Abstract
Rapid environmental change is predicted to compromise population survival, and the resulting strong selective pressure can erode genetic variation, making evolutionary rescue unlikely. Non-genetic inheritance may provide a solution to this problem and help explain the current lack of fit between purely genetic evolutionary models and empirical data. We hypothesize that epigenetic modifications can facilitate evolutionary rescue through ‘epigenetic buffering’. By facilitating the inheritance of novel phenotypic variants that are generated by environmental change—a strategy we call ‘heritable bet hedging’—epigenetic modifications could maintain and increase the evolutionary potential of a population. This process may facilitate genetic adaptation by preserving existing genetic variation, releasing cryptic genetic variation and/or facilitating mutations in functional loci. Although we show that examples of non-genetic inheritance are often maladaptive in the short term, accounting for phenotypic variance and non-adaptive plasticity may reveal important evolutionary implications over longer time scales. We also discuss the possibility that maladaptive epigenetic responses may be due to ‘epigenetic traps’, whereby evolutionarily novel factors (e.g. endocrine disruptors) hack into the existing epigenetic machinery. We stress that more ecologically relevant work on transgenerational epigenetic inheritance is required. Researchers conducting studies on transgenerational environmental effects should report measures of phenotypic variance, so that the possibility of both bet hedging and heritable bet hedging can be assessed. Future empirical and theoretical work is required to assess the relative importance of genetic and epigenetic variation, and their interaction, for evolutionary rescue.
Keywords: climate change, evolutionary traps, evolutionary tracking, plasticity, epimutation, transgenerational epigenetic inheritance
Introduction
We are living through a period of human-induced rapid environmental change, with a concurrent loss of biodiversity that is of major concern [ 1–4 ]. Organisms are adapted to live and reproduce within the range of environmental conditions experienced by their ancestors. If the environment changes outside these conditions, then population fitness (i.e. the average fitness of individuals in the population) is predicted to decline [ 5 , 6 ]. In the face of such rapid environmental change, populations could go extinct, migrate to more suitable environments or stay and adapt to the novel conditions [ 7 ]. Understanding the processes that lead to adaptation in changed environments is vitally important, both for theoretical insights into organismal evolution and practical attempts to conserve biodiversity.
The process by which genetic adaptation saves a population from otherwise inevitable extinction is termed ‘evolutionary rescue’ (hereafter, for phrases in single inverted commas, see Table 1 ), and it has received much attention in recent years [ 7 , 18–22 ]. Evolutionary rescue relies on the traditional tools of evolution: genetic variation that already exists within the population, new mutations and gene flow [ 7 ]. Having more genetic variance within a population increases its ‘evolutionary potential’, making rescue from extinction more likely [ 23–25 ]. However, there are few documented examples of evolutionary rescue, and it is unclear whether genetic evolution is sufficient for a population to cope with rapid environmental change [ 7 ]. Phenotypic change does not necessarily require changes in gene frequency: it can also be the result of ‘phenotypic plasticity’, and there is an increasing appreciation of heritable non-genetic sources of phenotypic variation (e.g. parental effects, cultural inheritance and epigenetic variation) [ 15 , 18 , 26 ].
Table 1.
Glossary of terms
| Term | Definition |
|---|---|
| Adaptive epigenetic response | An adaptive phenotypic response to selection brought about by environmental change that is mediated through epigenetic inheritance, resulting in the population reaching a fitness optimum in the new environment |
| Cryptic genetic variation | Genetic variability that is not translated into phenotypic variability under normal environmental conditions, but that is exposed under atypical environmental conditions generating heritable phenotypic variation [ 8 ] |
| Epigenetic buffering | Epigenetic modifications which provide phenotypic resilience against fluctuating environmental change, facilitating the persistence of a population through rapid environmental change over ecological timescales |
| Epigenetic inheritance | Inheritance of phenotypic variations that do not stem from differences in the DNA sequences [ 9 ]. With this term, we do not mean cellular epigenetic inheritance (i.e. within-generation maintenance of epigenetic states) but intergenerational or transgenerational epigenetic inheritance (see below) |
| Epigenetic trap | Any change in the environment which causes the existing epigenetic machinery of an organism to produce a maladapted phenotype, with no increase in phenotypic variance within the population |
| Evolutionary potential | Ability of a population to respond to future selection pressures, taking into account currently existing (visible and cryptic) genetic variation [ 10 ]. Note that Le Rouzic and Carlborg [ 10 ] use this definition to describe “evolvability”; however, we use this to describe evolutionary potential of a population |
| Evolutionary rescue | Genetic adaptation that allows population recovery from environmentally induced demographic effects that would normally cause extinction [ 7 ]. Increases in beneficial mutations and/or standing genetic variation may prevent negative population growth and extinction |
| Genetic assimilation | Process by which selection converts an environmentally responsive phenotype into a phenotype that no longer requires the environmental stimuli for its production [ 8 ] |
| Heritable bet hedging | Process in which phenotypic variation is increased due to environmental factors and importantly, induced phenotypic values are heritable. In contrast, traditional bet hedging is a process in which evolved phenotypic variability buffer unpredictable environmental changes but heritability of phenotypic values are usually not assumed ( Fig. 2 ; see also [ 11 , 12 , 13 ]) |
| Intergenerational (epigenetic) inheritance (i.e. parental effects) | Effect of a parental phenotype on their offspring’s phenotype that cannot be attributed to the parental or offspring genome, non-parental components of the environment or their interaction [ 14 ]. Effects occur across a single generation (F 0 –F 1 ). See also the definition of ‘epigenetic inheritance” above |
| Non-genetic inheritance | The transmission to offspring of parental phenotypic or environment variation that does not include the inheritance of DNA sequences (i.e. genes) [ 15 ] |
| Phenotypic plasticity | Changed phenotypic expression of a genotype/individual under different environmental conditions. Two forms of plasticity have recently been defined by Snell-Rood [ 16 ]: (i) developmental plasticity where a genotype/individual expresses different phenotypes in different environments by taking different developmental trajectories early in life that are often established during a sensitive period and (ii) activational plasticity, which is an immediate phenotypic change by a genotype/individual in response to the environment and can occur throughout an organism’s life |
| Standing genetic variation | Genetic variation that is present in the population as opposed to new mutations [ 8 ] |
| Transgenerational epigenetic inheritance | Transmission from parents to offspring of phenotypic traits resulting from different methylation patterns or chromatin structure that affects gene expression, generally over two or more generations (F 0 –F N , where N ≥ 2) [ 15 ]. See also the definition of ‘epigenetic inheritance’ above |
| Transposable elements | Mobile DNA segments in the genome. Two major types exist: (i) DNA transposon that do not use reverse transcriptase to integrate into the genome and (ii) retrotransposon that uses reverse transcriptase to integrate into the genome [ 17 ] |
Non-genetic variation is now recognized as an underappreciated component of evolution [ 27 ] and there is accumulating evidence that, by altering gene expression, variation in epigenetic marks can cause heritable phenotypic variation that is independent of genetic variation [ 28–31 ]. Interestingly, it appears that epigenetic modifications can also affect the probability that a region of the genome will mutate (e.g., single-base and transposon-mediated mutations and translocation) [ 32 ]. Therefore, not only may epigenetic modification promote heritable phenotypic variation, but it could also facilitate genetic evolution by modulating mutation rates across the genome [ 33 ]. It is now conceivable that epigenetic variation (and the resulting novel genetic variation) could be a key mechanism by which organisms adapt to rapid environmental change.
