigene phylogenies of Ophiostomataceae associated with Monterey pine bark beetles in Spain reveal three new fungal species. Mycologia 106: 119–132. https://doi.org/10.3852/13-073 [DOI] [PubMed] [Google Scholar]
- Romón P, Zhou XD, Iturrondobeitia JC, Wingfield MJ, Goldarazena A. (2007) Ophiostoma species (Ascomycetes: Ophiostomatales) associated with bark beetles (Coleoptera: Scolytinae) colonizing Pinus radiata in northern Spain. Canadian Journal of Microbiology 53: 756–767. https://doi.org/10.1139/w07-001 [DOI] [PubMed] [Google Scholar]
- Rumbold CT. (1931) Two blue-staining fungi associated with bark-beetle infestation of pines. Journal of Agricultural Research 43: 847–873. http://books.google.co.za/books?id=HjzMJAAACAAJ [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, Wingfield MJ, Aime MC, An KD, Bai FY, Barreto RW, Begerow D, Bergeron MJ, Blackwell M, Boekhout T, Bogale M, Boonyuen N, Burgaz AR, Buyck B, Cai L, Cai Q, Cardinali G, Chaverri P, Coppins BJ, Crespo A, Cubas P, Cummings C, Damm U, De Beer ZW, De Hoog GS, Del-Prado R, B D, Dieguez-Uribeondo J, Divakar PK, Douglas B, Duenas M, Duong TA, Eberhardt U, Edwards JE, Elshahed MS, Fliegerova K, Furtado M, Garcia MA, Ge ZW, Griffith GW, Griffiths K, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Guo LD, Hagen F, Hambleton S, Hamelin RC, Hansen K, Harrold P, Heller G, Herrera G, Hirayama K, Hirooka Y, Ho HM, Hoffmann K, Hofstetter V, Hognabba F, Hollingsworth PM, Hong SB, Hosaka K, Houbraken J, Hughes K, Huhtinen S, Hyde KD, James T, Johnson EM, Johnson JE, Johnston PR, Jones EB, Kelly LJ, Kirk PM, Knapp DG, Koljalg U, GM K, Kurtzman CP, Landvik S, Leavitt SD, Liggenstoffer AS, Liimatainen K, Lombard L, Luangsa-Ard JJ, Lumbsch HT, Maganti H, Maharachchikumbura SS, Martin MP, May TW, McTaggart AR, Methven AS, Meyer W, Moncalvo JM, Mongkolsamrit S, Nagy LG, Nilsson RH, Niskanen T, Nyilasi I, Okada G, Okane I, Olariaga I, Otte J, Papp T, Park D, Petkovits T, Pino-Bodas R, Quaedvlieg W, Raja HA, Redecker D, T R, Ruibal C, Sarmiento-Ramirez JM, Schmitt I, Schussler A, Shearer C, Sotome K, Stefani FO, Stenroos S, Stielow B, Stockinger H, Suetrong S, Suh SO, Sung GH, Suzuki M, Tanaka K, Tedersoo L, Telleria MT, Tretter E, Untereiner WA, Urbina H, Vagvolgyi C, Vialle A, Vu TD, Walther G, Wang QM, Wang Y, Weir BS, Weiss M, White MM, Xu J, Yahr R, Yang ZL, Yurkov A, Zamora JC, Zhang N, Zhuang WY, Schindel D, Consortium FB. (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. https://doi.org/10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA. (1993) Sapstain of commercial lumber by species of Ophiostoma and Ceratocystis. In: Wingfield MJ, Seifert KA, Webber JF. (Eds) Ceratocystis and Ophiostoma: Taxonomy, Ecology, and Pathogenicity. American Phytopathological Society, St. Paul, Minnesota, USA, 141–151.
