Abstract.
Atypical pathogens including Mycoplasma pneumoniae and Legionella pneumophila are increasingly recognized as important causes of community-acquired pneumonia (CAP). Mycoplasma pneumoniae accounts for 20–40% of all CAP and L. pneumophila is responsible for 3–15% of cases. The paucity of data from India in this regard prompted us to conduct this prospective multicentric analysis to detect the prevalence of M. pneumoniae and L. pneumophila in our geographical region. A total of 453 patients with symptoms of pneumonia and 90 controls with no history of lower respiratory tract infections were included in the study. A duplex polymerase chain reaction (PCR) targeting 543 bp region of P1 adhesin gene of M. pneumoniae and 375 bp region of macrophage infectivity potentiator (mip) gene of L. pneumophila was standardized for simultaneous detection of these atypical pathogens. Respiratory secretions, blood, and urine samples were collected from each patient and control and were subjected to duplex PCR, culture and serology for M. pneumoniae and L. pneumophila. Urine samples were subjected for detecting L. pneumophila antigen. Among the 453 patients investigated for M. pneumoniae, 52 (11.4%) were positive for IgM antibodies, 17 were positive by culture, and seven tested positive by PCR (P1 gene). Similarly for L. pneumophila, 50 cases (11%) were serologically positive for IgM antibodies, one was positive by PCR (mip gene) and urine antigen detection. A total of eight samples were positive by duplex PCR for M. pneumoniae P1 gene (N = 7) and L. pneumophila mip gene (N = 1). Of the 90 controls, two samples (2.2%) showed IgM positivity, and 15 (16.7%) showed IgG positivity for M. pneumoniae. For L. pneumophila, three samples (3.3%) tested positive for IgM, and 12 (13.3%) tested positive for IgG antibodies. The study findings indicate the presence of M. pneumoniae and L. pneumophila in our geographical region, and a combination of laboratory approaches including PCR, culture, and serology is required for effective detection of these agents.
INTRODUCTION
Atypical pathogens including Mycoplasma pneumoniae and Legionella pneumophila are increasingly recognized as the common causes of community-acquired pneumonia (CAP). Despite the wide spread use of effective antibiotics, respiratory diseases due to these nonzoonotic, bacterial, respiratory pathogens remain an important cause of morbidity and mortality. It is estimated that M. pneumoniae accounts for 20–40% of all CAP cases in certain populations.1,2 Legionella pneumophila was first described in 1976, and the bacteria are responsible for 2–15% of cases of CAP worldwide.3,4 The rates of respiratory infections in relation to these pathogens are grossly underestimated because of difficulty in identifying them.
M. pneumoniae infections can occur in upper and lower respiratory tract, but extra pulmonary involvement can also be seen without prominent respiratory disease. Infections are generally self-limiting, seen in children and adults of all age. These infections may progress to severe pneumonia that requires hospitalization especially in elderly population and immunocompromised patients. In complicated cases, death can happen due to neurological diseases, such as encephalitis.5,6 Legionnaire’s disease (LD) is a fatal pneumonia with multisystem involvement caused by breathing in small water droplets contaminated with gram negative bacteria of the genus Legionella. The bacteria mainly affect susceptible individuals as a result of age, underlying medical conditions, or immunosuppression.7 Globally, more than 90% of infections are caused by L. pneumophila which has 15 serogroups. Legionella pneumophila serogroup 1 (Lp1) is involved in ∼84% of cases.8–10 LD is associated with greater CAP severity and higher case fatality rate up to 30%.11
Outbreaks of LD have been reported throughout the world, and a recent outbreak of infection in Bronx, New York had sickened more than 120 people and claimed the lives of 13 patients. Community outbreaks of M. pneumoniae infections have been reported to occur in 3- to 7-year intervals.12–14 This evidence reinforces the need for an efficient diagnostic assay for early detection and therefore executing effective antibiotic treatment.
Presently, the laboratory tests for detecting these pathogens are fraught with limitations. Culture is time consuming; need specially formulated media and technical expertise. Serological tests using serum samples from acute and convalescent phases offer retrospective diagnosis, but specificity and sensitivity of results are questionable. Hence, nucleic acid amplification tests have been developed for rapid and sensitive detection of these pathogens.6,13,15,16
There is a paucity of data on infections due to M. pneumoniae and Legionella species from India. It can be due to lack of clinical awareness, nonclassical presentations of illness, extra pulmonary manifestations, and delayed seroconversion. Serology-based prevalence studies were conducted previously, but molecular tests such as polymerase chain reaction (PCR) are not frequently used in Indian population. To address these issues, we conducted a large scale, prospective, multicentric analysis to detect the prevalence of M. pneumoniae and L. pneumophila infections in our specific geographical region. Our study is the first multicentric large-scale study from India that included a battery of all possible diagnostic tests including PCR, culture, serology, and antigen detection for diagnosis of these infections.
METHODS
Origin of samples.
The study was a prospective multicentric analysis involving three major centers in New Delhi, India which included All India Institute of Medical Sciences, Vardhaman Mahavir Medical College and Safdarjung Hospital and Vallabhbhai Patel Chest Institute. The duration of the study was 3 years (March 2011 to March 2014). The study protocol was approved by the Institute’s ethics committee, and patients were recruited based on the following criteria.
Inclusion criteria.