In this article, we hypothesize that epigenetic-mediated variation can provide populations with the resilience to persist through periods of environmental change, and we name this phenomena ‘epigenetic buffering’. First, we provide a brief overview of epigenetic mechanisms that have so far been discovered, discuss how they may increase heritable phenotypic variance and outline how the interaction between genetic and epigenetic sources can maintain, and possibly increase, a population’s evolutionary potential. We then provide an extensive list of examples for both ‘intergenerational’ and ‘transgenerational epigenetic inheritance’, and discuss whether, during rapid environmental change, increased phenotypic variance caused by epigenetic mechanisms is likely to be adaptive. We speculate that bet hedging, in combination with transgenerational inheritance (termed ‘heritable bet hedging’), may be a common adaptive mechanism in response to rapid environmental change. We also discuss how maladaptive outcomes of transgenerational epigenetic inheritance could occur when organisms fall into an ‘epigenetic trap’. However, seemingly maladaptive responses may prove beneficial over a longer time period. We then reach our main thesis: that over ecological timescales, epigenetic change provides populations with the phenotypic and genetic variability required to persist during sudden environmental change, and possibly facilitates genetic adaptation over evolutionary timescales. Finally, we present predictions based on our proposed phenomena, and suggest avenues for future empirical and theoretical research.
Evolution and Epigenetic Inheritance
Evolution and the Epigenetic Mechanisms Underlying Phenotypic Variation
Adaptive phenotypic evolution (or adaptive tracking) relies on selection, variation and heritability, and the properties of these variables will influence evolutionary dynamics [ 32 ]. Although genetic mechanisms provide the most faithful mode of phenotypic transmission, less stable epigenetic mechanisms, such as DNA methylation (see below), may be selected to persist if a plastic phenotype is adaptive. The inclusion of ‘non-genetic inheritance’ mechanisms into evolutionary thinking is a major goal of the extended evolutionary synthesis [ 34 , 35 ]. Three alternative classes of epigenetic variation have now been recognized [ 36 ], each having different and important roles in the evolutionary process. These include (i) obligate epigenetic variation: epigenetic variation is completely associated with genetic variation; (ii) facilitated epigenetic variation: the genotype probabilistically determines the epigenotype and (iii) pure epigenetic variation: epigenetic variation is driven by stochastic events that are independent of the genotype [ 36 ]. Importantly, because obligate epigenetic variation is indistinguishable from genetic variation, in this article we exclude it from our discussions of epigenetic variation and evolutionary implications.
The molecular mechanisms underlying epigenetic variation are now beginning to be understood ([ 9 , 37–39 ]; for an accessible and extensive overview, see [ 40 ]) and have been shown to have transgenerational effects on phenotypic development. Three major categories of molecular epigenetic mechanisms are now known to regulate gene expression [ 41 ]: (i) DNA methylation (usually methylation of cytosine) which can silence genes by blocking transcription factors from binding to promoter sites; (ii) histone modifications (post-translational modification of histone tails by different chemical compounds) which can up-regulate, down-regulate and silence genes; and (iii) processes mediated by non-coding RNAs (ncRNAs) which can provide sophisticated gene regulation in both plants and animals. For example, mild heat stress in the nematode ( Caenorhabditis elegans ) produces a heritable change in gene expression due to RNA interference from small interference RNA (one type of ncRNA), which may involve interactions with chromatin modifications [ 42 ]. DNA methylation, histone modifications and ncRNAs can interact to influence gene regulation in a variety of ways. They can, for example, change chromatin configuration (chromatin remodelling) [ 43 ]. These molecular mechanisms contribute substantially to phenotypic development and thus have important consequences that influence phenotypic variation.
Phenotypic plasticity is likely mediated by epigenetic mechanisms, which can lead to permanent or transient changes in the phenotype that can influence the amount of phenotypic variation in a population [ 16 , 44 ]. Epigenetic processes translate environmental cues to changes in gene expression, leading to both permanent (developmental plasticity, sensu [ 16 ]) and transient phenotypic effects (activational plasticity, sensu [ 16 ]). Although phenotypic plasticity is recognized as an intra-generational phenomenon, the recognition that epigenetic processes have the capacity to be transgenerational suggests that epigenetic mechanisms responsible for influencing the development of the phenotype in one generation can affect phenotypic variation in subsequent generations. While theoretical models treat phenotypically plastic responses as a genetic phenomenon [ 45 ], it is important to recognize that, for many plastic responses, we lack a good understanding of how genetic and epigenetic mechanisms interact when responding to environmental cues, or how much genetic variation exists in the epigenetic machinery. More empirical work exploring the genetic and epigenetic processes of phenotypically plastic responses will inform our understanding of these important questions.
Increasing Evolutionary Potential: Three Pathways Via Epigenetic Mechanisms
Epigenetic mechanisms can enable heritable changes in gene expression that can increase both phenotypic and genetic variance. Epigenetic mutations, or epimutations, can change gene expression independent of DNA sequence change [ 43 ], and heritable epimutations and associated phenotypes can be selected and maintained [ 46 ]. Furthermore, an allele carrying an epimutation can be considered an allelic variant that is distinct from other genetic variants at a particular locus, and thus it can promote heterozygosity (or epiheterozygosity) with another allele [ 30 ]. A special type of epimutation is paramutation, which is well known in plants and has now been described in animals such as mice [ 47 ]. Paramutation at a heterozygous locus causes the other allele to adopt its epigenetic state [ 30 ]. Therefore, paramutation, if stable through generations, can quickly spread, changing the epigenetic landscape of a population. Epigenetic changes can also promote genetic mutations, such as when epigenetic regulation of ‘transposable elements’ (TEs) is destabilized. It is hypothesized that epigenetic mechanisms originally evolved as a defence mechanism against parasitic viral TEs, but have now been co-opted to regulate gene expression in eukaryotes [ 48 ]. For example, small RNAs (which affect gene regulation) probably evolved to disable invading RNAs from retroviruses (by RNA interference) [ 49 , 50 ]. Now, disruption of TEs by epigenetic changes could dramatically increase the occurrence of genomic mutations and rearrangements [ 17 , 51 , 52 ]. Mutations mediated by TEs are extremely frequent for two reasons. First, TEs constitute a large portion of eukaryote genomes (e.g., half of the human genome and 90% of the maze genome; [ 53 ]). Second, TEs can cause numerous types of genetic mutations; TEs can ‘jump’ around the genome by inserting or copying themselves, and they can also duplicate, translocate and invert a portion of the genome (although these mutations are mostly deleterious) [ 17 , 48 , 51 ].
Although evolutionary change driven by epimutations is probably short lived (due to their transient heritability [ 54 ]), their effects on genetic variation could have lasting evolutionary implications. In addition to the aforementioned ability of epigenetic factors to affect TE-mediated mutations, epigenetic changes also affect chromatin structure. Chromatin configuration affects the probability of a region of the genome mutating [ 55 ]; epimutations that affect chromatin structure could therefore influence mutation rates at these sites. More directly, there is experimental evidence that epimutations can lead to biased mutation rates [ 33 , 56 ]. Methylated cytosine, for example, has a higher probability of being replaced by thymine than non-methylated cytosine [ 56 ]. Methylated regions have also been associated with mutations involving copy number variation [ 33 ]. Far more speculatively, epiheterozygosity could also facilitate mutations, as it has recently been shown that genetic heterozygosity is associated with increased mutation rates [ 57 ]. This increased mutation rate is presumably due to mismatches between paternal and maternal alleles. It is unknown if epiheterozygosity similarly facilitates mutations, but it is an intriguing possibility. Indeed most literature on this topic is fairly speculative, and it remains unknown how prevalent epigenetic effects on mutation rates are in nature [ 9 , 58 ].
In addition to epimutations, epigenetic mechanisms could increase evolutionary potential in response to environmental change via two additional pathways ( Fig. 1 ). First, environmental change can expose ‘cryptic genetic variation’, which is part of the ‘standing genetic variation’ [ 8 ]. This might occur by demethylation or chromatin remodelling to turn genes ‘on’ which had previously been unexpressed. Second, environmental stress could create new genetic variation, by inducing random and/or biased genetic mutations (as described above).
Figure 1.
