Abstract
Numerous studies have examined the association between pharmacogenetic effects and the response to inhaled corticosteroids (ICS) in patients with asthma. In fact, several single nucleotide polymorphisms of a number of candidate genes have been identified that might influence the clinical response to ICS in children with asthma. Their direct or indirect effects depend on their role in the inflammatory process in asthma or the anti-inflammatory action of corticosteroids, respectively. Among the genes identified, variants in T-box 21 (TBX21) and Fc fragment of IgE receptor II (FCER2) contribute indirectly to the variability in the response to ICS by altering the inflammatory mechanisms in asthma, while other genes such as corticotropin releasing hormone receptor 1 (CRHR1), nuclear receptor subfamily 3 group C member 1 (NR3C1), stress induced phosphoprotein 1 (STIP1), dual specificity phosphatase 1 (DUSP1), glucocorticoid induced 1 (GLCCI1), histone deacetylase 1 (HDAC), ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3), and vascular endothelial growth factors (VEGF) directly affect this variability through the anti-inflammatory mechanisms of ICS. The results to date indicate various potential genetic factors associated with the response to ICS, which could be utilized to predict the individual therapeutic response of children with asthma to ICS. Clinical trials are underway and their results are greatly anticipated. Further pharmacogenetic studies are needed to fully understand the effects of genetic variation on the response to ICS in children with asthma.
Keywords: Corticoids, inhaled corticosteroid, children, gene, asthma
Introduction
Asthma is a common chronic disease in children, which makes them absent from school and sometimes requires emergency hospitalization. About 50% of patients with asthma have the initial symptoms from childhood.1 One of the key pathogenic characteristics of asthmatic children is chronic inflammation in the airways. Anti-inflammatory drugs are therefore the usual treatment of choice and they are recommended by many international asthma management guidelines.2 Children with asthma are recommended to use inhaled corticosteroids (ICS), bronchodilators (short- or long-acting beta-agonists), antileukotrienes, or anticholinergics for those more than 12 years old;2 and immunotherapy is recommended in some severe cases.3 To date, corticosteroids are known to be one of the most effective ways to treat inflammation for children with persistent asthma and for preventing airway remodelling.4 However, there is evidence that some asthmatic children do not respond when using ICS.5
Previous studies have suggested that 60–80% of asthma patients have different responses due to genetic factors.6 The results from pharmacogenetic studies have demonstrated the correlation between candidate genes and drug responses, for example adrenoceptor beta 2 (ADRB2), corticotropin releasing hormone receptor 2, and arginase 1 encoding for different responses to beta-2 agonists; arachidonate 5-lipoxygenase, leukotriene A4 hydrolase, leukotriene C4 synthase, and cysteinyl leukotriene receptor 2 for affecting the response to leukotriene antagonists; and corticotropin releasing hormone receptor 1 (CRHR1), T-box 21 (TBX21), and Fc fragment of IgE receptor II (FCER2) for response to ICS.5,7–9 This review summarizes the results of studies on genetic factors in the modulating corticotherapy effects in children with asthma.
Overview of airway inflammation in asthmatic children
Asthmatic children are particularly recognized by inflammation in the airway, bronchial hyperresponsiveness (BHR), and reversibility of airway obstruction.10 Many inflammatory and structural cells and inflammatory mediators are involved in the disease processes in asthmatic children.11 Negative impacts on the airways caused by pathogens induce structural changes of the airways or remodeling.12 These changes are responsible for the persistent clinical symptoms such as a chronic refractory cough and dyspnoea. It is also a common cause for bronchoconstriction and severe dyspnoea during and after exercise.
