Abstract
South China and Indochina host striking species diversity and endemism. Complex tectonic and climatic evolutions appear to be the main drivers of the biogeographic patterns. In this study, based on the geologic history of this region, we test 2 hypotheses using the evolutionary history of Microhyla fissipes species complex. Using DNA sequence data from both mitochondrial and nuclear genes, we first test the hypothesis that the Red River is a barrier to gene flow and dispersal. Second, we test the hypothesis that Pleistocene climatic cycling affected the genetic structure and population history of these frogs. We detect 2 major genetic splits that associate with the Red River. Time estimation suggests that late Miocene tectonic movement associated with the Red River drove their diversification. Species distribution modeling (SDM) resolves significant ecological differences between sides of the Red River. Thus, ecological divergence also probably promoted and maintained the diversification. Genogeography, historical demography, and SDM associate patterns in southern China with climate changes of the last glacial maximum (LGM), but not Indochina. Differences in geography and climate between the 2 areas best explain the discovery. Responses to the Pleistocene glacial–interglacial cycling vary among species and regions.
Keywords: demographic histories, ecological divergence, genogeography, LGM, pigmy narrow-mouth frog
Southern China and Indochina harbor extraordinary high levels of species diversity and share a zoogeographic fauna in the Oriental Region (Myers et al. 2000; Holt et al. 2013). The region includes China, Vietnam, Laos, Cambodia, Thailand, and Myanmar. This biodiversity has attracted much attention (e.g., Stuart et al. 2006; Matsui et al. 2010; McLeod 2010). High species richness in these areas has been attributed in large part to its complex geological and climatic history (e.g., Pleistocene climatic oscillations) and resulting changes in topography and environment (Woodruff 2010; de Bruyn et al. 2014). Advances in speciation and diversification, conservation, biogeography, and geology depend on understanding how these historic processes shaped the spatial patterns of genetic diversity, i.e., the distributions of species.
Tectonic movement and orogenesis can drive habitat fragmentation and create barriers to gene flow. Such events can drive genetic diversification and speciation (Che et al. 2010). Probably, the most famous geological formation of this area is the Red River shear zone, a major geological strike-slip fault zone that demarks the boundary between South China and Indochina tectonic plates (Leloup et al. 1995; Hall 1998). This shear zone runs southward from the southeastern corner of Tibet to the Gulf of Tonkin along the Red River (Figure 1). The Red River shear zone has a complex geological history (Hall 1998; Replumaz et al. 2001). For example, left-lateral Red River shearing occurred in the Oligocene and early Miocene (30∼15 Ma; Tapponnier et al. 1990; Searle 2006). This was followed several times by right-lateral strike-slips during late Miocene (e.g., 12.17∼9.19, 8.14∼6.18, Zhang et al. 2009). These events caused habitat changes along the Red River, which could act as potential geographic barriers for species dispersal and, thus, promote speciation. Many taxa, including both plants and animals, occur in 1 side of the Red River only (reviewed in Li and Li 1992, 1997; Bain and Hurley 2011; Fan et al. 2013; and references therein), or show genetic differences between populations on either side of the Red River (Zhang et al. 2010a, 2010b).
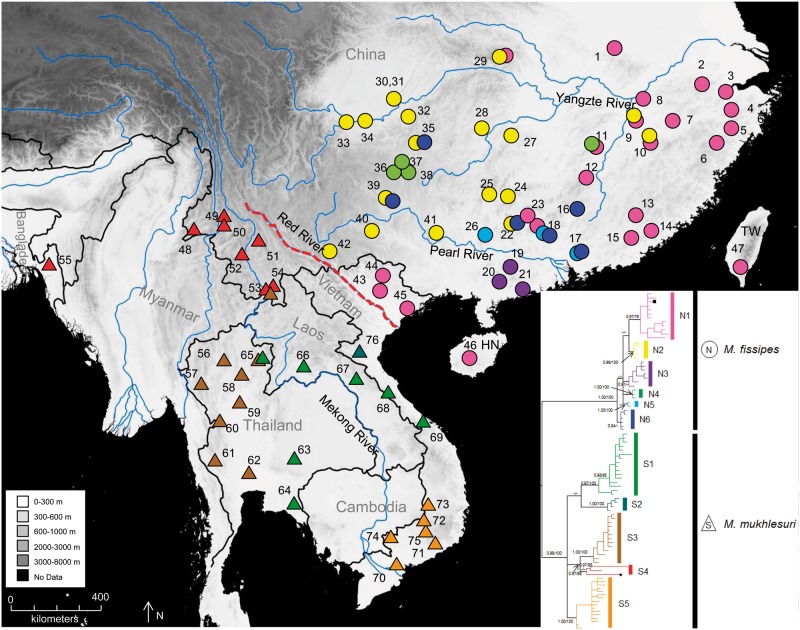
Figure 1.
Geographic distribution of samples of the M. fissipes complex. Localities are detailed in Supplementary Table S1 (supporting information). Colored triangles and circles correspond to the major matrilines in Figure 2. Dotted line denotes the Red River. Inset shows a simplified genealogy with major matrilines corresponding to Figure 2. The abbreviations HN and TW refer to Hainan and Taiwan, China, respectively.
Climatic cycling, especially during the Late Pleistocene, shaped the current distributions, genetic diversification, and demographic dynamics of many species (Hewitt 2000, 2004). Although southern China and Indochina were not glaciated, they experienced cooler and possibly drier climates during the Pleistocene (Williams et al. 1998; Li et al. 2004). In pace with the cyclical cooling–warming climate, many species experienced periodic habitat expansions and contractions to meet their ecological requirements. For example, during glacial times, species retracted into small refugia and during postglacial periods their ranges expanded. In southern China, both some plants (Qi et al. 2012; Tian et al. 2015) and animals (Zhang et al. 2008; Zhong et al. 2008; Blair et al. 2012) exhibit this pattern, as do several Indochinese forest-dwelling animals (e.g., Luo et al. 2004; Fuchs et al. 2008; Patou et al. 2010; Morgan et al. 2011). By comparison, several recent studies on southern China montane species have detected unusual expansions during the glacial periods, with demographic contractions and genetic divergence occurring after the glacial periods. For example, this occurs in tits (Dai et al. 2011; Wang et al. 2012), Chinese bamboo partridge (Huang et al. 2010), and Elliot’s laughing thrush (Qu et al. 2011). Several studies have questioned if the glacial climate adversely affected population sizes and genetic structures. For example, several forest-dwelling taxa from southern China (e.g., Song et al. 2009; Lei et al. 2012; Yan et al. 2012; Yan et al. 2013; Wu et al. 2013) and Indochina (Latinne et al. 2015) presented long-term stable demographic histories and deep genetic diversification that predated the Pleistocene. Most of these controversies likely owe to studying different parts of these areas and being based on species with different ecological requirements (e.g., lowland vs. montane species). Thus, studies involving additional widely distributed taxa can contribute to understanding how Pleistocene climate fluctuations affected the distributions and genetic patterns among species and regions.
Pigmy narrow-mouth frogs Microhyla fissipes complex (Microhylidae) constitute an ideal model group for testing hypotheses on how tectonic and climatic processes shape the patterns of organisms in southern China and Indochina. Similar to most other amphibians, they have low vagility and low physiological tolerance to extremely cold temperatures and dry environmental conditions. These characteristics can lead to isolation, which, in turn, makes them ideal taxa for carrying footprints of historical processes (Zeisset and Beebee 2008). These frogs occur in open lowlands (lowland scrub forest, grassland, agricultural land, pastureland, and some urban areas), and they range from western Myanmar eastward through Indochina and northward into southern China including Hainan and Taiwan (AmphibiaWeb 2016; Frost 2016). Because the populations show subtle differences in morphological characters and occur in similar habitats, the group was long considered to constitute the single species M. fissipes only (Poyarkov et al. 2014). Recently, Hasan et al. (2014) combined mitochondrial DNA (mtDNA) analyses and morphological comparisons to reveal 2 species within this complex. Populations from Bangladesh were assigned to M. mukhlesuri, and populations from China (1 locality from Anhui, 2 localities from Taiwan) were considered “true” M. fissipes (Hasan et al. 2014). However, the absence of extensive sampling leaves the Hasan et al. (2014) hypothesis with 2 possible scenarios of Indochinese species: “M. fissipes” from Thailand and Laos might be same as M. mukhlesuri, or they correspond to an undescribed and/or cryptic species. Presently, their ranges are unclear. The widespread distribution of this complex not only makes it as good model for testing the hypothesis that the Red River is a geographic barrier to dispersal, but also the hypothesis that the Pleistocene climate changes impacted both species, which occur in similar habitats, equally.
