Abstract
Assisted reproductive technologies are fertility treatments used by subfertile couples to conceive their biological child. Although generally considered safe, these pregnancies have been linked to genomic imprinting disorders, including Beckwith–Wiedemann and Silver–Russell Syndromes. Silver–Russell Syndrome is a growth disorder characterized by pre- and post-natal growth retardation. The Mest imprinted domain is one candidate region on chromosome 7 implicated in Silver–Russell Syndrome. We have previously shown that maintenance of imprinted methylation was disrupted by superovulation or embryo culture during pre-implantation mouse development. For superovulation, this disruption did not originate in oogenesis as a methylation acquisition defect. However, in comparison to other genes, Mest exhibits late methylation acquisition kinetics, possibly making Mest more vulnerable to perturbation by environmental insult. In this study, we present a comprehensive evaluation of the effects of superovulation and in vitro culture on genomic imprinting at the Mest gene. Superovulation resulted in disruption of imprinted methylation at the maternal Mest allele in blastocysts with an equal frequency of embryos having methylation errors following low or high hormone treatment. This disruption was not due to a failure of imprinted methylation acquisition at Mest in oocytes. For cultured embryos, both the Fast and Slow culture groups experienced a significant loss of maternal Mest methylation compared to in vivo-derived controls. This loss of methylation was independent of development rates in culture. These results indicate that Mest is more susceptible to imprinted methylation maintenance errors compared to other imprinted genes.
Keywords: genomic imprinting, DNA methylation, mouse, assisted reproduction, Mest
Introduction
Assisted reproductive technologies (ARTs) are fertility treatments used by infertile/subfertile couples to conceive their biological child. Although generally considered safe, ART pregnancies exhibit increased risk for preterm birth, low birth weight, intrauterine growth restriction [1] and have been linked to genomic imprinting disorders including Beckwith–Wiedemann Syndrome [2–10], Angelman Syndrome [11–13], and Silver–Russell Syndrome [7, 14–20].
Silver–Russell Syndrome (SRS) is an imprinting disorder characterized by pre- and post-natal growth retardation, relative macrocephaly at birth, protruding forehead, body asymmetry, and low body mass index and/or feeding difficulties [21]. Up to 44% of SRS cases observed in the general population are associated with hypomethylation at the H19 imprinted domain, which is located within the 11p15 region. By comparison, from the limited number of SRS patients conceived by ARTs that were studied, 92% possess H19 hypomethylation, indicating a higher incidence of imprinted methylation perturbations in SRS patients in ART versus the general population [7, 14–19]. Maternal uniparental disomy of chromosome 7 (two maternal and no paternal copies of chromosome 7) has been implicated in 5–10% of SRS cases, indicating that the absence of chromosome 7 imprinted genes of paternal origin and/or double the number of chromosome 7 imprinted genes of maternal origin leads to SRS [22]. The MEST (mesoderm-specific transcript; also known as paternally expressed gene 1) imprinted domain is one of the primary SRS candidate regions on chromosome 7 (7q32) [15, 23–26]. In mice, paternal inheritance of a Mest targeted deletion results in severe intrauterine growth restriction [27].
The Mest imprinted domain is located on mouse chromosome 6 in a region syntenic to human chromosome 7 [28, 29]. A gametic differentially methylated region (gDMR) spans the Mest putative promoter region and exon 1. The Mest gDMR is methylated on the maternal allele while the paternal allele is unmethylated [30–32]. During oogenesis, DNA methylation acquisition at imprinted, maternal gDMRs occurs in an oocyte diameter/size-, days postpartum-, and/or follicular stage-dependent manner, with imprinted gDMRs showing differential acquisition kinetics [33–36]. Compared to other imprinted gDMRs, the Mest gDMR methylation exhibits the latest acquisition kinetics [34, 37].
Late acquisition of de novo methylation has led to the suggestion that the Mest gDMR may be more vulnerable to perturbation by environmental insult [38]. To investigate the requirement for methyl donors during follicle development, Anckaert et al. [38] cultured preantral follicles in medium with low methyl donors. While acquisition of DNA methylation at the Snrpn and Igf2r gDMRs was not impeded, there was a reduced level of DNA methylation at the Mest gDMR. We also analyzed de novo methylation in connexin 43 (Gja4) deficient oocytes under the premise that gap junction communication provides important metabolites for DNA methylation acquisition. In contrast to the Snrpn and Peg3 gDMRs, we observed a loss or delay in methylation acquisition at the Mest gDMR, possibly due to its late methylation acquisition [36].
Additional consideration should be given to the grand-parental alleles with regard to environmental insult. Acquisition of Mest gDMR methylation occurs differentially with the grand-maternal allele acquiring methylation prior to the grand-paternal Mest allele [33, 34, 36]. This may place the grand-paternal Mest gDMR at a higher risk for methylation acquisition errors.
In this study, we characterize the effects of superovulation or embryo culture on the acquisition and maintenance of genomic imprinting at the Mest locus. Superovulation, also known as ovarian stimulation, is used to recover large numbers of mature oocytes at one time to increase the chances of generating diploid fertilized zygotes. Embryo culture allows the identification and selection of developmental competent in vitro produced embryos for transfer to the patient as a strategy to increase the chances of implantation. To provide a comprehensive allelic analysis of the response of the Mest gene to superovulation and embryo culture, and to avoid confounding factors such as intrinsic patient sub-fertility and sample pooling, our analysis was performed on individual oocytes and embryos in a mouse model. Here, we demonstrate that the Mest gDMR was hypermethylated on the grand-maternal and grand-paternal alleles in MII oocytes following low or high doses of superovulation similar to control oocytes, indicating that acquisition of DNA methylation at the Mest gDMR in the growing oocyte was not affected by hormonal stimulation. By comparison, significant methylation loss occurred at the maternal Mest gDMR in blastocysts following superovulation regardless of hormone dosage. For in vitro culture, both fast and slow developing embryos experienced a significant loss of maternal Mest gDMR methylation compared to in vivo-derived controls. These results contrast with our previous studies, where greater numbers of embryos in the high hormone treatment group and in the fast-developing culture group showed loss of maternal Snrpn and paternal H19 gDMR methylation compared to the high hormone treatment and slow-developing culture groups [39, 40]. These results indicate that Mest gDMR is more susceptible to imprinted methylation maintenance errors.