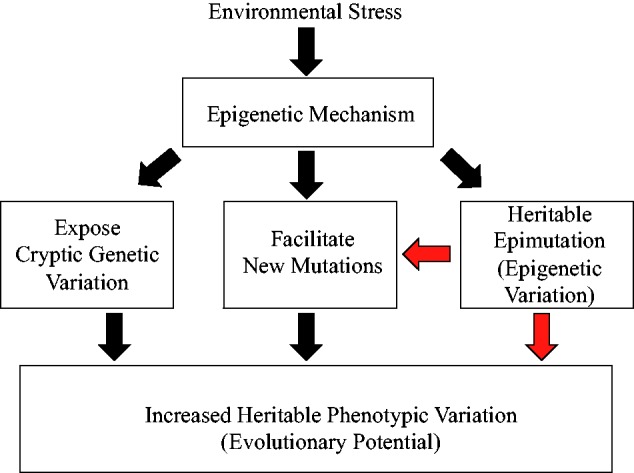
Three different ways in which epigenetic modification can increase heritable phenotypic variation and thus evolutionary potential. Red arrows indicate transient nature of these effects
In summary, we envisage three different ways in which environmental change could increase heritable phenotypic variation: (i) exposing cryptic genetic variation; (ii) generating genetic variation and (iii) creating more heritable epigenetic variation. Each of these paths, which we summarize in Fig. 1 , could maintain, and eventually enhance, the evolutionary potential of a population when faced with environmental change. In this article, our focus is on the third pathway, which is the basis of transgenerational epigenetic inheritance.
Adaptive Value of Transgenerational Epigenetic Inheritance
An Overview of Transgenerational Epigenetic Inheritance Examples
There is now growing evidence demonstrating that non-genetically transmitted phenotypes [ 9 , 15 , 28 , 37 , 59 ] can be generated by diverse environmental effects, affect a wide range of offspring traits (both positively and negatively) and be transmitted by both parental lines for several generations (i.e. transgenerational effects; Table 2 ). Numerous studies have reported non-genetic inheritance across one (F 0 –F 1 ) generation (i.e. intergenerational effects; see references within Supplementary Table S1 ), but documenting unequivocal transgenerational effects requires assessing the first generation not directly exposed to the environmental stress. For postnatal or adult F 0 exposure, this requires assessing the F 2 generation. For in utero embryonic exposure (i.e. gestating F 0 female exposure), however, assessment of the F 3 generation is required [ 37 , 84 ]. Currently, transgenerational inheritance research ( Table 2 ) is dominated by studies investigating effects of either nutrition (e.g. altered dietary composition, undernourishment) or pollution (particularly endocrine disrupting chemicals). Most endocrine disrupting chemicals have been documented to have negative effects on offspring reproduction, social behaviours, disease onset and even mate preference ( Table 2 ). At least one study identified a potential positive effect—first generation progeny exposed to bisphenol A in utero displayed fewer social interactions as compared with control mice, but increased social interactions and decreased non-social behaviours (grooming and cage exploration) were observed in later generations (F 2 and F 4 ) [ 66 ]. Likewise, parental malnutrition generally has negative effects on size, growth, longevity and disease ( Table 2 ), although one study found that parental habitat quality (i.e. food source quality) positively affects the foraging strategy of F 2 and F 3 generations of a flour beetle [ 78 ]. A recent study found that predation pressure results in earlier maturation and larger clutch size in three subsequent generations of Daphnia [ 79 ]. Finally, studies have documented transgenerational tolerance or increased sensitivity to abiotic factors like heavy metals or odours [ 72 , 73 ], the latter of which is a stunning example of how learned fear response, through odour fear conditioning in F 0 mice, can be transmitted via sperm [ 73 ]. As illustrated in Supplementary Table S1 , many more biotic and abiotic effects have been revealed in studies that assess non-genetic inheritance in the first generation; such effects need to be explored in subsequent generations.
Table 2.
Examples of environmental factors that can have transgenerational (≧F 2 ; F 3 if F 1 offspring were in utero during exposure of F 0 mother) effects, the nature of those effects on offspring and their consequences for offspring fitness [ 15 ]
| Environmental manipulation experienced by parental generation (F 0 ) a | Effect on offspring | Consequences for offspring fitness | Offspring generations affected | Species | Reference |
|---|---|---|---|---|---|
| Abiotic | |||||
| Chemicals: endocrine disrupters | |||||
| Fungicide (vinclozolin) | Behaviour: mate selection | Negative | F 3 | Mus musculus (mice) | [ 60 ] |
| Fungicide (vinclozolin) | Disease: testicular, prostate, kidney, ovarian | Negative | F 3 | M. musculus (mice; strain specific) | [ 61 ] |
| Fungicide (vinclozolin) and pesticide (methoxychor) | Male infertility | Negative | F 1 –F 4 | Rattus norvegicus (brown rat) | [ 62–64 ] |
| Hydrocarbons (jet fuel) | Disease: ovarian | Negative | F 1 –F 3 | R. norvegicus (brown rat) | [ 65 ] |
| Plastic (bisphenol A) | Social interaction and social behaviours | Positive or negative | F 1 –F 4 | M. musculus (mice) | [ 66 ] |
| Plastic (DEHP (di-2-ethylhexyl phthalate)) | Reproductive: sperm counts, motility and testis organization | Negative | F 1 –F 4 | M. musculus (mice) | [ 67 ] |
| Dioxin (TCDD (2,3,7,8-Tetrachlorodibenzo-p-dioxin)) | Sex ratio, skeletal abnormalities, fertility | Negative | F 1 –F 2 | Danio rerio (zebrafish) | [ 68 ] |
| Pesticides, plastics, dioxin, jet fuel | Reproductive: ovarian and spermatogenic | Negative | F 3 | R. norvegicus (brown rat) | [ 69 ] |
| 17α-ethinylestradiol | Fertilization rate, embryo survival | Negative | F 2 –F 3 | Oryzias latipes (medaka) | [ 70 ] |
| Tributyltin | Metabolic: obesity | Negative | F 2 –F 3 | M. musculus (mice) | [ 71 ] |
| Other abiotic | |||||
| Heavy metal exposure | Increased tolerance | Positive | F 2 | Oryza sativa L. (rice) | [ 72 ] |
| Exposure to odour (fear conditioning) | Sensitivity to odour | Positive | F 2 | M. musculus (mice) | [ 73 ] |
| Biotic | |||||
| Nutrition | |||||
| Undernourishment | Body weight and obesity | Negative | F 2 | Homo sapiens (human) | [ 74 ] |
| Dietary composition (food restricted) | Sex ratio and growth | Negative | F 2 | Mesocricetus auratus (golden hamster) | [ 75 ] |
| Dietary composition (over nutrition) | Longevity, disease | Negative | F 2 | H. sapiens (human) | [ 76 , 77 ] |
| Dietary composition (habitat quality) | Foraging strategy, population growth rate | Positive | F 2 –F 3 | Tribolium castaneum (flour beetle) | [ 78 ] |
| Other biotic | |||||
| Predation | Maturation and clutch size | Positive | F 1 –F 3 | Daphnia ambigua (water flea) | [ 79 ] |
| Maternal separation/maternal stress | Depressive-like behaviour, behavioural response to aversive environment | Negative | F 2 –F 3 | M. musculus (mice) | [ 80 ] |
| Traumatic stress (unpredictable maternal separation) | Avoidance and fear, depressive behaviours, abnormal metabolism | Negative and positive | F 1 –F 2 | M. musculus (mice) | [ 81 ] |
Adaptive Plasticity: Predictability and Stability
There is scant empirical evidence of an ‘adaptive epigenetic response’ to environmental change [ 54 ]. Two conditions need to be met to demonstrate that an environmentally induced phenotype is adaptive [ 85 ]. First, when exposed to the environmental trigger, individuals who express the modified phenotype are fitter than those who do not. Second, when the environmental trigger is absent, individuals who nevertheless express the modified phenotype are less fit than their ‘normal’ counterparts. Although simple in theory, the empirical reality of demonstrating adaptive plasticity is often cumbersome [ 26 ]. Where an adaptive transgenerational plastic response to an environmental trigger has been reported, there has not been a concurrent demonstration that transgenerational epigenetic mechanisms were the cause [ 54 ]. Conversely, examples of environmentally induced transgenerational epigenetic inheritance have not been shown to be adaptive [ 54 ]. Of course, absence of evidence is not evidence of absence, and examples of adaptive epigenetic transgenerational plasticity may well emerge as research methods improve and interest intensifies.