Herein, we use both mitochondrial DNA and nuclear DNA (nuDNA) markers to dissect the impact of geologic events and Pleistocene climatic cycling on the current geographical patterns of the distribution and genetic diversity of M. fissipes complex. More specifically, we test 2 following hypotheses. First, we test the null hypothesis (H0) that the Red River is not a barrier to dispersal and gene flow for the M. fissipes complex. Rejection of the H0 will require investigations into the potential mechanisms of isolation that promote diversification. Second, considering ice sheets did not cover both southern China and Indochina during the Pleistocene, we hypothesize that Pleistocene climatic changes had little effect on populations. This predicts that populations will have persisted in situ, exhibit a high level of divergence, and keep stable population demography throughout the Pleistocene. Alternatively, Pleistocene climatic cycling affected regions differently because of different landscapes. Signals of population shrinkages or expansions associated with cyclical Pleistocene glaciations should occur in regions where the climate experiences extreme changes.
Materials and Methods
Sampling, sequencing, and alignment
For the ingroup, we obtained 324 individuals from the M. fissipes complex representing 76 localities from southern China and Indochina (Supplementary Table S1; Figure 1). For the outgroups, 1 individual each of M. heymonsi, M. mixtura, M. okinavensis and M. mymensinghensis was included based on the studies of Matsui et al. (2005) and Hasan et al. (2014). Totally, new sequences from 319 individuals were analyzed, along with 9 individuals from GenBank (Supplementary Table S1). The Animal Ethics Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences approved the protocols for collection of specimens of this study. Toe samples were collected mostly from adults and preserved in 95% ethanol; several adult individuals from each population were euthanized using benzocaine and preserved as voucher specimens (Supplementary Table S1).
Total genomic DNA was extracted using standard phenol–chloroform extraction protocol (Sambrook et al. 1989). A partial fragment of the mitochondrial gene encoding cytochrome b (CYTB) was amplified for 319 individuals. Partial sequences of the nuclear gene encoding recombinase activating 2 protein (RAG2) were amplified for 201 individuals representing each matriline and most localities (Supplementary Table S1). Primers were summarized in Supplementary Table S2. Standard polymerase chain reactions (PCR) were conducted in a 25 uL volume reaction using the following cycling conditions: an initial denaturing step at 95°C for 5 min; 35 cycles of denaturing at 94°C for 1 min, annealing at 55°C (Supplementary Table S2) for 1 min, and extension at 72°C for 1 min; and a final extension step of 72°C for 10 min. PCR products were purified with Gel Extraction Mini Kit (Watson BioTechnologies, Shanghai, China) or a modification of the Exo-SAP method (Werle et al. 1994). Cleaned products were sequenced directly with the corresponding PCR primers using the BigDye Terminator Cycle Sequencing Kit (v.2.0, Applied Biosystems) and an ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Foster City, CA).
Sequences from each gene were aligned using mega 5.05 (Tamura et al. 2011). For CYTB, identical haplotypes were collapsed using dnasp 5.10 (Librado and Rozas 2009), and the overall value of nucleotide diversity (π) and haplotype diversity (H) were also estimated using the same program.
Phylogenetic analyses
Matrilineal relationships among mitochondrial haplotypes of CYTB and a nuclear gene tree based on RAG2 gene were independently constructed using Bayesian inference (BI) and maximum likelihood (ML) methods. Best-fit nucleotide substitution models were selected for the 3 codon position partitions using Akaike information criterion (AIC) in jmodeltest 2.1.6 (Guindon and Gascue 2003; Darriba 2012). BI analyses were performed using mrbayes 3.1.2 (Ronquist and Huelsenbeck 2003) with 10 million generations, and 4 metropolis-coupled Markov chain Monte Carlo (MCMC) chains (temp = 0.2) runs starting with a random tree. Trees were sampled every 100 generation with a burn-in of 25000. Within each run, the average standard deviation of split frequencies (ASDSF) and the potential scale reduction factor (PSRF) statistics from mrbayes were used to evaluate topological and branch-length convergence, respectively. We monitored effective sample size (ESS) values with tracer 1.4 (Rambaut and Drummond 2007). Support was assessed by Bayesian posterior probabilities (BPP). ML analyses were conducted using RAxML 7.0.4 (Stamatakis et al. 2008) but with only the GTR + I + G model for same data partition as BI analyses. Support values were estimated from 1000 non-parametric bootstrap replicates. Uncorrected pair-wise genetic distances (p distances) between major lineages and sublineages were calculated for CYTB using MEGA 5.0 (Tamura et al. 2011).
Divergence time estimation
Time to the most recent common ancestor (TMRCA) of matrilines in the M. fissipes complex was estimated using a Bayesian approach implemented in beast 1.7.5 (Drummond and Rambaut 2007). We employed a likelihood ratio test (LRT) to assess if our CYTB data followed a constant rate of molecular evolution (Huelsenbeck and Crandall 1997). The test was based on unconstrained and clock-enforced matrilineal genealogies estimated using phylip 3.6.9 (Felsenstein 2004). Due to the absence of a fossil record, we assumed a range of substitution rate of 0.65–1.00% (mean = 0.8%) per Ma for CYTB based on evolutionary rates commonly proposed for anurans (Macey et al. 1998, 2001; Monsen and Blouin 2003; Yan et al. 2013) and generally for mtDNA (Brown et al. 1979). Five tree-priors (Bayesian skyline, constant size, expansion growth, exponential growth, and logistic growth) were performed with a different model of nucleotide substitution for each of the 3 codon positions. The best prior was determined using a Bayes factor test with tracer 1.4 (Rambaut and Drummond 2007). For each process, Markov chains were run for 30 million generations. Trees were sampled every 1000 generations. Burn-in and convergence of the chains were determined with tracer 1.4. Further, ESSs were required to have values greater than 200.
Historical demography
Two analyses of the CYTB data served to assess historical changes in effective population size. Considering that population subdivision could have influenced the effect of expansion (Grant et al. 2012; Heller et al. 2013), we analyzed well-supported (BPP >0.95) matrilines N1, N2, S1, S2, S3, and S5, which harbored more than 10 samples and contained 3 haplotypes. First, we used non-genealogical summary statistics including Tajima’s D (Tajima 1989) and Fu’s Fs statistics (Fu 1997) to detect past demographical expansions. Summary statistics were calculated using arlequin 3.5 (Excoffier and Lischer 2010) with 10,000 coalescent simulations. Second, coalescent-based Bayesian skyline plots (BSPs) (Drummond et al. 2005) were generated using beast 1.75 (Drummond and Rambaut 2007). BSPs provided a temporal reference to demographical events such as bottlenecks and expansions. This analysis was performed using the same settings as above for the divergence time estimation, except the coalescent tree prior was specified as the Bayesian skyline with 4 groups. Plots for each analysis were visualized using tracer 1.4 (Rambaut and Drummond 2007).
SDM
We compared ecological niche models (ENMs) for the current and LGM habitats to approximate the distributions of M. fissipes and M. mukhlesuri; this approach assumed niche conservatism over time for the species (Peterson et al. 1999; Wiens and Graham 2005). Separate modeling was conducted for each species considering their relatively high level of divergence in both matrilineal relationships and nuclear genes. Their potential geographic ranges were constructed using the maximum entropy algorithm implemented in maxent 3.3.1 (Phillips et al. 2006; Phillips and Dudík 2008). Current and LGM (21 ka) climatic conditions were downloaded from the global climate database WorldClim (Hijmans et al. 2005). Considering the controversy about whether correlated variables should be included or not, we took the conservative approach and used all of the 19 bioclimatic variables. All layers were cropped to span from 91°E to 122°E and from 7.0°N to 33°N with a spatial resolution of 2.5 arc-minutes. We built ENMs using 75 localities for M. fissipes, and 57 localities for M. mukhlesuri. These data included 52 localities with GPS information downloaded from VertNet (http://vertnet.org; 24 localities for M. fissipes from north of the Red River and 28 localities for M. mukhlesuri from south of the Red River), and coordinates for the other localities were taken by us in the field.
We projected the present-day models on the climatic reconstructions of the LGM under the assumption that the climatic niche of each lineage remained conserved in these periods (Elith et al. 2010). Data were randomly partitioned into training (75%) and testing (25%) datasets. Bootstrapping with 100 replicates was used to generate the pseudoreplicate datasets. We compared area under the curve (AUC) to evaluate the performance of the models.