Methods
B6(CAST7p6) Mice
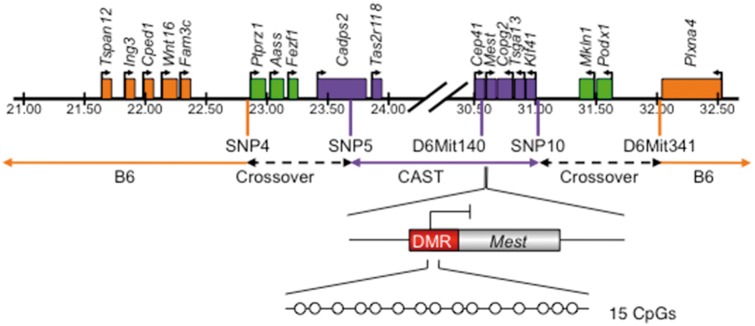
Previous studies from our lab utilized a mouse model suited for imprinting analyses, namely C57BL/6(CAST7) [B6(CAST7)] mice, which contains two Mus musculus castaneus chromosome 7 s on a B6 background [41]. Since Mest is located on chromosome 6, we screened our B6(CAST7) colony using satellite markers, or single nucleotide polymorphisms (SNPs) plus restriction digestion (refer to Table 1 for PCR primers and annealing temperatures) to identify a subset of mice that harbored a partial region of Mus musculus castaneus chromosome 6. The proximal crossover was mapped between SNP4 (rs3090864) at 22.8 MB and SNP5 (rs3088527) at 23.7 MB (Fig. 1). SNP4 is a polymorphic site between B6 (A) and CAST (G), which distinguished the parental alleles via HpyCH4III restriction digestion (B6, 181 and 12 bp; CAST, 101, 80, and 12 bp). SNP5, a polymorphic site between B6 (C) and CAST (A), identified parental alleles though CviKI-1 restriction digestion (B6, 74, 54, and 38 bp; CAST, 112 and 54 bp). The distal crossover was first mapped to a region between D6Mit140 (30.60 MB) and D6Mit341 (32.05 MB), and then narrowed to a region between SNP10 (rs6183467) (31.02 MB) and D6Mit341 (32.05 MB). For SNP10, a polymorphic site distinguished B6 (T) and CAST (G) alleles though HincII restriction digestion (B6, 316 bp; CAST, 232 and 84 bp). Thus, crossover events were mapped to 22.8–23.7 and 31.02–32.05 MB, which encompassed a 9.25 MB region containing the Mest imprinted domain (Fig. 1). All polymorphisms were confirmed by sequencing of PCR products. B6(CAST7p6) intercrosses were used to generate a B6(CAST7p6) mouse colony. This B6(CAST7p6) mouse model was used for all subsequent experiments. Experiments were performed in compliance with the guidelines set by the Canadian Council for Animal Care, and the policies and procedures approved by the Western University Council on Animal Care.
Table 1.
Mest primers
| Region | Accession | Primer/probe | Primer sequence (5'–3') | Annealing Temperature (°C) |
|---|---|---|---|---|
| Imprinted methylation analysis | ||||
| D6Mit140 | MGI: 702660 | F | TGCCAACTAAGGTACATCTATAGCC | 55 |
| R | TGGTTCAAAAAATAAGATTCTGAGC | |||
| D6Mit34 | MGI: 705219 | F | TGTGTGTGTTGCCTCCTCTC | 58 |
| R | ACCAGTTTTTACCTTTCAAAATAATG | |||
| SNP4 | rs3090864 | F | GTGCCAGATTGTCTTCCC | 55 |
| R | ACCCTCAGGACAGTTCG | |||
| SNP5 | rs3088527 | F | ATGCCTCATTTGGAGTCTG | 55 |
| R | AGCATCCTCTGGGAGTGTA | |||
| SNP10 | rs6183467 | F | CAGGATGGGTCTGGAGTGA | 55 |
| R | CTTAGTAGCAACTGGGTGGTG | |||
| Imprinted expression analysis | ||||
| 1380–1920 | NM_008590 | F | CACATTGGTGAACAAACTACAGG | 50 |
| R | AGAGTGCTGGGAACTGAACC | |||
| Imprinted methylation analysis | ||||
| 1309–1651 | AF017994 | OF | MestB TTTTAGATTTTGAGGGTTTTAGGTTG | 50 |
| OR | MestE TCATTAAAAACACAAACCTCCTTTAC | |||
| IF | MestC GGTGTTGGTATTTTTAGTGTTAGTTG | 57.5 | ||
| IR | MestD AATCCCTTAAAAATCATCTTTCACAC | |||
F, forward; R, reverse; OF, outer forward, OR, outer reverse; IF, inner forward; IR, inner reverse.