It is unlikely that novel and rapid environmental change would induce an adaptive phenotypic response. For an organism to exhibit adaptive plasticity in response to an environmental cue it requires pre-existing genetic or epigenetic architecture, presumably selected by evolution during past periods of similar environmental conditions. In the context of human-induced rapid environmental change, it is assumed that population fitness declines because the environmental conditions are beyond those previously experienced. Furthermore, adaptive transgenerational plasticity requires the environment to change predictably (either consistent change in the same direction or periodic fluctuations), so that an environmental cue experienced by one generation predicts the environment of future generations (detection-based effects [ 86 ]). For example, models have shown that non-genetic inheritance can be adaptive in fluctuating environments, provided that those fluctuations are predictable [ 87 , 88 ], and environmental stability can promote the evolution of partial epigenetic inheritance [ 89 ]. Therefore, both the instability and unpredictability of the environment during periods of human-induced rapid environmental change render it unlikely that transgenerational epigenetic effects could induce an adaptive phenotypic change. Given that the majority of genetic mutations are inconsequential or deleterious with respect to fitness [ 33 , 90 ], this will also likely be true for epimutations.
Maladaptive Plasticity: Epigenetic Traps or Adaptively Maladaptive?
Most examples of transgenerational epigenetic inheritance seem to lead to a reduction in fitness (but see Table 2 for a few positive effects); however, if heritable phenotypic variation is increased, then an adaptive response could evolve over time. If there is no increase in evolutionary potential, then negative transgenerational epigenetic inheritance could only be maladaptive, so why do such effects occur? We see similarities to the concept of an ‘evolutionary trap’, where the environmental cue that an organism uses to assess the quality of a resource becomes inappropriate due to environmental change, causing individuals to make the “wrong decision” [ 5 , 6 ]. In the case of epigenetic inheritance, the molecular machinery presumably evolved for adaptive purposes [ 86 ]. It is only when encountering something recent in evolutionary terms, such as an obesogenic environment or novel toxins, that the pre-existing epigenetic mechanisms produce inappropriate responses that can have maladaptive consequences for average population fitness. It is therefore the environmental mismatch that causes the epigenetic machinery of an organism to produce a heritable maladaptive phenotype—we term this an epigenetic trap. Although we recognize that this term is somewhat synonymous with evolutionary traps, we specifically use this to more effectively define what part of the molecular machinery is being monopolized to lead to maladaptive outcomes. A potential example of an epigenetic trap comes from a series of studies on epimutations in mammalian sperm. Glucocorticoid receptors are induced by stressful experiences, but they are also induced by evolutionarily novel stimuli such as endocrine disruptors and alcohol (presumably by “hacking” into the same or similar epigenetic pathways) [ 38 ]. Although epigenetic traps at first seem problematic to our thesis, it is important to recognize that, analogous to most mutations being deleterious, evolutionary rescue only requires a few beneficial mutations to rapidly spread through a population. Similarly, although many epigenetic responses may be maladaptive, the increased phenotypic variation may generate a small frequency of beneficial epimutations, enabling populations to explore new phenotypic space produced by environmental stressors. Furthermore, maladaptive phenotypic plasticity, mediated by epigenetic mechanisms, may itself facilitate adaptive evolutionary responses by increasing the strength of directional selection and allowing populations to reach the phenotypic optimum more quickly. For example, Ghalambor et al. [ 91 ] showed that the direction of plastic responses in gene expression was opposite to the direction of adaptive evolution, suggesting that adaptive plasticity may constrain evolutionary responses, while maladaptive plasticity allows populations to adapt more quickly to environmental change.
Heritable Bet Hedging
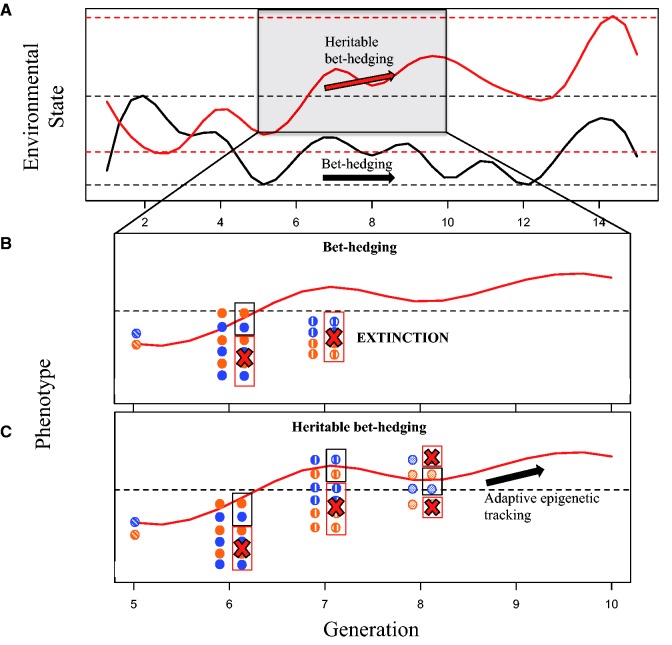
We speculate that where epigenetic mechanisms can provide an adaptive response to novel and unpredictable environmental change, it will most likely be through a random increase in heritable phenotypic variability combined with adaptive evolution (tracking), i.e. heritable bet hedging ( Fig. 2 ; cf. [ 54 ]). Bet hedging is often conceptualized as an intergenerational effect (such as a maternal effect on offspring size variation), where responses are driven by plastic allocation strategies in the parents that do not result in heritable effects in subsequent generations (i.e. no shifts in the offspring phenotypic mean values or frequencies; Fig. 2 B; Table 1 ). In fluctuating environments experienced by a population in its evolutionary past, bet hedging strategies can yield greater geometric mean fitness—despite a reduction in arithmetic mean fitness—by reducing the variance in fitness across fluctuating environments [ 11 ]. Nonetheless, under environmental conditions not encountered in a species’ evolutionary history, directional environmental change may lead to extinction when only bet hedging strategies are relied upon ( Fig. 2 B). We predict that heritable epigenetic mechanisms can lead to phenotypic variation generated by bet hedging strategies being transgenerationally inherited, facilitating adaptive evolution (i.e. changes in the offspring phenotypic mean values or frequencies; Fig. 2 C). When a population has not evolved an adaptive response to a particular environmental change, it is likely to be knocked off its fitness peak, and heritable phenotypic variation is then required for recovery through evolutionary processes [ 54 , 92 ]. Theoretically, heritable bet hedging could allow populations to move between different peaks in the adaptive landscape [ 93 ], which might lead to recovery from rapid (possibly transient) environmental change. To date, heritable bet hedging has not been demonstrated in multicellular organisms [ 54 ], but research that pays heed to phenotypic variance of offspring traits after parental exposure to an environmental change could tell a different story. This will be a necessary first step in assessing the importance of heritable bet hedging in epigenetic buffering [ 12 ].
Figure 2.