Point-based analysis of environmental variables
A point-based method of analysis was performed to compare ecological niche differences between the 2 species. The values for each of the 19 present bioclimatic variables were extracted for all sampling sites using diva-gis (Hijmans et al. 2005). Principal component analysis (PCA) was conducted in spss 16.0 to convert the 19 variables to a set of values that accounted for most of the variance. Principal components with eigenvalues >1 that explained >10% of the variation were retained. We used independent sample T test for the 19 bioclimatic variables to determine significant differences in environmental conditions between the populations from each side of the Red River.
Results
Sequence characteristics
For CYTB, we obtained sequences for 324 individuals of the M. fissipes complex, and the 4 outgroup samples (Supplementary Table S1). For this dataset, we sequenced 319 individuals de novo and retrieved 9 from the previous studies (Matsui et al. 2005; Zhang et al. 2005; Hasan et al. 2014). After trimming the ends, 588 base pairs (bp) were retained for downstream analyses. All nucleotide sequences successfully translated to amino acids without premature stop codons and heterozygotes. A total of 136 potentially parsimony-informative sites resulted in 89 haplotypes for the ingroup. The overall nucleotide diversity (π) was 0.054; haplotype diversity (H) was 0.948. The best-fit models for the first, second, and third codon positions of CYTB were K80 + G, HKY + I, and GTR + G, respectively.
One nuclear gene fragment, RAG2, was obtained from a subset of our samples representing matrilines of M. fissipes complex and 1 individual of M. mixtura (Supplementary Table S1). The data included 201 sequences. After trimming the ends, a 587-bp fragment was resolved, of which 42 sites were potentially parsimony informative. The best-fit models for the first, second, and third codon positions were all GTR + G. All newly obtained sequences were deposited in GenBank (Supplementary Table S1).
Matrilineal genealogy and nuclear gene tree
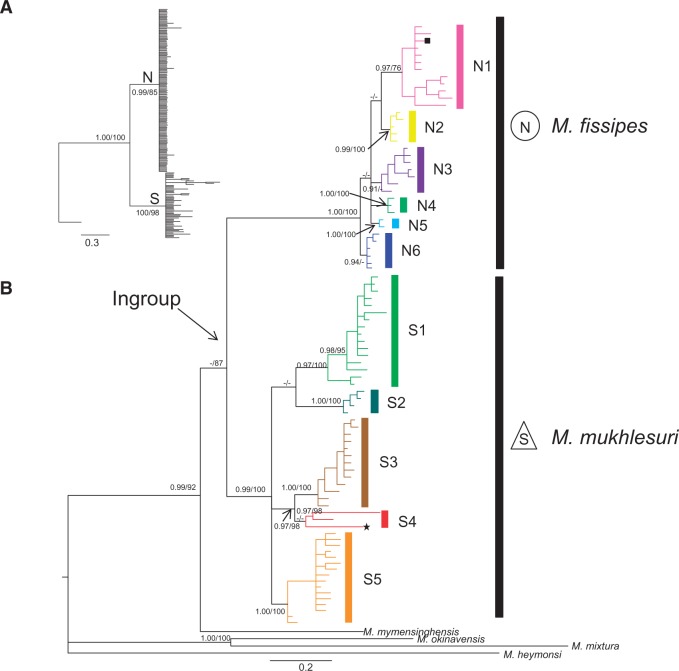
The BI and ML analyses produced extremely similar matrilines based on the mtDNA data. The BI tree was presented in Figure 2B. Distinctive matrilines N and S corresponded to M. fissipes and M. mukhlesuri, respectively. Microhyla fissipes (Lineage N) was broadly distributed mainly north of the Red River where it covered most regions of southern China including Taiwan and Hainan (circles, Figure 1; Figure 2). It contained 6 sublineages (N1–N6) whose relationships were unresolved. Only N1, N2, N4, and N5 were highly supported. Sublineage N1 occurred in coastal southern China, Hainan, and Taiwan (type locality of M. fissipes). Sublineage N2 mainly occurred from west of N1 from eastern Sichuan to Jiangxi. Sublineage N3 occurred at localities 19–21 (Figure 1). Sublineage N4 occurred at localities 11 and 36–38. Sublineage N5 occurred at localities 17, 18, and 26. Finally, sublineage N6 occurred at localities 16, 17, 18, 22, 35, and 39. Genetically distinct sympatric pairs of sublineages occurred at 9 localities in southern China (localities 9, 10, 11, 17, 18, 22, 29, 35, and 39; Figure 1 and Figure 2B). Haplotypes 9 (localities 1–13, 31) and 10 (localities 9, 10, 22, 25, 27–34) from sublineage N1 and N2, respectively, were broadly distributed (Supplementary Table S1).
Figure 2.
Bayesian nuclear gene tree (A) and maternal genealogy (B) for the M. fissipes complex. Vertical color bars show lineage/sublineage assignment. Bootstrap proportions ≥70% and BPP ≥ 90% shown, and lower values were treated as “-”.
Microhyla mukhlesuri (Lineage S) hosted 5 matrilines (Figure 2, S1–S5). The species occurred mainly in Indochina and extreme southwestern China south of Red River (triangles, Figure 1). Sublineage S1, the sister group of S2, consisted of individuals from central Vietnam to eastern Thailand. Sublineage S2 occurred at locality 76 in northern Vietnam only. Sublineage S3 was distributed in northwestern Thailand and 1 site in Yunnan (locality 53). Two haplotypes from southern Yunnan and 1 haplotype from Bangladesh (the type locality of M. mukhlesuri) formed sublineage S4 but without high support. Sublineages S3 and S4 clustered together with strong support. Sublineage S5 occurred mainly in southern Vietnam. Locality 53 contained sympatric sublineages S3 and S4, and locality 65 hosted both S1 and S3. No single haplotype occurred across the distribution of the species (Figure 1, Supplementary Table S1). The p distances for CYTB between matrilines N and S were 9.1%; distances between the 6 sublineages of matriline N ranged from 1.0% to 2.4% and the 5 sublineages of matriline S ranged from 3.4% to 6.8% (Supplementary Table S3).
BI and ML analyses of the RAG2 data (Figure 2A) also resolved well-supported matrilines N and S (M. fissipes and M. mukhlesuri, respectively). Owing to a limited number of potentially parsimony-informative sites, no strong historical patterning was recovered within either group. Within each species, the nuclear gene did not form geographic units, but were mixed together.
Estimated divergence times
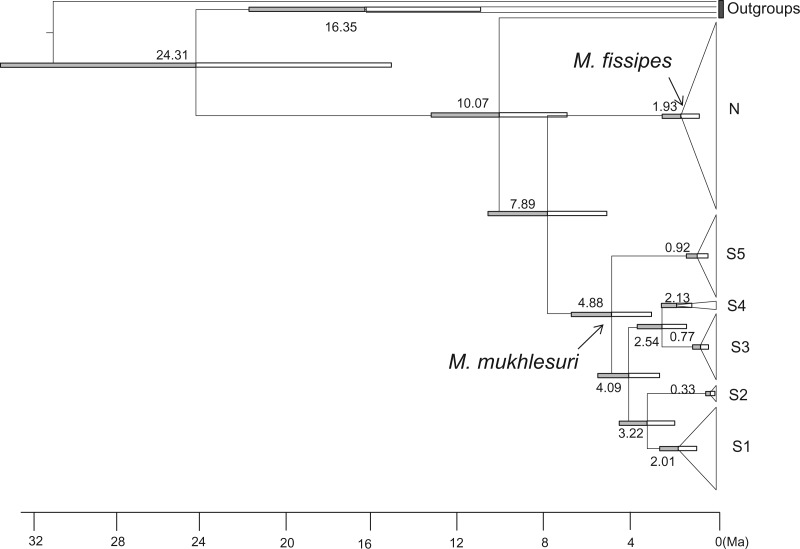
The null hypothesis of clock-like evolution for CYTB was not rejected by the LRT (v2 = 37.22, df = 63, P > 0.05). Therefore, a strict molecular clock was used. The Bayes factor test suggested Bayesian skyline as being the best tree prior process (Supplementary Table S4). The estimated times were presented in Figure 3. The MRCA of the M. fissipes complex and M. mymensinghensis dated to 10.07 Ma (95% CI: 6.95–13.22 Ma). Microhyla fissipes and M. mukhlesuri diverged about 7.89 Ma (95% CI: 5.13–10.53 Ma). Sublineages N1–N6 subsequently diverged starting from 1.93 Ma (95% CI: 0.9–2.60 Ma) to 0.5 Ma (95% CI: 0.20–1.51 Ma). Sublineage S5 diverged first within M. mukhlesuri about 4.88 Ma (95% CI: 3.04–6.81 Ma). Sublineages S3 and S4 split at about 2.54 Ma (95% CI: 1.87–3.63 Ma), and S1 and S2 about 3.22 Ma (95% CI: 2.05–2.57 Ma).