Figure 1:
crossover sites in the B6(CAST7p6) mouse model. Graphical representation of chromosome 6 in the B6(CAST7p6) mouse model. Genes in orange are located within the B6 region, genes in purple are located within the Mus musculus castaneus region, and genes in green fall within the crossover region. Orange and purple vertical lines represent satellite markers (D6Mit140 and D6Mit341) and single nucleotide polymorphisms (SNPs)/restriction digestion that were used to determine genotypes. The Mest imprinted domain was found to reside in a Mus musculus castaneus 9.25 MB region between 22.8–23.7 and 31.02–32.05 MB. The region analyzed for imprinted Mest methylation is indicated. Red box, maternally methylated Mest gDMR; blunted-ended arrow, repressed transcription start site; white circles, CpG dinucleotides
Oocyte and Embryo Collection, and Embryo Culture
Ovulated oocytes were collected from B6(CAST7p6)XB6 F1 females following spontaneous ovulation or superovulation as previously prescribed [42, 43]. Briefly, for superovulation, females were injected with either 6.25 IU or 10 IU equine chorionic gonadotropin (eCG, Intervet Canada) followed 40–44 h later by the same dose of human chorionic gonadotropin (hCG, Intervet Canada). Females in the estrus stage of spontaneous ovulation cycles were used as unstimulated controls. Oocyte-cumulus cell complexes were flushed from the oviducts at approximately 12 PM the following day (22 h post-hCG) into M2 media (Sigma). For the spontaneously ovulated group, we obtained 9.7 ± 1.6 oocytes/female (n = 15). For the 6.25 IU and 10 IU hormone-treated groups, 25.6 ± 5.7 and 29.7 ± 7.1 oocytes/female were obtained, respectively (n = 7–8 females). MII stage oocytes were dissociated from surrounding cumulus cells using 0.3 mg/ml hyaluronidase (Sigma) and washed three times in M2 media. Oocytes were treated with acidic tyrode’s solution (Sigma) at room temperature to remove the zona pellucida, washed twice more in M2 media, and placed individually on a glass slide in minimal media for agarose embedding.
To obtain B6(CAST7p6)XB6 F1 embryos following superovulation, B6(CAST7p6) females were mated with B6 males (Charles River, St Constant, Canada) the same day as the hCG injection as previously described [39]. Mating was determined by the presence of a vaginal plug at 0.5 days post-coitum (dpc). For embryo culture experiments, 2-cell embryos were flushed from the oviducts at 1.5 dpc, washed twice and cultured in Whitten’s medium. Embryo culture drops (10–20 μl) with a filter-sterilized mineral oil overlay (Sigma) were prepared prior to 9 AM and allowed to equilibrate. Embryos were cultured at a concentration of 1 embryo per microliter. Embryos were separated into four groups based on rate of development over the course of the 3-day culture period as previously described [40]. For in vivo-derived embryos, B6(CAST7p6) females were checked for estrus, and then mated with B6 males. Blastocyst stage embryos were flushed from uteri in M2 medium (Sigma) at 3.5 dpc. Blastocysts from control and experimental groups were placed in individual tubes, snap frozen on dry ice and stored at −80 °C.
Analysis of Imprinted Mest Methylation on Single Oocytes
Processing, agarose embedding, and bisulfite mutagenesis of individual oocytes was performed as previously described [42, 43]. Each oocyte sample was directly added as a solid agarose bead to an Illustra ready-to-go PCR bead (GE) containing 0.2 μM final concentration of Mest outer primers and 9.6 ng/ml final concentration of tRNA in a 15 μl solution, with 25 μl of mineral oil overlay. Negative controls (agarose bead without oocyte) were processed alongside each sample. For the second round, 5 μl of first round was added to a second 25 μl ready-to-go PCR bead containing 0.2 μM final concentration of Mest inner primers, with 25 μl of mineral oil overlay. Primers were designed within a previously described region [38], allowing for the analysis of 15 CpGs within the Mest gDMR (Fig. 1) (Accession number, AF017994; primer positions outer, MestBE 1088–1744, inner, MestCD 1309–1651; nucleotide 1343, A in B6, C in CAST). Refer to Table 1 for primers and annealing temperatures. For each oocyte, 5 clones were sequenced. Samples having two or more clones with different methylation patterns and different non-CpG conversion rates were excluded from analysis, as cumulus cell contamination could not be ruled out. Sequences with conversion rates <85% were not included. Following the bisulfite mutagenesis and PCR amplification process, 36% (10/28) of spontaneous, 38% (17/45) of 6.25 IU, and 40% (20/50) of 10 IU ovulated oocytes successfully amplified. Of these, 0, 12, and 25%, respectively, were excluded from analysis due to a conversion rate below 85%, or having more than two methylation patterns suggestive of cumulus cell contamination.
Analysis of Imprinted Mest Expression and Methylation in Blastocysts
The combined analysis of imprinted expression and methylation in individual blastocysts was performed as previously described [40]. For the analysis of imprinted Mest expression, amplification of a 541 bp fragment was tested using SYBR green to allow determination of the range of cycles located in log-phase amplification. PCR on subsequent embryos was performed to ensure that amplification was in the log-phase upon completion of the PCR program. Following PCR amplification using ready-to-go PCR Beads (GE; 0.2 μM final concentration Mest primers), embryos were digested with the BsiHKA1 restriction enzyme to determine allelic identity. Densitometry was performed using the Opticon Monitor Software.
For imprinted methylation analysis, bisulfite mutagenesis, nested PCR (0.2 μM final concentration Mest outer and inner primers; first round PCR was split in half to allow for two independent PCR reactions), cloning and sequencing was performed as described previously [40]. At least 40 clones per embryo were sequenced. Sequences with conversion rates <85% were not included. Identical clones (identical location and number of unconverted CpG-associated cytosines and identical location and number of unconverted non-CpG-associated cytosines) were included only once. Percent methylation was calculated as number of methylated CpGs over the total number of CpGs. Refer to Table 1 for primers and annealing temperature.