-
1.
Suspected cases of CAP.
-
2.
Presence of at least one of the major clinical criteria (cough, sputum production, or fever > 37.8°C) or two of the minor criteria (pleuritic chest pain, dyspnea, altered mental state, sign of pulmonary consolidation on examination, or total leukocyte count of > 12,000/cu mm.
-
3.
Presence of a new pulmonary infiltrate/shadow on chest X-ray suggestive of pneumonia at or within 24 hours of hospitalization.
-
4.
Patient residing in a community.
Exclusion criteria.
-
1.
Hospital-acquired pneumonia i.e., pneumonia not incubating at the time of hospital admission and occurring 48 hours or more after hospitalization.
-
2.
Cases not willing to give consent.
-
3.
Patients diagnosed with other established organisms causing pneumonia.
Study group.
A total of 453 subjects including 365 adults and 88 children were enrolled in the study collectively from all the three centers. Respiratory secretions (sputum, nasopharyngeal aspirates (NPA), bronchoalveolar lavage (BAL), endotracheal aspirate etc.), throat swabs, blood, and clean catch mid-stream urine samples were collected from each patient after obtaining informed consent. Serum was extracted from blood specimens, and all samples were stored at −20°C until processed.
Control group.
A total of 90 subjects, adults (above 15 years of age) were enrolled as control group. Controls were taken from hospital and laboratory staff and individuals attending out patient department with no history of lower respiratory tract infections. Because of technical difficulties, control samples from pediatric groups were not included.
Standard strains.
Experiments were conducted with the standard strains M. pneumoniae M129-B7 and L. pneumophila American Type Culture Collection (ATCC) 33153. Mycoplasma pneumoniae was maintained by serial subcultures in pleruropneumonia-like organism (PPLO) broth (BD Difco™, New Delhi, India) and PPLO agar (BD Difco, New Delhi, India) up to 4–5 weeks at 37°C under 5% CO2. Legionella pneumophila strain was grown on buffered charcoal yeast extract (BCYE) agar (BD BBL™, New Delhi, India) medium for 3–7 days at 37°C under 5% CO2.
Genomic DNA extraction.
For L. pneumophila strains grown on BCYE agar medium, colonies were resuspended in 200 μL of phosphate-buffered saline, pH 7.2 before extraction of genomic DNA. For M. pneumoniae strains grown in PPLO broth, 200 μL of liquid culture was used for extraction. Throat swabs were resuspended in 200 μL of PPLO broth, sputum samples, BAL, NPA, and other respiratory secretions were extracted without prior treatments. Genomic DNA was extracted from 200 μL of each sample by using QIAamp DNA blood extraction kit protocol (Qiagen, Hilden, Germany). Manufacturer’s instructions were followed, and DNA was eluted in a final volume of 200 μL, stored as aliquots at −20°C before being subjected to PCR.
Duplex PCR assay targeting M. pneumoniae P1 gene and L. pneumophila mip gene.
Two sets of primers which were previously described, each specific for P1 adhesin gene of M. pneumoniae and macrophage infectivity potentiator (mip) gene of L. pneumophila were used to develop the duplex PCR assay.17,18 Primers were initially tested, and PCR conditions were optimized in single plex format. The 543 bp PCR product from M. pneumoniae M129- B7 and 375 bp PCR product from L. pneumophila ATCC 33153 standard strains were cloned in pGEM-T Easy (Promega, Madison, WI) vector according to manufactures instructions. Positive clones were confirmed by restriction digestion and sequencing. Cloned Plasmids for P1 gene and mip gene were diluted (1 in 50) and used as the positive control for standardization of duplex PCR. Reaction mixture for duplex PCR was prepared in a final volume of 25 μL containing 2.5 μL of 10 × PCR buffer (Bangalore Genei, Bangalore, India), 0.5 μL of dNTPs (Thermo scientific, Vilnius, Lithuania), 0.5 μL of 10 pmol/μL of forward and reverse primers (Sigma, Bangalore, India) of each gene target, and nuclease-free water (Thermo Scientific, Vilnius, Lithuania) to achieve the desired final volume. The reaction was performed in a thermal cycler (Applied biosystems, Carlsbad, CA) under the following conditions. 94°C for 5 minutes, followed by 35 cycles of amplification each at 94°C for 1 minute denaturation, 55°C for 1 minute annealing and 72°C for 2 minute extension, and a final elongation step of 72°C for 10 minutes. A negative control was systematically run in parallel. Sensitivity and specificity of PCR reaction were checked. Clinical specimens for the detection of M. pneumoniae and L. pneumophila were tested with the same assay. Details of primers and gene targets for detection of M. pneumoniae and L. pneumophila are shown in Table1.
Table 1.
Primers and gene targets for detection of Mycoplasma pneumoniae and Legionella pneumophila
| Primer | Sequence (5′to 3′) | Gene target | Product (bp) | Reference |
|---|---|---|---|---|
| Forward primer | 5′-CAAGCCAAACACGAGCTCCGGCC-3′ | P1 | 543 | 17 |
| Reverse primer | 5′-GGGGAAGGACAAACAGCTGACACTGG-3′ | |||
| Forward primer | 5′-GACAAGGATAAGTTGTCTTATAGC-3′ | mip | 375 | 18 |
| Reverse primer | 5′-ACGACCAGTGTATTCCACAG-3′ |
Culture.