Mechanism of airway inflammation in asthmatic children
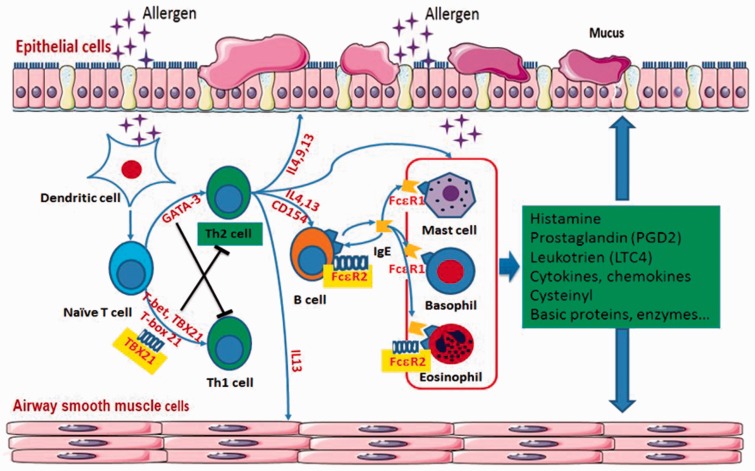
In children with asthma, the inflammation is initiated by infiltration of allergens into the lower airway, which are taken up by dendritic cells (DC).13 DC process allergens to peptides and present the peptides to naïve T cells, and in a suitable environment the naïve T cells develop into type 2 helper T (Th2) cells.11 Th2 cells produce cytokines such as interleukin (IL)-4 and IL-13, stimulating B lymphocytes to produce immunoglobulin (Ig)E; IL-3 and IL-5, attracting eosinophils to the lungs; and IL-4 and IL-9, which stimulate mast cell hyperplasia.14 Under the influence of Th2 cells, B lymphocytes secrete IgE, which is a key factor in allergic asthma because it activates mast cells (Figure 1). With exposure to the specific allergens that individuals are sensitized to, mast cells, secondary to binding of allergens to IgE, release histamine and start to produce prostaglandin D2 and cysteinyl-leukotrienes (LTC4, LTD4, and LTE4), which attract inflammatory cells to the lungs.11 The early phase of asthma is secondary to the effects of histamine and other mediators released from mast cells, while the delayed effect is secondary to other inflammatory cells and the release of mediators.
Figure 1.
Mechanism of airway inflammation in asthma. GATA-3, GATA binding protein 3; IL, interleukin; CD, cluster of differentiation; TBX21, T-box 21; Th, T helper; FcɛR, Fc fragment of IgE receptor. The colour version of this figure is available at: http://imr.sagepub.com.
Molecular mechanism of inflammation in asthmatic children
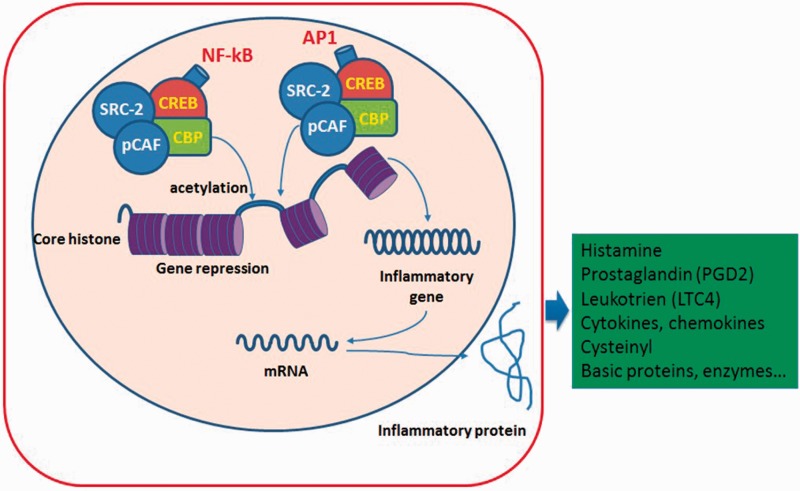
As in adults, the molecular mechanism of inflammation in asthmatic children is characterized by increasing various inflammatory genes controlled by proinflammatory transcription factors, such as nuclear factor-kB (NF-kB) and activator protein-1 (AP1).15 Both NF-kB and AP1 are activated by mediators, including cytokines, tumour necrosis factor-α, IL-1β, and other factors (Figure 2).14 There are many coactivators that also participate in the activation and repression of inflammatory genes through acetylating the core histones.4 As a result, inflammatory proteins or enzymes and other proteins are synthesized and their production can influence inflammation in asthma.16
Figure 2.
Molecular mechanism of inflammation in asthma. NF-kB, nuclear factor-kB; AP1, activator protein-1; CREB, cAMP response element-binding protein; CBP, CREB binding protein; pCAF, p300/CBP-associated factor; SRC, steroid receptor co-activator. The colour version of this figure is available at: http://imr.sagepub.com.