- Six DL. (2003) Bark beetle-fungus symbioses. In: Bourtzis K, Miller T. (Eds) Insect Symbiosis. CRC Press, Boca Raton, Florida, USA, 77–114. https://doi.org/10.1201/9780203009918.ch7
- Six DL, Wingfield MJ. (2011) The role of phytopathogenicity in bark beetle-fungus symbioses: a challenge to the classic paradigm. Annual Review of Entomology 56: 255–272. https://doi.org/10.1146/annurev-ento-120709-144839 [DOI] [PubMed] [Google Scholar]
- Stielow JB, Lévesque CA, Seifert KA, Meyer W, Irinyi L, Smits D, Renfurm R, Verkley GJM, Groenewald M, Chaduli D, Lomascolo A, Welti S, Lesage-Meessen L, Favel A, Al-Hatmi AMS, Damm U, Yilmaz N, Houbraken J, Lombard L, Quaedvlieg W, Binder M, Vaas LAI, Vu D, Yurkov A, Begerow D, Roehl O, Guerreiro M, Fonseca A, Samerpitak K, van Diepeningen AD, Dolatabadi S, Moreno LF, Casaregola S, Mallet S, Jacques N, Roscini L, Egidi E, Bizet C, Garcia-Hermoso D, Martín MP, Deng S, Groenewald JZ, Boekhout T, De Beer ZW, Barnes I, Duong TA, Wingfield MJ, De Hoog GS, Crous PW, Lewis CT, Hambleton S, Moussa TAA, Al-Zahrani HS, Almaghrabi OA, Louis-Seize G, Assabgui R, McCormick W, Omer G, Dukik K, Cardinali G, Eberhardt U, De Vries M, Robert V. (2015) One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35: 242–263. https://doi.org/10.3767/003158515X689135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone C, Simpson JA. (1990) Species associations in Ips grandicollis galleries in Pinus taeda. New Zealand Journal of Forestry Science 20: 75–96. [Google Scholar]
- Taerum SJ, Duong TA, De Beer ZW, Gillette N, Sun JH, Owen DR, Wingfield MJ. (2013) Large shift in symbiont assemblage in the invasive red turpentine beetle. PLoS ONE 8: e78126. http://dx.doi.org/10.1371/journal.pone.0078126 [DOI] [PMC free article] [PubMed]
- Upadhyay HP. (1981) Monograph of Ceratocystis and Ceratocystiopsis. University of Georgia Press, Athens, Georgia, USA.
- Uzunovic A, Seifert KA, Kim SH, Breuil C. (2000) Ophiostoma setosum, a common sapwood staining fungus from western North America, a new species of the Ophiostoma piceae complex. Mycological Research 104: 486–494. https://doi.org/10.1017/S0953756299001446 [Google Scholar]
- Van der Linde JA, Six DL, De Beer ZW, Wingfield MJ, Roux J. (2016) Novel ophiostomatalean fungi from galleries of Cyrtogenius africus (Scolytinae) infesting dying Euphorbia ingens. Antonie van Leeuwenhoek 109: 589–601. https://doi.org/10.1007/s10482-016-0661-1 [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238–4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego (California), 315–322.
- Wingfield MJ, Barnes I, De Beer ZW, Roux J, Wingfield BD, Taerum SJ. (2017) Novel associations between ophiostomatoid fungi, insects and tree hosts: current status – future prospects. Biological Invasions: 1–14. https://doi.org/10.1007/s10530-017-1468-3
- Yamaoka Y, Masuya H, Chung WH, Goto H, To-Anun C, Tokumasu S. (2007) Two species of Leptographium isolated from blue-stained sapwood of Pinus khasya and bark beetles in Thailand. Mycoscience 48: 100–103. https://doi.org/10.1007/s10267-006-0337-1 [Google Scholar]
- Yamaoka Y, Masuya H, Chung WH, Goto H, To-Anun C, Tokumasu S, Zhou X, Wingfield MJ. (2008) The teleomorph of Leptographium yunnanense, discovered in crosses among isolates from Thailand, China, and Japan. Mycoscience 49: 233–240. https://doi.org/10.1007/s10267-008-0412-x [Google Scholar]
- Yang YM, Tian K, Hao JM, Pei SJ, Yang YX. (2004) Biodiversity and biodiversity conservation in Yunnan, China. Biodiversity & Conservation 13: 813–826. https://doi.org/10.1023/B:BIOC.0000011728.46362.3c [Google Scholar]
- Yin HF, Huang F, Li Z. (1984) Economic Insect Fauna of China. Science Press, Beijing, China.