Figure 3.
beast time-tree for the M. fissipes complex. Branch lengths are proportional to divergence times. Bars on the nodes are 95% confidence intervals. Matrilines N and S1–S5 correspond to Figure 2. Numbers at nodes are the average ages.
Historical demography
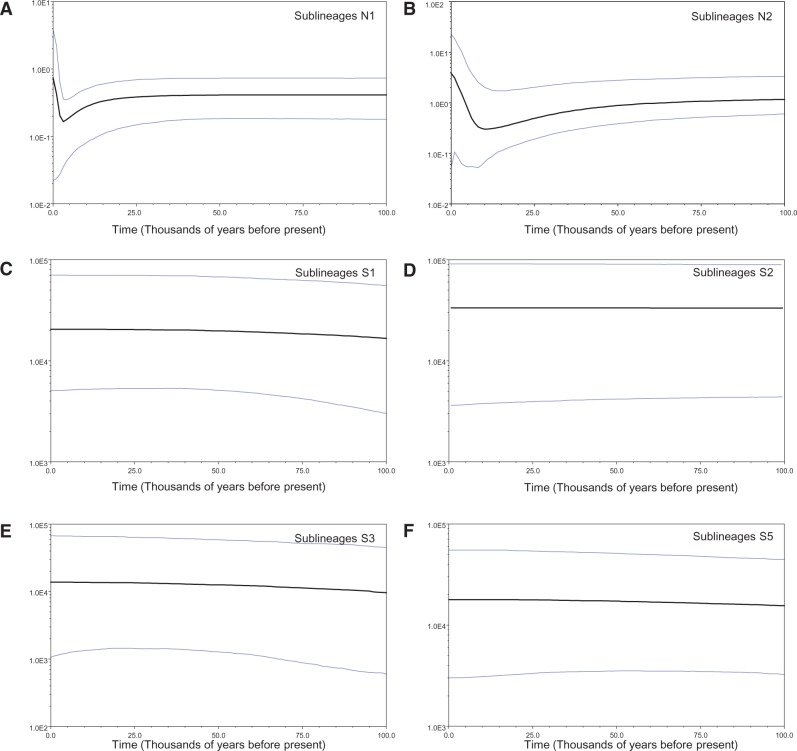
Based on the CYTB data, ESSs values were generally high (> 200) for all parameters in all BSPs analyses, indicating good MCMC mixing in the combined chains. Sublineages N1 and N2 from north of the Red River had similar demographic histories. Significant negative values of Tajima’s D and Fu’s Fs indicated population expansions (Table 1). The most prominent feature of the BSPs was a slight decline followed by a sharp increase in effective population size. As shown in Figure 4A and Figure 4B, the population sizes of sublineages N1 and N2 were relatively stable until approximately 25,000 years near the LGM. The decline spanned from 25,000 to 5,000 years ago with expansion starting about 5,000 years ago.
Table 1.
Statistics of neutrality tests for matrilines of the M. fissipes complex
| Tajima’s D | Tajima’s D P value | Fu’s Fs test | FsP value | |
|---|---|---|---|---|
| N1 | −2.127 | 0.002 | −3.488 | 0.013 |
| N2 | −1.706 | 0.031 | −6.136 | 0.002 |
| S1 | −1.809 | 0.019 | −4.279 | 0.025 |
| S2 | 0.755 | 0.799 | −0.780 | 0.193 |
| S3 | −1.369 | 0.078 | −3.300 | 0.095 |
| S5 | −1.563 | 0.043 | −6.804 | 0.004 |
Figure 4.
BSP estimated by beast for each matriline of the M. fissipes complex; y axis corresponds to effective population size (Neτ, the product of effective population size and generation length in years); x axis corresponds to time. (A–F) BSPs for matrilines N1, N2, S1, S2, S3, and S5, respectively. The mean estimate and both 95% HPD limits are indicated.
According to the coalescent-based BSPs analyses, the effective population sizes of sublineages S1–S3 and S5 were stable during LGM (Figure 4C–F). Significant negative values (P < 0.05) of Tajima’s D and Fu’s Fs were also revealed for sublineages S1 and S5 (Table 1). However, compared with BSP, which directly quantified demography from gene genealogies (Drummond et al. 2005), these summary statistics did not take into account tree structure and they were not based on all available information (Pybus et al. 2000).
Historical dynamics of distributions
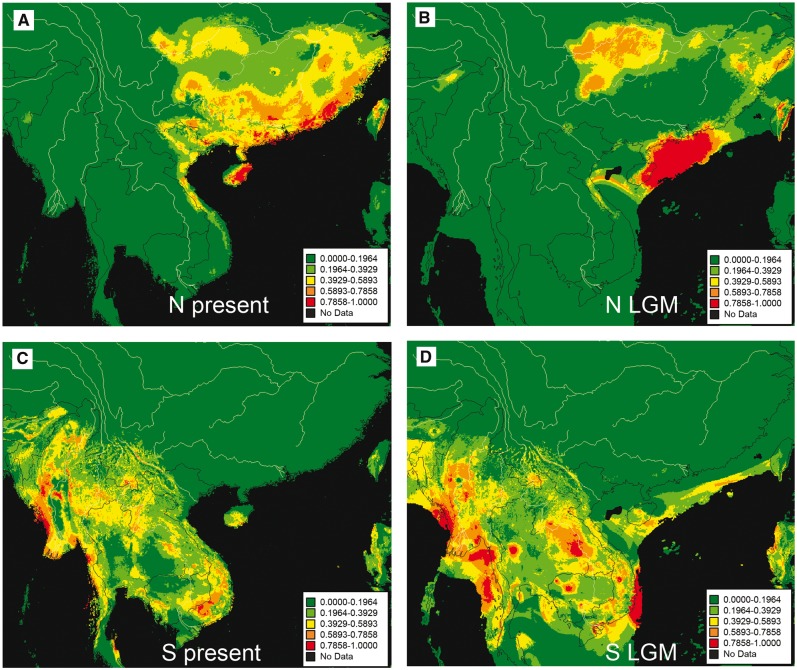
The ENMs had excellent predictive power for occurrences under conditions of both today and the LGM. The AUC had values higher than 0.85 in all analyses, which implied that the results greatly differed from random prediction (AUC = 0.5). For M. fissipes, suitable habitats occurred presently north of the Red River in southern China and coastal northern Vietnam (Figure 5A). In contrast, the LGM MAXENT projection predicted a more narrow distribution and shift to 4 major areas including coastal southeastern China, Taiwan, Zhejiang and northern Fujian, and eastern Sichuan, Chongqing and Guizhou (Figure 5B). For M. mukhlesuri, suitable habitats occurred presently south of the Red River including Hainan (Figure 5C). The resulting LGM MAXENT projection predicted a similar continuous distribution compared with the current range, although some subtle differences existed for the most suitable areas (Figure 5D).
Figure 5.
ENMs for lineages N (A, B) and S (C, D). SDMs at present (A, C) and LGM (∼21 ka; B, D) were presented. Color scale refers to probability of occurrence (habitat suitability) from maxent.
Point-based analysis
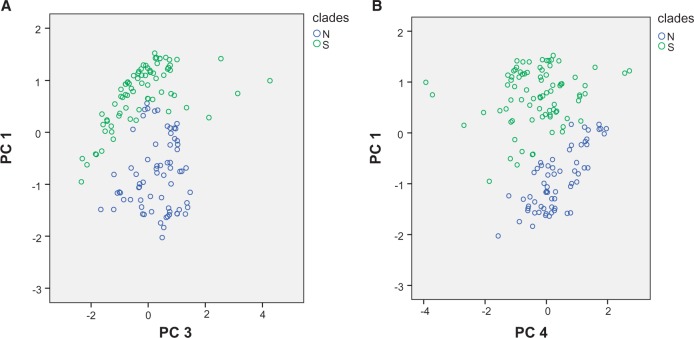
Four principal components (PC1 = 49.86%, PC2 = 17.34%, PC3 = 15.13%, PC4 = 7.80%) explained 90.13% of the total variation for the present ecological niche differences (Table 2). Plots from PC1 and PC3 (Figure 6A), PC1 and PC4 (Figure 6B) indicated that M. fissipes and M. mukhlesuri were well differentiated across the Red River. The independent sample T test indicated that the habitats of these 2 species differed significantly (P < 0.05) between the Red River for 12 of the 19 bioclimatic variables (Table 3).
Table 2.