Statistical Analysis
Statistical analysis was performed comparing maternal Mest methylation between in vivo-derived embryos, and embryos generated via superovulation (6.25 IU, low; 10 IU, high) or in vitro culture (Fast, Slow, FF, FS, SF, and SS). To account for variability among blastocysts within a given condition, random and mixed effects logistic regression models with random blastocyst effects were used. Models were fit through maximum pseudo-likelihood in SAS v9, estimated average group maternal methylation levels and odds ratios (OR) between groups were reported, 95% Wald based confidence intervals (CI) were computed, and two-sided level 5% Wald tests were used to test significance. To generate the box plot in Fig. 4, we used BoxPlotR; http://shiny.chemgrid.org/boxplotr/ (1 June 2017, date last accessed).
Figure 4:

box plot of maternal Mest gDMR methylation levels in control and experimental blastocysts. Each red circle represents the maternal Mest gDMR methylation levels for one embryo. Spon, spontaneous; and FF, fast–fast; FS, fast–slow; SF, slow–fast; SS, slow–slow developmental rates
Results
Effects of Superovulation at the Mest gDMR in Ovulated Oocytes
To determine the methylation status of the Mest gDMR in spontaneously ovulated and superovulated oocytes, we performed the single oocyte bisulfite mutagenesis and clonal sequencing assay developed by our group [42, 43]. Imprinted methylation was assessed at 15 CpGs within the Mest gDMR (Fig. 1). Individual oocytes from spontaneously ovulating B6(CAST7p6)XB6 females displayed 93–100% methylation at the Mest gDMR (Fig. 2). Similar to untreated controls, oocytes in the low and high dosage groups possessed 93–100% methylation at the Mest gDMR (Fig. 2), with mean methylation levels of 98.0, 97.8, and 98.2% in the spontaneous, 6.25 and 10 IU groups, respectively. Thus, superovulation did not alter acquisition of imprinted methylation at the Mest gDMR, at either the grand-maternal [B6(CAST7p6)] or the grand-paternal (B6) alleles during oogenesis.
Figure 2:
methylation of the Mest gDMR in individual oocytes derived from spontaneously ovulated and superovulated (6.25 and 10 IU) B6(CAST7p6)XB6 F1 females. Unmethylated CpGs are represented as white circles, while methylated CpGs are depicted as black circles. Each line denotes an individual strand of DNA from a single oocyte. Oocyte designations are indicated on the left of each DNA strand, and the grand-parental allele is indicated on the right of each strand (B, B6 grand-paternal; C, CAST grand-maternal)
Imprinted Mest gDMR Methylation in In Vivo-Derived Blastocyst-Stage Embryos
Prior to our investigation of superovulated or cultured embryos, we set out to determine the normal levels of DNA methylation at the Mest gDMR in our mouse model. Using our combined imprinted expression and methylation assay [40] to obtain information for individual blastocysts, we determined the methylation patterns of 10 in vivo-derived B6(CAST7p6)XB6 embryos. The mean maternal Mest methylation level in in vivo-derived blastocysts was 73.2%. The first quartile of the in vivo group (70%) was used as a cut-off, such that 70% and above was classified as normal (N) and below 70% was categorized as abnormal (A) methylation levels. Using this cut-off, 8/10 in vivo-derived blastocysts displayed normal methylation levels (70–93%), while 2/10 in vivo-derived embryos (46%; 61%) displayed abnormal methylation levels (Figs 3 and 4).
Figure 3:
methylation of the maternal Mest gDMR in embryos derived from spontaneously ovulating females. Each group of DNA strands represents one blastocyst. Unmethylated CpGs are represented as white circles while methylated CpGs are depicted as black circles. Each line denotes an individual strand of DNA, and each group of strands represents an individual blastocyst. Blastocyst designations are indicated at the top left, percent maternal methylation is indicated at the top center, and normal (N) or abnormal (A) methylation levels are indicated at the top right of each group. Percentage methylation was calculated as the number of methylated CpGs/total number of CpGs
Effects of Superovulation at the Imprinted Mest gDMR in Blastocyst-Stage Embryos
Next, we investigated imprinted methylation at the Mest gDMR in 20 blastocysts from superovulated females, using 6.25 IU (low) or 10 IU (high) hormone dosages. The estimated mean maternal methylation level in the low hormone group was 56.2% (CI= 42.3–69.2%) and was significantly less than that of in vivo-derived blastocysts (OR = 0.47, CI = 0.23–0.94, P = 0.03) (Figs 4 and 5). Of these embryos, 6/9 (25%; 33%; 38%; 47%; 54%; 68%) displayed loss of maternal Mest gDMR methylation. The estimated mean maternal methylation level in the high hormone group was 56.3% (CI= 44.8–67.2%) and was also significantly less than that of the in vivo-derived blastocysts (OR = 0.47, CI = 0.24–0.91, P = 0.02) (Figs 4 and 6 ). Of these blastocysts, 7/11 (20%; 33%; 33%; 46%; 61%; 61%; 61%) exhibited loss of maternal Mest gDMR methylation. No significant difference was found in maternal methylation levels between the low and high hormone groups (OR = 1.00, CI = 0.51–1.97, P = 0.99), indicating that methylation loss was not dose-dependent.