Anti-inflammatory mechanisms of corticosteroids
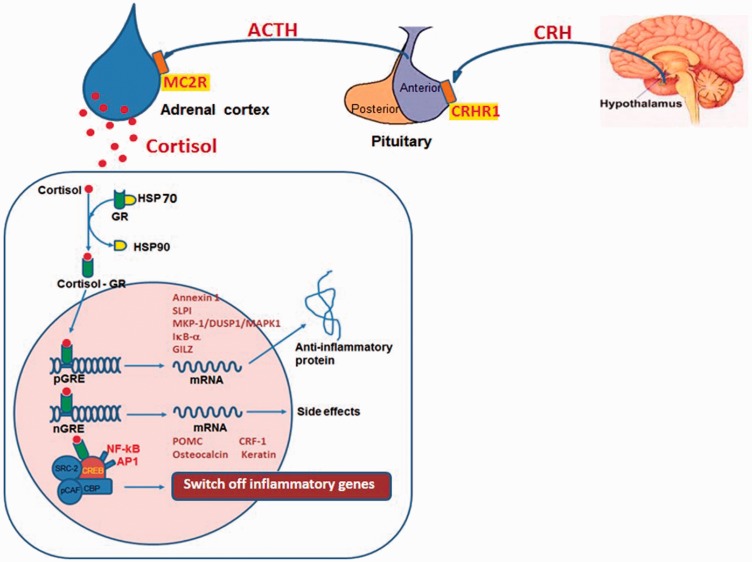
Corticosteroids are synthesized and secreted by the cortex of the adrenal gland as a result of stimulation by the hypothalamus–pituitary–adrenal (HPA) axis. The HPA axis is responsible for the adaptation to stress and inflammatory stimuli. This response is characterized by a hypothalamic release of corticotrophin-releasing hormone (CRH), which is important in regulating the secretion of adrenocorticotropic hormone (ACTH) and consequently in the catecholaminergic response to CRH. Hypothalamic CRH acts by combining with the CRH receptor (CRHR), predominantly CRH receptor 1 (CRHR1) (Figure 3). When CRH combines with CRHR1 on the anterior pituitary gland, ACTH is released and then binds to melanocortin 2 receptors on the adrenal cortex to stimulate cortisol secretion.17
Figure 3.
Anti-inflammatory mechanism of corticosteroids. CRH, corticotropin-releasing hormone; ACTH, adrenocorticotropic hormone; CRHR, CRH receptor; MC2R, melanocortin 2 receptors; HSP, heat shock protein; GR, glucocorticoid receptor; STIP1, stress induced phosphoprotein 1; NR3C1, nuclear receptor subfamily 3 group C member 1; DUSP1, dual specificity phosphatase 1; GRE, glucocorticoid response elements; HDAC1, histone deacetylase 1; NF-kB, nuclear factor-kB; AP1, activator protein-1; ORMDL3, ORMDL sphingolipid biosynthesis regulator 3; CREB, cAMP response element-binding protein; CBP, CREB binding protein; pCAF, p300/CBP-associated factor; SRC, steroid receptor co-activator; POMC, proopiomelanocortin; CRF, corticotrophin releasing factor; SLP, secretory leukoprotease inhibitor; GILZ, glucocorticoid-induced leucine zipper protein; MKP-1, mitogen-activated kinase phosphatase-1; IkB-α, inhibitor of NF-kB alpha. The colour version of this figure is available at: http://imr.sagepub.com.
Both endogenous and exogenous corticosteroids act effectively by combining with a cytoplasmic receptor known as the glucocorticoid receptor (GR). Two isoforms of the GR exist, GRα and GRβ, of which only GRα can bind ligand.18 GRs are expressed in most types of cells with different modular structures.19 A protein complex including heat shock protein (HSP)70 and HSP90 binds the inactive GR. While HSP70 inactivates GR through partial unfolding, HSP90 reverses this inactivation, and is required for activation of the GR. The nuclear localization of the activated GR–steroid complex takes place when the ligand combines with GR-HSP90 complexes. GR homodimers bind to glucocorticoid response elements (GREs) in the promoter region of steroid-sensitive genes, by which the anti-inflammatory proteins, such as annexin-1, secretary leucocyte protease inhibitor, MAPK phosphatase-1 (MKP-1/DUSP1/MAPK1),4 NF-kB, inhibitor of kappa B alpha, IL-10 and glucocorticoid-induced leucine zipper can be encoded.19 Less frequently, GR homodimers prevent genes from being encoded by combining with negative GREs, especially those relating to corticosteroid side-effects.4 Proinflammatory transcription factors like NF-kB or AP1 activate coactivator molecules, like cAMP-response-element-binding-protein binding protein when combining with nuclear GRs, thus inactivating the inflammatory genes activated by these transcription factors.4
Pharmacogenetic response to inhaled corticosteroids in the treatment of asthmatic children
Inhaled corticosteroids are one of the most effective anti-inflammatory medications for asthma and they are the first-line treatment of choice for persistent asthma in children.20 However, 5–15% of asthmatic children fail to respond to ICS and they are often treated with high doses of ICS, which then has the potential to cause significant side-effects.19,21 Although ICS non-responsive patients are uncommon, their treatment is associated with clinical challenges because alternative treatment choices are limited and the cost of treating this small population of patients accounts for more than 50% of all asthma cases.19 Gene polymorphisms are one of the causes for the inter-individual variability in a patient’s response to medication. With regard to the response to ICS, the genetic factors in question may be categorized into three groups: (i) the proinflammatory mechanisms of asthma; (ii) the anti-inflammatory mechanisms of corticosteroids; and (iii) a group with unclear mechanisms.