Epigenetic mechanisms can lead to heritable bet hedging that buffers populations from extinction. ( A ) A population experiences stochastic environmental variation across generations (black solid line) that falls within the zone experienced by the population over its evolutionary history (dotted black line). This is contrasted with directional stochastic variation (solid red line) caused by a rapid environmental shift across the generations that drives the population mean environment beyond normal environmental variation. For example, considering temperature, while we might assume that temperature fluctuations remain constant over time (i.e. they vary randomly within ±3°) the mean temperature might move from 23°C to 25°C over 10 years (what we consider directional stochastic variation). Populations can be maintained through bet hedging strategies within the dotted black lines, but require the presence of heritable phenotypic variation to cope with novel directional environmental changes (red dotted lines). ( B ) A bet hedging strategy fails to allow populations to cope with directional stochastic environmental change (red solid line). Bet hedging results in plastic allocation strategies (e.g. maternal effects) in the parental generation that leads to increased phenotypic variation in the subsequent generation. Selection favours individuals most closely matching the environmental optimum at the time of selection (black square) while selecting against individuals too far from the phenotypic optimum (red square with red X). The next generation, however, will on average exhibit similar phenotypes to generation 5, because these plastic responses are not heritable. ( C ) A bet hedging strategy where phenotypic variation is heritable (heritable bet hedging) allows a population to adaptively track an environmental optimum outside the range experienced in its evolutionary history. This is achieved by recruiting epigenetic mechanisms to ‘convert’ non-heritable phenotypic variability, generated through a bet hedging strategy, to heritable phenotypic variability (i.e. adaptive epigenetic tracking). In both (B) and (C) similar coloured circles (blue or orange) represent the phenotypes of two family lineages while different patterned circles represent each unique generation. Two columns of circles within a given generation represent the phenotypes before selection and the phenotypes left after selection (i.e. circles within the black square). Only 3–4 generations are shown for simplicity
Responding to Rapid Environmental Change
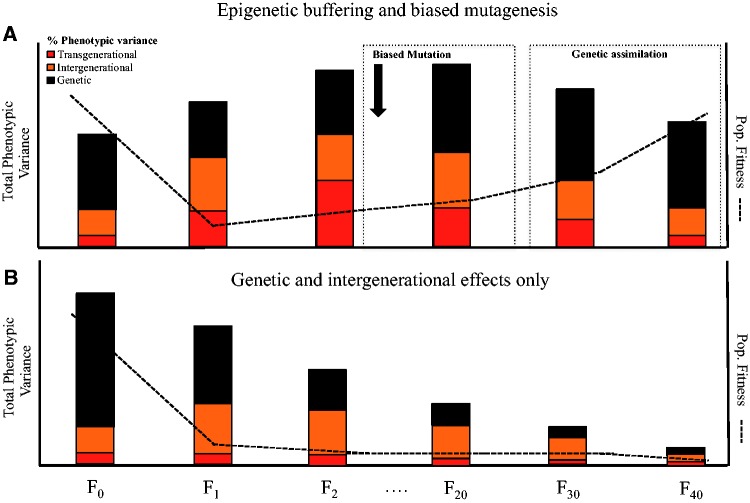
Epigenetic modifications could be the mechanism by which populations buffer against rapid, fluctuating environmental change ( Fig. 3 )—hereafter we will refer to this process as epigenetic buffering. If organisms depended on genetic adaptation alone, then a shift in the environment would decrease the populations’ genetic variance, reducing its future evolutionary potential. By decoupling the relationship between the phenotype under selection and the genotype, epigenetic inheritance could facilitate phenotypic adaptation, while both reducing genetic loss and providing the heritable phenotypic variation necessary for changes in trait distributions across generations. For example, consider a large population that experiences a sharp reduction in fitness due to a rapid environmental change ( Fig. 3 A). The environmental change increases genome-wide epigenetic variation causing a random increase in heritable phenotypic variance (i.e. heritable bet hedging). Positive selection increases the proportion of adaptive phenotypes, as well as their epigenetic creators, in subsequent generations. It is possible that epigenetic markers at genomic sites increase the probability of mutations at these locations (e.g. biased mutations—see ‘Increasing evolutionary potential: three pathways via epigenetic mechanisms’). When the epigenetic inheritance that promoted the adaptive phenotype degrades, some of these phenotypes may be maintained through the new genetic variants. Evolutionary rescue may then occur. In contrast, if heritable epigenetic mechanisms did not lead to some of the phenotypic variation being transmissible, we would predict a rapid decrease in genetic variation that may lead to population extinction ( Fig. 3 B). This simplified hypothetical scenario ( Fig. 3 ) necessitates some contentious assumptions based on the extended evolutionary synthesis [ 34 , 35 ], and whether such processes could actually lead to evolutionary rescue requires much further investigation. Nonetheless, this concept has been suggested in other papers without being directly named. For example, Bonduriansky et al. [ 15 ] suggested that non-genetic inheritance might allow the phenotype to match the environment when environmental change is too rapid or stochastic for genetic adaptation to keep up, and Harrisson et al. [ 24 ] claimed that “nongenetic inheritance may serve to buffer a population against rapid environmental change”.
Figure 3.
( A ) Epigenetic buffering helps retain genetic variance in response to a rapid decline in population fitness (dotted line). In response to an environmental stressor, we predict that total phenotypic variance should increase. In F 1 , intergenerational non-genetic/epigenetic effects (i.e. bet hedging) initially generate most phenotypic variance (orange bar), which shelters genetic variance (black bar) because this process dissociates the genotype from phenotype. In F 2 , phenotypic variation continues to increase, but a larger proportion of variance is attributed to transgenerational epigenetic inheritance, facilitating the heritability of a portion of the phenotypic variants. Over longer time scales, if the population remains in a stressful environment, it might begin to re-acquire genetic variants (replenishing genetic variance) through biased mutation rates (or through increased rates of mutation). Over longer time periods, we may get genetic assimilation as the population converges on the new fitness optima. ( B ) Depletion of genetic variation in a population when transgenerational epigenetic mechanisms (red bar) comprise a very low, non-significant proportion of the total phenotypic variance. In response to an environmental stressor, we see strong selection on phenotypic variation that slowly depletes genetic variation
Epigenetic buffering could occur by influencing each of the three modes of evolutionary potential depicted in Fig. 1 , thereby facilitating evolutionary rescue over evolutionary timescales. The scenario of epigenetic buffering we describe above focuses on epigenetic variation because this is a less familiar case, traditionally however, the interaction between epigenetic and genetic variation could also increase evolutionary potential. When a population experiences detrimental environmental change we would expect a reduction in genetic variance—due to negative selection and a declining population size—but epigenetic buffering may reduce this loss. This conservation of a populations’ genetic variance would provide greater scope for rescue. Furthermore, epigenetic buffering could increase the probability of new genetic variation. By preventing extinction in the short term, epigenetic buffering buys time for random mutations to occur, which could then aid evolution in the long term. An exciting possibility is that in addition to this random mechanism, epigenetic buffering could also facilitate biased mutations, which might hasten ‘genetic assimilation’ of the environmentally induced phenotypes. Genetic assimilation of phenotypic variants caused by environmental change is a common idea which is discussed in depth elsewhere [ 94 ]. Therefore, epigenetic buffering could provide population resilience to environmental change over ecological timescales, and paths towards population persistence over evolutionary timescales.
In summary, epigenetic buffering could facilitate evolutionary rescue through two basic phenomena which increase evolutionary potential: (i) heritability of environmentally induced phenotypes and (ii) reducing genetic loss and increasing the probability of novel genetic variation.
Outstanding Questions and Future Directions
Many factors should affect the likelihood of a population undergoing evolutionary rescue via epigenetic buffering. Below we present some outstanding questions. Ultimately, deeper insights into the mechanisms of evolutionary rescue—and the potential importance of epigenetic buffering—could provide creative solutions to conservation problems. However, we also stress the need for more empirical studies on epigenetic inheritance that test for adaptive or maladaptive phenotypic plasticity within the context of ecologically relevant environmental conditions. This will be informative in comparing and contrasting the importance of phenotypic plasticity to adaptive evolution under novel environmental conditions.
Which phyla are more likely to experience epigenetic buffering? We predict that there will be differences between phyla in the importance of epigenetic buffering. For example, there is mounting empirical evidence that plants possess more sophisticated mechanisms for genome evolution via epigenetic mechanisms than animals do [ 39 , 95–97 ]. There is a simple ecological hypothesis for this: plants are less able to move away from environmental disturbances or exhibit other types of behavioural plasticity, so they are more reliant on genetic adaptation and phenotypic plasticity through epigenetic pathways [ 51 ]. Furthermore, there is likely to be within-phyla variation in the magnitude of epigenetic buffering. Mammals, for example, undergo extensive epigenetic reprogramming between generations [ 98 ], but in zebrafish ( Danio rerio ) the paternal methylation pattern is faithfully inherited [ 99 , 100 ]. Studies from a diversity of species will be required to predict ecological impacts of environmental change, and these differences should be taken into account when evaluating empirical evidence.