Figure 5:
methylation of the maternal Mest gDMR in embryos derived from superovulated females treated with a low hormone dosage (6.25 IU). See Fig. 3 for details
Figure 6:
methylation of the maternal Mest gDMR in embryos derived from superovulated females treated with a high hormone dosage (10 IU). See Fig. 3 for details
Effects of In Vitro Culture at the Imprinted Mest gDMR in Blastocyst-Stage Embryos
In a previous study [40] and here, we utilized Whitten’s, a suboptimal culture medium, since it produces imprinting methylation defects, allowing us to investigate the relationship between developmental rates and imprint methylation maintenance. To evaluate the effects of embryo culture on imprinted methylation at the Mest gDMR in relation to rates of development, we analyzed 23 individual embryos cultured from the 2-cell to the blastocyst stage in Whitten’s medium. Embryos were separated based on rates of pre-implantation development [40]. Two-cell embryos were cultured for 24 h, after which embryos with 8 or more cells were placed in the Fast group, while those with less than 8 cells were placed in the Slow group. After another 24 h of culture, a second separation was performed with embryos in the Fast group split into fast–fast (FF) and fast–slow (FS) groups based on whether they were cavitating or compacted, respectively. Embryos in the Slow group were divided based on whether or not they had compacted [slow–fast (SF) or slow–slow (SS) groups, respectively]. Embryos were cultured for another 24 h to the expanded blastocyst-stage. At the first separation, the Fast (FF and FS) group displayed a mean maternal methylation level of 56.5% (CI = 50.4–62.5%), which was significantly less than in vivo-derived embryos (OR = 0.48, CI = 0.31–0.73, P < 0.01), while the Slow (SF and SS) group displayed a mean maternal methylation level of 59.5% (CI = 53.7–65.6%), which was also significantly less than in vivo-derived embryos (OR = 0.55, CI = 0.35–0.84, P < 0.01). The Fast (FF and FS) group displayed 11/13 embryos with loss of maternal Mest methylation, while the Slow (SF and SS) group had 8/11 blastocysts with abnormal maternal Mest methylation levels. At the second separation, the estimated mean maternal methylation levels in the FF and FS groups were 53.8% (CI = 46.6–60.8%) and 61.1% (CI = 51.1–70.2%), respectively, while in the SF and SS groups, the estimated mean maternal methylation levels were 57.1% (CI = 50.7–63.3%) and 63.1% (CI = 52.5–72.6%), respectively (Figs 7 and 8). In the FF group, 7/8 embryos (40%; 41%; 44%; 54%; 56%; 59%; 66%) displayed loss of maternal Mest gDMR methylation levels, as did 4/5 blastocysts (42%; 59%; 63%; 64%) in the FS group. In the SF and SS groups, 5/6 blastocysts (47%; 53%; 54%; 56%; 62%) and 3/5 embryos (47%; 58%; 60%) experienced a loss of maternal Mest gDMR methylation. We did not observe any significant difference in maternal Mest gDMR methylation between the Fast and Slow (OR = 0.87, CI = 0.58–1.31, P = 0.51), or FF and FS (OR = 0.74, CI = 0.43–1.29, P = 0.29), and SF and SS groups (OR = 0.78, CI = 0.43–1.40, P = 0.40).
Figure 7:
methylation of the maternal Mest gDMR in blastocysts from the fast (FF and FS) culture groups. See Fig. 3 for details
Figure 8:
methylation of the maternal Mest gDMR in blastocysts from the slow (SF and SS) culture groups. See Fig. 3 for details
Effects of Superovulation and Embryo Culture on Imprinted Mest Expression
Embryos in the in vivo-derived, 6.25 and 10 IU superovulated, and FF, FS, SF, and SS cultured groups, which were assayed for methylation analysis, were also analyzed for imprinted expression. Additional embryos in each group were also analyzed. Of the 102 blastocysts, Mest was expressed primarily from the paternal B6 allele (95–100%) in all embryos, where Mest expression was detectable, except for FF 2053 where expression was from the maternal B6(CAST7p6) allele (Fig. 9). Expression of Snrpn was also analyzed in these samples as a control for generation of the cDNA library, and exhibited normal paternal-specific expression in all samples (data not shown). These results are consistent with our previous observations, where paternal-specific Snrpn and Peg3 expression was not altered in superovulated or cultured blastocysts with imprinted methylation loss [41, 44, 45].
Figure 9:
imprinted Mest expression in blastocysts produced spontaneously, or through superovulation and embryo culture. Imprinted expression was classified as >90% expression from the paternal allele. Blue, paternal B6 allele; red, maternal CAST allele
Discussion
In this study, we present a comprehensive evaluation of the effects of superovulation and embryo culture on genomic imprinting at the Mest gene. Superovulation resulted in disruption of imprinted methylation at the maternal Mest gDMR in blastocyst-stage embryos compared to in vivo-derived controls, with a roughly equal loss in methylation in the low and high hormone treated groups. This disruption was not due to a failure of imprinted methylation acquisition at the Mest gDMR in the oocyte, on either grand-parental alleles. Cultured embryos also experienced a significant loss of maternal Mest methylation compared to in vivo-derived controls, with roughly an equal amount of methylation loss in the Fast and Slow culture groups.
To date, investigations of the effects of ARTs on imprinted methylation at gDMRs in mouse oocytes indicate that imprinted methylation acquisition is not perturbed by superovulation [42, 46], in vitro oocyte maturation [38, 47–49], or vitrification [50, 51]. Consistent with these reports, we found that acquisition of imprinted methylation at the Mest gDMR was not affected by superovulation. By comparison, Sato et al. [35] found that individual human germinal vesicle and metaphase I oocytes from women undergoing multiple hormone stimulations possessed aberrant imprinted methylation at MEST. It is important to acknowledge that human oocytes may be more susceptible to imprinted methylation errors following multiple ART procedures including ovarian stimulation and advanced maternal age [49, 52, 53]. Future investigations are required to assess methylation acquisition of Mest and other imprinted gDMRs in natural aged oocytes in mice.
By comparison, maintenance of imprinted methylation in pre-implantation embryos was disrupted by superovulation [39, 54], in vitro embryo culture [40, 41, 44, 45, 55], and vitrification [51]. In our previous study on the effects of superovulation at other imprinted gDMRs, loss of methylation was observed at both hormone dosages, with a greater loss of methylation at the high hormone treatment [39]. Here, the maternal Mest gDMR experienced methylation loss at roughly equal amounts at the low (56.2%) and high (56.3%) hormone dosage compared to in vivo-derived controls (73.2%), indicating that methylation loss was not hormone dose-dependent.