Culture methods were standardized using standard strains of M. pneumoniae and L. pneumophila according to ATCC guidelines (www.atcc.org).
Throat swabs collected in liquid media were processed for culture on PPLO broth for M. pneumoniae. PPLO broth was incubated at 37°C under 5% CO2 for 4–5 weeks. The indication of growth of M. pneumoniae was determined by color change in PPLO broth from orange red to yellow. Further confirmation was done by subculturing from broth to PPLO agar media and observing typical fried egg colonies under inverted microscope. For Legionella culture, respiratory samples were subjected to brief heat treatment at 50°C for 30 minutes and plated on BCYE agar containing BMPA-α (cefamandole, polymyxin, and anisomycin) selective supplements (Oxoid, United Kingdom). Plates were incubated at 37°C under 5% CO2 for 7 days. Gram negative bacilli recovered on BCYE agar with no growth after subculture to blood agar were presumptively identified as L. pneumophila and confirmed by PCR.
Urine antigen detection for L. pneumophila.
Legionella pneumophila urine antigen was detected using BinaxNOW Legionella urinary antigen ICT kit (Alere) which is specific for Lp1. The assay was performed according to manufacturer’s instructions for detecting the antigen.
Serology.
Commercial enzyme-linked immunosorbent assay (ELISA) kits (EUROIMMUN Medizinische Labordiagnostika AG, United Kingdom) were used for detection of serum IgM and IgG antibodies against M. pneumoniae and L. pneumophila according to the manufacturer’s instructions. The assays used single 1:101 dilutions of serum in sample buffer and included cutoff calibrators to score samples as negative or positive. Samples with borderline results were retested, and if same result obtained, it was scored as uncertain.
Statistical analysis.
Data analysis was done by using Fisher exact/Pearson chi-squared test.
RESULTS
A total of 453 cases and 90 controls were enrolled in the study. Among the cases, 365 (80.6%) were adults, and 88 (19.4%) were pediatric patients. Details of enrollment of cases and controls collectively from all three centers are shown in the Table 2.
Table 2.
Details of cases and controls enrolled from three centers
| Center | AIIMS | VMMC and Safdarjung Hospital | Vallabhbhai Patel Chest Institute | Total enrollment (cases + control) | |||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||
| Adults | 215 | 56 | 108 | 20 | 42 | 14 | 455 (365 + 90) |
| Pediatric | 87 | – | 1 | – | – | – | 88 (88 + 0) |
| Total | 302 | 56 | 109 | 20 | 42 | 14 | 543 (453 + 90) |
AIIMS = All India Institute of Medical Sciences; VMMC = Vardhaman Mahavir Medical College.
Standardization of PCR for M. pneumoniae P1 gene and L. pneumophila mip gene.
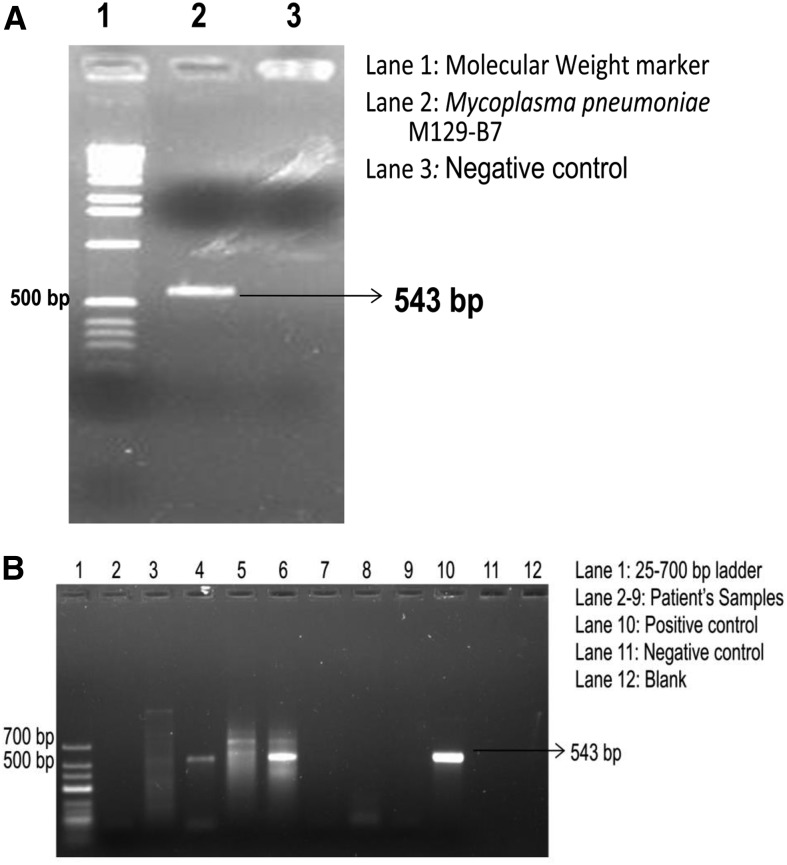
PCR reactions for P1 gene of M. pneumoniae and mip gene of L. pneumophila were standardized in singleplex formats. Standardized PCR reactions were used for detecting M. pneumoniae and L. pneumophila from patient samples. DNA extracted from the standard strain of M. pneumoniae M129-B7 grown in PPLO broth was used for PCR amplification of 543 bp fragment of P1 gene (Figure 1A). Among the 453 samples tested, a total of seven samples were positive for P1 gene of M. pneumoniae (Figure 1B).