Genetic factors and inflammation in asthma
The TBX21 gene
The transcription factor T-bet (also known as T-box 21 or Tbx21 protein) encoded by the TBX21 gene acts as a type 1 helper T cell development regulator by inducing interferon-γ production and inhibiting Th2 cytokines (Figure 1).22 Previous research has shown that knockout mice lacking T-bet developed BHR and other changes consistent with asthma, including peribronchial inflammation and collagen deposited in the basement membrane.23 ICS also modify BHR in asthma patients and it’s efficacy might be altered by TBX21 mutation.22
A correlation between the single nucleotide polymorphism (SNP) rs9910408 (c.-7947) in the TBX21 gene with BHR was reported in children,23 which was also seen in another study in adults.22 Changes in TBX21 expression in asthmatic children was previously demonstrated, in which the minor G allele in the TBX21 SNP rs2240017, which codes for glutamine (33Q), was associated with a dramatic improvement in the PC(20) (a measure of airway responsiveness) in the ICS-treated group (Table 1).24 However, this was not replicated in a study that genotyped four genes (ADRB2, adenylate cyclase 9, neurokinin receptor 2, and TBX21) in 53 adult Korean patients with mild-to-moderate asthma; after 4 weeks of treatment with ICS, there was a correlation between the control of asthma and the high frequency of the major C allele in TBX21 H33Q C>G.25
Table 1.
Interaction between single nucleotide polymorphisms (SNPs) of four candidate genes and corticosteroid response in patients with asthma.
| Gene | Variant | Population | Outcomes | Results |
|---|---|---|---|---|
| TBX21 | TBX21 H33Q (SNP rs2240017)24 | 311 children enrolled in the CAMP clinical trial who were treated with inhaled budesonide | BHR and FEV1 | Improvement of BHR in ICS group with minor allele G encoding for glutamine (33Q), but no correlation with FEV1 |
| FCER2 | 10 FCER2 SNPs resequenced and genotyped26 | 311 children from the CAMP clinical trial who were treated with inhaled budesonide over 4-years | IgE levels Severe exacerbations | FCER2 SNP (T2206C) associated with elevated IgE level and severe exacerbations |
| 17 phenotypic variables and polymorphisms in FCER2 and CRHR133 | 311 children from the CAMP clinical trial who were treated with inhaled budesonide | Poor lung function response (improvement of FEV1 < 7.5%) Recurrent asthma exacerbations | Minor allele of T2206C in FCER2 associated with both poor lung function and recurrent exacerbations | |
| FCER2 T2206C variant (rs28364072)34 | Two cohorts of asthmatic children: PACMAN study (n = 386) and BREATHE study (n = 939) | IgE levels Severe exacerbations Poor lung function response (improvement of FEV1 < 7.5%) Recurrent asthma exacerbations Asthma exacerbations ACQ and medication use | FCER2 SNP (T2206C) associated with elevated IgE level and severe exacerbations Minor allele of T2206C in FCER2 associated with both poor lung function and recurrent exacerbations T2206C variant increased risk of asthma exacerbations and uncontrolled asthma and associated with increased daily ICS dose | |
| CRHR1 | Screening 131 SNPs in 14 genes37 | CAMP (replicate): 311 childhood asthmatics used budesonide | Variation of FEV1 from baseline to 8 weeks | SNPs rs242941 in CRHR1 correlated with enhanced lung function GAT/GAT haplotype in CRHR1 demonstrated an improvement in FEV1 |
| 17 phenotypic variables and polymorphisms in FCER2 and CRHR1 33 | 311 asthmatic children used budesonide during the CAMP | Poor lung function response Recurrent asthma exacerbations | The minor allele (T) of rs242941 in CRHR1 gene associated with poor lung response, contrary to the above study | |
| GLCCI1 | rs37972; rs37973 7bib6 | FBAT screening cohort in the CAMP clinical trial population | FEV1 | Both rs37972 and rs37973 are associated with decrements in GLCCI1 expression, and reduced lung function in response to ICS |
CAMP, Childhood Asthma Management Program; BHR, bronchial hyperresponsiveness; FEV1, forced expiratory volume in 1 s; ICS, inhaled corticosteroids; PACMAN, Pharmacogenetics of Asthma medication in Children: Medication with ANti-inflammatory effects; IgE, immunoglobulin E; ACQ, Asthma Control Questionnaire; FBAT, family-based screening test.