Embryo culture has also been shown to cause perturbation of imprinted methylation and imprinted expression of several imprinted genes [41, 44, 45, 56–58]. Consistent with these previous studies, we reported loss of maternal methylation at the Mest gDMR following in vitro culture to the blastocyst stage in Whitten’s medium. Moreover, we assessed imprinted methylation loss based on rates of pre-implantation development during in vitro culture. In our previous study, a greater number of embryos lost methylation at the H19 and Snrpn gDMRs in the Fast developing group compared to the Slow developing group. Here, we found that loss of maternal methylation at the Mest gDMR was roughly equal in the Fast (56.5%) and Slow (59.5%) developing embryos, with no significant difference at the first separation between the Fast and Slow groups, or at the second separation between the FF and FS groups, and the SF and SS groups. Therefore, unlike Snrpn and H19 gDMRs, methylation at the Mest gDMR does not correlate with rates of development. Instead, methylation loss may occur at stages earlier than the first separation. These results together with those from superovulation indicate that Mest imprinted methylation is more susceptible to perturbations than other imprinted gDMRs during pre-implantation development.
During oogenesis, gDMRs are thought to transition from protective to permissive chromatin states that enable de novo methylation in growing oocytes [59]. This transition may occur in a two-step process involving gene transcription and H3K36 methylation followed by removal of H3K4 di/trimethylation (H3K4me2/3) [59, 60]. It was speculated that de novo methylation kinetics at early and late acquiring imprinted gDMRs may be the result of initial H3K4me2/3 levels and the timing/rate of H3K4me2/3 demethylation [59]. Furthermore, the idea was proposed that reiterative cycles of transcription/H3K36 methylation and H3K4me2/3 demethylation may be required until de novo methylation has been completed. Consistent with this notion, Mest had a protracted transcription profile in growing oocytes compared to other imprinted genes [37]. This leads to the question of whether methylation acquisition kinetics at various gDMRs in growing oocytes produces different stabilities in the pre-implantation embryo under environmental insult. In a recent study, shifting methylation acquisition at gDMRs earlier in growing oocytes by precocious Dnmt3a2 and Dnmt3l expression resulted in loss of gDMR methylation in embryonic day 9.5 embryos [61]. These data were interpreted to mean that “functional” imprinted methylation is attained late in oocyte-growth. However, an alternative explanation is that a longer acquisition period produces more stable imprinted methylation in the developing embryo. Thus, Mest with the shortest and latest acquisition period may render imprinted methylation less stable in the pre-implantation embryo and thus, more susceptible to perturbations under certain environmental conditions.
Overall, we observed similar amounts of Mest gDMR methylation loss following superovulation with low and high hormone treatment as well as in embryos with fast and slow developmental rates in culture. This suggests that Mest gDMR methylation is less stable in ART-produced pre-implantation embryos than other imprinted gDMRs. Studies targeting known regulators of epigenetic phenomena will be invaluable in pinpointing the specific factors involved in maintenance of imprinted methylation during oogenesis and at each stage of pre-implantation development, as well as how these factors are disrupted by superovulation and embryo culture. On the other hand, continued investigations of mechanisms regulating specific imprinted gDMRs will be equally important, and will allow a more detailed explanation of the differential responses of individual imprinted loci to similar environmental insults.
Acknowledgements
We thank William MacDonald for helpful comments on the manuscript. This work was supported by grants from the Canadian Institutes of Health Research (MOP 111210), Western University, the Magee-Womens Research Institute/University of Pittsburgh, and the Richard King Mellon Foundation to M.R.W.M., and the National Institutes of Health (UL1TR001857) to R.T.K. M.R.W.M. is a Magee Auxiliary Research Scholar. B.A.M.V. was supported by a NSERC Canada Graduate Scholarship, and M.M.D. was supported by a CIHR Training Program in Reproduction, Early Development and the Impact on Health (REDIH) Graduate Scholarship, and an Obstetrics and Gynecology, Western University Graduate Research Scholarship.
Author Contributions
M.R.W.M. conceived the study. M.R.W.M., B.A.M.V., and M.M.D. designed the study. B.A.M.V. and M.M.D. performed the experiments and acquired the data. All authors analyzed the data. R.T.K. performed the statistical analysis. All authors wrote, edited, and approved the manuscript.
Conflict of interest statement. None declared.