Figure 1.
Polymerase chain reaction (PCR) for detection of Mycoplasma pneumoniae: (A) Standardization of PCR targeting P1 adhesin gene 543 bp. (B) PCR analysis of DNA from patient samples for P1 adhesin gene of M. pneumoniae.
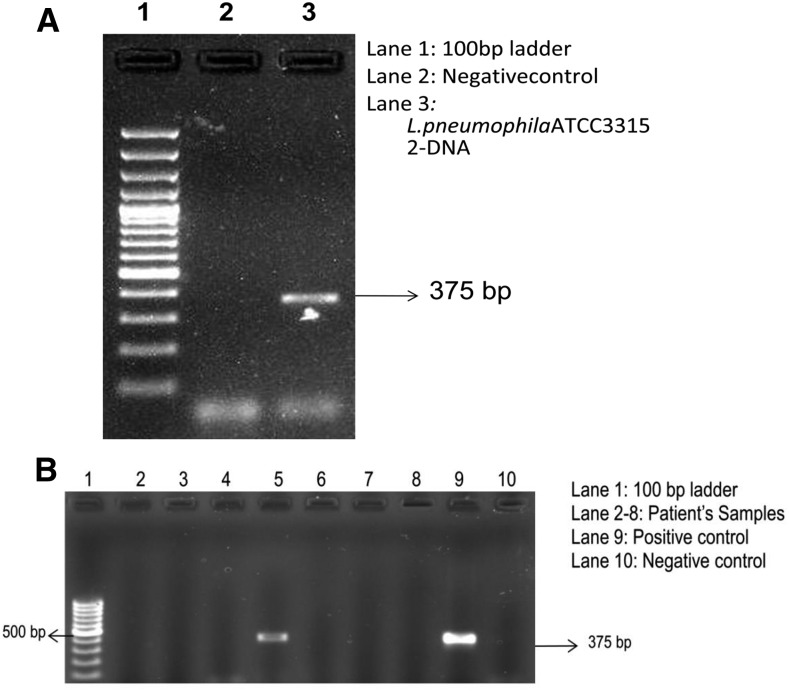
For L. pneumophila specific amplification, DNA isolated from L. pneumophila ATCC 33153 grown on BCYE media was used. The target for amplification was 375 bp segment of mip gene (Figure 2A). For L. pneumophila mip gene, only 1 of 453 tested samples was positive (Figure 2B). Among the controls tested, all were found to be negative by PCR.
Figure 2.
Polymerase chain reaction (PCR) for detection of Legionella pneumophila: (A) Standardization of PCR targeting mip gene 375 bp. (B) PCR analysis of DNA from patient samples for mip gene of L. pneumophila.
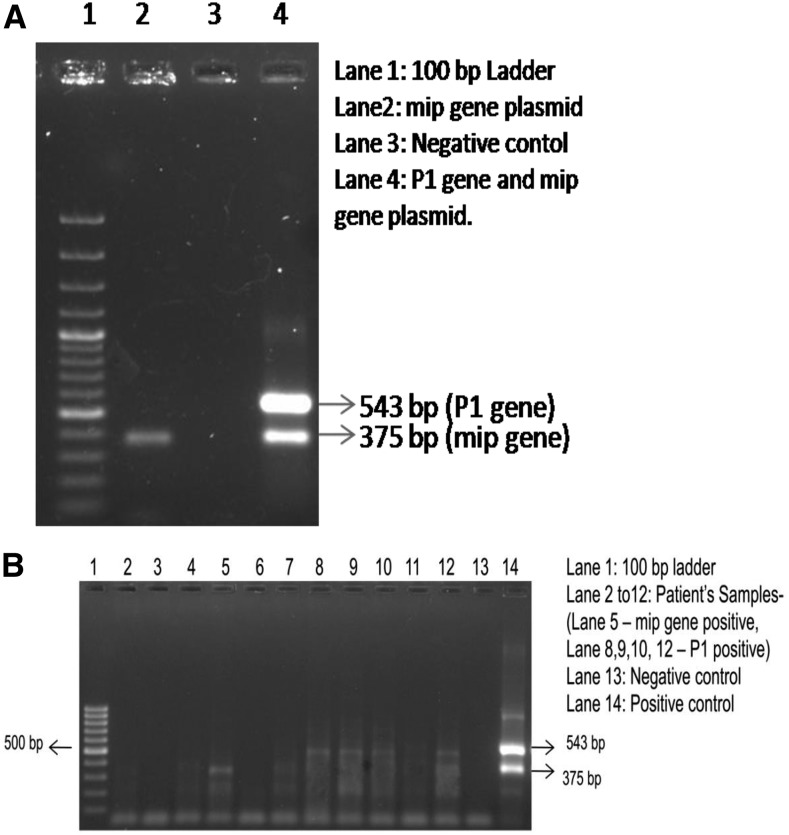
Standardization of duplex PCR for M. pneumoniae and L. pneumophila.