Nineteen biological environmental variables used in Point-based analysis and the PCA on these variables used in a comparison of climatic conditions between occurrence locations for matrilines N and S of the M. fissipes complex
| Bio-variable | Component |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| BIO1 = Annual Mean Temperature | 0.389 | −0.238 | 0.211 | 0.036 |
| BIO2 = Mean Diurnal Range | 0.883 | 0.012 | −0.586 | −0.237 |
| BIO3 = Isothermality | −0.945 | 0.122 | −0.137 | −0.392 |
| BIO4 = Temperature Seasonality | 0.436 | −0.101 | 0.104 | 0.268 |
| BIO5 = Max Temperature of Warmest Month | 0.938 | −0.664 | 0.403 | 0.203 |
| BIO6 = Min Temperature of Coldest Month | −0.879 | −0.118 | 0.252 | −0.087 |
| BIO7 = Temperature Annual Range | 0.655 | −0.171 | −0.104 | 0.195 |
| BIO8 = Mean Temperature of Wettest Quarter | 0.952 | −0.395 | 0.218 | 0.479 |
| BIO9 = Mean Temperature of Driest Quarter | 0.382 | −0.103 | 0.188 | −0.127 |
| BIO10 = Mean Temperature of Warmest Quarter | 0.981 | −0.596 | 0.573 | 0.378 |
| BIO11 = Mean Temperature of Coldest Quarter | 0.418 | −0.095 | 0.09 | −0.112 |
| BIO12 = Annual Precipitation | 0.541 | 0.712 | 0.477 | 0.021 |
| BIO13 = Precipitation of Wettest Month | −0.764 | 0.724 | 0.254 | 0.181 |
| BIO14 = Precipitation of Driest Month | 0.755 | 0.123 | 0.564 | −0.09 |
| BIO15 = Precipitation Seasonality | 0.561 | 0.22 | −0.452 | 0.269 |
| BIO16 = Precipitation of Wettest Quarter | −0.699 | 0.748 | 0.222 | 0.168 |
| BIO17 = Precipitation of Driest Quarter | −0.124 | 0.175 | 0.626 | −0.146 |
| BIO18 = Precipitation of Warmest Quarter | −0.199 | 0.702 | −0.076 | 0.577 |
| BIO19 = Precipitation of Coldest Quarter | 0.94 | 0.141 | 0.734 | −0.463 |
| Initial eigenvalues | 9.473 | 3.294 | 2.875 | 1.481 |
| % of variance | 49.857 | 17.337 | 15.129 | 7.796 |
Figure 6.
PCA of climatic niche differences between matrilines N and S framed by blue and green circles, respectively; (A) x axis indicates PC3, and y axis indicates PC1; (B) x axis indicates PC4, and y axis indicates PC1.
Table 3.
Results of T test for the 19 biological environmental variables, and 4 principal components derived from the PCA
| Bio-variable | F | P value |
|---|---|---|
| BIO1 = Annual Mean Temperature | 0.383 | 0.537 |
| BIO2 = Mean Diurnal Range* | 69.435 | 0.000 |
| BIO3 = Isothermality | 1.963 | 0.163 |
| BIO4 = Temperature Seasonality* | 12.092 | 0.001 |
| BIO5 = Max Temperature of Warmest Month* | 6.935 | 0.009 |
| BIO6 = Min Temperature of Coldest Month | 0.091 | 0.764 |
| BIO7 = Temperature Annual Range | 3.122 | 0.079 |
| BIO8 = Mean Temperature of Wettest Quarter* | 7.692 | 0.006 |
| BIO9 = Mean Temperature of Driest Quarter | 1.637 | 0.203 |
| BIO10 = Mean Temperature of Warmest Quarter* | 4.357 | 0.039 |
| BIO11 = Mean Temperature of Coldest Quarter* | 4.217 | 0.035 |
| BIO12 = Annual Precipitation* | 15.186 | 0.000 |
| BIO13 = Precipitation of Wettest Month* | 8.284 | 0.005 |
| BIO14 = Precipitation of Driest Month* | 11.975 | 0.001 |
| BIO15 = Precipitation Seasonality | 0.974 | 0.325 |
| BIO16 = Precipitation of Wettest Quarter* | 10.311 | 0.002 |
| BIO17 = Precipitation of Driest Quarter* | 8.746 | 0.004 |
| BIO18 = Precipitation of Warmest Quarter* | 18.085 | 0.000 |
| BIO19 = Precipitation of Coldest Quarter | 0.841 | 0.361 |
“*” denotes significant differences (P < 0.05) between the habitats of M. fissipes and M. mukhlesuri at the Red River.
Discussion
Species boundary
Hasan et al. (2014) hypothesized the existence of 2 or more species within M. fissipes complex based on morphology and mtDNA sequences, though with limited sampling. Analyses of our nuDNA sequences also resolve 2 major clades (N and S) and these correspond with the matrilines. This discovery supports of the recognition of both M. fissipes and M. mukhlesuri. Further, our extensive sampling clearly defines the distributions of both species. Microhyla fissipes occurs north of the Red River throughout southern China, including Taiwan (type locality) and Hainan Island (Figure 1). Samples of M. mukhlesuri are from Bangladesh (type locality), southern Yunnan, Thailand, Laos, and Vietnam south of the Red River. Although M. mukhlesuri has 5 divergent matrilines, the nuclear gene tree shows no sign of further division. Despite the absence samples from Myanmar and Cambodia, the widespread occurrence of M. mukhlesuri from Bangladesh to Thailand, Laos, and Vietnam implies that it also occurs in these areas. Further studies in these regions can fine-tune the distribution of M. mukhlesuri.
Red River barrier
Our study supports the allopatric occurrence of M. fissipes and M. mukhlesuri along the Red River shear zone (Figure 1). No locality examined hosts both species. This finding implies the absence of gene flow across the Red River. Two other species of frogs exhibit the same pattern: the spiny frog Nanorana yunnanensis (Zhang et al. 2010a) and the megophryid frog Leptobrachium ailaonicum (Zhang et al. 2010b). Both of these species occur around the Red River in Yunnan, which suggests that the river serves as an effective geographical barrier to contemporary dispersal. The Red River forms a major geological strike-slip fault zone that marks the boundary between South China and Indochina tectonic plates (Leloup et al. 1995; Hall 1998). The complex geology of the Red River was suggested to divide once continuous populations (Hall 1998; Zhang et al. 2010a, 2010b). The divergence of M. fissipes and M. mukhlesuri dates to 7.89 Ma, which broadly coincides with 1 period of the extrusion of the Indochina block shearing along the Red River fault in the Late Miocene (Hall 1998; Xiang et al. 2004; Zhang 2009). These geological events coincided with extremely dry, hot climatic conditions along the Red River basin and were likely responsible for the initial fragmentation of habitats.
Differing environmental conditions between north and south of the Red River likely promoted and maintained the divergence of M. fissipes and M. mukhlesuri. The Red River largely demarks the location of change between subtropical and tropical climates (Peel et al. 2007; Chen and Chen 2013) where temperature and precipitation can differ greatly. Differing climate and landscape features can promote local adaptations (Sobel et al. 2010) especially for frogs whose reproductive periods are strongly influenced by environmental variables. Both the ENMs and T tests indicate significant ecological differences between M. fissipes and M. mukhlesuri both at present and during the LGM (Figure 5). The PCAs based on the present environment data are consistent with these findings. Bioclimatic variables for the populations on either side of Red River generally form 2 clusters (Figure 6). Twelve of the 19 bioclimatic variables involved in temperature and precipitation differ significantly (T test, P < 0.05) across the Red River (Table 3). Ecological differences between populations could generate localized adaptations (Zhou et al. 2012). This process probably likely plays the key role in speciation within the M. fissipes complex.
Previous studies were limited to the narrow confines of geographic regions (e.g., Yunnan; Zhang et al. 2010a, 2010b) or involved either side of the Red River (e.g., reviewed in Li and Li 1992; Bain and Hurley 2011; Fan et al. 2013). Thus, they focused on the Red River as being a geographical barrier to dispersal alone; they did not identify environmental differences as the driver of the pattern. Future studies with detail biogeographic analyses should consider and test for alternative mechanisms of speciation.
Effect of the LGM
The effects of Pleistocene climatic cycling varied with regions, despite the M. fissipes complex having similar habitat requirements (lowland scrub forest and grassland, agricultural land and pastureland). Our results demonstrate that late Pleistocene climatic cycling had little influence on the distribution of M. mukhlesuri in Indochina. By comparison, it greatly affected the current genetic structure and population demography of M. fissipes in southern China.