- Yin ML, Duong TA, Wingfield MJ, Zhou XD, De Beer ZW. (2015) Taxonomy and phylogeny of the Leptographium procerum complex, including Leptographium sinense sp. nov. and Leptographium longiconidiophorum sp. nov. Antonie van Leeuwenhoek 107: 1–17. https://doi.org/10.1007/s10482-014-0351-9 [DOI] [PubMed] [Google Scholar]
- Yin ML, Wingfield MJ, Zhou XD, De Beer ZW. (2016) Multigene phylogenies and morphological characterization of five new Ophiostoma spp. associated with spruce-infesting bark beetles in China. Fungal Biology 120: 454–470. http://dx.doi.org/10.1016/j.funbio.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Zhou XD, Burgess TI, De Beer ZW, Lieutier F, Yart A, Klepzig K, Carnegie A, Portales JM, Wingfield BD, Wingfield MJ. (2007) High intercontinental migration rates and population admixture in the sapstain fungus Ophiostoma ips. Molecular Ecology 16: 89–99. https://doi.org/10.1111/j.1365-294X.2006.03127.x [DOI] [PubMed] [Google Scholar]
- Zhou XD, De Beer ZW, Wingfield MJ. (2006) DNA sequence comparisons of Ophiostoma spp., including Ophiostoma aurorae sp. nov., associated with pine bark beetles in South Africa. Studies in Mycology 55: 269–277. http://dx.doi.org/10.3114/sim.55.1.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XD, De Beer ZW, Wingfield MJ. (2013) Ophiostomatoid fungi associated with conifer-infecting bark beetles in China. In: Seifert KA, De Beer ZW, Wingfield MJ. (Eds) Ophiostomatoid fungi: Expanding frontiers. CBS, Utrecht, The Netherlands, 91–98.
- Zhou XD, Jacobs K, Kirisits T, Chhetri D, Wingfield MJ. (2008) Leptographium bhutanense sp. nov., associated with the root collar weevil Hylobitelus chenkupdorjii on Pinus wallichiana in Bhutan. Persoonia 21: 1–8. https://doi.org/10.3767/003158508X332435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XD, Jacobs K, Morelet M, Ye H, Lieutier F, Wingfield MJ. (2000) A new Leptographium species associated with Tomicus piniperda in south-western China. Mycoscience 41: 573–578. https://doi.org/10.1007/BF02460923 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1–S3
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Runlei Chang, Tuan A. Duong, Stephen J. Taerum, Michael J. Wingfield, Xudong Zhou, Z. Wilhelm de Beer
Data type: species data
Explanation note:
Table S1. Numbers of ophiostomatoid fungal isolates obtained from different mite species in this study.
Table S2. Haplotypes of Ophiostoma quercus.
Table S3. Haplotypes of Ophiostoma tsotsi.
Figure S1–S11
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Runlei Chang, Tuan A. Duong, Stephen J. Taerum, Michael J. Wingfield, Xudong Zhou, Z. Wilhelm de Beer
Data type: occurence
Explanation note:
Figure S1. Map of Yunnan Province in China showing the sites and tree hosts from which samples were collected during the three surveys in 2001, 2002 and 2010.
Figure S2. ML trees of the genus Graphium generated from DNA sequences of the ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S3. ML trees of the genus Sporothrix generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S4. ML trees of the O. ips complex generated from DNA sequences of ITS and BT regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S5. ML trees of the O. piceae complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S6. ML tree of the O. quercus complex generated from DNA sequences of ITS region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S7. ML tree of the O. quercus complex generated from DNA sequences of BT region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S8. ML tree of the O. quercus complex generated from DNA sequences of EF region. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S9. ML trees of the L. procerum complex generated from DNA sequences of BT, EF and CAL regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S10. ML trees of the G. galeiformis complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.
Figure S11. ML trees of the L. lundbergii complex generated from DNA sequences of BT and EF regions. Bold branches indicate posterior probabilities values ≥ 0.95. Bootstrap values ≥ 70 % are recorded at nodes as ML/MP. T = ex-type isolates.