PCR products from the above mentioned singleplex reactions were cloned in pGEM-T Easy vector and confirmed by restriction digestion and sequencing. Diluted clones were used as positive controls for standardization of duplex PCR for simultaneous detection and identification of M. pneumoniae and L. pneumophila. Standardization was done with the same primers that can amplify a 543 bp fragment for M. pneumoniae and a 375 bp fragment for L. pneumophila (Figure 3A). A total of 543 clinical samples including 453 cases and 90 controls were tested with the duplex PCR assay. A total of eight samples were positive by duplex PCR for M. pneumoniae P1 gene (N = 7) and L. pneumophila mip gene (N = 1). All the samples which were positive by singleplex PCR reactions were found to be positive for duplex PCR reaction (Figure 3B).
Figure 3.
Duplex polymerase chain reaction (PCR) for detection of Mycoplasma pneumoniae and Legionella pneumophila: (A) Standardization of Duplex PCR targeting M. pneumoniae P1 adhesin gene 543 bp and L. pneumophila mip gene 375 bp. (B) Analysis of DNA from patient samples for P1 gene of M. pneumoniae and mip gene of L. pneumophila using duplex PCR.
Culture.
A total of 17 of 429 tested samples (3.9%) were positive by culture for M. pneumoniae, and none of the tested samples (N = 334) were positive for Legionella culture on BCYE agar. A sample which was positive by mip gene PCR, and urine antigen detection for L. pneumophila did not show culture positivity. The culture results of clinical samples for M. pneumoniae and L. pneumophila are shown in Table 3.
Table 3.
Results of culture of patient samples for Mycoplasma pneumoniae and Legionella pneumophila
| Center | Cases/Controls | Total samples | Culture positive for M. pneumoniae (samples tested) | Culture positive for L. pneumophila (samples tested) |
|---|---|---|---|---|
| AIIMS | Cases | 302 | 13 (287) | 0 (226) |
| Control | 56 | 0 | – | |
| VMMC and Safdarjung Hospital | Cases | 109 | 3 (100) | 0 (72) |
| Control | 20 | 0 | – | |
| Vallabhbhai Patel Chest Institute | Cases | 42 | 1 (42) | 0 (36) |
| Control | 14 | 0 | – | |
| Total | 543 | 17 (429) | 0 (334) | |
AIIMS = All India Institute of Medical Sciences; VMMC = Vardhaman Mahavir Medical College.
Serology.
Among the 453 cases tested, 25.6% were scored serologically positive for M. pneumoniae. A total of 52 (11.5%) samples showed IgM positivity, and 64 (14.1%) samples showed IgG positivity for M. pneumoniae. Both IgM and IgG were detected in two patients. For L. pneumophila, there was an overall seropositivity of 27.2%. Of 453 patients tested, 50 (11%) samples showed IgM positivity and 73 (16.11%) samples showed IgG positivity by ELISA. Both IgM and IgG were detected in seven patients. A total of 11 samples were positive for IgM antibodies for both of the atypical pathogens.
Of the 90 controls tested, two samples (2.2%) showed IgM positivity and 15 samples (16.7%) showed IgG positivity for M. pneumoniae. For L. pneumophila 3 of 90 samples (3.3%) were tested positive for IgM antibodies, and 12 samples (13.3%) were positive for IgG antibodies.
For M. pneumoniae and L. pneumophila IgM ELISA results, difference between cases and controls was statistically significant (P < 0.05). Results of IgM and IgG positivity for M. pneumoniae and L. pneumophila in cases (N = 453) and controls (N = 90) are shown in Table 4.
Table 4.
Results of IgM and IgG positivity for Mycoplasma pneumoniae and Legionella pneumophila in patients having CAP and hospital-acquired pneumonia
| Center | Cases/Control | Total subjects | ELISA | |||
|---|---|---|---|---|---|---|
| M. pneumoniae | L. pneumophila | |||||
| IgM | IgG | IgM | IgG | |||
| AIIMS | Cases | 302 | 43 | 21 | 39 | 60 |
| Control | 56 | 2 | 9 | 3 | 7 | |
| VMMC and Safdarjung Hospital | Cases | 109 | 8 | 33 | 8 | 9 |
| Control | 20 | 0 | 4 | 0 | 3 | |
| Vallabhbhai Patel Chest Institute | Cases | 42 | 1 | 10 | 3 | 4 |
| Control | 14 | 0 | 2 | 0 | 2 | |
| Total | 543 | 54 | 79 | 53 | 85 | |
AIIMS = All India Institute of Medical Sciences; CAP = community-acquired pneumonia; ELISA = enzyme-linked immunosorbent assay; VMMC = Vardhaman Mahavir Medical College.
Urine antigen detection.
Among the 453 tested cases, L. pneumophila urine antigen was detected in one patient. The patient, who tested positive by mip gene PCR for L. pneumophila was found to be positive by urine antigen detection also.
Clinical signs and symptoms.
Fever (83.6%), cough (70.1%), dyspnea (45.2%), pleuritic chest pain (33.1%) and sore throat (32.2%) were the most common clinical features present in patients under investigation. Extra pulmonary manifestations including abdominal pain (7.2%) and diarrhea (5.9%) were present in a small population of patients. Chest X-ray findings suggestive of pneumonia were seen in 67.9% of patients. Clinical signs and symptoms of all patients and those tested positive for M. pneumoniae and L. pneumophila by serology- IgM ELISA are shown in Table 5. Fever, cough, dyspnea, pleuritic chest pain, sore throat, and chest X-ray findings suggestive of pneumonia were equally present in both groups of patients diagnosed by serology. Of the clinical symptoms, fever and confusion were found to be statistically significant (P < 0.05) for both groups.