LGM climatic change does not appear to have affected the population size and genetic structuring of M. mukhlesuri from Indochina. Demographic reconstructions do not detect any bottlenecking for any sublineage but rather predict stable demographic histories (Figure 4C–F). Further, the absence of widely distributed haplotypes rejects a hypothesis of recent population expansion. Divergence estimates for the 5 sublineages predate Pleistocene climatic changes, which implies that population structuring does not associate with climatic cycling. However, analyses of the nuDNA data do not obtain the same pattern. This is not surprising given the slower divergence rates of the nuDNA markers and usually higher effective population size, which could result in incomplete lineage sorting (Zhang and Hewitt 2003; Brito and Edwards 2009).
Large areas of suitable habitat during the LGM explain the stable demographic history of M. mukhlesuri. ENMs analyses reveal widespread suitable climatic conditions for M. mukhlesuri during the LGM (Figure 5C, D). In tropical Indochina, temperatures lowered during the LGM, but the consequences of this on humidity and types of vegetation are a hot topic of debate. Some palaeoclimatic studies suggested that most of the tropical rain forest was eliminated and replaced by pine grassland savannah during the LGM (Hope et al. 2004; Bird et al. 2005; Wurster et al. 2010). Consistent with this, several studies of forest-dwelling species in these regions identified population shrinkages during the LGM followed by drastic expansions, such as for Asian colobine monkeys (Brandon-Jones 1996), stone oaks (Cannon and Manos 2003), mosquito Anopheles dirus (O’Loughlin et al. 2008; Morgan et al. 2010), and the black fly Simulium tani (Pramual et al. 2005). In contrast, records of pollen and phytoliths indicated the existence of a large and continuous tract of tropical lowland rain forest during the LGM (Cannon et al. 2009; Wang et al. 2009b). Latinne et al. (2015) compared the suitable habitats for 3 Southeast Asian forest-dwelling murine rodents (Leopoldamys) in the present and LGM. They reported that these species did not experience significant range contractions during the LGM. However, little is known about how lowland species responded to these climate changes, especially in mainland Indochina. Our study documents a stable demographic history for 1 species of lowland frog.
Microhyla fissipes from southern China underwent demographic and range contractions in response to climate changes during the late Pleistocene, especially sublineages N1 and N2. Our coalescent-based BSP analyses suggest both population sizes of the sublineages N1 and N2 experienced declines during the LGM followed by recent, rapid expansions (Figure 4A, B). Further, widely distributed haplotypes (Hap 9, 10) occur across large areas of southern China, which possibly indicates recent population expansions. These 2 sublineages cover most of the distribution of M. fissipes, which suggests the climatic change possibly affected most of the populations of M. fissipes. The occurrence of several recently divergent sublineages within southern China reflects a pattern of range fragmentation into multiple refugia during the LGM. Moreover, the 9 or more pairs of sympatric haplotypes (Figure 1) may indicate secondary contact owing to recent population expansions from different refugia.
Drastic historic climatic change in southern China possibly contributed to the genetic pattern and population history of M. fissipes. Although no glacier covered these regions, the temperature of southern China was reduced by 6–7°C and precipitation decreased by 400–600 mm/year during the LGM (Zhou et al. 1991). Under this climate, drastic population size changes are likely to have happened, especially for the lowland frogs that are sensitive to the environment oscillations. Consistent with this scenario, our ENMs analyses suggest that the predicted range of M. fissipes from southern China shrink extensively during the LGM compared with today (Figure 5A, B).
Considering previous studies in southern China, we suggest that species with different ecological requirements vary in their responses to the same climatic changes even in similar regions. For example, the lowland and montane species in southern China show different responses to Late Pleistocene climatic changes. Similar to M. fissipes, population contractions and postglacial range expansions were also found in the lowland dwelling cricket frogs of southern China (Fejervarya multistriata; Zhong et al. 2008). In contrast, montane spiny frogs (Quasipaa boulengeri; Yan et al. 2013) and stream newts (Pachytriton; Wu et al. 2013) in southern China appear to have deep population divergences and stable demographic histories. The opposing patterns also correspond with differing types of vegetation in these regions, such as plants growing in thickets at forest margins (e.g., Qi et al. 2012; Tian et al. 2015) vs. those limited to montane forests of mixed evergreen and deciduous broadleaf trees (e.g., Wang and Ge 2006; Wang et al. 2009a). Montane species can shift their habitats along elevational gradients and survive environmental perturbations intact, but not lowland species (Hewitt 2000, 2004; Wu et al. 2013). These studies indicate that it is necessary to consider the ecological requirements of species when investigating how historic changes in climate shape genetic patterns.
Supplementary Material
Acknowledgments
We thank Sheng-guo Fang, Hai-tao Shi, Li-jun Wang, Li-min Ding, Yun-yu Wang, Bao-ling Zhang, Hai-peng Zhao, Jin-ming Chen, Jun-xiao Yang, Jia-jun Zhou, Gang Yao, Nikolai L. Orlov, Son Nguyen Truong, Tao Thien Nguyen, Truong Quang Nguyen, Tran Ngoc Ninh, Ba Dinh Vo, Andrei N. Kuznetsov, Leonid P. Korzoun, Anna B. Vassilieva, Eduard A. Galoyan, Igor V. Palko, Suwat Seaya, and Prakayjit Saengham for helping with sample collections. We thank the local Forestry Department and National reserve helped for the fieldworks in China. We thank the Forestry Department of Thailand and Vietnam for permissions to undertake field survey, specimen collections and local national park for helping. The Institute of Animal for Scientific Purposes Development (IAD) issued permission (No.U1-01205-2558) in Thailand. The Department of Forestry, Ministry of Agriculture and Rural Development of Vietnam issued permission (permits No. 170/ TCLN–BTTN, 982/ TCLN–BTTN and 831/ TCLN–BTTN) in Vietnam. For loans of tissues, we thank Amy Lathrop from the Royal Ontario Museum (ROM), Toronto, Canada. We gratefully acknowledge Wei-wei Zhou and Yong-jie Wu for providing thoughtful comments during preparation of the previous version of this manuscript.
Funding
This work was supported by the program of Chinese Academy of Sciences (2015CASEABRI002), the Ministry of Science and Technology of China (2011FY120200), the National Natural Science Foundation of China (31090250), and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences (the Large Research Infrastructure Funding) to J. C.; the Chinese Academy of Sciences Visiting Fellowship for Researchers (Postdoc) from Developing Countries (2013FFS130015) to C. S.; the National Natural Science Foundation of China (31501843) to H. M. C.; the Russian Foundation of Basic Research (Grant No. RFBR Taiwan No. 14-04-92000, 15-29-02771 and 15-04-02029) and the Russian Science Foundation (RSF grant No 14-50-00029) to N. A. P.; NSERC Discovery Grant (3148), the ROM Foundation, and the ROM Members Volunteer Committee to R. W. M.
Supplementary material
Supplementary material can be found at http://www.cz.oxfordjournals.org/.