How does the timing of environmental change affect the likelihood of epigenetic buffering? The life-history stage in which a species experiences an environmental disturbance could affect the likelihood of epigenetic inheritance, and we predict an interaction with taxa. For example, for mammals, an environmental disturbance early in development is far more likely to cause transgenerational inheritance than a disturbance that occurs in adulthood [ 101 ]. This is due to gamete differentiation (i.e. the Weismann barrier [ 102 ]). For species with different forms of reproduction this variable may be less important. We predict that the length of environmental fluctuations relative to the generation time of a species is another key factor. If the environment fluctuates too rapidly, then epigenetic inheritance might be eroded due to the instability of epigenetic markers. Overall, there will be limits on epigenetic buffering and some types of environmental change will inevitably lead to extinction. These dynamics are undoubtedly complex and require theoretical models.
Does heritable bet hedging occur in response to environmental change, and can bet hedging evolve? We urge empiricists to place greater importance on reporting variance in phenotypic traits. We predict that parental generations encountering environmental stress should produce offspring with more variable phenotypic traits, given the potential role of heritable bet hedging in epigenetic buffering. Understanding the adaptive significance of bet hedging has been difficult and most of the existing evidence is non-rigorous [ 12 ]. There are undoubtedly pragmatic reasons for this—measuring geometric mean fitness is difficult and requires multi-generational studies. As a result, environmental effects on trait variation are often unreported, with the focus primarily being on mean phenotypic outcomes. Nevertheless, reporting how phenotypic variance changes in response to environmental stressors will enable future meta-analytic tests of these theoretical ideas, and evaluation of the importance of transgenerational epigenetic inheritance. From a theoretical perspective, it would be interesting to consider the evolution of bet hedging itself. So far we have assumed that each individual in a population will generate the same variance in their offspring’s traits in response to environmental change, but this need not be the case. A population could undergo positive selection for increased offspring variance during periods of environmental instability, which may further facilitate rapid shifts across the fitness landscape.
What is the relative importance of genetic variation, new mutations and epigenetic variation for evolutionary rescue? Molecular biology can be used to investigate how evolutionary potential is affected by environmental stress. The three mechanisms shown in Fig. 1 —standing genetic variation, new genetic variation created by biased mutation and epigenetic variation—could all be tested empirically. Admittedly, this research will be limited by current knowledge of epigenetic mechanisms, choice of study species, and time and costs. However, there is still potential for important insights. Heritable epigenetic variation may be more important for populations with less standing genetic variation, as these populations will have lower evolutionary potential to begin with. The utility of epigenetic variation in evolutionary rescue may be constrained if epigenetic variation is correlated with genetic variation (of which there is some evidence; see [ 103 ]). It is, therefore, worth measuring the correlation between genetic and epigenetic variation to determine how independent these mechanisms are. In other words, it is important to distinguish among obligatory, facilitated and pure epigenetic variation ( sensu [ 36 ]). We would also predict the importance of new genetic variation (both random and biased) to increase with successive generations. Quantifying the magnitude and stability of heritable epigenetic variation is vital. This detail, combined with measurements of biased mutation rates, can inform evolutionary models.
Supplementary Material
Supplementary Data
Acknowledgements
We thank Losia Lagisz and three anonymous reviewers for commenting on earlier versions of this manuscript.
Supplementary data
Supplementary data are available at EnvEpig online.
Conflict of interest: None declared.
Funding
S.N. is funded by an Australian Research Council Future Fellowship (FT130100268) and D.W.A.N. is funded by an Australian Research Council Discovery Early Career Research Award (DE150101774). D.H. is supported by National Health and Medical Research Council (APP1063981).
References
- 1. Barnosky AD, Matzke N, Tomiya S, et al. . Has the Earth’s sixth mass extinction already arrived? . Nature 2011. ; 471 : 51 – 7 . [DOI] [PubMed] [Google Scholar]
- 2. Ceballos G, Ehrlich PR, Barnosky AD, et al. . Accelerated modern human–induced species losses: entering the sixth mass extinction . Sci Adv 2015. ; 1 : e1400253 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mace GM, Norris K, Fitter AH . Biodiversity and ecosystem services: a multilayered relationship . Trends Ecol Evol 2012. ; 27 : 19 – 25 . [DOI] [PubMed] [Google Scholar]
- 4. Sih A, Ferrari MCO, Harris DJ . Evolution and behavioural responses to human-induced rapid environmental change . Evol Appl 2011. ; 4 : 367 – 87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson BA, Rehage JS, Sih A . Ecological novelty and the emergence of evolutionary traps . Trends Ecol Evol 2013. ; 28 : 552 – 60 . [DOI] [PubMed] [Google Scholar]
- 6. Schlaepfer MA, Runge MC, Sherman PW . Ecological and evolutionary traps . Trends Ecol Evol 2002. ; 17 : 474 – 80 . [Google Scholar]
- 7. Carlson SM, Cunningham CJ, Westley PAH . Evolutionary rescue in a changing world . Trends Ecol Evol 2014. ; 29 : 521 – 30 . [DOI] [PubMed] [Google Scholar]
- 8. Paaby AB, Rockman MV . Cryptic genetic variation: evolution’s hidden substrate . Nat Rev Genet 2014. ; 15 : 247 – 58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jablonka E, Raz G . Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution . Q Rev Biol 2009. ; 84 : 131 – 76 . [DOI] [PubMed] [Google Scholar]
- 10. Le Rouzic A, Carlborg O . Evolutionary potential and hidden genetic variation . Trends Ecol Evol 2007. ; 23 : 33 – 7 . [DOI] [PubMed] [Google Scholar]
- 11. Starrfelt J, Kokko H . Bet-hedging: a triple trade-off between means, variances and correlations . Biol Rev 2012. ; 87 : 742 – 55 . [DOI] [PubMed] [Google Scholar]
- 12. Simons AM . Modes of response to environmental change and the elusive empirical evidence for bet hedging . Proc R Soc B Biol Sci 2011. ; 278 : 1601 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tufto J . Genetic evolution, plasticity, and bet-hedging as adaptive responses to temporally autocorrelated fluctuating selection: a quantitative genetic model . Evolution 2015. ; 69 : 2034 – 49 . [DOI] [PubMed] [Google Scholar]
- 14. Uller T . Developmental plasticity and the evolution of parental effects . Trends Ecol Evol 2008. ; 23 : 432 – 8 . [DOI] [PubMed] [Google Scholar]
- 15. Bonduriansky R, Crean AJ, Day T . The implications of nongenetic inheritance for evolution in changing environments . Evol Appl 2012. ; 5 : 192 – 201 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Snell-Rood EC . An overview of the evolutionary causes and consequences of behavioural plasticity . Anim Behav 2015. ; 85 : 1004 – 11 . [Google Scholar]
- 17. Slotkin RK, Martienssen R . Transposable elements and the epigenetic regulation of the genome . Nat Rev Genet 2007. ; 8 : 272 – 85 . [DOI] [PubMed] [Google Scholar]