Table 5.
Clinical features and signs of all patients and serologically positive cases for Mycoplasma pneumoniae and Legionella pneumophila
| Clinical symptoms and signs | All patients (N = 453) | M. pneumoniae IgM positive (N = 52) | L. pneumophila IgM positive (N = 50) | M. pneumoniae IgM+ L. pneumophila IgM positive (N = 11) |
|---|---|---|---|---|
| Fever | 379 (83.6%) | 34 (65.3%) | 36 (72%) | 9 (81.8%) |
| Cough | 318 (70.1%) | 33 (63.5%) | 37 (74%) | 11 (100%) |
| Dyspnea | 205 (45.2%) | 24 (46.1%) | 28 (56%) | 8 (72.7%) |
| Pharyngitis/Sore throat | 146 (32.2%) | 12 (23%) | 13 (24%) | 4 (36.4%) |
| Chest pain | 150 (33.1%) | 11 (21.1%) | 14 (28%) | 3 (27.3%) |
| Confusion | 34 (7.5%) | 8 (15.4%) | 8 (16%) | 2 (18.2%) |
| Chills | 80 (17.6%) | 8 (15.4%) | 7 (14%) | 2 (18.1%) |
| Myalgia | 33 (7.3%) | 4 (7.6%) | 3 (6%) | 1 (9.1%) |
| Hemoptysis | 40 (8.8%) | 4 (7.6%) | 4 (8%) | – |
| Diarrhea | 27 (5.9%) | 6 (11.5%) | 2 (4%) | 1 (9.1%) |
| Head ache | 44 (9.7%) | 2 (3.8%) | 3 (6%) | – |
| Abdominal pain | 33 (7.3%) | 7 (13.4%) | 3 (6%) | 1 (9.1%) |
| Relative bradycardia | 2 (0.4%) | 1 (1.9%) | – | – |
| Chest X-ray findings | 308 (67.9%) | 34 (65.3%) | 37 (74%) | 9 (81.8%) |
Comorbid conditions.
Chronic obstructive pulmonary disease (COPD) was the most common comorbid condition seen in 152 (33.5%) patients followed by diabetes mellitus (13.5%), hypertension (12.1%), bronchial asthma (10.1%) and malignancy (5.7%). The co-morbid conditions of all patients and those tested positive for M. pneumoniae and L. pneumophila by IgM ELISA are shown in Table 6. Of the co morbid conditions, COPD was found to be statistically significant (P < 0.05) for both groups.
Table 6.
Comorbid conditions of all patients and serologically positive cases for Mycoplasma pneumoniae and Legionella pneumophila
| Comorbid conditions | All patients (N = 453) | M. pneumoniae IgM positive (N = 52) | L. pneumophila IgM positive (N = 50) | M. pneumoniae IgM+ L. pneumophila IgM positive (N = 11) |
|---|---|---|---|---|
| COPD | 142 (31.3%) | 5 (9.6%) | 8 (16%) | 1 (9%) |
| Diabetes mellitus | 61 (13.4%) | 3 (5.8%) | 7 (14%) | 1 (9%) |
| Hypertension | 55 (12.1%) | 3 (5.8%) | 3 (6%) | – |
| Malignancy | 26 (5.7%) | 4 (7.7%) | 2 (4%) | 2 (17.5%) |
| Bronchial asthma | 46 (10.1%) | 4 (7.7%) | 2 (4%) | – |
| Tuberculosis | 21 (4.6%) | 1 (1.9%) | 1 (2%) | – |
| HIV | 3 (0.6%) | 1 (1.9%) | – | – |
COPD = chronic obstructive pulmonary disease; HIV = human immunodeficiency virus.
DISCUSSION
Together Legionella spp. Mycoplasma pneumoniae and Chlamydophila pneumoniae are responsible for around 22% of cases of CAP in United States and Canada and up to 28% of cases worldwide.19 According to the Centers for Disease Control and Prevention it is estimated that there may be 8,000 to 18,000 persons hospitalized with community-acquired LD in United States in 1990, and the reported cases of Legionellosis more than tripled between 2001 and 2012. Extrapolation from study incidence data showed estimates of the annual number of cases due to M. pneumoniae in United States are 18,700 to 108,000.6
Because of the lack of common clinical signs and symptoms, available diagnostic tests and active surveillance programs, usually these infections are overtreated and/or underdiagnosed more so in developing countries with resource-limited settings. Culture is rather time consuming and insensitive, serological assays are not promising for immediate clinical management.
Detection of these atypical bacterial pathogens by a duplex PCR provided rapid, sensitive, and specific diagnosis. Primers specific for M. pneumoniae allowed amplification of P1 adhesin gene similarly, those specific for L. pneumophila were targeted against mip gene. Our study is the first report of a duplex PCR assay from India, capable of simultaneous detection of both M. pneumoniae and L. pneumophila from respiratory specimens. Duplex PCR was performed along with serology and culture for improved detection of cases.