References
- AmphibiaWeb, 2016. Information on Amphibian Biology and Conservation. Berkley: University of California; Available from: http://amphibiaweb.org/. [Google Scholar]
- Bain RH, Hurley MM, 2011. A biogeographic synthesis of the amphibians and reptiles of Indochina. Bull Am Mus Nat Hist 360:1–138. [Google Scholar]
- Bird MI, Taylor D, Hunt C, 2005. Palaeoenvironments of insular Southeast Asia during the Last Glacial Period: a savanna corridor in Sundaland? Quat Sci Rev 24:2228–2242. [Google Scholar]
- Blair C, Davy CM, Ngo A, Orlov NL, Shi HT, et al. , 2012. Genealogy and demographic history of a widespread amphibian throughout Indochina. J Heredity 104:72–85. [DOI] [PubMed] [Google Scholar]
- Brandon-Jones D, 1996. The Asian Colobinae (Mammalia: Cercopithecidae) as indicators of Quaternary climatic change. Biol J Linn Soc 59:327–350. [Google Scholar]
- Brito PH, Edwards SV, 2009. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica 135:439.–. [DOI] [PubMed] [Google Scholar]
- Brown WM, George MJ, Wilson AC, 1979. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA 76:1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon CH, Manos PS, 2003. Phylogeography of the Southeast Asian stone oaks (Lithocarpus). J Biogeogr 30:211–226. [Google Scholar]
- Cannon CH, Morley RJ, Bush ABG, 2009. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA 106:11188–11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che J, Zhou WW, Hu JS, Yan F, Papenfuss TJ, et al. , 2010. Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc Natl Acad Sci USA 107:13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Chen HW, 2013. Using the Köppen classification to quantify climate variations and change: an example for 1901–2010. Environ Dev 6:69–79. [Google Scholar]
- Dai CY, Zhao N, Wang WJ, Lin CT, Gao B, et al. , 2011. Profound climatic effects on two East Asian black-throated tits (Aves: Aegithalidae), revealed by ecological niche models and phylogeographic analysis. PLoS ONE 6:e29329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D, 2012. JMODELTEST 2: more models, new heuristics and parallel computing. Nat Meth 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyn M, Stelbrink B, Morley RJ, Hall R, Carvalho GR, et al. , 2014. Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Sys Biol 63:879–901. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG, 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22:1185–1192. [DOI] [PubMed] [Google Scholar]
- Elith J, Kearney M, Phillips S, 2010. The art of modelling range-shifting species. Methods Ecol Evol 1:330–342. [Google Scholar]
- Excoffier L, Lischer H, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567. [DOI] [PubMed] [Google Scholar]
- Fan DM, Yue JP, Nie ZL, Li ZM, Comes HP, et al. , 2013. Phylogeography of Sophora davidii (Leguminosae) across the “Tanaka-Kaiyong Line”, and important phytogeographic boundary in Southwest China. Mol Ecol 22:4270–4288. [DOI] [PubMed] [Google Scholar]
- Felsenstein J, 2004. PHYLIP (Phylogeny Inference Package). Version 3.6. Distributed by the author. Seattle, Washington: Department of Genome Sciences, University of Washington. [Google Scholar]
- Frost DR, 2016. Amphibian species of the World: An Online Reference. Version 6.0. New York: American Museum of Natural History. Available from: http://research.amnh.org/herpetology/amphibia/index.html. [Google Scholar]
- Fu YX, 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:9145–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J, Ericson PGP, Pasquet E, 2008. Mitochondrial phylogeographic structure of the white-browed piculet Sasia ochracea: cryptic genetic differentiation and endemism in Indochina. J Biogeogr 35:565–575. [Google Scholar]
- Grant WS, Liu M, Gao TX, Yanagimoto TG, 2012. Limits of Bayesian skyline plot analysis of mtDNA sequences to infer historical demographies in Pacific herring (and other species). Mol Phylogen Evol 65:203–212. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O, 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Sys Biol 52:696–704. [DOI] [PubMed] [Google Scholar]
- Hall R, 1998. The plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In: Hall R, Holloway JD, editors. Biogeography and Geological Evolution of SE Asia. Leiden: Backhuys, 99–131. [Google Scholar]
- Hasan M, Islam MM, Kuramoto M, Kurabayashi A, Sumida M, et al. , 2014. Description of two new species of Microhyla (Anura: Microhylidae) from Bangladesh. Zootaxa 3755:401–408. [DOI] [PubMed] [Google Scholar]
- Heller R, Chikhi L, Siegismund HR, 2013. The confounding effect of population structure on Bayesian skyline plot inferences of demographic history. PLoS ONE 8:e62992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt GM, 2000. The genetic legacy of the Quaternary ice ages. Nature 405:907–913. [DOI] [PubMed] [Google Scholar]
- Hewitt GM, 2004. Genetic consequences of climatic oscillations in the Quaternary. Proc R Soc B 359:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A, et al. , 2005. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978. [Google Scholar]
- Hope G, Kershaw AP, van der Kaars S, Sun XJ, Liew PM, et al. , 2004. History of vegetation and habitat change in the Austral-Asian region. Quatern Int 118:103–126. [Google Scholar]
- Holt BG, Lessard JP, Borregaard MK, Fritz SA, Araújo MB, et al. , 2013. An update of Wallace’s zoogeographic regions of the world. Science 339: 4–78. [DOI] [PubMed] [Google Scholar]
- Huang ZH, Liu NF, Liang W, Zhang YY, Liao XJ, et al. , 2010. Phylogeography of Chinese bamboo partridge Bambusicola thoracica thoracica (Aves: Galliformes) in south China: inference from mitochondrial DNA control-region sequences. Mol Phylogen Evol 56:273–280. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Crandall KA, 1997. Phylogeny estimation and hypothesis testing using maximum likelihood. Ann Rev Ecol Sys 28:437–466. [Google Scholar]
- Latinne A, Meynard CN, Herbreteau V, Waengsothorn S, Morand S, et al. , 2015. Influence of past and future climate changes on the distribution of three Southeast Asian murine rodents. J Biogeogr 42:1714–1726. [Google Scholar]
- Lei M, Wang Q, Wu ZJ, López-Pujol J, Li DZ, et al. , 2012. Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: limited admixture among multiple refugia. Tree Genet Genom 8:1203–1212. [Google Scholar]
- Leloup PH, Lacassin R, Tapponnier P, Schärer U, Zhong DL, et al. , 1995. The Ailao Shan-Red River shear zone (Yunnan, China), Tertiary transform boundary of Indochina. Tectonophysics 251:3–84. [Google Scholar]
- Li JJ, Shu Q, Zhou SZ, Zhang J, 2004. Review and prospects of Quaternary glaciation research in China. J Glaciol Geocryol 26:235–243. [Google Scholar]
- Li XW, Li J, 1992. On the validity of Tanaka line and its significance viewed from the distribution of eastern Asiatic genera in Yunnan. Plant Div Res 14:1–12. [Google Scholar]
- Li XW, Li J, 1997. The Tanaka-Kaiyong line: an important floristic line for the study of the flora of East Asia. Ann Mo Bot Gard 84:888–892. [Google Scholar]
- Librado P, Rozas J, 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Luo SJ, Kim JH, Johnson WE, van der Walt J, Martenson J, et al. , 2004. Phylogeography and genetic ancestry of tigers Panthera tigris. PLoS Biol 2:e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macey JR, Schulte JA, Larson A, Fang ZL, Wang YZ, et al. , 1998. Phylogenetic relationships of toads in the Bufo bufo species group from the eastern escarpment of the Tibetan Plateau: a case of vicariance and dispersal. Mol Phylogen Evol 9:80–87. [DOI] [PubMed] [Google Scholar]
- Macey JR, Strasburg JL, Brisson JA, Vredenburg VT, Jennings M, et al. , 2001. Molecular phylogenetics of western North American frogs of the Rana boylii species group. Mol Phylogenet Evol 19:131–143. [DOI] [PubMed] [Google Scholar]
- Matsui M, Ito H, Shimada T, Ota H, Saidapur SK, et al. , 2005. Taxonomic relationships within the Pan–Oriental narrow-mouth toad Microhyla ornata as revealed by mtDNA analysis (Amphibia, Anura, Microhylidae). Zool Sci 22:489–495. [DOI] [PubMed] [Google Scholar]
- Matsui M, Hamidy A, Murphy RW, Khonsue W, Yambun P, et al. , 2010. Phylogenetic relationships of megophryid frogs of the genus Leptobrachium (Amphibia, Anura). Mol Phylogen Evol 56:259–272. [DOI] [PubMed] [Google Scholar]
- McLeod DS, 2010. Of least concern? Systematics of a cryptic species complex: Limnonectes kuhlii (Amphibia: Anura: Dicroglossidae). Mol Phylogen Evol 56:991–1000. [DOI] [PubMed] [Google Scholar]
- Monsen KJ, Blouin MS, 2003. Genetic structure in a montane ranid frog, restricted gene flow and nuclear-mitochondrial discordance. Mol Ecol 12:3275–3286. [DOI] [PubMed] [Google Scholar]
- Morgan K, Linton Y-M, Somboon P, Somboon P, Saikia P, et al. , 2010. Inter-specific gene flow dynamics during the Pleistocene-dated speciation of forest-dependent mosquitoes in Southeast Asia. Mol Ecol 19:2269–2285. [DOI] [PubMed] [Google Scholar]