References
- 1. Schieve LA, Meikle SF, Ferre C, Peterson HB, Jeng G, Wilcox LS.. Low and very low birth weight in infants conceived with use of assisted reproductive technology. N Engl J Med 2002;346:731–7. [DOI] [PubMed] [Google Scholar]
- 2. DeBaun MR, Niemitz EL, Feinberg AP.. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet 2003;72:156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y.. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet 2003;72:1338–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM.. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART). J Med Genet 2003;40:62–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang AS, Moley KH, Wangler M, Feinberg AP, Debaun MR.. Association between Beckwith-Wiedemann syndrome and assisted reproductive technology: a case series of 19 patients. Fertil Steril 2005;83:349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rossignol S, Steunou V, Chalas C, Kerjean A, Rigolet M, Viegas-Pequignot E, Jouannet P, Le Bouc Y, Gicquel C.. The epigenetic imprinting defect of patients with Beckwith-Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet 2006;43:902–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azzi S, Rossignol S, Steunou V, Sas T, Thibaud N, Danton F, Le Jule M, Heinrichs C, Cabrol S, Gicquel C, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet 2009;18:4724–33. [DOI] [PubMed] [Google Scholar]
- 8. Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, Clayton-Smith J, Brueton LA, Bannister W, Maher ER.. Assisted reproductive therapies and imprinting disorders—a preliminary British survey. Hum Reprod 2006;21:1009–11. [DOI] [PubMed] [Google Scholar]
- 9. Lim D, Bowdin SC, Tee L, Kirby GA, Blair E, Fryer A, Lam W, Oley C, Cole T, Brueton LA, et al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod 2009;24:741–7. [DOI] [PubMed] [Google Scholar]
- 10. Lennerz JK, Timmerman RJ, Grange DK, DeBaun MR, Feinberg AP, Zehnbauer BA.. Addition of H19 ‘loss of methylation testing‘ for Beckwith-Wiedemann syndrome (BWS) increases the diagnostic yield. J Mol Diagn 2010;12:576–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox GF, Burger J, Lip V, Mau UA, Sperling K, Wu BL, Horsthemke B.. Intracytoplasmic sperm injection may increase the risk of imprinting defects. Am J Hum Genet 2002;71:162–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Orstavik KH, Eiklid K, van der Hagen CB, Spetalen S, Kierulf K, Skjeldal O, Buiting K.. Another case of imprinting defect in a girl with Angelman syndrome who was conceived by intracytoplasmic semen injection. Am J Hum Genet 2003;72:218–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ludwig M, Katalinic A, Gross S, Sutcliffe A, Varon R, Horsthemke B.. Increased prevalence of imprinting defects in patients with Angelman syndrome born to subfertile couples. J Med Genet 2005;42:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bliek J, Terhal P, van den Bogaard MJ, Maas S, Hamel B, Salieb-Beugelaar G, Simon M, Letteboer T, van der Smagt J, Kroes H, Mannens M.. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am J Hum Genet 2006;78:604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kagami M, Nagai T, Fukami M, Yamazawa K, Ogata T.. Silver-Russell syndrome in a girl born after in vitro fertilization: partial hypermethylation at the differentially methylated region of PEG1/MEST. J Assist Reprod Genet 2007;24:131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chopra M, Amor DJ, Sutton L, Algar E, Mowat D.. Russell-Silver syndrome due to paternal H19/IGF2 hypomethylation in a patient conceived using intracytoplasmic sperm injection. Reprod Biomed Online 2010;20:843–7. [DOI] [PubMed] [Google Scholar]
- 17. Hiura H, Okae H, Miyauchi N, Sato F, Sato A, Van De Pette M, John RM, Kagami M, Nakai K, Soejima H, et al. Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproduction technologies. Hum Reprod 2012;27:2541–8. [DOI] [PubMed] [Google Scholar]
- 18. Chiba H, Hiura H, Okae H, Miyauchi N, Sato F, Sato A, Arima T.. DNA methylation errors in imprinting disorders and assisted reproductive technology. Pediatr Int 2013;55:542–9. doi:10.1111/ped.12185. [DOI] [PubMed] [Google Scholar]
- 19. Cocchi G, Marsico C, Cosentino A, Spadoni C, Rocca A, De Crescenzo A, Riccio A.. Silver-Russell syndrome due to paternal H19/IGF2 hypomethylation in a twin girl born after in vitro fertilization. Am J Med Genet A 2013;161A:2652–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vermeiden JP, Bernardus RE.. Are imprinting disorders more prevalent after human in vitro fertilization or intracytoplasmic sperm injection? Fertil Steril 2013;99:642–51. [DOI] [PubMed] [Google Scholar]
- 21. Wakeling EL, Brioude F, Lokulo-Sodipe O, O'Connell SM, Salem J, Bliek J, Canton AP, Chrzanowska KH, Davies JH, Dias RP, et al. Diagnosis and management of Silver-Russell syndrome: first international consensus statement. Nat Rev Endocrinol 2017;13:105–24. [DOI] [PubMed] [Google Scholar]
- 22. Eggermann T. Russell-Silver syndrome. Am J Med Genet C Semin Med Genet 2010;154C:355–64. [DOI] [PubMed] [Google Scholar]
- 23. Kotzot D, Schmitt S, Bernasconi F, Robinson WP, Lurie IW, Ilyina H, Mehes K, Hamel BC, Otten BJ, Hergersberg M, et al. Uniparental disomy 7 in Silver-Russell syndrome and primordial growth retardation. Hum Mol Genet 1995;4:583–7. [DOI] [PubMed] [Google Scholar]
- 24. Kobayashi S, Kohda T, Miyoshi N, Kuroiwa Y, Aisaka K, Tsutsumi O, Kaneko-Ishino T, Ishino F.. Human PEG1/MEST, an imprinted gene on chromosome 7. Hum Mol Genet 1997;6:781–6. [DOI] [PubMed] [Google Scholar]
- 25. Hannula K, Lipsanen-Nyman M, Kontiokari T, Kere J.. A narrow segment of maternal uniparental disomy of chromosome 7q31-qter in Silver-Russell syndrome delimits a candidate gene region. Am J Hum Genet 2001;68:247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eggermann T, Spengler S, Begemann M, Binder G, Buiting K, Albrecht B, Spranger S.. Deletion of the paternal allele of the imprinted MEST/PEG1 region in a patient with Silver-Russell syndrome features. Clin Genet 2012;81:298–300. [DOI] [PubMed] [Google Scholar]
- 27. Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA.. Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet 1998;20:163–9. [DOI] [PubMed] [Google Scholar]
- 28. Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA.. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 1995;11:52–9. [DOI] [PubMed] [Google Scholar]