- 18. Gomulkiewicz R, Shaw RG . Evolutionary rescue beyond the models . Philos Trans R Soc B Biol Sci 2013. ; 368 : 20120093 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez A, Ronce O, Ferriere R, et al. . Evolutionary rescue: an emerging focus at the intersection between ecology and evolution . Philos Trans R Soc B Biol Sci 2013. ; 368 : 20120404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hao YQ, Brockhurst MA, Petchey OL, et al. . Evolutionary rescue can be impeded by temporary environmental amelioration . Ecol Lett 2015. ; 18 : 892 – 8 . [DOI] [PubMed] [Google Scholar]
- 21. Lindsey HA, Gallie J, Taylor S, et al. . Evolutionary rescue from extinction is contingent on a lower rate of environmental change . Nature 2013. ; 494 : 463 – 7 . [DOI] [PubMed] [Google Scholar]
- 22. Whiteley AR, Fitzpatrick SW, Funk WC, et al. . Genetic rescue to the rescue . Trends Ecol Evol 2015. ; 30 : 42 – 9 . [DOI] [PubMed] [Google Scholar]
- 23. Hansen TF, Pélabon C, Houle D . Heritability is not evolvability . Evol Biol 2011. ; 38 : 258 – 77 . [Google Scholar]
- 24. Harrisson KA, Pavlova A, Telonis-Scott M, et al. . Using genomics to characterize evolutionary potential for conservation of wild populations . Evol Appl 2014. ; 7 : 1008 – 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Rouzic A, Carlborg Ö . Evolutionary potential of hidden genetic variation . Trends Ecol Evol 2008. ; 23 : 33 – 37 . [DOI] [PubMed] [Google Scholar]
- 26. Merila J, Hendry AP . Climate change, adaptation, and phenotypic plasticity: the problem and the evidence . Evol Appl 2014. ; 7 : 1 – 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danchin E . Avatars of information: towards an inclusive evolutionary synthesis . Trends Ecol Evol 2013. ; 28 : 351 – 8 . [DOI] [PubMed] [Google Scholar]
- 28. Burton T, Metcalfe NB . Can environmental conditions experienced in early life influence future generations? . Proc R Soc Lond B Biol Sci 2014. ; 281 : 20140311 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Champagne FA . Maternal imprints and the origins of variation . Horm Behav 2011. ; 60 : 4 – 11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geoghegan JL, Spencer HG . The evolutionary potential of paramutation: a population-epigenetic model . Theor Popul Biol 2013. ; 88 : 9 – 19 . [DOI] [PubMed] [Google Scholar]
- 31. Skinner MK, Gurerrero-Bosagna C, Haque MM, et al. . Epigenetics and the evolution of Darwin’s Finches . Genome Biol Evol 2014. ; 6 : 1972 – 89 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jablonka EL, Lamb MJ . Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life . Cambridge, MA: : The MIT Press; , 2005. . [Google Scholar]
- 33. Skinner MK . Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-Lamarckian concept that facilitates neo-Darwinian evolution . Genome Biol Evol 2015. ; 7 : 1296 – 302 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laland K, Uller T, Feldman M, et al. . Does evolutionary theory need a rethink? . Nature 2014. ; 514 : 161 – 4 . [DOI] [PubMed] [Google Scholar]
- 35. Laland KN, Uller T, Feldman MW, et al. . The extended evolutionary synthesis: its structure, assumptions and predictions . Proc R Soc B Biol Sci 2015. ; 282 : 20151019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richards EJ . Inherited epigenetic variation—revisiting soft inheritance . Nat Rev Genet 2006. ; 7 : 395 – 401 . [DOI] [PubMed] [Google Scholar]
- 37. Schaefer S, Nadeau JH . The genetics of epigenetic inheritance: modes, molecules, and mechanisms . Q Rev Biol 2015. ; 90 : 381 – 415 . [DOI] [PubMed] [Google Scholar]
- 38. Bohacek J, Mansuy IM . Molecular insights into transgenerational non-genetic inheritance of acquired behaviours . Nat Rev Genet 2015. ; 16 : 641 – 52 . [DOI] [PubMed] [Google Scholar]
- 39. Heard E, Martienssen RA . Transgenerational epigenetic inheritance: myths and mechanisms . Cell 2014. ; 157 : 95 – 109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moore DS . The Developing Genome . Oxford: : Oxford University Press; , 2015. . [Google Scholar]
- 41. Rosenfeld CS . Chapter 11—Animal models of transgenerational epigenetic effects . In: Tollefsbol T. (ed.), Transgenerational Epigenetics . Oxford: : Academic Press; , 2014. , 123 – 45 . [Google Scholar]
- 42. Schott D, Yanai I, Hunter CP . Natural RNA interference directs a heritable response to the environment . Sci Rep 2014. ; 4 : 7387 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Geoghegan JL . Chapter 15—Inheritance of epigenome variants across generations and its implications on the emergence of phenotypic novelties during evolution . In: Tollefsbol T. (ed.), Transgenerational Epigenetics . Oxford: : Academic Press; , 2014. , 187 – 94 . [Google Scholar]
- 44. West-Eberhard MJ . Developmental Plasticity and Evolution . New York: : Oxford University Press Inc. , 2003. . [Google Scholar]
- 45. Lande R . Evolution of phenotypic plasticity in colonizing species . Mol Ecol 2015. ; 24 : 2038 – 45 . [DOI] [PubMed] [Google Scholar]
- 46. Cropley JE, Dang THY, Martin DIK, et al. . The penetrance of an epigenetic trait in mice is progressively yet reversibly increased by selection and environment . Proc R Soc B Biol Sci 2012. ; 279 : 2347 – 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rassoulzadegan M, Grandjean V, Gounon P, et al. . RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse . Nature 2006. ; 441 : 469 – 74 . [DOI] [PubMed] [Google Scholar]
- 48. Burt A, Trivers R . Genes in Conflict: The Biology of Selfish Genetic Elements . Cambridge, MA: : Belknap Press of Harvard University Press; , 2006. . [Google Scholar]
- 49. Mello CC, Conte D . Revealing the world of RNA interference . Nature 2004. ; 431 : 338 – 42 . [DOI] [PubMed] [Google Scholar]
- 50. Wilson RC, Doudna JA . Molecular mechanisms of RNA interference . Annu Rev Biophys 2013. ; 42 : 217 – 39 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fedoroff NV . Presidential address. Transposable elements, epigenetics, and genome evolution . Science 2012. ; 338 : 758 – 67 . [DOI] [PubMed] [Google Scholar]
- 52. McClintock B . The significance of responses of the genome to challenge . Science 1984. ; 226 : 792 – 801 . [DOI] [PubMed] [Google Scholar]
- 53. SanMiguel P, Tikhonov A, Jin YK, et al. . Nested retrotransposons in the intergenic regions of the maize genome . Science 1996. ; 274 : 765 – 8 . [DOI] [PubMed] [Google Scholar]
- 54. Herman JJ, Spencer HG, Donohue K, et al. . How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging . Evolution 2014. ; 68 : 632 – 43 . [DOI] [PubMed] [Google Scholar]
- 55. Makova KD, Hardison RC . The effects of chromatin organization on variation in mutation rates in the genome . Nat Rev Genet 2015. ; 16 : 213 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ruden DM, Rasouli P, Lu X . Epignenetics, environment, and evolution . In: Esteller M. (ed.), Epigenetics in Biology and Medicine . Boca Raton, FL: : CRC; , 2009. , 241 – 60 . [Google Scholar]
- 57. Yang SH, Wang L, Huang J, et al. . Parent-progeny sequencing indicates higher mutation rates in heterozygotes . Nature 2015. ; 523 : 463 – 7 . [DOI] [PubMed] [Google Scholar]
- 58. Klironomos FD, Berg J, Collins S . How epigenetic mutations can affect genetic evolution: model and mechanism . Bioessays 2013. ; 35 : 571 – 8 . [DOI] [PubMed] [Google Scholar]
- 59. Skinner MK, Manikkam M, Guerrero-Bosagna C . Epigenetic transgenerational actions of environmental factors in disease etiology . Trends Endocrinol Metab 2010. ; 21 : 214 – 22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Crews D, Gore AC, Hsu TS, et al. . Transgenerational epigenetic imprints on mate preference . Proc Natl Acad Sci U S A 2007. ; 104 : 5942 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guerrero-Bosagna C, Covert TR, Haque MM, et al. . Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers . Reprod Toxicol 2012. ; 34 : 694 – 707 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anway MD, Leathers C, Skinner MK . Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease . Endocrinology 2006. ; 147 : 5515 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Anway MD, Cupp AS, Uzumcu M, et al. . Epigenetic transgenerational actions of endocrine disruptors and mate fertility . Science 2005. ; 308 : 1466 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schneider S, Kaufmann W, Buesen R, et al. . Vinclozolin—the lack of a transgene rational effect after oral maternal exposure during organogenesis . Reprod Toxicol 2008. ; 25 : 352 – 60 . [DOI] [PubMed] [Google Scholar]