The FCER2 gene
The Fc fragment of IgE receptor II (FCER2) gene, which encodes the low-affinity receptor for IgE (CD23),26 decreases IgE-mediated immune responses when activated (Figure 1).27 In patients with asthma, high IgE levels cause acute exacerbations,28 emergency consultations,29,30 and hospitalizations (Table 1).31,32 A novel variant (T2206C) of the FCER2 gene was shown to be associated with higher IgE levels and increased severe exacerbations in children with asthma treated with ICS (Table 1).26 A study that aimed to identify predictors of a poor response to ICS during long-term therapy, evaluated 17 phenotypic variables and polymorphisms of the FCER2 and CRHR1 genes in 311 asthmatic children treated with budesonide during the Childhood Asthma Management Program (CAMP) clinical trial by comparing recurrent exacerbations and lung function.33 The results showed that both a poor lung function response and repeated exacerbations were caused only by the FCER2 gene mutation.33 Children that were homozygous for the FCER2 gene T2206C mutant allele had a 3.3-times higher risk of recurrent exacerbations and a 3.9-times higher risk of poor lung function than the wild-type homozygous children (Table 1).33
Data from two cohort studies undertaken in the Netherlands, which included 386 children in the Pharmacogenetics of Asthma medication in Children: Medication with ANti-inflammatory effects study and 939 children in the BREATHE study, were used to analyse the effect of the FCER2 gene T2206C variant on acute asthma exacerbations, symptoms and treatment.34 The results showed that the T2206C variant was associated with a higher risk of asthma-related hospitalization (odds ratio [OR] 1.91, 95% confidence interval [CI] 1.08, 3.40), an increased risk of uncontrolled asthma (OR 2.64, 95% CI 1.00, 6.98), and a higher daily steroid dose (OR 2.46, 95% CI 1.38, 4.39).34
The CRHR1 gene
Corticotropin releasing hormone receptor 1, encoded by the CRHR1 gene, is one of the key receptors of the pituitary gland and it plays an intermediary role in releasing ACTH (Figure 3). CRHR1 has been shown to be important in the pathogenesis of asthma in children and adults.35 The absence of CRHR1 leads to enhanced airway inflammation and dysfunction.36 A study that evaluated 14 candidate genes that are relevant to the entire corticosteroid pathway demonstrated that the rs242941 polymorphism of the CRHR1 gene was associated with a positive treatment response in both children from the CAMP clinical trial and a population of adults with asthma (known as the Adult Study) (Table 1).37 However, this result was not replicated in adults with asthma from the Asthma Clinical Research Network population who were also evaluated in this study.37
The NR3C1 gene
The nuclear receptor subfamily 3 group C member 1 (NR3C1) gene encodes the glucocorticoid receptor, which acts as a transcription factor that binds to the promoter area of the glucocorticoid responsive genes (Figure 3).38 Mutations in this gene are associated with generalized glucocorticoid resistance.39
The STIP1 gene
Stress induced phosphoprotein 1 (STIP1) is a protein that coordinates the function of HSP70 and HSP90 (Figure 3).40 Research suggests that STIP1 gene mutations may take part in the regulation of the corticosteroid response of severely asthmatic patients with lung dysfunction.41 Four STIP1 gene SNPs (rs4980524, rs6591838, rs2236647, and rs2236648) were closely associated with a decrease in the baseline percentage predicted forced expiratory volume in 1 s (FEV1) and the largest difference was measured for SNP rs6591838.41 The rare GG genotype of rs6591838 increased the percentage change in FEV1 at both 4 and 8 weeks of treatment.41
The GLCCI1 gene
Corticosteroids induce the expression of the glucocorticoid induced 1 (GLCCI1) gene.7 GLCCI1 gene polymorphisms are associated with a decreased ICS response in asthmatic patients.7 In the CAMP clinical trial, the minor (T) rs37972 allele was demonstrated to be significantly associated with a poor response to ICS (Table 1).7
The HDAC genes
Members of the histone deacetylase (HDAC) family of enzymes are involved in regulating the inflammatory genes by removing acetyl groups from histones (Figure 3).42,43 Conditional gene targeting that resulted in the T cell-specific loss of HDAC1 lead to an inflamed airway and Th2 cytokine production in an in vivo allergic airway inflammation model in mice.44 In addition, HDAC is related to the corticosteroid activity mechanism.45 For example, HDAC2 recruitment is an important step in the process by which corticosteroids repress inflammatory genes; and HDAC2 is less active in some diseases where a proportion of patients respond poorly to corticosteroids.46 Therefore, HDAC1 and HDAC2 can be studied as target mediators for corticosteroid-response prediction.47
A study on 70 children and 35 adults with asthma showed that there was a significant correlation between the rs1741981 polymorphism in the HDAC1 gene and asthma severity, while the rs58677352 polymorphism in the HDAC2 gene showed no correlation with asthma severity.47 Children with the rs1741981 CC genotype had the lower increase of percentage FEV1 in response to corticosteroid therapy in comparison with children with the CT and TT genotypes.47