Of a total 453 patients, six (1.3%) tested positive for M. pneumoniae by duplex PCR. Of the six positive cases, three were positive by culture and four were seropositive (IgG antibodies). PCR, culture, and serology were positive in two patients. All of the PCR positive patients tested negative for IgM antibodies. Among the 17 culture positive patients, only three cases were positive by duplex PCR. Possible reason can be the presence of PCR inhibitors or a low copy number of bacteria in the respiratory secretions of patients.
Chaudhry et al.20 in 2011, reported an overall seroprevalence of 19%. IgG, IgM, and IgA antibodies were positive in 14.92%, 4.47%, and 5.22% of the tested population, respectively. In the present study, overall seropositivity was found to be slightly elevated (25.6%) with an increased IgM (11.5%) positivity. IgG (14.1%) positivity was in consistent with the above mentioned study.
Of 453 patients tested for L. pneumophila, only one tested positive by duplex PCR. Patient tested positive by duplex PCR was found to be positive for urinary antigen, IgG and IgM antibodies as well. There was no culture positivity. Overall seropositivity was found to be 27.2% with 16.11% IgG and 11% IgM positivity.
LD was first recognized as a cause of fatal pneumonia more than three decades ago and was reported for the first time from India in 1991.21 A study that included 45 clinical specimens and 17 environmental samples showed the presence of Legionella in four (9%) clinical specimens and 13 (76%) environmental specimens. A prospective study conducted by Bahl et al.22 in 1997 reported low antibody titers of Legionella in 21 of 100 patients. Chaudhry et al.23 in 2000 conducted a study to estimate the incidence of L. pneumophila infections in patients having CAP and reported 15% seropositivity for IgM antibodies. Another study by the same investigators in 2010, reported an overall seropositivity of 27.43%. Anti-Legionella IgG, IgM and IgA antibodies were positive in 7.96%, 15.92%, and 11.5% patients, respectively. Urinary antigen was detected in 17.69% of the tested patients.24 In the present study, IgM positivity (11%) is slightly decreased compared with the above mentioned study, but IgG positivity (16.11%) was found to be elevated considerably. There was no isolation by culture; this can probably be due to empirical antibiotic treatment. Urine antigen detection can provide a presumptive diagnosis within a short time frame but the assays uniquely target the predominant serogroup, Lp1. Hence, a total reliance on this diagnostic test may result in significant numbers of undetected cases of LD.8,25,26 It is important to be noted that PCR can be an attractive tool for rapid diagnosis in the early phase and the test is not affected by prior antibiotic therapy. Duplex PCR can be used as a potential tool for detection of M. pneumoniae and L. pneumophila DNA under routine conditions in diagnostic laboratories.
Analysis of clinical signs and symptoms among seropositive patients for M. pneumoniae and L. pneumophila showed no significant difference. It is well established that there is a significant overlap in the clinical manifestations of Legionellosis and M. pneumoniae infection, and it is very difficult to distinguish from other pulmonary infections based on clinical signs and symptoms. Kunha et al. suggested that rapid clinical diagnosis of Legionellosis can be made by diagnostic triad which included signs and symptoms of CAP along with a new infiltration on chest radiograph, fever more than 102°F with relative bradycardia and any three of the key laboratory features: hypophosphatemia, highly increased serum ferritin levels (> 2 × n), increased serum transaminases, and relative lymphopenia .Clinical suspicion of LD is a major factor and once infection is suspected, effective diagnosis can be done by implementing proper laboratory tools.
In our study, Legionella infection was diagnosed in a patient who was a known case of sarcoidosis with a previous history of pulmonary tuberculosis. The patient had clinical symptoms such as fever (39.6°C), dry cough, and bilateral infra-axillary and infrascapular crepitations. Chest X-ray findings showed bilateral lower zone infiltrations. Laboratory examinations showed hyponatremia, hypophosphatemia, and elevated serum creatinine levels.
Of the 90 controls tested, all were negative by duplex PCR and culture for M. pneumoniae and L. pneumophila. Urine antigen detection was negative for L. pneumophila. IgM antibodies for M. pneumoniae and L. pneumophila were seen in 2/90 and 3/90 tested controls, respectively. Mostly, cases that showed IgM positivity were health care professionals who were in constant exposure to these pathogens in laboratory. Similarly, IgG antibodies for M. pneumoniae were seen in 15/90 controls, and those for L. pneumophila were present in 12/90 controls. This may be due to prior exposure of the individuals to these pathogens. Patients showed significantly higher seropositivity for the tested pathogens as compared with controls.
Our results indicate the presence of M. pneumoniae and L. pneumophila in this geographical region that can create a greater awareness and reporting of these diseases. Clinicians should have a high index of suspicion for these agents while treating patients with CAP. As mortality in a percentage of population in our country is due to respiratory illness without any specific microbial etiology, the impact of M. pneumoniae and L. pneumophila among these patients is suspected to be significant. Hence, for determining the true significance of Legionellosis and M. pneumoniae infections in our country, large scale studies are required from various geographical regions.
As a conclusion, a convergence of laboratory approaches including PCR, culture, and serology may be required for the effective identification of M. pneumoniae and L. pneumophila in patients having CAP. Proper monitoring of these respiratory pathogens are mandatory and implementation of rapid and more sensitive assays especially real-time PCR can improve the detection of cases and implementation of specific therapeutic options.