- 65. Tracey R, Manikkam M, Guerrero-Bosagna C, et al. . Hydrocarbons (jet fuel JP-8) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations . Reprod Toxicol 2013. ; 36 : 104 – 16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wolstenholme JT, Edwards M, Shetty SRJ, et al. . Gestational exposure to bisphenol A produces transgenerational changes in behaviors and gene expression . Endocrinology 2012. ; 153 : 3828 – 38 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Doyle TJ, Bowman JL, Windell VL, et al. . Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice . Biol Reprod 2013. ; 88 : 112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baker TR, Peterson RE, Heideman W . Using zebrafish as a model system for studying the transgenerational effects of dioxin . Toxicol Sci 2014. ; 138 : 403 – 11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Manikkam M, Guerrero-Bosagna C, Tracey R, et al. . Transgenerational actions of environmental compounds on reproductive disease and identification of epigenetic biomarkers of ancestral exposures . PLoS One 2012. ; 7 : e31901 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bhandari RK, vom Saal FS, Tillitt DE . Transgenerational effects from early developmental exposures to bisphenol A or 17 alpha-ethinylestradiol in medaka, Oryzias latipes . Sci Rep 2015. ; 5 : 9303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chamorro-Garcia R, Sahu M, Abbey RJ, et al. . Transgenerational inheritance of increased fat depot size, stem cell reprogramming, and hepatic steatosis elicited by prenatal exposure to the obesogen tributyltin in mice . Environ Health Perspect 2013. ; 121 : 359 – 66 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ou X, Zhang Y, Xu C, et al. . Transgenerational inheritance of modified DNA methylation patterns and enhanced tolerance induced by heavy metal stress in rice ( Oryza sativa L.) . PLoS One 2012. ; 7 : e41143 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dias BG, Ressier KJ . Parental olfactory experience influences behavior and neural structure in subsequent generations . Nat Neurosci 2014. ; 17 : 89 – 96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Veenendaal MVE, Painter RC, de Rooij SR, et al. . Transgenerational effects of prenatal exposure to the 1944–45 Dutch famine . BJOG 2013. ; 120 : 548 – 54 . [DOI] [PubMed] [Google Scholar]
- 75. Huck UW, Labov JB, Lisk RD . Food-restricting first generation juvenile female hamsters ( Mesocricetus auratus ) affects sex ratio and growth of third generation offspring . Biol Reprod 1987. ; 37 : 612 – 7 . [DOI] [PubMed] [Google Scholar]
- 76. Bygren LO, Kaati G, Edvinsson S . Longevity determined by paternal ancestors’ nutrition during their slow growth period . Acta Biotheor 2001. ; 49 : 53 – 9 . [DOI] [PubMed] [Google Scholar]
- 77. Kaati G, Bygren LO, Edvinsson S . Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period . Eur J Hum Genet 2002. ; 10 : 682 – 8 . [DOI] [PubMed] [Google Scholar]
- 78. Van Allen BG, Rudolf VHW . Ghosts of habitats past: environmental carry-over effects drive population dynamics in novel habitat . Am Nat 2013. ; 181 : 596 – 608 . [DOI] [PubMed] [Google Scholar]
- 79. Walsh MR, Cooley F, Biles K, et al. . Predator-induced phenotypic plasticity within- and across-generations: a challenge for theory? . Proc R Soc B Biol Sci 2015. ; 282 : 20142205 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Franklin TB, Russig H, Weiss IC, et al. . Epigenetic transmission of the impact of early stress across generations . Biol Psychiatry 2010. ; 68 : 408 – 15 . [DOI] [PubMed] [Google Scholar]
- 81. Gapp K, Jawaid A, Sarkies P, et al. . Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice . Nat Neurosci 2014. ; 17 : 667 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Holeski LM, Jander G, Agrawal AA . Transgenerational defense induction and epigenetic inheritance in plants . Trends Ecol Evol 2012. ; 27 : 618 – 26 . [DOI] [PubMed] [Google Scholar]
- 83. Ho DH, Burggren WW . Parental hypoxic exposure confers offspring hypoxia resistance in zebrafish ( Danio rerio ) . J Exp Biol 2012. ; 215 : 4208 – 16 . [DOI] [PubMed] [Google Scholar]
- 84. Skinner MK . What is an epigenetic transgenerational phenotype? F3 or F2 . Reprod Toxicol 2008. ; 25 : 2 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nettle D, Bateson M . Adaptive developmental plasticity: what is it, how can we recognize it and when can it evolve? . Proc R Soc B Biol Sci 2015. ; 282 : 23 – 31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shea N, Pen I, Uller T . Three epigenetic information channels and their different roles in evolution . J Evol Biol 2011. ; 24 : 1178 – 87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jablonka E, Lachmann M, Lamb MJ . Evidence, mechanisms and models for the inheritance of acquired characters . J Theor Biol 1992. ; 158 : 245 – 68 . [Google Scholar]
- 88. Lachmann M, Jablonka E . The inheritance of phenotypes: an adaptation to fluctuating environments . J Theor Biol 1996. ; 181 : 1 – 9 . [DOI] [PubMed] [Google Scholar]
- 89. Uller T, English S, Pen I . When is incomplete epigenetic resetting in germ cells favoured by natural selection? . Proc R Soc B Biol Sci 2015. ; 282 : 20150682 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Peck JR . A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex . Genetics 1994. ; 137 : 597 – 606 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ghalambor GK, Hoke KL, Ruell EW, et al. . Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature . Nature 2015. ; 525 : 372 – 5 . [DOI] [PubMed] [Google Scholar]
- 92. Pal C . Plasticity, memory and the adaptive landscape of the genotype . Proc R Soc B Biol Sci 1998. ; 265 : 1319 – 23 . [Google Scholar]
- 93. Pál C, Miklós I . Epigenetic inheritance, genetic assimilation and speciation . J Theorl Biol 1999. ; 200 : 19 – 37 . [DOI] [PubMed] [Google Scholar]
- 94. Crispo E . The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity . Evolution 2007. ; 61 : 2469 – 79 . [DOI] [PubMed] [Google Scholar]
- 95. Feng S, Jacobsen SE, Reik W . Epigenetic reprogramming in plant and animal development . Science 2010. ; 330 : 622 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Henderson IR, Jacobsen SE . Epigenetic inheritance in plants . Nature 2007. ; 447 : 418 – 24 . [DOI] [PubMed] [Google Scholar]
- 97. Ito H . Chapter 12—Plant models of transgenerational epigenetic inheritance . In: Tollefsbol T. (ed.), Transgenerational Epigenetics . Oxford: : Academic Press; , 2014. , 147 – 61 . [Google Scholar]
- 98. Morgan HD, Santos F, Green K, et al. . Epigenetic reprogramming in mammals . Hum Mol Genet 2005. ; 14 : R47 – 58 . [DOI] [PubMed] [Google Scholar]
- 99. Jiang L, Zhang J, Wang JJ, et al. . Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos . Cell 2013. ; 153 : 773 – 84 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Potok ME, Nix DA, Parnell TJ, et al. . Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern . Cell 2013. ; 153 : 759 – 72 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Feil R, Fraga MF . Epigenetics and the environment: emerging patterns and implications . Nat Rev Genet 2012. ; 13 : 97 – 109 . [DOI] [PubMed] [Google Scholar]
- 102. Sabour D, Schöler HR . Reprogramming and the mammalian germline: the Weismann barrier revisited . Curr Opin Cell Biol 2012. ; 24 : 716 – 23 . [DOI] [PubMed] [Google Scholar]
- 103. Pecinka A, Abdelsamad A, Vu GTH . Hidden genetic nature of epigenetic natural variation in plants . Trends Plant Sci 2013. ; 18 : 624 – 32 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data