The ORMDL3 gene
The ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) gene at locus 17q21 is ubiquitously expressed and encodes a protein that is a crucial mediator in decreasing the synthesis of sphingolipids.48 In asthma, sphingolipids play an important role in synthesizing many inflammatory proteins (Figure 3).49 The ORMDL3 gene increases the risk of asthma in children.50 The SNP rs2872507 that modulates ORMDL3 gene expression has been shown to have a significant correlation with asthma.51,52 In a study of 311 children with persistent asthma and 276 healthy control subjects, the rs2872507 polymorphism was demonstrated to be associated with ORMDL3 gene expression and with ICS treatment response in children with atopic asthma.53
The VEGF genes
Members of the family of vascular endothelial growth factors (VEGFs) regulate angiogenesis and vascular permeability.54 In mouse models, VEGF (now known as VEGF-A) has been observed to increase mucus production, collagen deposition, as well as smooth muscle hyperplasia.55 Empirical evidence suggested that increased VEGF levels were inversely correlated with FEV1 and positively associated with airway hyperresponsiveness.56 However, there is a lack of evidence about the effects of the VEGF genes on medication responsiveness. A study that investigated the effects of the VEGFA gene in 131 asthmatic children being treated with different therapies (the ICS fluticasone propionate or the leukotriene receptor antagonist [LTRA] montelukast) for 12 months, demonstrated that the polymorphism rs2146323 A>C was associated with ICS response.57 Specifically, patients with the AA genotype had a greater improvement in FEV1 when compared with those with the AC and CC genotypes.57 However, among children treated with the LTRA, the AA genotype was associated with uncontrolled asthma and a worse FEV1/forced vital capacity ratio compared with the other genotypes.57 Another polymorphism associated with the LTRA response was rs833058 C>T, in which patients with the TT genotype had an improvement in the percentage predicted FEV1 in comparison with no improvement in patients with the CT or CC genotypes.57
Other genes
The role of other genes such as dual specificity phosphatase 1 (DUSP1) or allantoicase (ALLC) in response to corticosteroids in children with asthma has not been well demonstrated.58,59 DUSP1 protein (also known as mitogen-activated protein kinase phosphatase-1) is important in the human cellular response to stress caused by the environment and to cell proliferation.60 In asthmatic patients, DUSP1 seems to mediate corticosteroid activity (Figure 3).58 The ALLC gene, an uricolysis enzyme involved in uric acid degradation, has an unclear role in ICS response in asthmatic patients.
Conclusion
In conclusion, despite the use of ICS and other therapies, childhood asthma causes significant morbidity worldwide. Understanding the pathogenesis of asthma and the impact of genetic polymorphisms on the ICS treatment response could lead to personalized therapy and improved quality of care for children with asthma in the future.
Declaration of conflicting interests
The authors declare that there are no conflicts of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Simpson CR, Sheikh A. Trends in the epidemiology of asthma in England: a national study of 333,294 patients. J R Soc Med 2010; 103: 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Initiative for Asthma, http://ginasthma.org/wp-content/uploads/2016/01/GINA_Report_2015_Aug11-1.pdf (accessed 12 April 2016).
- 3.Leung TF, Tang MF, Sy HY, et al. Novel asthma therapeutics: insights from whole-genome studies. J Pharmacogenomics Pharmacoproteomics 2013; 4: 115–115. [Google Scholar]
- 4.Barnes PJ. Corticosteroid effects on cell signalling. Eur Respir J 2006; 27: 413–426. [DOI] [PubMed] [Google Scholar]
- 5.Tantisira KG, Damask A, Szefler SJ, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med 2012; 185: 1286–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baye TM, Abebe T, Wilke RA. Genotype–environment interactions and their translational implications. Per Med 2011; 8: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tantisira KG, Lasky-Su J, Harada M, et al. Genome wide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 2011; 365: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung LP, Waterer G, Thompson PJ. Pharmacogenetics of β2 adrenergic receptor gene polymorphisms, long-acting β-agonists and asthma. Clin Exp Allergy 2011; 41: 312–326. [DOI] [PubMed] [Google Scholar]
- 9.Tantisira KG, Drazen JM. Genetics and pharmacogenetics of the leukotriene pathway. J Allergy Clin Immunol 2009; 124: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Asthma Education and Prevention Program, Third Expert Panel on the Diagnosis and Management of Asthma, www.ncbi.nlm.nih.gov/books/NBK7223/ (accessed 13 May 2016).