Acknowledgments:
We acknowledge Mohanlal Sharma, Puran Ram Pramod Kumar and Anil Kheriwal for their technical supports.
REFERENCES
- 1.Marston BJ, Plouffe JF, File TM, Jr, Hackman BA, Salstrom SJ, Lipman HB, Kolczak MS, Breiman RF, 1997. Incidence of community-acquired pneumonia requiring hospitalization. Results of a population-based active surveillance study in Ohio. The Community-Based Pneumonia Incidence Study Group. Arch Intern Med 157: 1709–1718. [PubMed] [Google Scholar]
- 2.Winchell JM, 2013. Mycoplasma pneumoniae—a national public health perspective. Curr Pediatr Rev 9: 324–333. [Google Scholar]
- 3.Muder RR, Yu VL, Fang GD, 1989. Community-acquired Legionnaires disease. Semin Respir Infect 4: 32–39. [PubMed] [Google Scholar]
- 4.Berger P, Papazian L, Drancourt M, La Scola B, Auffray JP, Raoult D, 2006. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg Infect Dis 12: 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waites KB, Talkington DF, 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17: 697–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS, 2008. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 46: 3116–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha BA, Burillo A, Bounza E, 2016. Legionnaires’ disease. Lancet 387: 376–385. [DOI] [PubMed] [Google Scholar]
- 8.Fields BS, Benson RF, Besser RE, 2002. Legionella and Legionnaires’ disease: 25 years of investigation. Clin Microbiol Rev 15: 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak NA, Benson RF, Brown E, Alexander NT, Taylor TH, Shelton BG, Fields BS, 2009. Distribution of lag-1 alleles and sequence-based types among Legionella pneumophila serogroup 1 clinical and environmental isolates in the United States. J Clin Microbiol 47: 2525–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muder RR, Yu VL, 2002. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis 35: 990–998. [DOI] [PubMed] [Google Scholar]
- 11.Mercante JW, Winchell JM, 2015. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 28: 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klement E, et al. 2006. Identification of risk factors for infection in an outbreak of Mycoplasma pneumoniae respiratory tract disease. Clin Infect Dis 43: 1239–1245. [DOI] [PubMed] [Google Scholar]
- 13.Waites KB, Talkington DF, 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17: 697–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter ND, et al. 2008. Community outbreak of Mycoplasma pneumoniae infection: school‐based cluster of neurologic disease associated with household transmission of respiratory illness. J Infect Dis 198: 1365–1374. [DOI] [PubMed] [Google Scholar]
- 15.Gullsby K, Storm M, Bondeson K, 2008. Simultaneous detection of Chlamydophila pneumoniae and Mycoplasma pneumoniae by use of molecular beacons in a duplex real-time PCR. J Clin Microbiol 46: 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loens K, Ursi D, Goossens H, Ieven M, 2003. Molecular diagnosis of Mycoplasma pneumoniae respiratory tract infections. J Clin Microbiol 41: 4915–4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson J, Marmion BP, Worswick DA, Kok TW, Tannock G, Herd R, Harris RJ, 1992. Laboratory diagnosis of Mycoplasma pneumoniae infection. 4. Antigen capture and PCR-gene amplification for detection of the Mycoplasma: problems of clinical correlation. Epidemiol Infect 109: 519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welti M, Jaton K, Altwegg M, Sahli R, Wenger A, Bille J, 2003. Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn Microbiol Infect Dis 45: 85–95. [DOI] [PubMed] [Google Scholar]
- 19.Arnold FW, et al. 2007. A worldwide perspective of atypical pathogens in community-acquired pneumonia. Am J Respir Crit Care Med 175: 1086–1093. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhry R, Sharma S, Javed S, Passi K, Dey AB, Malhotra P, 2013. Molecular detection of Mycoplasma pneumoniae by quantitative real-time PCR in patients with community acquired pneumonia. Indian J Med Res 138: 244–251. [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal L, Dhunjibhoy KR, Nair KG, 1991. Isolation of Legionella pneumophila from patients of respiratory tract disease & environmental samples. Indian J Med Res 93: 364–365. [PubMed] [Google Scholar]
- 22.Bahl S, Wali JP, Handa R, Rattan A, Aggarwal P, Kindo AJ, 1997. Legionella as a lower respiratory pathogen in north India. Indian J Chest Dis Allied Sci 39: 81–86. [PubMed] [Google Scholar]
- 23.Chaudhry R, Dhawan B, Dey AB, 2000. The incidence of Legionella pneumophila: a retrospective study in a tertiary care hospital in India. Trop Doct 30: 197–200. [DOI] [PubMed] [Google Scholar]
- 24.Javed S, Chaudhry R, Passi K, Sharma S, Padmaja K, Dhawan B, 2010. Sero diagnosis of Legionella infection in community acquired pneumonia. Indian J Med Res 131: 92–96. [PubMed] [Google Scholar]
- 25.Benin AL, Robert FB, Richard EB, 2002. Trends in Legionnaires disease, 1980–1998: declining mortality and new patterns of diagnosis. Clin Infect Dis 35: 1039–1046. [DOI] [PubMed] [Google Scholar]
- 26.St-Martin G, Uldum S, Molbak K, 2013. Incidence and Prognostic Factors for Legionnaires’ disease in Denmark 1993–2006. ISRN. Epidemiol 2013: 1–8. [Google Scholar]