- Morgan K, O’Loughlin SM, Chen B, Linton YM, Thongwat D, et al. , 2011. Comparative phylogeography reveals a shared impact of Pleistocene environmental change in shaping genetic diversity within 9 Anopheles mosquito species across the Indo-Burma biodiversity hotspot. Mol Ecol 20:4533–4549. [DOI] [PubMed] [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J, et al. , 2000. Biodiversity hotspots for conservation priorities. Nature 403:854–858. [DOI] [PubMed] [Google Scholar]
- O’Loughlin SM, Okabayashi T, Honda M, Kitazoe Y, Kishino H, et al. , 2008. Complex population history of two Anopheles dirus mosquito species in Southeast Asia suggests the influence of Pleistocene climate change rather than human-mediated effects. J Evol Biol 21:1555–1569. [DOI] [PubMed] [Google Scholar]
- Patou ML, Wilting A, Gaubert P, Esselstyn JA, Cruaud C, et al. , 2010. Evolutionary history of the Paradoxurus palm civets: a new model for Asian biogeography. J Biogeogr 37:2077–2097. [Google Scholar]
- Peel MC, Finlayson BL, McMahon TA, 2007. Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Sys Sci 11:1633–1644. [Google Scholar]
- Peterson A, Soberón J, Sánchez-Cordero V, 1999. Conservatism of ecological niches in evolutionary time. Science 285:1265–1267. [DOI] [PubMed] [Google Scholar]
- Phillips SJ, Dudík M, 2008. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175. [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE, 2006. Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259. [Google Scholar]
- Poyarkov NA, Vassilieva AB, Orlov NL, Galoyan EA, Tran TAD, et al. , 2014. Taxonomy and distribution of narrow-mouth frogs of the genus Microhyla Tschudi, 1838 (Anura: Microhylidae) from Vietnam with descriptions of five new species. Russ J Herpetolo 21:89–148. [Google Scholar]
- Pramual P, Kuvangkadilok C, Baimai V, Walton C, 2005. Phylogeography of the black fly Simulium tani (Diptera: Simuliidae) from Thailand as inferred from mtDNA sequences. Mol Ecol 14:3989–4001. [DOI] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A, Harvey PH, 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, et al. , 2012. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytolo 196:617–630. [DOI] [PubMed] [Google Scholar]
- Qu Y, Luo X, Zhang R, Song G, Zou F, et al. , 2011. Lineage diversification and historical demography of a montane bird Garrulax elliotii: implications for the Pleistocene evolutionary history of the eastern Himalayas. BMC Evol Biol 11:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond A, 2007. Tracer v1.4. Available from: http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Replumaz A, Lacassin R, Tapponnier P, Leloup PH, 2001. Large river offsets and Plio-Quaternary dextral slip on the Red River fault (Yunnan, China). J Geophys Resear 106:819–836. [Google Scholar]
- Ronquist F, Huelsenbeck JP, 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T, 1989. Molecular Cloning: A Laboratory Manual. 2nd edn New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Searle MP, 2006. Role of the Red River Shear zone, Yunnan and Vietnam, in the continental extrusion of SE Asia. J Geol Soc 163:1025–1036. [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW, et al. , 2010. The biology of speciation. Evolution 64:295–315. [DOI] [PubMed] [Google Scholar]
- Song G, Qu YH, Yin ZH, Li S, Liu NF, et al. , 2009. Phylogeography of the Alcippe morrisonia (Aves: Timaliidae): long population history beyond late Pleistocene glaciations. BMC Evol Biol 9:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J, 2008. A rapid bootstrap algorithm for the RAXML Web servers. Sys Biol 57:758–771. [DOI] [PubMed] [Google Scholar]
- Stuart BL, Inger RF, Voris HK, 2006. High level of cryptic species diversity revealed by sympatric lineages of Southeast Asian forest frogs. Biol Lett 2:470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapponnier P, Lacassin R, Leloup PH, Schärer U, Zhong DL, et al. , 1990. The Ailao Shan/Red River metamorphic belt: Tertiary left-lateral shear between Indochina and South China. Nature 343:431–437. [Google Scholar]
- Tian S, Lei SQ, Hu W, Deng LL, Li B, et al. , 2015. Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Mol Phylogen Evol 8:238–246. [DOI] [PubMed] [Google Scholar]
- Wang HW, Ge S, 2006. Phylogeography of the endangered Cathaya argyrophylla (Pinaceae) inferred from sequence variation of mitochondrial and nuclear DNA. Mol Ecol 15:4109–4122. [DOI] [PubMed] [Google Scholar]
- Wang J, Gao P, Kang M, Lowe AJ, Huang HW, et al. , 2009a. Refugia within refugia: the case study of a canopy tree Eurycorymbus cavaleriei in subtropical China. J Biogeogr 36:2156–2164. [Google Scholar]
- Wang W, McKay BD, Dai C, Zhao N, Zhang RY, et al. , 2012. Glacial expansion and diversification of an East Asian montane bird, the green-backed tit Parus monticolus. J Biogeogr 40:1156–1169. [Google Scholar]
- Wang X, Sun X, Wang P, Stattegger K, 2009b. Vegetation on the Sunda Shelf, South China Sea, during the Last Glacial Maximum. Palaeogeog Palaeoclim Palaeoecol 278:88–97. [Google Scholar]
- Werle E, Schneider C, Renner M, Völker M, Fiehn W, et al. , 1994. Convenient single-step, one tube purification of PCR products for direct sequencing. Nuc Aci Res 22:4354–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ, Graham CH, 2005. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Sys 36:519–539. [Google Scholar]
- Williams MAJ, Dunkerley DL, de Deckker P, Kershaw AP, Chappell JA, et al. , 1998. Quaternary Environments. London: Arnold. [Google Scholar]
- Woodruff DS, 2010. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today’s patterns and the future of the remaining refugial-phase biodiversity. Biodiv Conserv 19:919–941. [Google Scholar]
- Wu YK, Wang YZ, Jiang K, Hanken J, 2013. Significance of pre-Quaternary climate change for montane species diversity: insights from Asian salamanders (Salamandridae: Pachytriton). Mol Phylogen Evol 66:380–390. [DOI] [PubMed] [Google Scholar]
- Wurster CM, Bird MI, Bull ID, Creed F, Bryant C, et al. , 2010. Forest contraction in north equatorial Southeast Asia during the Last Glacial Period. Proc Natl Acad Sci USA 107:15508–15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang HF, Han ZJ, Guo SM, Chen LC, Zhang WX, et al. , 2004. Several questions about the quantitative study of the large scale right-lateral strike-slip movement of the Red River fault zone. Adv Earth Sci 19:56–59. [Google Scholar]
- Yan HF, Zhang CY, Wang FY, Hu CM, Ge XJ, et al. , 2012. Population expanding with the phalanx model and lineages split by environmental heterogeneity: a case study of Primula obconica in subtropical China. PLoS ONE 7:e41315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Zhou WW, Zhao HT, Yuan ZY, Wang YY, et al. , 2013. Geological events play a larger role than Pleistocene climatic fluctuations in the genetic structure of the montane stream-dwelling frog Quasipaa boulengeri (Anura: Dicroglossidae) in southern China. Mol Ecol 22:1120.–. [DOI] [PubMed] [Google Scholar]
- Zeisset I, Beebee TJC, 2008. Amphibian phylogeography: a model for understanding historical aspects of species distributions. Heredity 101:109–119. [DOI] [PubMed] [Google Scholar]
- Zhang BL, Liu RX, Xiang HF, Wan JL, Huang XN, et al. , 2009. FT dating of fault rocks in the central-southern section of the Red River Fault zone and its geological implications. Seismol Geol 31:44–56. [Google Scholar]
- Zhang D, Hewitt GM, 2003. Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects .Mol Ecol 12:563.–. [DOI] [PubMed] [Google Scholar]
- Zhang DR, Chen MY, Murphy RW, Che J, Pang JF, et al. , 2010a. Genealogy and palaeodrainage basins in Yunnan Province: phylogeography of the Yunnan spiny frog Nanorana yunnanensis (Dicroglossidae). Mol Ecol 19:3406–3420. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yan J, Zhang GQ, Zhou KY, 2008. Phylogeography and demographic history of Chinese black-spotted frog populations Pelophylax nigromaculata: evidence for independent refugia expansion and secondary contact. BMC Evol Biol 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MW, Rao DQ, Yang JX, Yu GH, Wilkinson JA, et al. , 2010b. Molecular phylogeography and population structure of a mid-elevation montane frog Leptobrachium ailaonicum in a fragmented habitat of southwest China. Mol Phylogen Evol 54:47–58. [DOI] [PubMed] [Google Scholar]
- Zhang P, Zhou H, Chen YQ, Liu YF, Qu LH, et al. , 2005. Mitogenomic perspectives on the origin and phylogeny of living amphibians. Sys Biol 54:391–400. [DOI] [PubMed] [Google Scholar]
- Zhong J, Liu ZQ, Wang YQ, 2008. Phylogeography of the rice frog Fejervarya multistriata (Anura: Ranidae), from China based on mtDNA D-loop Sequences. Zool Sci 25:811.–. [DOI] [PubMed] [Google Scholar]
- Zhou WW, Yan F, Fu JZ, Xu YB, Jin JQ, et al. , 2012. Speciation in the Rana chensinensis species complex and its relationship to the uplift of the Qinghai-Tibetan Plateau. Mol Ecol 21:960–973. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Qiu G, Guo D, 1991. Changes of permafrost in China during Quaternary. In: Liu TS, editor. Quaternary Geology and Environment in China. Beijing: Science Press, 86–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.