- 11.Vijverberg SJ, Hilvering B, Raaijmakers JA, et al. Clinical utility of asthma biomarkers: from bench to bedside. Biologics 2013; 7: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes PJ. Pathophysiology of asthma. Eur Respir Mon 2003; 23: 84–113. [Google Scholar]
- 13.Brugha R, Mushtaq N, McCarthy NE, et al. Respiratory tract dendritic cells in paediatric asthma. Clin Exp Allergy 2015; 45: 624–631. [DOI] [PubMed] [Google Scholar]
- 14.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest 2008; 118: 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alangari AA. Genomic and non-genomic actions of glucocorticoids in asthma. Ann Thorac Med 2010; 5: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes PJ, Adcock IM. How do corticosteroids work in asthma? Ann Intern Med 2003; 139(5 Pt 1): 359–370. [DOI] [PubMed] [Google Scholar]
- 17.Priftis KN, Papadimitriou A, Nicolaidou P, et al. The hypothalamic-pituitary-adrenal axis in asthmatic children. Trends Endocrinol Metab 2008; 19: 32–38. [DOI] [PubMed] [Google Scholar]
- 18.Fenech AG, Ellul-Micallef R. Pharmacogenetics of asthma therapeutics (respiratory supplement). The Chronic Ill 2005; 9: 24–29. [Google Scholar]
- 19.Adcock IM, Lane SJ. Corticosteroid-insensitive asthma: molecular mechanisms. J Endocrinol 2003; 178: 347–355. [DOI] [PubMed] [Google Scholar]
- 20.van Aalderen WM, Sprikkelman AB. Inhaled corticosteroids in childhood asthma: the story continues. Eur J Pediatr 2011; 170: 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss ST, Litonjua AA, Lange C, et al. Overview of the pharmacogenetics of asthma treatment. Pharmacogenomics J 2006; 6: 311–326. [DOI] [PubMed] [Google Scholar]
- 22.Lopert A, Rijavec M, Zavbi M, et al. Asthma treatment outcome in adults is associated with rs9910408 in TBX21 gene. Sci Rep 2013; 3: 2915–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raby BA, Hwang ES, Van Steen K, et al. T-bet polymorphisms are associated with asthma and airway hyperresponsiveness. Am J Respir Crit Care Med 2006; 173: 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tantisira KG, Hwang ES, Raby BA, et al. TBX21: a functional variant predicts improvement in asthma with the use of inhaled corticosteroids. Proc Natl Acad Sci U S A 2004; 101: 18099–18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye YM, Lee HY, Kim SH, et al. Pharmacogenetic study of the effects of NK2R G231E G>A and TBX21 H33Q C>G polymorphisms on asthma control with inhaled corticosteroid treatment. J Clin Pharm Ther 2009; 34: 693–701. [DOI] [PubMed] [Google Scholar]
- 26.Tantisira KG, Silverman ES, Mariani TJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol 2007; 120: 1285–1291. [DOI] [PubMed] [Google Scholar]
- 27.Sharma V, Michel S, Gaertner V, et al. A role of FCER1A and FCER2 polymorphisms in IgE regulation. Allergy 2014; 69: 231–236. [DOI] [PubMed] [Google Scholar]
- 28.Wever-Hess J, Kouwenberg JM, Duiverman EJ, et al. Risk factors for exacerbations and hospital admissions in asthma of early childhood. Pediatr Pulmonol 2000; 29: 250–256. [DOI] [PubMed] [Google Scholar]
- 29.Duff AL, Pomeranz ES, Gelber LE, et al. Risk factors for acute wheezing in infants and children: viruses, passive smoke, and IgE antibodies to inhalant allergens. Pediatrics 1993; 92: 535–540. [PubMed] [Google Scholar]
- 30.Pollart SM, Chapman MD, Fiocco GP, et al. Epidemiology of acute asthma: IgE antibodies to common inhalant allergens as a risk factor for emergency room visits. J Allergy Clin Immunol 1989; 83: 875–882. [DOI] [PubMed] [Google Scholar]
- 31.Zieg G, Lack G, Harbeck RJ, et al. In vivo effects of glucocorticoids on IgE production. J Allergy Clin Immunol 1994; 94(2 Pt 1): 222–230. [DOI] [PubMed] [Google Scholar]
- 32.Fuhlbrigge AL, Weiss ST, Kuntz KM, et al. Forced expiratory volume in 1 second percentage improves the classification of severity among children with asthma. Pediatrics 2006; 118: e347–e355. [DOI] [PubMed] [Google Scholar]
- 33.Rogers AJ, Tantisira KG, Fuhlbrigge AL, et al. Predictors of poor response during asthma therapy differ with definition of outcome. Pharmacogenomics 2009; 10: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koster ES, Maitland-van der Zee AH, Tavendale R, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid-treated asthmatic children. Allergy 2011; 66: 1546–1552. [DOI] [PubMed] [Google Scholar]
- 35.Tse SM, Tantisira K, Weiss ST. The pharmacogenetics and pharmacogenomics of asthma therapy. Pharmacogenomics J 2011; 11: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maitland-van der Zee AH and Daly AK. Pharmacogenetics and individualized therapy. Wiley, 2012.