- 29. MacIsaac JL, Bogutz AB, Morrissy AS, Lefebvre L.. Tissue-specific alternative polyadenylation at the imprinted gene Mest regulates allelic usage at Copg2. Nucleic Acids Res 2012;40:1523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lefebvre L, Viville S, Barton SC, Ishino F, Surani MA.. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum Mol Genet 1997;6:1907–15. [DOI] [PubMed] [Google Scholar]
- 31. Riesewijk AM, Hu L, Schulz U, Tariverdian G, Hoglund P, Kere J, Ropers HH, Kalscheuer VM.. Monoallelic expression of human PEG1/MEST is paralleled by parent-specific methylation in fetuses. Genomics 1997;42:236–44. [DOI] [PubMed] [Google Scholar]
- 32. Nishita Y, Sado T, Yoshida I, Takagi N.. Effect of CpG methylation on expression of the mouse imprinted gene Mest. Gene 1999;226:199–209. [DOI] [PubMed] [Google Scholar]
- 33. Lucifero D, Mann MR, Bartolomei MS, Trasler JM.. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 2004;13:839–49. [DOI] [PubMed] [Google Scholar]
- 34. Hiura H, Obata Y, Komiyama J, Shirai M, Kono T.. Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells 2006;11:353–61. [DOI] [PubMed] [Google Scholar]
- 35. Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T.. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod 2007;22:26–35. [DOI] [PubMed] [Google Scholar]
- 36. Denomme MM, White CR, Gillio-Meina C, Macdonald WA, Deroo BJ, Kidder GM, Mann MR.. Compromised fertility disrupts Peg1 but not Snrpn and Peg3 imprinted methylation acquisition in mouse oocytes. Front Genet 2012;3:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Obata Y, Kono T.. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem 2002;277:5285–829. [DOI] [PubMed] [Google Scholar]
- 38. Anckaert E, Romero S, Adriaenssens T, Smitz J.. Effects of low methyl donor levels in culture medium during mouse follicle culture on oocyte imprinting establishment. Biol Reprod 2010;83:377–86. [DOI] [PubMed] [Google Scholar]
- 39. Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR.. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet 2010;19:36–51. [DOI] [PubMed] [Google Scholar]
- 40. Market Velker BA, Denomme MM, Mann MR.. Loss of genomic imprinting in mouse embryos with fast rates of preimplantation development in culture. Biol Reprod 2012;86:143, 141-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS.. Selective loss of imprinting in the placenta following preimplantation development in culture. Development 2004;131:3727–35. [DOI] [PubMed] [Google Scholar]
- 42. Denomme MM, Zhang L, Mann MR.. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil Steril 2011;96:734–8. [DOI] [PubMed] [Google Scholar]
- 43. Denomme MM, Zhang L, Mann Mr.. Single oocyte bisulfite mutagenesis. J Vis Exp 2012;64: e4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Doherty AS, Mann MRW, Tremblay KD, Bartolomei MS, Schultz RM.. Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod 2000;62:1526–35. [DOI] [PubMed] [Google Scholar]
- 45. Market-Velker BA, Fernandes AD, Mann MRW.. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod 2010;83:938–50. [DOI] [PubMed] [Google Scholar]
- 46. Imamura T, Kerjean A, Heams T, Kupiec JJ, Thenevin C, Paldi A.. Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. J Biol Chem 2005;280:20171–5. [DOI] [PubMed] [Google Scholar]
- 47. Anckaert E, Adriaenssens T, Romero S, Dremier S, Smitz J.. Unaltered imprinting establishment of key imprinted genes in mouse oocytes after in vitro follicle culture under variable follicle-stimulating hormone exposure. Int J Dev Biol 2009;53:541–8. [DOI] [PubMed] [Google Scholar]
- 48. Geuns E, De RM, Van Steirteghem A, Liebaers I.. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet 2003;12:2873–9. [DOI] [PubMed] [Google Scholar]
- 49. Geuns E, Hilven P, Van Steirteghem A, Liebaers I, De Rycke M.. Methylation analysis of KvDMR1 in human oocytes. J Med Genet 2007;44:144–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trapphoff T, El Hajj N, Zechner U, Haaf T, Eichenlaub-Ritter U.. DNA integrity, growth pattern, spindle formation, chromosomal constitution and imprinting patterns of mouse oocytes from vitrified pre-antral follicles. Hum Reprod 2010;25:3025–42. [DOI] [PubMed] [Google Scholar]
- 51. Cheng KR, Fu XW, Zhang RN, Jia GX, Hou YP, Zhu SE.. Effect of oocyte vitrification on deoxyribonucleic acid methylation of H19, Peg3, and Snrpn differentially methylated regions in mouse blastocysts. Fertil Steril 2014;102:1183–90. [DOI] [PubMed] [Google Scholar]
- 52. El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet 2001;27:341–4. [DOI] [PubMed] [Google Scholar]
- 53. Borghol N, Lornage J, Blachère T, Sophie Garret A, Lefèvre A.. Epigenetic status of the H19 locus in human oocytes following in vitro maturation. Genomics 2006;87:417–26. [DOI] [PubMed] [Google Scholar]
- 54. El Hajj N, Trapphoff T, Linke M, May A, Hansmann T, Kuhtz J, Reifenberg K, Heinzmann J, Niemann H, Daser A, et al. Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single early mouse embryos and bovine oocytes. Epigenetics 2011;6:1176–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, Hu JF, Behr B, Giudice L, Hoffman AR.. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod 2005;11:631–40. [DOI] [PubMed] [Google Scholar]
- 56. de Waal E, Mak W, Calhoun S, Stein P, Ord T, Krapp C, Coutifaris C, Schultz RM, Bartolomei MS.. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol Reprod 2014;90:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Waal E, Vrooman LA, Fischer E, Ord T, Mainigi MA, Coutifaris C, Schultz RM, Bartolomei MS.. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum Mol Genet 2015;24:6975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rivera RM, Stein P, Weaver JR, Mager J, Schultz RM, Bartolomei MS.. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet 2008;17:1–14. [DOI] [PubMed] [Google Scholar]
- 59. Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, Smallwood SA, Chen T, Kelsey G.. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev 2015;29:2449–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T.. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009;461:415–8. [DOI] [PubMed] [Google Scholar]
- 61. Hara S, Takano T, Fujikawa T, Yamada M, Wakai T, Kono T, Obata Y.. Forced expression of DNA methyltransferases during oocyte growth accelerates the establishment of methylation imprints but not functional genomic imprinting. Hum Mol Genet 2014;23:3853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]