- 37.Tantisira KG, Lake S, Silverman ES, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet 2004; 13: 1353–1359. [DOI] [PubMed] [Google Scholar]
- 38.HUGO Gene Nomenclature Committee. NR3C1 nuclear receptor subfamily 3 group C member 1 [Homo sapiens (human)]. Available from: http://www.ncbi.nlm.nih.gov/gene/2908.
- 39.Mohamed NA, Abdel-Rehim ASM, Farres MN, et al. Influence of glucocorticoid receptor gene NR3C1 646 C>G polymorphism on glucocorticoid resistance in asthmatics: a preliminary study. Cent Eur J Immunol 2015; 40: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song Y, Masison DC. Independent regulation of Hsp70 and Hsp90 chaperones by Hsp70/Hsp90-organizing protein Sti1 (Hop1). J Biol Chem 2005; 280: 34178–34185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawkins GA, Lazarus R, Smith RS, et al. The glucocorticoid receptor heterocomplex gene STIP1 is associated with improved lung function in asthmatic subjects treated with inhaled corticosteroids. J Allergy Clin Immunol 2009; 123: 1376–83. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adcock IM, Ford P, Ito K, et al. Epigenetics and airways disease. Respir Res 2006; 7: 21–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhavsar P, Ahmad T, Adcock IM. The role of histone deacetylases in asthma and allergic diseases. J Allergy Clin Immunol 2008; 121: 580–584. [DOI] [PubMed] [Google Scholar]
- 44.Grausenburger R, Bilic I, Boucheron N, et al. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J Immunol 2010; 185: 3489–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: importance in inflammatory lung diseases. Eur Respir J 2005; 25: 552–563. [DOI] [PubMed] [Google Scholar]
- 46.Barnes PJ, Ito K, Adcock IM. Corticosteroid resistance in chronic obstructive pulmonary disease: inactivation of histone deacetylase. Lancet 2004; 363: 731–733. [DOI] [PubMed] [Google Scholar]
- 47.Kim MH, Kim SH, Kim YK, et al. A polymorphism in the histone deacetylase 1 gene is associated with the response to corticosteroids in asthmatics. Korean J Intern Med 2013; 28: 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breslow DK, Collins SR, Bodenmiller B, et al. Orm family proteins mediate sphingolipid homeostasis. Nature 2010; 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan JJ, Spiegel S. The role of sphingosine-1-phosphate and its receptors in asthma. Drug News Perspect 2008; 21: 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007; 448: 470–473. [DOI] [PubMed] [Google Scholar]
- 51.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010; 363: 1211–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sleiman PM, Flory J, Imielinski M, et al. Variants of DENND1B associated with asthma in children. N Engl J Med 2010; 362: 36–44. [DOI] [PubMed] [Google Scholar]
- 53.Berce V, Kozmus CE, Potocnik U. Association among ORMDL3 gene expression, 17q21 polymorphism and response to treatment with inhaled corticosteroids in children with asthma. Pharmacogenomics J 2013; 13: 523–529. [DOI] [PubMed] [Google Scholar]
- 54.Shibuya M. Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: a crucial target for anti- and pro-angiogenic therapies. Genes Cancer 2011; 2: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee KS, Kim SR, Park HS, et al. Cysteinyl leukotriene receptor antagonist regulates vascular permeability by reducing vascular endothelial growth factor expression. J Allergy Clin Immunol 2004; 114: 1093–1099. [DOI] [PubMed] [Google Scholar]
- 56.Hoshino M, Takahashi M, Aoike N. Expression of vascular endothelial growth factor, basic fibroblast growth factor, and angiogenin immunoreactivity in asthmatic airways and its relationship to angiogenesis. J Allergy Clin Immunol 2001; 107: 295–301. [DOI] [PubMed] [Google Scholar]
- 57.Balantic M, Rijavec M, Skerbinjek Kavalar M, et al. Asthma treatment outcome in children is associated with vascular endothelial growth factor a (VEGFA) polymorphisms. Mol Diagn Ther 2012; 16: 173–180. [DOI] [PubMed] [Google Scholar]
- 58.Jin Y, Hu D, Peterson EL, et al. Dual specificity phosphatase-1 as a pharmacogenetic modifier of inhaled steroid response among asthma patients. J Allergy Clin Immunol 2010; 126: 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jindal SK. Genetic basis of asthma. Indian J Med Res 2015; 142: 640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu YX, Wang J, Guo J, et al. DUSP1 is controlled by p53 during the cellular response to oxidative stress. Mol Cancer Res 2008; 6: 624–633. [DOI] [PubMed] [Google Scholar]